Abstract
Deregulation of the c-Myc oncoprotein (Myc) is implicated in many types of cancer. Myc is a sequence-specific transcription factor that regulates transcription of genes involved in the control of cell proliferation and apoptosis via mechanisms that are still poorly understood. Cell transformation by Myc involves its association with the transformation-transactivation domain-associated protein (TRRAP) and the human histone acetyltransferase (HAT) GCN5. TRRAP and GCN5 are components of a variety of shared and distinct multiprotein HAT complexes with diverse functions. Myc induces TRRAP recruitment and histone hyperacetylation at specific Myc-activated genes in vivo. However, the identity of the HAT complexes recruited by Myc and the roles of TRRAP and GCN5 in Myc function are still unclear. Here we show that Myc co-recruits TRRAP and GCN5 via direct physical interactions of its N-terminal activation/transformation domain with the human STAGA (SPT3-TAF-GCN5 acetylase) coactivator complex. We demonstrate that GCN5 and TRRAP cooperate to enhance transcription activation by the N-terminal activation domain of Myc in vivo and that this synergy requires both the SPT3/GCN5 interaction domain of TRRAP and the HAT activity of GCN5. Thus, TRRAP might function as an adaptor within the STAGA complex, which helps recruit GCN5 HAT activity to Myc during transcription activation.
The c-Myc protein (Myc) is the most widely expressed member of a small family of highly related cellular oncoproteins that includes L-Myc and N-Myc. Myc is essential for early embryo-genesis and regulates cell growth, proliferation, differentiation, and apoptosis. Deregulated Myc expression contributes to tumorigenesis in animal models and is associated with many types of cancer in humans. Most of the biological effects of Myc, including its cellular transformation properties, result from its gene-specific transcription regulatory functions. Myc has a complex N-terminal transcription regulatory domain with both transcription activating and repressive functions and a C-terminal basic helix-loop-helix leucine zipper (bHLHZip)1 domain, which is required for heterodimerization with its obligatory bHLHZip partner Max and for binding to E-box DNA sequences (consensus CACGTG). The domains of Myc that are essential for cell transformation correspond to domains also required for transcription activation by Myc. These include the bHLHZip domain and two essential amino acid regions within the Myc N-terminal regulatory domain: amino acids 1–110 and 129–145, which contain, respectively, the two phylogenetically conserved Myc box 1 (MB1) and Myc box 2 (MB2) sequences (reviewed in Refs. 1 and 2).
Several different proteins that associate with the N-terminal regulatory domain of Myc have been identified (3). However, generally the precise role(s) of these interactions in Myc functions is still unclear. Of the various Myc-interacting proteins identified so far, the so-called transformation-transactivation domain-associated protein (TRRAP), the human histone acetyltransferase (HAT) GCN5, TIP48, TIP49, and BAF53 proteins appear to play essential roles in Myc-dependent cell transformation (4–7). These proteins are part of a variety of both shared and distinct multiprotein complexes involved in regulation of chromatin structure via either ATP-dependent nucleosome remodeling (e.g. BAF53-containing SWI/SNF-like complexes) or histone acetylation. In particular, TRRAP is a subunit of the TIP48/TIP49-containing TIP60 HAT complex, which preferentially acetylates histone H4 (and H2A) within nucleosomes (8), a related p400 complex that appears to lack TIP60 and HAT activity (9), a novel TRRAP-BAF53 HAT complex that acetylates preferentially nucleosomal histones H4 and H2A (7), and at least three distinct human complexes related to the yeast SAGA coactivator complex that preferentially acetylate nucleosomal histone H3: the PCAF (p300/CBP-associated factor) complex (10), and the two distinct human GCN5-containing complexes STAGA (11) and TFTC (12). Some of these TRRAP-containing complexes have been implicated in transcription regulation and/or DNA repair within chromatin, although the detailed mechanisms are still unclear. Thus, the essential role of TRRAP in transformation by Myc may result from its possible function as an adaptor linking Myc to one or several of the above HAT complexes and cognate activities. Indeed, TRRAP has been shown to bind directly to N-Myc in vitro (13) and TRRAP association with Myc in vivo is dependent on the integrity of both the MB2 (amino acids 129–145) and MB1-containing (amino acids 1–110) sequences (4). In addition, the C-terminal ATM-related domain of TRRAP is required for in vivo association of TRRAP with human SPT3 (hSPT3) and GCN5 (two subunits of STAGA and TFTC complexes) and for Myc-dependent oncogenic transformation (13). A role for TRRAP-HAT complexes in mediating transcription activation by Myc is suggested by the observed recruitment of TRRAP to certain Myc target genes during Myc-dependent activation in vivo and by a corresponding Myc-dependent increase in histone H3 and H4 hyperacetylation at target promoters/regulatory DNA regions, which depend on both MB2 and MB1-containing sequences (14, 15, 16). However, transcription functions of Myc at steps after histone acetylation and RNA polymerase II recruitment and independently of MB2 and TRRAP have also been reported (16–19).
Here we demonstrate an association of Myc:Max heterodimers with the human STAGA coactivator complex in vivo. We show that Myc N-terminal transformation/activation domain recruits the STAGA complex via direct physical interactions and that a Myc N-terminal subdomain (amino acids 1–110) is necessary and sufficient for STAGA recruitment in vivo and in vitro. We further provide the first evidence for synergistic functions of human GCN5 and TRRAP and a requirement for GCN5 HAT activity in mediating the transcription activating functions of the transformation/activation domain of Myc. Thus, our results strongly suggest a role for the human STAGA complex and associated GCN5 HAT activity in transcription activation by Myc.
EXPERIMENTAL PROCEDURES
Plasmids
Plasmid pG5-E4-Luc (kindly provided by X. Liu) contains 5 Gal4 binding sites upstream of the adenovirus E4 gene (TATA) box and the luciferase reporter gene. The plasmids pSV-Gal4(1–147), pSV-Gal4-Myc(1–262), pSV-Gal4-Myc(1–103), and pSV-Gal4-Myc(41–143) were kindly provided by C. V. Dang and described elsewhere (20). The plasmids pCbS, pCbS/FLAG-c-Myc, pCbS/FLAG-c-Myc(Δ1–110), pCβF, pCβF-TRRAPwt, pCβF-TRRAP(1–3713), and pCβF-TRRAP(1899–2401) were kindly provided by M. Cole and were described previously (4, 13). pFH:hGCN5L-IRESneo was constructed by inserting the MluI/EcoRI Klenow-filled cDNA fragment encoding full-length human GCN5L (human GCN5L cDNA kindly provided by P. Moore) into the Klenow-filled NotI site of pFH-IRESneo vector (11) and was verified by DNA sequencing (more detailed information is available upon request). The pcDNA-mGCN5 vector was generated by inserting mouse GCN5 cDNA into pcDNA3.1B (Invitrogen). The expression vectors for catalytically inactive mGCN5 mut.HAT-1 and mut.HAT-2: pcDNA-mGCN5(E568A) and pcDNA-mGCN5(GYG→AYA/582Y584), respectively, were generated by site-directed mutagenesis (with the Quick-Change site-directed mutagenesis kit, Stratagene) and verified by DNA sequencing. The plasmid pcDNA-AS-GCN5 contains the hGCN5L cDNA in an antisense orientation (21) and was kindly provided by S. Kato. The bacterial GST-Max expression vector (pGEX2TK-Max125) encodes a fusion of GST with Max amino acids 36–160 (i.e. lacks 35 residues at the Max N terminus including the basic region). The GST-Myc expression vectors were obtained from R. Bernards and described previously (22).
Cell Culture and Transfection
Hela cells and HEK293 cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% (v/v) fetal bovine serum at 37 °C with 5% CO2. For transient transfection Hela cells at ~80% confluence were co-transfected with 0.2 μg of luciferase reporter vector, 0.1 μg of Myc expression plasmids, and the indicated amount of cofactors expression plasmids using LipofectAMINE reagent (Invitrogen) in 6-well plates. An internal control pCMV-β-galactosidase (0.2 μg) was included in all transfections, and total amount of DNA was kept constant by supplementing with corresponding empty vectors. After 48 h, cell extracts were prepared and luciferase assays were performed as previously described (23). Since β-galactosidase activity was moderately increased (less than 2-fold) by transfection of TRRAP, luciferase activities were normalized to protein concentration in extracts, and results are the means ± S.D. from at least three independent experiments each performed in duplicate.
Immunoprecipitations and GST Pull-down Assays
HEK293 cells were seeded on 10-cm plates in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (without antibiotics) at a density of 6 × 106 24 h before transfection. Cells at ~90% confluence were transfected with 10 μg of CMV expression vectors encoding FLAG-tagged mouse Myc full-length or Δ1–110 using LipofectAMINE 2000 reagent (Invitrogen). After 48 h, cells were washed with phosphate-buffered saline twice and lysed with a buffer containing 50 mM Hepes, pH 7.0, 250 mm NaCl, 0.1% Nonidet P-40, 0.2 mm EDTA, 0.2 mm phenylmethylsulfonyl fluoride. Lysates were adjusted to IP buffer (179 mm NaCl, 0.08% Nonidet P-40, 40 mm Hepes, pH 7.0, 6% glycerol, 0.2 mm EDTA, 1.4 mm β-mercaptoethanol, 0.2 mm phenylmethylsulfonyl fluoride) and subjected to immunoprecipitation by incubation with anti-FLAG resin (M2-agarose) (Sigma) overnight at 4 °C. Myc:Max complexes in lysates of untransfected HEK293 cells were immunoprecipitated with a Max polyclonal antibody (C-17, Santa Cruz Biotechnology) and protein-A-agarose (Pierce). After washing with IP buffer three times, proteins were eluted with SDS gel loading buffer, separated by SDS-PAGE, transferred to nitrocellulose membranes, and immunostained with specific antibodies: FLAG (M2, Sigma), Myc (C-33, Santa Cruz Biotechnology), Max (clone 2.6, BD Biosciences), TRRAP (14), GCN5 (N-18, Santa Cruz Biotechnology), hSPT3 (24), PAF65β/TAF5L (gift from Y. Nakatani), hTBP, hTAF31/TAF9, and hTAF100/TAF5 (gifts from R. G. Roeder), with the enhanced chemiluminesence (ECL) kit (Amersham Biosciences). Nuclear extracts and purification of the STAGA complex were performed as described previously (11). GST-Myc fusion proteins expressed in bacteria were immobilized on glutathione-agarose and used in pull-down assays essentially as previously described (11).
RESULTS
Intracellular Association of Myc with Several STAGA Subunits Depends on Myc Amino Acids 1–110
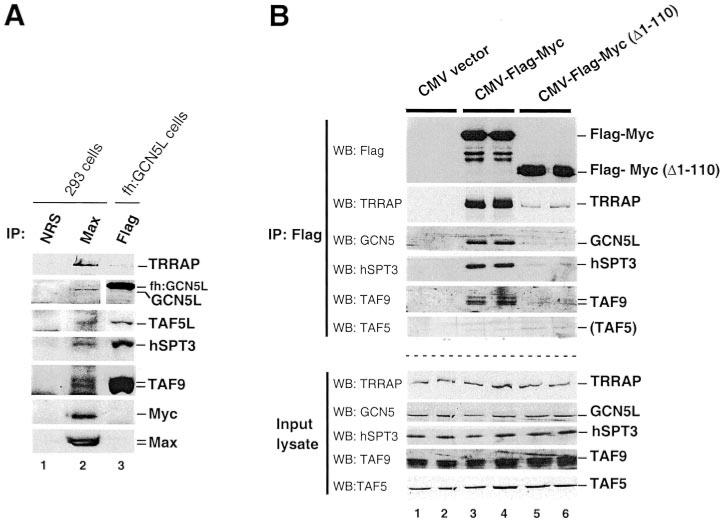
TRRAP and GCN5 co-exist in human STAGA and TFTC complexes and interact with c-Myc in vivo (see Introduction). Thus, TRRAP and GCN5 might be co-recruited by Myc as part of a single complex. However, TRRAP and GCN5 are also found separately in different TRRAP and ADA/GCN5 complexes (see Introduction, reviewed in Ref. 25). Thus, it is also possible that TRRAP and GCN5 interact with Myc separately, as part of two (or more) distinct complexes, different from STAGA/TFTC. To specifically address whether Myc associates with human STAGA and/or TFTC in vivo, we analyzed in co-immunoprecipitation experiments Myc interaction with components specific of these complexes (i.e. SPT3 and TAFs) that are not found in other known TRRAP or ADA/GCN5 complexes. Endogenous Myc:Max heterodimers and associated proteins in human HEK293 cells were immunoprecipitated with a Max antibody and analyzed by SDS-PAGE and Western blotting with specific antibodies to Myc and STAGA subunits. As shown in Fig. 1A, Myc and the STAGA/TFTC subunits TRRAP, the 97-kDa “long” form of GCN5 (GCN5L), TAF5L (formerly PAF65β, Ref. 26), hSPT3, and TAF9 (formerly TAF31) co-precipitated with Max (lane 2). To further address whether STAGA subunits interact with Myc and to test the requirement for Myc-N-terminal transformation domain, FLAG-tagged Myc full-length and truncated (Δ1–110) proteins were expressed in HEK293 cells by transient transfection, immunoprecipitated with a FLAG antibody, and analyzed as above. Control immunoprecipitations were performed with cells transfected with the empty expression vector. Consistent with the above results, TRRAP, GCN5L, hSPT3, and TAF9 were also co-immunoprecipitated with ectopic full-length FLAG-Myc in this assay (Fig. 1B, lanes 3 and 4). Interestingly, TAF5 (formerly hTAF100, Ref. 26), a subunit of TFTC (and TFIID) that is absent in STAGA (11, 12, 24), was not detected in the immune precipitates, although it was abundant in HEK293 cell lysates. Significantly, when compared with full-length Myc (lanes 3 and 4), Myc(Δ1–110) deleted protein did not as efficiently interact with TRRAP, GCN5L, hSPT3, and TAF9 (lanes 5 and 6). Note a residual TRRAP binding to Myc(Δ1–110). Thus, we conclude that Myc requires residues 1–110 within its N-terminal transformation/activation domain to efficiently interact with components of the human STAGA complex in vivo.
Fig. 1. In vivo interaction of Myc with various subunits of the human STAGA complex requires its N-terminal amino acids 1-110.
A, lysates of HEK293 cells (lanes 1 and 2) were immunoprecipitated with non-immune rabbit serum (NRS, lane 1) or with rabbit polyclonal antibodies to Max (C-17) (lane 2), and immunoprecipitated proteins were detected by Western blot with specific antibodies for the indicated proteins. Lane 3 contains STAGA subunits immunoprecipitated with a FLAG antibody from cells expressing FLAG/HA-GCN5L (fh:GCN5L). Different strips of the same blot are shown. B, lysates of HEK293 cells transfected with pCbS (CMV vector; lanes 1 and 2), pCbS/FLAG-c-Myc (lanes 3 and 4), and pCbS/FLAG-c-Myc(Δ1–110) (lanes 5 and 6) were immunoprecipitated (IP) with an anti-FLAG antibody and bound proteins were analyzed by Western blot (WB). The upper panels show the immunoprecipitated proteins in different strips of the same blot probed with the indicated antibodies. Similarly, the bottom panels show the corresponding proteins in the input lysates used for immunoprecipitation. The strips probed with GCN5 antibody were reprobed (after stripping) with TAF5 antibodies.
Selective and Direct Interaction of Myc N-terminal Domain with the Human STAGA Complex in Vitro
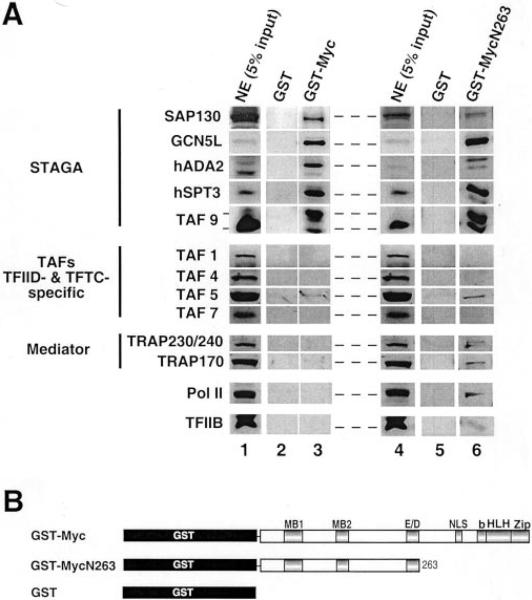
Since Myc interacted specifically with several human STAGA subunits in vivo, we further analyzed this interaction in vitro by GST pull-down assays (Fig. 2). A chimeric GST-Myc full-length protein immobilized on glutathione-agarose beads was able to efficiently recruit various STAGA components from HeLa cell nuclear extracts (Fig. 2A, lane 3). The N-terminal 1–263 regulatory domain of Myc fused to GST (GST-MycN263, Fig. 2B) was sufficient for this interaction (Fig. 2A, lane 6), whereas GST protein alone did not bind STAGA subunits (lanes 2 and 5). Consistent with the above in vivo results, TAF5 and other TAFs (TAF1, TAF4, and TAF7) that are only found in TFTC and TFIID complexes were either undetectable or present at very low concentration in the GST-Myc-bound fractions (compare TAFs panels input in lane 1 with bound proteins in lane 3). This was also observed in a pull-down assay with a 3-fold higher concentration of GST/GST-MycN263 (lanes 5 and 6). Similarly, several subunits of the human Mediator/TRAP coactivator complex (TRAPs 240, 230, and 170), RNA polymerase II, and the general transcription factor TFIIB bound relatively poorly to GST-Myc and GST-MycN263 (i.e. binding was either undetectable or well below 5% of the input in corresponding panels in lanes 3 and 6). Thus, Myc interacts specifically in cells and in cell-free extracts with several components of the human STAGA complex.
Fig. 2. Selective interaction of STAGA components with Myc N-terminal activation domain in vitro.
A, GST pull-down with HeLa cell nuclear extracts (NE) and GST, GST-Myc, and GST-MycN263 resins. Input and bound fractions were analyzed by Western blot with antibodies against the indicated general transcription factors and cofactors. GST pull-down assays in lanes 5 and 6 were performed with 3 times higher concentration of GST fusion protein as compared with lanes 2 and 3. All reactions were run on the same SDS-PAGE gel. STAGA panels (except GCN5L and hSPT3) in lanes 1–3 are longer exposures than the corresponding panels in lanes 4–6 (compare signals of the duplicated input, lanes 1 and 4). B, structure of GST-Myc full-length and GST-MycN263 fusion proteins. Myc boxes 1 and 2 (MB1 and MB2), acidic region (E/D), nuclear localization signal (NLS), and basic helix-loop-helix leucine zipper domain (bHLHZip) are indicated.
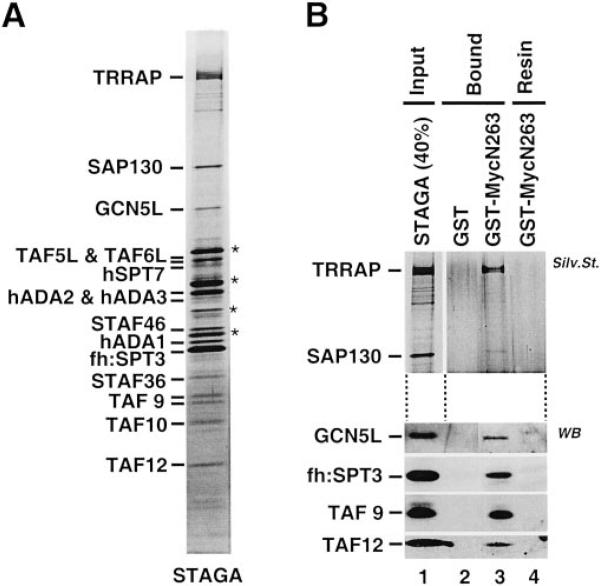
The above results establish that Myc interacts in vivo and in vitro via its N-terminal regulatory domain with human SPT3, ADA2, TAF5L, TAF9, and SAP130 in addition to TRRAP and GCN5 and suggest the possible co-recruitment of these proteins as part of a pre-assembled STAGA complex. To test this, purified human STAGA complex (Fig. 3A) was used in pull-down assays with the GST-MycN263 resin. As shown in Fig. 3B, the purified STAGA complex interacted efficiently with GST-MycN263 (lane 3) but not with GST (lane 2). This demonstrates a direct physical recruitment of the whole STAGA complex by Myc N-terminal regulatory domain.
Fig. 3. Direct interaction of purified human STAGA complex with Myc activation domain in vitro.
A, purified STAGA complex in a silver-stained SDS-PAGE. Asterisks indicate nonspecific proteins. B, GST pull-down assay with purified STAGA (Input, lane 1) and GST (lane 2) or GST-MycN263 (lane 3). Lane 4 contains the GST-MycN263 resin before incubation with STAGA. The upper panel is a silver-stained SDS-PAGE, and the bottom panels are Western blot analyses with antibodies to the indicated proteins on different strips from the same blot.
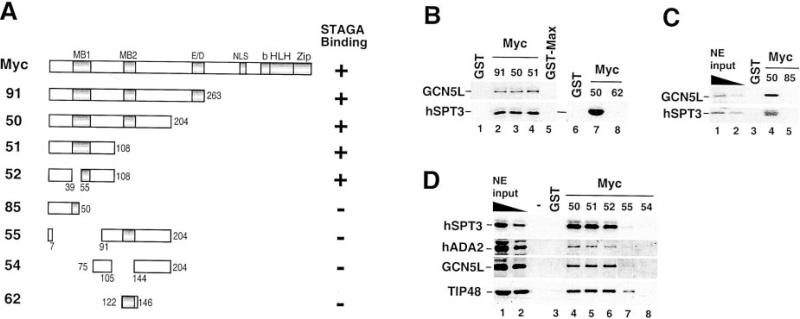
The STAGA-binding domain of Myc was further mapped in GST pull-down assays by using various GST-Myc deletion mutants (Fig. 4). Interestingly, amino acids 1–108 were sufficient for STAGA binding (mutant 51), and neither the conserved MB2 motif nor the N-terminal half of MB1 was required for STAGA interaction (compare mutants 50, 51, and 52). However, deletion of amino acids 51–108 abolished STAGA binding (mutant 85). MB2 was also not sufficient for efficient STAGA binding in the absence of N-terminal amino acids 8–90 (mutants 55 and 62). Thus, the STAGA-binding domain is located within amino acids 1–108, and important sequences reside between amino acids 50 and 108, which include the C-terminal half of MB1. Interestingly, TIP48, a subunit of various complexes distinct from STAGA/TFTC, bound to both the MB1-containing 1–108 region (mutants 51 and 52, Fig. 4D, lanes 5 and 6) and the MB2-containing 91–204 region (mutant 55, Fig. 4D, lane 7). TIP48 binding to the latter region was abolished by deletion of residues 106–143 (mutant 54) suggesting a role for MB2 in this interaction. Thus TIP48 and STAGA components have overlapping but also distinct Myc interaction domains.
Fig. 4. Mapping of the STAGA-binding domain on Myc.
A, structure of GST-Myc deletion mutants used and summary of interaction results: +, corresponds to a binding activity similar to that of the full-length Myc protein (arbitrary 100%); –, indicates less than 30% of that activity. B–D, representative pull-down analyses with HeLa nuclear extracts (NE, input) and the GST-Myc fusion proteins depicted in A.
Human GCN5 and TRRAP Are Required for Transcription Activation by the N-terminal Activation Domain of Myc
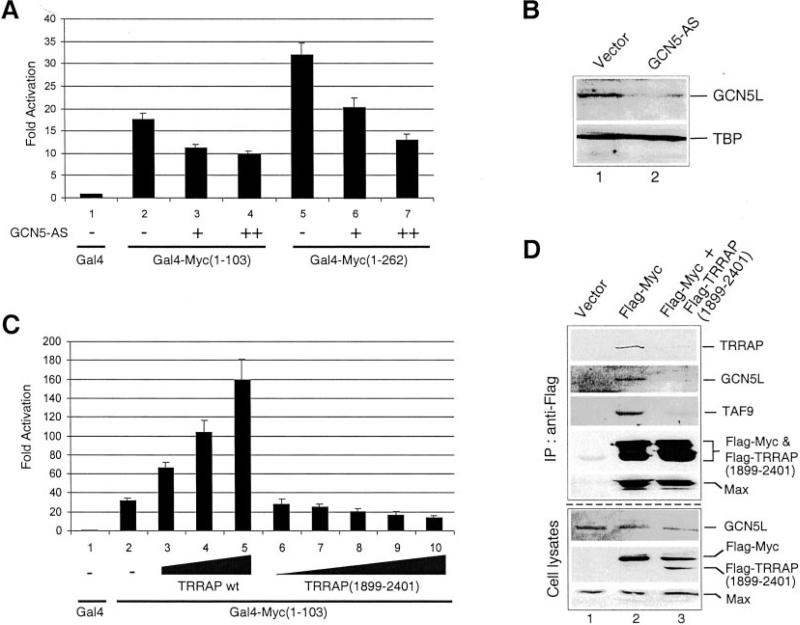
To address the role of the STAGA subunits TRRAP and GCN5 in transactivation by Myc N-terminal regulatory domain in vivo, independently of the activity of other E-box-binding proteins, transient transfection experiments were performed in HeLa cells with a luciferase reporter gene containing 5 Gal4 binding sites and expression vectors for Gal4-Myc fusion proteins consisting of the DNA-binding domain of yeast Gal4 fused to Myc N-terminal domain (1–262 or 1–103). Consistent with the above interaction assays, both Gal4-Myc(1–262) and Gal4-Myc(1–103) interacted with TRRAP in transfected cells (data not shown, see also Ref.4) and, as expected from previous studies (20), activated transcription of the Gal4 site-containing reporter gene (Fig 5A, lanes 2 and 5). To address the role of GCN5 in this assay, Gal4-Myc expression vectors were co-transfected with an antisense GCN5 (GCN5-AS) expression plasmid, which decreased endogenous GCN5 protein levels in transfected cells (about 60% reduction; Fig. 5B, lane 2) as previously reported (21). Co-transfection of GCN5-AS resulted in decreased activation by both activators (Fig. 5A, lanes 3, 4, and 6, 7) suggesting that endogenous GCN5 is required for the activity of the MB2-containing (1–262) and MB2-lacking (1–103) Myc domains (see also below).
Fig. 5. Requirement for TRRAP and GCN5 in transcription activation by Myc N-terminal activation domain.
A and C, HeLa cells were transiently transfected with pG5-E4-luciferase reporter and either pSV-Gal4(1–147) (Gal4), pSV-Gal4-Myc(1–262), or pSV-Gal4-Myc(1–103) and either an antisense GCN5 expression plasmid (GCN5-AS) or expression vectors for TRRAP wild type (wt) and TRRAP(1899–2401) as indicated. Luciferase activity is represented as “Fold Activation” (±S.D.) relative to the activity obtained by cotransfection with Gal4 alone (lane 1), which was arbitrarily set to 1. A, the antisense GCN5 expression vector pcDNA-AS-GCN5 (0.5 μg, and 1.0 μg, + and ++, respectively) was cotransfected with Gal4 and Gal4-Myc expression vectors as indicated. The corresponding empty pcDNA3 vector was added to transfections in lanes 1, 2, 3, 5, and 6 to keep total DNA constant. B, Western blot analysis of endogenous GCN5L and TBP in HEK293 cells transfected with equal amounts (2 μg in 6-cm plates) of pcDNA (vector, lane 1) and pcDNA-AS-GCN5 (lane 2). C, pCβF-TRRAPwt (0.3, 0.4, and 0.5 μg; lanes 3–5) or pCβF-TRRAP(1899–2401) (0.1, 0.2, 0.3, 0.4, and 0.5 μg; lanes 6–10) were cotransfected with Gal4 and Gal4-Myc(1–103) expression vectors as indicated. The corresponding empty vector pCβF was included (lanes 1–4 and 6–9) to keep total DNA constant. D, HEK293 cells were transfected with pCβF (18 μg, lane 1), pCbS/FLAG-Myc (8 μg, lane 2) and with both pCbS/FLAG-Myc (8 μg), and pCβF-TRRAP(1899–2401) (10 μg, lane 3). Total DNA was adjusted to 18 μg (per 10-cm dish) in all transfections by complementing with pCβF. Whole cell extracts (Cell lysates) were immunoprecipitated with a FLAG antibody. Both the input cell lysates (bottom panels) and the immunoprecipitates (top panels) were analyzed by Western blot with antibodies to the indicated proteins. FLAG-Myc and FLAG-TRRAP(1899–2401) were detected with the FLAG antibody.
To analyze the role of TRRAP in activation by the minimal Myc(1–103) domain, transfection experiments were performed (as above) with expression vectors for TRRAP wild type and, as a control, a truncated TRRAP mutant (1899–2401) that interacts with Myc but not with GCN5 in vivo and has dominant-negative effects on growth of neuroblastoma cells (13). As shown in Fig. 5C, expression of wild-type TRRAP stimulated transactivation by Gal4-Myc(1–103) in a dose-dependent manner (lanes 3–5). In contrast, TRRAP(1899–2401) had no stimulatory effect but rather repressed the activity of Gal4-Myc(1–103) (lanes 6–10). Consistent with this, overexpression of TRRAP(1899–2401) diminished (by about 60%) the intracellular association of TRRAP, GCN5, and TAF9 with Myc in transfected cells (Fig. 5D). These results thus establish a role for TRRAP in mediating at least part of the transcription activating functions of Myc(1–103) domain.
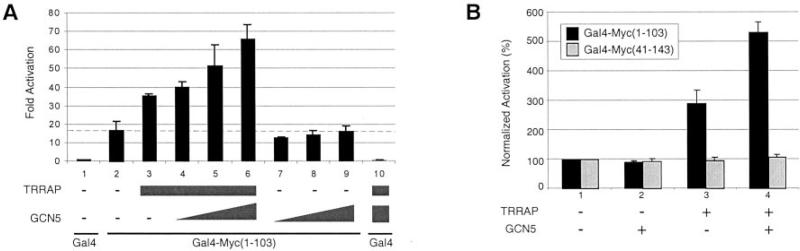
Since Myc N-terminal domain co-recruits TRRAP and GCN5 as part of the STAGA complex we tested whether TRRAP and GCN5 cooperate in stimulating activation by Gal4-Myc(1–103). Whereas ectopic TRRAP stimulated activation (see above and Fig. 6A, lane 3), expression of GCN5 alone did not stimulate activation by Gal4-Myc(1–103) under these conditions (Fig. 6A, lanes 7–9). However, co-expression of TRRAP with increasing amounts of GCN5 resulted in a dose-dependent activation by GCN5 (lanes 4–6), which was not observed with the Gal4 DNA-binding domain alone (lane 10). Interestingly, activation by a Gal4-Myc(41–143) fusion protein, which contains MB2 but lacks the first 40 N-terminal residues of Myc that are important for efficient TRRAP recruitment in vivo (4) was not stimulated by ectopic TRRAP and GCN5 (Fig. 6B). Note that this is not due to an inefficient activating function of Gal4-Myc(41–143) since this activator is about 2–3-fold more active than Gal4-Myc(1–103) in this assay (data not shown, see also Ref. 20). Thus, TRRAP and GCN5 specifically stimulate the transcription activation functions of Myc(1–103) domain in a synergistic manner.
Fig. 6. TRRAP and GCN5 stimulate transcription activation by Gal4-Myc(1–103) in a cooperative manner.
A, transient transfections were performed as in Fig. 5, A and C with the pG5-E4-luciferase reporter, Gal4, or Gal4-Myc(1–103) expression vectors, and pCβF-TRRAPwt (TRRAP; 0.5 μg, lanes 3–6 and 10), pFH:hGCN5L-IRESneo (GCN5; 0.1, 0.2, and 0.25 μg; lanes 4–6 and 7–10) as indicated. The corresponding empty vectors pCβF and pFH:IRESneo were used to keep total vector DNA constant. B, TRRAP and GCN5 enhance transcription activation by Gal4-Myc(1–103) but not by Gal4-Myc(41–143). Transfections (as above) were performed with either pSV-Gal4-Myc(1–103) (black bars) or pSV-Gal4-Myc(41–143) (gray bars) and with pCβF-TRRAPwt (0.5 μg, TRRAP) and/or pcDNA-mGCN5 (0.25 μg, GCN5), as indicated. Corresponding empty vectors were used (in lanes labeled –) to keep total DNA constant. Activation values without transfected TRRAP and GCN5 were arbitrarily set to 100% (lane 1).
The hSPT3-/GCN5-interacting Domain of TRRAP and GCN5 HAT Activity Contribute to Transcription Activation by the N-terminal Activation Domain of Myc
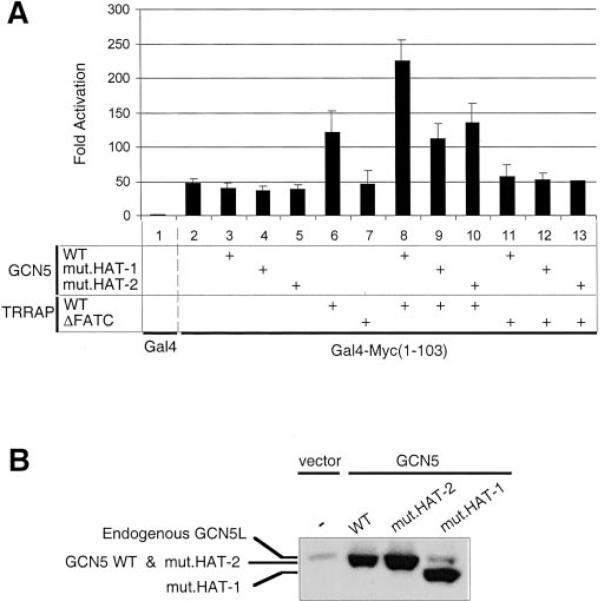
A central region within TRRAP (residues 1591–2401) is sufficient for Myc interaction, while TRRAP C-terminal residues (3714–3830) containing the FATC (FRAP/ATM/TRRAP C-terminal) domain are necessary for TRRAP interaction with GCN5 and SPT3 in vivo (13, 27). Thus, the coactivator function of TRRAP and its synergy with GCN5 (above) might reflect the recruitment by TRRAP of coactivator-HAT activities within GCN5/STAGA that are important for Myc transactivation. To address this, a TRRAP deletion mutant (1–3713) that lacks the FATC domain (TRRAP-ΔFATC) was tested for stimulation of transcription by Gal4-Myc(1–103) in HeLa cells (Fig. 7). In contrast to full-length TRRAP (WT, lane 6), TRRAP-ΔFATC (lane 7) did not stimulate activation by Gal4-Myc(1–103). Moreover, GCN5 did not synergize with TRRAP-ΔFATC (compare lanes 8 and 11) suggesting that interaction of GCN5 (and/or SPT3) with TRRAP (directly or indirectly) is required for GCN5-mediated transactivation in this assay.
Fig. 7. A functional GCN5 HAT domain and the FATC domain of TRRAP are required for activation by Gal4-Myc(1–103).
Top panel, transfections in HeLa cells were performed as described in the legend to Fig. 5 and under “Experimental Procedures” with pSV-Gal4 (lane 1) or pSV-Gal4-Myc(1–103) (lanes 2–13) and with the indicated combinations (+ signs) of pcDNA-mGCN5 (GCN5 WT, 0.25 μg), pcDNA-mGCN5(E568A) (mut.HAT-1, 0.25 μg), pcDNA-mGCN5 (GYG→AYA/582Y584) (mut.HAT-2, 0.25 μg), pCβF-TRRAPwt (TRRAP WT, 0.5 μg), or pCβF-TRRAP(1–3713) (ΔFATC, 0.5 μg). The corresponding empty expression vectors (0.25 μg of pcDNA; 0.5 μg of pCβF) were transfected in empty lanes to keep total DNA constant. B, lysates (40 μg of protein) from HEK293 cells transfected with either the empty pcDNA3 plasmid (vector) or the indicated GCN5 expression vectors (WT and mut.HAT-1/2) were analyzed by Western blot with polyclonal GCN5 (N-18) antibodies. Endogenous and ectopic GCN5 proteins are indicated.
To further test whether GCN5 HAT activity is important for transcription activation by Gal4-Myc(1–103), cotransfection experiments (as above) were performed with two GCN5 mutants, mut.HAT-1 and mut.HAT-2, having, respectively, one and two amino acid substitutions in the catalytic site of the GCN5 HAT domain (see legend to Fig. 7) that abolish HAT activity in vitro (data not shown). As shown in Fig. 7 both of these HAT-deficient GCN5 mutants failed to synergize with TRRAP (compare lane 8 with lanes 9 and 10). We verified that wild-type GCN5 and the HAT mutants are expressed to comparable levels in transfected cells (Fig. 7B). Thus, altogether these results strongly suggest that TRRAP-dependent recruitment of GCN5 and GCN5-mediated acetylation are important for the transactivating function of Myc N-terminal activation domain in vivo.
DISCUSSION
The previously reported association of Myc with TRRAP and GCN5 in vivo (4, 6, 13) and the increased acetylation of histones H3 and H4 during Myc-dependent transcription activation of specific genes within cellular chromatin (14–16) suggested a model for Myc-dependent transcription activation on chromatin involving the direct recruitment by Myc of TRRAP- and/or GCN5-containing human HAT complexes (2, 28). In this report we identify specifically the human STAGA coactivator complex as a TRRAP- and GCN5-containing HAT complex that interacts with Myc in vivo and in vitro. We show (i) an intracellular interaction of Myc (in association with Max) with various STAGA components (TRRAP, GCN5, SPT3, TAF5L, and TAF9) that requires Myc N-terminal amino acids 1–110 within its activation/transformation domain, (ii) a direct physical interaction of purified STAGA complex with Myc N-terminal activation/transformation domain in vitro, and (iii) that a small Myc N-terminal region (residues 1–108) lacking Myc box 2 (MB2) is sufficient for efficient recruitment of STAGA in vitro. We further demonstrate that TRRAP and GCN5 synergistically stimulate transcription activation by the N-terminal TRRAP-binding domain of Myc (residues 1–103) in vivo, in a manner that is dependent on the integrity of both the SPT3/GCN5 interaction domain of TRRAP (i.e. the FATC domain-containing C-terminal region 3714–3830) and a functional GCN5 HAT domain.
Previous analyses of Myc deletion mutants have shown that both the MB1-containing region (residues 1–110) and MB2 (residues 129–145) are necessary for Myc-dependent transformation and for efficient interaction with TRRAP in vivo (4, 32). Our results showing that the Myc 1–110 region is necessary for Myc to interact with STAGA in vivo and sufficient as a GST fusion to recruit STAGA in vitro are therefore consistent with these earlier studies on the essential role of Myc residues 1–110 and further identify the STAGA complex as a human TRRAP-containing complex that binds to this region. The fact that Gal4-Myc(1–103) bound TRRAP and activated transcription in a TRRAP- and GCN5-dependent manner in vivo further indicates that MB2 is not necessary for functional STAGA recruitment under these conditions (see also below). Taken together these results suggest that specific residues within the Myc 1–110 region are likely to contact directly TRRAP and/or other components of the STAGA complex. Similar observations were made recently for the interaction of Myc with the yeast SAGA, NuA4, and SWI/SNF complexes (29).
The fact that the isolated Myc 1–110 region is sufficient for STAGA binding in the assays described above but that both the 1–110 region and MB2 in the context of full-length Myc are required for efficient recruitment of TRRAP and histone H3- and H4-specific HAT activities (14–16) suggests that MB2 either makes additional direct contacts with STAGA (and other TRRAP-HAT complexes), which are not revealed in our GST pull-down assays or, alternatively, can indirectly facilitate interactions between the flanking 1–110 region and the STAGA complex in the context of the full-length Myc protein. Indeed, we cannot exclude that in our assays the fusion of the Myc 1–110 region to GST and to the Gal4 DNA-binding domain might have bypassed the need for MB2. In this respect we also note that a possible structural role for MB2 in the proper folding of Myc activation domain was proposed previously (3, 29).
Interestingly, we found that in contrast to components of STAGA, TIP48 in HeLa nuclear extracts interacts with both the 1–108 and the 91–204 regions of Myc and binding to the latter domain requires MB2 sequences. Since TIP48 has been reported to assemble with TIP49 in TRRAP-containing complexes distinct from STAGA (8, 9) and in a TIP48/49-BAF53 complex that lacks TRRAP and HAT activity (7), MB2 might thus play a more direct/important role than the 1–110 region in recruiting one or more of these complexes. This indeed seems to be the case for Myc interaction with the TIP48/49-BAF53 complex and is thus consistent with our observations (7). In addition, these results also suggest the possibility that a specific TIP48 complex could bind directly to the 1–110 region.
The biological importance of the N-terminal residues of Myc is further underscored by the observation that Myc-S, a naturally occurring short form of Myc (derived from alternative translation initiation), which lacks the first N-terminal 100 amino acids (but retains MB2), has a reduced transcription activation potential (30), and is deficient in promoting apoptosis and transformation of primary fibroblasts (4, 31, 32). Interestingly, however, Myc-S can stimulate cell proliferation and induce transformation and apoptosis of immortalized cells (33). Thus, it is tempting to speculate that STAGA and/or other cofactors that preferentially bind to this N-terminal region, including P-TEFb (19) and perhaps a specific TIP48 complex (see above), might be essential for Myc to activate specific genes required for transformation (or apoptosis) in primary cells by stimulating a step in the activation pathway that is not rate-limiting and/or is constitutively active in immortalized cell lines.
Our results further show that ectopic TRRAP and GCN5 cooperate in stimulating activation by the STAGA-binding domain of Myc in vivo, suggesting that these factors might be limiting and/or readily exchangeable within STAGA complexes in HeLa cells. Recent studies have also shown that transfected TRRAP and GCN5 stimulate activation by E2F4 and the estrogen receptor α and that TRRAP stimulates p53-dependent activation of the MDM2 promoter (34, 35, 36). However, in these cases whether TRRAP functions by recruiting GCN5 HAT activity remains to be determined. Consistent with a possible variable STAGA subunit composition in vivo, STAGA complexes having different amounts of associated SAP130 and GCN5 have been observed in HeLa cells (11) and the subunit composition of the homologous yeast SAGA complex has been shown to vary with growth conditions (37–39). Thus, the composition of yeast SAGA and human STAGA complexes might be dynamic and/or regulated in the cell.
The essential role of the SPT3/GCN5-interacting FATC region of TRRAP in activation by the STAGA-binding domain of Myc in HeLa cells strongly suggests a function for TRRAP as an adaptor that mediates the recruitment of a STAGA/TFTC-like complex to Myc during activation of target genes in vivo. Since the binding of TFTC-/TFIID-specific TAFs to Myc in vivo and in vitro was very weak (if at all detectable), our results suggest that Myc preferentially associates with the STAGA complex. Similarly, STAGA rather than a TFTC-like complex has been proposed recently to interact with acetylated p53 (40). It is important, however, to emphasize that these results, do not exclude the involvement of TFTC/TFIID-specific TAFs during the activation process on Myc target promoters.
We have also shown that, in addition to TRRAP, a functional catalytic site within GCN5 HAT domain is required for stimulation of transcription activation by Myc(1–103). This strongly suggests that the acetyltransferase activity of GCN5 can mediate at least part of the stimulatory function of a minimal Myc activation domain and is consistent with a role of the GCN5 HAT domain in Myc-dependent cell transformation (6). These results also suggest that GCN5 could be involved in the previously reported Myc-dependent histone H3 acetylation at certain Myc target genes in vivo (15, 16). However, this remains to be demonstrated. We note also that a function of GCN5-mediated acetylation of general transcription factors instead of (or in addition to) histone H3 cannot be excluded. Since GCN5/STAGA is a histone H3-specific HAT in vitro and histone H4 is also acetylated during Myc-dependent activation of target genes in vivo (14–16), either GCN5 HAT activity has a broader specificity in vivo or, more likely, Myc recruits additional H4-specific HATs during transcription activation. Indeed, several TRRAP/BAF53-containing H4-specific HAT complexes have been described and are good candidates for this activity (7, 8).
Finally, since STAGA is a multiprotein complex, Myc-STAGA interactions might also regulate specific genes via mechanisms that do not require GCN5 and/or its HAT activity in a manner perhaps similar to the GCN5-independent function of yeast SAGA in recruitment of TBP to certain promoters (41–43). Clearly, further characterization of the exact role of STAGA and GCN5 in Myc-dependent transcription activation is required. Specifically, identification of physiological Myc target genes that require STAGA, including the analysis of the role of other STAGA components in Myc-dependent gene regulation and cell proliferation should provide valuable new insights into the mechanisms involved in Myc-dependent cell transformation.
Acknowledgments
We thank Bob Roeder for reagents and support. We also thank Bruno Amati, Shelley Berger, Rene Bernards, Mike Cole, Chi.V. Dang, Shigeaki Kato, Xuan Liu, Paul Moore, Yoshi Nakatani, and Laszlo Tora for reagents.
Footnotes
The abbreviations used are: bHLHZip, basic helix-loop-helix leucine zipper; HAT, histone acetyltransferase; STAGA, SPT3-TAF-GCN5 acetyltransferase; TBP, TATA-binding protein; TAF, TBP-associated factor; TFTC, TBP-free TAF complex; GST, glutathione S-transferase; TRRAP, transformation-transactivation domain-associated protein; FATC, FRAP/ATM/TRRAP C-terminal; MB, Myc box.
REFERENCES
- 1.Grandori C, Cowley SM, James LP, Eisenman RN. Annu. Rev. Cell Dev. Biol. 2000;16:653–699. doi: 10.1146/annurev.cellbio.16.1.653. [DOI] [PubMed] [Google Scholar]
- 2.Amati B, Frank SR, Donjerkovic D, Taubert S. Biochim. Biophys. Acta. 2001;1471:M135–145. doi: 10.1016/s0304-419x(01)00020-8. [DOI] [PubMed] [Google Scholar]
- 3.Sakamuro D, Prendergast GC. Oncogene. 1999;18:2942–2954. doi: 10.1038/sj.onc.1202725. [DOI] [PubMed] [Google Scholar]
- 4.McMahon SB, Van Buskirk HA, Dugan KA, Copeland TD, Cole MD. Cell. 1998;94:363–374. doi: 10.1016/s0092-8674(00)81479-8. [DOI] [PubMed] [Google Scholar]
- 5.Wood MA, McMahon SB, Cole MD. Mol. Cell. 2000;5:321–330. doi: 10.1016/s1097-2765(00)80427-x. [DOI] [PubMed] [Google Scholar]
- 6.McMahon SB, Wood MA, Cole MD. Mol. Cell. Biol. 2000;20:556–562. doi: 10.1128/mcb.20.2.556-562.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park J, Wood MA, Cole MD. Mol. Cell. Biol. 2002;22:1307–1316. doi: 10.1128/mcb.22.5.1307-1316.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ikura T, Ogryzko VV, Grigoriev M, Groisman R, Wang J, Horikoshi M, Scully R, Qin J, Nakatani Y. Cell. 2000;102:463–473. doi: 10.1016/s0092-8674(00)00051-9. [DOI] [PubMed] [Google Scholar]
- 9.Fuchs M, Gerber J, Drapkin R, Sif S, Ikura T, Ogryzko V, Lane WS, Nakatani Y, Livingston DM. Cell. 2001;106:297–307. doi: 10.1016/s0092-8674(01)00450-0. [DOI] [PubMed] [Google Scholar]
- 10.Vassilev A, Yamauchi J, Kotani T, Prives C, Avantaggiati ML, Qin J, Nakatani Y. Mol. Cell. 1998;2:869–875. doi: 10.1016/s1097-2765(00)80301-9. [DOI] [PubMed] [Google Scholar]
- 11.Martinez E, Palhan VB, Tjernberg A, Lymar ES, Gamper AM, Kundu TK, Chait BT, Roeder RG. Mol. Cell. Biol. 2001;21:6782–6795. doi: 10.1128/MCB.21.20.6782-6795.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brand M, Yamamoto K, Staub A, Tora L. J. Biol. Chem. 1999;274:18285–18289. doi: 10.1074/jbc.274.26.18285. [DOI] [PubMed] [Google Scholar]
- 13.Park J, Kunjibettu S, McMahon SB, Cole MD. Genes Dev. 2001;15:1619–1624. doi: 10.1101/gad.900101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frank SR, Schroeder M, Fernandez P, Taubert S, Amati B. Genes Dev. 2001;15:2069–2082. doi: 10.1101/gad.906601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bouchard C, Dittrich O, Kiermaier A, Dohmann K, Menkel A, Eilers M, Luscher B. Genes Dev. 2001;15:2042–2047. doi: 10.1101/gad.907901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nikiforov MA, Chandriani S, Park J, Kotenko I, Matheos D, Johnsson A, McMahon SB, Cole MD. Mol. Cell. Biol. 2002;22:5054–5063. doi: 10.1128/MCB.22.14.5054-5063.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eberhardy SR, D'Cunha CA, Farnham PJ. J. Biol. Chem. 2000;275:33798–33805. doi: 10.1074/jbc.M005154200. [DOI] [PubMed] [Google Scholar]
- 18.Eberhardy SR, Farnham PJ. J. Biol. Chem. 2001;276:48562–48571. doi: 10.1074/jbc.M109014200. [DOI] [PubMed] [Google Scholar]
- 19.Eberhardy SR, Farnham PJ. J. Biol. Chem. 2002;277:40156–40162. doi: 10.1074/jbc.M207441200. [DOI] [PubMed] [Google Scholar]
- 20.Kato GJ, Barrett J, Villa-Garcia M, Dang CV. Mol. Cell. Biol. 1990;10:5914–5920. doi: 10.1128/mcb.10.11.5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hardy S, Brand M, Mittler G, Yanagisawa J, Kato S, Meisterernst M, Tora L. J. Biol. Chem. 2002;277:32875–32882. doi: 10.1074/jbc.M205860200. [DOI] [PubMed] [Google Scholar]
- 22.Hateboer G, Timmers HT, Rustgi AK, Billaud M, van't Veer LJ, Bernards R. Proc. Natl. Acad. Sci. U. S. A. 1993;90:8489–8493. doi: 10.1073/pnas.90.18.8489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brasier AR, Tate JE, Habener JF. BioTechniques. 1989;7:1116–1122. [PubMed] [Google Scholar]
- 24.Martinez E, Kundu TK, Fu J, Roeder RG. J. Biol. Chem. 1998;273:23781–23785. doi: 10.1074/jbc.273.37.23781. [DOI] [PubMed] [Google Scholar]
- 25.Roth SY, Denu JM, Allis CD. Annu. Rev. Biochem. 2001;70:81–120. doi: 10.1146/annurev.biochem.70.1.81. [DOI] [PubMed] [Google Scholar]
- 26.Tora L. Genes Dev. 2002;16:673–675. doi: 10.1101/gad.976402. [DOI] [PubMed] [Google Scholar]
- 27.Bosotti R, Isacchi A, Sonnhammer EL. Trends Biochem. Sci. 2000;25:225–227. doi: 10.1016/s0968-0004(00)01563-2. [DOI] [PubMed] [Google Scholar]
- 28.Eisenman RN. Genes Dev. 2001;15:2023–2030. doi: 10.1101/gad928101. [DOI] [PubMed] [Google Scholar]
- 29.Flinn EM, Wallberg AE, Hermann S, Grant PA, Workman JL, Wright AP. J. Biol. Chem. 2002;277:23399–23406. doi: 10.1074/jbc.M201704200. [DOI] [PubMed] [Google Scholar]
- 30.Spotts GD, Patel SV, Xiao Q, Hann SR. Mol. Cell. Biol. 1997;17:1459–1468. doi: 10.1128/mcb.17.3.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hirst SK, Grandori C. Oncogene. 2000;19:5189–5197. doi: 10.1038/sj.onc.1203904. [DOI] [PubMed] [Google Scholar]
- 32.Stone J, DeLange T, Ramsay G, Jakobovitz E, Bishop JM, Varmus H, Lee W. Mol. Cell. Biol. 1987;7:1697–1709. doi: 10.1128/mcb.7.5.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiao Q, Claassen G, Shi J, Adachi S, Sedivy J, Hann SR. Genes Dev. 1998;12:3803–3808. doi: 10.1101/gad.12.24.3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lang SE, McMahon SB, Cole MD, Hearing P. J. Biol. Chem. 2001;276:32627–32634. doi: 10.1074/jbc.M102067200. [DOI] [PubMed] [Google Scholar]
- 35.Yanagisawa J, Kitagawa H, Yanagida M, Wada O, Ogawa S, Nakagomi M, Oishi H, Yamamoto Y, Nagasawa H, McMahon SB, Cole MD, Tora L, Takahashi N, Kato S. Mol. Cell. 2002;9:553–562. doi: 10.1016/s1097-2765(02)00478-1. [DOI] [PubMed] [Google Scholar]
- 36.Ard PG, Chatterjee C, Kunjibettu S, Adside LR, Gralinski LE, McMahon SB. Mol. Cell. Biol. 2002;22:5650–5661. doi: 10.1128/MCB.22.16.5650-5661.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Belotserkovskaya R, Sterner DE, Deng M, Sayre MH, Lieberman PM, Berger SL. Mol. Cell. Biol. 2000;20:634–647. doi: 10.1128/mcb.20.2.634-647.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sterner DE, Belotserkovskaya R, Berger SL. Proc. Natl. Acad. Sci. U. S. A. 2002;99:11622–11627. doi: 10.1073/pnas.182021199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu PY, Winston F. Mol. Cell. Biol. 2002;22:5367–5379. doi: 10.1128/MCB.22.15.5367-5379.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barlev NA, Liu L, Chehab NH, Mansfield K, Harris KG, Halazonetis TD, Berger SL. Mol. Cell. 2001;8:1243–1254. doi: 10.1016/s1097-2765(01)00414-2. [DOI] [PubMed] [Google Scholar]
- 41.Dudley AM, Rougeulle C, Winston F. Genes Dev. 1999;13:2940–2945. doi: 10.1101/gad.13.22.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Larschan E, Winston F. Genes Dev. 2001;15:1946–1956. doi: 10.1101/gad.911501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bhaumik SR, Green MR. Mol. Cell. Biol. 2002;22:7365–7371. doi: 10.1128/MCB.22.21.7365-7371.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]