SUMMARY
Abalone, a broadcast spawning marine mollusk, is an important model for molecular interactions and positive selection in fertilization, but the focus has previously been on only two sperm proteins, lysin and sp18.We used genomic and proteomic techniques to bring new insights to this model by characterizing the testis transcriptome and sperm proteome of the Red abalone Haliotis rufescens. One pair of homologous, testis-specific proteins contains a secretion signal and is small, abundant, and associated with the acrosome. Comparative analysis revealed that homologs are extremely divergent between species, and show strong evidence for positive selection. The acrosomal localization and rapid evolution of these proteins indicates that they play an important role in fertilization, and could be involved in the species-specificity of sperm-egg interactions in abalone. Our genomic and proteomic characterization of abalone fertilization resulted in the identification of interesting, novel peptides that have eluded detection in this important model system for 20 years.
INTRODUCTION
Fertilization is a cascade of events for which surprisingly few molecular interactions are known. A detailed molecular understanding of its mechanisms and evolution begins with the identification of sperm and egg proteins, because sperm-egg interaction is a major source of diversity in sperm proteins. Broadcast spawning marine invertebrates provide an excellent system to study how selective pressures influence the evolution of reproductive proteins. Rapid evolution in sperm proteins has been found in sea urchins (Strongylocentrotus), top snails (Tegula), ascidian (Ciona), and abalone (Haliotis). In Strongylocentrotus species, the sperm protein bindin, which is involved in sperm-egg recognition and binding, is under positive selection (Pujolar and Pogson, 2011). In the top snail Tegula, a protein with no homology to bindin or lysin, TMAP, is the major acrosomal protein under positive selection (Hellberg et al., 2000). In addition, three potential gamete recognition proteins from the sperm of ascidian Ciona intestinalis are under positive selection between two closely related lineages (Nydam and Harrison, 2011).
Abalone has a long history as a model system for the biochemical events of fertilization (Swanson and Vacquier, 2002), and provides an excellent opportunity for further study. California abalone exhibit species specificity in fertilization as a result of sequence diversity in sperm-egg interacting proteins (Swanson and Vacquier, 1998). Abalone sperm have an unusually large acrosome granule (Lewis et al., 1980), which allowed the early purification of the proteins lysin and sp18. Sp18is implicated in sperm-egg fusion (Swanson and Vacquier, 1995) whereas lysin dimers create a hole in the egg vitelline envelope, allowing sperm to swim through, by interacting with the vitelline envelope receptor for lysin (VERL) (Lewis et al., 1982; Kresge et al., 2001). The species-specificity of vitelline envelope dissolution by lysin is mediated by two highly variable regions in the amino acid sequence (Lyon and Vacquier, 1999). The rapid evolution of lysin and VERL may be driven by coevolution between the two proteins (Clark et al., 2009). Lysin in particular has become a textbook example of rapidly evolving fertilization proteins (Kresge et al., 2001), and was an early focus of study due to its abundance and acrosomal localization. Surprisingly, it is not the most abundant acrosomal protein.
The discovery of new abalone sperm-egg interaction proteins is of interest for studying positive selection and is important to the elucidation of general mechanisms of fertilization. Most animal egg coats are made of glycosylated zona pellucida (ZP)-domain proteins (Jovine et al., 2005). Thirty ZP-domain proteins have been identified as components of the abalone egg vitelline envelope in addition to VERL, several of which show evidence of positive selection (Aagaard et al., 2006, 2010). Despite rapid evolution in sperm-egg interaction proteins, there is shared ancestry in the structural homology between distant species. For example, abalone VERL repeats are structurally homologous to the ZP-N domain of human egg ZP3 (Swanson et al., 2011). ZP3, like VERL, is under positive selection in mammals (Swanson et al., 2001) and is thought to be involved in sperm-egg coat interactions. Interestingly, the sites subject to positive selection in both molecules fall on the same face, in a region implicated in species-specificity, suggesting that selection is related to sperm-egg interaction. Lysin and VERL’s interactions and evolution are well studied, but there are many other steps in the fertilization cascade that are not characterized at the molecular level. For example, chemoattraction is also species specific between the closely related Red and Green abalone (respectively, Haliotis rufescens and H. fulgens), but no sperm receptor is known (Riffell et al., 2004).
We are interested in finding new abalone sperm proteins and candidate sperm-egg interaction proteins. Of particular interest to us are proteins that are abundant, and have a transmembrane domain, a signal sequence, or a carbohydrate- interaction domain. Proteins with a transmembrane domain may be involved in the initial egg-recognition event or in membrane fusion, like the gamete fusion protein HAP2-GCS1 (Wong and Johnson, 2010). A signal sequence indicates that a protein is secreted, either outside the sperm cell or into the acrosome; we are interested in identifying acrosome proteins other than lysin and sp18. Egg coats are made up of glycoproteins, so a carbohydrate-interaction domain could be part of a protein involved in binding to and recognition of the egg coat. In sea urchins, species-specific induction of the acrosome reaction is mediated by sulfated polysaccharides in the egg jelly (Vilela-Silva et al., 2008). Unknown proteins might also be of interest: if they are divergent, they may be evolving under positive selection. In this study we assembled a testis RNA-seq library with no reference sequence, and identified hundreds of sperm proteins with mass spectrometry in the Red abalone H. rufescens. The most interesting result was the discovery of a small, abundant, rapidly evolving pair of homologous proteins. Abalone sperm proteins have been studied for over 20 years, but we have observed these important proteins for the first time by applying new genomic and proteomic technologies to a non-traditional model system.
RESULTS AND DISCUSSION
We identified 975 sperm proteins in H. rufescens by shotgun mass spectrometry. The most striking result was the discovery of a pair of homologous proteins that appear to be as or more abundant than the major abalone acrosomal proteins lysin and sp18. We can approximate the relative transcript abundance with read coverage of RNA-seq data and protein abundance with the normalized spectral abundance factor (NSAF) (Zybailov et al., 2006) from the proteomic mass spectrometry data. As expected, lysin and sp18 had high coverage in both RNA-seq and mass spectrometry, but the combined coverage of the two novel proteins was higher. One form is approximately 6 kDa, so we call the protein sperm protein 6, or sp6. Among the differences between these two forms is the length of an aspartic acid stretch, so we refer to those as sp6_4D and sp6_8D for the 4 or 8 aspartic acids, respectively. Both sequences have an N-terminal signal peptide, which is predicted by SignalP and is supported by mass spectra coverage starting at amino acid 20. Mature sp6_8D has a predicted pI of 4.13 and molecular weight of 6.86 kDa while mature sp6_4D has a predicted pI of 3.93 and molecular weight of 6.28 kDa. The highly acidic nature of these novel peptides is in stark contrast to the strongly basic nature of lysin and sp18.
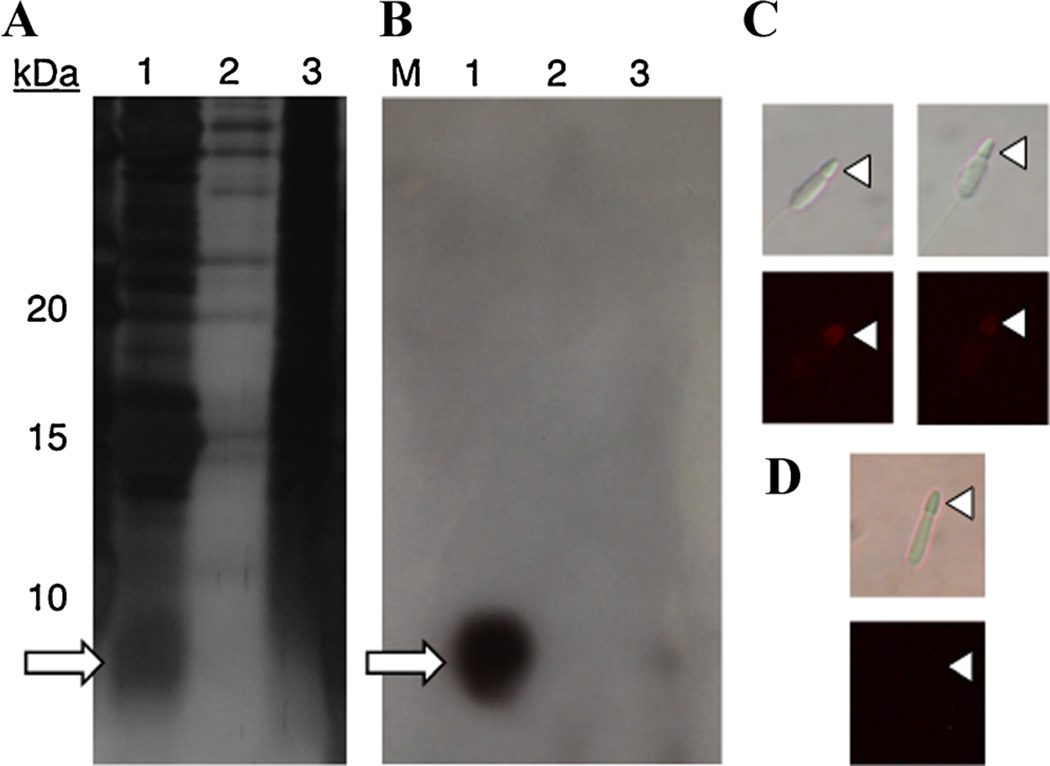
We purified sp6_4D and sp6_8D, which had previously not been observed by SDS protein gel electrophoresis. As they are very small proteins, sp6_4D and sp6_8D typically migrate out of standard single-percentage gels. In addition, they do not stain well with Coomassie due to their acidic nature (Tal et al., 1985). When sperm or testis proteins were separated by SDS-PAGE using 15% acrylamide and silver stained, a dark band appeared around the predicted sizes of the two sp6 proteins (Fig. 1A; the first lane shows acrosome-enriched testis proteins, the second acrosome-depleted, and the third ovary proteins). To confirm that the band contains sp6 proteins, we excised the gel bands and performed mass spectrometry and found that sp6_8D is the dominant protein. We purified sp6 proteins from sperm-protein extracts with an anion exchange column for further characterization, and used the product to develop an antibody. Purification and antibody generation were done with a fraction that includes both sp6 proteins, so subsequent antibody work refers to sp6 in the broad sense. By Western blot and sperm immunofluorescence, sp6 is not present in ovary protein extracts, but is associated with the acrosome of abalone sperm (Fig. 1). The high abundance and acrosomal localization strongly suggests sp6 plays an important role in fertilization.
Figure 1.
Sp6 resides in the acrosome. In (A) and (B), sp6 is indicated with an arrow. In (C) and (D), the acrosome is indicated with a triangle. A: Silver-stained 15% SDS-PAGE, with sizes marked on far left. Lane 1: acrosome-enriched testis proteins, 2: acrosome-depleted testis proteins, 3: ovary proteins. B: Western blot of the same proteins probed with sp6 serum. C: Sperm stained with sp6 serum and a rhodamine-conjugated secondary antibody show fluorescence in the acrosome. D: No acrosome staining is seen in sperm incubated with pre-immune serum.
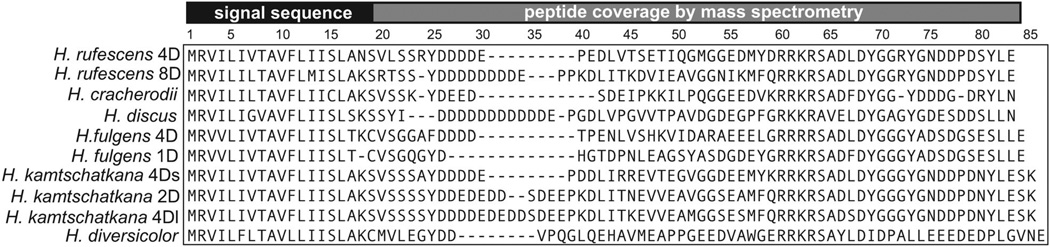
We identified sp6 forms expressed in the testis of four other Haliotis species by PCR using primers from the known H. rufescens sequences (Fig. 2). We also looked for sp6 in two available Haliotis whole-body expressed sequence tag (EST) databases using BLAST, with our sequenced sp6 sequences as queries. We found one homolog in the H. diversicolor database (Jiang et al., 2011), but not in the South African abalone H. midae. For H. rufescens, H. discus, and H. kamtchaskana, agarose gels of PCR products showed bands at multiple sizes, which correspond to multiple forms based on sequencing results of these clones. In contrast, we only amplified one form of sp6 for H. cracherodii and H. discus. In total, sp6 sequences are highly divergent, between forms and between closely related species. BLAST searches against the National Center for Biotechnology Information (NCBI) protein database and the gastropod mollusk L. gigantea EST database revealed no homologous proteins. Sp6 proteins vary in length, which is largely determined by a region of variable-length aspartic acid repeats. We do not know the mechanism and consequences of this variation, but it is interesting to note that these features (tandem repeats and sequence variability) are present in other rapidly evolving sperm proteins. For example, oyster bindin’s F-lectin domain repeats are both polymorphic between individuals and variable within individuals, as a result of alternative splicing and recombination (Moy et al., 2008). In addition, insertion–deletion variation appears to be prevalent in Drosophila accessory gland proteins, which are delivered to the female upon mating (Schully and Hellberg, 2006).
Figure 2.
Amino acid sequences of sp6 from Haliotis species. The gray bar shows sequence that is covered by peptide sequences resulting from shotgun mass spectrometry. The black bar indicates the signal sequence, both as predicted by SignalP and supported by the absence of mass spectra coverage.
The high level of sequence diversity made it difficult to align sp6 sequences. High confidence alignments are necessary to analyze substitution rates to detect positive selection, so we created two separate alignments for subsets of sp6_4D and sp6_8D, removing the signal sequence and any gaps, and calculated dN/dS with codeml. Both forms of sp6 show evidence of positive selection among species (Table 1). In fact, sp6 appears to evolve more rapidly than either lysin or sp18, suggesting it may play a role in species-specificity.
TABLE 1.
dN/dS Analysis on Abalone Sperm Proteins, Using Codeml in PAML
| Contig name | Species | dN/dS | 2ΔInL M7 v. M8 | 2ΔInL pairwise | Feature |
|---|---|---|---|---|---|
| sp6_8D | H. rufescens | 5.461 | 12.335* | sp6_8D | Abundant, no BLAST match |
| H. discus | |||||
| H. kamtschatkana 2D | |||||
| H. kamtschatkana 4Ds | |||||
| sp6_4D | H. rufescens | 10.725 | 17.844* | sp6_4D | Abundant, no BLAST match |
| H. fulgens 4D | |||||
| H. kamtschatkana 4Ds | |||||
| comp6_seq1 | H. rufescens | 0.417 | — | 2.187 | Abundant, no BLAST match |
| H. fulgens | |||||
| comp81_seq1 | H. rufescens | 0.722 | 0.518 | Calcium binding | |
| H. fulgens | |||||
| comp230_seq1 | H. rufescens | 1.172 | — | 0.779 | Transmembrane |
| H. discus | |||||
| comp266_seq4 | H. rufescens | 2.669 | 0.996 | Abundant | |
| H. discus | |||||
| H. walallensis | |||||
| H. fulgens | |||||
| comp694_seq1 | H. rufescens | 3.173 | 4.99 | Glycosyltransferase | |
| H. discus | |||||
| H. fulgens | |||||
| comp1804_seq3 | H. rufescens | 1.640 | 12.597* | Abundant | |
| H. discus | |||||
| H. walallensis |
Alignments used for sp6 sequences are available in Figure S2.
P > 0.005, two degrees of freedom.
Sp6 is abundant, evolves rapidly, and may be abalone-specific, based on BLAST queries of other species databases. Given its location and rapid evolution, one possibility is that sp6 interacts with lysin and/or sp18. Lysin and sp18 are both positively charged, so sp6 may help them remain soluble while tightly packed in the acrosome. Or it may interact with an egg protein to facilitate species-specific fertilization. Alternatively, akin to the anti-bacterial peptides previously isolated from whole abalone (Park et al., 2012), sp6 may act as an anti-microbicide, protecting the fertilized egg or the developing embryo from pathogens.
While sp6 is the most striking example of rapid evolution in the list of identified sperm proteins, we also found other proteins that are under positive selection (Table 1). We used RNA-seq to generate a transcriptome assembly for H. rufescens testis, whose de novo assembly yielded 41,300 contigs. We added capillary-sequenced cDNA clones from a testis library to the assembly file for a final contig number of 41,576. We identified 975 total sperm proteins with shotgun mass spectrometry of whole sperm, acrosome enriched fractions, and extracellular digestions. The final assembly and the protein list are available at http://depts.washington.edu/swansonw/Swanson_Lab/Data.html. In a BLAST search against the NCBI non-redundant protein database, 676 of the proteins have a significant match. Our sperm protein numbers are similar to those found in mouse: 858 (Baker et al., 2008), fruit fly: 1108 (Wasbrough et al., 2010), and human: 1056 (Baker, 2007), but greater than seen so far in ascidian: 304 (Nakachi et al., 2011).
Sp6 was not discovered until we used new genomic and proteomic technologies for protein discovery to learn more about abalone fertilization. The combined use of testis transcriptome and sperm proteome gives us an excellent list of candidate fertilization proteins, in addition to sp6. This is particularly useful for abalone, because it contributes more information to an already well-studied system. We will perform more detailed studies of the functions of sp6 and other proteins that may play an important role in abalone fertilization, and combine this information with what we know about the egg coat proteome (Aagaard et al., 2010). This will advance our understanding of the entire fertilization cascade in a model system, which also provides the opportunity to study how selection can affect evolutionary rates at different steps of fertilization.
MATERIAL AND METHODS
Strand-specific cDNA was prepared from RNA from the testes of 5 H. rufescens individuals. Sequencing was performed on an Illumina Genome Analyzer with 76-base pair paired-end reads. Raw reads are available at the NCBI Sequence Read Archive, accession number SRR770266. The paired reads were filtered with a phred quality cutoff of 5. Assembly was performed with the Trinity package (Grabherr et al., 2011), then contigs were added from Sanger sequencing of a cDNA library for a total of 41,576 predicted transcripts. Read coverage of contigs was estimated with the BWA and Samtools packages.
Spawning was induced in H. rufescens males in 0.2-µm filtered seawater with the Tris/H2O2 protocol (Morse et al., 1977). Intact sperm were treated with trypsin or elastase (Promega, Madison, WI) to extract extracellular proteins, or induced to acrosome react by addition of a calcium ionophore (Sigma A23187, St. Louis, MO) to a final concentration of 0.2 mg/ml, and incubation on ice until most sperm were acrosome-reacted in a subsample viewed with a microscope, or spun down and homogenized intact. All protein preparations were digested with trypsin or elastase. Peptides were eluted over a 4-hour gradient through an in-line HPLC column and into an LTQ ion-trap mass spectrometer (Thermo) for tandem mass spectrometry. Mass spectra were searched against a six-frame translation of the transcriptome assembly for protein identification, with a decoy database of reversed sequences to determine the false discovery rate, using Sequest (Eng et al., 1994). Data were visualized and summarized with the University of Washington’s Yeast Resource Center’s Mass Spectrometry Data Platform: MSDaPl (Sharma et al., 2012). Relative abundance of proteins in a sample was estimated by the normalized spectral abundance factor (NSAF) (Paoletti et al., 2006).
To determine homology to known proteins, Blast2GO (Götz et al., 2008) was run against the non-human, nonmouse EST database, and the non-redundant protein database in NCBI. The protein and transcript sequences were also searched for transmembrane domains and signal sequences, using HMM (THMM Server v. 2.0, CGS Technical University of Denmark) and SignalP (Petersen et al., 2011). Protein molecular weights and charges were estimated with tools on the ExPASy Bioinformatics Resource Portal. From H. rufescens candidate sequences, primer sequences were designed to PCR amplify the transcripts from testis cDNA of other species. dN/dS analyses were done with the codeml package in PAML (Yang, 1997). These sequences were deposited in GenBank (accession numbers KC752594_KC752618).
Sp6 was purified with anion-exchange chromatography and confirmed with SDS-PAGE. Polyclonal antibodies were raised against purified sp6 from H. rufescens testis in rabbits by R&R Research, LLC (Stanwood, WA), and diluted serum was used for Western blots and sperm immunofluorescence.
Supplementary Material
ACKNOWLEDGMENTS
We thank Gennifer Merrihew for mass spectrometry assistance, Joe Hiatt and Choli Lee for sequencing assistance, Renee George for RNA-seq data analysis assistance, and Jan Aagaard for experimental advice. We also thank all members of the Swanson Lab and three anonymous reviewers for helpful comments on the manuscript. This work was supported by the National Institute of Health (5R01HD057974 and 5R01HD042563 to W.J.S., and P41 GM103533 to M.J.M.); the National Science Foundation (DEB-0918106 and DEB-0716761 to W.J.S.); and a Bridge Fund Award from the University of Washington, School of Medicine to W.J.S.
Abbreviations
- EST
expressed sequence tag
- NCBI
National Center for Biotechnology Information
- VERL
vitelline envelope receptor for lysin
Footnotes
SUPPORTING INFORMATION
Additional supporting information may be found in the online version of this article at the publisher’s web-site.
REFERENCES
- Aagaard JE, Vacquier VD, MacCoss MJ, Swanson WJ. ZP domain proteins in the abalone egg coat include a paralog of VERL under positive selection that binds lysin and 18-kDa sperm proteins. Mol Biol Evol. 2010;27:193–203. doi: 10.1093/molbev/msp221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aagaard JE, Yi X, MacCoss MJ, Swanson WJ. Rapidly evolving zona pellucida domain proteins are a major component of the vitelline envelope of abalone eggs. Proc Natl Acad Sci U S A. 2006;103:17302–17307. doi: 10.1073/pnas.0603125103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker MA, Hetherington L, Reeves GM, Aitken RJ. The mouse sperm proteome characterized via IPG strip pre-fractionation and LC-MS/MS identification. Proteomics. 2008;8:1720–1730. doi: 10.1002/pmic.200701020. [DOI] [PubMed] [Google Scholar]
- Baker MA. Identification of gene products present in Triton X-100 soluble and insoluble fractions of human spermatozoa lysates using LC-MS/MS analysis. Proteomic Clin Appl. 2007:1–9. doi: 10.1002/prca.200601013. [DOI] [PubMed] [Google Scholar]
- Clark NL, Gasper J, Sekino M, Springer SA, Aquadro CF, Swanson WJ. Coevolution of interacting fertilization proteins. PLoS Genet. 2009;5:e1000570. doi: 10.1371/journal.pgen.1000570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng JK, McCormack AL, Yates JR., III An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- Götz S, García-Gómez JM, Terol J, et al. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008;36:3420–3435. doi: 10.1093/nar/gkn176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellberg M, Moy G, Vacquier V. Positive selection and propeptide repeats promote rapid interspecific divergence of a gastropod sperm protein. Mol Biol Evol. 2000;17:458–466. doi: 10.1093/oxfordjournals.molbev.a026325. [DOI] [PubMed] [Google Scholar]
- Jiang J-Z, Zhang W, Guo Z-X, Cai C-C, Su Y-L, Wang R-X, Wang J-Y. Functional annotation of an expressed sequence tag library from Haliotis diversicolor and analysis of its plant-like sequences. Mar Genomics. 2011;4:189–196. doi: 10.1016/j.margen.2011.05.001. [DOI] [PubMed] [Google Scholar]
- Jovine L, Darie CC, Litscher ES, Wassarman PM. Zona pellucida domain proteins. Annu Rev Biochem. 2005;74:83–114. doi: 10.1146/annurev.biochem.74.082803.133039. [DOI] [PubMed] [Google Scholar]
- Kresge N, Vacquier V, Stout C. Abalone lysin: The dissolving and evolving sperm protein. Bioessays. 2001;23:95–103. doi: 10.1002/1521-1878(200101)23:1<95::AID-BIES1012>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Lewis CA, Leighton D, Vacquier V. Morphology of abalone spermatozoa before and after the acrosome reaction. J Ultrastruct Res. 1980;72:39–46. doi: 10.1016/s0022-5320(80)90133-1. [DOI] [PubMed] [Google Scholar]
- Lewis CA, Talbot CF, Vacquier VD. A protein from abalone sperm dissolves the egg vitelline layer by a nonenzymatic mechanism. Dev Biol. 1982;92:227–239. doi: 10.1016/0012-1606(82)90167-1. [DOI] [PubMed] [Google Scholar]
- Lyon J, Vacquier V. Interspecies chimeric sperm lysins identify regions mediating species-specific recognition of the abalone egg vitelline envelope. Dev Biol. 1999;214:151–159. doi: 10.1006/dbio.1999.9411. [DOI] [PubMed] [Google Scholar]
- Morse DE, Duncan H, Hooker N, Morse A. Hydrogen peroxide induces spawning in mollusks, with activation of prostaglandin endoperoxide synthetase. Science. 1977;196:298–300. doi: 10.1126/science.403609. [DOI] [PubMed] [Google Scholar]
- Moy GW, Springer SA, Adams SL, Swanson WJ, Vacquier VD. Extraordinary intraspecific diversity in oyster sperm bindin. Proc Natl Acad Sci U S A. 2008;105:1993–1998. doi: 10.1073/pnas.0711862105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakachi M, Nakajima A, Nomura M, Yonezawa K, Ueno K, Endo T, Inaba K. Proteomic profiling reveals compartment-specific, novel functions of ascidian sperm proteins. Mol Reprod Dev. 2011;78:529–549. doi: 10.1002/mrd.21341. [DOI] [PubMed] [Google Scholar]
- Nydam ML, Harrison RG. Reproductive protein evolution in two cryptic species of marine chordate. BMC Evol Biol. 2011;11:18. doi: 10.1186/1471-2148-11-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paoletti AC, Parmely TJ, Tomomori-Sato C, Sato S, Zhu D, Conaway RC, Conaway JW, Florens L, Washburn MP. Quantitative proteomic analysis of distinct mammalian Mediator complexes using normalized spectral abundance factors. Proc Natl Acad Sci U S A. 2006;103:18928–18933. doi: 10.1073/pnas.0606379103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S-C, Kim J-Y, Lee J-K, Hahm K-S, Park Y. Antibacterial action of new antibacterial peptides, Nod1 and Nod2, isolated from Nordotis discus discus. J Agric Food Chem. 2012 doi: 10.1021/jf3006646. [DOI] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, Heijne von G, Nielsen H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- Pujolar JM, Pogson GH. Positive Darwinian selection in gamete recognition proteins of Strongylocentrotus sea urchins. Mol Ecol. 2011;20:4968–4982. doi: 10.1111/j.1365-294X.2011.05336.x. [DOI] [PubMed] [Google Scholar]
- Riffell JA, Krug PJ, Zimmer RK. The ecological and evolutionary consequences of sperm chemoattraction. Proc Natl Acad Sci U S A. 2004;101:4501–4506. doi: 10.1073/pnas.0304594101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schully SD, Hellberg ME. Positive selection on nucleotide substitutions and indels in accessory gland proteins of the Drosophila pseudoobscura subgroup. J Mol Evol. 2006;62:793–802. doi: 10.1007/s00239-005-0239-4. [DOI] [PubMed] [Google Scholar]
- Sharma V, Eng JK, MacCoss MJ, Riffle M. A mass spectrometry proteomics data management platform. Mol Cell Proteomics. 2012;11:824–831. doi: 10.1074/mcp.O111.015149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson W, Vacquier V. Liposome fusion induced by a M(R)-18000 protein localized to the acrosomal region of acrosome-reacted abalone spermatozoa. Biochemistry-US. 1995;34:14202–14208. doi: 10.1021/bi00043a026. [DOI] [PubMed] [Google Scholar]
- Swanson W, Vacquier V. Concerted evolution in an egg receptor for a rapidly evolving abalone sperm protein. Science. 1998;281:710–712. doi: 10.1126/science.281.5377.710. [DOI] [PubMed] [Google Scholar]
- Swanson W, Vacquier V. Reproductive protein evolution. Annu Rev Ecol Syst. 2002;33:161–179. [Google Scholar]
- Swanson WJ, Aagaard JE, Vacquier VD, Monné M, Sadat Al Hosseini H, Jovine L. The molecular basis of sex: Linking yeast to human. Mol Biol Evol. 2011;28:1963–1966. doi: 10.1093/molbev/msr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson WJ, Yang Z, Wolfner MF, Aquadro CF. Positive Darwinian selection drives the evolution of several female reproductive proteins in mammals. Proc Natl Acad Sci U S A. 2001;98:2509–2514. doi: 10.1073/pnas.051605998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tal M, Silberstein A, Nusser E. Why does Coomassie brilliant blue R interact differently with different proteins? A partial answer. J Biol Chem. 1985;260:9976–9980. [PubMed] [Google Scholar]
- Vilela-Silva A-CES, Hirohashi N MouraoPAS. The structure of sulfated polysaccharides ensures a carbohydrate-based mechanism for species recognition during sea urchin fertilization. Int J Dev Biol. 2008;52:551–559. doi: 10.1387/ijdb.072531av. [DOI] [PubMed] [Google Scholar]
- Wasbrough ER, Dorus S, Hester S, Howard-Murkin J, Lilley K, Wilkin E, Polpitiya A, Petritis K, Karr TL. The Drosophila melanogaster sperm proteome-II (DmSP-II) J Proteomics. 2010;73:2171–2185. doi: 10.1016/j.jprot.2010.09.002. [DOI] [PubMed] [Google Scholar]
- Wong JL, Johnson MA. Is HAP2-GCS1 an ancestral gamete fusogen? Trends Cell Biol. 2010;20:134–141. doi: 10.1016/j.tcb.2009.12.007. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML:A program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Zybailov B, Mosley AL, Sardiu ME, Coleman MK, Florens L, Washburn MP. Statistical analysis of membrane proteome expression changes in Saccharomyces cerevisiae. J Proteome Res. 2006;5:2339–2347. doi: 10.1021/pr060161n. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.