SUMMARY
Botch promotes embryonic neurogenesis through inhibition of Notch-1 signaling through inhibition of the initial S1 furin-like cleavage step of Notch maturation. The biochemical process by which Botch inhibits Notch maturation is not known. Here we show that Botch has γ-glutamyl cyclotransferase (GGCT) activity that deglycinates Notch, which prevents the S1 furin-like cleavage. Moreover, Notch is mono-glycinated on the γ-glutamyl carbon of glutamate 1669. The deglycinase activity of Botch is required for inhibition of Notch signaling, both in vitro and in vivo. When the γ-glutamyl-glycine at position 1669 of Notch is degylcinated it is replaced by 5-oxy-proline. These results reveal that Botch regulates Notch signaling through deglycination and identify a novel posttranslational modification of Notch that plays an important role in neurogenesis.
INTRODUCTION
Botch promotes embryonic neurogenesis through inhibition of Notch-1 signaling (Chi et al., 2012). Notch processing and subsequent signaling, requires a S1 furin-like cleavage event (Kopan and Ilagan, 2009; Louvi and Artavanis-Tsakonas, 2006). This cleavage generates the mature processed form of Notch in which one polypeptide becomes divided into the Notch1 extracellular domain (NECD) and the trans-membrane intracellular domain (TMIC). The NECD is the ligand binding domain through which the ligands, Delta or Jagged-like activate Notch. The TMIC upon ligand binding to NECD will undergo S2 and S3 cleavage to generate the intracellular domain (NICD). NICD translocates to the nucleus to direct target gene expression (Ilagan and Kopan, 2013), where it converts C-promoter binding factor-1 (CBF-1) complex from a transcriptional repressor to a transcriptional activator resulting in Notch1 target gene expression. Botch prevents Notch signaling by interfering with the initial S1 furin-like cleavage step of Notch maturation (Chi et al., 2012). This maintains Notch in an immature unprocessed form. Botch interferes with the S1 furin-like cleavage of Notch by physical binding of Botch to the NECD. The mechanism of how Botch controls the S1 furin-like cleavage of Notch is not known. Here we show that Botch has γ-glutamyl cyclotransferase (GGCT) activity that deglycinates Notch, which prevents the S1 furin-like cleavage. The deglycinase activity of Botch is required for inhibition of Notch signaling, both in vitro and in vivo. These results reveal that Botch regulates Notch signaling through deglycination and identify a novel posttranslational modification of Notch that plays an important role in neurogenesis.
RESULTS
Enzymatic Activity of Botch is γ-Glutamyl-Cyclotransferase (GGCT)
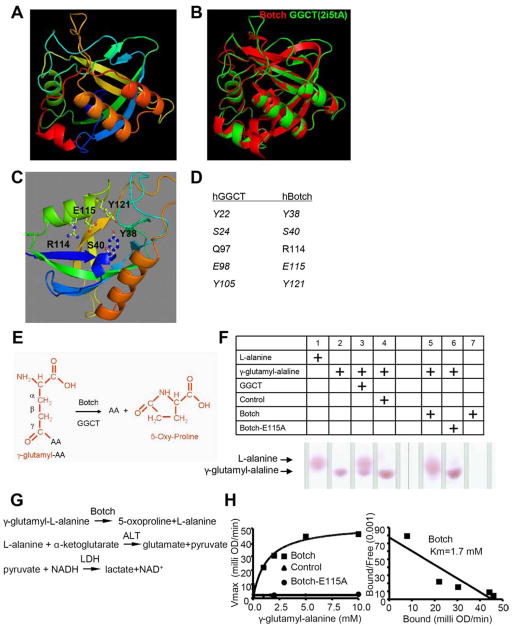
The structure of Botch was modeled on Swiss-Model website and Botch was found to closely resemble the structure of human γ-glutamyl cyclotransferase (GGCT) (Figure 1A). There is a very high structural similarity between Botch and GGCT (Figure 1B), even though there is little amino acid homology. The Swiss-Model search provided a putative catalytic domain in Botch, which aligned with GGCT indicating it may function in a similar manner. The model also predicted that E115 may be a key amino acid for Botch enzymatic function (Figure 1C). All the key amino acids within the catalytic domain of GGCT could be aligned with Botch (Figure 1D). To determine if Botch has GGCT activity thin layer chromatography (TLC) assays were performed. In this TLC assay the γ-amino acid (γ-glutamyl-AA) is converted to the free amino acid (AA) by GGCTs and in the process generates a 5-oxy-proline (Figure 1E). The free AA is detected by an upward shift on the TLC plate. A typical GGCT substrate γ-glutamyl-alanine was incubated in the presence of GGCT, Botch or Botch E115A. Both GGCT and Botch release alanine by cleavage of γ-glutamyl-alanine whereas Botch E115A is inactive (Figure 1F). Importantly Botch does not self cleave (Figure 1F). Similar observations are made for γ-glutamyl-methionine and γ-glutamyl-glutamate (Figure S1A and S1B). In order to determine the kinetics of enzymatic function a well-established quantitative biochemical assay to monitor GGCT activity was used with γ-glutamyl-L-alanine as a substrate (Figure 1G) (Oakley et al., 2008). Botch is active and has a Km of 1.7 mM similar to GGCT (Oakley et al., 2008), whereas Botch E115A is inactive (Figure 1H). These results indicate that Botch has GGCT-like activity.
Figure 1. Botch has γ-Glutamyl-Cyclotransferase (GGCT) activity.
(A) Botch structure generated by Swiss-model.
(B) Super-imposition of Botch (Red) structure with human GGCT structure (Green, PDB code 2i5tA) generated by Swiss-model.
(C) Predicted Botch putative active site residues.
(D) Alignment of human Botch putative active site residues with corresponding active site residues of human GGCT.
(E) Schematic representation of GGCT activity. Glutamate that is modified by an amino acid (AA) on the γ carbon is removed by the GGCT activity of Botch liberating and AA and forming 5-oxy-proliine.
(F) Thin layer chromatography of γ-glutamyl-Alanine cleavage by either recombinant GGCT or Botch, but not Botch-E115A.
(G) A schematic diagram of the serial reactions by using oxidation of NADH as the final readout for Botch’s GGCT like activity.
(H) Scatchard plot of quantitative activity of control or Botch or Botch-E115A.
F–H, Experiments were repeated three times with similar results.
Botch GGCT like activity is required for regulation of embryonic neurogenesis in vivo
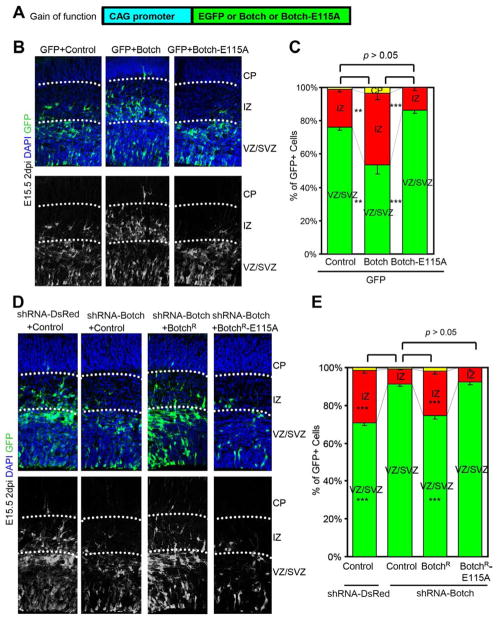
Gain and loss of function studies were performed as previously described to explore the role of Botch’s GGCT-like activity in neocortical development (Chi et al., 2012). In utero co-electroporation of pCAG-EGFP and pCAG-Botch or pCAG-Botch E115A into E13.5 CD1 mouse brain was performed (Figure 2). Embryos were harvested at E15.5 and immunostained for GFP to identify Botch overexpressing cells and counterstained with DAPI to identify all cells. As previously described Botch overexpression results in fewer GFP positive cells in the ventricular (VZ) and subventricular (SVZ) zones and more cells in the cortical plate (CP) and intermediate zone (IZ) when compared to co-electroporation with control (pCAG empty vector) (Figure 2B and 2C) (Chi et al., 2012). Botch E115A has no effect and is similar to control (Figure 2B and 2C).
Figure 2. Botch GGCT like activity is required for regulation of embryonic neurogenesis in vivo.
(A) A schematic diagram of pCAG constructs for overexpression (gain of function) for in utero injection and electroporation.
(B–E) Distribution of GFP+ cells 2 days after in utero injection and electroporation.
(B) Representative confocal images of cortex immunostained for GFP with and without the DAPI channel with Botch expression. Abbreviations: CP, cortical plate; IZ, intermediate zone; VZ, ventricular zone; SVZ, subvetricular zone.
(C) Quantification of distribution of GFP+ cells in (B). Values represent the mean ± SEM (n ≥ 3; ** p < 0.01; *** p < 0.001; n.s. p > 0.05, one-way ANOVA, post-test: Tukey’s multiple comparison tests).
(D) Representative confocal images of cortex immunostained for GFP with and without the DAPI channel following knockdown of Botch and rescue either with BotchR or BotchR-E115A.
(E) Quantification of distribution of GFP+ cells in (D). Values represent the mean ± SEM (n ≥ 3; *** p < 0.001; n.s. p > 0.05; one-way ANOVA, post-test: Tukey’s multiple comparison test).
To explore the role of Botch’s GGCT-like activity in neurogenesis in utero electroporation of shRNA DsRed, shRNA Botch, shRNA Botch and shRNA resistant Botch, and shRNA Botch with shRNA resistant Botch E115A (Figure S2A) into E13.5 CD1 mouse brain was performed. Embryos were harvested at E15.5 (Figure 2D and 2E). Knockdown of Botch greatly increases the percentage of cells in the VZ and SVZ while significantly decreasing the percentage of GFP positive cells in the CP and IZ (Figure 2D and 2E). Co-expression of shRNA resistant Botch (BotchR), which is not susceptible to shRNA Botch (Chi et al., 2012) rescues the knock down phenotype whereas shRNA resistant Botch E115A (BotchR E115A) has no effect (Figure 2D and 2E). Co-immunoprecipitation of Botch-E115A-myc with SP-NECD-GFP confirms this mutant can bind Notch1 (Figure S2B) and supports the notion that inactivity of Botch-E115A during neurogenesis is due to a lack of catalytic activity. These results taken together indicate that Botch’s GGCT-like activity is required for Botch’s promotion of neurogenesis.
Botch blocks Notch signaling in vitro through GGCT like activity
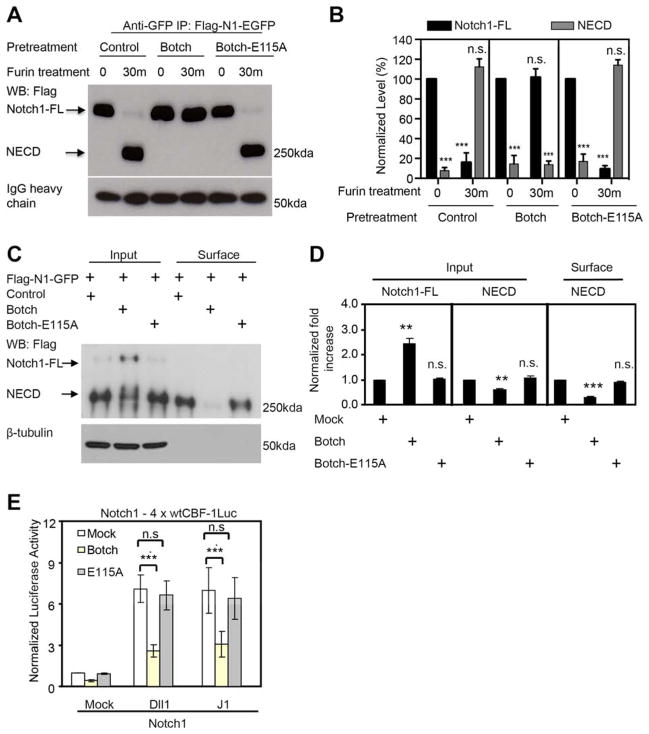
Botch promotes neurogenesis by preventing the cell surface presentation of Notch by inhibiting the S1-furin-like cleavage of Notch, maintaining Notch in the immature full-length form (Chi et al., 2012). To determine whether the GGCT like activity of Botch is required for the regulation of S1 cleavage of Notch1, Flag-Notch1-EGFP (Flag-N1-GFP) was treated with furin in the presence or absence of Botch or Botch E115A. As previously reported, wild type Botch completely prevents the furin cleavage of Notch1 (Chi et al., 2012), whereas Botch E115A is devoid of activity (Figure 3A and 3B). To determine if Botch acts generally on proteins that are furin substrates we investigated whether Botch can inhibit the cleavage of proBMP10 (Susan-Resiga et al., 2011). Botch fails to block the furin cleavage proBMP10 to BMP10 (Figure S3).
Figure 3. The GGCT activity of Botch is required to block Notch1 signaling in vitro.
(A) Immunoblots of Notch1 cleavage by furin and inhibition of furin cleavage by, Botch or Botch-E115A. Abbreviations: FL, Full Length; ECD, Extracellular domain.
(B) Optical densitometry quantification of a normalized to IgG heavy chain. Values represent the mean ± SEM (n ≥ 3; *** p < 0.001; n.s. p > 0.05; Student’s t-test).
(C) Immunoblot analysis of Notch1 in whole cell lysate and surface expression following overexpression of Botch or Botch-E115A and Flag-Notch1-GFP. Abbreviations: FL, Full Length; ECD, Extracellular domain.
(D) Optical densitometry quantification of (C) normalized to β-tubulin. Values represent the mean ± SEM (n = 3; ** p < 0.01; *** p < 0.001; n.s. p > 0.05; one-way ANOVA, post-test: Tukey’s multiple comparison test).
(E) wtCBF-1 luciferase reporter assays of HeLa cells expressing Notch1 and either Botch or Botch-E115A co-cultured with NIH 3T3 cells expressing Notch ligands. wtCBF-1 luciferase activity was normalized to β-galatacosidase activity and then to the Mock group without Botch. Values represent the mean ± SEM (n ≥ 3; *** p < 0.001; n.s. p > 0.05, one-way ANOVA, post-test: Tukey’s multiple comparison test).
To further investigate the actions of Botch on Notch, Botch and Notch1 GFP tagged constructs were expressed in HEK293 cells to monitor Notch1 processing. Overexpression of Botch leads to an approximate 2-fold increase in unprocessed full-length Notch1 (Notch1-FL) with a significant decrease in NECD (Figure 3c and 3D). Botch E115A has no effect (Figure 3C to D). HeLa cells were transfected with full-length Notch1 (Notch1-FL) and 4X wtCBF-1-response element luciferase reporter, and β-galatosidase reporter (β-Gal) and Botch or Botch E115A. HeLa cells were co-cultured 24 hours later with NIH3T3 cells expressing the ligand Delta-like 1 (DII1) or Jagged 1 (JI). β-Gal and luciferase activity were determined 24 hours after co-culture. Botch significantly inhibits the activation of CBF-luciferase activity whereas Botch E115A has no effect (Fig 3E). These results indicate that Botch blocks Notch1 signaling through inhibition of the furin-like cleavage of Notch1 via its GGCT-like activity.
Botch deglycinates Notch1 and Notch E1669 is required for Botch to block Notch signaling
Since Botch liberates amino acids from γ-carbon-modified glutamate (γ-glutamyl-AA) (see Figure 1 and Figure S1) experiments were performed to determine whether Notch1 has γ-carbon-modified glutamates. Since Botch binds to the full-length unprocessed Notch1 at the extracellular domain containing the EGF repeat 32–36 plus LNR domains corresponding to amino acids 1224–1723 (Chi et al., 2012), Notch1 1224–1723 was fused downstream to the 20 amino acid N-terminus signal peptide from Notch1 to insure proper subcellular localization, and was overexpressed, immunoprecipitated and purified from HEK293 cells (Figure S4A). A Notch1 peptide spanning 832–1310 also containing the 20 amino acid N-terminus signal peptide from Notch1 served as a control (Figure S4A). The bands corresponding to Notch 1224–1723 and 832–1310 were excised and subjected to trypsin digestion and mass spectrometry analysis. A total of 31 non-redundant tryptic peptides that contain 72% glutamate (21/29) were observed and mapped to 63% of amino acids of Notch 1225–1723. Using a targeted approach focused on glutamate residues in Notch1, mass spectrometry reveals that three glutamates including glutamate 1669 are modified by glycine (Figure S4B). If glycine is removed from γ-glutamyl-glycine, via the GGCT-like activity of Botch, a 5-oxy-proline, which has a unique mass different from all other amino acids, should be formed (Figure S4C). As such, the mass spectrometry data were analyzed for 5-oxy-proline and it was determined that the γ-glutamyl-glycine at position 1669 is replaced by 5-oxy-proline (Figure S4D). A 5-oxy-proline substitution was not detected at positions 1595 or 1655. Glutamate 1669 is conserved between the mammalian Notch’s as well as Drosophila (Figure S4E). These results suggest that Notch glutamate 1669 is modified via glycine on the γcarbon and undergoes removal to create a 5-oxy-proline.
Botch deglycinates Notch1
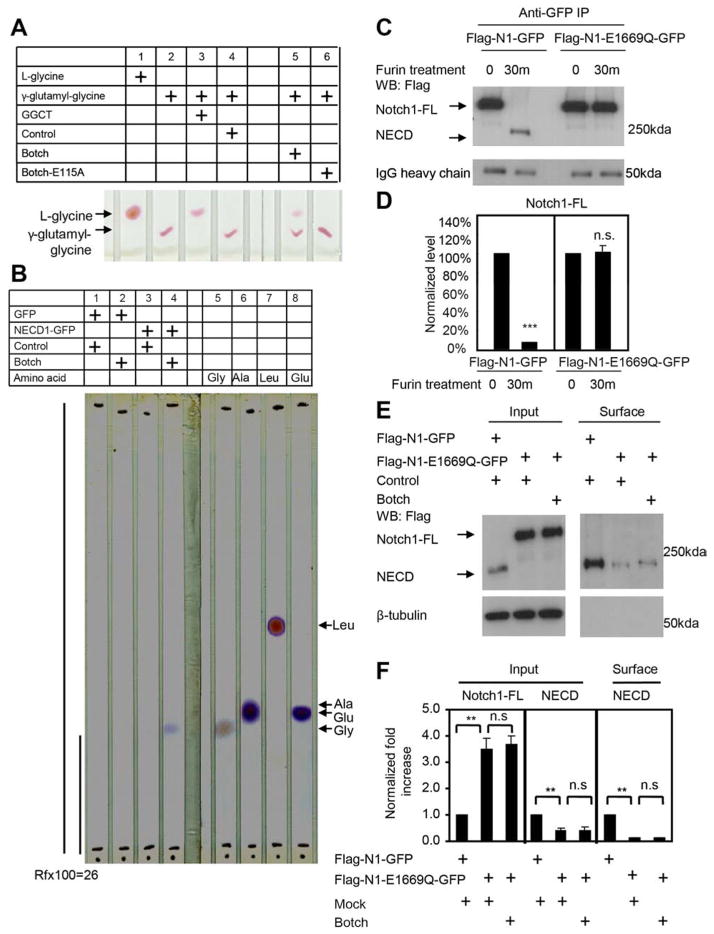
To determine if Botch has GGCT activity against γ-glutamyl-glycine, TLC assays were performed. The substrate γ-glutamyl-glycine was incubated in the presence of GGCT, Botch or Botch E115A. Both GGCT and Botch release glycine by cleavage of γ-glutamyl-glycine whereas Botch E115A is inactive (Figure 4A). To ascertain whether Botch is able to release glycine from Notch1, the Notch1 extracellular domain that binds to Botch (NECD1-GFP) was expressed and purified and incubated with purified Botch. TLC analysis reveals a band at the correct migration for glycine, but not glutamate, alanine or leucine (Figure 4B). A migration factor (Rfx100) was calculated at 26 and confirms that the band detected by TLC migrates identically to glycine (Sleckman and Sherma, 1982b) (Figure 4B).
Figure 4. Botch deglycinates Notch1 and Notch1 E1669 is required for Botch to block Notch1 signaling.
(A) Thin layer chromatography of γ-glutamyl-Glycine cleavage by either recombinant GGCT or Botch, but not Botch-E115A. (n = 3).
(B) Thin layer chromatography of NECD liberation of glycine by Botch. Four reference amino acids were run along with the cleavage product. Rfx100 for NECD cleavage product is 26. (n = 3).
(C) Immunoblots of Notch1 or Notch1-E1669Q cleavage by furin. Abbreviations: FL, Full Length; ECD, Extracellular domain.
(D) Optical densitometry quantification of a normalized to IgG heavy chain. Values represent the mean ± SEM (n ≥ 3; *** p < 0.001; n.s. p > 0.05; Student’s t-test).
(E) Immunoblot analysis of Notch1 in whole cell lysate and surface expression following overexpression of Botch and Flag-Notch1-GFP or Flag-Notch1-E1669Q-GFP.
(F) Optical densitometry quantification of (E) normalized to β-tubulin. Values represent the mean ± SEM (n = 3; ** p < 0.01; n.s. p > 0.05; one-way ANOVA, post-test: Tukey’s multiple comparison tests).
Notch E1669 is required for furin-like cleavage of Notch and Botch dependent regulation of Notch signaling
To determine if E1669 is required for the furin-like cleavage of Notch1, a conservative amino acid substitution from glutatmate to glutamine was made at 1669 in full-length Notch1 (Flag-N1-E1669Q-GFP). Flag-N1-GFP or Flag-N1-E1669Q-GFP was treated with furin in the presence or absence of Botch. Wild type Notch1 is cleaved by furin, whereas Flag-N1-E1669Q-GFP is not (Figure 4C to D). Botch is without effect on Flag-N1-E1669Q-GFP (Figure S4F and S4G). Botch-myc binds to SP-NECD-E1669Q-GFP Botch (Figure S4H and S4I). To further investigate the impact of the E1669Q mutation on the processing of Notch, wild type Flag-N1-GFP or Flag-N1-E1669Q-GFP in the presence or absence of Botch were expressed in HEK293 cells to monitor Notch1 processing. Similar to the effects of Botch overexpression, Notch1-E1669Q leads to an approximate 3-fold increase in unprocessed Notch1-FL with a significant decrease in NECD (Figure 4E and 4F) but has no effect on Flag-N1-E1669Q-GFP processing.
DISCUSSION
The major finding of this study is that Botch functions as a GGCT to remove glycine from a γ-carbon modified glutamate on Notch1. This results in the loss of the furin-like cleavage of full-length Notch1 leading to decreased cell surface expression of Notch1 and inhibition of Notch1 signaling and regulation of neurogenesis. Botch’s GGCT-like activity is required for the promotion of neurogenesis and the inhibition of Notch1 signaling. By mass spectrometry and mutational analysis a critical glutamate was found in human Notch1 that is required for Notch1’s furin-like cleavage. Post-translation modification via mono-glycination on glutamate 1669 appears to be required for the furin-like cleavage of Notch1 since a conservative substitution of glutamate 1669 to glutamine in Notch1 prevents its furin-like cleavage similar to the effect of Botch removing the glycine from the γ-glutamyl modified glutamate in Notch1.
In mammalian cells biochemical studies indicate that S1 cleavage of the extracellular domain by furin protease, a proprotein convertase, leads to maturation of the Notch receptor. This cleavage and creation of the heterodimeric form of Notch occurs in the Golgi apparatus, where Notch and Botch interact (Chi et al., 2012). While there is substantial evidence in mammalian systems for the importance of S1 cleavage the generality of this event across species is controversial. In Drosophila, in which Notch was first discovered, the furin cleavage of Drosophila Notch has long been controversial (Kidd and Lieber, 2002). However, recent investigations with new reagents and approaches provide strong evidence that the Drosophila Notch requires S1 cleavage (Lake et al., 2009). To explore the generality of Botch regulation of Notch, S2 drosophila cells were transfected with full-length drosophila Notch-VP16 (pANLV) (Saj et al., 2010b), a Notch responsive NRE-luciferase reporter or a Notch unresponsive NRE mutant luciferase reporter, β-Gal and a Drosophila Botch wild type expression construct and a Botch GGCT mutant construct with an E135A mutation, which corresponds to the E115A mutation (Chi et al., 2012). In pANLV transfected S2 cells Botch significantly inhibits the activation of Notch whereas Botch E135A has no effect (data not shown) suggesting that Botch blocks Notch1 signaling through inhibition of the furin-like cleavage of Notch1 via its GGCT-like activity and that this activity appears to be evolutionarily conserved. Future studies are required to clarify the role of Botch’s potential regulation of Drosophila Notch similar to those outlined here for mammalian Notch1.
Our study focuses on the regulation of Notch1 by Botch but it is notable that the Notch isoforms have been observed to have differential requirement for S1 cleavage. While S1 cleavage appears important for the maturation of Notch1, mutation of the S1 cleavage site in Notch2 has no apparent effect on transport of Notch2 to the cell surface or subsequent signaling events (Gordon et al., 2009). Thus, although Botch can potentially block the S1 cleavage of all Notch receptors, it might inhibit Notch signaling in a paralog-specific manner through interference with Notch1 signaling while sparing Notch2 signaling. Given that the number and isoform type of cell surface Notch receptors is critical for the functional outcome of Notch signaling the control of processing events provides an important layer of control that can have important biological and therapeutic implications.
When γ-glutamyl-glutamate is cleaved to remove the amino acid modification it creates a unique 5-oxy-proline. Consistent with this idea we observed a 5-oxy-proline modification at position 1669 in Notch1, the same amino acid we found by mass spectrometry to be modified by glycine. Mutations in human Notch that flank this conserved E1669, prevent furin cleavage resulting in T cell acute lymphoblastic leukemias, supporting the idea that this site is important for Notch function. (Van Vlierberghe and Ferrando, 2012).
The post-translational modification of proteins on γ-glutamyl-glutamate by amino acids has been described for tubulin, 14-3-3, nucleosome assembly protein1 (NAP1) and others (Lalle et al., 2011; van Dijk et al., 2008; Wloga et al., 2010; Wloga and Gaertig, 2010). Tubulin ligase like (TLL) 3 and 4 ligate glycine or glutamate to tubulin on the γ-carbon of glutamate where they are then elongated via TLL10 in Drosophila to form poly-glycine or poly-glutamate side chains that modify tubulin function (Rogowski et al., 2009). The mechanism by which Notch1 is glycinated is not known but TLL’s are likely candidates. Notch1 appears to be mono-glycinated since we were unable to detect poly-glycine chains on Notch1 via mass spectrometry. This is consistent with the observation in humans that TLL10, which is required for the poly-glycination is inactive (Rogowski et al., 2009). Carboxypeptidase 5 (CCP5) acts as a tubulin deglutamylase (Kimura et al., 2010). The mechanism by which γ-glutamyl-glycines are removed is not known. The finding that Botch acts as a deglycinase suggests that Botch represents the first known deglycinase. Consistent with this notion is the observation that YER63C, the yeast homologue of Botch, was recently shown to possess GGCT activity where it converts glutathione to 5-oxy-proline and the dipeptide cysteine-glycine (Kumar et al., 2012). Thus, Botch may regulate other proteins that are post-translationally modified with the addition of glycine to the γ-carbon of glutamate.
In summary, we identify a previously unknown post-translational modification in which the γ-glutamyl-glycine of Notch1 at position 1669, is removed by the GGCT activity of Botch. This prevents the S1-furin-like ‘cleavage’ of Notch1, maintaining it in an inactive state. These findings expand the repertoire of proteins that undergo post-translational modification on the γ-carbon of glutamate. Since there are other GCCT-like proteins there are likely to be other proteins that are regulated by post-translation amino acid addition and removal. These findings have the potential to open up a new area of investigation into this understudied post-translational modification and the biologic consequences of regulation of this modification.
EXPERIMENTAL PROCEDURES
Plasmid constructs and Vector-based shRNA constructs
Botch cDNA from rat brain (GenBank NM_001173437.1) was cloned into pcDNA3.1/myc/His (Invitrogen) and APTag5 (GenHunter) as previous described (Chi et al., 2012; Saj et al., 2010a). For the rescue experiments shRNA resistant Botch (BotchR) and shRNA-DsRed was generated as previous described (Chi et al., 2012). The extracellular domain of Notch1 (NECD1) contains a cDNA fragment from 227 bp to 5395 bp of rat Notch1 cDNA (NM_001105721.1). The NECD1 was further divided into four fragments including EGF repeats 1–12, 11–24, 22–33 and 32–36 with LNR, which contain cDNA fragments from 227 bp to 1705 bp, from 1472 bp to 2956 bp, from 2723 bp to 4156 bp and from 3899 bp to 5395 bp, respectively. To ensure proper processing the cDNA encoding the 20 amino-acid signal peptide from Notch1 was inserted to the N-terminal of each NECD1 fragment by PCR. Botch-E115A mutant and Notch1-E1669Q mutant were generated by site-directed mutagenesis (Stratagene) and verified by sequencing. Human GGCT cDNA (GenBank C7orf24) was subcloned into pGEX-6p-1 vector (GE Healthcare).
In utero injection and electroporation
DNA transfer into E13.5 CD1 mouse brains in utero was performed using a Nepagene CUY21EDIT electroporator. 2.5 μg of DNA constructs in PBS were injected into the lateral ventricle using an orally controlled capillary micropipette. Five electric pulses (33V, 50ms pulse followed by 950ms gap) were delivered. Embryos were harvested at E15.5 fixed in 4% paraformaldehyde with 30% sucrose dehydration. 0.5 μg shRNA-Botch-pCAG-EGFP or shRNA-DsRed-pCAG-EGFP was co-injected with 2 μg either Mock (pCAG empty vector) or pCAG-BotchR or pCAG-BotchR-E115A or pCAG-Botch plasmids. 1 μg pCAG-EGFP was co-injected with 1 μg of either Mock (pCAG empty vector) or pCAG-Botch or pCAG-Botch-E115A plasmid.
Thin layer chromatography (TLC) assay
TLC assays were performed as described (Sleckman and Sherma, 1982a) on silica gel 60A plates (Whatman) with n-butanol/acetic acid/water, 3:1:1. Chromatography plates were dried and developed with Ninhydrin (Sigma) to produce blue or purple zones of amino acids. GGCT-GST fusion protein cloned into pGEX-6P-1 vector was expressed in E. coli. Botch protein was purified with Glutathione-Sepharose 4B and the GST tag was cleaved by Precision protease (GE healthcare). Alkaline phosphatase (AP) or Botch tagged with Botch-AP or Botch-E115A-AP constructs were transiently transfected into HEK293 cells (Chi et al., 2012). The conditioned medium containing the Botch-AP fusion protein was centrifuged, pH adjusted, concentrated and filtered. Concentration was determined by p-Nitrophenyl phosphate (GenHunter) at 405 nm.
Characterization of Botch-AP enzymatic activity
Enzymatic activity assay was performed (Oakley et al., 2008). The activity of either AP or Botch-AP or Botch-E115A-AP was measured with γ-glutamyl-L-Alanine as a substrate by using oxidation of NADH as the final readout.
Cell cultures
HEK293, HeLa, and NIH 3T3 cells were obtained from the American Type Culture Collections.
CBF-1RE and NRE Luciferase reporter assay
The co-culture reporter assay was performed as described (Chi et al.). Wild-type CBF-1RE reporter constructs were provided by D. Hayward (Johns Hopkins University School of Medicine, Baltimore, MD). Wild-type and mutant NRE luciferase reporter constructs were made by S. Bray (University of Cambridge, UK).
Immunoprecipitation assays
The immunoprecipitation protocols were as described (Zhang et al., 2000). Protein G Sepharose beads, 5 μg anti-GFP antibody (Rockland) or 5 μg anti-Notch1 antibody (C20, Santa-Cruz) was used. In HEK293 cell lysates, heterodimeric Notch1 is dissociated by 1% TX-100.
Immunoblot analysis and immunostaining
Primary antibodies used were rabbit anti-Notch1 (Millipore), mouse anti-Botch (Neuromab, 75–181), mouse anti-myc (Sungene, Tianjin), goat anti-GFP (Rockland) and mouse anti-β-tubulin (Sigma-Aldrich). Immunostainings were imaged with a Zeiss LSM 510. Images were processed with Adobe Photoshop.
Cell surface biotinylation labeling
Biotin was used to label and isolate cell surface protein as described (Chi et al., 2012).
Furin cleavage assay
As previous described (Chi et al., 2012), HEK293 cells were transfected with Flag-Notch1-GFP construct. 24 hours later, the cell lysates were subjected to anti-GFP antibody immunoprecipitation. The Flag-Notch1-GFP protein binding on protein G beads was first pre-treated with AP or Botch-AP or Botch-E115A-AP, and then treated with furin (New England Biolabs) at room temperature in total volume of 1 ml using 20 units of recombinant furin in 100mM HEPES 7.5, 0.5% Triton and 1 mM CaCl2.
Statistical analysis
Statistical analysis was performed using Prism or Excel software and specific tests are noted in the text and figure legends. Unless otherwise noted, all error bars represent ± SEM. and significance was assessed as p < 0.05.
HIGHLIGHTS.
Botch has γ-glutamyl cyclotransferase (GGCT) activity.
Notch is mono-glycinated on the γ-glutamyl carbon of glutamate 1669.
Botch deglycinates Notch preventing S1 furin-like cleavage and thus Notch signaling.
Botch creates a 5-oxy-proline post-translational modification at Notch glutamate 1669.
Acknowledgments
This work was supported by McKnight Foundation Neuroscience of Brain Disorders Award, USPHS NS40809, DA00266, and MSCRFII-0429 to V.L.D. T.M.D. is the Leonard and Madlyn Abramson Professor in Neurodegenerative Diseases.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Chi Z, Zhang J, Tokunaga A, Harraz MM, Byrne ST, Dolinko A, Xu J, Blackshaw S, Gaiano N, Dawson TM, et al. Botch promotes neurogenesis by antagonizing Notch. Dev Cell. 22:707–720. doi: 10.1016/j.devcel.2012.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi Z, Zhang J, Tokunaga A, Harraz MM, Byrne ST, Dolinko A, Xu J, Blackshaw S, Gaiano N, Dawson TM, et al. Botch promotes neurogenesis by antagonizing Notch. Dev Cell. 2012;22:707–720. doi: 10.1016/j.devcel.2012.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon WR, Vardar-Ulu D, L’Heureux S, Ashworth T, Malecki MJ, Sanchez-Irizarry C, McArthur DG, Histen G, Mitchell JL, Aster JC, et al. Effects of S1 cleavage on the structure, surface export, and signaling activity of human Notch1 and Notch2. PloS one. 2009;4:e6613. doi: 10.1371/journal.pone.0006613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilagan MX, Kopan R. Selective Blockade of Transport via SERCA Inhibition: The Answer for Oncogenic Forms of Notch? Cancer Cell. 2013;23:267–269. doi: 10.1016/j.ccr.2013.02.020. [DOI] [PubMed] [Google Scholar]
- Kidd S, Lieber T. Furin cleavage is not a requirement for Drosophila Notch function. Mechanisms of development. 2002;115:41–51. doi: 10.1016/s0925-4773(02)00120-x. [DOI] [PubMed] [Google Scholar]
- Kimura Y, Kurabe N, Ikegami K, Tsutsumi K, Konishi Y, Kaplan OI, Kunitomo H, Iino Y, Blacque OE, Setou M. Identification of tubulin deglutamylase among Caenorhabditis elegans and mammalian cytosolic carboxypeptidases (CCPs) J Biol Chem. 2010;285:22936–22941. doi: 10.1074/jbc.C110.128280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Tikoo S, Maity S, Sengupta S, Kaur A, Bachhawat AK. Mammalian proapoptotic factor ChaC1 and its homologues function as gamma-glutamyl cyclotransferases acting specifically on glutathione. EMBO Rep. 2012;13:1095–1101. doi: 10.1038/embor.2012.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake RJ, Grimm LM, Veraksa A, Banos A, Artavanis-Tsakonas S. In vivo analysis of the Notch receptor S1 cleavage. PloS one. 2009;4:e6728. doi: 10.1371/journal.pone.0006728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalle M, Camerini S, Cecchetti S, Blasetti Fantauzzi C, Crescenzi M, Pozio E. Giardia duodenalis 14-3-3 protein is polyglycylated by a tubulin tyrosine ligase-like member and deglycylated by two metallocarboxypeptidases. J Biol Chem. 2011;286:4471–4484. doi: 10.1074/jbc.M110.181511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louvi A, Artavanis-Tsakonas S. Notch signalling in vertebrate neural development. Nat Rev Neurosci. 2006;7:93–102. doi: 10.1038/nrn1847. [DOI] [PubMed] [Google Scholar]
- Oakley AJ, Yamada T, Liu D, Coggan M, Clark AG, Board PG. The identification and structural characterization of C7orf24 as gamma-glutamyl cyclotransferase. An essential enzyme in the gamma-glutamyl cycle. J Biol Chem. 2008;283:22031–22042. doi: 10.1074/jbc.M803623200. [DOI] [PubMed] [Google Scholar]
- Rogowski K, Juge F, van Dijk J, Wloga D, Strub JM, Levilliers N, Thomas D, Bre MH, Van Dorsselaer A, Gaertig J, et al. Evolutionary divergence of enzymatic mechanisms for posttranslational polyglycylation. Cell. 2009;137:1076–1087. doi: 10.1016/j.cell.2009.05.020. [DOI] [PubMed] [Google Scholar]
- Saj A, Arziman Z, Stempfle D, van Belle W, Sauder U, Horn T, Durrenberger M, Paro R, Boutros M, Merdes G. A combined ex vivo and in vivo RNAi screen for notch regulators in Drosophila reveals an extensive notch interaction network. Dev Cell. 2010a;18:862–876. doi: 10.1016/j.devcel.2010.03.013. [DOI] [PubMed] [Google Scholar]
- Saj A, Arziman Z, Stempfle D, van Belle W, Sauder U, Horn T, Durrenberger M, Paro R, Boutros M, Merdes G. A Combined Ex Vivo and In Vivo RNAi Screen for Notch Regulators in Drosophila Reveals an Extensive Notch Interaction Network. Developmental Cell. 2010b;18:862–876. doi: 10.1016/j.devcel.2010.03.013. [DOI] [PubMed] [Google Scholar]
- Sleckman B, Sherma J. J Liquid Chromatography. 1982a;5 [Google Scholar]
- Sleckman B, Sherma J. A Comparison of Amino-Acid Separations on Silica-Gel, Cellulose, and Ion-Exchange Thin-Layers. J Liquid Chromatography. 1982b;5:1051–1068. [Google Scholar]
- Susan-Resiga D, Essalmani R, Hamelin J, Asselin MC, Benjannet S, Chamberland A, Day R, Szumska D, Constam D, Bhattacharya S, et al. Furin is the major processing enzyme of the cardiac-specific growth factor bone morphogenetic protein 10. J Biol Chem. 2011;286:22785–22794. doi: 10.1074/jbc.M111.233577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk J, Miro J, Strub JM, Lacroix B, van Dorsselaer A, Edde B, Janke C. Polyglutamylation is a post-translational modification with a broad range of substrates. J Biol Chem. 2008;283:3915–3922. doi: 10.1074/jbc.M705813200. [DOI] [PubMed] [Google Scholar]
- Van Vlierberghe P, Ferrando A. The molecular basis of T cell acute lymphoblastic leukemia. The Journal of clinical investigation. 2012;122:3398–3406. doi: 10.1172/JCI61269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wloga D, Dave D, Meagley J, Rogowski K, Jerka-Dziadosz M, Gaertig J. Hyperglutamylation of tubulin can either stabilize or destabilize microtubules in the same cell. Eukaryot Cell. 2010;9:184–193. doi: 10.1128/EC.00176-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wloga D, Gaertig J. Post-translational modifications of microtubules. Journal of cell science. 2010;123:3447–3455. doi: 10.1242/jcs.063727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Gao J, Chung KK, Huang H, Dawson VL, Dawson TM. Parkin functions as an E2-dependent ubiquitin- protein ligase and promotes the degradation of the synaptic vesicle-associated protein, CDCrel-1. Proc Natl Acad Sci U S A. 2000;97:13354–13359. doi: 10.1073/pnas.240347797. [DOI] [PMC free article] [PubMed] [Google Scholar]