Abstract
The objectives of this study were to determine if global DNA methylation, as reflected in LINE-1 and Alu elements, is associated with telomere length and whether it modifies the rate of telomeric change. A repeated-measures longitudinal study was performed with a panel of 87 boilermaker subjects. The follow-up period was 29 months. LINE-1 and Alu methylation was determined using pyrosequencing. Leukocyte relative telomere length was assessed via real-time qPCR. Linear-mixed models were used to estimate the association between DNA methylation and telomere length. A structural equation model (SEM) was used to explore the hypothesized relationship between DNA methylation, proxies of particulate matter exposure, and telomere length at baseline. There appeared to be a positive association between both LINE-1 and Alu methylation levels, and telomere length. For every incremental increase in LINE-1 methylation, there was a statistically significant 1.0 × 10−1 (95% CI: 4.6 × 10−2, 1.5 × 10−1, P < 0.01) unit increase in relative telomere length, controlling for age at baseline, current and past smoking status, work history, BMI (log kg/m2) and leukocyte differentials. Furthermore, for every incremental increase in Alu methylation, there was a statistically significant 6.2 × 10−2 (95% CI: 1.0 × 10−2, 1.1 × 10−1, P = 0.02) unit increase in relative telomere length. The interaction between LINE-1 methylation and follow-up time was statistically significant with an estimate −9.8 × 10−3 (95% CI: −1.8 × 10−2, −1.9 × 10−3, P = 0.02); suggesting that the rate of telomeric change was modified by the degree of LINE-1 methylation. No statistically significant association was found between the cumulative PM exposure construct, with global DNA methylation and telomere length at baseline.
Keywords: telomere, LINE-1, Alu, DNA methylation, longitudinal
Background
Telomere length has emerged as a prominent molecular marker of adverse health outcomes such as cancer and cardiovascular disease in epidemiological studies [Nilsson et al., 2013;Wentzensen et al., 2011]. Telomeres are characterized as hexameric (TTAGGG)n repeats, spanning thousands of base pairs at the ends of eukaryotic chromosomes. Furthermore, they interact with an assortment of DNA-binding proteins to form the protective telosome complex [Liu et al., 2004]. Telomeres erode by approximately 30–100 bp after each cellular division in somatic tissue due to limitations in lagging strand DNA synthesis at chromosomal ends and inadequate telomerase expression [Blackburn, 1991]. As a result, telomeres are analogous to a molecular clock that is reflective of mitotic history and degree of cellular aging. Additionally, telomeres have the pivotal role as a vanguard against induced nucleolytic decay, fusion of chromosomal ends, and atypical recombination [De Vivo et al., 2009]. Therefore, telomere length may also reflect the accumulation of past instances of environmental and biochemical trauma to the genome. In the event that telomeres truncate to critically short lengths, subsequent chromosomal fusions may result in adverse cellular abnormalities including neoplasia. Indeed, truncated telomeres have been found to be associated with increased risk of cancers of the bladder, lung, prostate, esophagus, and stomach [Ma et al., 2011].
DNA methylation is a reversible epigenetic augmentation of the genome that is forged in utero and during early stages of postnatal development. This modification is achieved through the addition of methyl groups to cytosine by methyl-transferases and is one of the primary mechanisms regulating expression of human genes [Suva et al., 2013]. Furthermore, DNA methylation exhibits cell-specific plasticity across the life span, with various environmental and lifestyle factors known to modify epigenetic signatures [Herceg, 2007; Slattery et al., 2007]. A substantial proportion of methylation sites are located in repetitive sequences such as short-interspersed nuclear elements (SINEs) and long-interspersed nuclear elements (LINEs) [Ehrlich et al., 1982; Lange et al., 2012]. There are over 500,000 LINEs and 1,500,000 SINEs across the human genome [Cordaux and Batzer, 2009]. Alu elements are the most common SINE; comprising 11% the human genome and containing a third of all methylation sites [Deininger and Batzer, 1999; Wilson et al., 2007]. The substantial representation of these repetitive elements throughout the genome makes their effectiveness as proxies for global DNA methylation highly evident. Indeed, methylation levels in Alu and LINE-1 repeats have been shown in previous studies to be associated with total genomic methylation content [Weisenberger et al., 2005;Yang et al., 2004].Moreover, global DNA hypomethylation has been shown to be associated with greater genomic instability and deregulation of gene expression [Gravina and Vijg, 2010]. Several studies have implicated the role of global DNA methylation in the variability of telomere length, which is biomarker of cancer. Maintenance and de novo DNA methyltransferases have been previously found to negatively regulate telomere length through global methylation of cytosine, including those located in subtelomeric regions [Gonzalo et al., 2006].
Particulate matter with an aerodynamic diameter less than 2.5 µm (PM2.5) is the primary size fraction suspected of toxicity [Xu and Zhang, 2004]. Inhalation of PM2.5 poses a substantial environmental health concern because the particles are deeply deposited into the alveoli, the primary lung compartment responsible for gas exchange [Wang et al., 2002]. The heavy metal components of PM2.5 produced from combustion processes can then readily diffuse into the blood stream and subsequently deposit into bone marrow, liver, kidneys, and other target organs [Laohaudomchok et al., 2010]. Metal particles such as lead, arsenic, cadmium, nickel, and manganese can disrupt tissue function and impact health outcomes through mechanisms involving oxidative stress, lipid peroxidation of cell membranes, and DNA adduct formation [Nuernberg et al., 2008]. Indeed, exposure to PM2.5 has been found in epidemiological studies to be associated with increased risk of cancer and cardiopulmonary outcomes [Pope et al., 2002]. In vitro studies have suggested that reactive oxygen species (ROS), one of the main cellular stressors produced via particulate matter exposure, may contribute to global DNA hypomethylation [Bollati et al., 2010]. PM2.5 exposure has been shown in numerous studies to be associated with both telomere length and global DNA methylation. In a study of steel workers, investigators found a significant short-term increase in leukocyte telomere length 3 days after particulate matter exposure compared with baseline samples [Dioni et al., 2011]. Furthermore, a short-term study of truck drivers in Beijing, China found that telomere length increased in association with personal PM2.5 and ambient PM10 [Hou et al., 2012]. With respect to epigenetic changes, surrogates for long-term cumulative PM2.5 exposure such as biological levels of heavy metals, have been found to be correlated with DNA methylation. In particular, arsenic concentration in toenails has been previously found to be negatively associated with LINE-1 methylation, whereas both iron and nickel were positively associated [Tajuddin et al., 2013].
Although telomeres are an established biomarker of chronic health outcomes, few if any studies have examined epigenetic factors affecting their length and rate of change in a longitudinal setting; leaving substantial gaps in knowledge to be addressed. In order to overcome past methodological limitations, the Harvard Boilermakers Longitudinal Study was used to assess the relationship between proxies of global DNA methylation, long-term particulate matter exposure, and telomere length dynamics. We hypothesize that epigenetic dysregulation in the form of global DNA hypomethylation, is associated with shorter telomeres and accelerates its rate of change. Therefore, the primary aim of this study was to determine if LINE-1 and Alu methylation levels, as established surrogates of global DNA methylation, are associated with telomere length and modifies the rate of telomeric change over a 2-year follow-up. Variables related to particulate matter exposure such as work history and toenail metal concentrations were included as potential confounders in the longitudinal and baseline analyses, respectively. Further understanding of the dynamics of telomere length regulation would shed light on the role these structures play in the etiology of chronic diseases and may ultimately support the development of preventative measures.
Materials and Methods
Study Design
The Harvard Boilermakers Longitudinal Study is a prospective open-cohort comprised of a series of periodic short-term panel studies. The source population consists of members of the International Brotherhood of Boilermakers Union, Local 29 in Quincy, MA. There are currently 400 active union members, 190 of which have been prospectively followed since 1999. Their primary occupational activity is assembling and welding boilers that provide high-pressure steam to drive electrical turbines in power plants. The inclusion criteria for this study were being male, unionized apprentices or journeymen, and over 18 years of age at the time of recruitment. The follow-up period for the current longitudinal study was from January 29th 2010 to June 16th 2012. The current study population consisted of 87 subjects at baseline. The timescale of interest was months of follow-up, while repeated measures for the outcome and predictors were obtained at irregularly spaced time intervals. There were a total of 451 blood draws (observations) between all the subjects. There was a median of 4 and mean of 5.9 ± 4.3 SD repeated measures. The range of repeated measures was from 1 to 18. This study was approved by the Institutional Review Board of Harvard School of Public Health. The authors declare no conflicts of interest.
Exposure Assessment—Pyrosequencing
Whole blood samples were extracted from subjects by a certified phlebotomist. Buffy coat was extracted from whole blood and stored in cell lyses solution at −20°C until DNA extraction. Peripheral blood leukocyte DNA was extracted using the QI Amp DNA blood kits (Qiagen). DNA methylation was assessed using the Pyromark MA System (Pyrosequencing Inc., Westborough, MA) by the Andrea Baccarelli laboratory. Briefly, 1 µg of genomic DNA at a concentration of 50 ng/µL was bisulfite treated using an EZ DNA Methylation-Gold Kit (Zymo Research, Orange, CA) according to the manufacturer’s protocol. Non-CpG cytosine residues were used as internal controls to verify bisulfite conversion. A 30-µL PCR reaction was then performed in 15 µL of GoTaq Green Master mix (Promega), 10 pmol forward primer, 10 pmol reverse primer, and 50 ng of bisulfite-treated genomic DNA. PCR cycling conditions for LINE-1 were 95°C for 30 sec, 56.3°C for 30 sec, and 72°C for 30 sec for 45 cycles. PCR cycling conditions for Alu were 95°C for 30 sec, 50°C for 30 sec, and 72°C for 30 sec for 45 cycles. Primers for the LINE-1 assay were: forward-(TTTTGAGTTAGGTGTGGGATATA), reverse-(biotin-AAAATCAAAAAATTCCCTTTC), sequencing primer-(AGTTAGGTGTGGGATATAGT), and sequencing entry-(TTC/TGT GGTGC/TGTC/TGTTTTTTAAGTC/TGGTTTGAAAAGC/TGTA). Primers for Alu assay were: forward-(biotin-TTTTTATTAAAAATATAAAAATT), reverse-(CCCAAACTAAAATACAATAA), sequencing primer-(AATAACTAAAATTACAAAC), and sequencing entry-(A/GCA/GCA/GCCAC/TCACA/GCCCA/GACT). PCR products were purified and sequenced via pyrosequencing as previously described [Bollati et al., 2007]. The degree of methylation was expressed for each CpG site as percent of methylated cytosines (%5 mC); calculated by dividing the number of methylated cytosines by the sum of methylated and unmethylated cytosines. Each sample was performed in duplicate to confirm reproducibility. The grand mean average degree of methylation was calculated for 5 random Alu regions and 4 random LINE-1 regions as proxies for global DNA methylation.
Outcome Assessment–Real-Time Quantitative PCR
Average relative telomere length was assessed using real-time quantitative PCR [Cawthon, 2002]. This assay determines the copy-number ratio between telomeric repeats and a single-copy (36B4) reference gene (T/S Ratio, −dCt). The T/S ratio is reflective of the average telomere length across all chromosomes in a population of cells and was calculated for each participant by subtracting the average 36B4 threshold cycle (Ct) value from the average telomere Ct value. The T/S ratio value for all experimental samples was then compared to the T/S ratio of a reference sample, consisting of a pooled genomic DNA sample in order to normalize for batch variations. The relative T/S ratio (−ddCt) was determined by subtracting the T/S ratio of the reference sample from the T/S ratio of each unknown sample, and then exponentiating (2− −ddCt). A modified version of the real-time PCR telomere assay was performed in a 384-well format with an Applied Biosystems 7900HT PCR System. Briefly, 5 ng of buffy-coat derived genomic DNA was dried down in a 384-well plate and resuspended in 10 µL of either the telomere or 36B4 reaction mixture for 2 hr at 4°C. The telomere reaction mixture consisted of 1× Quantitect SYBR Green Master Mix (Qiagen, Valencia, CA), 2.5 mM of DTT, 270 nM of Tel-1 primer-(GGTTTTTGAGGGTGAGGGTGAGGGTGAGGGTGAGGGT), and 900 nM of Tel-2 primer-(TCCCGACTATCCCTATCCCTATCCCTATCCCTATCCCTA). The reaction proceeded for 1 cycle at 95°C for 5 min, followed by 40 cycles at 95°C for 15 sec, and 54°C for 2 min. The 36B4 reaction consisted of 1× Quantitect SYBR Green Master Mix, 300 nM of 36B4U primer-(CAGCAAGTGGGAAGGTGTAATCC), and 500 nM of 36B4D primer- (CCCATTCTATCATCAACGGGTACAA). The 36B4 reaction proceeded for 1 cycle at 95°C for 5 min, followed by 40 cycles at 95°C for 15 sec, and 58°C for 1 min 10 sec. All samples for both the telomere and 36B4 reactions were performed in triplicate on different plates. Each 384-well plate contained a 6-point standard curve from 0.625 ng to 20 ng to assess PCR efficiency. The slope of the standard curve for both the telomere and 36B4 reactions was −3.40 ± 0.15. Quality control samples were interspersed throughout the plates in order to assess interplate and intraplate variability of Ct values. The interplate percent coefficient of variation for both the telomere Ct and 36B4 Ct values was under 0.75%. Multiple relative telomere length measurements for each subject within a 4-week timeframe were averaged to obtain a mean relative T/S ratio for that monthly unit of follow-up time.
Covariate, White Blood Cell Differential, and Toenail Metal Assessment
Self-reported questionnaires were used to collect demographic data such as birth date, sex, race, current smoking status, past smoking status, and work history. Number of years as a boilermaker was used as a proxy for long-term particulate matter exposure from welding fumes [Kile et al. 2013; Ozdemir et al., 1995]. Anthropometric data such as height and weight used to derive body mass index were measured by research personnel at the study site.
In order to control for variability in average leukocyte telomere length due to changing proportions of subtypes, complete blood counts including leukocyte differentials were assessed using a XE-2100™ Automated Hematology System (Sysmex, Kobe, Japan) by Laboratory Corporation of America Holdings (LabCorp) (Newton, MA). Proportions of neutrophils, lymphocytes, monocytes, eosinophils, and basophils were obtained.
Toenail samples were collected at baseline to assess biological accumulation of heavy metals. Toenail heavy metal concentrations were used as surrogate biological markers for long-term cumulative particulate matter exposure from welding fumes. Subjects with sufficient toenail growth clipped all 10 toenails at the study site. Subjects who provided toenails 21 or more days after questionnaire information collection by mail were excluded. Toenail concentrations of lead, manganese, cadmium, nickel, and arsenic were assessed at the Harvard School of Public Health Trace Metals Laboratory using a dynamic reaction cell-inductively coupled plasma mass spectrometer (DRC-ICP-MS, Elan 6100, Perkin Elmer, Norwalk, CT). Toenail clippings from all ten toes were combined for each subject and analyzed as previously described [Mordukhovich et al., 2012]. Five replicate reactions were performed for each subject. The net average concentration for each metal was calculated by subtracting detectable laboratory blank concentrations within each batch. Previously published studies have shown statistically significant correlations between toenail manganese, lead, and cadmium levels and exposure towelding fumes in the prior 7–9 months and 10–12 months [Grashow et al., 2013; Laohaudomchok et al., 2011]. Furthermore, other metals such as selenium have been found to be useful markers of exposure in a 26–52 week period prior to toenail collection [Longnecker et al., 1993].
Analysis
Structural Equation Model at Baseline
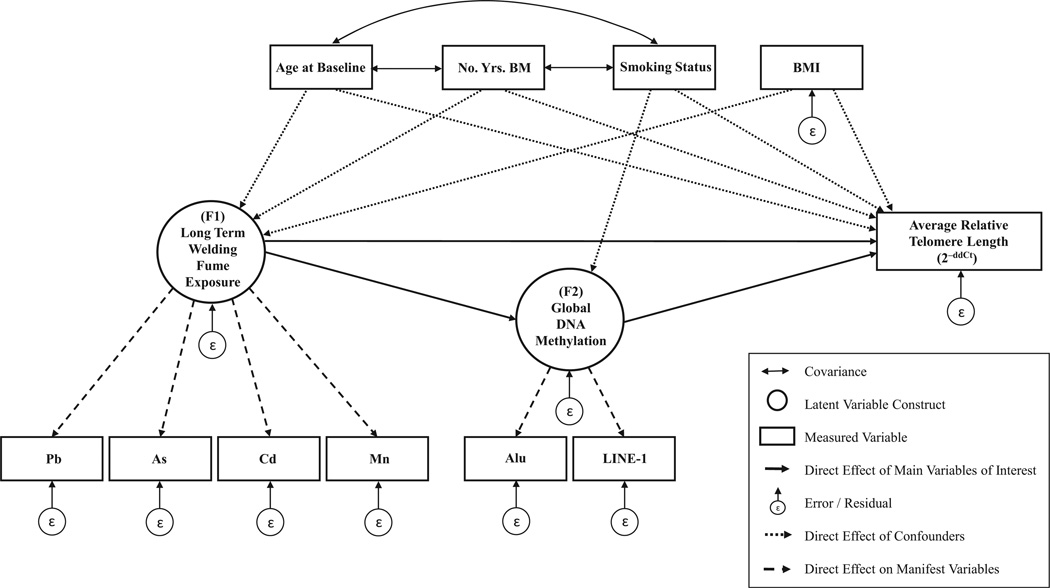
A cross-sectional structural equation model (SEM) was constructed to evaluate the hypothesized causal structure between global DNA methylation, long-term particulate matter exposure, and telomere length at baseline (Fig. 1). Structural equation modeling is a versatile framework that enables modeling of complex multivariate data and multiple predictors in unison [Sanchez et al., 2009]. Furthermore, SEMs allow for the evaluation of total, path-specific, direct, and indirect effects between all specified variables [VanderWeele, 2012]. Latent variables are singular, immeasurable concepts that are linearly represented by measured manifest variables. The measurement portion of the model contained latent variable constructs for cumulative particulate matter exposure in the past 7–12 months (F1) and global DNA methylation (F2). The latent variable for cumulative particulate matter was loaded onto measured surrogate biomarkers: log-transformed toenail levels (µg/g) of arsenic, cadmium, manganese, and lead. The latent variable for global DNA methylation was loaded onto the average degree of methylation (%5 mC) of LINE-1 and Alu repetitive elements. The structural portion of the model contained relative telomere length as the outcome, along with age at baseline blood draw, log-transformed number of years as a boilermaker, current smoking status, and log-transformed body mass index (BMI, kg/m2) as measured confounders. Overall model fit was assessed using a χ2 test, comparative fit index (CFI), root mean square error of approximation (RMSEA), and Tucker–Lewis Index (TLI). SEM analysis was performed using SPSS AMOS v.21.0 (IBM). Pearson correlation coefficients, simple linear regression, and scatterplots were used to assess linear associations between the outcome and continuous predictors. Mardia’s Kurtosis test was used to evaluate multivariate normality. Normality of residuals was checked with QQ-plots. Homoscedasticity was checked by plotting the residuals against their predicted values.
Figure 1.
Hypothesized cross-sectional causal structure between long-term welding fume exposure, global DNA methylation, and relative telomere length.
Linear-Mixed Models for Longitudinal Analysis
Linear-mixed effects regression models were used to analyze the association between repeated measures of relative telomere length as the outcome, and both time-varying LINE-1 and Alu %5mC as continuous exposures. The linear-mixed models also controlled for time-varying confounders such as log-transformed number of years as a boilermaker (continuous), smoking status (smoker/nonsmoker), past smoking (ever/never), log-transformed BMI (continuous), time-invariant age at baseline blood draw (continuous), white blood cell count, neutrophil%, lymphocyte%, monocyte%, and eosinophil%. Basophil proportions were not controlled for due to miniscule values. Interaction terms between the main effect and time were included to assess effect modification of the rate of telomeric change. The main effects for LINE-1 and Alu methylation were centered on their baseline mean values. Linear-mixed models with random intercept were implemented in SAS v9.3 using PROC MIXED. In order to obtain unbiased estimates with a limited sample size, restricted maximum likelihood (REML) was used for estimation.
Missing Data
The study population was an open cohort and therefore subject to staggered entry and loss of follow-up. Furthermore, data collection occurred at irregularly spaced time intervals. As a result, data was unbalanced across the time points. Linear-mixed models do not require data to be balanced nor collected at regularly spaced intervals because they use maximum likelihood (ML) or REML to estimate the coefficients and covariance structures [Cnaan et al., 1997]. The estimates are valid under the assumption that data are missing at random(MAR) and distributional assumptions are correct [Cnaan et al., 1997]. However, verification of MAR assumptions is rarely possible in epidemiological studies. Missing data in the structural equation model were handled using full-information maximum likelihood (FIML) estimation.
Results
Baseline Study Population Characteristics
The study population was composed of 87 male subjects. The subjects were 85.1% (74/87) self-identified white, whereas the remaining subjects were identified as African-American, Hispanic, and Asian (Table 1). One-way ANOVA analysis indicated no statistical difference in telomere length between categories of self-reported race (F-statistic 0.49, P value = 0.69). Race was not controlled for in the analysis because there were no statistical differences between groups and the study population was predominantly white. Current smokers composed 36.8% of the sample and ever smokers composed 54.0% of the sample. Two-sample Student’s t-tests indicated no statistical difference in telomere length between smokers and nonsmokers (P = 0.48), as well as between ever-and never smokers (P = 0.80). However, past and current smoking status was included as confounders in subsequent analysis because of its established causal relationship with telomere length [Cassidy et al., 2010; Liu et al., 2013; Sun et al., 2012]. The average age at baseline was 42.7 ± 13.1 SD years; having worked an average of 10.8 ± 10.5 SD years as a boilermaker. The average body mass index was 29.2 ± 6.3 SD kg/m2. A statistically significant positive linear association was found between mean LINE-1 and Alu methylation levels, as reflected by a Pearson coefficient of 0.36, P < 0.01.
Table 1.
Demographic and anthropometric characteristics of the Harvard boilermaker longitudinal study population at baseline
| Variable | Total n = 87 frequency |
(%) | Mean relative telomere length |
SD | Test statistica | P-value | |
|---|---|---|---|---|---|---|---|
| Current smoking status | |||||||
| Nonsmoker | 55 | 63.2 | 0.62 | 0.18 | − 0.71 | 0.48 | |
| Smoker | 32 | 36.8 | 0.65 | 0.17 | |||
| Past smoking status | |||||||
| Never | 17 | 19.5 | 0.63 | 0.17 | − 0.25 | 0.80 | |
| Ever | 47 | 54.0 | 0.65 | 0.19 | |||
| Self-reported race | |||||||
| White | 74 | 85.1 | 0.63 | 0.17 | 0.49 | 0.69 | |
| African-American | 8 | 9.2 | 0.64 | 0.23 | |||
| Hispanic | 3 | 3.4 | 0.56 | 0.24 | |||
| Asian | 2 | 2.3 | 0.76 | 0.11 | |||
| Mean | SD | Median | |||||
| Age | 42.7 | 13.1 | 40.5 | ||||
| Number of years working as a boilermaker | 10.8 | 10.4 | 9.0 | ||||
| Body mass index (BMI), kg/m2 | 29.2 | 6.3 | 27.6 | ||||
| Relative telomere length (2− −ddCt) | 0.63 | 0.17 | 0.61 | ||||
| DNA methylation | |||||||
| Grand mean Alu methylation (%5 mC) | 29.8 | 1.0 | 29.7 | ||||
| Average Alu (%5 mC) position 1 | 35.8 | 1.2 | 35.9 | ||||
| Average Alu (%5 mC) position 2 | 28.5 | 1.6 | 28.3 | ||||
| Average Alu (%5 mC) position 3 | 15.0 | 0.6 | 15.0 | ||||
| Average Alu (%5 mC) position 4 | 36.4 | 1.3 | 36.4 | ||||
| Average Alu (%5 mC) position 5 | 32.9 | 1.5 | 32.8 | ||||
| Grand mean LINE-1 methylation (%5 mC) | 83.1 | 1.0 | 83.2 | ||||
| Average LINE-1 (%5 mC) position 1 | 83.1 | 1.9 | 82.9 | ||||
| Average LINE-1 (%5 mC) position 2 | 80.7 | 1.1 | 80.7 | ||||
| Average LINE-1 (%5 mC) position 3 | 80.1 | 1.5 | 80.2 | ||||
| Average LINE-1 (%5 mC) position 4 | 88.8 | 2.0 | 88.8 | ||||
| Toenail metal concentrations | |||||||
| Pb (µg/g) | 0.67 | 1.03 | 0.31 | ||||
| As (µg/g) | 0.18 | 0.14 | 0.14 | ||||
| Cd (µg/g) | 0.01 | 0.06 | 0.01 | ||||
| Mn (µg/g) | 1.35 | 2.14 | 0.78 | ||||
A two-sample Student’s t-test was used to compare telomere length in covariates with two categories (t-statistic), whereas a one-way ANOVA was used to evaluate covariates with three or more categories (F-statistic). P-values < 0.05 were considered statistically significant.
Discrepancies in counts due to missing data.
Structural Equation Model: The Association Between Global DNA Methylation, Long-term Cumulative Fine Particulate Matter Exposure, and Telomere Length at Baseline
A structural equation model was used to evaluate the hypothesized causal structure between global DNA methylation, long-term fine particulate matter exposure, and telomere length at baseline (Fig. 1). The direct effect of global DNA methylation on average relative telomere length at baseline was statistically significant with a regression weight of 0.22 ± 0.08 SE P < 0.01 (Table 2). No statistically significant association was found between the cumulative PM exposure construct, with global DNA methylation and telomere length at baseline. Log-transformed body mass index was highly negatively associated with the latent variable for cumulative PM exposure with a regression weight of −0.92 ± 0.34 SE P < 0.01. The χ2 statistic of 38.99 (P = 0.18) was above the threshold of α = 0.05 which was indicative of adequate overall model fit. Furthermore, the comparative fit index (CFI) was 0.95 which also corroborates a well-fitted model. The normalized χ2 test (CFMIN/DF), which minimizes the impact of sample size on the χ2 statistic was 1.22; below the threshold of 2.00 that indicated an adequate fit. The root mean square error of approximation (RMSEA) was 0.05 which indicated a fair fitted model. The Mardia’s Kurtosis test statistic of 0.12, P = 0.90 indicated that there was no statistically significant departure from multivariate normality.
Table 2.
Structural equation model relating global DNA methylation and long-term welding fume exposure to telomere length at baseline (n = 87)
| Regression weight |
Standardized estimate |
Standard error |
Critical ratio |
P-value | ||
|---|---|---|---|---|---|---|
| Path from welding fume exposure in the past year (F1) to: | ||||||
| Toenail arsenic (As) | 1.00a | 1.23 | ||||
| Toenail lead (Pb) | 0.27 | 0.23 | 0.18 | 1.52 | 0.13 | |
| Toenail manganese (Mn) | 0.46 | 0.39 | 0.25 | 1.84 | 0.07 | |
| Toenail cadmium (Cd) | 0.77 | 0.85 | 0.22 | 3.46 | <0.001b | |
| Global DNA methylation (F2) | − 0.07 | − 0.16 | 0.07 | − 1.01 | 0.31 | |
| Average relative telomere length | 0.03 | 0.13 | 0.03 | 1.09 | 0.28 | |
| Path from global DNA methylation (F2) to: | ||||||
| Mean Alu methylation (%5mC) | 1.00a | 0.37 | ||||
| Mean LINE-1 methylation (%5 mC) | 2.73 | 0.97 | 1.57 | 1.74 | 0.08 | |
| Average relative telomere length | 0.22 | 0.46 | 0.08 | 2.62 | <0.01b | |
| Path from Age at blood draw (years) to: | ||||||
| Welding fume exposure in the past year (F1) | − 0.02 | − 0.29 | 0.01 | − 2.54 | 0.01b | |
| Average relative telomere length | 0.00 | − 0.16 | 0.00 | − 1.04 | 0.30 | |
| Path from current smoking status to: | ||||||
| Welding fume exposure in the past year (F1) | 0.22 | 0.13 | 0.14 | 1.56 | 0.12 | |
| Global DNA methylation (F2) | − 0.05 | − 0.06 | 0.10 | − 0.46 | 0.64 | |
| Average relative telomere length | 0.03 | 0.09 | 0.04 | 0.80 | 0.42 | |
| Path from log-transformed body mass index (BMI) to: | ||||||
| Welding fume exposure in the past year (F1) | − 0.92 | − 0.22 | 0.34 | − 2.74 | <0.01b | |
| Average relative telomere length | − 0.14 | − 0.16 | 0.09 | − 1.50 | 0.13 | |
| Path from log-transformed number of years as a boilermaker to: | ||||||
| Welding fume exposure in the past year (F1) | 0.04 | 0.05 | 0.10 | 0.42 | 0.67 | |
| Average relative telomere length | 0.02 | 0.09 | 0.03 | 0.59 | 0.56 | |
In order to establish a measurement scale for the latent variable, the first manifest variable loading must be fixed at 1.00 and not estimated. Model Fit Indices: Chi-square = 38.99, P = 0.18, CMIN/DF = 1.22, CFI = 0.95, TLI = 0.89, RMSEA = 0.05.
P-values < 0.05 were considered statistically significant.
Linear-Mixed Models: Longitudinal Analysis of the Association Between DNA Methylation and Telomere Length
LINE-1 methylation was found to be positively associated with telomere length in the linear-mixed models (Table 3). The coefficient for LINE-1%5mC was highly significant with an estimate of 1.0 × 10−1 (95% CI: 4.6 × 10−2, 1.5 × 10−1, P < 0.01) controlling for all measured covariates at any month of follow-up. Furthermore, for every unit increase in Alu methylation, there was a statistically significant 6.2 × 10−2 (95% CI: 1.0 × 10−2, 1.1 × 10−1, P = 0.02) unit increase in relative telomere length, controlling for measured covariates at any month of follow-up. The interaction term between LINE-1 methylation and follow-up months was statistically significant with an estimate of −9.8 × 10−3 (95% CI: −1.8 × 10−2, −1.9 × 10−3, P = 0.02); indicating that LINE-1 DNA methylation modified the rate of change of telomere length. Furthermore, number of years as a boilermaker was not significantly associated with telomere length in both the Alu and LINE-1 models, in addition to the model treating number of years as a boilermaker as the main effect (Tables 3 and 4).
Table 3.
Longitudinal analysis of the association between global DNA methylation and telomere length (2010−2012)
|
aModel 1 |
bModel 2 |
cModel 3 |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Main effect of LINE-1 methylation | Estimate | 95%CI lower | 95%CI upper | P-value | Estimate | 95%CI lower | 95%CI upper | P-value | Estimate | 95%CI lower | 95%CI upper | P-value |
| Grand mean LINE-1 (%5 mC) | 7.5 × 10−2 | 4.1 ×10−2 | 1.1 × 10−1 | <0.0001d | 8.0 × 10−2 | 4.1 × 10−2 | 1.2 × 10−1 | <0.001d | 1.0 × 10−1 | 4.6 × 10−2 | 1.5 × 10−1 | <0.01d |
| Time (months of follow-up) | − 6.7 × 10−3 | − 1.2 × 10−2 | − 1.5 × 10−3 | <0.01d | − 7.5 × 10−3 | − 1.3 × 10−2 | − 1.6 × 10−3 | 0.01d | − 7.8 × 10−3 | − 1.7 × 10−2 | 1.6 × 10−3 | 0.10 |
| LINE-1 × Time interaction | − 9.5 × 10−3 | − 1.5 × 10−2 | − 4.3 × 10−3 | <0.001d | − 1.0 × 10−2 | − 1.6 × 10−2 | − 4.2 × 10−3 | <0.01d | − 9.8 × 10−3 | − 1.8 × 10−2 | − 1.9 × 10−3 | 0.02d |
| Main effect of Alu methylation | ||||||||||||
| Grand mean Alu (%5 mC) | 2.9 × 10−2 | − 9.3 × 10−3 | 6.7 × 10−2 | 0.14 | 2.9 × 10−2 | − 1.2 × 10−2 | 7.1 × 10−2 | 0.16 | 6.2 × 10−2 | 1.0 × 10−2 | 1.1 × 10−1 | 0.02d |
| Time (months of follow-up) | − 5.4 × 10−3 | − 1.1 × 10−2 | − 7.0 × 10−5 | <0.05d | − 6.6 × 10−3 | − 1.3 × 10−2 | − 4.0 × 10−4 | 0.04d | − 4.3 × 10−3 | − 1.3 × 10−2 | 4.7 × 103 | 0.32 |
| Alu × time interaction | − 3.5 × 10−3 | − 9.3 × 10−3 | 2.4 × 10−3 | 0.24 | − 3.5 × 10−3 | − 1.0 × 10−2 | 3.1 × 10−3 | 0.29 | − 6.0 × 10−3 | − 1.3 × 10−2 | 1.4 × 10−3 | 0.11 |
Model 1: Random intercept linear-mixed model with main effect, time, and main effect × time interaction term.
Model 2: Random intercept linear-mixed model additionally controlling for age at baseline blood draw, current and past smoking status, log-transformed No. of years as a boilermaker, and log-transformed BMI (kg/m2).
Model 3: Random intercept linear-mixed model additionally controlling for age at baseline blood draw, current and past smoking status, log-transformed No. of years as a boilermaker, and log-transformed BMI (kg/m2), white blood cell count, neutrophil%, lymphocyte%, monocyte%, and eosinophil%.
P-values < 0.05 were considered statistically significant.
Table 4.
Longitudinal analysis of the association between work history and telomere length (2010−2012)
|
aModel 4 |
bModel 5 |
cModel 6 |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Main effect of work history | Estimate | 95%CI lower | 95%CI upper | P-value | Estimate | 95%CI lower | 95%CI upper | P-value | Estimate | 95%CI lower | 95%CI upper | P-value |
| Log number of years as BM | − 3.8 × 10−2 | − 7.8 × 10−2 | 1.3 × 10−3 | 0.06 | 3.0 × 10−3 | − 5.4 × 10−2 | 6.0 × 10−2 | 0.92 | − 3.6 × 10−2 | − 1.3 × 10−1 | 5.5 × 10−2 | 0.40 |
| Time (months of follow-up) | − 3.2 × 10−3 | − 1.6 × 10−2 | 9.9 × 10−3 | 0.63 | − 1.0 × 10−2 | − 2.7 × 10−2 | 6.2 × 10−3 | 0.22 | − 9.3 × 10−4 | − 2.8 × 10−2 | 2.6 × 10−2 | 0.94 |
| Log number of years BM × time interaction | − 1.6 × 10−3 | − 8.9 × 10−3 | 5.7 × 10−3 | 0.66 | 2.3 × 10−3 | − 6.9 × 10−3 | 1.1 × 10−2 | 0.62 | − 3.6 × 10−3 | − 1.7 × 10−2 | 9.9 × 10−3 | 0.57 |
Model 4: Random intercept linear-mixed model with main effect, time, and main effect × time interaction term.
Model 5: Random intercept linear-mixed model additionally controlling for age at baseline blood draw, current and past smoking status, and log-transformed BMI (kg/m2).
Model 6: Random intercept linear-mixed model additionally controlling for age at baseline blood draw, current and past smoking status, and log-transformed BMI (kg/m2), white blood cell count, neutrophil%, lymphocyte%, monocyte%, and eosinophil%.
P-values < 0.05 were considered statistically significant.
Discussion
This study is one of the few longitudinal investigations into the relationship between global DNA methylation and telomere length. Furthermore, this study incorporates a recent fundamental shift in approaching the study of telomeres; evaluating the rate of change as opposed to viewing telomeres only as static entities. The most significant finding of this study was that LINE-1 and Alu methylation was positively associated with telomere length in peripheral blood leukocytes (Table 3). Furthermore, LINE-1 methylation, but not Alu methylation, was found to modify the rate of telomeric change over the follow-up. The longitudinal main effects were corroborated by the structural equation model analysis at baseline. Taken together, the direction of these findings suggests that global DNA hypomethylation may be associated with shorter telomere length in white blood cells in our target population.
The discrepancy between LINE-1 and Alu methylation with respect to their modification of rates of telomeric change may have a biological basis. Although this study focused on global DNA methylation, previous investigations have shown that methylation status of subtelomeric regions are associated with telomere length and may be important regions of epigenetic regulation of telomere dynamics [Vera et al., 2008; Yehezkel et al., 2008]. LINE-1 and Alu elements have nonhomogeneous densities and distributions throughout the genome. SINEs tend to accumulate in regions of the genome with high GC content, whereas LINEs tend to incorporate into AT-rich regions [Hackenberg et al., 2005; Sellis et al., 2007; Soriano et al., 1983]. Given these characteristics, LINEs may be more represented in subtelomeric regions of the genome compared to Alu elements. Indeed, subtelomeric mapping studies of chromosomal assemblies have found that LINEs composed approximately 25% of the number of bases in the subtelomere compared to 10% for SINEs [Riethman et al., 2004]. Conversely, LINEs composed about 20% of the number of bases across the genome compared to 12% for SINEs. Moreover, the study also found very large strand and subtelomere-specific biases in LINE, SINE, LTR, and DNA repeat content [Riethman et al., 2004]. The overrepresentation of LINEs in the subtelomeric region may explain why there appeared to be a significant statistical association between LINE-1 methylation and telomere length, in addition to an interaction with follow-up time (Table 3). Although increasing amounts of LINE-1%5 mC appeared to modify the rate of telomeric change, a significant interaction between Alu methylation and follow-up time was not observed. This result may be due to the underrepresentation of Alu elements in subtelomeric regions. Conversely, the disparate findings with respect to effect modification of rates between LINE-1 and Alu may be attributed to chance from limited sample sizes. However, the use of repeated-measures greatly increases statistical power to observe small effects.
The cross-sectional relationship between global DNA methylation and telomere length was evaluated at baseline using a structural equation model. The SEM indicated a significant positive association between the construct for global DNA methylation and telomere length; which was in concordance with the positive associations between LINE-1 and Alu %5mC with telomere length from the longitudinal analysis. SEMs are a powerful framework for evaluating the relationship between multiple predictors, latent variables, and outcomes simultaneously; thereby alleviating false discovery rate issues inherent in multiple testing. However, SEMs only test the associations of the assumed causal structure as specified by the investigator. The presence or absence of arrows must be specified a priori based on expert subject matter knowledge and assumptions [Bollen and Noble, 2011; Greenland et al., 1999]. Although temporality is difficult to assess with SEMs in cross-sectional situations, the purpose of using a SEM was to confirm and disconfirm associations, rather than to infer causation. Fitting of SEMs is sensitive to both model specification and sample size [Bollen and Noble, 2011].With 87 subjects in the current study, the sample size was on the lower limits of acceptability for use in SEMs [Bagozzi et al., 2012]. However, the proposed model was not overly complex with only 2 latent and 11 measured variables. Even with a limited sample size, the effect of global DNA methylation on telomere length was highly significant. A study of this sample size and number of parameters has 80% power to detect a minimum effect size of 0.13 [Westland, 2010]. Furthermore, all relevant fit statistics including χ2 test, CMIN/DF, CFI, RMSEA, and TLI indicated an adequately fitted model despite assessing different criteria. In the measurement portion of the SEM, the standardized estimate for arsenic was above a value of 1, which may indicate a high degree of multicollinearity or an unreasonable model constraint may have been imposed. However, if the factors are correlated and the factor loadings are regression coefficients but not correlations, they can be validly larger than 1 in magnitude [Jöreskog, 1999].
The most prominent strength of this study was its prospective nature that permits the temporality between exposure, covariates, and outcomes to be established; allowing greater control of confounding. Moreover, the prospective cohort design imparts robustness against differential misclassification. Although the sample size was limited, the incorporation of repeated measurements of telomere length over the follow-up period substantially increases statistical power to detect subtle effects [Bernal-Rusiel et al., 2012;Vickers, 2003]. Furthermore, considering the entire source population was 400 union members, the study population actually constituted a sizable proportion of the totality.
This study addresses gaps in knowledge pertaining to epigenetic factors affecting telomere length and its rate of change. There have been few if any studies in the literature that have examined this relationship. The majority of studies focusing on epigenetic regulation of telomere length have been laboratory experiments in cell lines and animal models; limiting their generalizability to human populations. This is one of the few epidemiological studies that has attempted to use SEMs to characterize long-term particulate matter exposure and global DNA methylation. Both cumulative PM2.5 exposure and global methylation do not have singular accepted quantitative measures. In particular, long-term cumulative PM2.5 is difficult to assess due to contribution from multiple emission sources and its complex composition of particles. SEMs permit estimation of the effect these immeasurable latent constructs on an outcome. The use of heavy metal biomarkers of particulate matter exposure as manifest variables in the construct was advantageous because they allowed assessment of cumulative biological intake in specific exposure windows. The use of metal biomarkers circumvents external factors affecting person-specific intake of air particles such as respiratory rate, particle concentration, lifestyle factors, and multiple emission sources.
As with all scientific investigations, this study was not without limitations. Both telomere length and DNA methylation were laboratory measurements on a continuous scale; therefore the presence of random misclassification (mismeasurement) is unavoidable and would increase variance of the estimates in linear models. However, even in the presence of random misclassification and increased variance, statistically significant associations were still found between DNA methylation and telomere length. The presence of differential misclassification of the exposures and outcome was unlikely because DNA methylation and telomere length were independently assessed via laboratory assays. The use of real-time qPCR to assess telomere length has various trade-offs. It is currently the most cost effective method for high-throughput measurement of telomere length and requires only a fraction of genomic DNA compared to Southern blot and quantitative fluorescent in situ hybridization. However, the T/S ratio is a relative, not an absolute metric of telomere length. Although a previous technical study described the use of oligomer standard curves to quantify absolute telomere length, the method was not used because small variances while working in the picomolar scale can result in substantial systematic bias [O’Callaghan et al., 2008; O’Callaghan and Fenech, 2011]. However, previous studies have established a high correlation between relative T/S ratio and absolute telomere length [Cawthon, 2002]. The T/S ratio reflects the average telomere length across a population of cells but does not assess chromosome-specific telomere length [Britt-Compton et al., 2006]. Despite this drawback, relative average telomere length is still a highly informative metric as exemplified in its association with numerous chronic health outcomes [Fyhrquist et al., 2013;Wentzensen et al., 2011]. There was the possibility of unmeasured confounding in this study. However, the most pertinent time varying and time invariant confounders related to both global DNA methylation and telomere length, including white blood cell differentials, were controlled for in the analysis. Although controlling for time-varying confounders in traditional models may result in selection bias (conditioning on a collider), the degree of bias would depend on the strength of association of the measured and unmeasured covariates [Hernan and Robins, 2006].
Conclusions
In summary, significant positive associations were found between surrogates of global DNA methylation and telomere length in a prospective occupational cohort. These findings suggest that global DNA hypomethylation may be related to decreased telomere length in peripheral blood leukocytes in our source population. Furthermore, a significant interaction was observed between LINE-1 %5mC and follow-up time, which suggests that LINE-1 methylation signatures may modify the rate of telomeric change. The apparent rate of change in telomere length over the 2-year follow-up was modified by increased global DNA methylation as reflected in LINE-1. Findings from this study are consistent with the concept that deregulation of epigenetic signatures throughout the genome may affect telomere length and the rate in which it changes over time. Although the sample size was limited, this exploratory study may potentially stimulate further research into the nuances behind epigenetic and environmental factors affecting telomere length regulation and its temporal dynamics. Future longitudinal studies would benefit from focusing on gene-specific methylation signatures potentially related to telomere length. Moreover, future studies involving larger cohorts, balanced repeated measures of telomere length, and extended follow-up periods would aid in unraveling the role telomeres play in the etiological tapestry of chronic diseases.
Acknowledgments
This project was made possible by funding from the National Institute of Environmental Health Sciences (NIEHS) grants R01ES009860 and P50ES00002, in addition to the CPWR: The Center for Construction Research and Training through NIOSH cooperative agreement OH009762. We would like to thank Jinming Zhang, Hyang-Min Byun, Shona Fang, Li Su, Zhonghua Liu, Joel Schwartz, and Donna Spiegelman for their support.
References
- Bagozzi RP, Bergami M, Marzocchi GL, Morandin G. Customer-organization relationships: development and test of a theory of extended identities. J Appl Psychol. 2012;97(1):63–76. doi: 10.1037/a0024533. [DOI] [PubMed] [Google Scholar]
- Bernal-Rusiel JL, Greve DN, Reuter M, Fischl B, Sabuncu MR. Statistical analysis of longitudinal neuroimage data with linear mixed effects models. Neuroimage. 2012;66C:249–260. doi: 10.1016/j.neuroimage.2012.10.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn EH. Structure and function of telomeres. Nature. 1991;350(6319):569–573. doi: 10.1038/350569a0. [DOI] [PubMed] [Google Scholar]
- Bollati V, Baccarelli A, Hou L, Bonzini M, Fustinoni S, Cavallo D, Byun HM, Jiang J, Marinelli B, Pesatori AC, et al. Changes in DNA methylation patterns in subjects exposed to low-dose benzene. Cancer Res. 2007;67(3):876–880. doi: 10.1158/0008-5472.CAN-06-2995. [DOI] [PubMed] [Google Scholar]
- Bollati V, Marinelli B, Apostoli P, Bonzini M, Nordio F, Hoxha M, Pegoraro V, Motta V, Tarantini L, Cantone L, et al. Exposure to metal-rich particulate matter modifies the expression of candidate microRNAs in peripheral blood leukocytes. Environ Health Perspect. 2010;118(6):763–768. doi: 10.1289/ehp.0901300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollen KA, Noble MD. Structural equation models and the quantification of behavior. Proc Natl Acad Sci USA. 2011;108(Suppl 3):15639–15646. doi: 10.1073/pnas.1010661108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britt-Compton B, Rowson J, Locke M, Mackenzie I, Kipling D, Baird DM. Structural stability and chromosome-specific telomere length is governed by cisacting determinants in humans. Hum Mol Genet. 2006;15(5):725–733. doi: 10.1093/hmg/ddi486. [DOI] [PubMed] [Google Scholar]
- Cassidy A, De Vivo I, Liu Y, Han J, Prescott J, Hunter DJ, Rimm EB. Associations between diet, lifestyle factors, and telomere length in women. Am J Clin Nutr. 2010;91(5):1273–1280. doi: 10.3945/ajcn.2009.28947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cawthon RM. Telomere measurement by quantitative PCR. Nucleic Acids Res. 2002;30(10):e47. doi: 10.1093/nar/30.10.e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cnaan A, Laird NM, Slasor P. Using the general linear mixed model to analyse unbalanced repeated measures and longitudinal data. Stat Med. 1997;16(20):2349–2380. doi: 10.1002/(sici)1097-0258(19971030)16:20<2349::aid-sim667>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nat Rev Genet. 2009;10(10):691–703. doi: 10.1038/nrg2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Counter CM, Gupta J, Harley CB, Leber B, Bacchetti S. Telomerase activity in normal leukocytes and in hematologic malignancies. Blood. 1995;85(9):2315–2320. [PubMed] [Google Scholar]
- De Vivo I, Prescott J, Wong JY, Kraft P, Hankinson SE, Hunter DJ. A prospective study of relative telomere length and postmenopausal breast cancer risk. Cancer Epidemiol Biomarkers Prev. 2009;18(4):1152–1156. doi: 10.1158/1055-9965.EPI-08-0998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deininger PL, Batzer MA. Alu repeats and human disease. Mol Genet Metab. 1999;67(3):183–193. doi: 10.1006/mgme.1999.2864. [DOI] [PubMed] [Google Scholar]
- Dioni L, Hoxha M, Nordio F, Bonzini M, Tarantini L, Albetti B, Savarese A, Schwartz J, Bertazzi PA, Apostoli P, et al. Effects of short-term exposure to inhalable particulate matter on telomere length, telomerase expression, and telomerase methylation in steel workers. Environ Health Perspect. 2011;119(5):622–627. doi: 10.1289/ehp.1002486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich M, Gama-Sosa MA, Huang LH, Midgett RM, Kuo KC, McCune RA, Gehrke C. Amount and distribution of 5-methylcytosine in human DNA from different types of tissues of cells. Nucleic Acids Res. 1982;10(8):2709–2721. doi: 10.1093/nar/10.8.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fyhrquist F, Saijonmaa O, Strandberg T. The roles of senescence and telomere shortening in cardiovascular disease. Nat Rev Cardiol. 2013;10(5):274–283. doi: 10.1038/nrcardio.2013.30. [DOI] [PubMed] [Google Scholar]
- Gonzalo S, Jaco I, Fraga MF, Chen T, Li E, Esteller M, Blasco MA. DNA methyltransferases control telomere length and telomere recombination in mammalian cells. Nat Cell Biol. 2006;8(4):416–424. doi: 10.1038/ncb1386. [DOI] [PubMed] [Google Scholar]
- Grashow R, Zhang J, Fang SC, Weisskopf MG, Christiani DC, Cavallari JM. Toenail metal concentration as a biomarker of occupational welding fume exposure. J Occupational Environ Hygiene. 2013 doi: 10.1080/15459624.2013.875182. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravina S, Vijg J. Epigenetic factors in aging and longevity. Pflugers Arch. 2010;459(2):247–258. doi: 10.1007/s00424-009-0730-7. [DOI] [PubMed] [Google Scholar]
- Greenland S, Pearl J, Robins JM. Causal diagrams for epidemiologic research. Epidemiology. 1999;10(1):37–48. [PubMed] [Google Scholar]
- Hackenberg M, Bernaola-Galvan P, Carpena P, Oliver JL. The biased distribution of Alus in human isochores might be driven by recombination. J Mol Evol. 2005;60(3):365–377. doi: 10.1007/s00239-004-0197-2. [DOI] [PubMed] [Google Scholar]
- Herceg Z. Epigenetics and cancer: towards an evaluation of the impact of environmental and dietary factors. Mutagenesis. 2007;22(2):91–103. doi: 10.1093/mutage/gel068. [DOI] [PubMed] [Google Scholar]
- Hernan MA, Robins JM. Estimating causal effects from epidemiological data. J Epidemiol Community Health. 2006;60(7):578–586. doi: 10.1136/jech.2004.029496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou L, Wang S, Dou C, Zhang X, Yu Y, Zheng Y, Avula U, Hoxha M, Diaz A, McCracken J, et al. Air pollution exposure and telomere length in highly exposed subjects in Beijing, China: a repeated-measure study. Environ Int. 2012;48:71–77. doi: 10.1016/j.envint.2012.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jöreskog K. How large can a standardized coefficient be? Scientific Software Central. 1999 Available at: http://www.ssicentral.com/lisrel/techdocs/HowLargeCanaStandardizedCoefficientbe.pdf. [Google Scholar]
- Kile ML, Fang S, Baccarelli AA, Tarantini L, Cavallari J, Christiani DC. A panel study of occupational exposure to fine particulate matter and changes in DNA methylation over a single workday and years worked in boilermaker welders. Environ Health. 2013;12(1):47. doi: 10.1186/1476-069X-12-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange NE, Sordillo J, Tarantini L, Bollati V, Sparrow D, Vokonas P, Zanobetti A, Schwartz J, Baccarelli A, Litonjua AA, et al. Alu and LINE-1 methylation and lung function in the normative ageing study. BMJ Open. 2012;2(5) doi: 10.1136/bmjopen-2012-001231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laohaudomchok W, Cavallari JM, Fang SC, Lin X, Herrick RF, Christiani DC, Weisskopf MG. Assessment of occupational exposure to manganese and other metals in welding fumes by portable X-ray fluorescence spectrometer. J Occup Environ Hyg. 2010;7(8):456–465. doi: 10.1080/15459624.2010.485262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laohaudomchok W, Lin X, Herrick RF, Fang SC, Cavallari JM, Christiani DC, Weisskopf MG. Toenail, blood, and urine as biomarkers of manganese exposure. J Occup Environ Med. 2011;53(5):506–510. doi: 10.1097/JOM.0b013e31821854da. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, O’Connor MS, Qin J, Songyang Z. Telosome, a mammalian telomere-associated complex formed by multiple telomeric proteins. J Biol Chem. 2004;279(49):51338–51342. doi: 10.1074/jbc.M409293200. [DOI] [PubMed] [Google Scholar]
- Liu JJ, Prescott J, Giovannucci E, Hankinson SE, Rosner B, De Vivo I. Onecarbon metabolism factors and leukocyte telomere length. Am J Clin Nutr. 2013;97(4):794–799. doi: 10.3945/ajcn.112.051557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longnecker MP, Stampfer MJ, Morris JS, Spate V, Baskett C, Mason M, Willett WC. A 1-y trial of the effect of high-selenium bread on selenium concentrations in blood and toenails. Am J Clin Nutr. 1993;57(3):408–413. doi: 10.1093/ajcn/57.3.408. [DOI] [PubMed] [Google Scholar]
- Ma H, Zhou Z, Wei S, Liu Z, Pooley KA, Dunning AM, Svenson U, Roos G, Hosgood HD, 3rd, Shen M, et al. Shortened telomere length is associated with increased risk of cancer: a meta-analysis. PLoS One. 2011;6(6):e20466. doi: 10.1371/journal.pone.0020466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mordukhovich I, Wright RO, Hu H, Amarasiriwardena C, Baccarelli A, Litonjua A, Sparrow D, Vokonas P, Schwartz J. Associations of toenail arsenic, cadmium, mercury, manganese, and lead with blood pressure in the normative aging study. Environ Health Perspect. 2012;120(1):98–104. doi: 10.1289/ehp.1002805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson PM, Tufvesson H, Leosdottir M, Melander O. Telomeres and cardiovascular disease risk: an update 2013. Transl Res. 2013;162(6):371–380. doi: 10.1016/j.trsl.2013.05.004. [DOI] [PubMed] [Google Scholar]
- Nuernberg AM, Boyce PD, Cavallari JM, Fang SC, Eisen EA, Christiani DC. Urinary 8-isoprostane and 8-OHdG concentrations in boilermakers with welding exposure. J Occup Environ Med. 2008;50(2):182–189. doi: 10.1097/JOM.0b013e31815cf6cc. [DOI] [PubMed] [Google Scholar]
- O’Callaghan N, Dhillon V, Thomas P, Fenech M. A quantitative real-time PCR method for absolute telomere length. Biotechniques. 2008;44(6):807–809. doi: 10.2144/000112761. [DOI] [PubMed] [Google Scholar]
- O’Callaghan NJ, Fenech M. A quantitative PCR method for measuring absolute telomere length. Biol Proced Online. 2011;13:3. doi: 10.1186/1480-9222-13-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozdemir O, Numanoglu N, Gonullu U, Savas I, Alper D, Gurses H. Chronic effects of welding exposure on pulmonary function tests and respiratory symptoms. Occup Environ Med. 1995;52(12):800–803. doi: 10.1136/oem.52.12.800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope CA, 3rd, Burnett RT, Thun MJ, Calle EE, Krewski D, Ito K, Thurston GD. Lung cancer, cardiopulmonary mortality, and long-term exposure to fine particulate air pollution. JAMA. 2002;287(9):1132–1141. doi: 10.1001/jama.287.9.1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riethman H, Ambrosini A, Castaneda C, Finklestein J, Hu XL, Mudunuri U, Paul S, Wei J. Mapping and initial analysis of human subtelomeric sequence assemblies. Genome Res. 2004;14(1):18–28. doi: 10.1101/gr.1245004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez BN, Houseman EA, Ryan LM. Residual-based diagnostics for structural equation models. Biometrics. 2009;65(1):104–115. doi: 10.1111/j.1541-0420.2008.01022.x. [DOI] [PubMed] [Google Scholar]
- Sellis D, Provata A, Almirantis Y. Alu and LINE 1 distributions in the human chromosomes: evidence of global genomic organization expressed in the form of power laws. Mol Biol Evol. 2007;24(11):2385–2399. doi: 10.1093/molbev/msm181. [DOI] [PubMed] [Google Scholar]
- Shlush LI, Skorecki KL, Itzkovitz S, Yehezkel S, Segev Y, Shachar H, Berkovitz R, Adir Y, Vulto I, Lansdorp PM, et al. Telomere elongation followed by telomere length reduction, in leukocytes from divers exposed to intense oxidative stress—implications for tissue and organismal aging. Mech Ageing Dev. 2011;132(3):123–130. doi: 10.1016/j.mad.2011.01.005. [DOI] [PubMed] [Google Scholar]
- Slattery ML, Curtin K, Sweeney C, Levin TR, Potter J, Wolff RK, Albertsen H, Samowitz WS. Diet and lifestyle factor associations with CpG island methylator phenotype and BRAF mutations in colon cancer. Int J Cancer. 2007;120(3):656–663. doi: 10.1002/ijc.22342. [DOI] [PubMed] [Google Scholar]
- Soriano P, Meunier-Rotival M, Bernardi G. The distribution of interspersed repeats is nonuniform and conserved in the mouse and human genomes. Proc Natl Acad Sci USA. 1983;80(7):1816–1820. doi: 10.1073/pnas.80.7.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenstrup T, Hjelmborg JV, Kark JD, Christensen K, Aviv A. The telomere lengthening conundrum—artifact or biology? Nucleic Acids Res. 2013;41(13):e131. doi: 10.1093/nar/gkt370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Shi L, Prescott J, Chiuve SE, Hu FB, De Vivo I, Stampfer MJ, Franks PW, Manson JE, Rexrode KM. Healthy lifestyle and leukocyte telomere length in U.S. women. PLoS One. 2012;7(5):e38374. doi: 10.1371/journal.pone.0038374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suva ML, Riggi N, Bernstein BE. Epigenetic reprogramming in cancer. Science. 2013;339(6127):1567–1570. doi: 10.1126/science.1230184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajuddin SM, Amaral AF, Fernandez AF, Rodriguez-Rodero S, Rodriguez RM, Moore LE, Tardon A, Carrato A, Garcia-Closas M, Silverman DT, et al. Genetic and non-genetic predictors of LINE-1 methylation in leukocyte DNA. Environ Health Perspect. 2013;121(6):650–656. doi: 10.1289/ehp.1206068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanderWeele TJ. Invited commentary: structural equation models and epidemiologic analysis. Am J Epidemiol. 2012;176(7):608–612. doi: 10.1093/aje/kws213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vera E, Canela A, Fraga MF, Esteller M, Blasco MA. Epigenetic regulation of telomeres in human cancer. Oncogene. 2008;27(54):6817–6833. doi: 10.1038/onc.2008.289. [DOI] [PubMed] [Google Scholar]
- Vickers AJ. Underpowering in randomized trials reporting a sample size calculation. J Clin Epidemiol. 2003;56(8):717–720. doi: 10.1016/s0895-4356(03)00141-0. [DOI] [PubMed] [Google Scholar]
- Wang G, Huang L, Gao S, Wang L. Measurements of PM10 and PM2.5 in urban area of Nanjing, China and the assessment of pulmonary deposition of particle mass. Chemosphere. 2002;48(7):689–695. doi: 10.1016/s0045-6535(02)00197-2. [DOI] [PubMed] [Google Scholar]
- Weisenberger DJ, Campan M, Long TI, Kim M, Woods C, Fiala E, Ehrlich M, Laird PW. Analysis of repetitive element DNA methylation by MethyLight. Nucleic Acids Res. 2005;33(21):6823–6836. doi: 10.1093/nar/gki987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wentzensen IM, Mirabello L, Pfeiffer RM, Savage SA. The association of telomere length and cancer: ameta-analysis. Cancer Epidemiol Biomarkers Prev. 2011;20(6):1238–1250. doi: 10.1158/1055-9965.EPI-11-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westland JC. Lower bounds on sample size in structural equation modeling. Electronic Commerce Research and Applications. Electronic Commerce Res Appl. 2010;9(6):476–487. [Google Scholar]
- White LP. Theintravascular life span of transfused leukocytes tagged with atabrine. Blood. 1954;9(1):73–82. [PubMed] [Google Scholar]
- Wilson AS, Power BE, Molloy PL. DNA hypomethylation and human diseases. Biochim Biophys Acta. 2007;1775(1):138–162. doi: 10.1016/j.bbcan.2006.08.007. [DOI] [PubMed] [Google Scholar]
- Xu DQ, Zhang WL. Monitoring of pollution of air fine particles (PM2.5) and study on their genetic toxicity. Biomed Environ Sci. 2004;17(4):452–458. [PubMed] [Google Scholar]
- Yang AS, Estécio MRH, Doshi K, Kondo Y, Tajara EH, Issa JP. A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucleic Acids Res. 2004;32:e38. doi: 10.1093/nar/gnh032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yehezkel S, Segev Y, Viegas-Pequignot E, Skorecki K, Selig S. Hypomethylation of subtelomeric regions in ICF syndrome is associated with abnormally short telomeres and enhanced transcription from telomeric regions. Hum Mol Genet. 2008;17(18):2776–2789. doi: 10.1093/hmg/ddn177. [DOI] [PubMed] [Google Scholar]