Abstract
ABCA1 is a major regulator of cellular cholesterol efflux and plasma HDL biogenesis. Even though the transcriptional activation of ABCA1 is well established, the posttranscriptional regulation of ABCA1 expression is poorly understood. Here, we investigate the potential contribution of the RNA binding protein (RBP) human antigen R (HuR) on the posttranscriptional regulation of ABCA1 expression. RNA immunoprecipitation assays demonstrate a direct interaction between HuR and ABCA1 mRNA. We found that HuR binds to the 3′ untranslated region of ABCA1 and increases ABCA1 translation, while HuR silencing reduces ABCA1 expression and cholesterol efflux to ApoA1 in human hepatic (Huh-7) and monocytic (THP-1) cells. Interestingly, cellular cholesterol levels regulate the expression, intracellular localization, and interaction between HuR and ABCA1 mRNA. Finally, we found that HuR expression was significantly increased in macrophages from human atherosclerotic plaques, suggesting an important role for this RBP in controlling macrophage cholesterol metabolism in vivo. In summary, we have identified HuR as a novel posttranscriptional regulator of ABCA1 expression and cellular cholesterol homeostasis, thereby opening new avenues for increasing cholesterol efflux from atherosclerotic foam macrophages and raising circulating HDL cholesterol levels.
Keywords: ATP binding cassette transporter A1, human antigen R, posttranscriptional regulation, lipid homeostasis, cholesterol efflux
Lipid dysregulation is a critical factor associated with the development of several pathologies, such as Alzheimer’s disease, type 2 diabetes, and atherosclerosis (1, 2). Early stages of atherosclerosis are characterized by the accumulation of lipid-loaded macrophages in the artery wall. To avoid massive accumulation of cholesterol, macrophages can efflux cholesterol to lipid-poor circulating ApoAI and HDL through the ABCA1 and ABCG1 transporters, respectively (3). ABCA1 also plays an important role in the biogenesis of HDL, and mutations in this gene are associated with low plasma HDL-cholesterol (HDL-C) levels and a high risk for developing atherosclerotic vascular disease (4). ABCA1-mediated cholesterol efflux represents the first step in reverse cholesterol transport (RCT), a process that results in the net cholesterol removal from peripheral tissues back to the liver for excretion into bile and feces (5). The expression of ABCA1 is regulated at the transcriptional level primarily by the liver X receptors (LXRs). During cellular cholesterol excess, LXRs induce cholesterol efflux through the transcriptional regulation of ABCA1 expression, keeping cellular cholesterol balanced (6). Although the classical transcriptional regulation of ABCA1 expression is well established, its regulation at the posttranscriptional level is just starting to be revealed. In this regard, micro-RNAs (miRNAs) and RNA binding proteins (RBPs) have been shown to regulate gene expression by controlling mRNA stability and/or translation. miRNAs are a class of small (19–23 nt) noncoding RNAs that bind to the 3′ untranslated regions (3′UTRs) of target mRNAs promoting degradation and/or translational repression by forming incomplete base pairing with its target mRNA (7–9). Recently, we and others have shown that ABCA1 expression is regulated by several miRNAs including miR-33, miR-758, miR-106b, and miR-144, thus revealing a novel layer of regulation of cholesterol homeostasis (10–16).
In addition to miRNAs, RBPs (especially turnover- and translation-regulatory RBPs) are known to regulate all aspects of mRNA metabolism including processing, transport, translation, and turnover via different RNA interaction motifs generally present in the 3′UTR of the target mRNA (17–20). Human antigen R (HuR), also known as embryonic lethal, abnormal vision Drosophila-like 1 (ELAVL1), is a ubiquitously expressed RBP belonging to the Hu/Elav family that modulates the stability and translational efficiency of mRNAs by interacting with uridylate (U)-rich or adenylate-uridylate (AU)-rich elements in the 3′UTR (21, 22). HuR is constitutively expressed and localized predominantly in the nucleus of most unstimulated cells but can translocate to the cytoplasm upon cellular stimulation, a process that has been linked to the stabilization of many targets (23). While HuR has been reported to be involved in many pathologic processes, particularly cancer and inflammation (24), the role of HuR in atherosclerosis has not yet been well characterized. Previous genome-wide studies in HuR identified two preferred individual-nucleotide resolution cross-linking and immunoprecipitation (iCLIP) sites associated with two U-rich segments in the 3′UTR of ABCA1 (25). Building on this information, we decided to study the role of HuR in the regulation of ABCA1 expression and function. Our results reveal a direct interaction between HuR and ABCA1 mRNA and demonstrate that HuR controls ABCA1 protein expression levels and cholesterol efflux in human macrophages and hepatic cells. Interestingly, cellular cholesterol levels in turn regulate the expression, intracellular localization, and interaction between HuR and ABCA1 mRNA. Finally, we found that HuR expression was significantly increased in macrophages accumulated in human atherosclerotic plaques, suggesting a role for HuR in controlling macrophage lipid homeostasis in vivo. Altogether, these results demonstrate that HuR can be potentially considered a therapeutic target for increasing cholesterol efflux from atherosclerotic foam macrophages and raising circulating HDL-C levels.
MATERIALS AND METHODS
Chemicals
Chemicals were obtained from Sigma unless otherwise noted. Human lipoproteins [acetylated LDL (Ac-LDL)] were obtained from Biomedical Technologies Inc. The synthetic LXR ligand T0901317 (T090) was purchased from Cayman Chemical. Human ApoA1 and HDL were obtained from Meridian Life Sciences. A mouse monoclonal antibody against ABCA1 was purchased from Abcam, a mouse monoclonal heat shock protein 90 (HSP90) antibody was purchased from BD Bioscience, a mouse monoclonal p84 antibody was from GeneTex, and mouse monoclonal HuR and α-tubulin antibodies were from Santa Cruz Biotechnology. Goat polyclonal T-cell restricted intracellular antigen (TIA-1), heterogeneous nuclear ribonucleoprotein C, and glycine-tryptophan protein of 182 KDa (GW-182) antibodies were from Santa Cruz. Mouse monoclonal T-cell intracellular antigen-1 related protein (TIAR) antibody was from BD Bioscience. Antibody recognizing AU-rich element RNA-binding protein 1 (AUF1) was from Millipore. The LDL receptor (LDLR) polyclonal antibody was from Cayman Chemical, and a mouse monoclonal antibody recognizing Renilla luciferase (RLuc) was from Abcam. Secondary fluorescently labeled antibodies were from Molecular Probes (Invitrogen).
Cell culture and transfection
Human monocytic (THP-1) and human hepatic (Huh-7) cells were obtained from American Type Tissue Collection. THP-1 cells were maintained in RPMI 1640 media (Sigma) supplemented with 10% FBS and 2% penicillin-streptomycin in 10 cm2 dishes at 37°C and 5% CO2. THP-1 differentiation into macrophages was induced using 100 nM PMA for 72 h. Huh-7 cells were maintained in DMEM containing 10% FBS and 2% penicillin-streptomycin. The siRNAs against HuR (HuR siRNA) and control siRNA (Ctrl siRNA) were obtained from Dharmacon (Lafayette, CO). THP-1 and Huh-7 cells were transfected with 60 nM siRNA utilizing RNAiMax (Invitrogen) and analyzed 72 h after transfection. For HuR overexpression, Huh-7 cells were transfected with 1 μg of HuR fused to a tandem affinity purification (TAP) tag (TAP-HuR) or control TAP (TAP) utilizing Lipofectamine 2000 (Invitrogen) and analyzed 48 h after transfection. For mRNA stability assays, Huh-7 cells were treated with actinomycin D (2.5 μg/ml) to inhibit de novo transcription.
RNA isolation and quantitative real-time PCR
Total RNA was isolated using TRIzol reagent (Invitrogen) according to the manufacturer’s protocol. For mRNA quantification, cDNA was synthesized using iScript RT Supermix (Bio-Rad), following the manufacturer’s protocol. Quantitative real-time PCR (qPCR) was performed in triplicate using iQ SYBR Green Supermix (BioRad) on a Real-Time Detection System (Eppendorf). The mRNA levels were normalized to the levels of 18S rRNA. The human primer sequences used were the following: ABCA1, 5′-GGTTTGGAGATGGTTATACAATAGTTGT-3′ and 5′-CCCGGAAACGCAAGTCC-3; ABCG1, 5′-TCACCCAGTTCTGCATCCTCTT-3′ and 5′-GCAGATGTGTCAGGACCGAGT-3′ 18S, 5′-GCTTAATTTGACTCAACACGGGA-3′ and 5′-AGCTATCAATCTGTCAATCCTGTC-3′ HuR (ELAVL1), 5′-GCGCAGAGATTCAGGTTCTCCC-3′ and 5′-GGCCATCGCGGCTTCTTCAT-3′ LDLR, 5′-AGTTGGCTGCGTTAATGTGAC-3′ and 5′-TGATGGGTTCATCTGACCAGT-3′.
Western blot analysis
Cells were lysed in ice-cold buffer containing 50 mM Tris-HCl, pH 7.5, 125 mM NaCl, 1% NP-40, 5.3 mM NaF, 1.5 mM NaP, 1 mM orthovanadate, 1 mg/ml of protease inhibitor cocktail (Roche), and 0.25 mg/ml 4-benzenesulfonyl fluoride hydrochloride (AEBSF; Roche). Cell lysates were rotated at 4°C for 1 h before the insoluble material was removed by centrifugation at 12,000 g for 10 min. After normalizing for equal protein concentration, cell lysates were resuspended in SDS sample buffer before separation by SDS-PAGE. Following overnight transfer of the proteins onto nitrocellulose membranes, the membranes were probed with the indicated antibodies, and protein bands were visualized using the Odyssey Infrared Imaging System (LI-COR Biotechnology). Densitometry analysis of the gels was carried out using ImageJ software from the National Institutes of Health (http://rsbweb.nih.gov/ij/).
Nuclear and cytosolic extract preparation
Huh-7 and THP-1 cells (average 4 × 106) were incubated in 100 μl of buffer A [10 mM HEPES pH 7.6, 10 mM KCl, 0.1 mM EDTA, 0.2 mM EGTA, 0.75 mM spermidine, 0.15 mM spermine, 1% NP-40, 5.3 mM NaF, 1.5 mM NaP, 1 mM DTT, 1 mM orthovanadate, 1 mg/ml of protease inhibitor cocktail (Roche), and 0.25 mg/ml AEBSF (Roche)] for 15 min at 4°C followed by vortexing for 15–20 s. Cell suspension was then centrifuged at 12,000 rpm for 1 min. After collecting the supernatants (cytosolic extracts), the pellets were washed several times with 1 ml buffer A and resuspended in 50 μl buffer C [20 mM HEPES pH 7.6, 400 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, 1 mM orthovanadate, 1 mg/ml of protease inhibitor cocktail (Roche), and 0.25 mg/ml AEBSF (Roche)]. Cell lysates were rotated at 4°C for 30 min before the insoluble material was removed by centrifugation at 12,000 rpm for 5 min, and the supernatants were used as nuclear fraction (NF).
Immunofluorescence and confocal microscopy
THP-1 macrophages and Huh-7 cells were fixed with 4% paraformaldehyde (PFA) in PBS and washed with PBS. Cells were permeabilized with 0.1% Triton and blocked with 3% BSA and then were incubated with primary antibodies against HuR in 3% BSA for 1 h, washed, and incubated with antibodies conjugated to Alexa Fluor 488 (A-21202 at 1:250; Invitrogen) for 1 h. After samples were washed, cells were mounted in the presence of ProLong Gold Antifade Reagent (P36390, Invitrogen) with Topro-3 for nuclear staining (T3604, Invitrogen). Images were collected with a Leica TCS SP5 confocal laser-scanning microscope with a 63×oil objective and a 1.4 numerical aperture (63×/1.4). The frame size was 1,024 by 1,024 pixels.
3′UTR luciferase reporter assays
The cDNA fragments corresponding to the entire 3′UTR of human ABCA1 mRNAs were amplified by RT-PCR using the following primers: 5′-AGCGGCCGCTTTCTGTAGACCAACAGAACTGTCA-3′ (NotI) and 5′- AACTCGAGAGAATCCTGTTCATACGGGG-3′ (XhoI) from total RNA extracted from cells. The PCR product was directionally cloned downstream of the RLuc open reading frame of the psiCHECK2TM vector (Promega) that also contains a constitutively expressed firefly luciferase gene, which is used to normalize transfections (ABCA1 3′UTR). ABCA1 3′UTR Δ1, Δ2 construct was obtained by deletion of specific HuR site 1 and site 2 in the 3′UTR of ABCA1 using QuikChange II XL Site-Directed Mutagenesis Kit (Agilent Technologies). All constructs were confirmed by sequencing. COS-7 cells were plated into 12-well plates and cotransfected with 1 μg of the indicated 3′UTR luciferase reporter vectors and the mammalian vector that expresses HuR fused to a TAP tag (TAP-HuR) or control TAP (TAP) utilizing Lipofectamine 2000 (Invitrogen). Luciferase activity was measured using the Dual-Glo Luciferase Assay System (Promega). RLuc activity was normalized to the corresponding firefly luciferase activity and plotted as a percentage of the control (cells cotransfected with the corresponding concentration of control TAP). Experiments were performed in triplicate wells of 12-well plates and repeated at least three times.
Immunoprecipitation assays
Immunoprecipitation (IP) of endogenous ribonucleoprotein (RNP) complexes was performed from whole-cell extracts (WCEs) as previously described (26, 27). Briefly, cells were lysed in 20 mM Tris-HCl at pH 7.5, 100 mM KCl, 5 mM MgCl2, and 0.5% NP-40 for 10 min on ice and centrifuged at 10,000 g for 15 min at 4°C. The supernatants were incubated with protein A-Sepharose beads coated with antibodies that recognized HuR (Santa Cruz Biotechnology) or control IgG (Santa Cruz Biotechnology) for 1 h at 4°C. After the beads were washed with NT2 buffer (50 mM Tris-HCl at pH 7.5, 150 mM NaCl, 1 mM MgCl2, and 0.05% NP-40), the complexes were incubated with 20 units of RNase-free DNase I (15 min at 37°C) and further incubated with 0.1% SDS/0.5 mg/ml Proteinase K (15 min at 55°C) to remove DNA and proteins, respectively. The RNPs isolated from the IP materials were further assessed by RT-qPCR analysis. In the ribonucleoprotein immunoprecipitation (RIP) experiment in cells transfected with ABCA1 3′UTR or ABCA1 3′UTR Δ1, Δ2, mRNA enrichment was assessed using primers that recognized the RLuc coding region: 5′-TCCAGAACAAAGGAAACGGA-3′ and 5′-ATAATACACCGCGCTACTGG-3′.
Polysome analysis
Forty-eight hours after transfection with siRNAs, Huh-7 cells were preincubated with cycloheximide (Sigma; 100 μg/ml for 15 min), and cytoplasmic lysates were prepared and fractionated by ultracentrifugation through 15–60% linear sucrose gradients; 12 fractions were collected, and RNA extracted from each fraction was used for RT-qPCR analysis, as described (28).
Cholesterol efflux assays
THP-1 macrophages (1 × 106/well) were transfected with either an HuR siRNA, or a Ctrl siRNA 48 h prior to loading with 0.5 μCi/ml 3H-cholesterol for 24 h with or without T090 (3 μM) for 12 h. Then, cells were washed twice with PBS and incubated in RPMI supplemented with 2 mg/ml fatty-acid free BSA (FAFA media) in the presence of ACAT inhibitor Sandoz (2 μM) for 4 h prior to the addition of 50 μg/ml of human ApoA1 in FAFA media. Supernatants were collected after 6 h, and radioactivity content was expressed as a percentage of 3H-cholesterol in the media/total cell 3H-cholesterol content (total effluxed 3H-cholesterol + cell-associated 3H-cholesterol).
Immunohistochemistry of carotid endarterectomy plaques
The study was approved by the hospital’s ethics committee (IIS-Fundación Jiménez Díaz) according to the institutional and the Good Clinical Practice guidelines and was performed in accordance with the Declaration of Helsinki. Written informed consent was obtained from patients undergoing carotid endarterectomy. The carotid atherosclerotic plaques were fixed with PFA and embedded in paraffin for histological analysis and were then cross-sectioned into 4 mm thick pieces, dewaxed, and rehydrated. Immunohistochemical staining was assessed using anti-CD68 (clone kp1, M0814, Dako), anti-HuR (sc-5261, Santa Cruz Biotechnology), or anti-ABCA1 (ab18180, Abcam) antibodies, followed by mouse secondary antibody. ABComplex/HRP (Vector Laboratories) was added, and sections were stained with 3,3′-diaminobenzidine (Dako). Corresponding hematoxylin staining was used for nucleus identification. For colocalization studies in plaques, double immunohistochemistry/immunofluorescence with anti-HuR or anti-ABCA1 antibodies was carried out in the same sections followed by CD68 staining, as previously described (29). After ABComplex/HRP for HuR or ABCA1, sections were stained with 3,3′-diaminobenzidine (Dako) followed by immunofluorescence for CD68 and mounted in ProLong Gold Antifade Reagent (P36390, Invitrogen). Negative controls using the corresponding IgG were included for checking nonspecific staining.
Statistical analysis
All data are expressed as ± SEM. Statistical differences were measured by either a Student t-test or two-way ANOVA with Bonferroni correction for multiple comparisons when appropriate. A value of P ≤ 0.05 was considered statistically significant. Data analysis was performed using GraphPad Prism 5.0a software (GraphPad, San Diego, CA).
RESULTS
HuR interacts with the 3′UTR of ABCA1
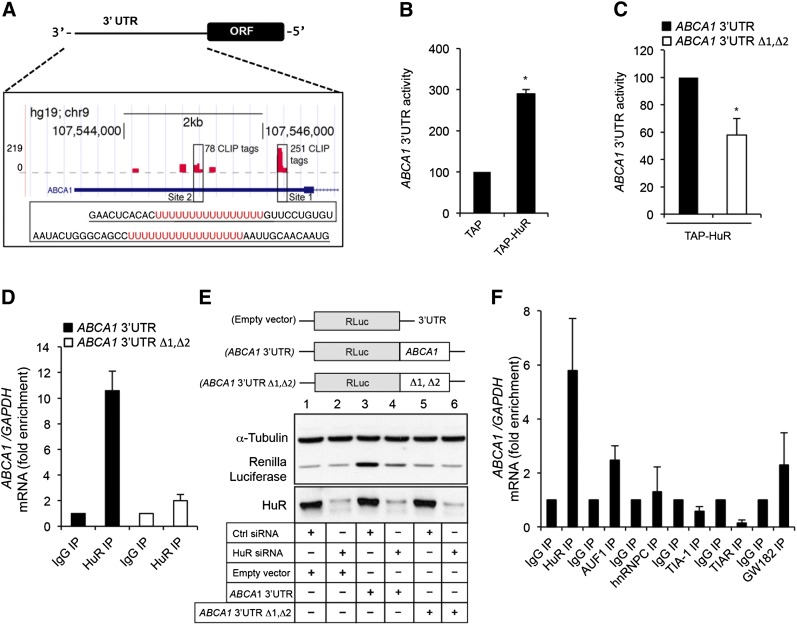
Initial analysis of the ABCA1 3′UTR revealed several U/AU-rich elements similar to the bona fide HuR binding sites previously described (30). In agreement with this observation, a recent throughput study from Penalva’s laboratory (25) mapped these sites via iCLIP in HeLa cells (Fig. 1A and supplementary Fig. I). HuR regulates gene expression by increasing mRNA stability and/or protein translation. Thus, we evaluated whether HuR overexpression influences the ABCA1 3′UTR activity using a luciferase reporter assay. As seen in Fig. 1B, HuR overexpression (TAP-HuR) markedly increased the 3′UTR activity compared with cells transfected with control vector (TAP), suggesting that HuR enhances the stability and/or translation rate of ABCA1 mRNA. As expected, specific deletion of the two major HuR binding sites (ABCA1 3′UTR Δ1, Δ2) significantly reduced the ABCA1 3′UTR activity (Fig. 1C). To confirm the direct interaction between HuR and the 3′UTR of ABCA1, we performed RIP analysis in Huh-7 cells using a specific antibody against HuR. The results showed a significant enrichment of ABCA1 mRNA relative to the levels in IP samples obtained by using a control antibody (IgG1) and normalized to the levels of GAPDH mRNA (a transcript that is not a target of HuR) (Fig. 1D, solid bars). Of note, specific deletion of HuR binding sites (ABCA1 3′UTR Δ1, Δ2) abolished the interaction between HuR and the 3′UTR of ABCA1 (Fig. 1D, open bars). To confirm the influence of endogenous HuR levels on ABCA1 expression, we analyzed the RLuc protein levels in cells expressing endogenous (Ctrl siRNA) or reduced (HuR siRNA) HuR levels and transfected with RLuc empty vector, RLuc vector fused to the 3′UTR of ABCA1 (ABCA1 3′UTR construct), and RLuc fused to ABCA1 3′UTR Δ1, Δ2 (ABCA1 3′UTR Δ1, Δ2). As shown in Fig. 1E, we observed significantly higher expression of RLuc in Huh-7 cells transfected with the RLuc vector fused to the 3′UTR of ABCA1 (third column) compared with cells transfected with RLuc empty vector (first column) or RLuc vector fused ABCA1 3′UTR Δ1, Δ2 (fifth column). Importantly, RLuc expression was significantly reduced in Huh-7 cells transfected with the RLuc vector fused to the 3′UTR of ABCA1 when we silenced the expression of HuR (Fig. 1E, fourth column vs. third column). As expected, HuR inhibition did not affect RLuc levels in cells transfected with the RLuc empty vector or ABCA1 3′UTR Δ1, Δ2 construct (Fig. 1E). Finally, we determined the specificity of HuR in regulating ABCA1 expression compared with other RBPs implicated in mRNA stabilization and/or translation such as AUF1, hnRNP, TIA-1, TIAR, and GW-182 by RIP assays. The results revealed that ABCA1 mRNA associated preferentially to HuR compared with other RBPs assessed (Fig. 1F). Taken together, these results demonstrate a direct interaction between HuR and ABCA1 mRNA and suggest that HuR positively regulates ABCA1 expression posttranscriptionally through the ABCA1 3′UTR.
Fig. 1.
HuR associates with ABCA1 mRNA. A: Graphical representation of the HuR binding sites in the 3′UTR of ABCA1 (red bars) mapped by iCLIP. Sequences for major binding sites (Site 1 and Site 2) with 251 and 78 iCLIP tags, respectively. B: 3′UTR activity of human ABCA1 mRNA in COS-7 cells transfected with a control plasmid that expresses the TAP tag (TAP) or with TAP-HuR plasmids, which express TAP-tagged HuR. Data are expressed as relative luciferase activity to control samples cotransfected with TAP-empty vector (normalized to 100) and are the mean ± SEM of three independent experiments in triplicate. * P < 0.05 versus untreated group. C: 3′UTR activity of human ABCA1 mRNA and ABCA1 mRNA with specific deletions in the HuR binding sites (ABCA1 3′UTR Δ1, Δ2) in COS-7 cells transfected with with TAP-HuR plasmid. Data are expressed as relative luciferase activity to cells cotransfected with the 3′UTR ABCA1 (normalized to 100) and are the mean ± SEM of three independent experiments in triplicate. * P < 0.05 versus untreated group. D: RIP analysis of Huh-7 cells overexpressing ABCA1 3′UTR (solid bars) or ABCA1 3′UTR Δ1, Δ2 (open bars) constructs using IgG and an anti-HuR antibody. ABCA1 and GAPDH mRNAs were quantified using RT-qPCR and were represented as fold enrichment compared with IgG RIP analysis. E: Representative Western blot analysis of the RLuc protein expression in Huh-7 cells overexpressing RLuc empty vector (Empty vector), RLuc vector fused to the 3′UTR of ABCA1 (ABCA1 3′UTR), and RLuc fused to ABCA1 3′UTR Δ1, Δ2 (ABCA1 3′UTR Δ1, Δ2), treated with Ctrl siRNA versus HuR siRNA. F: Analysis of the association between the ABCA1 mRNA and different RBPs by RIP using specific antibodies. ABCA1 and GAPDH mRNAs were quantified using RT-qPCR and were represented as fold enrichment compared with IgG RIP.
HuR regulates ABCA1 expression by enhancing protein translation
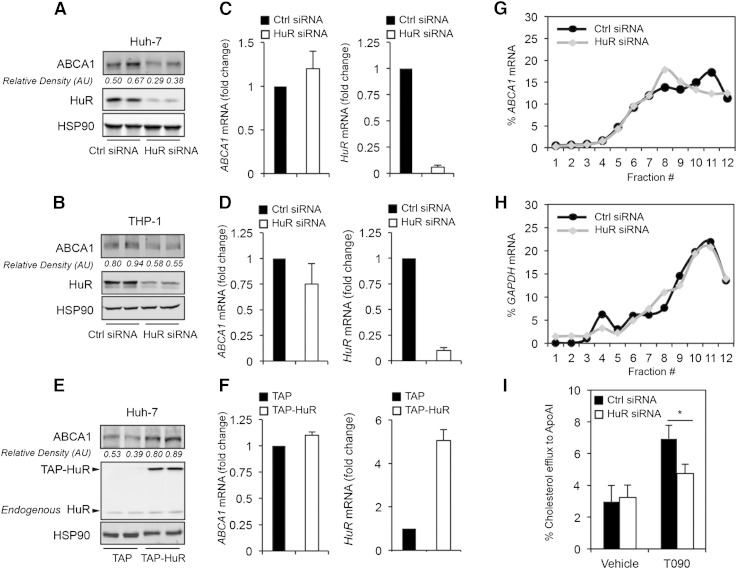
To analyze the contribution of HuR in regulating ABCA1 expression, we performed experiments in which HuR expression levels were modified. As shown in Fig. 2A, B, siRNA-mediated inhibition of HuR in Huh-7 and THP-1 cells using a specific HuR-directed siRNA resulted in a significant decrease in ABCA1 protein expression without changes in ABCA1 mRNA levels (Fig. 2C, D, left panels). As expected, transfection of HuR-directed siRNA resulted in the knockdown of the mRNA levels of HuR in both cells types (Fig. 2C, D, right panels). Conversely, HuR overexpression in Huh-7 cells using a mammalian vector that expresses HuR fused to a TAP tag (TAP-HuR) boosted ABCA1 protein expression (Fig. 2E). In both sets of experiments, ABCA1 mRNA levels were not affected by either HuR silencing or overexpression (Fig. 2C, D, F, left panels), suggesting that HuR does not influence ABCA1 mRNA stability. Indeed, ABCA1 mRNA half-life was similar in Huh-7 cells transfected with HuR siRNA compared with Ctrl siRNA (supplementary Fig. IIA). GAPDH and sirtuin 1 (SIRT1) mRNAs expression were studied as negative and positive controls of HuR-regulated mRNA stability, respectively (supplementary Fig. IIB, C). Because HuR did not influence ABCA1 mRNA stability, we asked whether HuR could affect ABCA1 translation. To test this hypothesis, we performed polysome analysis in Huh-7 cells transfected with a control siRNA or an HuR-directed siRNA for 48 h. The results show that in control siRNA-transfected cells the distribution of ABCA1 mRNA along the sucrose gradient shifted toward heavier polysome fractions (Fig. 2G) while HuR silencing abrogated the shift in translation of ABCA1 mRNA. The distribution of the housekeeping GAPDH mRNA did not show this pattern (Fig. 2H), indicating that silencing HuR specifically affected ABCA1 mRNA translation. These results support the notion that HuR enhances ABCA1 mRNA association with actively translating polysomes and that HuR promotes ABCA1 translation.
Fig. 2.
Role of HuR in the regulation of ABCA1 protein expression in Huh-7 and THP-1 cells and cholesterol efflux. A, B: Representative Western blot analysis of ABCA1 and HuR expression in Huh-7 (A) and THP-1 (B) cells transfected with a nontargeted control siRNA (Ctrl siRNA) or HuR siRNA (HuR siRNA). HSP90 was used as loading control. C, D: qRT-PCR analysis of ABCA1 and HuR expression in Huh-7 (C) and THP-1 (D) cells transfected with a nontargeted Ctrl siRNA or HuR siRNA. Data are represented as mRNA fold change compared with cells transfected with Ctrl siRNA (normalized to 1) and are the means ± SEM of three independent experiments performed in triplicate. E: Representative Western blot analysis of ABCA1 and HuR expression in Huh-7 cells transfected with empty vector (TAP) and TAP-HuR. HSP90 was used as loading control. F: qRT-PCR analysis of ABCA1 and HuR expression in Huh-7 cells transfected with empty vector (TAP) and TAP-HuR. Data are represented as mRNA fold change compared with cells transfected with Ctrl siRNA (normalized to 1) and are the means ± SEM of three independent experiments performed in triplicate. G, H: Polysome analysis performed in Huh-7 cells transfected with Ctrl siRNA or HuR siRNA. The relative levels of ABCA1 (G) and GAPDH (H) mRNAs in each fraction were analyzed by RT-qPCR and represented as percentage of total RNA in the gradient. I: Cholesterol efflux analysis to ApoA1 performed in THP-1-derived macrophages transfected with Ctrl siRNA or HuR siRNA and treated or not with T090. Data are the means ± SEM of three independent experiments in triplicate. * P < 0.05 Ctrl siRNA compared with HuR siRNA.
HuR controls ABCG1 expression
In addition to ABCA1, ABCG1 regulates cellular cholesterol efflux. The expression of both ABC transporters is regulated by similar regulatory mechanisms. Therefore, we asked whether HuR also regulates ABCG1 expression. Similarly to ABCA1, HuR overexpression markedly increased ABCG1 3′UTR activity (supplementary Fig. IIIA). This effect was specific for HuR because other RBPs assessed did not interact directly with the 3′UTR of ABCG1 (supplementary Fig. IIIB). We next evaluated the effect of HuR silencing on ABCG1 expression in Huh-7 and THP-1 cells. As seen in supplementary Fig. IIIC, D, inhibition of HuR resulted in a significant decrease of ABCG1 expression in both cell types. Conversely, overexpression of HuR in Huh-7 cells increased ABCG1 protein levels (supplementary Fig. IIIE). Taken together these results demonstrate that HuR controls ABCG1 expression.
HuR regulates cholesterol efflux to ApoAI
ABCA1 plays a critical role in promoting cellular cholesterol efflux to ApoA1. To determine whether HuR affects ABCA1 function, we performed cholesterol efflux assays in THP-1 cells transfected with a control siRNA or HuR-targeted siRNA. In keeping with the enhancement in ABCA1 expression by HuR, HuR silencing attenuated cholesterol efflux to ApoA1 in cells stimulated with a synthetic LXR agonist T090 (Fig. 2I), indicating that HuR affects cellular cholesterol efflux, a critical step in the RCT pathway to remove cholesterol excess to the liver.
Cholesterol content affects HuR expression and localization
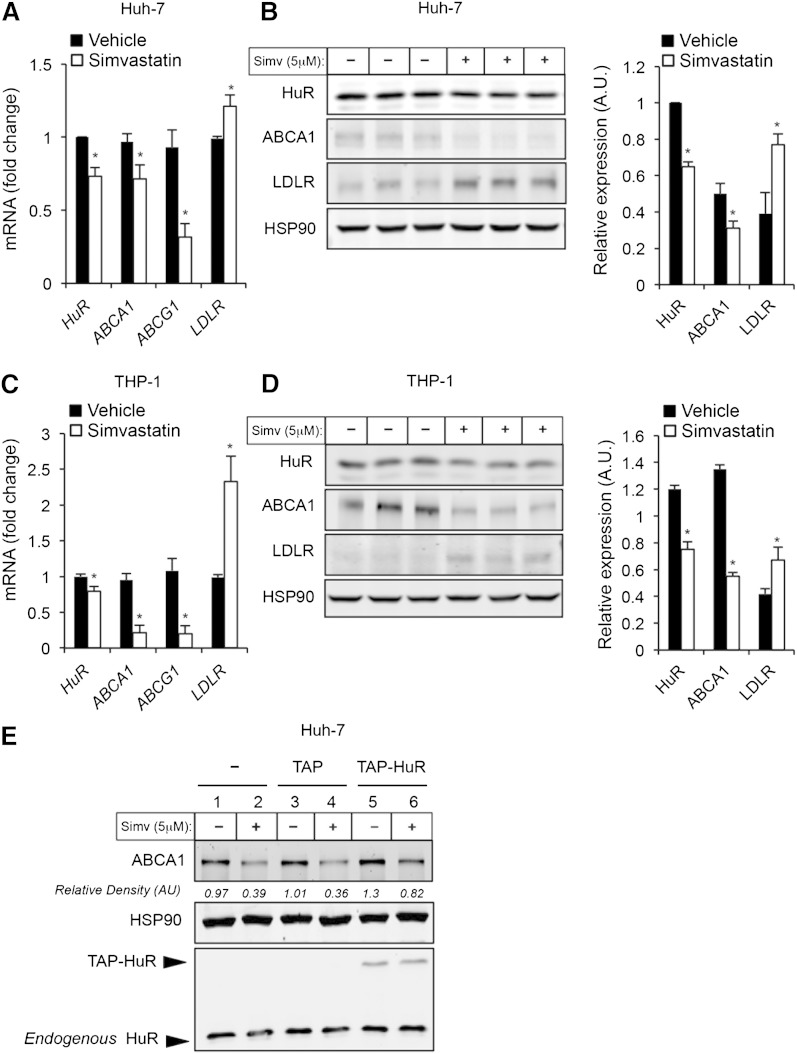
The regulation of ABCA1 expression is crucial to maintain lipid homeostasis and is determined by cellular cholesterol content. Given the role of HuR in the regulation of ABCA1 expression and function, we sought to analyze the expression and subcellular localization of HuR under different cholesterol content conditions. To this end, we depleted cholesterol in Huh-7 and THP-1 cells by treating them with simvastatin and then analyzed HuR expression levels by RT-qPCR and Western blotting. Interestingly, Huh-7 and THP-1 cells treated with simvastatin showed significantly lower levels of the HuR mRNA protein (Fig. 3A–D). In addition to analyzing HuR expression levels, we also assessed ABCA1 and LDLR expression levels as internal controls for cholesterol depletion in cells treated with simvastatin. As expected, simvastatin treatment resulted in a significant reduction of ABCA1 expression and increased LDLR mRNA and LDLR protein levels (Fig. 3A–D). The fact that ABCA1 expression levels correlate positively with HuR levels suggests a possible coregulation of both proteins in response to cellular cholesterol content. To test whether HuR influences ABCA1 expression under cholesterol-depleting conditions, we overexpressed HuR in Huh-7 cells treated with simvastatin. As shown in Fig. 3E, HuR overexpression rescued partially ABCA1 expression in cells treated with simvastatin (sixth column vs. second and forth columns). These results demonstrated that both HuR and ABCA1 expression might be regulated in a similar manner to control cellular cholesterol efflux.
Fig. 3.
Cholesterol depletion decreases HuR expression in Huh-7 and THP-1 cells. A, C: qRT-PCR analysis of HuR, ABCA1, ABCG1, and LDLR expression in Huh-7 (A) and THP-1 (C) cells treated or not with simvastatin. Data are the mean ± SEM of three independent experiments in triplicate. * P < 0.05 versus untreated group. B, D: Representative Western blot analysis of HuR, ABCA1, ABCG1, and LDLR expression in Huh-7 (B) and THP-1 (D) cells treated or not with simvastatin (5 μM) for 24 h. Right panels show the densitometric analysis of the Western blots. E: Representative Western blot analysis of HuR, ABCA1, and HSP90 in Huh-7 cells transfected or not with empty vector (TAP) or TAP-HuR and treated or not with simvastatin (5 μM) for 24 h.
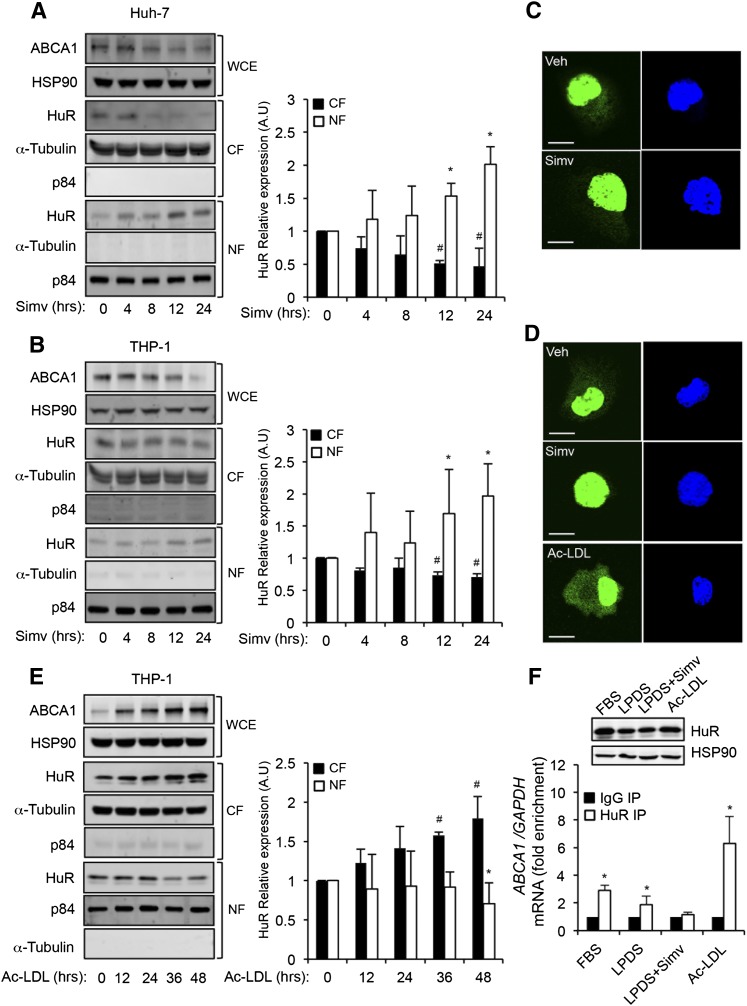
HuR regulates mRNA stabilization and protein translation in the cytoplasm. Therefore, we further explored whether cholesterol content may affect the subcellular localization of HuR. Treatment of Huh-7 and THP-1 cells with simvastatin at different time points followed by analysis of HuR levels in the cytosolic fraction (CF) and NF by Western blot revealed that simvastatin treatment promoted nuclear localization of HuR in a time-dependent manner (Fig. 4A, B). Similar results were observed when we analyzed HuR intracellular localization by immunofluorescence (Fig. 4C, D). We also assessed the HuR localization in cholesterol-loaded THP-1 cells. In contrast to the subcellular distribution of HuR observed in THP-1 cells treated with simvastatin, loading of cholesterol using Ac-LDL promoted cytosolic translocation of HuR (Fig. 4E, left panel). These results were also confirmed by immunofluorescence analysis in THP-1 cells loaded with cholesterol during 48 h (Fig. 4D, bottom panel). In agreement with these data, RIP experiments showed a reduction in the levels of ABCA1 mRNA associated with HuR when THP-1 cells were incubated in lipoprotein-deficient serum (LPDS) (Fig. 4F). This effect was even more pronounced when cells were treated with simvastatin for 24 h (Fig. 4F). In contrast, we observed a significant enrichment of ABCA1 mRNA in THP-1 cells that were loaded with cholesterol for 48 h (Fig. 4F). Interestingly, the increased interaction between HuR and ABCA1 was independent of changes in HuR protein levels (Fig. 4F, upper panels). Overall, these results demonstrate that cholesterol content in the cells not only regulates HuR expression but also controls its subcellular localization and its association with ABCA1 mRNA.
Fig. 4.
Translocation of HuR is regulated by simvastatin and Ac-LDL. A, C: Representative Western blot analysis of ABCA1, HSP90, HuR, α-tubulin, and p84 expression in Huh-7 (A) and THP-1-derived macrophages (C) treated with 5 μM of simvastatin at different time points. HuR abundance in CF (50 μg protein) and NF (10 μg protein) was analyzed. ABCA1 levels in the WCEs (50 μg protein) were measured in parallel as a control of cholesterol-depletion conditions. HSP90, α-tubulin, and p84 were used as housekeeping controls in WCE, CF, and NF, respectively. Right panels show the band densitometry analysis of the Western blots. Data are expressed as relative expression levels compared with untreated cells (time = 0 h) and normalized to 1 and correspond to the mean ± SEM of three independent experiments in triplicate. #,* P < 0.05 versus untreated group. C, D: Immunofluorescence analysis of HuR intracellular localization (green) in Huh-7 cells treated with vehicle (C, upper panel) or simvastatin (C, bottom panel) and THP-1 cells treated with vehicle (D, upper panel), simvastatin (D, middle panel), or Ac-LDL (D, bottom panel). Nuclei were stained with DAPI (blue). E: Representative Western blot analysis of ABCA1, HSP90, HuR, α -tubulin, and p84 expression in THP-1-derived macrophages treated with 120 μg/ml of Ac-LDL at different time points. HuR abundance in CF (50 μg protein) and NF (10 μg protein) was analyzed. ABCA1 levels in the WCEs (50 μg protein) were measured in parallel as a control of cholesterol-loading conditions. HSP90, α-tubulin, and p84 were used as housekeeping controls in WCE, CF, and NF, respectively. Right panels show the band densitometry analysis of the Western blots. Data are expressed as relative expression levels compared with untreated cells (time = 0 h) and normalized to 1 and correspond to the mean ± SEM of three independent experiments in triplicate. #,* P < 0.05 versus untreated group. F: RIP analysis of ABCA1 mRNA binding to HuR in Huh-7 cells cultured in presence of FBS, LPDS, LPDS with simvastatin (5 μM), or Ac-LDL (120 μg/ml). ABCA1/GAPDH mRNA ratio during RIP using anti-HuR antibody were quantified by RT-qPCR and were represented as fold enrichment compared with ABCA1/GAPDH mRNA ratio in the IgG RIP. Data are mean ± SEM of three independent experiments in triplicate. * P < 0.05 versus HuR expression was evaluated under these conditions by Western blot (F, upper panel). HSP90 expression was used as internal control of protein loading.
HuR and ABCA1 colocalize in macrophages in human atherosclerotic plaques
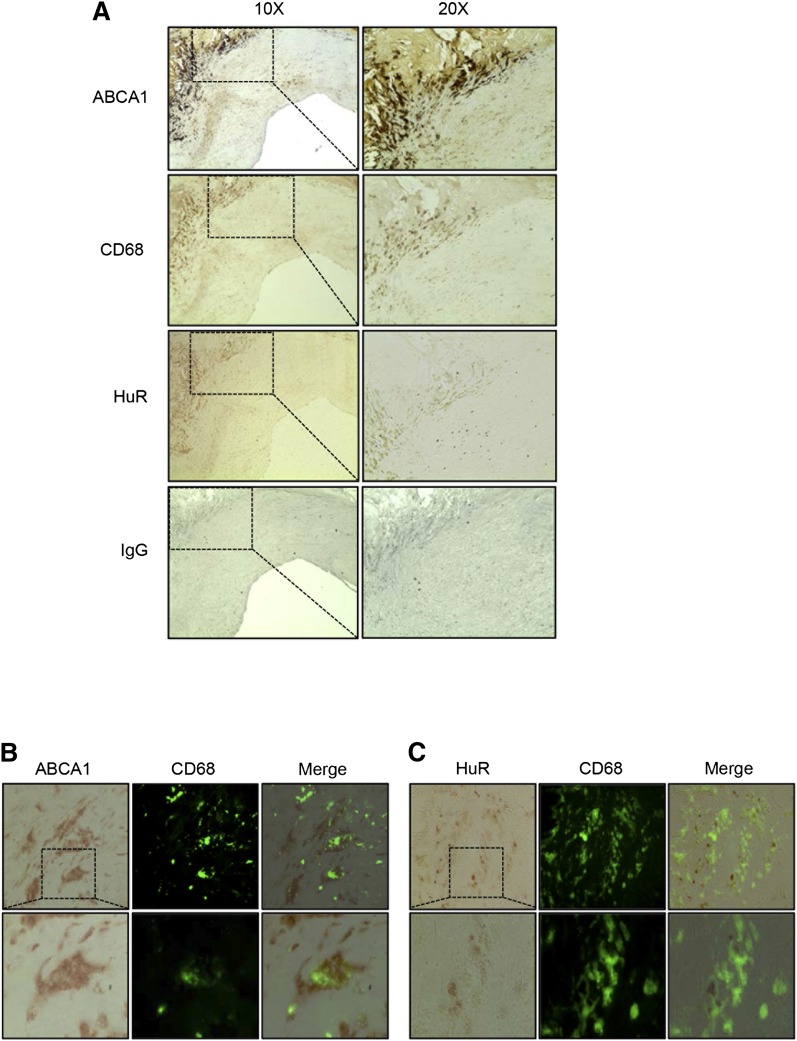
A number of studies have demonstrated that ABCA1 exerts a major atheroprotective role by raising plasma HDL levels, which is directly linked to the attenuation of the progression and the enhancement of the regression of atherosclerosis (31–33). The lack of conservation of HuR binding sites in the murine ABCA1 3′UTR makes it difficult to develop in vivo studies to extrapolate our results using mouse models of atherosclerosis. Therefore, we decided to explore the potential relevance of our previous data in human atherosclerosis. To this end, we performed several immunohistochemical analyses of HuR and ABCA1 expression in human carotid atherosclerotic plaques. These experiments revealed that HuR and ABCA1 expression mostly localized within similar regions in atherosclerotic plaques. Moreover, HuR was markedly expressed in plaque areas characterized by an abundant macrophage accumulation (CD68 positive area) (Fig. 5A–C). These results suggest that HuR might play a role in controlling ABCA1 expression in atherosclerotic plaque macrophages.
Fig. 5.
HuR and ABCA1 expression in human carotid atherosclerotic plaques. A: Immunohistochemical analysis of ABCA1 and HuR expression in human carotid plaques. CD68 was used as a marker for macrophages in the lesion area. Corresponding hematoxylin staining was used for nuclei identification. Right panels represent enlarged pictures from the same regions. Nonspecific immunostaining was tested using an IgG immunoglobulin as a negative control (bottom panel). B, C: Representative pictures of HuR and CD68 (B) or ABCA1 and CD68 (C) expression in the same section assessed by immunohistochemistry. Lower panel represents enlarged pictures in the indicated area.
DISCUSSION
The role of RBPs, including HuR, in regulating cell differentiation and growth, apoptosis, and inflammatory responses has been extensively investigated (24). However, the posttranscriptional regulation of cholesterol-related genes by RBPs and their association with cardiovascular disorders remains poorly understood. Here, we describe for the first time that HuR, one of the most studied members of the ELAVL family of RBPs, acts as a novel posttranscriptional regulator of ABCA1 expression. HuR interacts directly with the ABCA1 3′UTR, increases its expression, and enhances cellular cholesterol efflux in human macrophages and hepatic cell lines. Although the lack of conservation of HuR binding sites in the murine Abca1 3′UTR limited our in vivo studies, we found that HuR was highly expressed in human atherosclerotic plaques. Importantly, we found a strong correlation between the expression of HuR and ABCA1, which became more evident in macrophage-enriched areas of the lesions. This observation agrees with the hypothesis that HuR regulates cholesterol efflux through the transcriptional regulation of ABCA1 in atherosclerotic plaque macrophages. To our knowledge, this is the first evidence of a direct influence of an RBP on ABCA1 expression.
In addition to the many biological functions in which HuR is implicated, such as cell differentiation, cell growth, DNA repair, cell activation, and inflammatory response, growing evidence suggests that HuR, as well as other RBPs, can also impact upon metabolic homeostasis by affecting mRNAs involved in glucose and lipid metabolism (34). In this regard, RBPs have been shown to participate in the formation of lipid droplets during adipogenesis (35), as well as insulin signaling and secretion (36). The mechanisms that control HuR function and its influence on target mRNAs have been investigated extensively. Most of these studies have shown a role for HuR in the stabilization of mRNAs. However, HuR and other Hu/ELAV members have also been found to promote the translation of a growing number of target transcripts. In addition to enhancing ABCA1 translation, as described here, HuR was shown to stimulate the translation of mRNAs such as those that encode ProTα, p53, and Glut-1 (26, 37, 38). The mechanism by which HuR enhances translation of target mRNAs is unclear but may be linked to a mechanism of recruitment of these mRNAs to translationally active polysomes or HuR-elicited exclusion of translational repressors like miRNAs or other RBPs. In the present investigation, no differences in ABCA1 mRNA abundance or stability were observed, and only an HuR-mediated promotion of ABCA1 translation was observed.
Given HuR’s influence on the expression of ABCA1, we explored whether the cellular cholesterol content, known to regulate the expression of this transporter, could also determine cellular HuR levels. Interestingly, we found that cholesterol depletion using simvastatin, an inhibitor of HMG-CoA reductase that reduces cholesterol levels and ABCA1 expression, induced downregulation of HuR mRNA and protein expression in macrophages and hepatic cells. Although the transcriptional regulation of HuR is poorly understood, several pieces of evidence indicate that HuR mRNA and protein are subject to multiple posttranscriptional and posttranslational regulatory mechanisms (mainly involving HuR phosphorylation) that may affect HuR binding to mRNAs and potentially influence its function (39). The observation that simvastatin reduces HuR expression, as well as its translocation from the nucleus, suggests that HuR and ABCA1 levels and function are coregulated. In agreement with this idea, our RIP experiments showed that the interaction between HuR and ABCA1 mRNA is also impaired under cholesterol-depletion conditions and, in contrast, is induced after loading cells with cholesterol. This enhanced interaction is not due to an increase of total protein levels of HuR, but it might be due to its accumulation in the cytosol (by the induction of the translocation from the nucleus) found in macrophages after Ac-LDL treatment. Similarly, the translocation of HuR has also been described after oxidative stress or other cellular stimuli, and it is considered a general phenomenon linked to HuR’s ability to stabilize and modulate the translation of target mRNAs (40). The results presented in this study suggest that cholesterol loading increases ABCA1 by activating the transcription of the gene and by enhancing its translation via HuR. Thus, a plausible scenario is one in which cholesterol loading promotes, in a coordinate manner, transcriptional and posttranscriptional mechanisms to ensure the precise and timely regulation of ABCA1. This is particularly the case of the negative feedback loop that represents the posttranscriptional regulation of ABCA1 by miR-33. Further analysis is needed to determine the mechanism by which cholesterol modulates the expression, subcellular localization, and function of HuR.
To add to the complexity of HuR posttranscriptional actions, studies over the past 5 years have revealed that the influence of HuR on many bound transcripts is dependent on HuR’s interplay with some miRNAs, which are associated with the same mRNAs. This fact could be especially interesting in the case of the ABCA1 mRNA, which has been shown to be highly susceptible to posttranscriptional regulation by several miRNAs and now by the RBP HuR. MiR-33, miR-758, miR-106b, and miR-144 have been shown to regulate ABCA1 expression in hepatocytes, macrophages, and neuronal cells (41). The differential contribution of these miRNAs to the posttranscriptional regulation of ABCA1 expression may be determined by their abundance under different physiological conditions but also by the competition or cooperation among themselves, with other regulatory RNAs like competing endogenous RNAs, and/or with other RBPs like HuR. Importantly, interference between HuR and miRNAs has been proposed to be one of the possible mechanisms that could explain the increased translational activity of HuR on mRNA targets, as demonstrated here for ABCA1. Future studies should examine directly whether HuR functionally competes or cooperates with these miRNAs.
In conclusion, this study shows that the RBP HuR binds and regulates the expression and function of ABCA1 in human hepatic cells and macrophages. This novel posttrancriptional regulation of ABCA1 by HuR, together with miRNAs, may be part of the complex mechanisms developed by the cell to ensure the tight regulation of cholesterol homeostasis.
Supplementary Material
Acknowledgments
The authors thank Jennifer L. Martindale for help with the polysome analysis.
Footnotes
Abbreviations:
- Ac-LDL
- acetylated LDL
- AEBSF
- 4-benzenesulfonyl fluoride hydrochloride
- CF
- cytosolic fraction
- HSP90
- heat shock protein 90
- HuR
- human antigen R
- iCLIP
- individual-nucleotide resolution cross-linking and immunoprecipitation
- IP
- immunoprecipitation
- LDLR
- LDL receptor
- LPDS
- lipoprotein-deficient serum
- LXR
- liver X receptor
- miRNA
- micro-RNA
- NF
- nuclear fraction
- qPCR
- quantitative real-time PCR
- RBP
- RNA binding protein
- RIP
- ribonucleoprotein immunoprecipitation
- RLuc
- Renilla luciferase
- TAP
- tandem affinity purification
- T090
- T0901317
- UTR
- untranslated region
- WCE
- whole-cell extract
This work was supported by National Institutes of Health Grants R01HL107953 and R01HL106063 (C.F-H.) and 1F31AG043318-01 (L.G.), and American Heart Association Grant 12POST9780016 (C.M.R.). C. S. Lin was supported by Tri-Service General Hospital (TSGH-C103-027). K. Abdelmohsen, J-H. Yoon, and M. Gorospe were supported by the National Institute on Aging-Intramural Research Program, the National Institutes of Health.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of three figures.
REFERENCES
- 1.Liu J. P., Tang Y., Zhou S., Toh B. H., McLean C., Li H. 2010. Cholesterol involvement in the pathogenesis of neurodegenerative diseases. Mol. Cell. Neurosci. 43: 33–42. [DOI] [PubMed] [Google Scholar]
- 2.Del Prato S., Enzi G., Vigili de Kreutzenberg S., Lisato G., Riccio A., Maifreni L., Iori E., Zurlo F., Sergi G., Tiengo A. 1990. Insulin regulation of glucose and lipid metabolism in massive obesity. Diabetologia. 33: 228–236. [DOI] [PubMed] [Google Scholar]
- 3.Gelissen I. C., Harris M., Rye K. A., Quinn C., Brown A. J., Kockx M., Cartland S., Packianathan M., Kritharides L., Jessup W. 2006. ABCA1 and ABCG1 synergize to mediate cholesterol export to apoA-I. Arterioscler. Thromb. Vasc. Biol. 26: 534–540. [DOI] [PubMed] [Google Scholar]
- 4.Tall A. R. 2008. Cholesterol efflux pathways and other potential mechanisms involved in the athero-protective effect of high density lipoproteins. J. Intern. Med. 263: 256–273. [DOI] [PubMed] [Google Scholar]
- 5.Wang X., Collins H. L., Ranalletta M., Fuki I. V., Billheimer J. T., Rothblat G. H., Tall A. R., Rader D. J. 2007. Macrophage ABCA1 and ABCG1, but not SR-BI, promote macrophage reverse cholesterol transport in vivo. J. Clin. Invest. 117: 2216–2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Venkateswaran A., Laffitte B. A., Joseph S. B., Mak P. A., Wilpitz D. C., Edwards P. A., Tontonoz P. 2000. Control of cellular cholesterol efflux by the nuclear oxysterol receptor LXR alpha. Proc. Natl. Acad. Sci. USA. 97: 12097–12102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ambros V. 2004. The functions of animal microRNAs. Nature. 431: 350–355. [DOI] [PubMed] [Google Scholar]
- 8.Bartel D. P. 2009. MicroRNAs: target recognition and regulatory functions. Cell. 136: 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Filipowicz W., Bhattacharyya S. N., Sonenberg N. 2008. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 9: 102–114. [DOI] [PubMed] [Google Scholar]
- 10.Rayner K. J., Suarez Y., Davalos A., Parathath S., Fitzgerald M. L., Tamehiro N., Fisher E. A., Moore K. J., Fernandez-Hernando C. 2010. MiR-33 contributes to the regulation of cholesterol homeostasis. Science. 328: 1570–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marquart T. J., Allen R. M., Ory D. S., Baldan A. 2010. miR-33 links SREBP-2 induction to repression of sterol transporters. Proc. Natl. Acad. Sci. USA. 107: 12228–12232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Najafi-Shoushtari S. H., Kristo F., Li Y., Shioda T., Cohen D. E., Gerszten R. E., Naar A. M. 2010. MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science. 328: 1566–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramirez C. M., Davalos A., Goedeke L., Salerno A. G., Warrier N., Cirera-Salinas D., Suarez Y., Fernandez-Hernando C. 2011. MicroRNA-758 regulates cholesterol efflux through posttranscriptional repression of ATP-binding cassette transporter A1. Arterioscler. Thromb. Vasc. Biol. 31: 2707–2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim J., Yoon H., Ramirez C. M., Lee S. M., Hoe H. S., Fernandez-Hernando C., Kim J. 2012. MiR-106b impairs cholesterol efflux and increases Abeta levels by repressing ABCA1 expression. Exp. Neurol. 235: 476–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramírez C. M., Rotllan N., Vlassov A. V., Dávalos A., Li M., Goedeke L., Aranda J. F., Cirera-Salinas D., Araldi E., Salerno A., et al. 2013. Control of cholesterol metabolism and plasma high-density lipoprotein levels by microRNA-144. Circ. Res. 112: 1592–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Aguiar Vallim T. Q., Tarling E. J., Kim T., Civelek M., Baldan A., Esau C., Edwards P. A. 2013. MicroRNA-144 regulates hepatic ATP binding cassette transporter A1 and plasma high-density lipoprotein after activation of the nuclear receptor farnesoid X receptor. Circ. Res. 112: 1602–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keene J. D. 2007. RNA regulons: coordination of post-transcriptional events. Nat. Rev. Genet. 8: 533–543. [DOI] [PubMed] [Google Scholar]
- 18.Moore M. J. 2005. From birth to death: the complex lives of eukaryotic mRNAs. Science. 309: 1514–1518. [DOI] [PubMed] [Google Scholar]
- 19.Pullmann R., Jr, Kim H. H., Abdelmohsen K., Lal A., Martindale J. L., Yang X., Gorospe M. 2007. Analysis of turnover and translation regulatory RNA-binding protein expression through binding to cognate mRNAs. Mol. Cell. Biol. 27: 6265–6278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abdelmohsen K., Kuwano Y., Kim H. H., Gorospe M. 2008. Posttranscriptional gene regulation by RNA-binding proteins during oxidative stress: implications for cellular senescence. Biol. Chem. 389: 243–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hinman M. N., Lou H. 2008. Diverse molecular functions of Hu proteins. Cell. Mol. Life Sci. 65: 3168–3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.López de Silanes I., Lal A., Gorospe M. 2005. HuR: post-transcriptional paths to malignancy. RNA Biol. 2: 11–13. [DOI] [PubMed] [Google Scholar]
- 23.Kim H. H., Abdelmohsen K., Lal A., Pullmann R., Jr, Yang X., Galban S., Srikantan S., Martindale J. L., Blethrow J., Shokat K. M., et al. 2008. Nuclear HuR accumulation through phosphorylation by Cdk1. Genes Dev. 22: 1804–1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Srikantan S., Gorospe M. 2012. HuR function in disease. Front. Biosci. (Landmark Ed.) 17: 189–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Uren P. J., Burns S. C., Ruan J., Singh K. K., Smith A. D., Penalva L. O. 2011. Genomic analyses of the RNA-binding protein Hu antigen R (HuR) identify a complex network of target genes and novel characteristics of its binding sites. J. Biol. Chem. 286: 37063–37066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mazan-Mamczarz K., Galban S., Lopez de Silanes I., Martindale J. L., Atasoy U., Keene J. D., Gorospe M. 2003. RNA-binding protein HuR enhances p53 translation in response to ultraviolet light irradiation. Proc. Natl. Acad. Sci. USA. 100: 8354–8359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tenenbaum S. A., Lager P. J., Carson C. C., Keene J. D. 2002. Ribonomics: identifying mRNA subsets in mRNP complexes using antibodies to RNA-binding proteins and genomic arrays. Methods. 26: 191–198. [DOI] [PubMed] [Google Scholar]
- 28.Vo D. T., Abdelmohsen K., Martindale J. L., Qiao M., Tominaga K., Burton T. L., Gelfond J. A., Brenner A. J., Patel V., Trageser D., et al. 2012. The oncogenic RNA-binding protein Musashi1 is regulated by HuR via mRNA translation and stability in glioblastoma cells. Mol. Cancer Res. 10: 143–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martín-Ventura J. L., Ortego M., Esbrit P., Hernández-Presa M. A., Ortega L., Egido J. 2003. Possible role of parathyroid hormone-related protein as a proinflammatory cytokine in atherosclerosis. Stroke. 34: 1783–1789. [DOI] [PubMed] [Google Scholar]
- 30.López de Silanes I., Zhan M., Lal A., Yang X., Gorospe M. 2004. Identification of a target RNA motif for RNA-binding protein HuR. Proc. Natl. Acad. Sci. USA. 101: 2987–2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Castelli W. P., Doyle J. T., Gordon T., Hames C. G., Hjortland M. C., Hulley S. B., Kagan A., Zukel W. J. 1977. HDL cholesterol and other lipids in coronary heart disease. The cooperative lipoprotein phenotyping study. Circulation. 55: 767–772. [DOI] [PubMed] [Google Scholar]
- 32.Wilson P. W. 1990. High-density lipoprotein, low-density lipoprotein and coronary artery disease. Am. J. Cardiol. 66: 7A–10A. [DOI] [PubMed] [Google Scholar]
- 33.Plump A. S., Scott C. J., Breslow J. L. 1994. Human apolipoprotein A-I gene expression increases high density lipoprotein and suppresses atherosclerosis in the apolipoprotein E-deficient mouse. Proc. Natl. Acad. Sci. USA. 91: 9607–9611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim W., Kyung Lee E. 2012. Post-transcriptional regulation in metabolic diseases. RNA Biol. 9: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kawagishi H., Wakoh T., Uno H., Maruyama M., Moriya A., Morikawa S., Okano H., Sherr C. J., Takagi M., Sugimoto M. 2008. Hzf regulates adipogenesis through translational control of C/EBPalpha. EMBO J. 27: 1481–1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee E. K., Kim W., Tominaga K., Martindale J. L., Yang X., Subaran S. S., Carlson O. D., Mercken E. M., Kulkarni R. N., Akamatsu W., et al. 2012. RNA-binding protein HuD controls insulin translation. Mol. Cell. 45: 826–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lal A., Kawai T., Yang X., Mazan-Mamczarz K., Gorospe M. 2005. Antiapoptotic function of RNA-binding protein HuR effected through prothymosin alpha. EMBO J. 24: 1852–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gantt K. R., Cherry J., Richardson M., Karschner V., Atasoy U., Pekala P. H. 2006. The regulation of glucose transporter (GLUT1) expression by the RNA binding protein HuR. J. Cell. Biochem. 99: 565–574. [DOI] [PubMed] [Google Scholar]
- 39.Srikantan S., Tominaga K., Gorospe M. 2012. Functional interplay between RNA-binding protein HuR and microRNAs. Curr. Protein Pept. Sci. 13: 372–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gorospe M. 2003. HuR in the mammalian genotoxic response: post-transcriptional multitasking. Cell Cycle. 2: 412–414. [PubMed] [Google Scholar]
- 41.Aranda J. F., Madrigal-Matute J., Rotllan N., Fernandez-Hernando C. 2013. MicroRNA modulation of lipid metabolism and oxidative stress in cardiometabolic diseases. Free Radic. Biol. Med. 64: 31–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.