The current antibiotic crisis stems from two distinct phenomena – drug resistance, and drug tolerance. Resistance mechanisms such as drug efflux or modification prevent antibiotics from binding to their targets1, allowing pathogens to grow. Antibiotic tolerance is the property of persister cells, phenotypic variants of regular bacteria2. Antibiotics kill by corrupting targets, but these are inactive in dormant persisters, leading to tolerance. Persisters were first identified by Joseph Bigger in 1944, when he discovered a surviving sub-population of Staphylococcus following treatment with penicillin3. Persisters are largely responsible for recalcitrance of chronic diseases such as tuberculosis, and various infections associated with biofilms - endocarditis, osteomyelitis, infections of catheters and indwelling devices, and deep-seated infections of soft tissues4. There are a number of redundant pathways involved in persister formation5,6 precluding development of drugs inhibiting their formation. The acyldepsipeptide antibiotic (ADEP 4) has been shown to activate the ClpP protease resulting in death of growing cells7. Here we show that ADEP4 activated ClpP becomes a fairly non-specific protease and kills persister cells by degradation of over 400 intracellular targets. clpP mutants are resistant to ADEP47, but we find that they display increased susceptibility to killing by a range of conventional antibiotics. Combining ADEP4 with rifampicin leads to eradication of persisters, stationary and biofilm populations of Staphylococcus aureus in vitro and in a deep-seated murine infection. Target corruption/activation provides an approach to killing persisters and eradicating chronic infections.
ADEP4 is a semi-synthetic derivative of the natural product acyldepsipeptide, factor A, with increased potency against gram-positive organisms7. ADEP4 binds to ClpP and keeps the catalytic chamber open, allowing access to peptides and proteins, normally too large to access the chamber8,9. Normally, peptides are delivered to ClpP by ATP-dependent ClpX, C or A subunits9. In the presence of ADEP, proteolysis by ClpP no longer depends on ATP10. ADEP4 shows efficacy in a lethal systemic murine infection of Enterococcus faecalis and S. aureus and lethal sepsis caused by Streptococcus pneumoniae in the rat7. Nascent polypeptides emerging from the ribosome, rather than mature folded proteins, were proposed to be the primary target of ADEP/ClpP10. This would suggest that ADEP has activity against growing cells with active protein synthesis10. A particular mature protein, FtsZ, has also been reported to be the main target of ADEP/ClpP11. FtsZ forms the cell division ring, and this would also suggest that ADEP is active against growing cells. Experiments with nascent peptides and FtsZ were performed with short exposure times and with rapidly growing cells, and we reasoned that longer incubation with ADEP may result in non-specific degradation of proteins in non-growing cells. A stationary phase population of S. aureus was chosen to test this, as cells are not dividing and synthesis of nascent polypeptides is strongly down-regulated12. Stationary cells of methicillin resistant S. aureus (MRSA) were exposed to ADEP4 for 24 hours and the resulting proteome was compared with that of an untreated control (Fig 1).
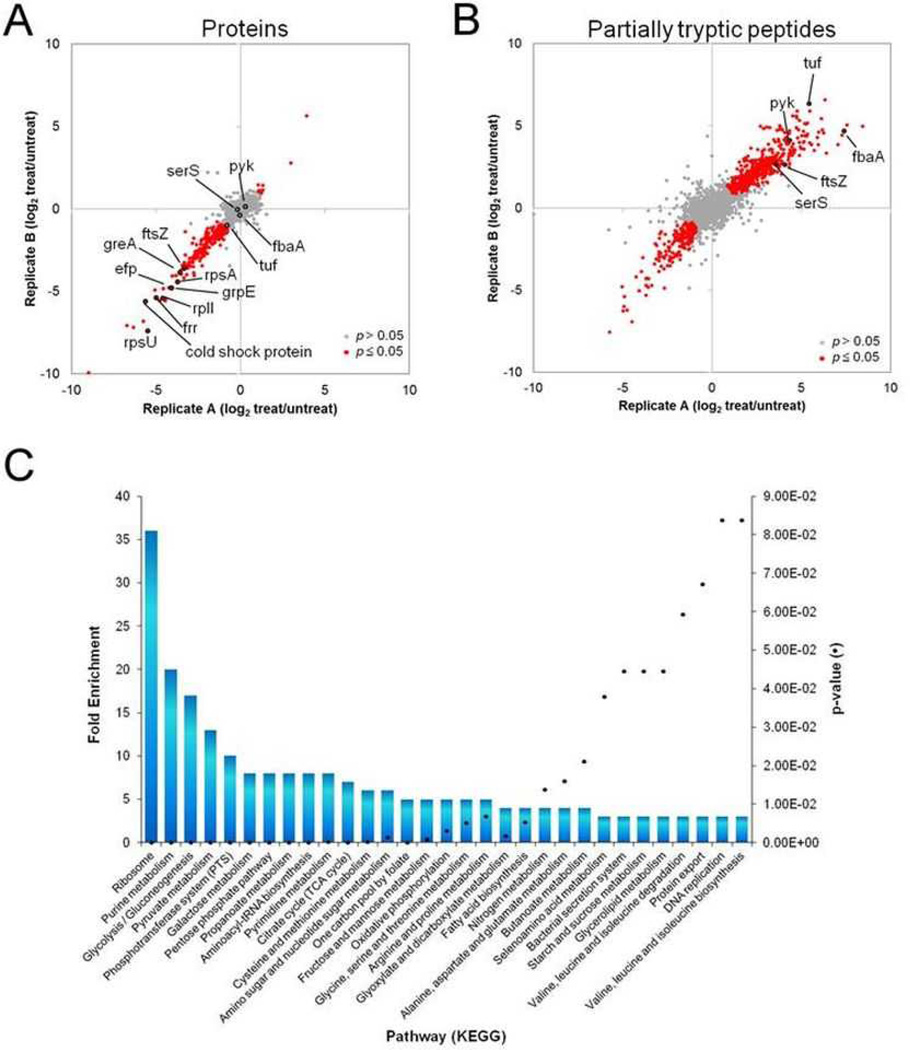
Figure 1. Quantitative proteomic analysis of S. aureus cells treated with ADEP4 reveals extensive protein degradation.
S. aureus cells were treated with ADEP4 in biological duplicates and submitted toa global quantitative proteomic analysis. The dispersion graphs show the relative abundances (treated/untreated) of a, total proteins and b, partiallytryptic peptides in different biological replicates. The significant changes in abundances (p≤0.05 and >2 fold) are represent in red circles. c, Function-enrichment analysis of proteins degraded by ADEP4. Functions overrepresented among proteins degraded by ADEP4 were annotated using Database for Annotation, Visualization and Integrated Discovery (DAVID) and the overrepresented pathways compared to the genome background are shown as columns, whereas their p-values are represented by the black dots.
Proteomic analysis of untreated stationary cells led to the detection of 1712 proteins (65% of the predicted ORFs). Treatment with ADEP4 resulted in decreased abundance of 243 proteins (p≤0.05 and 2-fold decrease) (Fig 1A) (Extended Data Table 1). This however is likely an underestimate. The proteome reports changes in the relative abundance of peptides produced by trypsin cleavage. A protein that was only cleaved once, for example, by ADEP4/ClpP and was not further degraded prior to trypsin treatment would still generate several tryptic peptides and not show an overall decrease in protein abundance. To address this, we analyzed partially-tryptic peptides to uncover additional ADEP/ClpP targets. Partially tryptic peptides exist at certain abundance in untreated cells due to natural degradation, followed by trypsin treatment. However, the levels of these peptides increases or decreases markedly due to degradation induced by addition of ADEP; as is obvious from the abundance of red spots. An increase of partially tryptic peptides indicates ADEP degradation of a protein resulting in partially tryptic peptides. This analysis revealed 174 additional ADEP/ClpP targets (peptides of increased abundance; Fig 1B; Extended Data Table 2). A decrease on the other hand indicates that a particular degradation product, present at the time of ADEP addition, can be further degraded by ADEP/ClpP and are of less relevance to the study.
Combined analysis of both data sets brings the total of detected degraded proteins to 417. Interestingly, essential ribosomal proteins were among the most strongly diminished by ADEP4-ClpP, with proteins S21, L9, S1 and ribosomal recycling factor all showing between 17 and 64-fold reduction in the ADEP4 treated sample. Elongation factor Tu, pyruvate kinase and fructose bi-phosphate aldolase were among the proteins with the largest increase in non-trypsin cleavage sites (Fig 1B). FtsZ was also one of the many strongly degraded proteins, displaying a marked reduction in protein levels. It is feasible that the disordered C-terminus of FtsZ13 is responsible for the degradation of this protein. Other than the ribosome, degraded proteins belonged to various functional types, including purine metabolism, glycolysis and aminoacyl-tRNA biosynthesis, among others (Fig 1C).
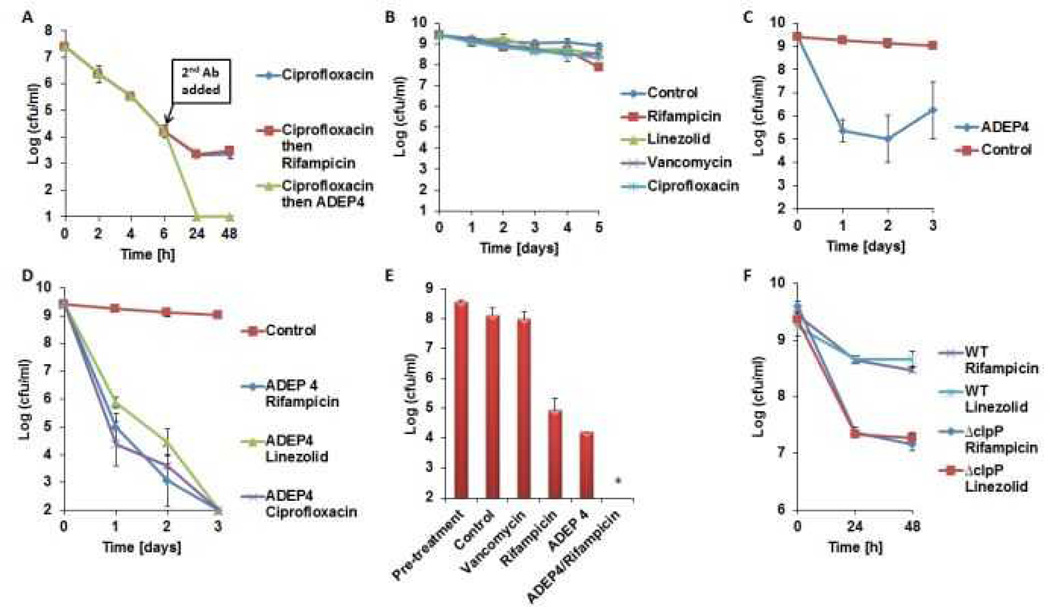
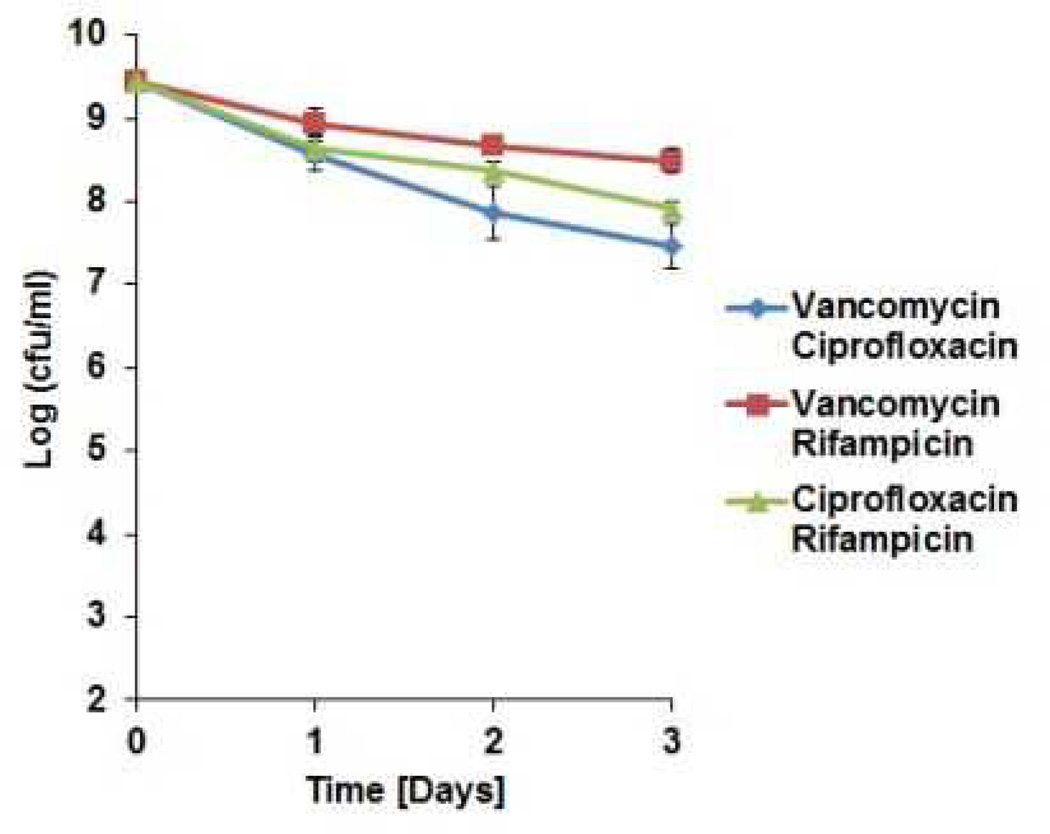
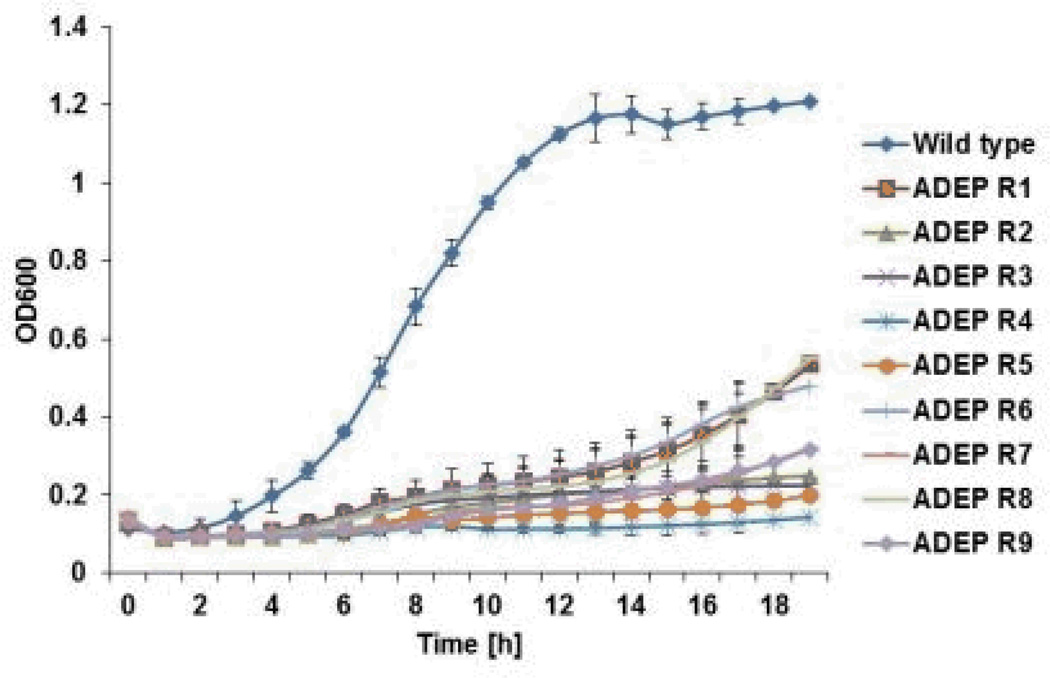
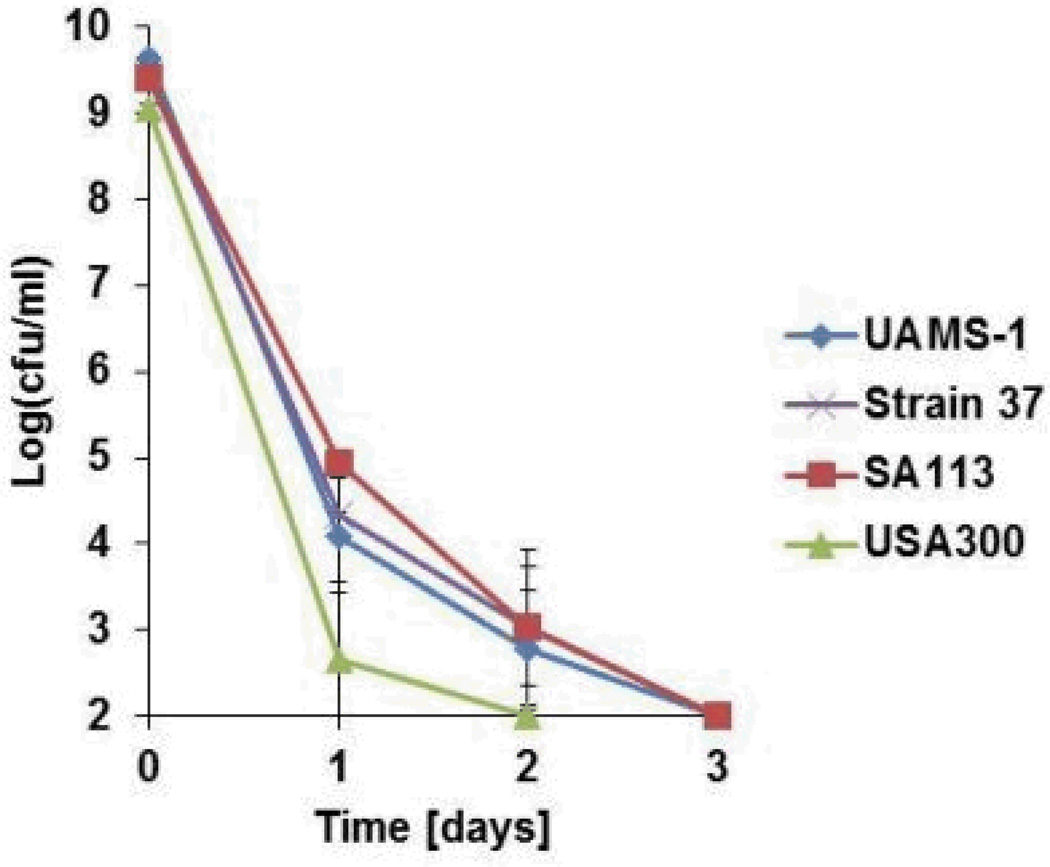
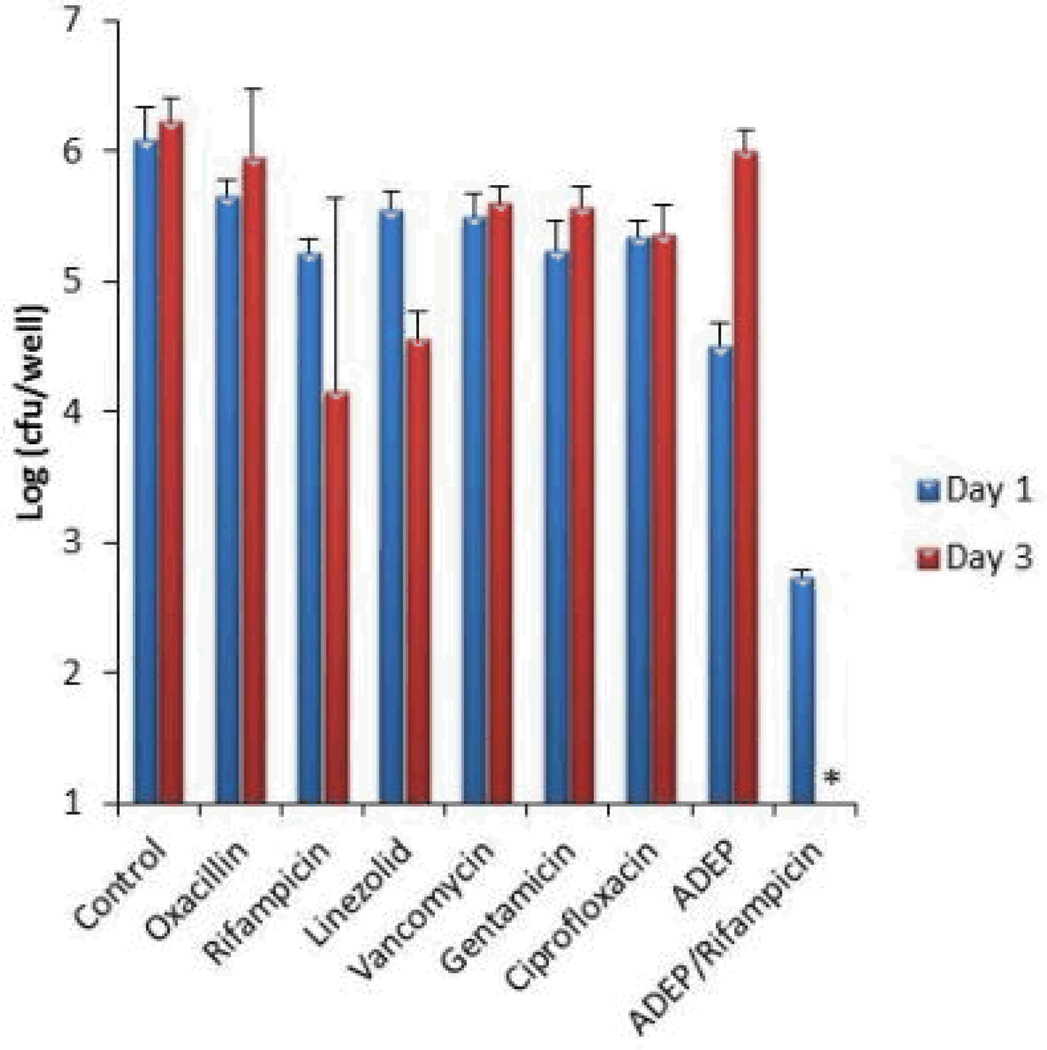
The proteomic data suggests that ADEP4 forces the cell to self-digest, and may be effective in killing dormant cells. Importantly, ADEP4 uncouples ClpP from the requirement to use ATP, which would help kill persisters with low energy levels14. In order to test the action of ADEP4, ciprofloxacin was added to an exponentially growing culture of S. aureus, which produced a typical biphasic killing pattern with surviving persisters (Fig 2A). Addition of rifampicin to surviving persisters had no effect on their viability, in agreement with previous observations on the multidrug tolerant nature of these cells15,16. By contrast, addition of ADEP4 led to eradication of persisters below the limit of detection (Fig 2A). Next, we examined the ability of ADEP4 to kill stationary cells of S. aureus. Stationaryphase S. aureus behave as persisters and are extremely difficult to kill with antibiotics17, even over a 5 day period (Fig 2B). Furthermore, combinations of vancomycin, rifampicin and ciprofloxacin had limited activity against this population (Extended Data Fig. 4) ADEP4 showed excellent killing, decreasing the cells count of a stationary culture by 4 log in two days (Fig 2C), but the population rebounded after day 3. Null mutants of clpP are resistant to ADEP47and arise with high frequency because ClpP is not essential in S. aureus. No cross-resistance to marketed antibiotics was identified for ADEP47. Sequencing of 9 isolates of this culture showed mutations in clpP, and all of them displayed the temperature sensitive phenotype characteristic of null clpP mutants18 (Extended Data Fig. 1). To suppress resistant mutants, ADEP4 was paired with either rifampicin or linezolid, which are used to treat chronic MRSA infections and ciprofloxacin. The ADEP4/antibiotic combination eradicated a stationary population of S. aureus to the limit of detection (Fig 2D). This shows that ADEP4, unlike conventional antibiotics, has a remarkable ability to kill drug-tolerant persister cells. The rich Mueller-Hinton broth (MHB) in which these experiments were performed probably does not reflect conditions in vivo where pathogens experience nutrient limitation. We therefore tested susceptibility to killing of stationary cells in a chemically defined medium19. Killing in the minimal medium by ADEP4/rifampicin was even more effective than in MHB, eradicating the population in 24 hours (Fig 2E). Complete sterilization in this experiment was unexpected, particularly for the usually bacteriostatic antibiotic linezolid – the frequency of clpP mutants is 10−6, and in a population of 109 cells, there should have been 103 survivors. To investigate this eradication, a clpP mutant was examined for its susceptibility to linezolid and rifampicin (Fig 2F). The ΔclpP strain had the same MIC as the wild type, but stationary phase counts were reduced 10–100 fold more than the wild type by linezolid or rifampicin in stationary state. Apparently, a mutation in clpP diminishes the fitness of cells and makes them vulnerable to certain antibiotics. In agreement with this, clpP mutants were reported to be avirulent in vivo18. We then tested the eradicating potential of the ADEP4/rifampicin combination against a variety of S. aureus strains. These included laboratory strain SA113, as well as clinical isolates USA300, UAMS-1 and strain 37, a S. aureus strain isolated from a patient undergoing vancomycin therapy who succumbed to infection20 (Extended Data Fig. 2). No colonies were detected after 72 hours of incubation with ADEP4/rifampicin. Biofilms produced by the osteomyelitis associated strain UAMS-1 displayed a similar tolerance to antibiotics as stationary phase cultures (Fig 3). ADEP4 showed considerable killing following 24 hours of treatment, but the population rebounded after 72 hours. Again, a combination of ADEP4 with rifampicin resulted in eradication of living cells in the biofilm below the limit of detection (Fig 3). The addition of fresh media to the biofilm did not result in re-growth after 3 days of ADEP/rifampicin treatment, confirming the eradication of living cells. An elimination of a biofilm is unprecedented for such low, clinically achievable concentrations of compounds.
Figure 2. ADEP4 kills persisters and in combination with rifampicin or linezolid eradicates stationary phase S. aureus.
a, ADEP4 kills persisters isolated following ciprofloxacin treatment. b, Conventional antibiotics are inactive against stationary phase S. aureus. c, ADEP4 activity against stationary S. aureus. d, ADEP4 in combination with rifampicin or linezolid eradicates stationary phase S. aureus to the detection limit in 72h in MHB and e, 24h in chemically defined media. f, ADEP4 resistant mutants are less tolerant to rifampicin and linezolid than the parent wild type strain. The X axis is the limit of detection.* represents eradication to the limit of detection.
Extended data Figure 4.
Extended data Figure 1.
ADEP4 resistant strains are heat sensitive. Wild type ATCC 33591 and 9 ADEP4 resistant isolates with mutations in clpP were grown for 20 hours in MHB at 44°C in 96-well polystyrene plates.
Extended data Figure 2.
ADEP4/rifampicin eradicates a variety of S. aureus strains. S. aureus was grown in MHB for 16 hours and challenged with 10 × MIC of ADEP4 and rifampicin. Colony counts were performed every 24 hours. Limit of detection = 2.
Figure 3. ADEP4 kills S. aureus biofilm and in combination with rifampicin eradicates the population.
The X axis is the limit of detection.* represents eradication to the limit of detection.
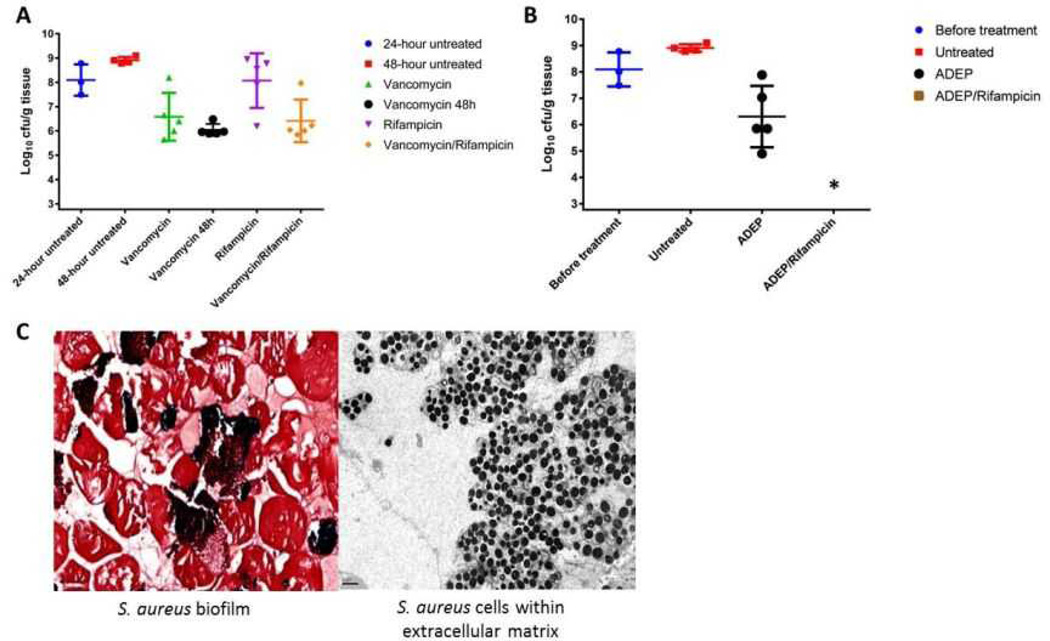
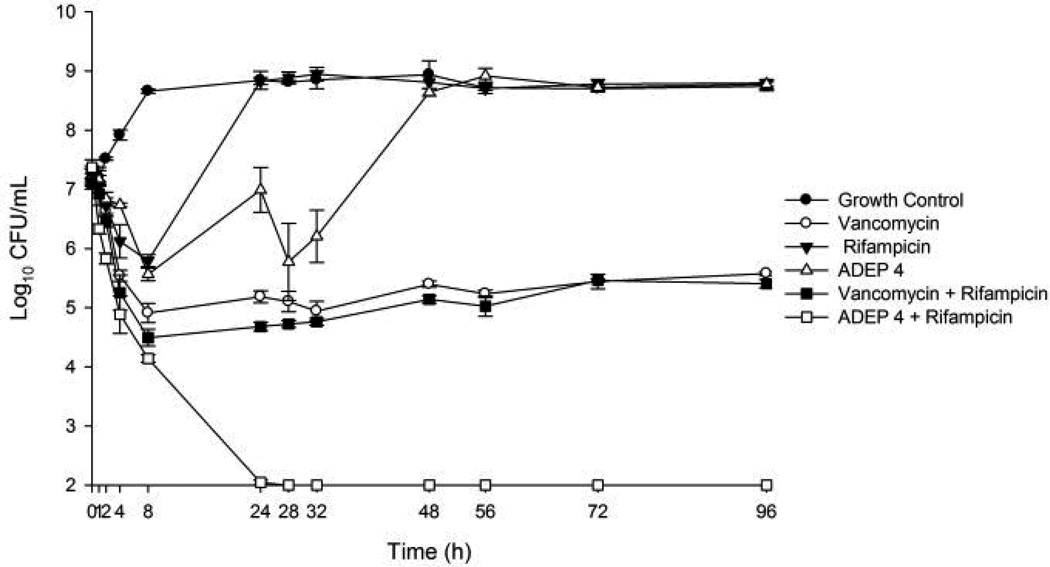
This eradication of stationary and biofilm populations was an encouraging sign that ADEP4 would be a very useful antibiotic against untreatable chronic infections. To test this we employed a deep-seated mouse thigh infection model. In a standard thigh model, a mouse is infected with a low dose of pathogen and antibiotic therapy begins within a few hours of infection. Under these conditions, conventional antibiotics are very effective. In the deep-seated model, the mouse is made neutropenic by treatment with cyclophosphamide, a large dose of pathogen is delivered and the infection is allowed to develop for 24 hours prior to therapy, leading to a severe, recalcitrant, deep-seated infection. This model emulates a difficult to treat human deep-seated chronic infection in immunocompromised patients. Administration of vancomycin, rifampicin, or a combination of both decreased the viable counts, but did not clear the infection (Fig 4A). Furthermore, no significant difference was observed between mice treated for 24 or 48 hours with vancomycin in this model, suggesting the presence of a persister subpopulation surviving the antibiotic treatment (Fig 4A). Remarkably, an ADEP4-rifampicin combination led to sterilization of the infected tissue to the limit of detection within 24 hours (Fig 4B). Based on this efficacious dose and the mouse pharmacokinetics data7, we performed a hollow fiber experiment, which also resulted in complete eradication of the infection to the limit of detection (Extended Data Fig. 3). Gram-stained histopathology and electron microscopy of cross-sections of the infected tissue from an untreated mouse revealed S. aureus growing in biofilms adhered to muscle cells (Fig 4C).
Figure 4. ADEP4 in combination with rifampicin eradicates a deep-seated mouse thigh infection.
a, Single day treatments with rifampicin and vancomycin reveal the deep-seated nature of the mouse thigh lesion. A second day of vancomycin treatment (vancomycin 48h) reveals an antibiotic tolerant subpopulation. b, Single day ADEP4 rifampicin combination eradicates S. aureus in the deep-seated infection. * represents eradication to the limit of detection c, Histopathology of S. aureus infected thighs reveals the presence of a biofilm. Left panel, Gram staining 80 × magnification; right panel, electron micrograph 8000 × magnification.
Extended data Figure 3.
ADEP4/rifampicin eradicates S. aureus in a hollow fiber infection model. The concentration of ADEP was varied over time to match the pharmacokinetics in the mouse model. Limit of detection = 2.
Our results show that persister cells can be killed with the protease activating antibiotic ADEP4. Without an eradicating treatment, it had been unclear whether conventional antibiotics fail due to their ineffective killing of persisters, or simply because they do not penetrate to the site of infection. Eradication of a deep-seated infection by ADEP4/rifampicin settles this important question. The ability to efficiently eradicate an infection greatly reduces the chance of antibiotic resistance evolving during prolonged treatments, an enormous problem associated with chronic infections21. ADEP4 is remarkable in its ability to kill dormant cells. Activation of a protease suggests a general approach to effectively kill persisters. Apart from ADEP4, other activators of ClpP22 may be developed into therapeutics, and additional bacterial proteases such as Lon may be used as targets for killing specialized survivor cells.
Methods
Bacterial strains, plasmids, media and growth conditions
Methicillin resistant Staphylococcus aureus strain ATCC 33591 was used for proteome analysis, antibiotic killing assays and in vivo infections. USA300, SA113, UAMS-1 and strain 37 were also used in antibiotic killing assays. Biofilm experiments were carried out with UAMS-1. Stationary phase populations were prepared as follows: bacteria from frozen stock were grown in 20 ml of Mueller-Hinton broth (MHB) or a chemically defined media19 in 250 ml conical flasks with aeration at 225rpm at 37°C overnight. Exponential phase cultures were prepared as follows: A stationary overnight culture was diluted 1:1000 in MHB and incubated at 37°C with aeration at 225rpm until an OD of 0.5 was reached. Biofilms were grown in brain heart infusion (BHI) broth. Mueller-Hinton agar (MHA) and BHI agar were used for colony counts.
Antibiotic susceptibility assays
Bacteria were incubated in the presence of antibiotics at 37°C with aeration at 225rpm. Antibiotic concentrations, corresponding to 10 × the minimum inhibitory concentration were as follows: vancomycin 10 µgml−1, ADEP4 5 µgml−1, rifampicin 0.4 µgml−1, linezolid 10 µgml−1 and ciprofloxacin 3 µgml−1. Live cell numbers at a given timepoint were determined as follows: 100µl of culture was removed from the flask and centrifuged at 10,000g for 1 minute. The resulting pellet was resuspended in PBS. Serial dilutions were performed and 10 µl of each dilution was spotted onto Mueller-Hinton agar plates. Plates were allowed to dry and then incubated overnight at 37°C.
Biofilm assays
Overnight, stationary cultures of UAMS-1 were diluted 1:20 in BHI broth. 100 µl of this culture was added to each well of a tissue-culture treated polystyrene 96-well plate. Plates were incubated at 37°C, static, for 24 hours. Medium was carefully removed and wells were gently washed twice with PBS. 100 µl of fresh media containing 10 × MIC of antibiotics was carefully added to each well. Plates were incubated for either 24 or 72 hours. Medium was carefully removed and wells were gently washed twice with PBS. 100 µl of PBS was then added to each well and biofilm was solubilized by sonication. Serial dilutions of each well were performed and 10 µl of each dilution was spotted onto BHI plates and incubated overnight at 37°C.
Proteomic analysis
Stationary phase cultures of MRSA cells were treated with 10 × MIC of ADEP4 for 24 h at 37°C. Biological duplicates of untreated control and ADEP4-treated cells were harvested and lysed in 100 mM NH4HCO3, 1 mM PMSF, 2 mM N-ethylmaleimide (NEM) and 5 mM ETDA, by mechanical disruption by vigorous vortexing in the presence of 0.1 mm silica/zirconia beads. A buffer exchange was performed on the cell lysates through an Amicon 10-kDa MWCO filter into 100 mM NH4HCO3. The lysate was denatured/reduced in 100 mM NH4HCO3,8 M urea, 5 mM DTT for 30 min at 60 °C, and then diluted to obtain a final concentration of 1 M urea, and digested with trypsin for 3 h at 37 °C. The resulting peptides were desalted using C18 SPE cartridges (Discovery C18, 1 mL, 50 mg, Sulpelco), labeled with 4-plex iTRAQ reagent (Applied Biosystems) following the manufacturer recommendations, and each of the labeled samples was mixed in equal amounts based on total peptide concentrations measured by BCA assay (Thermo Scientific). The peptide mix was then fractionated into 96 fractions by high pH reverse phase chromatography and concatenated into 24 fractions as previously described23, and submitted to liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis in a LTQ OrbitrapVelos mass spectrometer (Thermo Fisher Scientific). Peptides were loaded into capillary columns (75 µm × 65 cm, Polymicro) packed with C18 beads (3 µm particles, Phenomenex) connected to a custom-made 4-column LC system24. The elution was performed in an exponential gradient from 0–100% B solvent (solvent A: 0.1% formic acid; solvent B: 90% acetonitrile/0.1% formic acid) with a constant pressure of 10,000 psi and flow rate of ~300 nL/min. Full-MS scans were collected in a 400–2000 m/z, and the six most intense ions were submitted for HCD fragmentation using a 2 m/z isolation width and 45% normalized collision energy.
Raw mass spectrometry data were converted to peak lists (DTA files) using the DeconMSn25(version 2.2.2.2, http://omics.pnl.gov/software/DeconMSn.php) and searched with MSGF+26 against the S. aureus COL NC 002951 (2615 sequences), bovine trypsin and human keratin sequences (all in correct and reverse orientations, 5362 total sequences). Searching parameters included tryptic digestion in at least one of the peptide termini (partially tryptic), 10 ppm peptide mass tolerance, methionine oxidation as variable modification, and cysteine alkylation with NEM and N-terminus and lysine labeling with iTRAQ reagent as fixed modifications. Peptides were filtered with an MSGF score ≤ 1e-9, resulting ≤ 1% false-discovery rate at protein level. For the quantitative analysis, the iTRAQ report ion intensities were extracted with MASIC27 (MS/MS Automated Selected Ion Chromatogram Generator, version v2.5.3923, http://omics.pnl.gov/software/MASIC.php). Peptides that were fragmented multiple times had their iTRAQ reporter ions intensities summed to remove redundancy and to improve signal to noise ratio. For protein quantification, the reporter intensities of different fully tryptic peptides belonging to the same proteins were also summed. Peptides and proteins with missing data were excluded from the analysis. Since only 2 replicates were analyzed, a Bayesian moderated t-test (available through “limma” BioConductor package28) was applied to determine the differentially abundant proteins.
Mouse thigh infection
Groups of 5, 6 week old Swiss-Webster mice (Taconic) were rendered neutropenic by cyclophosphamide therapy29. A stationary culture of S. aureus ATCC 33591 was centrifuged and resuspended in PBS. 100 µl of a 1:100 dilution (2 × 106 cells) was injected to the right thigh of each mouse. Infection was allowed to progress for 24 hours and mice displayed measurable increase in thigh diameter. Mice were then treated with vancomycin (Hospira), rifampicin (Pfizer), or ADEP4 (WuXiAppTec). ADEP4 and rifampicin were solubilized in 100% PEG400. Vancomycin was solubilized in water. Vancomycin was dosed intraperitoneally at 110 mg/kg every 12 hours. Rifampicin was dosed intraperitoneally at 30 mg/kg every 24 hours. ADEP4 was dosed intraperitoneally at 25 mg/kg followed by a second 35mg/kg dose 4 hours later. Control mice were sacrificed 24 hours after infection (pretreatment) and 48 hours after infection (post-treatment). Thighs were aseptically removed and homogenized in PBS using a Bullet Blender™ homogenizer. Homogenates were serially diluted and samples were plated on BHI agar and incubated at 37°C overnight.
Microscopy
Histopathology was performed at the Boston University School of Medicine Experimental Pathology Laboratory Service Core.
Gram Stain
Infected thigh tissues were aseptically dissected and fixed overnight at 4°C in 10% formalin. Samples were dehydrated using a graded alcohol series from 70–100%, cleared with xylene to remove the dehydrant, and infiltrated with paraffin. Processed tissue was embedded in paraffin, cut in 5 µm sections, and placed on microscope slides. Slides were baked at 67°C for 36 minutes. After cooling, slides were washed twice with xylene for 5 minutes, twice with 100% alcohol for 5 minutes, twice with 95% alcohol for 2 minutes each, with 70% alcohol for 2 minutes, and left in distilled water until staining. Slides were stained using a Gram Stain Kit from Poly Scientific R&D Corp (cat# s205). Slides were stained with crystal violet for 1 minute and washed thoroughly with distilled water. Next, Gram’s iodine was applied for 30 seconds and the slides were washed thoroughly with distilled water. Slides were discolorized with Gram’s decolorizer until the crystal violet was washed away. Slides were rinsed with distilled water and counterstained with Gram’s safranin O counterstain. Slides were washed with distilled water and air dried before a coverslip was applied. Slides were digitized at 40× using VentanaiScanCoreo AU slide scanner and viewed using Image Viewer v. 3.1.
Electron Microscopy
2 mm cross sections of infected thigh were fixed overnight at 4°C in 2.5% glutaraldehyde/2.0% paraformaldehyde in 0.1 M sodium cacodylate buffer. Samples were post-fixed 1 hr in 1.0% osmium tetroxide in 0.15 M cacodylate buffer at room temperature, dehydrated through a graded acetone series, and embedded in epoxy resin. Sections were cut at 70 nm, stained with uranyl acetate and lead citrate, and examined in a JEOL electron microscope at 80 kV. Images were recorded using a Gatan side mounted 11 Megapixel digital camera.
In vitro Hollow Fiber Model
In vitro pharmacokinetic/pharmacodynamic modeling experiments were performed over a 96 hour period using a hollow fiber model (Fibercell Systems, Frederick, MD) with a culture of ~107 CFU/mL as a starting inoculum. Fresh MHB was continuously supplied via a peristaltic pump (Masterflex; Cole-Parmer Instrument Company, Chicago, IL) set to simulate the half-lives of the antibiotics. After inoculation of the bacteria into the extracapillary space of the hollow fiber cartridge, antibiotic was infused into the reservoir chamber via a dosing port. Free drug concentrations of vancomycin (1 g every 12 hours: fCmax: 30 µg/mL, fCmin: 7.5 µg/mL, half-life: 6 h; at 50% protein binding for vancomycin these levels correspond to a total Cmax of 60 µg/mL and Cmin of 15 µg/mL) and rifampicin (300 mg every 8 hours: fCmax: 0.8 µg/mL, half-life: 3 h; at 80% protein binding for rifampicin these levels correspond to a total Cmax of 4 µg/mL) were dosed to simulate human pharmacokinetics while ADEP4 (25 mg/kg followed by 35 mg/kg 4 hours later – repeated every 24 hours: Cmax for 25 mg/kg: 11.7 µg/mL, Cmax for 35 mg/kg: 16.4 µg/mL, half-life 1.5 h) was dosed to simulate mouse pharmacokinetics. Mouse pharmacokinetics were used for ADEP4 because there is no human pharmacokinetic data nor is there sufficient animal pharmacokinetic data for an allometric conversion. Antibiotic regimens tested included ADEP4 alone, vancomycin alone, rifampicin alone, ADEP4 combined with rifampicin, and vancomycin combined with rifampicin. Model simulations involving two drugs with different half-lives were performed using a previously validated method30. All experiments were performed at 37°C in triplicate to ensure reproducibility.
Samples (1 mL) were removed at 0, 1, 2, 4, 8, 24, 28, 32, 48, 56, 72, and 96 h, serially diluted, plated on BHI agar, and incubated at 37°C with a lower limit of detection of 2 log10 CFU/mL. Antibiotic concentrations were verified by bioassay utilizing antibiotic medium 19 and S. aureus ATCC 33591 for ADEP4, antibiotic medium 5 and B. subtilis for vancomycin, and Mueller Hinton Agar and K. rhizophila ATCC 9341for rifampicin. Only models using a single agent had pharmacokinetics verified while combination models were performed using the verified method described above. Pharmacokinetic parameters were analyzed using WinNonlin modeling software (Pharsight, Cary, NC). Pharmacokinetic values from the models were all within 10% of targets. Overall activity of regimens over the 96 hour period was compared by calculating the area under the bacterial kill curve for each regimen using SigmaPlot software (version 11.1, Systat Software Inc., San Jose, CA). The AUCs were then compared using analysis of variance (ANOVA) with Tukey’s post-hoc test with IBM SPSS Statistics (Version 19.0, SPSS Inc., Chicago, IL).
Supplementary Material
Acknowledgements
We thank Dr. Barry Wright and Christina Blinn of AstraZeneca for assisting with the establishment of the mouse infection model, Prof. Richard E. Lee, Prof. Michael Pollastri, and Dr. Zelkja Maglika for critical discussions and advice, Dr. Iris Keren and Dr. Sarah Rowe for reading of the manuscript, Heather Brewer and Dr. David Camp II for assistance with proteomics. This work was supported by NIH awards T-RO1AI085585 and 3R01 GM061162 to KL, by Arietis Corporation to ML and KC, by the NIH-NIAID IAA Y1-AI-8401 (to JNA) and P41 GM103493-10 (RDS). Work was performed in the EMSL, a DOE-BER national scientific user facility PNNL. PNNL is a multi-program national laboratory operated by Battelle Memorial Institute for the DOE under contract DE-AC05-76RLO 1830.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions B.C, M.L, K.C and K.L designed the study, analyzed results and wrote the manuscript. B.C performed in vitro antibiotic susceptibility assays, harvested samples for proteomics, performed biofilm susceptibility studies and mouse infection models. V.I assisted with in vitro susceptibility assays. E.N and J.A performed I-TRAQ proteomics and analyzed results. L.F participated in mouse infection model experiments. S.L performed hollow-fiber experiments. M.L was responsible for histopathology. R.S provided the proteomics measurement capabilities
The authors declare no competing financial interests.
References
- 1.Alekshun MN, Levy SB. Molecular mechanisms of antibacterial multidrug resistance. Cell. 2007;128:1037–1050. doi: 10.1016/j.cell.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 2.Lewis K. Persister cells. Annu Rev Microbiol. 2010;64:357–372. doi: 10.1146/annurev.micro.112408.134306. [DOI] [PubMed] [Google Scholar]
- 3.Bigger JW. Treatment of staphylococcal infections with penicillin. Lancet. 1944;ii:497–500. [Google Scholar]
- 4.Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: A common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 5.Hansen S, Lewis K, Vulić M. The role of global regulators and nucleotide metabolism in antibiotic tolerance in Escherichia coli. Antimicrob Agents Chemother. 2008 doi: 10.1128/AAC.00144-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maisonneuve E, Shakespeare LJ, Jorgensen MG, Gerdes K. Bacterial persistence by RNA endonucleases. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:13206–13211. doi: 10.1073/pnas.1100186108. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 7.Brotz-Oesterhelt H, et al. Dysregulation of bacterial proteolytic machinery by a new class of antibiotics. Nature medicine. 2005;11:1082–1087. doi: 10.1038/nm1306. [DOI] [PubMed] [Google Scholar]
- 8.Li DH, et al. Acyldepsipeptide antibiotics induce the formation of a structured axial channel in ClpP: A model for the ClpX/ClpA-bound state of ClpP. Chem Biol. 2010;17:959–969. doi: 10.1016/j.chembiol.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee BG, et al. Structures of ClpP in complex with acyldepsipeptide antibiotics reveal its activation mechanism. Nat Struct Mol Biol. 2010;17:471–478. doi: 10.1038/nsmb.1787. [DOI] [PubMed] [Google Scholar]
- 10.Kirstein J, et al. The antibiotic ADEP reprogrammes ClpP, switching it from a regulated to an uncontrolled protease. EMBO Mol Med. 2009;1:37–49. doi: 10.1002/emmm.200900002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sass P, et al. Antibiotic acyldepsipeptides activate ClpP peptidase to degrade the cell division protein FtsZ. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:17474–17479. doi: 10.1073/pnas.1110385108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Michalik S, et al. Proteolysis during long-term glucose starvation in Staphylococcus aureus COL. Proteomics. 2009;9:4468–4477. doi: 10.1002/pmic.200900168. [DOI] [PubMed] [Google Scholar]
- 13.Buske PJ, Levin PA. A flexible C-terminal linker is required for proper FtsZ assembly in vitro and cytokinetic ring formation in vivo. Molecular microbiology. 2013;89:249–263. doi: 10.1111/mmi.12272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dorr T, Vulic M, Lewis K. Ciprofloxacin causes persister formation by inducing the TisB toxin in Escherichia coli. PLoS biology. 2010;8:e1000317. doi: 10.1371/journal.pbio.1000317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keren I, Shah D, Spoering A, Kaldalu N, Lewis K. Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. J Bacteriol. 2004;186:8172–8180. doi: 10.1128/JB.186.24.8172-8180.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson PJ, Levin BR. Pharmacodynamics, population dynamics, and the evolution of persistence in Staphylococcus aureus. PLoS genetics. 2013;9:e1003123. doi: 10.1371/journal.pgen.1003123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keren I, Kaldalu N, Spoering A, Wang Y, Lewis K. Persister cells and tolerance to antimicrobials. FEMS Microbiol Lett. 2004;230:13–18. doi: 10.1016/S0378-1097(03)00856-5. [DOI] [PubMed] [Google Scholar]
- 18.Frees D, Qazi SN, Hill PJ, Ingmer H. Alternative roles of ClpX and ClpP in Staphylococcus aureus stress tolerance and virulence. Molecular microbiology. 2003;48:1565–1578. doi: 10.1046/j.1365-2958.2003.03524.x. [DOI] [PubMed] [Google Scholar]
- 19.Hussain M, Hastings JG, White PJ. A chemically defined medium for slime production by coagulase-negative staphylococci. J Med Microbiol. 1991;34:143–147. doi: 10.1099/00222615-34-3-143. [DOI] [PubMed] [Google Scholar]
- 20.Miyazaki M, et al. Vancomycin bactericidal activity as a predictor of 30-day mortality in patients with methicillin-resistant Staphylococcus aureus bacteremia. Antimicrobial agents and chemotherapy. 2011;55:1819–1820. doi: 10.1128/AAC.01536-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mwangi MM, et al. Tracking the in vivo evolution of multidrug resistance in Staphylococcus aureus by whole-genome sequencing. Proc Natl Acad Sci U S A. 2007;104:9451–9456. doi: 10.1073/pnas.0609839104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leung E, et al. Activators of cylindrical proteases as antimicrobials: identification and development of small molecule activators of ClpP protease. Chem Biol. 2011;18:1167–1178. doi: 10.1016/j.chembiol.2011.07.023. [DOI] [PubMed] [Google Scholar]
- 23.Wang Y, et al. Reversed-phase chromatography with multiple fraction concatenation strategy for proteome profiling of human MCF10A cells. Proteomics. 2011;11:2019–2026. doi: 10.1002/pmic.201000722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Livesay EA, et al. Fully automated four-column capillary LC-MS system for maximizing throughput in proteomic analyses. Analytical chemistry. 2008;80:294–302. doi: 10.1021/ac701727r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mayampurath AM, et al. DeconMSn: a software tool for accurate parent ion monoisotopic mass determination for tandem mass spectra. Bioinformatics. 2008;24:1021–1023. doi: 10.1093/bioinformatics/btn063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim S, et al. The generating function of CID, ETD, and CID/ETD pairs of tandem mass spectra: applications to database search. Mol Cell Proteomics. 2010;9:2840–2852. doi: 10.1074/mcp.M110.003731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Monroe ME, Shaw JL, Daly DS, Adkins JN, Smith RD. MASIC: a software program for fast quantitation and flexible visualization of chromatographic profiles from detected LC-MS(/MS) features. Comput Biol Chem. 2008;32:215–217. doi: 10.1016/j.compbiolchem.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smyth GK. In: Bioinformatics and Computational Biology Solutions using R and Bioconductor. Gentleman R, et al., editors. 2005. Springer. pp. 397–420. [Google Scholar]
- 29.Zuluaga AF, et al. Neutropenia induced in outbred mice by a simplified low-dose cyclophosphamide regimen: characterization and applicability to diverse experimental models of infectious diseases. BMC Infect Dis. 2006;6:55. doi: 10.1186/1471-2334-6-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blaser J. In-vitro model for simultaneous simulation of the serum kinetics of two drugs with different half-lives. The Journal of antimicrobial chemotherapy. 1985;15(Suppl A):125–130. doi: 10.1093/jac/15.suppl_a.125. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.