Abstract
Major histocompatibility complex (MHC) molecules play a central role in the immune response and in the recognition of non-self. Found in all jawed vertebrate species, including zebrafish and other teleosts, MHC genes are considered the most polymorphic of all genes. In this review we focus on the multi-faceted diversity of zebrafish MHC class I genes, which are classified into three sequence lineages: U, Z, and L. We examine the polygenic, polymorphic, and haplotypic diversity of the zebrafish MHC class I genes, discussing known and postulated functional differences between the different class I lineages. In addition, we provide the first comprehensive nomenclature for the L lineage genes in zebrafish, encompassing at least 15 genes, and characterize their sequence properties. Finally, we discuss how recent findings have shed new light on the remarkably diverse MHC loci of this species.
Keywords: immunity, multigene families, haplotype, polymorphism, histocompatibility, MHC
1. Introduction
Originally identified as a genetic locus responsible for transplant rejection between congenic mouse strains (Klein 2001; Snell et al. 1951), Major Histocompatibility Complex (MHC) loci have been well studied in many species. With a high gene density, the MHC locus is one of the most polymorphic regions in mammalian genomes (Klein 2001; Kumanovics et al. 2003;Takada et al. 2003). The human MHC locus comprises a 4 Mb section of chromosome 6 containing hundreds of genes (MHC Sequencing Consortium, 1999;Shiina et al. 2009) including the MHC class I and class II genes, which encode molecules that present peptide antigens to T cells and play a central role in distinguishing between self and non-self (Chaplin 2010). These transmembrane proteins have been found in all jawed vertebrate species examined including cartilaginous fishes (Kulski et al. 2002;Okamura et al. 1997;Trowsdale 1995).
Levels of sequence diversity found between MHC genes and their alleles are considered the highest of any genes in the vertebrate genome (Vandiedonck and Knight 2009). Evidence for overdominance has been found within MHC loci favoring heterozygosity and is likely associated with response to diverse pathogens (Hughes and Nei 1988). MHC genes also play a fundamental role in allograft recognition (Thorsby 2009), and matching at MHC loci between donors and recipients is important to ensure successful transplant outcomes by minimizing acute graft rejection and/or graft-versus-host disease after bone marrow transplantation (reviewed by Groth et al. 2000). Human donor and recipient matching at the most polymorphic classical MHC genes may have a disproportionate impact on transplantation success (Horan et al. 2012). MHC loci are also considered model regions for genomics research in large part due to the association of the MHC with a number of different diseases (Trowsdale and Knight 2013;Vandiedonck and Knight 2009).
As a vertebrate model species, the zebrafish (Danio rerio) has several advantages over the more traditional mouse model including higher fecundity, smaller size, more rapid development, external fertilization, and optical clarity at the embryonic and larval stages. Because of these advantages, zebrafish are routinely employed for studies of in vivo immune function (van der Vaart et al. 2012), host-pathogen interactions (Kanther and Rawls 2010;Tobin et al. 2012), and cell migration (Ignatius and Langenau 2011;Renaud et al. 2011), as well as transplantation assays, particularly for the study of hematopoietic stem cells (de Jong et al. 2011;Li et al. 2011;Taylor and Zon 2009). However their use in this type of assay highlights a current limitation: attempts at generating inbred zebrafish lines met with only limited success (Shinya and Sakai 2011), resulting in standard laboratory “lines” of zebrafish harboring a multitude of polymorphisms as well as haplotypic variation especially at immune loci (Howe et al. 2013;Patowary et al. 2013). As immunologically compatible zebrafish donors and recipients are not readily available, genotyping strategies have been and are being developed for identifying MHC-matched zebrafish. However, the number of described MHC class I loci in the zebrafish genome is growing (Dirscherl and Yoder 2014;McConnell et al. 2014). In this review we will summarize the current knowledge on the polygenic, polymorphic and haplotypic nature of the zebrafish MHC class I gene families. We will also provide an overview on the data implicating zebrafish MHC class I genes in immune function.
2. Classical and nonclassical MHC class I molecules
Classical function of MHC class I molecules is defined as presentation of peptide antigens to CD8+ T cells in order to initiate an immune response. In mammals, only a subset of MHC class I genes is associated with classical function, as some MHC molecules interact with other immune cells, present non-peptide antigens, or have non-immune functions (Parham 2005; Rodgers and Cook 2005). Classical MHC class I genes have extremely high levels of polymorphism, often found as specific patterns of substitutions in the MHC molecules particularly among residues associated with the peptide binding site (Bjorkman et al. 1987). Additional characteristics of classical MHC molecules include conservation of peptide anchor residues, conserved structural residues and conserved surface residues associated with binding sites with other molecules such as beta-2-microglobulin (β2M) and CD8 (Hee et al. 2013). Presentation of diverse processed intracellular peptides by classical MHC class I molecules at the cell surface allows their interrogation by the immune system, and thus T cells become activated upon recognizing these MHC-presented allo-antigens to help initiate an immune response (Bjorkman and Parham 1990;reviewed in Klein and Sato 2000).
The nonclassical MHC genes are frequently at least an order of magnitude less polymorphic than their classical counterparts and are often expressed in limited tissues (Shiina et al. 2009). Interestingly, some MHC molecules considered classical, such as HLA-C, also have nonclassical characteristics, and are thus capable of initiating an immune response by more than one mechanism (Colonna et al. 1993). It has been hypothesized that classical MHC genes are periodically duplicated within the genome (Klein et al. 2007;Nei and Rooney 2005) and thus over time may be free to be selected upon to acquire diverse roles (Flajnik and Kasahara 2001). Of note, a large fraction of the nonclassical MHC genes are not found within the core MHC locus but are instead scattered on additional chromosomes in humans (Horton et al. 2004) and many other vertebrate species. This scattering of class I genes outside of the core MHC locus also applies to zebrafish (Dirscherl and Yoder 2014;McConnell et al. 2014;Sambrook et al. 2005).
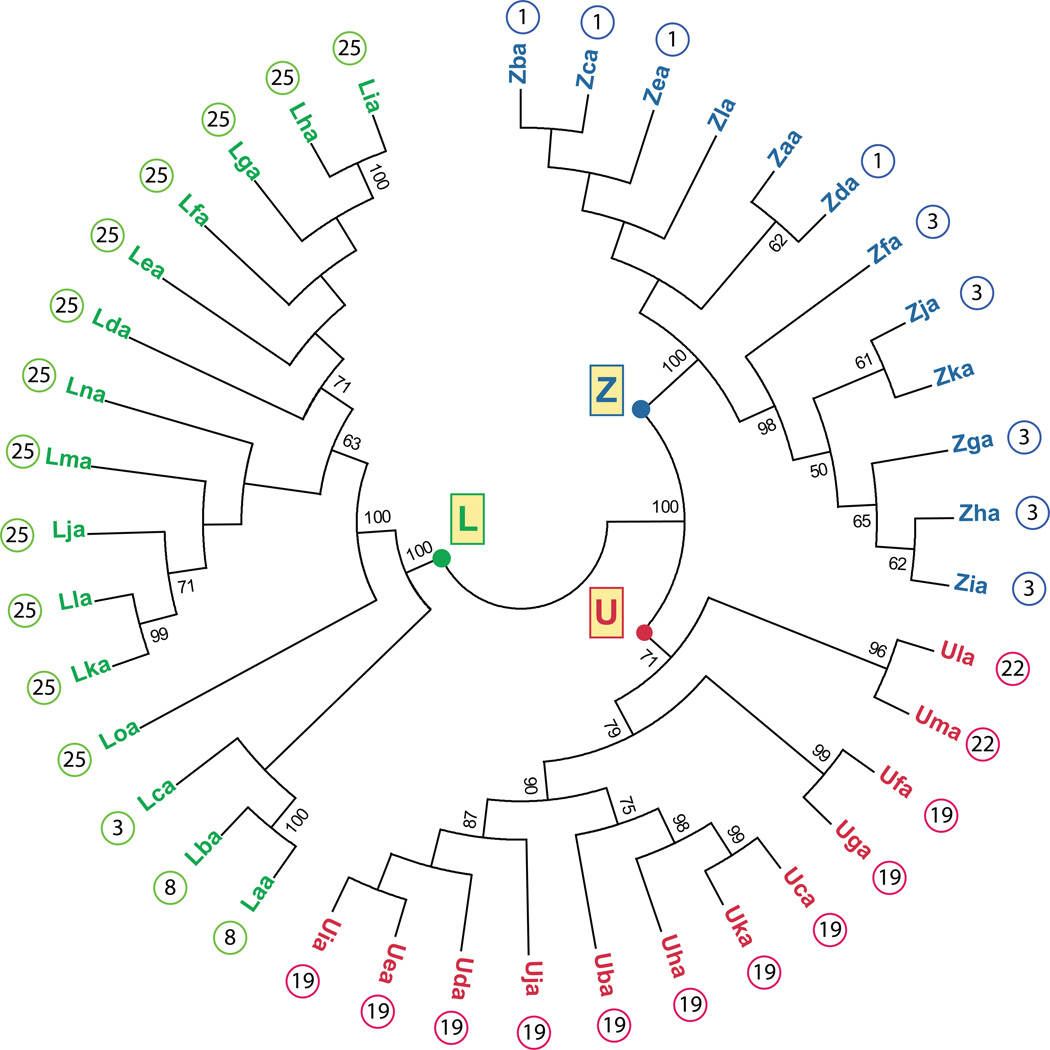
In zebrafish, three MHC class I lineages have been described: U, Z and L. The U lineage genes function as classical MHC class I genes in zebrafish whereas the L lineage genes likely represent nonclassical MHC class I genes. The zebrafish Z lineage genes cannot be currently classified as either classical or nonclassical MHC class I genes. A phylogenetic analysis of all identified zebrafish MHC class I proteins (α1 – α3 domains) illustrates their classification into these three lineages and highlights their subgrouping based on chromosomal location (Figure 1). The evidence supporting the classification of these genes is discussed below.
Figure 1. Phylogenetic analysis of zebrafish MHC class I proteins.
A neighbor-joining tree was constructed from an alignment of the α1 – α3 domains from MHC class I U, Z and L lineages. The analysis of U sequences included Uba through Ula as previously described (McConnell et al. 2014) as well as Uma (GenBank: XP_005161940.1). The analysis of Z sequences included Zaa through Zla described previously (Dirscherl and Yoder 2014). The analysis of L sequences included Laa through Loa as described in Table 1. Mhc1uaa and additional putative L sequences were excluded as they did not include complete α1-α3 domains. If known, the chromosomal assignment for each gene is shown next to the protein symbol. The consensus tree was constructed using MEGA5 and employed 2000 bootstrap replications (Tamura et al. 2011): bootstrap values less than 50 are not shown.
3. Nomenclature and history of MHC class I lineages
Based on guidelines proposed for all species (Klein et al. 1990), the current zebrafish MHC class I gene nomenclature reflects the nomenclature system in Atlantic salmon (Salmo salar; Lukacs et al. 2010) and has been approved by the zebrafish nomenclature committee (http://zfin.org/). If the zebrafish “mhc1uaa” gene symbol is dissected as an example, the “mhc1” is included to define the sequence as class I. The first letter “u” (or “z” or “l”) corresponds to the MHC class I lineage. The variable second letter “a” (or “b” or “c” etc.) designates the gene, named alphabetically in order of discovery. The third letter “a” indicates the alpha chain. For MHC class II genes that form heteromeric complexes of an alpha and a beta chain, the beta chain gene is designated with “b”. When zebrafish MHC class I genes are compared to sequences from other species, then the inclusion of a species identifier is crucial. In this case, the four letter prefix, Dare (which refers to the first two letters of the genus and species, Danio rerio), is added to the gene symbol (Dare-mhc1uaa).
3.1 The classical U lineage genes
The first description of a classical MHC class I transcript from any teleost was the cloning of salmon UBA (Grimholt et al. 1993). Named for “uno”, meaning “one”, other U lineage genes and transcripts have since been described in numerous teleost species including zebrafish (Clark et al. 2001;Figueroa et al. 2001;Lukacs et al. 2010;Malmstrom et al. 2013;Nonaka et al. 2011;Sato et al. 1997;Sato et al. 1998;Shum et al. 2001;Stet et al. 2003;Takeuchi et al. 1995;van Erp et al. 1996).
Most zebrafish U genes are located in the core MHC locus found on chromosome 19, a region syntenic with the mammalian MHC core locus including several additional class I pathway genes important for antigen processing and transport, such as abcb3/tap2, psmb8-11, and tapbp. Unlike the mammalian MHC core locus, teleost classical MHC class I and class II genes are unlinked, with the zebrafish class II genes present on chromosome 8 (Bingulac-Popovic et al. 1997;Dijkstra et al. 2013). This arrangement has been hypothesized to facilitate distinct forms of selection on class I and class II genes in teleosts (Shum et al. 2001). Low gene density of the class II locus on chromosome 8 and the presence of genes unlinked to the mammalian MHC locus have been interpreted as evidence of separation by chromosomal translocation rather than duplication (Kuroda et al. 2002), but additional analysis is needed to support this hypothesis (Dijkstra et al. 2007).
Recently we have demonstrated that the zebrafish core MHC locus at chromosome 19 exhibits extensive haplotypic variation, distributing at least ten distinct U genes among six divergent haplotypes (McConnell et al. 2014). Genomic sequence is available for only five of the ten zebrafish U genes identified: mhc1uda, mhc1uea and mhc1ufa on Haplotype A, and mhc1uba and mhc1uca on Haplotype B. The remaining five genes are unmapped to the current reference genome (Zv9), having been identified from BLAST searches of cDNA libraries. Furthermore, two additional MHC class I genes can be found on chromosome 22. While they are phylogenetically distinct from the chromosome 19 genes (Figure 1), they fall within the branch of the U lineage and have been named mhc1ula (GenBank XM_001336713.5) and mhc1uma (GenBank XM_005161883.1).
3.2 A brief history of the Z lineage genes
The Z lineage genes in zebrafish have historically been referred to as ZE lineage genes; however, the zebrafish nomenclature committee (http://zfin.org/) recently agreed to employ a nomenclature similar to that used for the zebrafish U genes as well as the Z gene nomenclature in Atlantic salmon (Dirscherl and Yoder 2014). In order to understand the history of the Z gene nomenclature in zebrafish one needs to first understand the origins of the nomenclature in carp. The first Z lineage genes to be reported were described from common carp (Cyprinus carpio) and ginbuna crucian carp (Carassius auratus langsdorfii) and named ZA, ZB, ZC, and ZD (e.g. Cyca-ZA1 etc.) (Hashimoto et al. 1990;Okamura et al. 1993). Approximately ten years later, an additional MHC class I gene was described from zebrafish, common carp, and barbus (Barbus intermedius), that is phylogenetically distinct from the U and ZA/ZB/ZC/ZD gene lineages. This gene shares more similarity with the ZA/ZB/ZC/ZD sequences than the U lineage sequences, and thus was named ZE (Kruiswijk et al. 2002). However, differences between ZE and the ZA/ZB/ZC/ZD genes soon became apparent, as the ZA/ZB/ZC/ZD genes appear to be restricted to carp species and are not present in the zebrafish genome (Hashimoto et al. 1990;Okamura et al. 1993). Also, while the ZA/ZB/ZC/ZD genes have properties consistent with nonclassical MHC genes, ZE genes are distinguished by having features of both classical and nonclassical MHC genes. Since the only MHC class I Z genes identified outside of carp species are the ZE genes, the zebrafish ZE genes are now referred to simply as Z genes in accordance with MHC class I nomenclature guidelines.
The initial full-length Z transcripts from zebrafish were named Dare-ZE*0101 and Dare-ZE*0102 not knowing if they represented different genes or different alleles of a single gene (Kruiswijk et al. 2002). However, it was clear then that these sequences were likely not derived from a single copy gene as 14 unique Z sequences were amplified from only five individual zebrafish (Kruiswijk et al. 2002). Further evidence of the Z lineage being a multi-gene family came in 2005 when a genome-wide survey of zebrafish MHC loci placed one full length Z gene on an unplaced scaffold as well as four similar genes on chromosome 1 (Zv4) but did not provide sequence data (Sambrook et al. 2005).
We recently reported that the zebrafish reference genome (Zv9) encodes nine unique Z genes on chromosomes 1 and 3 (Dirscherl and Yoder 2014). Three additional Z genes have been identified from other genomic sequences, including those corresponding to the two initial full-length transcripts Dare-ZE*0101 and Dare-ZE*0102 (Kruiswijk et al. 2002) that are now renamed Dare-mhc1zaa and Dare-mhc1zla, respectively (Dirscherl and Yoder 2014). Although the original carp ZA, ZB, ZC and ZD genes have been characterized as nonclassical MHC class I genes and zebrafish Z sequences share some of these nonclassical features (reviewed in detail by Stet et al. 1998), the zebrafish Z genes currently defy classification as either classical or nonclassical MHC class I genes. This is due to the fact that their extracellular α1 and α2 domains (which would be polymorphic for a diverse peptide-binding repertoire) are relatively well conserved yet they possess conserved residues thought to be important for peptide binding and generally exhibit ubiquitous expression. It has been suggested that the Z genes may represent an intermediate functional category of MHC class I molecules (Stet et al. 2003). However, functional data are needed to accurately determine their classification.
3.3 The nonclassical L lineage genes
A third lineage of MHC class I genes in teleosts was first reported in 2007 and designated the L lineage due to the linkage of at least one of its members with MHC class II (Dijkstra et al. 2007). This linkage of MHC class I and class II genes in zebrafish was in contrast to the previously described absence of linkage between these genes in teleosts (Bingulac-Popovic et al. 1997). While this lineage was most extensively characterized in rainbow trout (Oncorhynchus mykiss), L gene sequences were also identified in zebrafish, Atlantic salmon and fathead minnow (Pimephales promelas) but not in species outside the teleosts (Dijkstra et al. 2007).
The MHC class I L genes are characterized as being nonclassical primarily due to the fact that the molecules they encode lack most of the residues thought to be important for peptide-termini binding. Other features that define this lineage are the lack of two tryptophan residues (W51 and W60) in the α1 domain that are otherwise highly conserved in MHC class I molecules (although four of the twelve zebrafish U lineage genes have W60D substitutions) and a HINLTL motif in the α3 domain (see section 5.3) (Dijkstra et al. 2007). When this motif was used to identify potential MHC class I L lineage proteins from other species (via BLASTp), sequences from cichlid species were readily identified including Haplochromis burtoni (GenBank: XP_005935938.1), Oreochromis niloticus (GenBank: XP_005462427.1), and Maylandia zebra (GenBank: XP_004572211.1). Phylogenetic analysis of the alpha domains from these three genes verifies that they group with the zebrafish L lineage genes (Dirscherl and Yoder, data not shown).
Two full-length and four partial MHC class I L lineage transcripts have been reported from zebrafish (Dijkstra et al. 2007). A search of the zebrafish genome in 2007 (Zv6) with characteristic L gene α3 sequences identified 24 partial L genes: 20 that mapped to a 450 kb stretch of chromosome 25, three that mapped to a 30 kb stretch of chromosome 8, and one gene that mapped to chromosome 3; however, specific sequences were not reported for most of these genes (Dijkstra et al. 2007). A comparison of one full length zebrafish L sequence (GenBank: NM_001017904.1) to the current zebrafish reference protein database (BLASTp) identified fifteen unique L proteins that include α1-α3 domains: one on chromosome 3, two on chromosome 8, and 12 on chromosome 25. Table 1 summarizes the GenBank accession numbers for these proteins, the proposed nomenclature for the genes encoding the proteins, and the previous names of the protein products.
Table 1.
Zebrafish MHC class I L gene nomenclature
| Gene | Genomic Location |
Previous Name |
Genbank Accession Nos. |
|---|---|---|---|
| mhc1laa | Chr8 | zgc:113060 | XM_005167247.1; NM_001017904.1 |
| mhc1lba | Chr8 | LOC100150607 | XM_001920752.3 |
| mhc1lca | Chr3 | LOC100000140 | XM_001340377.4 |
| mhc1lda | Chr25 | LOC100149810 | XM_001920255.4 |
| mhc1lea | Chr25 | LOC100150713 | XM_001920310.3 |
| mhc1lfa | Chr25 | LOC100148359 | XM_001920284.3 |
| mhc1lga | Chr25 | LOC100150525 | XM_001920246.4 |
| mhc1lha | Chr25 | LOC100535946 | XM_005174295.1 |
| mhc1lia | Chr25 | LOC100151195 | XM_001920233.2 |
| mhc1lja | Chr25 | LOC100150018 | XM_001920322.3; XM_005161517.1 |
| mhc1lka | Chr25 | LOC100334758 | XM_002666795.2 |
| mhc1lla | Chr25 | LOC100536492 | XM_003201408.2 |
| mhc1lma | Chr25 | LOC101887116 | XM_005174300.1 |
| mhc1lna | Chr25 | LOC101883841 | XM_005161519.1 |
| mhc1loa | Chr25 | LOC101886720 | XM_005174294.1 |
Dijkstra et al. reported two overlapping zebrafish BAC clones showing the linkage of an L lineage MHC class I gene (GenBank: CAD56801.1) to several MHC class IIα and class IIβ genes on chromosome 8 (2007). Although this L gene doesn’t match any of the predicted L genes identified in the current draft of the reference genome, the two L genes encoded on chromosome 8 (mhc1laa and mhc1lba, Zv9_scaffold1130, GenBank: NW_001879369.3) are indeed adjacent to a class II histocompatibility gene (GenBank: XP_699949.2) that corresponds to one of the class II genes reported by Djikstra et al. (GenBank: CAD32273.1). While there is not sufficient evidence in the flanking sequences to conclude that these genomic sequences represent alternative haplotypes, this further supports the linkage of MHC class I L lineage genes to MHC class II genes on chromosome 8.
3.4 MHC class I S lineage genes are absent from zebrafish
A fourth MHC class I lineage in bony fish is referred to as the S lineage as it was first described in salmonid species, including Atlantic salmon and rainbow trout (Lukacs et al. 2010;Shum et al. 1999), although a related cDNA also has been identified from channel catfish (Ictalurus punctatus) which is a siluriforme species (Dijkstra et al. 2007). Atlantic salmon appear to encode a single S lineage gene, SAA (previously named UAA) (Lukacs et al. 2010). A BLASTp search of the non-redundant protein database using the α1-α3 domains of Atlantic salmon SAA as a query (GenBank: ACY30362) identified nearly identical sequences from other salmonids (95–100% identity; E values 6e-122 to 0.0). However, the sequences from a cyprinid species with the highest similarity to SAA are the grass carp UAA and UBA sequence (51% identity, E value 1e-42). In addition, tBLASTn searches of the zebrafish reference genome (Zv9) with SAA as a query identified the two MHC class I U genes on chr 22 (mhc1ula and mhc1uma) as the most similar to salmon SAA (30–31% identity; E values 4e-32 to 9e-33). These observations strengthen the argument that S lineage genes are absent in cyprinids such as zebrafish and carp.
4. MHC class I loci are in highly variable genomic regions
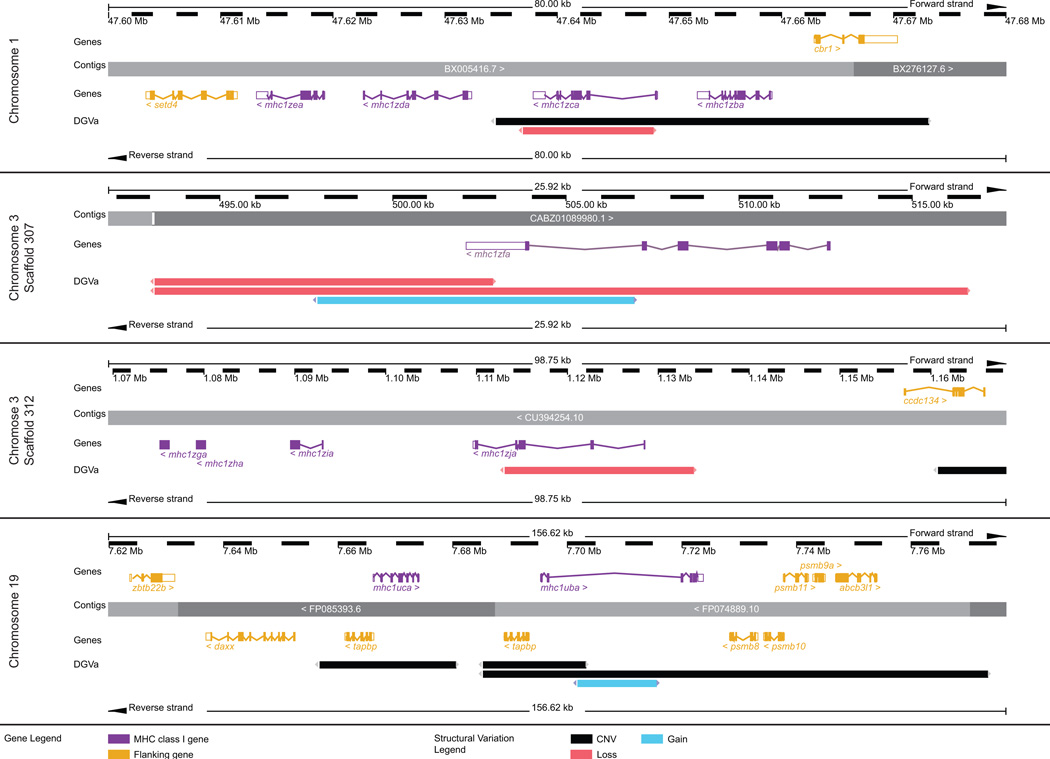
The different MHC class I lineages are encoded across multiple chromosomes: the U genes are encoded on chromosomes 19 and 22, the Z genes are encoded on chromosomes 1 and 3, and the L genes are encoded on chromosomes 3, 8, and 25. As shown in Figure 2, a common feature of these MHC class I loci is that they are encoded in regions of high copy number variation (CNV). While the zebrafish reference genome has been sequenced, analysis of its polymorphic and structural variation is only beginning. An evaluation of copy number variation (CNV) between the genomes of 80 individual zebrafish revealed 6,080 CNV elements encompassing 14.6% of the zebrafish reference genome (Brown et al. 2012). In addition, a comparison of the genomic sequence of a wild caught zebrafish to the reference genome revealed 5.2 million single nucleotide variations and over 1.6 million insertion-deletion variations (Patowary et al. 2013). This type of variation can significantly influence phenotypic responses so knowledge of this variation and the ability to define the variation between experimental animals is critical for accurately interpreting experimental findings, especially when these findings may be translated to human disease applications. Figure 3 shows that the zebrafish U and Z loci are encoded in genomic regions with increased levels of gene loss, gain, or copy number variation (as identified by Brown et al. 2012). As discussed further in Section 6, the high degree of CNV in the MHC class I loci is evidence that some of these genes may be encoded by only a subset of the different haplotypes.
Figure 2. Genomic loci of zebrafish MHC class I genes.

The location of each zebrafish MHC class I locus is shown in relation to a published chromosomal map of copy number variation (CNV). 31,749 CNVs found in 80 individual zebrafish were combined into a non-redundant dataset of 6,080 CNV elements (CNVEs) (Brown et al. 2012). Lengths of green (CNV gain) and red (CNV loss) horizontal lines reflect relative CNVE frequencies at respective chromosomal locations. CNV image courtesy of Charles Lee, The Jackson Laboratory Institute for Genomic Medicine.
Figure 3. Evidence for haplotypic variation at the MHC class I U and Z lineage loci.
The regions of chromosomes 1 and 3 that encode the Z genes and the region of chromosome 19 that encodes the U genes are shown in detail (adapted from the Ensembl Genome Browser, www.ensembl.org/Danio_rerio/). The MHC class I genes (purple) and relevant flanking genes (yellow) are annotated above and below the genomic contigs. The Database of Genomic Variants archive (DGVa) provides the genomic structural variants: regions of copy number variation (CNV) are indicated in black, regions of gene loss are indicated in red, and regions of gene gain are indicated in blue.
5. Sequence diversity
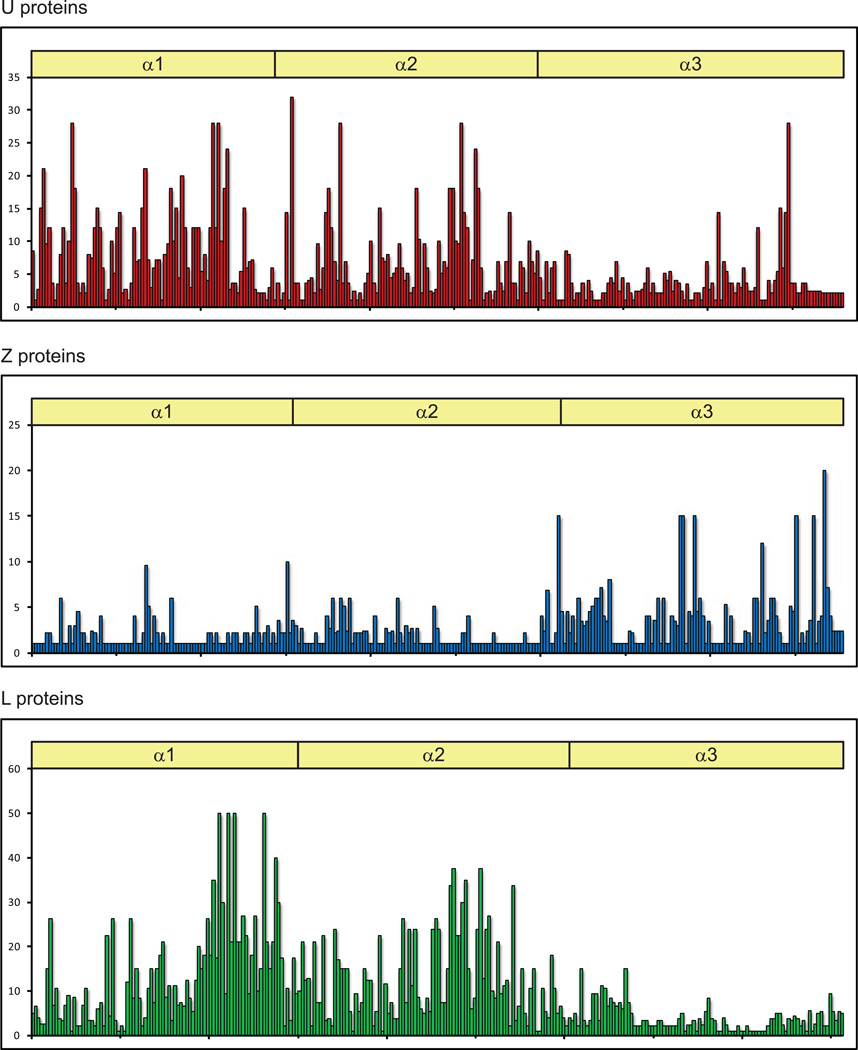
A characteristic that helps to define each MHC class I lineage is the differential variability of its α domains. Figure 4 shows variability plots based on the amino acid alignments of the α1 to α3 domains of the molecules associated with each MHC class I lineage, the same molecules incorporated into the phylogenetic tree seen in Figure 1. The classical U proteins have the majority of their variability concentrated in the α1 and α2 domains, with a few highly variable residues likely important for determining ligand specificity as is the case with mammalian classical MHC class I proteins. The L proteins also have α1 and α2 domains that are much more variable than their α3 domains, yet this variability is more evenly distributed across the two alpha domains suggesting that these molecules perform functions other than peptide presentation. The Z proteins show the opposite trend of having variable α3 domains and relatively well conserved α1 and α2 domains. This is difficult to reconcile with the fact that the Z proteins exhibit conserved residues important for binding peptide and calls for functional data to explain the effect of the α3 variability.
Figure 4. Variability plots for all three lineages of MHC class I proteins.
Variability plots are based on the ClustalW alignments of the α1 – α3 amino acid sequences (X axis) of the same MHC class I proteins included in Figure 1. The analysis is of representative zebrafish proteins excluding polymorphic data on individual protein sequences. Variability (y axis) was calculated using the Wu-Kabat method (Protein Variability Server, http://imed.med.ucm.es/PVS/).
5.1 Sequence diversity of U genes
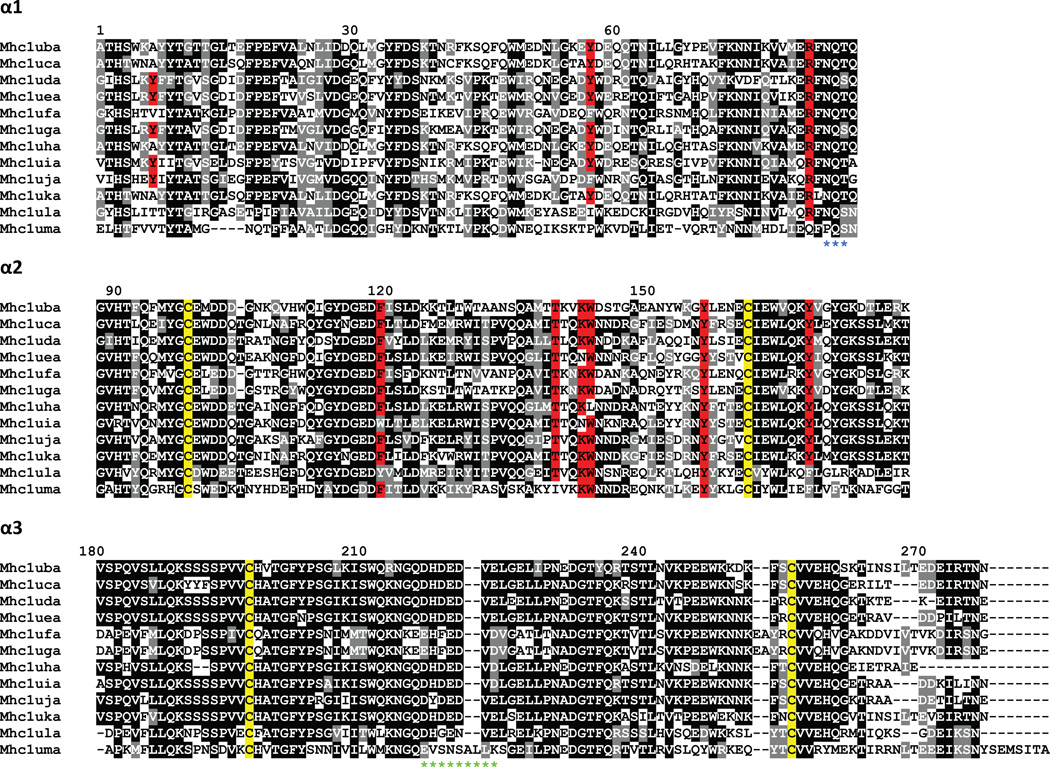
The level of sequence diversity between zebrafish class I U lineage genes (McConnell et al. 2014;Takeuchi et al. 1995) is similar to the level of diversity observed between alleles of the highly polymorphic UBA gene in salmonids (Aoyagi et al. 2002;Shum et al. 2001). Like the salmonid genes, the zebrafish U lineage genes have the most diversity in the α1 and α2 domains, but also maintain some diversity in the α3 domain (Figure 5; McConnell et al. 2014). Many of the presumed classical zebrafish class I U lineage genes have specific polymorphisms found within their otherwise well-conserved peptide anchor residues. For example, Dare-Uha has two predicted peptide anchor residue substitutions, Y7A and W147L. The Y7A substitution has been identified within a divergent sequence lineage found among salmonid classical sequences (Aoyagi et al. 2002;Kiryu et al. 2005;Lukacs et al. 2010). The W147L substitution has also been found in class I molecules from additional teleosts such as common carp and Atlantic salmon (Chen et al. 2010;Lukacs et al. 2010). Similarly, Dare-Ufa and Dare-Uja both have amino acid substitutions at a different putative peptide anchor residue, Y59F, a substitution that has also been observed in class I genes from medaka and Atlantic salmon (Lukacs et al. 2010;Nonaka et al. 2011). Furthermore, a substitution within another putative peptide anchor residue of Dare-Uea and Dare-Uia, K146N, has also been observed within common carp and medaka U genes (Chen et al. 2010;Nonaka et al. 2011).
Figure 5. Alignment of MHC class I U lineage proteins.
The proteins encoded by the U lineage genes were organized into domains and aligned. Numbering of amino acids is based on the Mhc1uba sequence starting with the first residue of α1. Positions that are at least 50% identical are shaded in black and similar residues are shaded in gray. Conserved cysteine residues likely involved in the Ig-fold are highlighted in yellow. Residues implicated in peptide binding are highlighted in red. A potential N-linked glycosylation site at the end of α1 is indicated by blue asterisks below the alignment. A stretch of acidic residues in α3 that may associate with CD8 is indicated by green asterisks below the alignment.
These observations, along with additional sequence changes, suggest trans-specific conservation of substitutions within the sequence lineages of species that are estimated to have diverged at least 100 million years ago (Takeuchi et al. 1995). Furthermore, these frequently observed substitutions indicate that many of the peptide anchor residues remain somewhat flexible to accommodate the function of class I molecules within teleosts. This finding is in contrast to the more strict conservation of peptide anchor residues within mammals. Therefore, conservation of the peptide anchor residues may not represent an absolute test to discriminate between classical and nonclassical molecules in teleosts.
Despite a large amount of sequence divergence among zebrafish U genes, many amino acids with presumed functional roles are well conserved (McConnell et al. 2014). These include structural residues such as those involved in disulfide or salt bridges to maintain the protein fold. This also includes residues likely to interact with additional molecules such as CD8 and β2M that have been conserved across most if not all vertebrate classical molecules. In summary, zebrafish U lineage molecules have remarkably high levels of sequence diversity largely confined to regions responsible for determining antigen specificity, in conjunction with many conserved residues important for contributing to classical function. Additional cell-based studies are necessary to evaluate the function of individual U genes in the immune response.
5.2 Sequence diversity of Z genes
In an attempt to classify the Z genes as classical or nonclassical, the features of their amino acid sequences were studied in detail (Figure 6; Dirscherl and Yoder 2014). Despite the unusually high level of sequence conservation across the α1 and α2 domains, the Z genes exhibit many features of classical MHC class I molecules. These features include conserved cysteine residues important for forming an Ig fold, conserved residues thought to be important for anchoring the ends of bound peptides, and a stretch of acidic residues in the α3 domain that potentially associate with CD8. Through RT-PCR and cloning strategies, it was also shown that the individual Z genes exhibit low to moderate levels of polymorphism in comparison to the reference genome with the sequence divergence ranging up to 11% on the amino acid level. The more polymorphic alleles of the Z genes were generally found in individuals that were just three generations away from their wild-caught founders while the alleles detected in the standard laboratory lines aligned more closely with the reference sequences (Dirscherl and Yoder 2014).
Figure 6. Alignment of MHC class I Z lineage proteins.
The proteins encoded by the Z lineage genes were organized into domains and aligned. Numbering of amino acids is based on the Mhc1zaa sequence starting with the first residue of α1. Sequence features are indicated as in Figure 5.
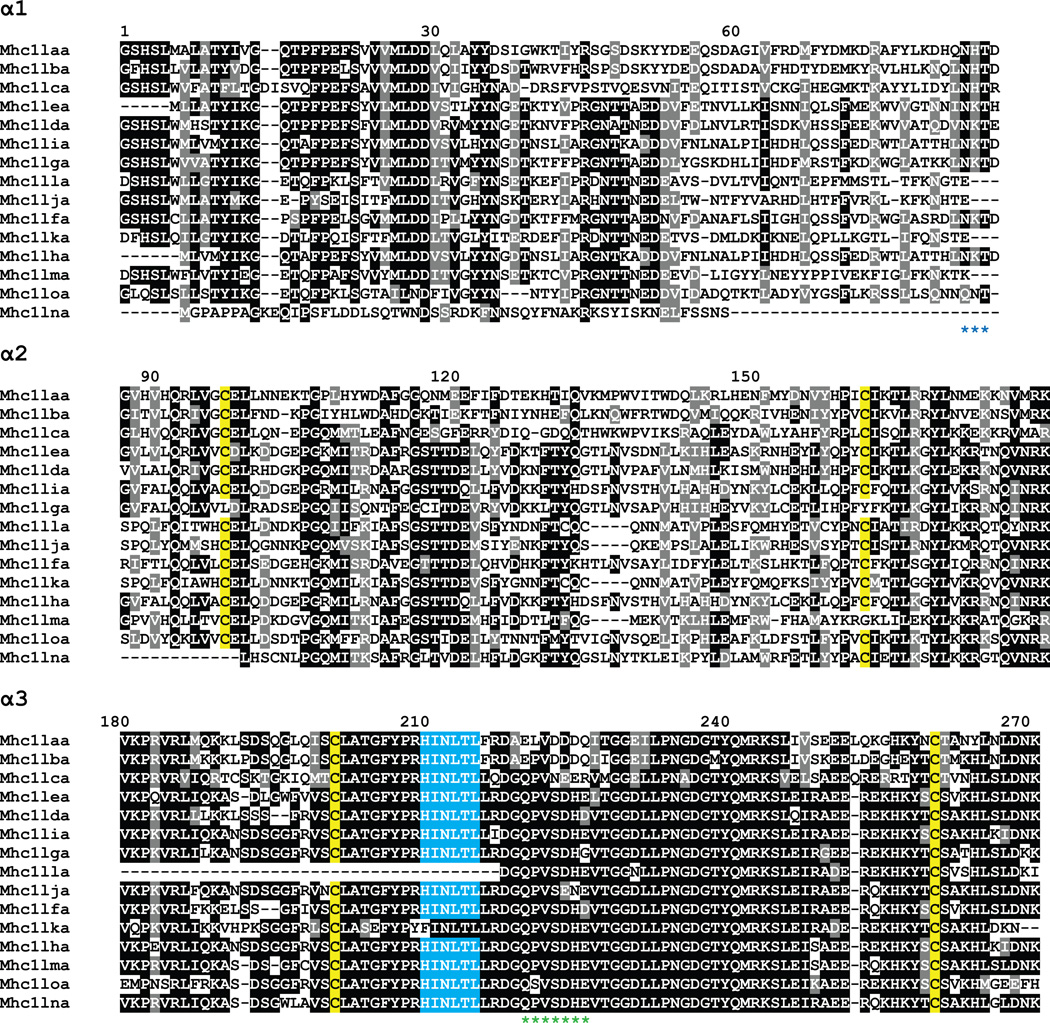
5.3 Sequence diversity of L genes
Aligning the amino acid sequences encoded by the α1 to α3 domains of the L genes reveals many interesting sequence features (Figure 7). The alignment highlights the variability of the α1 and α2 domains in contrast to the conservation of the α3 domain as mentioned above. The four cysteine residues important for the Ig fold of α2 and α3 are conserved in almost all cases (yellow highlight). However, the residues implicated in peptide binding are not conserved in this lineage. All but two of the genes have the HINLTL motif in the α3 domain that is specific to the L lineage (blue highlight) (Dijkstra et al. 2007). Nine of the fifteen genes encode a potential glycosylation site at the end of the α1 domain (marked by blue asterisks below the alignment). Finally, a stretch of acidic residues in α3 may indicate that these L molecules associate with CD8 (marked by green asterisks below the alignment).
Figure 7. Alignment of predicted MHC class I L lineage proteins.
The proteins encoded by the predicted L lineage genes (Table 1) were organized into domains and aligned. Numbering of amino acids is based on the Mhc1laa sequence starting with the first residue of α1. Positions that are at least 50% identical are shaded in black and similar residues are shaded in gray. Conserved cysteine residues likely involved in the Ig-fold are highlighted in yellow. The conserved HINLTL motif in α3 is highlighted in blue. A potential N-linked glycosylation site at the end of α1 is indicated by blue asterisks below the alignment. A stretch of acidic residues in α3 that may associate with CD8 is indicated by green asterisks below the alignment. The proteins lack the normally conserved tryptophan residues, W51 and W60, a characteristic feature of the L lineage molecules. They also lack the residues important for anchoring bound peptide, a feature that supports their nonclassical nature.
6. MHC class I haplotypic diversity
6.1 U gene haplotypes encode distinct sets of MHC class I genes
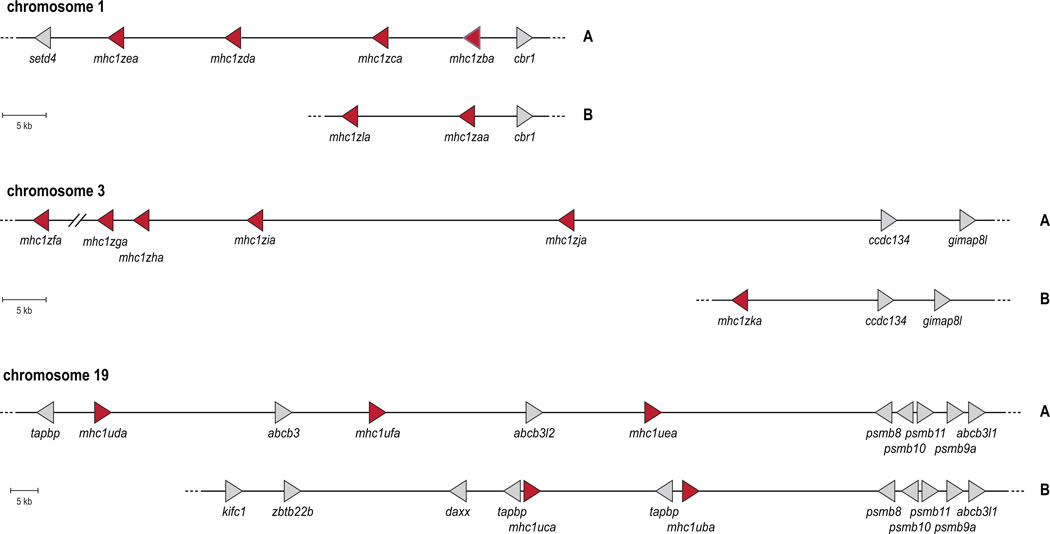
In zebrafish, haplotypic variation greatly contributes to the diversity of U gene loci. Haplotypic variation was first indicated via linkage studies mapping U lineage genes to chromosome 19 in zebrafish (Bingulac-Popovic et al. 1997), and then extended to encompass many other U lineage genes on several additional haplotypes (de Jong et al. 2011;McConnell et al. 2014). Genomic sequences have been assembled for two different haplotypes on zebrafish chromosome 19 (Michalova et al. 2000). Strikingly, these two haplotypes maintain different numbers of U lineage genes without any overlap of individual U lineage genes (Figure 8). Furthermore, six distinct zebrafish haplotypes have been described to date via specific polymorphisms in two linked flanking genes, with each distinct haplotype carrying one to three out of the ten unique U lineage genes (McConnell et al. 2014). Southern blot analyses of fish that are homozygous for each of the six haplotypes support the conclusion that each haplotype contains one to three U genes. Experiments to identify other haplotypes at this locus are underway, and genomic sequencing will provide insight into the extent of this diversity within the zebrafish.
Figure 8. Genomic maps of MHC class I U and Z gene haplotypes.
Genomic maps include the MHC class I genes as red triangles and relative flanking genes as gray triangles, both pointing in the direction of transcription. Alternate haplotypes (labeled “A” and “B” on the right) of the Z gene loci on chromosomes 1 and 3 and the U locus on chromosome 19 are summarized. Detailed descriptions of these haplotypes have been reported previously (Dirscherl and Yoder 2014;McConnell et al. 2014).
Two additional candidate U lineage genes have been identified on zebrafish chromosome 22 (Section 3.1). Although their polymorphic and haplotypic variation remains to be determined, Southern blot analysis provides evidence for some variation in this region (McConnell et al. 2014).
6.2 Evidence of Z gene haplotypes
Recent work investigating the MHC class I Z lineage revealed that the diverse Z transcripts that have been identified are members of a multigene family encoded on chromosomes 1 and 3, and these genes appear to be inherited on different haplotypes (Dirscherl and Yoder 2014). This is supported by genomic sequences in a BAC clone and an unplaced genomic scaffold that encode different sets of Z genes than the reference genome (Figure 8). Additionally, data mining of genomic assemblies from two double haploid homozygous zebrafish, one of the AB background and one of the Tübingen background, revealed that these individuals encode different haplotypes at the chromosome 3 Z locus (Dirscherl and Yoder 2014). A genotyping PCR strategy revealed that individual zebrafish of different genetic backgrounds express different sets of Z genes. Southern blot analyses provided additional evidence for haplotypic variation as the genomic DNA from these same individuals displayed different banding patterns when hybridized with DNA probes designed to detect all Z genes. Future genomic sequencing efforts should clarify the different Z gene haplotypes allowing for MHC matching across multiple lineages and improved success in transplantation experiments (Dirscherl and Yoder 2014).
6.3 Evidence of L gene haplotypes
Although the genomic organization of a single zebrafish MHC class I L lineage locus has been described (Dijkstra et al. 2007), to the extent of our knowledge, no sequence evidence has been reported supporting haplotypic variation of these genes in any species. However, Southern blot analyses from three clonal rainbow trout lines with three different probes (α2 and α3 domains) revealed different banding patterns suggestive of haplotypic variation (Dijkstra et al. 2007). It remains to be determined if the zebrafish L genes display this level of variation.
7. Evidence of functionality
7.1 U genes exhibit classical functions
MHC class I genes from the U lineage are conserved throughout diverse teleost families (Malmstrom et al. 2013;Pinto et al. 2013;Wegner 2008), including those that are considered young (neoteleosts such as medaka) and old (cyprinids such as zebrafish). Conservation of U genes across all teleost species may be considered one important prerequisite in order to maintain classical function. Importantly, cytotoxic T cell function also appears to be highly conserved in teleosts (Fischer et al. 2013;Laing and Hansen 2011;Nakanishi et al. 2011;Somamoto et al. 2002;Takizawa et al. 2011), which may rely on antigen presentation via U molecules to activate the immune response of CD8+ cells. Additionally, it has been demonstrated that rainbow trout UBA is expressed in similar cell types as mammalian classical MHC class I molecules (Dijkstra et al. 2003).
In contrast to the Z or L lineage genes, at the genomic level only the U lineage genes are consistently associated with the core MHC locus within teleosts, meaning that they maintain linkage to additional class I pathway genes such as psmb8, tapbp, and abcb3/tap2 (Clark et al. 2001;Matsuo et al. 2002;Michalova et al. 2000). These genes also maintain synteny within the MHC across other vertebrate species including mammals (Kulski et al. 2002;Matsuo et al. 2002). Furthermore, these genes (psmb8, tapbp, and abcb3/tap2) function specifically within the class I pathway for antigen presentation, providing a potential selective advantage for maintaining their ancient linkage to the U lineage genes.
Studies performed with other species including common carp and stickleback (Gasterosteus aculeatus) have shown 1) MHC class I U protein expression on the surface of cells (Scharsack et al. 2007), 2) binding of peptide to U molecules, 3) association of U molecules with β2M, and 4) the interaction of labeled U complexes with T cells (Chen et al. 2010), providing direct experimental evidence of classical peptide presentation and function for U molecules in a teleost species. Interferon response is conserved in bony fish (Zou et al. 2005), and upregulation of MHC class I transcripts (including U genes such as carp UBA) has been observed following immune stimulation (Chen et al. 2010;Martin et al. 2007;Rise et al. 2008). Transplantation experiments in zebrafish indicate a functional role for the polymorphic U gene loci in rejection of MHC-mismatched transplanted cells (de Jong et al. 2011;Sarder et al. 2003). Finally, specific alleles of U lineage genes are implicated in pathogen resistance (Grimholt et al. 2003;Kjoglum et al. 2008). Taken together, this evidence provides strong support that U genes are the class I genes associated with classical function in teleosts.
7.2 Evidence for Z gene function
To date, little has been done to characterize the function of the Z genes. However, some of the Z genes have been included in microarray and qRT-PCR panels used for quantifying the immune response. It is likely that the Z genes are important for zebrafish immune function as these methods have indicated altered levels of Z gene transcripts in response to different immune stimuli and disease states. For example mhc1zba transcript levels decreased in response to siRNA silencing of a bacterial recognition molecule (Chang et al. 2009). The host response induced by injecting transformed mesenchymal stem cells (MSCs) into zebrafish embryos included decreased transcript levels of mhc1zaa or mhc1zja when compared with injection of MSCs that had not been transformed (Mohseny et al. 2012). In addition, immunization of zebrafish with an attenuated vaccine caused levels of mhc1zea transcripts to increase 3-fold in the liver (Yang et al. 2012).
7.3 MHC class I L genes likely exhibit nonclassical functions
While structural evidence points to the L genes as being nonclassical, no experimental data exists to explain their function. It is possible that L gene function differs from chromosome to chromosome, as they are encoded at three different loci, or even within a single chromosomal locus.
8. Conclusions and future directions
Recent advances have provided much needed clarity to understand the MHC class I genes in zebrafish, yet much remains to be elucidated about these molecules. While analyses of the peptide sequences provide testable hypotheses about the function of the class I U, Z and L genes, transplantation assays and other experiments are needed to verify the role of each individual protein. Questions to be addressed include: Which MHC molecules are expressed on the cell surface? What peptides or other antigens do they present? What receptors on T cells and other effector cells recognize the class I molecules?
At present, peptides have been identified that bind to the common carp UBA protein (Chen et al. 2010), but these results have not yet been elicited for the zebrafish. Likewise, β2M has been proven to associate with common carp UBA, but β2M association with zebrafish MHC class I molecules remains to be determined. Presumably some of the zebrafish MHC class I molecules present non-peptide antigens, such as phospholipids, but these have not been identified to date. Finally we anticipate that we have only scratched the surface of the repertoire of MHC genes found in the zebrafish species. Searches for additional haplotypes at all MHC class I chromosomal regions are underway. Genomic sequencing of these haplotypes will provide the means for interspecies comparisons to ascertain the evolution and help predict the function of MHC genes within this important preclinical model organism.
Highlights.
The diversity of MHC class I genes in zebrafish is discussed.
Class I sequence diversity is polymorphic, polygenic, and haplotypic.
Zebrafish class I genes appear to contribute to a robust immune system.
Nomenclature for 15 zebrafish L lineage MHC class I genes is provided.
Acknowledgements
We are very grateful to Charles Lee (The Jackson Laboratory Institute for Genomic Medicine) for providing the chromosomal map of copy number variation. H.D. is supported in part by a National Institutes of Health Biotechnology Traineeship (T32 GM008776) and by a Joseph E. Pogue Fellowship through the UNC Royster Society of Fellows. J.d. is supported by the University of Chicago Cancer Research Foundation Auxiliary Board and by the National Institute of Diabetes and Digestive and Kidney Diseases (R03-DK091497).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- Complete sequence and gene map of a human major histocompatibility complex. The MHC sequencing consortium. Nature. 1999;401:921–923. doi: 10.1038/44853. [DOI] [PubMed] [Google Scholar]
- Aoyagi K, Dijkstra JM, Xia C, Denda I, Ototake M, Hashimoto K, Nakanishi T. Classical MHC class I genes composed of highly divergent sequence lineages share a single locus in rainbow trout (Oncorhynchus mykiss) J. Immunol. 2002;168:260–273. doi: 10.4049/jimmunol.168.1.260. [DOI] [PubMed] [Google Scholar]
- Bingulac-Popovic J, Figueroa F, Sato A, Talbot WS, Johnson SL, Gates M, Postlethwait JH, Klein J. Mapping of Mhc class I and class II regions to different linkage groups in the zebrafish, Danio rerio. Immunogenetics. 1997;46:129–134. doi: 10.1007/s002510050251. [DOI] [PubMed] [Google Scholar]
- Bjorkman PJ, Parham P. Structure, function, and diversity of class I major histocompatibility complex molecules. Annu. Rev. Biochem. 1990;59:253–288. doi: 10.1146/annurev.bi.59.070190.001345. [DOI] [PubMed] [Google Scholar]
- Bjorkman PJ, Saper MA, Samraoui B, Bennett WS, Strominger JL, Wiley DC. The foreign antigen binding site and T cell recognition regions of class I histocompatibility antigens. Nature. 1987;329:512–518. doi: 10.1038/329512a0. [DOI] [PubMed] [Google Scholar]
- Brown KH, Dobrinski KP, Lee AS, Gokcumen O, Mills RE, Shi X, Chong WW, Chen JY, Yoo P, David S, Peterson SM, Raj T, Choy KW, Stranger BE, Williamson RE, Zon LI, Freeman JL, Lee C. Extensive genetic diversity and substructuring among zebrafish strains revealed through copy number variant analysis. Proc. Natl. Acad. SciUSA. 2012;109:529–534. doi: 10.1073/pnas.1112163109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang MX, Wang YP, Nie P. Zebrafish peptidoglycan recognition protein SC (zfPGRP-SC) mediates multiple intracellular signaling pathways. Fish. Shellfish. Immunol. 2009;26:264–274. doi: 10.1016/j.fsi.2008.11.007. [DOI] [PubMed] [Google Scholar]
- Chaplin DD. Overview of the immune response. J Allergy Clin Immunol. 2010;125:S3–S23. doi: 10.1016/j.jaci.2009.12.980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Jia Z, Zhang T, Zhang N, Lin C, Gao F, Wang L, Li X, Jiang Y, Li X, Gao GF, Xia C. MHC class I presentation and regulation by IFN in bony fish determined by molecular analysis of the class I locus in grass carp. J. Immunol. 2010;185:2209–2221. doi: 10.4049/jimmunol.1000347. [DOI] [PubMed] [Google Scholar]
- Clark MS, Shaw L, Kelly A, Snell P, Elgar G. Characterization of the MHC class I region of the Japanese pufferfish (Fugu rubripes) Immunogenetics. 2001;52:174–185. doi: 10.1007/s002510000285. [DOI] [PubMed] [Google Scholar]
- Colonna M, Borsellion G, Falco M, Ferrara GB, Strominger JL. HLA-C is the inhibitory ligand that determines dominant resistance to lysis by NK1-and NK2-specific natural killer cells. Proc Natl Acad Sci USA. 1993;90:12000–12004. doi: 10.1073/pnas.90.24.12000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong JL, Burns CE, Chen AT, Pugach E, Mayhall EA, Smith AC, Feldman HA, Zhou Y, Zon LI. Characterization of immune-matched hematopoietic transplantation in zebrafish. Blood. 2011;117:4234–4242. doi: 10.1182/blood-2010-09-307488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijkstra JM, Grimholt U, Leong J, Koop BF, Hashimoto K. Comprehensive analysis of MHC class II genes in teleost fish genomes reveals dispensability of the peptide-loading DM system in a large part of vertebrates. BMC Evol. Biol. 2013;13:260. doi: 10.1186/1471-2148-13-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijkstra JM, Katagiri T, Hosomichi K, Yanagiya K, Inoko H, Ototake M, Aoki T, Hashimoto K, Shiina T. A third broad lineage of major histocompatibility complex (MHC) class I in teleost fish; MHC class II linkage and processed genes. Immunogenetics. 2007;59:305–321. doi: 10.1007/s00251-007-0198-6. [DOI] [PubMed] [Google Scholar]
- Dijkstra JM, Kollner B, Aoyagi K, Sawamoto Y, Kuroda A, Ototake M, Nakanishi T, Fischer U. The rainbow trout classical MHC class I molecule Onmy-UBA*501 is expressed in similar cell types as mammalian classical MHC class I molecules. Fish. Shellfish Immunol. 2003;14:1–23. doi: 10.1006/fsim.2001.0407. [DOI] [PubMed] [Google Scholar]
- Dirscherl H, Yoder JA. Characterization of the Z lineage Major histocompatability complex class I genes in zebrafish. Immunogenetics. 2014 doi: 10.1007/s00251-013-0748-z. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa F, Mayer WE, Sato A, Zaleska-Rutczynska Z, Hess B, Tichy H, Klein J. Mhc class I genes of swordtail fishes, Xiphophorus: variation in the number of loci and existence of ancient gene families. Immunogenetics. 2001;53:695–708. doi: 10.1007/s00251-001-0378-8. [DOI] [PubMed] [Google Scholar]
- Fischer U, Koppang EO, Nakanishi T. Teleost T and NK cell immunity. Fish. Shellfish. Immunol. 2013;35:197–206. doi: 10.1016/j.fsi.2013.04.018. [DOI] [PubMed] [Google Scholar]
- Flajnik MF, Kasahara M. Comparative genomics of the MHC: glimpses into the evolution of the adaptive immune system. Immunity. 2001;15:351–362. doi: 10.1016/s1074-7613(01)00198-4. [DOI] [PubMed] [Google Scholar]
- Grimholt U, Hordvik I, Fosse VM, Olsaker I, Endresen C, Lie O. Molecular cloning of major histocompatibility complex class I cDNAs from Atlantic salmon (Salmo salar) Immunogenetics. 1993;37:469–473. doi: 10.1007/BF00222473. [DOI] [PubMed] [Google Scholar]
- Grimholt U, Larsen S, Nordmo R, Midtlyng P, Kjoeglum S, Storset A, Saebo S, Stet RJ. MHC polymorphism and disease resistance in Atlantic salmon (Salmo salar); facing pathogens with single expressed major histocompatibility class I and class II loci. Immunogenetics. 2003;55:210–219. doi: 10.1007/s00251-003-0567-8. [DOI] [PubMed] [Google Scholar]
- Groth CG, Brent LB, Calne RY, Dausset JB, Good RA, Murray JE, Shumway NE, Schwartz RS, Starzl TE, Terasaki PI, Thomas ED, van Rood JJ. Historic landmarks in clinical transplantation: conclusions from the consensus conference at the University of California, Los Angeles. World J. Surg. 2000;24:834–843. doi: 10.1007/s002680010134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto K, Nakanishi T, Kurosawa Y. Isolation of carp genes encoding major histocompatibility complex antigens. Proc. Natl. Acad. Sci. USA. 1990;87:6863–6867. doi: 10.1073/pnas.87.17.6863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hee CS, Beerbaum M, Loll B, Ballaschk M, Schmieder P, Uchanska-Ziegler B, Ziegler A. Dynamics of free versus complexed beta2-microglobulin and the evolution of interfaces in MHC class I molecules. Immunogenetics. 2013;65:157–172. doi: 10.1007/s00251-012-0667-4. [DOI] [PubMed] [Google Scholar]
- Horan J, Wang T, Haagenson M, Spellman SR, Dehn J, Eapen M, Frangoul H, Gupta V, Hale GA, Hurley CK, Marino S, Oudshoorn M, Reddy V, Shaw P, Lee SJ, Woolfrey A. Evaluation of HLA matching in unrelated hematopoietic stem cell transplantation for nonmalignant disorders. Blood. 2012;120:2918–2924. doi: 10.1182/blood-2012-03-417758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton R, Wilming L, Rand V, Lovering RC, Bruford EA, Khodiyar VK, Lush MJ, Povey S, Talbot CC, Jr, Wright MW, Wain HM, Trowsdale J, Ziegler A, Beck S. Gene map of the extended human MHC. Nat. Rev. Genet. 2004;5:889–899. doi: 10.1038/nrg1489. [DOI] [PubMed] [Google Scholar]
- Howe K, Clark MD, Torroja CF, Torrance J, Berthelot C, Muffato M, Collins JE, Humphray S, McLaren K, Matthews L, McLaren S, Sealy I, Caccamo M, Churcher C, Scott C, Barrett JC, Koch R, Rauch GJ, White S, Chow W, Kilian B, Quintais LT, Guerra-Assuncao JA, Zhou Y, Gu Y, Yen J, Vogel JH, Eyre T, Redmond S, Banerjee R, Chi J, Fu B, Langley E, Maguire SF, Laird GK, Lloyd D, Kenyon E, Donaldson S, Sehra H, Almeida-King J, Loveland J, Trevanion S, Jones M, Quail M, Willey D, Hunt A, Burton J, Sims S, McLay K, Plumb B, Davis J, Clee C, Oliver K, Clark R, Riddle C, Eliott D, Threadgold G, Harden G, Ware D, Mortimer B, Kerry G, Heath P, Phillimore B, Tracey A, Corby N, Dunn M, Johnson C, Wood J, Clark S, Pelan S, Griffiths G, Smith M, Glithero R, Howden P, Barker N, Stevens C, Harley J, Holt K, Panagiotidis G, Lovell J, Beasley H, Henderson C, Gordon D, Auger K, Wright D, Collins J, Raisen C, Dyer L, Leung K, Robertson L, Ambridge K, Leongamornlert D, McGuire S, Gilderthorp R, Griffiths C, Manthravadi D, Nichol S, Barker G, Whitehead S, Kay M, Brown J, Murnane C, Gray E, Humphries M, Sycamore N, Barker D, Saunders D, Wallis J, Babbage A, Hammond S, Mashreghi-Mohammadi M, Barr L, Martin S, Wray P, Ellington A, Matthews N, Ellwood M, Woodmansey R, Clark G, Cooper J, Tromans A, Grafham D, Skuce C, Pandian R, Andrews R, Harrison E, Kimberley A, Garnett J, Fosker N, Hall R, Garner P, Kelly D, Bird C, Palmer S, Gehring I, Berger A, Dooley CM, Ersan-Urun Z, Eser C, Geiger H, Geisler M, Karotki L, Kirn A, Konantz J, Konantz M, Oberlander M, Rudolph-Geiger S, Teucke M, Osoegawa K, Zhu B, Rapp A, Widaa S, Langford C, Yang F, Carter NP, Harrow J, Ning Z, Herrero J, Searle SM, Enright A, Geisler R, Plasterk RH, Lee C, Westerfield M, de Jong PJ, Zon LI, Postlethwait JH, Nusslein-Volhard C, Hubbard TJ, Roest CH, Rogers J, Stemple DL. The zebrafish reference genome sequence and its relationship to the human genome. Nature. 2013;496:498–503. doi: 10.1038/nature12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL, Nei M. Pattern of nucleotide substitution at major histocompatibility complex class I loci reveals overdominant selection. Nature. 1988;335:167–170. doi: 10.1038/335167a0. [DOI] [PubMed] [Google Scholar]
- Ignatius MS, Langenau DM. Fluorescent imaging of cancer in zebrafish. Methods Cell Biol. 2011;105:437–459. doi: 10.1016/B978-0-12-381320-6.00019-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanther M, Rawls JF. Host-microbe interactions in the developing zebrafish. Curr. Opin. Immunol. 2010;22:10–19. doi: 10.1016/j.coi.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiryu I, Dijkstra JM, Sarder RI, Fujiwara A, Yoshiura Y, Ototake M. New MHC class Ia domain lineages in rainbow trout (Oncorhynchus mykiss) which are shared with other fish species. Fish. Shellfish. Immunol. 2005;18:243–254. doi: 10.1016/j.fsi.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Kjoglum S, Larsen S, Bakke HG, Grimholt U. The effect of specific MHC class I and class II combinations on resistance to furunculosis in Atlantic salmon (Salmo salar) Scand J. Immunol. 2008;67:160–168. doi: 10.1111/j.1365-3083.2007.02052.x. [DOI] [PubMed] [Google Scholar]
- Klein J. George Snell's first foray into the unexplored territory of the major histocompatibility complex. Genetics. 2001;159:435–439. doi: 10.1093/genetics/159.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein J, Bontrop RE, Dawkins RL, Erlich HA, Gyllensten UB, Heise ER, Jones PP, Parham P, Wakeland EK, Watkins DI. Nomenclature for the major histocompatibility complexes of different species: a proposal. Immunogenetics. 1990;31:217–219. doi: 10.1007/BF00204890. [DOI] [PubMed] [Google Scholar]
- Klein J, Sato A. The HLA system. First of two parts. N. Engl. J. Med. 2000;343:702–709. doi: 10.1056/NEJM200009073431006. [DOI] [PubMed] [Google Scholar]
- Klein J, Sato A, Nikolaidis N. MHC TSP, the origin of species: from immunogenetics to evolutionary genetics. Annu. Rev. Genet. 2007;41:281–304. doi: 10.1146/annurev.genet.41.110306.130137. [DOI] [PubMed] [Google Scholar]
- Kruiswijk CP, Hermsen TT, Westphal AH, Savelkoul HF, Stet RJ. A novel functional class I lineage in zebrafish (Danio rerio), carp (Cyprinus carpio), and large barbus (Barbus intermedius) showing an unusual conservation of the peptide binding domains. J. Immunol. 2002;169:1936–1947. doi: 10.4049/jimmunol.169.4.1936. [DOI] [PubMed] [Google Scholar]
- Kulski JK, Shiina T, Anzai T, Kohara S, Inoko H. Comparative genomic analysis of the MHC: the evolution of class I duplication blocks, diversity and complexity from shark to man. Immunol. Rev. 2002;190:95–122. doi: 10.1034/j.1600-065x.2002.19008.x. [DOI] [PubMed] [Google Scholar]
- Kumanovics A, Takada T, Lindahl KF. Genomic organization of the mammalian MHC. Annu. Rev. Immunol. 2003;21:629–657. doi: 10.1146/annurev.immunol.21.090501.080116. [DOI] [PubMed] [Google Scholar]
- Kuroda N, Figueroa F, O'hUigin C, Klein J. Evidence that the separation of Mhc class II from class I loci in the zebrafish, Danio rerio, occurred by translocation. Immunogenetics. 2002;54:418–430. doi: 10.1007/s00251-002-0473-5. [DOI] [PubMed] [Google Scholar]
- Laing KJ, Hansen JD. Fish T cells: recent advances through genomics. Dev. Comp Immunol. 2011;35:1282–1295. doi: 10.1016/j.dci.2011.03.004. [DOI] [PubMed] [Google Scholar]
- Li P, White RM, Zon LI. Transplantation in zebrafish. Methods Cell Biol. 2011;105:403–417. doi: 10.1016/B978-0-12-381320-6.00017-5. [DOI] [PubMed] [Google Scholar]
- Lukacs MF, Harstad H, Bakke HG, Beetz-Sargent M, McKinnel L, Lubieniecki KP, Koop BF, Grimholt U. Comprehensive analysis of MHC class I genes from the U-, S-, and Z-lineages in Atlantic salmon. Bmc Genomics. 2010;11:154. doi: 10.1186/1471-2164-11-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmstrom M, Jentoft S, Gregers TF, Jakobsen KS. Unraveling the evolution of the Atlantic cod's (Gadus morhua L.) alternative immune strategy. PLoS. One. 2013;8:e74004. doi: 10.1371/journal.pone.0074004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SA, Zou J, Houlihan DF, Secombes CJ. Directional responses following recombinant cytokine stimulation of rainbow trout (Oncorhynchus mykiss) RTS-11 macrophage cells as revealed by transcriptome profiling. Bmc Genomics. 2007;8:150. doi: 10.1186/1471-2164-8-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo MY, Asakawa S, Shimizu N, Kimura H, Nonaka M. Nucleotide sequence of the MHC class I genomic region of a teleost, the medaka (Oryzias latipes) Immunogenetics. 2002;53:930–940. doi: 10.1007/s00251-001-0427-3. [DOI] [PubMed] [Google Scholar]
- McConnell SC, Restaino AC, de Jong JL. Multiple divergent haplotypes express completely distinct sets of class I MHC genes in zebrafish. Immunogenetics. 2014 doi: 10.1007/s00251-013-0749-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalova V, Murray BW, Sultmann H, Klein J. A contig map of the Mhc class I genomic region in the zebrafish reveals ancient synteny. J Immunol. 2000;164:5296–5305. doi: 10.4049/jimmunol.164.10.5296. [DOI] [PubMed] [Google Scholar]
- Mohseny AB, Xiao W, Carvalho R, Spaink HP, Hogendoorn PC, Cleton-Jansen AM. An osteosarcoma zebrafish model implicates Mmp-19 and Ets-1 as well as reduced host immune response in angiogenesis and migration. J. Pathol. 2012;227:245–253. doi: 10.1002/path.3998. [DOI] [PubMed] [Google Scholar]
- Nakanishi T, Toda H, Shibasaki Y, Somamoto T. Cytotoxic T cells in teleost fish. Dev. Comp Immunol. 2011;35:1317–1323. doi: 10.1016/j.dci.2011.03.033. [DOI] [PubMed] [Google Scholar]
- Nei M, Rooney AP. Concerted and birth-and-death evolution of multigene families. Annu. Rev. Genet. 2005;39:121–152. doi: 10.1146/annurev.genet.39.073003.112240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka MI, Aizawa K, Mitani H, Bannai HP, Nonaka M. Retained orthologous relationships of the MHC Class I genes during euteleost evolution. Mol. Biol. Evol. 2011;28:3099–3112. doi: 10.1093/molbev/msr139. [DOI] [PubMed] [Google Scholar]
- Okamura K, Nakanishi T, Kurosawa Y, Hashimoto K. Expansion of genes that encode MHC class I molecules in cyprinid fishes. J. Immunol. 1993;151:188–200. [PubMed] [Google Scholar]
- Okamura K, Ototake M, Nakanishi T, Kurosawa Y, Hashimoto K. The most primitive vertebrates with jaws possess highly polymorphic MHC class I genes comparable to those of humans. Immunity. 1997;7:777–790. doi: 10.1016/s1074-7613(00)80396-9. [DOI] [PubMed] [Google Scholar]
- Parham P. MHC class I molecules and KIRs in human history, health and survival. Nat. Rev. Immunol. 2005;5:201–214. doi: 10.1038/nri1570. [DOI] [PubMed] [Google Scholar]
- Patowary A, Purkanti R, Singh M, Chauhan R, Singh AR, Swarnkar M, Singh N, Pandey V, Torroja C, Clark MD, Kocher JP, Clark KJ, Stemple DL, Klee EW, Ekker SC, Scaria V, Sivasubbu S. A sequence-based variation map of zebrafish. Zebrafish. 2013;10:15–20. doi: 10.1089/zeb.2012.0848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto RD, Randelli E, Buonocore F, Pereira PJ, dos Santos NM. Molecular cloning and characterization of sea bass (Dicentrarchus labrax, L.) MHC class I heavy chain and beta2-microglobulin. Dev. Comp Immunol. 2013;39:234–254. doi: 10.1016/j.dci.2012.10.002. [DOI] [PubMed] [Google Scholar]
- Renaud O, Herbomel P, Kissa K. Studying cell behavior in whole zebrafish embryos by confocal live imaging: application to hematopoietic stem cells. Nat. Protoc. 2011;6:1897–1904. doi: 10.1038/nprot.2011.408. [DOI] [PubMed] [Google Scholar]
- Rise ML, Hall J, Rise M, Hori T, Gamperl A, Kimball J, Hubert S, Bowman S, Johnson SC. Functional genomic analysis of the response of Atlantic cod (Gadus morhua) spleen to the viral mimic polyriboinosinic polyribocytidylic acid (pIC) Dev. Comp Immunol. 2008;32:916–931. doi: 10.1016/j.dci.2008.01.002. [DOI] [PubMed] [Google Scholar]
- Rodgers JR, Cook RG. MHC class Ib molecules bridge innate and acquired immunity. Nat. Rev. Immunol. 2005;5:459–471. doi: 10.1038/nri1635. [DOI] [PubMed] [Google Scholar]
- Sambrook JG, Figueroa F, Beck S. A genome-wide survey of Major Histocompatibility Complex (MHC) genes and their paralogues in zebrafish. Bmc Genomics. 2005;6:152. doi: 10.1186/1471-2164-6-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarder MR, Fischer U, Dijkstra JM, Kiryu I, Yoshiura Y, Azuma T, Kollner B, Ototake M. The MHC class I linkage group is a major determinant in the in vivo rejection of allogeneic erythrocytes in rainbow trout (Oncorhynchus mykiss) Immunogenetics. 2003;55:315–324. doi: 10.1007/s00251-003-0587-4. [DOI] [PubMed] [Google Scholar]
- Sato A, Figueroa F, O'hUigin C, Steck N, Klein J. Cloning of major histocompatibility complex (Mhc) genes from threespine stickleback, Gasterosteus aculeatus. Mol. Mar. Biol. Biotechnol. 1998;7:221–231. [PubMed] [Google Scholar]
- Sato A, Klein D, Sultmann H, Figueroa F, O'hUigin C, Klein J. Class I mhc genes of cichlid fishes: identification, expression, and polymorphism. Immunogenetics. 1997;46:63–72. doi: 10.1007/s002510050243. [DOI] [PubMed] [Google Scholar]
- Scharsack JP, Kalbe M, Schaschl H. Characterization of antisera raised against stickleback (Gasterosteus aculeatus) MHC class I and class II molecules. Fish. Shellfish Immunol. 2007;23:991–1002. doi: 10.1016/j.fsi.2007.03.011. [DOI] [PubMed] [Google Scholar]
- Shiina T, Hosomichi K, Inoko H, Kulski JK. The HLA genomic loci map: expression, interaction, diversity and disease. J. Hum. Genet. 2009;54:15–39. doi: 10.1038/jhg.2008.5. [DOI] [PubMed] [Google Scholar]
- Shinya M, Sakai N. Generation of highly homogeneous strains of zebrafish through full sib-pair mating. G3. (Bethesda.) 2011;1:377–386. doi: 10.1534/g3.111.000851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shum BP, Guethlein L, Flodin LR, Adkison MA, Hedrick RP, Nehring RB, Stet RJM, Secombes C, Parham P. Modes of salmonid MHC class I and II evolution differ from the primate paradigm. J Immunol. 2001;166:3297–3308. doi: 10.4049/jimmunol.166.5.3297. [DOI] [PubMed] [Google Scholar]
- Shum BP, Rajalingam R, Magor KE, Azumi K, Carr WH, Dixon B, Stet RJ, Adkison MA, Hedrick RP, Parham P. A divergent non-classical class I gene conserved in salmonids. Immunogenetics. 1999;49:479–490. doi: 10.1007/s002510050524. [DOI] [PubMed] [Google Scholar]
- Snell G, Higgins G. Alleles at the histocompatibility-2 locus in the mouse as determined by tumor transplantation. Genetics. 1951;36:306–310. doi: 10.1093/genetics/36.3.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somamoto T, Nakanishi T, Okamoto N. Role of specific cell-mediated cytotoxicity in protecting fish from viral infections. Virology. 2002;297:120–127. doi: 10.1006/viro.2002.1486. [DOI] [PubMed] [Google Scholar]
- Stet RJ, Kruiswijk CP, Dixon B. Major histocompatibility lineages and immune gene function in teleost fishes: the road not taken. Crit Rev. Immunol. 2003;23:441–471. doi: 10.1615/critrevimmunol.v23.i56.50. [DOI] [PubMed] [Google Scholar]
- Stet RJ, Kruiswijk CP, Saeij JP, Wiegertjes GF. Major histocompatibility genes in cyprinid fishes: theory and practice. Immunol. Rev. 1998;166:301–316. doi: 10.1111/j.1600-065x.1998.tb01271.x. [DOI] [PubMed] [Google Scholar]
- Takada T, Kumanovics A, Amadou C, Yoshino M, Jones EP, Athanasiou M, Evans GA, Fischer LK. Species-specific class I gene expansions formed the telomeric 1 mb of the mouse major histocompatibility complex. Genome Res. 2003;13:589–600. doi: 10.1101/gr.975303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi H, Figueroa F, O'hUigin C, Klein J. Cloning and characterization of class I Mhc genes of the zebrafish, Brachydanio rerio. Immunogenetics. 1995;42:77–84. doi: 10.1007/BF00178581. [DOI] [PubMed] [Google Scholar]
- Takizawa F, Dijkstra JM, Kotterba P, Korytar T, Kock H, Kollner B, Jaureguiberry B, Nakanishi T, Fischer U. The expression of CD8alpha discriminates distinct T cell subsets in teleost fish. Dev. Comp Immunol. 2011;35:752–763. doi: 10.1016/j.dci.2011.02.008. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor AM, Zon LI. Zebrafish tumor assays: the state of transplantation. Zebrafish. 2009;6:339–346. doi: 10.1089/zeb.2009.0607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorsby E. A short history of HLA. Tissue Antigens. 2009;74:101–116. doi: 10.1111/j.1399-0039.2009.01291.x. [DOI] [PubMed] [Google Scholar]
- Tobin DM, May RC, Wheeler RT. Zebrafish: a see-through host and a fluorescent toolbox to probe host-pathogen interaction. PLoS. Pathog. 2012;8:e1002349. doi: 10.1371/journal.ppat.1002349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trowsdale J. "Both man & bird & beast": comparative organization of MHC genes. Immunogenetics. 1995;41:1–17. doi: 10.1007/BF00188427. [DOI] [PubMed] [Google Scholar]
- Trowsdale J, Knight JC. Major histocompatibility complex genomics and human disease. Annu. Rev. Genomics Hum. Genet. 2013;14:301–323. doi: 10.1146/annurev-genom-091212-153455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Vaart M, Spaink HP, Meijer AH. Pathogen recognition and activation of the innate immune response in zebrafish. Adv. Hematol. 2012;2012:159807. doi: 10.1155/2012/159807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Erp SH, Dixon B, Figueroa F, Egberts E, Stet RJ. Identification and characterization of a new major histocompatibility complex class I gene in carp (Cyprinus carpio L.) Immunogenetics. 1996;44:49–61. doi: 10.1007/BF02602656. [DOI] [PubMed] [Google Scholar]
- Vandiedonck C, Knight JC. The human Major Histocompatibility Complex as a paradigm in genomics research. Brief. Funct. Genomic. Proteomic. 2009;8:379–394. doi: 10.1093/bfgp/elp010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegner KM. Historical and contemporary selection of teleost MHC genes: did we leave the past behind? J Fish Biol. 2008;73:2110–2132. [Google Scholar]
- Yang D, Liu Q, Yang M, Wu H, Wang Q, Xiao J, Zhang Y. RNA-seq liver transcriptome analysis reveals an activated MHC-I pathway and an inhibited MHC-II pathway at the early stage of vaccine immunization in zebrafish. Bmc Genomics. 2012;13:319. doi: 10.1186/1471-2164-13-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Carrington A, Collet B, Dijkstra JM, Yoshiura Y, Bols N, Secombes C. Identification and bioactivities of IFN-gamma in rainbow trout Oncorhynchus mykiss: the first Th1-type cytokine characterized functionally in fish. J. Immunol. 2005;175:2484–2494. doi: 10.4049/jimmunol.175.4.2484. [DOI] [PubMed] [Google Scholar]