Abstract
Osteoarthritis (OA) is characterized by remodeling and degradation of joint tissues. Microarray studies have led to a better understanding of the molecular changes that occur in tissues affected by conditions such as OA; however, such analyses are limited to the identification of a list of genes with altered transcript expression, usually at a single time point during disease progression. While these lists have identified many novel genes that are altered during the disease process, they are unable to identify perturbed relationships between genes and gene products. In this work, we have integrated a time course gene expression data set with network analysis to gain a better systems level understanding of the early events that occur during the development of OA in a mouse model. The subnetworks that were enriched at one or more of the time points examined (2, 4, 8, and 16 weeks after induction of OA) contained genes from several pathways proposed to be important to the OA process, including the extracellular matrix-receptor interaction and the focal adhesion pathways and the Wnt, Hedgehog and TGF-β signaling pathways. The genes within the subnetworks were most active at the 2 and 4 week time points and included genes not previously studied in the OA process. A unique pathway, riboflavin metabolism, was active at the 4 week time point. These results suggest that the incorporation of network-type analyses along with time series microarray data will lead to advancements in our understanding of complex diseases such as OA at a systems level, and may provide novel insights into the pathways and processes involved in disease pathogenesis.
Keywords: gene expression, systems biology, articular cartilage, arthritis, cell signaling, extracellular matrix
1. Introduction
In order to generate new hypotheses and obtain a better understanding of complex disease processes, investigators have used a systems biology approach to complement the more traditional reductionist approach. A systems approach has been facilitated by the availability of technologies which allow for interrogation of the entire genome, proteome and/or metabolome in an organism or tissue of interest. One of the most widely available and commonly used approaches is analysis of gene expression using microarrays. The analysis and interpretation of gene expression data generated by microarrays most commonly involves grouping or clustering genes based on the measured changes in expression, followed by identification of over-represented annotations in each of the gene clusters using various commercial or publically available bioinformatics tools. Although this methodology can elucidate processes and pathways that are differentially regulated in diseased cells or tissues, it does not incorporate information needed to define the relationships between specific genes or proteins to discover interacting networks and pathways important to a disease process.
In this work, we integrated microarray gene expression data into a network-based analysis to identify the signaling and metabolic pathways most highly regulated during the development of osteoarthritis (OA). OA is a condition of increasing public health interest. The most common form of arthritis affecting over 27 million people in the US1 with similar prevalence worldwide, OA results in significant pain and loss of function making it the most common cause of chronic disability in adults2. A major limitation in the treatment of OA is the lack of any intervention proven to directly impact the disease process, either in the early or late stage of the disease. A better understanding of the molecular changes that occur during the development of OA will improve our knowledge of how this disease progresses and will help identify new targets needed to develop therapies to improve clinical outcomes.
Microarray studies using RNA extracted from joint tissues affected by OA have been reported in the literature. Many of these studies have focused on a single tissue, like articular cartilage3, 4, subchondral bone5 or synovium6, 7 most often at single time points. While these approaches have identified many novel genes with altered expression in OA, there is no information on how the gene product interactions may be altered to influence the development of the disease process.
We recently generated a microarray dataset that assessed gene expression changes over time during the early stages of OA in a commonly used mouse model8. In this model, OA develops after surgical destabilization of the medial meniscus (DMM model). We evaluated changes in gene expression and histological evidence of OA at 2, 4, 8, and 16 weeks after DMM surgery with a sham surgery group serving as control. In order to be able to take a more systems level approach, we examined gene expression changes in the joint as an organ by extracting RNA from a pool of tissues including cartilage, subchondral bone, meniscus and the joint capsule with synovium. Using this dataset, we previously filtered a total of 371 genes into 27 clusters with various temporal gene expression patterns8. These clusters included many genes previously associated with OA—such as COMP, MMP14, TIMP2, SDC1, DDR2, TGFB2, MMP13, FMOD, BGN, LECT1, S100B and multiple collagen genes—and elucidated their expression kinetics during the onset of OA. However, the cluster analysis was unable to identify connections between the genes needed to better understand how particular pathways are regulated during the disease process.
Utilizing the data from this time course microarray experiment, in the present study we were able to discover how perturbed pathways change over time by observing which transcripts are active (differentially expressed between DMM and sham controls) or inactive at each time point. Subnetworks relevant to the OA disease process were found, along with novel genes within these networks that have not been previously studied in OA. The results of this network-based analysis demonstrate that an improved comprehensive systems level understanding of OA can be obtained by incorporating pathway-level information into the analyses of gene expression microarray data.
2. Methods
2.1 Normalization and processing of microarray data
The gene expression microarray dataset used for the present analysis was obtained from our recently published study8. In brief, 12 week-old male C57BL/6 mice underwent DMM surgery to induce OA or sham surgery as control. After sacrifice, RNA was isolated from the medial side of the joint (n=9 mice per surgical group per time point) pre-surgery (time 0) and at 2, 4, 8 and 16 weeks after surgery. RNA from 3 mice were pooled for each array performed using the Affymetrix Mouse 430 2.0 chips (3 independent arrays run for each time point and group). Microarray data was processed as previously described8. Briefly, microarrays were imaged and raw data was normalized using systematic variation normalization (SVN)9, 10 with the log2 intensity and detection p-value being reported. Relative gene expression changes due to the DMM-induced changes in the joint were calculated by using the time-matched average sham values as the control. The signal log ratio (SLR) of control versus each DMM replicate for each time point was then calculated and used as the DMM-induced gene expression changes.
2.2 Filtering and clustering of microarray data
The relative gene expression data was filtered and clustered as previously described8. Briefly, replicate time courses were first filtered individually on detection p-value (p-value ≤ 0.06) and SLR (SLR ≤0.5 or ≥0.5). The resulting lists of genes were then intersected and further filtered for consistency of gene expression over time by using the Euclidean distance (≤0.6) and Pearson correlation coefficient (≥0.7) scores between replicate time course profiles. The final set of differentially expressed genes was then clustered using the consensus clustering option provided by SC2ATmd11.
2.3 Network analysis
Network analysis was performed using the jActiveModules version 2.23 (JAM) plugin for Cytoscape12, 13. Prior to performing a network analysis using JAM, a known protein interaction and reaction network had to be obtained and gene expression data had to be converted for compatibility with JAM. Additionally, a consensus approach to the original JAM algorithm (cJAM) was implemented to obtain active subnetworks that appeared at a statistically significant number of times in multiple runs of JAM. Obtaining known interactions, processing of gene expression data, and the implementation of cJAM are detailed below.
2.4 Obtaining Known Network Interactions
The Cytoscape plugin BioNetBuilder v2.0 was used to query the Kyoto Encyclopedia of Genes and Genomes (KEGG)14 for all known mouse interactions and reactions present between any two genes represented on the Affymetrix Mouse 430 2.0 chip13, 15. The list of probe sets was imported into BioNetBuilder, and all associated nodes (representing gene products present on the chip) and edges (representing molecular interactions and reactions) for the Mus musculus genome were downloaded into Cytoscape version 2.8.1, and visualized as a network. Self-edges and edges labeled as “Shared Compound” were deleted from the network because self-edges resulted in JAM returning single-node results, which were not biologically meaningful, and Shared Compound edges were determined to be biologically irrelevant for the system under study. The resulting mouse network contained 2,393 nodes uniquely identified by Entrez ID that also had genes represented on the microarray chip with 32,342 edges (Cytoscape File available upon request). Interactions and reactions between gene products included 15,240 edges labeled as Enzyme-Compound interactions (EC), 440 edges labeled as Gene Expression interactions (GE) and 16,662 edges identified as Protein-Protein interactions (PP). JAM used this “global network” as the background for identifying active subnetworks.
2.5 Data Conversion and Mapping to the Global Network
Gene expression data was converted for compatibility with JAM12 using an in-house script written in MATLAB (MathWorks Inc.). Briefly, the DMM SLRs for each replicate and time point were independently filtered by detection p-value, where all 3 sham control p-values plus the respective DMM p-value had to be ≤0.06, to identify those genes that were significantly detected on the chip. At this point, the mean DMM SLR and standard deviation for each replicate and time point filtered list was calculated for future use. Next, the remaining Affymetrix probe set ID's for each replicate were mapped to the corresponding Entrez ID based on the Affymetrix annotations file downloaded from NetAffx (http://www.affymetrix.com). On occasion, several probe sets would map to a single Entrez ID, or one probe set would map to several Entrez IDs creating a many-to-many relationship. The later relationship was considered trivial; however, having several probe sets mapping to a single Entrez ID was a problem because only one SLR value could be represented in the network per Entrez ID. To resolve this issue a “summary SLR” was calculated as the average SLR value across all probe sets mapping to the same Entrez ID for a given replicate at a given time point. Finally, the SLR and summary SLR values were converted to the exclusive range of (0,1) by integrating the cumulative distribution function (Equation 1)16 over each replicate for each time point (a normal distribution was assumed). Integration was done relative to the respective mean and standard deviations calculated earlier for each replicate and time point for a given gene's SLR value. SLR values represented a z-score on the distribution where in Equation 1, Φ represents the fraction of SLR values below a given gene's SLR, and Equation 2 yields the fraction of genes with a greater absolute z-score than that of a given gene's SLR for the given replicate and time point.
| (1) |
| (2) |
In Equations 1 and 2, μ and σ are the mean and standard deviation, respectively, of all p-value filtered SLR values for each replicate i, and x is the SLR value for gene j under replicate i.
Once array data was converted into a format acceptable by JAM, data were mapped onto the global network obtained from KEGG using the Entrez IDs. Due to the p-value filter, not all genes represented in the network contained converted expression values for every time point and replicate. Thus, missing data was given a converted value of 0.5, which gave that gene a 50% chance of showing up in subnetwork results for the respective replicate and time point.
2.6 Consensus jActiveModules (cJAM) Analysis
Network analysis using JAM was performed using the described global mouse network and converted DMM gene expression data. Details of the original JAM algorithm have been described previously12, 17. Briefly, converted gene expression data is overlaid onto the known network, and an annealing search algorithm with a random initial start is used to search the network space by adding or removing nodes one at a time from the current selection. Subnetworks are scored by JAM using the average of the selected z-scores after each iteration. Once subnetwork scores no longer significantly improve, the search is halted and the current selection of subnetworks are those deemed as active (active subnetworks). A known issue with this approach is that it is impossible to search the entire subnetwork space12, so only part of the network space is searched with random initialization. The stochastic nature of this approach can result in different subnetworks each time it is run. Therefore, a consensus approach was implemented to identify those subnetwork nodes that appeared as active a significant number of times over multiple runs of JAM, which we have termed Consensus JAM (cJAM).
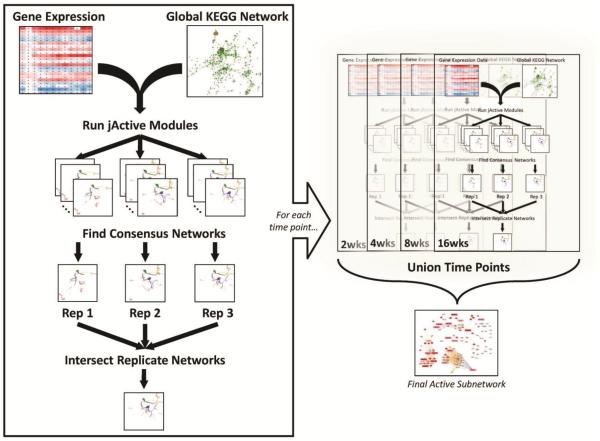
Consensus JAM was performed on the DMM time course as shown in Fig. 1. Each replicate DMM SLR data set was overlaid onto the global network individually and was run through JAM 10 times, using a different starting seed for each run (see Fig. 1 legend for JAM settings). Subnetwork results from each of the 10 runs were filtered to remove those with a score less than 3.018. The remaining subnetworks for each DMM SLR data set were analyzed using an in-house script that calculated the probability of each node appearing in the search results a given number of times (described below). Once the set of nodes that appeared a statistically significant number of times was determined, edges connecting those nodes were mined from the global network to create the consensus subnetwork. This was done for each replicate and each time point. To determine biological consensus, the consensus subnetworks for each replicate were intersected to obtain one subnetwork for each time point. These subnetworks were then unioned to generate an active subnetwork where any given node was deemed active in all replicates for at least one time point.
Fig. 1. Flow chart of the Consensus jActiveModules (cJAM) process.
cJAM was performed on the replicates of each time point independently. Processed gene expression data was imported into the JAM plugin and overlaid onto the global network extracted from KEGG. The annealing algorithm was run on each replicate 10 times using the following settings: number of modules = 20; Adjust score for size was turned OFF; Regional Scoring was turned ON; Iterations = 100,000; Start Temp = 2.0; End Temp = 0.0001; Quenching was turned OFF; Hubfinding = 10; and a random seed was chosen for each run based on time (used same seed for same run in all time points). Next, the consensus network for all runs was obtained for each replicate, and these were then intersected to identify the final subnetwork that showed up consistently in all replicates for a given time point. Lastly, the resulting active subnetworks were unioned to create one network that represented the activity of all time points (details of the nodes in this network can be seen in Fig. 2). Refer to the Methods section for specific process details. Disclaimer: All the network images, except the last one, are not from this analysis—they are just used to demonstrate the cJAM workflow.
2.7 Node significance calculation in cJAM
Statistical significance for each node returned by cJAM was calculated as follows. Assume that Gi = (Vi, Ei) represents the collection of graphs (G), made up of a set of nodes (vertices V) and edges (E), returned from the ith of k total runs of JAM over the global network with t total nodes. Let max be the number of nodes in the largest node set Vi returned by JAM for any one run. Let U be the union of all node sets, thereby defining the set of nodes seen across any run of JAM with its size represented as |U|. For each node u in the union U, a count of how many node sets the node appeared in was recorded. High values for this count would reveal if a node were seen in many of the repeated searches. To quantify how often a node should appear to be statistically significant, the following process is employed. Assume that max/t, the maximum number of nodes returned from any run divided by the total number of nodes searched in the global network represents the probability of a node being selected by chance. This value corresponds to the probability of a node being selected under the most “aggressive” searcher. Given this, the probability of a node being selected was computed using the binomial distribution, with parameters of k trials and probability, max/t, of success within a trial. The nodes in U can then be rank ordered by this probability to identify those that are statistically significant. Since hundreds or thousands of nodes are being evaluated under the binomial distribution, a Bonferroni correction for repeated tests is employed. To apply the Bonferroni correction, the cutoff probability for statistical significance is divided by the number of repeated tests being employed, which in this scenario is |U|, the number of nodes seen across all runs of the search algorithm.
2.8 Enrichment Analysis
The Database for Analysis, Visualization, Integration and Discovery (DAVID)19 was used for all gene-annotation enrichment analyses, which identify annotations that are significantly over-represented in an input list of genes. Briefly, a list of gene symbols (from clusters or active subnetworks) was imported into DAVID with the Mus musculus genome used as background. All default annotation-term sources were turned off except KEGG_PATHWAY. All other settings were left at their default value. The Annotation Chart tool was used to identify KEGG pathways14 that were significantly overrepresented in the input list. A Benjamini corrected p-value of 0.05 was used to determine significance.
3. Results and Discussion
3.1 Identification of subnetworks actively regulated during the time course of OA development
Our approach was to identify actively regulated subnetworks from a known background global network. We defined an active subnetwork as a collection of interacting gene products (proteins/nodes in the network) for which the majority of the genes are regulated across the time course. For this method, a threshold for significant gene expression is not required. This process is summarized in Fig. 1. Briefly, we constructed a background global network of 2,393 nodes, from the Kyoto Encyclopedia of Genes and Genomes (KEGG)14 as detailed in the Methods. The consensus JAM (cJAM) algorithm was then run using this global network and the converted time course gene expression data for each replicate dataset and each time point independently. Identified subnetworks were intersected to obtain one active subnetwork for each time point. The active subnetworks across the entire time course were unioned to obtain a single subnetwork where a node (gene product) was deemed active for at least one of the four time points (2, 4, 8 and 16 weeks after DMM surgery). The total number of nodes in this single “union” subnetwork was 116 (Fig. 2). The number of active nodes in the subnetworks at each time point varied considerably with the 2 and 4 week time points having the highest (65 and 72 nodes respectively) followed by a reduction to 29 nodes at 8 weeks and then an increase to 59 active nodes at 16 weeks (Fig. 3 and Supplemental File 1).
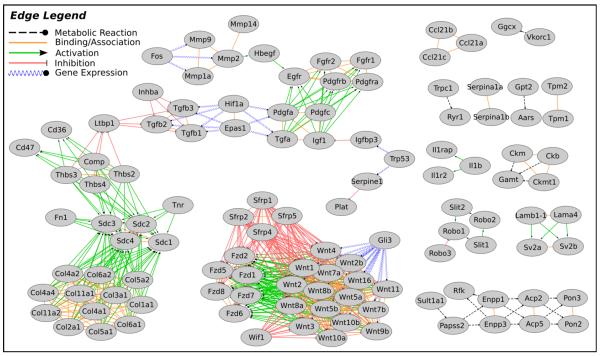
Fig. 2. Union of active subnetworks identified by cJAM.
The nodes present in the active subnetworks identified by cJAM for each time point were unioned to generate a composite network image. Edge types were obtained from KEGG and are described in the legend. Node labels are gene symbols.
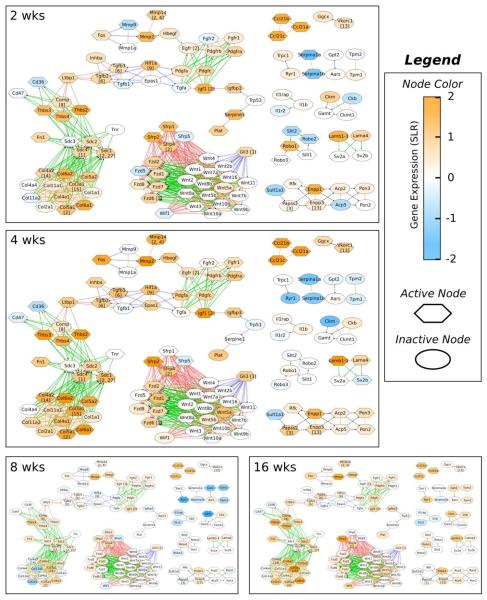
Fig. 3. Actively regulated subnetworks identified for each time point.
Active subnetwork, gene expression and cluster membership information is overlaid on the unioned, composite network from Fig. 2. Nodes considered active by cJAM (i.e. part of the active subnetwork) for each time point are indicated by hexagons. Node color represents gene expression as indicated by the color bar (orange is up-regulation, blue is down-regulation). Nodes that overlap with the previously described cluster analysis have the associated cluster numbers in square brackets after the gene symbol in the node label. Edge types are as described in the legend for Fig. 2.
The nodes included in an active subnetwork for a given time point were next analyzed using DAVID for gene-annotation enrichment in order to determine which specific pathways were actively regulated during the development of OA. Table 1 summarizes the primary active pathways for each time point, with specific gene names and the complete results of the DAVID analysis provided in Supplementary File 1. Most notably, the extracellular matrix (ECM)-receptor interaction pathway (KEGG_PATHWAY mmu04512) and the focal adhesion pathway (KEGG_PATHWAY mmu04510) were found to be enriched in the subnetworks identified in all four time points (Table 1). These pathways included the classic OA-associated gene COMP (cartilage oligomeric matrix protein), multiple collagen genes (COL1A1, COL2A1, COL3A1, COL4A1, COL4A2, COL4A4, COL5A1, COL5A2, COL6A1, COL6A2 and COL11A2), syndecans (SDC1, SDC2, SDC3, SDC4), thrombospondins (THBS2, THBS3, THBS4), FN1 (fibronectin 1), IGF1 (insulin-like growth factor-1), CD36, CD47, and TNR (tenascin R). These genes and/or members of these gene families have been implicated in the OA process in previous studies (for review see20–22) which helps to validate that this approach is finding relevant networks.
Table 1.
Summary of actively regulated pathways for each time point. Active genes for each pathway are detailed in Supplementary File 2.
| KEGG Pathway | Time Points | |||
|---|---|---|---|---|
| 2wks | 4wks | 8wks | 16wks | |
| ECM-receptor interaction | X | X | X | X |
| Focal adhesion | X | X | X | X |
| Wnt signaling pathway | X | X | X | |
| Hedgehog signaling pathway | X | X | X | |
| TGF-beta signaling pathway | X | X | X | |
| Riboflavin metabolism | X | |||
By examining the level of gene expression in the subnetworks at the various time points, it can be seen that many of the genes in the subnetworks were up-regulated in the DMM joints at the 2 and 4 week time points and then returned to levels similar to the sham control joints at 8 weeks (Fig. 3). At 8 weeks, the only two KEGG pathways that reached significance were the ECM-receptor interaction pathway and the focal adhesion pathway consistent with the importance of altered cell-matrix interactions in the OA process. Some of the genes in these pathways were down-regulated below the controls, including COL2A1 and COL11A1. These are both genes coding for fibrillar collagens found primarily in the articular cartilage. The decreased expression of these genes in the OA joints relative to controls could be due to a loss of articular cartilage as OA develops. However, at 16 weeks expression of COL2A1 along with COMP, THBS3, THBS4, COL3A1, COL5A1, COL5A2, COL6A1, and COL6A2 were increased, along with members of Wnt, Hedgehog and TGF-β signaling pathways, suggesting that a phasic process of active joint tissue remodeling was at play, as we had suggested previously8, rather than just a progressive loss of matrix as had been thought to occur in OA.
The Wnt signaling (KEGG_PATHWAY mmu04310), Hedgehog signaling (KEGG_PATHWAY mmu0430) and TGF-β signaling (KEGG_PATHWAY mmu04350) pathways were identified as actively regulated subnetworks at all time points except for 8 weeks (Table 1 and Fig. 3). Genes included in the Wnt signaling pathway were Wingless-type MMTV integration site family, member 5A (WNT5A), Frizzled family receptors (FZD1, FZD2, FZD6, FZD7) and secreted frizzled-related proteins (SFRP5, SFRP1, SFRP2, SFRP4). Genes in the Hedgehog signaling pathway included the transcriptional regulator Glioma-Associated Oncogene Family Zinc Finger 3 (GLI3) as well as WNT5A, and genes in the TGF-β signaling pathway including transforming growth factors (TGFB3, TGFB2), thrombospondins (THBS2, THBS3, THBS4), inhibin (INHBA), latent transforming growth factor beta binding protein (LTBP1) and COMP.
Although altered Wnt, Hedgehog, and TGF-β signaling has been previously implicated in the OA process21, 23–26, our analysis provided additional information on how genes in these pathways may interact during the development of OA. For example, there were multiple collagen genes in a subnetwork that connects to syndecans through the ability of collagens to activate syndecans which can also be activated by fibronectin, thrombospondins, tenascin and COMP that were found in the same subnetwork. COMP and thrombospondins can bind and inhibit latent TGF-binding protein 1 (Ltbp1) which in turn regulates activity of members of the TGF-β family. Although the Wnt and Hedgehog pathway members are in a separate subnetwork, studies have shown that Wnt signaling and cell-ECM adhesion pathways cross-talk via integrin and syndecan regulation of Wnt signaling (reviewed in27).
A small subnetwork that consisted of three isoforms of the same chemokine, CCL21A,CCL21B, and CCL21C, was unique in that it was up-regulated in DMM relative to sham control joints at all of the time points studied, even 8 weeks, but with the higher expression at 2 and 4 weeks (Fig. 3). We had previously reported up-regulation of CCL21 expression in a study where we compared expression of genes at 8 weeks after DMM surgery in young (12 week-old) and older adult (12 month-old) mice and localized CCL21 to chondrocytes and meniscal cells using immunohistochemistry10. CCL21 had been found in human OA synovial tissue and synovial fluid although at levels below that seen in rheumatoid arthritis where it is thought to promote angiogenesis28. The identification of the CCL21 subnetwork in the present analysis indicates that further studies on its role in OA are warranted.
3.2 Riboflavin subnetwork is active at 4 weeks
As noted above, the 4 week time point contained the largest number of nodes (72) of any time point included in the active subnetworks (Fig. 3). This time point included 5 genes in an active subnetwork identified by gene-annotation enrichment as the riboflavin metabolism pathway (KEGG_PATHWAY mmu00740). These genes included ENPP1 (Ectonucleotide Pyrophosphatase/Phosphodiesterase 1), which was the most upregulated of the 5 genes, ENPP3 (Ectonucleotide Pyrophosphatase/Phosphodiesterase 3), RFK (riboflavin kinase), ACP5 (Acid Phosphatase 5, Tartrate Resistant) and ACP2 (acid phosphatase 2, lysosomal). Although some of these genes, in particular ENPP1, have been individually associated with OA in previous studies29–31, the riboflavin metabolism pathway has not been specifically implicated in OA. However, supplements that include riboflavin have been found to reduce the severity of spontaneous OA in male STR/1N mice32 and C57 black mice33, and older adults with knee OA have been found to consume a diet that may be deficient in foods that contain riboflavin34. These reports, combined with the present network analysis, point to a possible role for the metabolism or deficiency of riboflavin in OA that deserves further investigation.
3.3 Overlap and comparison of active subnetworks with temporal gene expression clusters
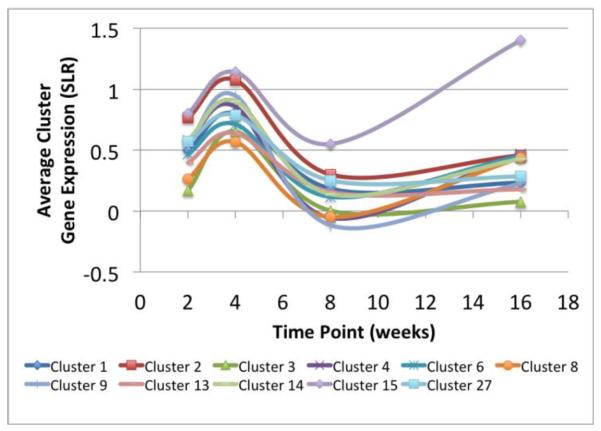
In our previous analysis of this gene expression dataset, we clustered the genes based on their temporal expression patterns into 27 gene clusters8. We intersected the previous clustering results with the set of genes identified in the active subnetworks in the present study to identify the genes that appeared in both analyses and to examine the relationship between the clusters and the active subnetworks. The genes appearing in both the subnetworks and the temporal expression clusters included COMP, SDC1, SDC4, COL4A2, COL3A1, COL5A1, SFRP4, FZD6, GLI3, MMP14, EGFR, TGFB3, TGFB2, VKORC1, HIF1A, ENPP3, PAPSS2 and IGF1 which were present in clusters 1–4, 6, 8, 9, 13–15 and 27 from the previous work8. Fig. 4 summarizes the temporal gene expression profiles of clusters relevant to this work. Genes present in the temporal clusters and also found in the actively regulated subnetworks are indicated in Fig. 3 by the cluster number shown in square brackets after the gene name.
Fig. 4. Summary of temporal profiles for relevant clusters.
Replicate gene expression values for each cluster at each time point were averaged and plotted for those clusters that overlapped with the network analysis.
With the exception of the Hedgehog signaling pathway, each of the identified pathways from the network analysis contained genes that had been previously placed in 2 or more clusters based on temporal gene expression. For example, the ECM-receptor interaction pathway in the subnetwork analysis contains genes from temporal clusters 1, 2, 8, 14, 15 and 27 while the TGF-β signaling pathway contains genes from clusters 6 (TGFB2,TGFB3) and 8 (COMP). The advantage to using the subnetwork analysis to identify KEGG pathways that were actively regulated at various time points during the OA time course can be seen when comparing the DAVID gene-annotation enrichment analysis results from the temporal clusters to that from the subnetworks. Analysis of temporal clusters 1, 2, 8, 14, 15 and 27 that included the genes identified as the ECM-receptor interaction pathway in the subnetwork analysis had revealed that clusters 8 and 15 had some ECM and cell adhesion-related enriched annotations based on gene ontology (GO) terms, but returned no significant KEGG pathways, while clusters 14 and 27 had no significant ECM annotations or pathways8. Cluster 2, the largest of the group, did contain significant annotations related to cell adhesion and the ECM, and the only significant KEGG_PATHWAY was that of Focal Adhesion—which is closely related to the ECM-receptor interaction pathway. It is not until you combine this group of clusters that you get the ECM-receptor interaction KEGG_PATHWAY as the most significantly enriched pathway (p-value=2.86E-4) for this set of genes (Supplemental File 1). This demonstrates that performing temporal clustering followed by enrichment analysis cannot reveal all the pathways regulated during the development of OA as each pathway has genes that are regulated differently over time, and thus will appear in several temporal clusters.
4. Conclusions
Network-based analysis of a time course gene expression dataset revealed the signaling and metabolic pathways that were actively regulated at one or more time points during the development of OA in a commonly used mouse model of the disease. The major pathways identified included the extracellular matrix-receptor interaction and the focal adhesion pathways along with the Wnt, Hedgehog and TGF-β signaling pathways. A newly identified pathway active at the 4 week time point was riboflavin metabolism. Members of these pathways have been examined for their role in OA in previous studies that have most often focused on one or a few genes in an individual pathway. However, the present study uniquely demonstrates how these pathways may interact with each other as subnetworks active at specific time points during the development of OA.
The predominance of ECM, focal adhesion and growth factor genes in the largest active subnetworks is consistent with an attempt of the joint tissues to repair or replace damaged and lost matrix as OA develops. This process appears to be driven by the Wnt, Hedgehog and TGF-β signaling pathways as well as by IGF-1. Although matrix degrading enzymes, such as the MMPs, and inflammatory mediators, such as cytokines and chemokines, are known to also be involved in the development of OA, only a few genes from these groups, including MMP-2, MMP-14, IL-1β and Ccl21, were found in the actively regulated subnetworks. This reflects a limitation of this type of network analysis which was mostly focused on identifying genes in signaling and metabolic pathways. Also, identification of active subnetworks requires that they contain genes already identified as part of that network in previous studies of known signaling or metabolic pathways. Despite these limitations, the network analysis enhanced and extended the information about the OA process gained from the previous cluster analysis of temporal gene expression and suggested additional genes within the key pathways that should be further studied.
Supplementary Material
Highlights
Network-based analysis of time course gene expression data identified pathways active during the development of osteoarthritis.
Active pathways included extracellular matrix-receptor interaction, focal adhesion, Wnt, Hedgehog and TGF-β signaling.
A unique pathway active early in the development of OA was riboflavin metabolism.
Acknowledgments
This work was supported by the National Institute of Arthritis Musculoskeletal and Skin Diseases (R37 AR049003-12) and by an Innovative Research Award from the Arthritis Foundation. The authors thank Brian Westwood for assistance with the network analysis.
Abbreviations
- COMP
cartilage oligomeric matrix protein
- DAVID
Database for Analysis, Visualization, Integration and Discovery
- DMM
destabilization of the medial meniscus
- EC
Enzyme-Compound interactions
- IGF-1
insulin-like growth factor-1
- JAM
jActiveModules
- cJAM
consensus jActiveModules
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- OA
Osteoarthritis
- PP
Protein-Protein interactions
- SLR
signal log ratio
- SVN
systematic variation normalization
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest There are no conflicts of interests.
Supplementary Files Supplementary File 1: Excel Sheet: DAVID gene-annotation enrichment analysis of selected clusters and of the cJAM results for each time point.
References
- 1.Lawrence RC, Felson DT, Helmick CG, Arnold LM, Choi H, Deyo RA, et al. Estimates of the prevalence of arthritis and other rheumatic conditions in the United States: Part II. Arthritis Rheum. 2008;58:26–35. doi: 10.1002/art.23176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prevalence of disabilities and associated health conditions among adults--United States, 1999. MMWR Morb Mortal Wkly Rep. 2001;50:120–125. [PubMed] [Google Scholar]
- 3.Appleton CT, Pitelka V, Henry J, Beier F. Global analyses of gene expression in early experimental osteoarthritis. Arthritis Rheum. 2007;56:1854–1868. doi: 10.1002/art.22711. [DOI] [PubMed] [Google Scholar]
- 4.Wei T, Kulkarni NH, Zeng QQ, Helvering LM, Lin X, Lawrence F, et al. Analysis of early changes in the articular cartilage transcriptisome in the rat meniscal tear model of osteoarthritis: pathway comparisons with the rat anterior cruciate transection model and with human osteoarthritic cartilage. Osteoarthritis Cartilage. 2010;18:992–1000. doi: 10.1016/j.joca.2010.04.012. [DOI] [PubMed] [Google Scholar]
- 5.Hopwood B, Tsykin A, Findlay DM, Fazzalari NL. Microarray gene expression profiling of osteoarthritic bone suggests altered bone remodelling, WNT and transforming growth factor-beta/bone morphogenic protein signalling. Arthritis Res Ther. 2007;9:R100. doi: 10.1186/ar2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kato H, Matsumine A, Wakabayashi T, Hasegawa M, Sudo A, Shintani K, et al. Large-scale gene expression profiles, differentially represented in osteoarthritic synovium of the knee joint using cDNA microarray technology. Biomarkers. 2007;12:384–402. doi: 10.1080/13547500601162482. [DOI] [PubMed] [Google Scholar]
- 7.Scanzello CR, McKeon B, Swaim BH, DiCarlo E, Asomugha EU, Kanda V, et al. Synovial inflammation in patients undergoing arthroscopic meniscectomy: molecular characterization and relationship to symptoms. Arthritis Rheum. 2011;63:391–400. doi: 10.1002/art.30137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Loeser RF, Olex AL, McNulty MA, Carlson CS, Callahan M, Ferguson C, et al. Disease progression and phasic changes in gene expression in a mouse model of osteoarthritis. PLoS One. 2013;8:e54633. doi: 10.1371/journal.pone.0054633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chou JW, Paules RS, Bushel PR. Systematic variation normalization in microarray data to get gene expression comparison unbiased. J Bioinform Comput Biol. 2005;3:225–241. doi: 10.1142/s0219720005001028. [DOI] [PubMed] [Google Scholar]
- 10.Loeser RF, Olex A, McNulty MA, Carlson CS, Callahan M, Ferguson C, et al. Microarray analysis reveals age-related differences in gene expression during the development of osteoarthritis in mice. Arthritis Rheum. 2012;64:705–717. doi: 10.1002/art.33388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Olex AL, Fetrow JS. SC(2)ATmd: a tool for integration of the figure of merit with cluster analysis for gene expression data. Bioinformatics. 2011;27:1330–1331. doi: 10.1093/bioinformatics/btr115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ideker T, Ozier O, Schwikowski B, Siegel AF. Discovering regulatory and signalling circuits in molecular interaction networks. Bioinformatics. 2002;18(Suppl 1):S233–240. doi: 10.1093/bioinformatics/18.suppl_1.s233. [DOI] [PubMed] [Google Scholar]
- 13.Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Avila-Campillo I, Drew K, Lin J, Reiss DJ, Bonneau R. BioNetBuilder: automatic integration of biological networks. Bioinformatics. 2007;23:392–393. doi: 10.1093/bioinformatics/btl604. [DOI] [PubMed] [Google Scholar]
- 16.Abramowitz M, Stegun IA. Handbook of Mathematical Functions With Formulas, Graphs, and Mathematical Tables. National Bureau of Standards; Washington, D.C.: 1972. [Google Scholar]
- 17.Chuang HY, Lee E, Liu YT, Lee D, Ideker T. Network-based classification of breast cancer metastasis. Mol Syst Biol. 2007;3:140. doi: 10.1038/msb4100180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cline MS, Smoot M, Cerami E, Kuchinsky A, Landys N, Workman C, et al. Integration of biological networks and gene expression data using Cytoscape. Nat Protoc. 2007;2:2366–2382. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dennis G, Jr., Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 20.Loeser RF, Goldring SR, Scanzello CR, Goldring MB. Osteoarthritis: A disease of the joint as an organ. Arthritis Rheum. 2012;64:1697–1707. doi: 10.1002/art.34453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loeser RF. Osteoarthritis year in review 2013: biology. Osteoarthritis Cartilage. 2013;21:1436–1442. doi: 10.1016/j.joca.2013.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goldring MB, Marcu KB. Cartilage homeostasis in health and rheumatic diseases. Arthritis Res Ther. 2009;11:224. doi: 10.1186/ar2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blom AB, Brockbank SM, van Lent PL, van Beuningen HM, Geurts J, Takahashi N, et al. Involvement of the Wnt signaling pathway in experimental and human osteoarthritis: prominent role of Wnt-induced signaling protein 1. Arthritis Rheum. 2009;60:501–512. doi: 10.1002/art.24247. [DOI] [PubMed] [Google Scholar]
- 24.Bush JR, Beier F. TGF-beta and osteoarthritis--the good and the bad. Nat Med. 2013;19:667–669. doi: 10.1038/nm.3228. [DOI] [PubMed] [Google Scholar]
- 25.van der Kraan PM, Blaney Davidson EN, van den Berg WB. A role for age-related changes in TGFbeta signaling in aberrant chondrocyte differentiation and osteoarthritis. Arthritis Res Ther. 2010;12:201. doi: 10.1186/ar2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin AC, Seeto BL, Bartoszko JM, Khoury MA, Whetstone H, Ho L, et al. Modulating hedgehog signaling can attenuate the severity of osteoarthritis. Nat Med. 2009;15:1421–1425. doi: 10.1038/nm.2055. [DOI] [PubMed] [Google Scholar]
- 27.Astudillo P, Larrain J. Wnt Signaling and Cell-Matrix Adhesion. Curr Mol Med. 2014 doi: 10.2174/1566524014666140128105352. in press. [DOI] [PubMed] [Google Scholar]
- 28.Pickens SR, Chamberlain ND, Volin MV, Pope RM, Mandelin AM, 2nd, Shahrara S. Characterization of CCL19 and CCL21 in rheumatoid arthritis. Arthritis Rheum. 2011;63:914–922. doi: 10.1002/art.30232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson K, Hashimoto S, Lotz M, Pritzker K, Goding J, Terkeltaub R. Up-regulated expression of the phosphodiesterase nucleotide pyrophosphatase family member PC-1 is a marker and pathogenic factor for knee meniscal cartilage matrix calcification. Arthritis Rheum. 2001;44:1071–1081. doi: 10.1002/1529-0131(200105)44:5<1071::AID-ANR187>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 30.Suk EK, Malkin I, Dahm S, Kalichman L, Ruf N, Kobyliansky E, et al. Association of ENPP1 gene polymorphisms with hand osteoarthritis in a Chuvasha population. Arthritis Res Ther. 2005;7:R1082–1090. doi: 10.1186/ar1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang R, Fang H, Chen Y, Shen J, Lu H, Zeng C, et al. Gene expression analyses of subchondral bone in early experimental osteoarthritis by microarray. PLoS One. 2012;7:e32356. doi: 10.1371/journal.pone.0032356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kurz B, Jost B, Schunke M. Dietary vitamins and selenium diminish the development of mechanically induced osteoarthritis and increase the expression of antioxidative enzymes in the knee joint of STR/1N mice. Osteoarthritis Cartilage. 2002;10:119–126. doi: 10.1053/joca.2001.0489. [DOI] [PubMed] [Google Scholar]
- 33.Wilhelmi G, Tanner K. Effect of riboflavin (vitamin B2) on spontaneous gonarthrosis in the mouse. Z Rheumatol. 1988;47:166–172. [PubMed] [Google Scholar]
- 34.White-O'Connor B, Sobal J, Muncie HL., Jr. Dietary habits, weight history, and vitamin supplement use in elderly osteoarthritis patients. J Am Diet Assoc. 1989;89:378–382. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.