Abstract
Importance
Infantile Hypertrophic Pyloric Stenosis (IHPS) is a serious condition in which hypertrophy of the pyloric sphincter muscle layer leads to gastric outlet obstruction. IHPS shows strong familial aggregation and heritability, but knowledge about specific genetic risk variants is limited.
Objective
To search the genome comprehensively for genetic associations with IHPS and validate findings in three independent sample sets.
Design, Setting, and Participants
In stage 1, we used reference data from the 1000 Genomes Project for imputation into a genome-wide dataset of 1001 Danish surgery-confirmed cases (diagnosed between 1987–2008) and 2371 disease-free controls. In stage 2, the five most significantly associated loci were tested in independent case-control sample sets from Denmark (cases diagnosed between 1983–2010), Sweden (cases diagnosed between 1958–2011), and the United States (cases diagnosed between 1998–2005) with a total of 1663 cases and 2315 controls.
Main Outcome Measure
Presence of Infantile Hypertrophic Pyloric Stenosis
Results
We found a new genomewide significant locus for IHPS at chromosome 11q23.3. The most significant SNP at the locus, rs12721025 (odds ratio [OR], 1.59; 95% confidence interval [CI], 1.38–1.83, P = 1.9×10−10), is located 301 bases downstream of the Apolipoprotein A-I (APOA1) gene and is correlated (r2 between 0.46 and 0.80) with SNPs previously found to be associated with levels of circulating cholesterol. For these SNPs, the cholesterol lowering allele consistently conferred increased risk of IHPS.
Conclusions and Relevance
We have identified a new genomewide significant locus for IHPS. Characteristics of this locus suggest the possibility of an inverse relationship between levels of circulating cholesterol in neonates and IHPS risk which warrants further investigation.
INTRODUCTION
Infantile Hypertrophic Pyloric Stenosis (IHPS) is the leading cause of gastrointestinal obstruction in the first months of life, with an incidence of 1–3 per 1000 live births in Western countries.1;2 It affects four to five times as many boys as girls3 and typically presents 2 – 8 weeks after birth4 with projectile vomiting, weight loss, and dehydration.
Although IHPS is a clinically well-defined entity, the etiology of the condition is complex and remains unclear. A genetic predisposition is well established; IHPS aggregates strongly in families and has an estimated heritability of more than 80%.2 However, environmental factors, such as erythromycin exposure5 and feeding practice6;7, have also been implicated in IHPS etiology. Moreover, sharp changes in incidence seen in several countries over the last decades8 underline the importance of modifiable environmental exposures.
Cases of IHPS sometimes occur as part of a syndrome of known genetic etiology9. For example, Smith-Lemli-Opitz syndrome is an autosomal recessive congenital disorder caused by mutations in the 7-dehydrocholesterol reductase (DHCR7; NCBI Entrez Gene 1717) gene.10 Affected individuals are unable to complete the final step in cholesterol biosynthesis causing a wide range of metabolic and developmental abnormalities, including IHPS in 10–15% of cases.10
Less is known about the genetic background of isolated IHPS. Several association studies have focused on genes involved in gastric contractility.11–13 However, these studies were relatively small and produced conflicting results. In contrast, we recently conducted the first genomewide association study of IHPS and identified three susceptibility loci near the muscleblind-like splicing regulator 1 (MBNL1; NCBI Entrez Gene 4154) gene and the NK2 homeobox 5 (NKX2-5; NCBI Entrez Gene 1482) gene.14 Still these variants only explain a small fraction of the variance in disease liability.
The present study followed a 2-stage approach with the aim to identify novel genetic variants for IHPS. In the first (discovery) stage, we used a hypothesis-free approach to identify variants associated with IHPS. In the second stage, we carried forward the most promising variants for validation in three independent case-control sample sets. Finally, we also aimed to investigate the biological relevance of novel genetic loci through follow-up experiments based on prospectively collected plasma samples from cases and controls.
METHODS
PARTICIPANTS
Eligible IHPS cases for the discovery sample were defined as singleton children of Danish ancestry who in their first year of life had a surgery code for pyloromyotomy in the Danish National Patient Registry and did not have any additional major malformations. Eligible controls for the discovery sample were non-affected Danish singleton children who did not have any major malformations. In addition, we excluded severe pregnancy complications from both cases and controls. All Danish samples were drawn from the Danish National Biobank; cases were sampled from dried blood spot samples and controls were sampled from dried blood spots or buffy coats (see the eAppendix for further details).
For the validation stage, we used IHPS case and unaffected control samples from three different countries. For the validation sample from Denmark, we used the same case and control definitions as for the discovery sample. The US sample was obtained from archived, residual newborn blood spots of participants of mostly non-Hispanic white descent delivered by New York State (NYS) residents between 1998–2005. Cases were identified from the population-based NYS Congenital Malformations Registry. Controls were a random sample of all NYS live births delivered during the same time frame. The Swedish validation cases were identified as IHPS patients who had undergone pyloromyotomy at pediatric surgery clinics. Available Swedish controls included healthy middle-aged anonymous blood donors, infants born in 2006, and unaffected relatives of IHPS cases. Swedish cases were sampled from whole blood; controls were sampled from whole blood or placenta.
The results of the genetic study suggested a possible relationship between low plasma lipid levels and risk of IHPS. To investigate this hypothesis, we set up a follow-up study comparing lipid levels in cases and controls. The dried blood spot samples used for genotyping were not suitable for lipid measurements. Instead, we used plasma from prospectively collected umbilical cord blood samples from the Danish National Birth Cohort (DNBC)15. We included all DNBC children who met our IHPS case definition and had adequate amounts of plasma available as cases. To increase statistical power, we sampled four controls for each case. Controls were matched to cases on sex and gestational age at birth and then selected randomly among the DNBC children who were already included as controls in the discovery sample of the genetic study.
The study was approved by the Scientific Ethics Committee for the Capital City Region (Copenhagen) and the Danish Data Protection Agency for the Danish sample. The Scientific Ethics Committee also granted exemption from obtaining informed consent from Danish participants (H-3-2009-093) since the study was based on biobank material. The Ethics Committee at Karolinska Institutet approved the study for the Swedish sample and informed consent was obtained from all Swedish participants. The New York State DOH IRB approved the study and did not require informed consent from the US participants because the samples were de-identified. The study was also approved by the National Institutes of Health Office of Human Subjects Research Protection for the US sample.
GENOTYPING
For the discovery phase, samples were genotyped using the Illumina Human 660W-Quadv1_A Bead Array. After quality control, 529128 SNPs remained available for association and imputation analyses. The Danish validation samples were genotyped at deCODE Genetics using the Centaurus platform (Nanogen) or TaqMan assays (Applied Biosystems). The US samples were genotyped at LGC Genomics using KASP assays, and the Swedish samples were genotyped at Karolinska Institutet using TaqMan assays. See the eAppendix for a detailed description of sampling, genotyping, and quality control.
PLASMA MEASUREMENTS
For the plasma samples, aliquots of 40 ul were prepared, diluted 1:1 with phosphate buffered saline and spectrophotometric measurements were done using the Roche Cobas c 111 Analyzer yielding measurements of circulating LDL, HDL, and Total Cholesterol, as well as Triglycerides. All measurements were conducted at the University of Iowa.
STATISTICAL ANALYSIS
We imputed unobserved genotypes using phased haplotypes from the integrated Phase I release of the 1000 Genomes Project16. (see the eAppendix for imputation details). We used logistic regression to test for differences in allele dosages between cases and controls under an additive genetic model. We carried out combined analysis of the discovery and validation data using the inverse variance method applying genomic control17 to the discovery stage results. We estimated heterogeneity between studies using the I2 statistic.18 We also assessed the robustness of the meta-analysis results by using hierarchical logistic regression as an alternative approach for combined analysis of discovery and replication data. We conducted the genetic analyses overall and stratified by sex and also tested for sex by genotype interaction19. We conditioned on the top SNP at associated loci to explore possible allelic heterogeneity. Family-based association testing was performed using an extended sib transmission/disequilibrium test.20
We evaluated the association between lipid levels in umbilical cord blood and the risk of IHPS by odds ratios (ORs) estimated in a conditional logistic regression using R (http://www.R-project.org/). The analysis took into account matching by sex and gestational age at birth using strata and was adjusted for gestational age. P values were obtained using likelihood ratio tests. We tested for possible nonlinear effects by adding a quadratic term to the model and conducted additional analyses based on lipid level quartiles to allow for nonlinear association.
We used SHAPEIT21, IMPUTE222, and SNPTEST23 software for imputation and association testing. METAL24 and R were used for meta-analysis, and we used PLINK20 for family-based association testing. We used a genomewide significance threshold of P<5×10−8 in the combined analyses of discovery and validation stage results. The lipid measurement study used a significance threshold of P<0.05. All statistical tests were two-sided.
RESULTS
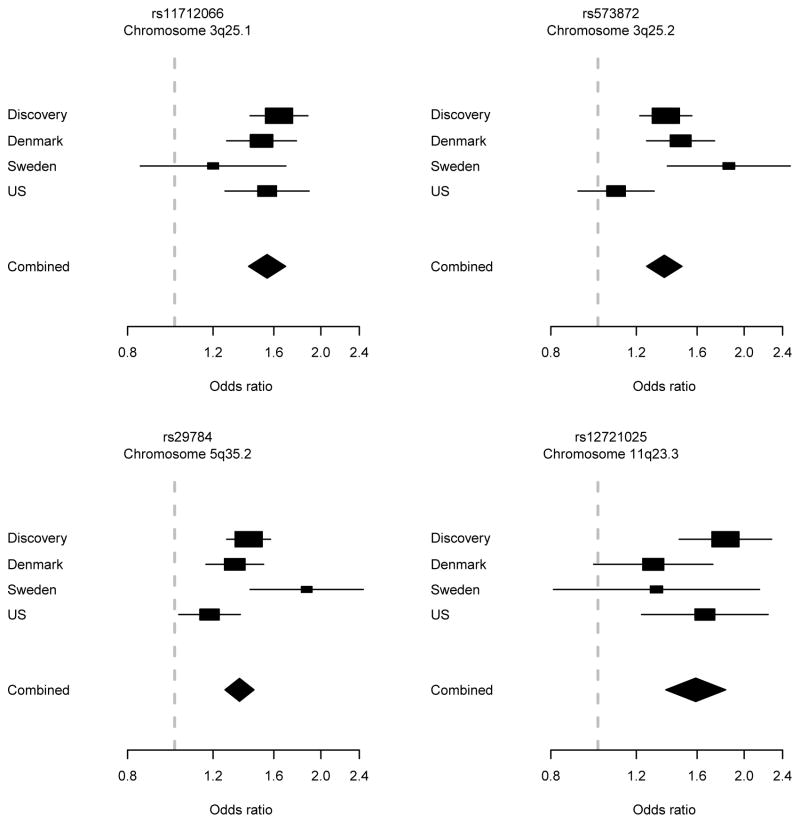
Table 1 shows sample characteristics of the participants contributing to the two stages of the genetic study. In the discovery stage, we analyzed the association between the disease and 9737928 imputed genetic variants in 1001 cases and 2371 controls. Genomic inflation factors were 1.05 in the complete discovery data, and 1.02 and 1.03 in the data restricted to boys and girls, respectively. Imputed SNPs at four loci showed P values <1×10−7 and were selected for further study. These included two novel loci on chromosomes 11q23.3 and 19p13.2, as well as two already confirmed loci on chromosomes 3q25.1 and 5q35.2. The third known locus on chromosome 3q25.2 was also selected for completeness. The chromosomal regions harboring the five selected loci were re-imputed with the original IMPUTE2 algorithm (i.e. without pre-phasing) for increased accuracy, and association tests were repeated for these regions. To confirm the associations at these loci, we genotyped a total of seven SNPs in validation samples from Denmark, Sweden, and the US with a total of 1663 cases and 2315 controls. One novel locus (11q23.3) was validated with genomewide significance (Table 2), the three known loci (3q25.1, 3q25.2, and 5q35.2) were confirmed, and one locus (19p13.2) could not be validated (eTable 1). Results based on hierarchical logistic regression were very similar (eTable 2). Figure 1 displays forest plots for the four genomewide significant loci.
Table 1.
Study Sample Characteristics
| Stage 1 Samples (Discovery)
|
Stage 2 Samples (Validation)
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Denmark | Denmark | United States | Sweden | |||||
| Cases (n=1001) | Controls (n=2371) | Cases (n=796) | Controls (n=879) | Cases (n=738) | Controls (n=697) | Cases (n=129) | Controls (n=742) | |
| Boys, No. (%) | 826 (83) | 1276 (54) | 666 (84) | 412 (47) | 619 (84) | 584 (84) | 110 (85) | 395 (54)a |
| Year of birth, range | 1987–2008 | 1986–2008 | 1983–2010 | 1998–2003 | 1998–2005 | 1998–2005 | 1958–2011 | NAb |
| Year of birth, mean (SD) | 1996 (6) | 2001 (3) | 1990 (7) | 2000 (1) | 2001 (2) | 2001 (2) | 1998 (6) | NAb |
| Maternal Age, mean (SD), years | 28 (5) | 29 (5) | 28 (5) | 24 (2) | 28 (6) | 30 (6) | 28 (5)c | NA |
| Season of birth, No. (%) | ||||||||
| Winter | 247 (25) | 562 (24) | 170 (21) | 192 (22) | 165 (22) | 193 (28) | 34 (26) | |
| Spring | 246 (25) | 566 (24) | 184 (23) | 207 (24) | 196 (27) | 178 (26) | 26 (20) | |
| Summer | 280 (28) | 635 (27) | 242 (30) | 249 (28) | 194 (26) | 170 (24) | 30 (23) | NA |
| Autumn | 228 (23) | 608 (26) | 200 (25) | 231 (26) | 183 (25) | 156 (22) | 39 (30) | |
| Age at diagnosis, mean (SD), days | 37 (18)d | - | 39 (22) | - | NA | - | 41 (19)e | - |
| Case definition | Surgery code for pyloromyotomy (ICD-8 codes 41840, 41841, 44100 up through December 1995; ICD-10 codes KJDH60, KJDH61 from January 1, 1996) in the Danish National Patient Register. | Surgery code for pyloromyotomy (ICD-8 codes 41840, 41841, 44100 up through December 1995; ICD-10 codes KJDH60, KJDH61 from January 1, 1996) in the Danish National Patient Register. | British Pediatric Association code for pyloric stenosis (750.510) in New York State Congenital Malformations Registry. | Pyloromyotomy registered in pediatric surgery clinic records. | ||||
| Sample Types (n) | DBSS (1001) | DBSS (961); Buffy coat (1410) | DBSS (796) | DBSS (879) | DBSS (738) | DBSS (697) | Whole blood (129) | Whole blood (380); Placenta (362) |
| Genotyping Platform | Illumina Human 660W-Quadv1_A Bead Array | Centaurus (Nanogen) or TaqMan (Applied Biosystems) | KASP (LGC Genomics) | TaqMan (Applied Biosystems) | ||||
Abbreviations: DBSS, Dried Blood Spot Samples; ICD-8, International Statistical Classification of Diseases, Eighth Revision; ICD-10, International Statistical Classification of Diseases, Tenth Revision; NA, data not available.
Sex was missing for 14 of the Swedish controls. The percentage is based on the 728 participants with sex data available.
362 of the Swedish controls were infants born in 2006; the remaining 380 were middle aged anonymous blood donors for whom year of birth was not available.
Based on 96 cases with maternal age information available.
Based on 996 cases with age at diagnosis information available.
Based on 101 cases with age at diagnosis information available.
Table 2.
Discovery stage, validation stage and combined results for the novel 11q23.3 locus associated with IHPS.
| chr | SNP | Position (bp) | Alleles | Frequency | Number | Odds Ratio (95% CI) | I2 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Eff | Alt | Sample Set | Cases | Controls | Cases | Controls | P value | (95% CI) | Het P | ||||
| 11 | rs12721025 | 116706047 | A | G | Discovery | 0.0911 | 0.0569 | 1001 | 2371 | 1.83 (1.47–2.28) | 1.0×10−7 | ||
| Denmark | 0.0741 | 0.0582 | 789 | 876 | 1.30 (0.98–1.70) | 0.06 | |||||||
| Sweden | 0.0820 | 0.0634 | 128 | 733 | 1.32 (0.81–2.16) | 0.27 | |||||||
| U.S. | 0.0860 | 0.0538 | 738 | 697 | 1.66 (1.23–2.33) | 7.4×10−4 | |||||||
| Combined | 2656 | 4677 | 1.59 (1.38–1.83) | 1.9×10−10 | 31 (0–75) | 0.23 | |||||||
| 11 | rs77349713 | 116765476 | C | T | Discovery | 0.0896 | 0.0533 | 1001 | 2371 | 1.76 (1.43–2.16) | 1.1×10−7 | ||
| Denmark | 0.0713 | 0.0554 | 793 | 876 | 1.31 (0.99–1.73) | 0.06 | |||||||
| Sweden | 0.0930 | 0.0760 | 129 | 730 | 1.25 (0.78–1.98) | 0.35 | |||||||
| U.S. | 0.0867 | 0.0603 | 738 | 697 | 1.48 (1.11–1.97) | 6.7×10−3 | |||||||
| Combined | 2661 | 4674 | 1.53 (1.33–1.75) | 1.2×10−9 | 21 (0–90) | 0.28 | |||||||
| 11 | rs150758276 | 117085874 | C | G | Discovery | 0.0392 | 0.0188 | 1001 | 2371 | 2.94 (2.00–4.31) | 4.0×10−8 | ||
| Denmark | 0.0269 | 0.0201 | 781 | 872 | 1.35 (0.86–2.12) | 0.19 | |||||||
| Sweden | 0.0310 | 0.0236 | 129 | 741 | 1.32 (0.61–2.89) | 0.48 | |||||||
| U.S. | 0.0373 | 0.0244 | 738 | 697 | 1.55 (1.00–2.39) | 0.05 | |||||||
| Combined | 2649 | 4681 | 1.86 (1.48–2.35) | 1.4×10−7 | 66 (50–88) | 0.03 | |||||||
Eff, effect allele; Alt, alternative allele; Frequency, effect allele frequency; CI, confidence interval; I2, heterogeneity estimate; Het P, P value for Cochran’s Q test of heterogeneity.
Figure 1. Forest plot.
Discovery stage, validation stage, and combined results for the most significant SNP at each of the four genomewide significant IHPS loci.
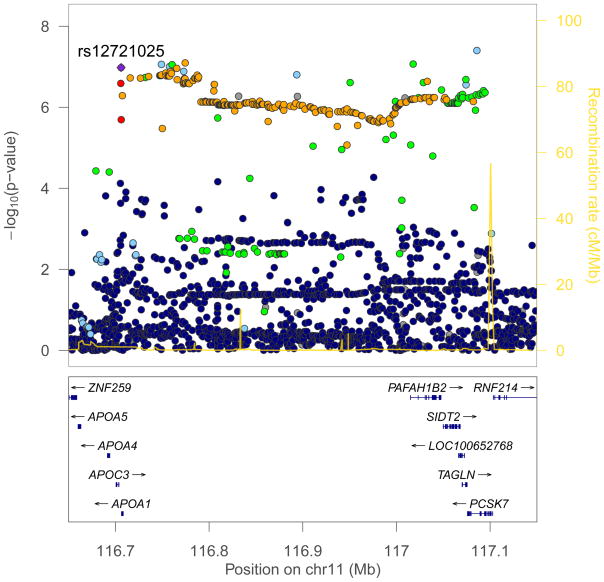
The variant rs12721025 yielded the lowest P value (OR, 1.59; 95% CI, 1.38–1.83; P=1.9×10−10) at the 11q23.3 locus. This SNP is located 301 bases downstream of the Apolipoprotein A-I (APOA1; NCBI Entrez Gene 335) gene with additional apolipoprotein genes APOC3 (NCBI Entrez Gene 345), APOA4 (NCBI Entrez Gene 337), and APOA5 (NCBI Entrez Gene 116519) within 50 kb centromeric (Figure 2). A region of strong linkage disequilibrium extends several hundred kb to the telomeric side; here the variant rs77349713, which is intronic in the salt-inducible kinase 3 (SIK3; NCBI Entrez Gene 23387) gene, was also associated with genomewide significance (OR, 1.53; 95% CI, 1.33–1.75; P=1.2×10−9).
Figure 2. Regional association plot showing imputed SNP results for the novel IHPS locus on chromosome 11q23.3.
SNPs are plotted by chromosomal location (x axis) and association with IHPS (−log10 P value; y axis). The colours reflect the linkage disequilibrium of each SNP with rs12721025 (based on pairwise r2 values from the 1000 genomes project; red, r2>0.8; orange, 0.6<r2<0.8; green, 0.4<r2<0.6; light blue, 0.2<r2<0.4; purple, r2<0.2; grey, r2 data not available). Estimated recombination rates (from HapMap) are plotted to reflect the local LD structure. Genes are indicated in the lower panel of the plot. The figure was generated using LocusZoom (http://csg.sph.umich.edu/locuszoom/).
We explored the possible functional impact of the 11q23.3 associations by considering all 222 genotyped or imputed variants (SNPs and indels) at the locus with P<1×10−6. These were all correlated with rs12721025 (r2 between 0.46 and 0.91) and we found no evidence for allelic heterogeneity (all of these SNPs had P>0.01 when conditioning on rs12721025). Most of the SNPs were intronic in the genes APOA1, SIK3, PAFAH1B2 (NCBI Entrez Gene 5049), SIDT2 (NCBI Entrez Gene 51092), TAGLN (NCBI Entrez Gene 6876), or PCSK7 (NCBI Entrez Gene 9159); see eTable 3. Two SNPs were in exons, but both were synonymous. Fifty out of the 222 SNPs were previously found to be associated with levels of total and/or HDL cholesterol25 with P values down to 5.7×10−7 (results retrieved from http://www.wikigwa.org). For all of these SNPs, the cholesterol lowering allele consistently conferred increased risk of IHPS (eTable 4). A search of the GWAS catalog (http://www.genome.gov/gwastudies) did not reveal any associations to other phenotypes and no associations to gene expression were found in a search of the expression quantitative trait loci (eQTL) browser (http://eqtl.uchicago.edu). However, chromatin immunoprecipitation sequencing (ChIP-seq) data from the ENCODE Consortium (explored using the UCSC genome browser, NCBI build 37, http://www.genome.ucsc.edu/cgi-bin/hgGateway) showed that small islands of histone modification involving mono- and trimethylation of histone H3 on lysine 4 (H3K4me1 and H3K4me3) directly cover rs12721025.
The functional characteristics of the 11q23.3 locus suggest the hypothesis that low levels of circulating lipids in newborns are associated with increased risk of IHPS. We addressed this hypothesis by measuring plasma levels of total, LDL, and HDL cholesterol as well as triglycerides in prospectively collected umbilical cord blood from a set of 46 IHPS cases and 189 controls of Danish ancestry, most of which were also in the discovery sample (see eTable 5 for sample characteristics). eFigure 1 summarizes the distribution of the four biomarkers in cases and controls. Table 3 shows levels of total cholesterol levels in umbilical cord blood plasma for cases and controls overall, and divided into quartiles. The mean total cholesterol levels for 46 cases and 189 matching controls were 65.22 mg/dL and 75.25 mg/dL, respectively. The risk of IHPS was inversely and significantly associated with total cholesterol level with an OR of 0.77 (95% CI, 0.64–0.92; P=0.005) per 10 mg/dL. An omnibus test of differences between quartiles was also significant (P=0.02). Results for LDL and HDL cholesterol and triglycerides are shown in eTable 6. For HDL cholesterol and triglycerides, adding a quadratic term to the analyses indicated non-linear effects (P=0.05 and P=0.004, respectively). For both of these biomarkers there were significant differences between quartiles (P=0.04 and P=0.01, respectively in the omnibus test), and for both quartile 3 had the lowest ORs.
Table 3.
Odds ratios for IHPS in infants according to total cholesterol levels in plasma from umbilical cord blood.
| Cases | Controls | OR (95% CI) | P | P nonlinearity | |||
|---|---|---|---|---|---|---|---|
|
| |||||||
| n | Mean | n | Mean | ||||
| All | 46 | 65.22 | 189 | 75.25 | 0.02b | 0.07d | |
| Quartile 1 (cholesterol level ≤ 58)a | 20 | 48.20 | 46 | 51.00 | 1.00 (reference) | ||
| Quartile 2 (58 < cholesterol level ≤ 70) | 10 | 64.20 | 48 | 65.33 | 0.54 (0.23–1.30) | ||
| Quartile 3 (70 < cholesterol level ≤ 84) | 11 | 76.55 | 45 | 78.31 | 0.51 (0.21–1.21) | ||
| Quartile 4 (84 < cholesterol level) | 5 | 110.4 | 50 | 104.32 | 0.21 (0.07–0.63) | ||
| OR per 10 mg/dL increase | – | – | – | – | 0.77 (0.64–0.92) | 0.005c | |
Quartiles of total cholesterol (mg/dL).
Omnibus test for no difference in ORs between quartiles using conditional logistic regression.
Test for no change in risk per 10 mg/dL increase using conditional logistic regression.
Test for nonlinearity using conditional logistic regression and comparing a model including cholesterol as a quadratic term to a model only including cholesterol as a linear term.
To explore the role of other lipid related variants on IHPS risk, we identified 247 SNPs representing 134 regions reported in the GWAS catalogue to be associated with lipid levels. Only one region on chromosome 19p13.2 was associated with IHPS below a Bonferroni adjusted threshold of P<3.8×10−4. For all SNPs in this region, the cholesterol lowering allele conferred increased risk of IHPS. The region was represented by rs2228671, a synonymous SNP in the LDL receptor gene (LDLR; NCBI Entrez Gene 3949), in our study, but the association was not seen in the validation cohorts (eTable 1).
A subset of 94 familial cases and 93 unaffected relatives in 37 pedigrees was excluded from the Swedish case-control analyses. These data were instead analyzed using family based association testing. This gave very limited statistical power, particularly for the rarer SNPs, and results did not reach statistical significance (eTable 7). All genomewide significant SNPs showed the same direction of effects in the two sexes and there was no interaction between sex and genotype (eTables 8 and 9).
COMMENT
We identified a novel genomewide significant locus for IHPS on chromosome 11q23.3 in a region harboring the apolipoprotein (APOA1/C3/A4/A5) gene cluster and we also confirmed three previously reported loci. The most significant SNP at the new locus, rs12721025, is located immediately downstream of APOA1 and is covered by several different histone modification regions.
APOA1 encodes apolipoprotein A-I which is the major protein component of high density lipoprotein (HDL) in plasma. Furthermore, rs12721025 is correlated with SNPs previously found to be associated with levels of circulating cholesterol. For these SNPs, the cholesterol lowering allele consistently conferred increased risk of IHPS. These findings suggest the hypothesis that low levels of plasma cholesterol in newborns are associated with increased risk of IHPS. We addressed this hypothesis experimentally using prospectively collected umbilical cord blood samples and found lower cholesterol levels at birth in infants who went on to develop IHPS compared with matched controls who did not develop the disease.
IHPS is a prominent clinical feature in many reports of the Smith-Lemli-Opitz syndrome (SLOS), an inborn defect of cholesterol biosynthesis in the gene DHCR7 associated with low cholesterol levels in infants at birth. In one large case series, six of 49 cases with proven DHCR7 mutations had IHPS.26 Also, a large epidemiological study found that SLOS is 150 times more prevalent in IHPS cases than in the general population.3 A study of 10 patients with isolated IHPS and 8 controls found no cholesterol metabolism anomalies, but did find that plasma cholesterol levels were lower in cases compared to controls.27 The study was however too small for firm statistical conclusions and appears not to have been followed up.
A number of previous findings would also be consistent with low lipid levels representing an important risk factor for IHPS. First, the protective effect of female sex could at least partly be due to higher cholesterol levels, since it is well-known that levels of LDL cholesterol and HDL cholesterol are on average higher in newborn girls compared to boys.28 Second, the IHPS risk associated with bottle-feeding6;7 could in part be caused by insufficient lipid levels since bottle-feeding is known to be associated with lower total and LDL cholesterol levels in infancy.29 Finally, the drop in IHPS incidence observed in the 1990s in several countries8;30–33 coincided temporally with increasing percentages of mothers breastfeeding their infants34, suggesting that better nutritional status in infants may have prevented IHPS from developing in a fraction of potential cases.
Other previously reported lipid related variants were not significantly associated with IHPS. However, different loci regulate different aspects of lipid metabolism at different stages over a lifetime, and most lipid genetics studies have used adult participants. Further functional study is clearly needed to illuminate the biological mechanisms underlying our findings. One approach that might be revealing would focus on the essential role of cholesterol in nervous system development35–37 given the deficiencies in enteric innervation seen in pyloric sphincter muscle tissue from IHPS patients38–40.
The previously identified IHPS loci point to candidate genes involved in alternative splicing, cardiac muscle development, and embryonic gut development14, and further investigation is needed of the interplay between these loci and the genetic and biochemical findings reported here.
Strengths of our study include the use of samples from three different populations with a total of more than 2600 cases and 4600 controls. Furthermore, by leveraging 1000 Genomes Project data, we were able to impute and analyze almost 10 million genetic variants in the discovery stage. Also, we were able to perform a follow-up study based on prospectively collected samples that investigated the potential biological relevance of the new IHPS locus.
The study also has limitations. First, our data permit no assertions about putative causal SNPs at the associated loci, something that would require further fine-mapping studies, e.g., through targeted sequencing of affected individuals in families41. Furthermore, it is important to emphasize that our results do not establish a causal link between cholesterol levels and IHPS. If some of the risk is mediated through cholesterol further study is required to assess the relative importance of other genetic and environmental risk factors. Finally, the lipid measurement study was limited by small numbers of cases and also only covered the time right at birth, i.e. several weeks before the condition typically developed in the cases.
In conclusion, we identified a novel genetic locus that associates with IHPS at genomewide significance. Characteristics of this locus suggest the possibility of an inverse relationship between levels of circulating cholesterol in neonates and IHPS risk. Further investigation is required to illuminate the functional significance of the association identified here.
Supplementary Material
Acknowledgments
Funding/Support: The study was supported by grants from the Lundbeck Foundation (R34-A3931), the Novo Nordisk Foundation, the Danish Medical Research Council (271-06-0628), the Swedish Research Council, the Centers for Disease Control and Prevention (5U01DD000492), and the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development. B.F. is supported by an Oak Foundation Fellowship. We thank the participants and their families, as well as the staff involved in recruiting and managing the study groups for their contributions to this study.
Role of the Sponsor: The funding agency had no role in the design and conduct of the study; in the collection, analysis, management, and interpretation of the data; or in the preparation, review, and approval of the manuscript.
Footnotes
Note: We have submitted an abstract based on results from this study to the American Society of Human Genetics meeting, October 22–26, 2013.
Conflict of Interest Disclosures: Statens Serum Institut has filed a priority patent application at the Danish Patent and Trademark Office on the use of genetic profiling to identify newborns at risk of IHPS, which contains subject matter drawn from the work also published here. Dr Feenstra, Mr Geller, and Dr Melbye are listed on the patent application. No other potential conflict of interest relevant to this article was reported.
Online-Only Material: The eAppendix, eFigure 1, and eTables 1 through 9 are available at http://www.jama.com.
Author Contributions: Dr Feenstra had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. Dr Feenstra and Mr Geller contributed equally to the study.
Study concept and design: Feenstra, Geller, Melbye.
Acquisition of data: Feenstra, Geller, Romitti, Baranowska Körberg, Bedell, Krogh, Svenningsson, Caggana, Nordenskjöld, Mills, Murray.
Analysis and interpretation of data: Feenstra, Geller, Carstensen, Romitti, Baranowska Körberg, Fan, Murray, Melbye.
Drafting of the manuscript: Feenstra, Geller, Melbye.
Critical revision of the manuscript for important intellectual content: Feenstra, Geller, Melbye, Carstensen, Romitti, Baranowska Körberg, Bedell, Krogh, Fan, Svenningsson, Caggana, Nordenskjöld, Mills, Murray.
Statistical analysis: Feenstra, Geller, Carstensen, Fan.
Obtained funding: Melbye, Feenstra, Geller, Romitti, Nordenskjöld, Mills, Murray.
Study supervision: Feenstra, Melbye.
Reference List
- 1.Mitchell LE, Risch N. The genetics of infantile hypertrophic pyloric stenosis. A reanalysis. Am J Dis Child. 1993;147:1203–1211. doi: 10.1001/archpedi.1993.02160350077012. [DOI] [PubMed] [Google Scholar]
- 2.Krogh C, Fischer TK, Skotte L, et al. Familial aggregation and heritability of pyloric stenosis. JAMA. 2010;303:2393–2399. doi: 10.1001/jama.2010.784. [DOI] [PubMed] [Google Scholar]
- 3.Schechter R, Torfs CP, Bateson TF. The epidemiology of infantile hypertrophic pyloric stenosis. Paediatr Perinat Epidemiol. 1997;11:407–427. doi: 10.1046/j.1365-3016.1997.d01-32.x. [DOI] [PubMed] [Google Scholar]
- 4.Ranells JD, Carver JD, Kirby RS. Infantile hypertrophic pyloric stenosis: epidemiology, genetics, and clinical update. Adv Pediatr. 2011;58:195–206. doi: 10.1016/j.yapd.2011.03.005. [DOI] [PubMed] [Google Scholar]
- 5.Honein MA, Paulozzi LJ, Himelright IM, et al. Infantile hypertrophic pyloric stenosis after pertussis prophylaxis with erythromcyin: a case review and cohort study. Lancet. 1999;354:2101–2105. doi: 10.1016/s0140-6736(99)10073-4. [DOI] [PubMed] [Google Scholar]
- 6.Pisacane A, de Luca U, Criscuolo L, et al. Breast feeding and hypertrophic pyloric stenosis: population based case-control study. BMJ. 1996;312:745–746. doi: 10.1136/bmj.312.7033.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krogh C, Biggar RJ, Fischer TK, Lindholm M, Wohlfahrt J, Melbye M. Bottle-feeding and the Risk of Pyloric Stenosis. Pediatrics. 2012;130:e943–e949. doi: 10.1542/peds.2011-2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pedersen RN, Garne E, Loane M, Korsholm L, Husby S. Infantile hypertrophic pyloric stenosis: a comparative study of incidence and other epidemiological characteristics in seven European regions. J Matern Fetal Neonatal Med. 2008;21:599–604. doi: 10.1080/14767050802214824. [DOI] [PubMed] [Google Scholar]
- 9.Peeters B, Benninga MA, Hennekam RC. Infantile hypertrophic pyloric stenosis--genetics and syndromes. Nat Rev Gastroenterol Hepatol. 2012;9:646–660. doi: 10.1038/nrgastro.2012.133. [DOI] [PubMed] [Google Scholar]
- 10.Kelley RI, Hennekam RC. The Smith-Lemli-Opitz syndrome. J Med Genet. 2000;37:321–335. doi: 10.1136/jmg.37.5.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Svenningsson A, Lagerstedt K, Omrani MD, Nordenskjold A. Absence of motilin gene mutations in infantile hypertrophic pyloric stenosis. J Pediatr Surg. 2008;43:443–446. doi: 10.1016/j.jpedsurg.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 12.Lagerstedt-Robinson K, Svenningsson A, Nordenskjold A. No association between a promoter NOS1 polymorphism (rs41279104) and Infantile Hypertrophic Pyloric Stenosis. J Hum Genet. 2009;54:706–708. doi: 10.1038/jhg.2009.101. [DOI] [PubMed] [Google Scholar]
- 13.Saur D, Vanderwinden JM, Seidler B, Schmid RM, De Laet MH, Allescher HD. Single-nucleotide promoter polymorphism alters transcription of neuronal nitric oxide synthase exon 1c in infantile hypertrophic pyloric stenosis. Proc Natl Acad Sci U S A. 2004;101:1662–1667. doi: 10.1073/pnas.0305473101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feenstra B, Geller F, Krogh C, et al. Common variants near MBNL1 and NKX2-5 are associated with infantile hypertrophic pyloric stenosis. Nat Genet. 2012;44:334–337. doi: 10.1038/ng.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Olsen J, Melbye M, Olsen SF, et al. The Danish National Birth Cohort--its background, structure and aim. Scand J Public Health. 2001;29:300–307. doi: 10.1177/14034948010290040201. [DOI] [PubMed] [Google Scholar]
- 16.A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 18.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21:1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 19.Magi R, Lindgren CM, Morris AP. Meta-analysis of sex-specific genome-wide association studies. Genet Epidemiol. 2010;34:846–853. doi: 10.1002/gepi.20540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Delaneau O, Marchini J, Zagury JF. A linear complexity phasing method for thousands of genomes. Nat Methods. 2012;9:179–181. doi: 10.1038/nmeth.1785. [DOI] [PubMed] [Google Scholar]
- 22.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marchini J, Howie B. Genotype imputation for genome-wide association studies. Nat Rev Genet. 2010;11:499–511. doi: 10.1038/nrg2796. [DOI] [PubMed] [Google Scholar]
- 24.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Teslovich TM, Musunuru K, Smith AV, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ryan AK, Bartlett K, Clayton P, et al. Smith-Lemli-Opitz syndrome: a variable clinical and biochemical phenotype. J Med Genet. 1998;35:558–565. doi: 10.1136/jmg.35.7.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hennekam RC, Waterham HR, Wanders RJ, Aronson DC. No cholesterol metabolism anomalies detectable in infants with hypertrophic pyloric stenosis. Am J Med Genet. 2001;99:256–257. doi: 10.1002/1096-8628(2001)9999:9999<::aid-ajmg1148>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 28.Carlson LA, Hardell LI. Sex differences in serum lipids and lipoproteins at birth. Eur J Clin Invest. 1977;7:133–135. doi: 10.1111/j.1365-2362.1977.tb01585.x. [DOI] [PubMed] [Google Scholar]
- 29.Owen CG, Whincup PH, Odoki K, Gilg JA, Cook DG. Infant feeding and blood cholesterol: a study in adolescents and a systematic review. Pediatrics. 2002;110:597–608. doi: 10.1542/peds.110.3.597. [DOI] [PubMed] [Google Scholar]
- 30.Nielsen JP, Haahr P, Haahr J. Infantile hypertrophic pyloric stenosis. Decreasing incidence. Dan Med Bull. 2000;47:223–225. [PubMed] [Google Scholar]
- 31.Persson S, Ekbom A, Granath F, Nordenskjold A. Parallel incidences of sudden infant death syndrome and infantile hypertrophic pyloric stenosis: a common cause? Pediatrics. 2001;108:E70. doi: 10.1542/peds.108.4.e70. [DOI] [PubMed] [Google Scholar]
- 32.Sommerfield T, Chalmers J, Youngson G, Heeley C, Fleming M, Thomson G. The changing epidemiology of infantile hypertrophic pyloric stenosis in Scotland. Arch Dis Child. 2008;93:1007–1011. doi: 10.1136/adc.2007.128090. [DOI] [PubMed] [Google Scholar]
- 33.de Laffolie J, Turial S, Heckmann M, Zimmer KP, Schier F. Decline in infantile hypertrophic pyloric stenosis in Germany in 2000–2008. Pediatrics. 2012;129:e901–e906. doi: 10.1542/peds.2011-2845. [DOI] [PubMed] [Google Scholar]
- 34.Yngve A, Sjostrom M. Breastfeeding in countries of the European Union and EFTA: current and proposed recommendations, rationale, prevalence, duration and trends. Public Health Nutr. 2001;4:631–645. doi: 10.1079/phn2001147. [DOI] [PubMed] [Google Scholar]
- 35.Mauch DH, Nagler K, Schumacher S, et al. CNS synaptogenesis promoted by glia-derived cholesterol. Science. 2001;294:1354–1357. doi: 10.1126/science.294.5545.1354. [DOI] [PubMed] [Google Scholar]
- 36.Fan QW, Yu W, Gong JS, et al. Cholesterol-dependent modulation of dendrite outgrowth and microtubule stability in cultured neurons. J Neurochem. 2002;80:178–190. doi: 10.1046/j.0022-3042.2001.00686.x. [DOI] [PubMed] [Google Scholar]
- 37.Funfschilling U, Jockusch WJ, Sivakumar N, et al. Critical time window of neuronal cholesterol synthesis during neurite outgrowth. J Neurosci. 2012;32:7632–7645. doi: 10.1523/JNEUROSCI.1352-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Langer JC, Berezin I, Daniel EE. Hypertrophic pyloric stenosis: ultrastructural abnormalities of enteric nerves and the interstitial cells of Cajal. J Pediatr Surg. 1995;30:1535–1543. doi: 10.1016/0022-3468(95)90151-5. [DOI] [PubMed] [Google Scholar]
- 39.Vanderwinden JM, Liu H, De Laet MH, Vanderhaeghen JJ. Study of the interstitial cells of Cajal in infantile hypertrophic pyloric stenosis. Gastroenterology. 1996;111:279–288. doi: 10.1053/gast.1996.v111.pm8690192. [DOI] [PubMed] [Google Scholar]
- 40.Piotrowska AP, Solari V, Puri P. Distribution of heme oxygenase-2 in nerves and interstitial cells of Cajal in the normal pylorus and in infantile hypertrophic pyloric stenosis. Arch Pathol Lab Med. 2003;127:1182–1186. doi: 10.5858/2003-127-1182-DOHOIN. [DOI] [PubMed] [Google Scholar]
- 41.Cirulli ET, Goldstein DB. Uncovering the roles of rare variants in common disease through whole-genome sequencing. Nat Rev Genet. 2010;11:415–425. doi: 10.1038/nrg2779. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.