Abstract
Lysosomes are acidic compartments in mammalian cells that are primarily responsible for the breakdown of endocytic and autophagic substrates such as membranes, proteins, and lipids into their basic building blocks. Lysosomal storage diseases (LSDs) are a group of metabolic disorders caused by genetic mutations in lysosomal hydrolases required for catabolic degradation, mutations in lysosomal membrane proteins important for catabolite export or membrane trafficking, or mutations in nonlysosomal proteins indirectly affecting these lysosomal functions. A hallmark feature of LSDs is the primary and secondary excessive accumulation of undigested lipids in the lysosome, which causes lysosomal dysfunction and cell death, and subsequently pathological symptoms in various tissues and organs. There are more than 60 types of LSDs, but an effective therapeutic strategy is still lacking for most of them. Several recent in vitro and in vivo studies suggest that induction of lysosomal exocytosis could effectively reduce the accumulation of the storage materials. Meanwhile, the molecular machinery and regulatory mechanisms for lysosomal exocytosis are beginning to be revealed. In this paper, we first discuss these recent developments with the focus on the functional interactions between lipid storage and lysosomal exocytosis. We then discuss whether lysosomal exocytosis can be manipulated to correct lysosomal and cellular dysfunction caused by excessive lipid storage, providing a potentially general therapeutic approach for LSDs.
Keywords: calcium, transient receptor potential mucolipin 1, transcription factor EB, lysosomal storage disease
LYSOSOMES AS THE DEGRADATION AND NUTRIENT-SENSING CENTER
Lysosomes are acidic compartments in mammalian cells that are primarily responsible for the breakdown of various biomaterials and macromolecules, such as membranes, proteins, polysaccharides, and complex lipids into their respective smaller building-block molecules: amino acids, monosaccharides, free fatty acids, or nucleotides (1–4). Hence, since their discovery by Christian De Duve in 1955, lysosomes have been viewed as the cell’s degradation center (3, 5). Lysosomes, several hundreds of them in each mammalian cell, are heterogeneous in size (100–1,000 nm) and morphology, and collectively constitute ∼5% of the cell volume (6). Lysosomes are filled with more than 50 different types of hydrolases: sulphatases, phosphatases, lipases, proteases, carbohydrases, and glycosidases, which effectively catabolize proteins and complex lipids into their building-block molecules (1, 4). The functions of most lysosomal hydrolytic enzymes require an acidic lumen, which is established and maintained by the vacuolar-ATPase (V-ATPase)... proton pumps located on the perimeter limited membrane (6). To protect the perimeter membrane and its resident membrane proteins from degradation, the inner leaflet of the lysosomal membrane is coated with a polysaccharide layer called the glycocalyx (7).
The biomaterials destined for degradation are delivered to lysosomes through endocytic/phagocytic and autophagic pathways (Fig. 1). Endocytosis or phagocytosis of extracellular particles or plasma membrane proteins begins with the invagination of the plasma membrane to form endocytic vesicles, which then undergo a series of maturation processes to first become early endosomes, and then late endosomes [excellently reviewed in (5)] (Fig. 1). In the late endosomes, the destined-to-be-degraded cargo is sorted into the intraluminal vesicles via the endosomal sorting complexes required for transport system (5). Intraluminal vesicle-containing late endosomes are thus called multi-vesicular bodies (MVBs). Mature late endosomes fuse with lysosomes to become endolysosome hybrids to mediate the bulk of the degradation (Fig. 1). In a parallel pathway, worn-out intracellular organelles and misfolded proteins are delivered to lysosomes through autophagy (Fig. 1). Double membrane autophagosomes fuse directly with lysosomes to form autolysosomes (8, 9), in which the autophagic substrates are broken down (Fig. 1). Although lysosomes receive cargo from both autophagic and endocytic pathways, there are no clear boundaries between them, as autophagosomes may merge directly with late endosomes, or even early endosomes (10). The products of lysosomal degradation, i.e., lysosome-derived catabolites, are transported out of lysosomes through specific exporters on the perimeter membrane, or through vesicular membrane trafficking (11, 12) (Fig. 1). Once reaching the cytoplasm or the Golgi apparatus, these catabolites are either further degraded to provide energy or reutilized by the biosynthetic pathways to produce complex molecules. Indeed, the biosynthesis of many complex lipids, such as the glycosphingolipids, mainly utilizes the lysosomal catabolites as its building blocks (4, 13). Therefore, the lysosome is the central platform for cellular recycling and energy metabolism.
Fig. 1.
Lysosomal membrane trafficking pathways. Lysosomes receive inputs from both endocytic and autophagic pathways. Endocytosed cargo including membranes, complex lipids, membrane proteins, and polysaccharides enter the endocytic pathway first to early endosomes and then intraluminal vesicle-containing late endosomes (MVBs). Late endosomes then fuse with lysosomes to form endolysosome hybrids. Damaged intracellular organelles enter the autophagic pathway in autophagosomes, which then fuse with lysosomes to form autolysosomes. Endocytosed and autophagocytosed cargo is degraded in endolysosomes and autolysosomes through lysosomal hydrolases. The resulting products are building-block precursor molecules for complex macromolecules. These catabolites are transported out of the lysosomes via specific exporters or vesicular trafficking. Insoluble catabolites, such as lipids, can be transported to the Golgi apparatus for reutilization via the retrograde trafficking pathway. Alternatively, degradation products can also be released into the extracellular medium via lysosomal exocytosis. Lysosomal ion homeostasis and membrane trafficking are regulated by lysosomal ion channels and transporters on the perimeter membrane.
Because the catabolic and anabolic pathways cross-talk with each other to regulate metabolic homeostasis, the endocytic and autophagic pathways that supply the catabolic cargos are highly regulated by cellular signaling and nutrient status (14). For example, autophagy is triggered by nutrient starvation but terminated upon completion of lysosomal degradation (15). Indeed, lysosomes contain the lysosomal nutrient sensing system (LYNUS) that consists of V-ATPase, Rag GTPases, and the mammalian target of rapamycin (mTOR) complex (14). Equipped with both degradation and signaling functions, lysosomes are now known to participate in a wide spectrum of basic cellular and physiological functions, including secretion, plasma membrane repair, cell growth, and cell death (3). Hence, lysosomes should not be simply viewed as a dead-end degradation compartment, but rather as a highly dynamic organelle that integrates multiple metabolic pathways to maintain cellular homeostasis.
LYSOSOMAL LIPID STORAGE DISORDERS
Lipids, such as sphingolipids, glycolipids, phospholipids, and cholesterol, are the essential structural constituents of the plasma membrane and the membranes of intracellular compartments (1, 4). In addition, lipids, especially phospholipids, also serve as signaling molecules (4). While lipid synthesis is initiated in the endoplasmic reticulum (ER) and modified in the Golgi apparatus, lipid degradation occurs mostly in the lysosomes, which are filled with various acid lipases and lipid phosphatases (4). Unlike water soluble substrates, such as proteins, that are readily accessible to lysosomal proteases and peptidases, the digestion of membrane-bound complexes takes place in intraluminal vesicles and requires both lipid-binding activator proteins and anionic lipids such as bis(monoacylglycero)phosphate (1). Complex lipids are usually delivered to lysosomes for degradation as parts of the autophagocytosed or endocytosed membranes (3). While the perimeter membrane is protected by glycocalyx, intraluminal vesicles in the MVBs are the main sites for lipid extraction and digestion (Fig. 1) (1). The membrane-stabilizing cholesterol is first sorted and extracted out of intraluminal vesicles via the luminal cholesterol-binding Niemann-Pick type C (NPC)2 proteins and then exported out of lysosomes via the NPC1 proteins on the limited membrane (16). Auxiliary lipid-binding activator proteins solubilize and present the complex lipids to the hydrolases for degradation (1). Complex lipids such as sphingoglycolipids require both lipases/phosphatases and glycosidases/carbohydrases for degradation (1). Because there are multiple steps in lipid digestion, lacking any of these lipid or carbohydrate hydrolases would result in accumulation of specific undigested precursor lipids in the lysosome (4).
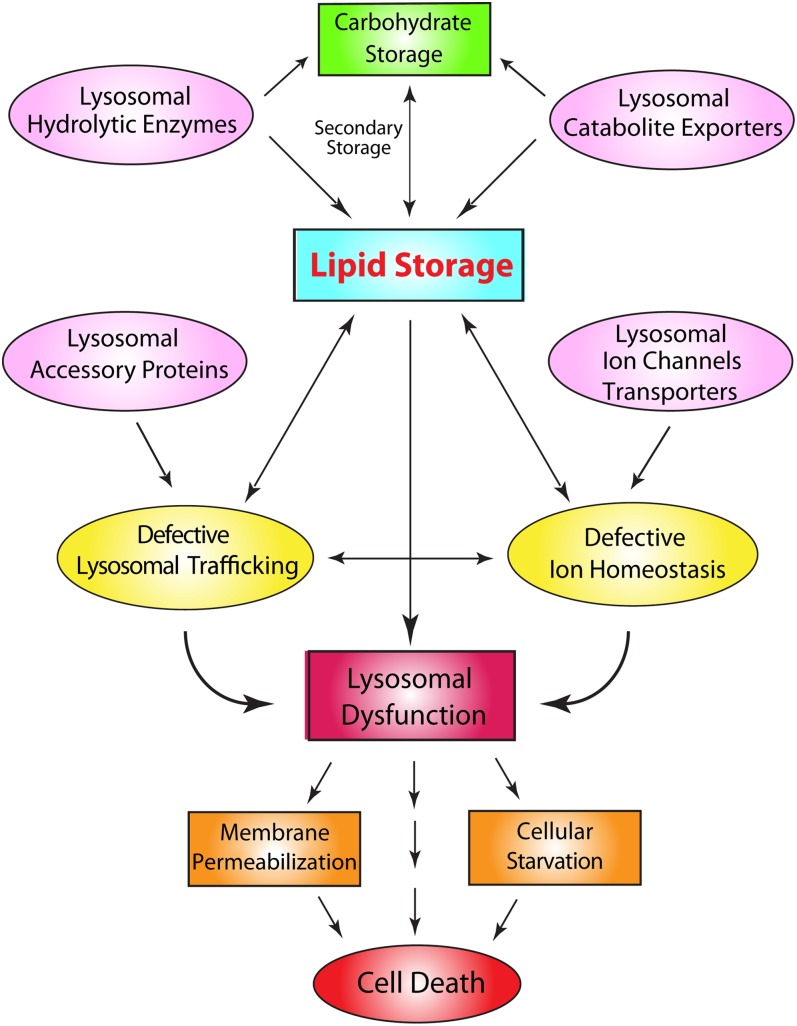
Accumulation of undigested lipids or other biomaterials in the lysosome would lead to a group of metabolic disorders collectively called lysosomal storage diseases (LSDs) (17). LSDs are traditionally classified by the nature of undigested materials within the lysosome. Hence, lipidoses are the most common LSDs. However, undigested lipids may also accumulate due to secondary mechanisms, for example, secondary to carbohydrate/protein accumulation or membrane trafficking (17, 18). Therefore, a more useful classification is based on the specific mutation for each LSD. Most LSDs are caused by mutations in the lysosomal hydrolases (4). However, defective membrane trafficking and catabolite export could cause a “traffic jam”, resulting in secondary lipid storage in the lysosome (Fig. 2) (17). In addition, a subset LSDs are caused by mutations in nonlysosomal proteins indirectly affecting degradation and transport (4). Furthermore, primary accumulation of undigested insoluble lipids may also cause a traffic jam, slowing down membrane trafficking and sorting, ultimately affecting the delivery of lysosomal hydrolases (3, 17). In addition, insoluble lipids may also precipitate in the lysosomes to reduce the activity of hydrolytic enzymes. Subsequently, secondary storage of ubiquitinated proteins, carbohydrates, or other lipids may occur. Hence, most LSDs manifest in the heterogeneity of storage lipids in the lysosome.
Fig. 2.
Molecular pathogenic mechanisms of LSDs. LSDs are characterized by the progressive build-up of undigested materials, such as lipids, carbohydrates, and proteins within the lysosomes. Mutations in lipases, lipid phosphatases, and carbohydrases lead to the accumulation of undigested lipids/carbohydrates/proteins in the lysosome. Likewise, mutations in the catabolite exporters also lead to the accumulation of lipids/carbohydrates/proteins in the lysosome. Mutations in lysosomal accessory proteins lead to lysosomal trafficking defects. Mutations in lysosomal ion channels and transporters lead to defective ion homeostasis, which in turn affects membrane trafficking to cause lipid accumulation. Lipid storage causes a traffic jam and secondary accumulation, resulting in lysosomal dysfunction, membrane permeabilization, and cellular starvation. Lysosomal membrane permeabilization may cause leakage of lysosomal enzymes into the cytoplasm to cause cell death.
Progressive accumulation of undigested lipids and protein aggregates, primarily or secondarily, leads to build-up of enlarged (>500 nm) but dysfunctional lysosomes. These enlarged “lysosomes” are presumably endolysosomes and autolysosomes, which under normal conditions are used to regenerate new lysosomes via the process of lysosome reformation, upon completion of lysosomal degradation (15). Hence, LSD is a state of endocytic and autophagic “block” or “arrest” (8). Subsequently, although the total number of lysosomes is not reduced in LSDs, the overall lysosomal function within a cell is compromised, and this can lead to severe cellular consequences. First, accumulation of undigested materials in the lysosomes could result in a deficiency of building block precursors for the biosynthetic pathways. Therefore, LSD cells must bear the consequence of cellular starvation (4) (Fig. 2). Second, accumulation of various membrane-bound lipids may affect the properties and integrity of the perimeter membrane (20). Third, lipid storage may alter the functionality of lysosomal membrane proteins, such as V-ATPase, lysosomal ion channels, or catabolite exporters, affecting the physiology and ionic composition of lysosomes. Altered heavy metal ion homeostasis, in turn, may increase oxidative stress to cause lipid peroxidation, affecting membrane integrity (21).
In an attempt to compensate for lysosomal dysfunction and reduced power in degradation, hence an increase of lysosomal substrates, most LSD cells exhibit an increase in the basal autophagy and in the expression levels of housekeeping lysosomal proteins such as Lamp-1 (8, 22). These compensatory changes may allow LSD cells to survive under normal nonstressed conditions. Under stressed conditions, however, altered luminal ion homeostasis and membrane integrity may cause lysosomal membrane permeabilization, leading to the leakage of lysosomal enzymes to the cytoplasm and triggering cell death (23). An increase in cell death would result in damage in tissues and organs with large amounts of postmitotic cells, including the brain, liver, eye, muscle, and spleen (18). Thus, the most common manifestations in LSDs are neurodegeneration, mental retardation, and motor disabilities (17).
Several therapeutic approaches have been developed for LSDs. These include substrate reduction therapy, bone marrow transplantation, gene therapy, and enzyme replacement therapy (ERT) (24). ERT and substrate reduction are the most common therapies, and are currently used effectively for the treatment of Gaucher and Fabry diseases (24, 25). In addition, chemical chaperones have been proven helpful for a subset of Gaucher, Fabry, and Tay-Sachs diseases that is caused by enzyme misfolding in the ER, (26). For most LSDs, however, an effective therapeutic approach is still lacking. For the currently available approaches, there are various intrinsic limitations, especially the problem of low efficacy. For instance, while the brain is the most commonly affected tissue in LSDs, it is very difficult for ERT to deliver enzymes across the blood-brain barrier. Most importantly, all the current available approaches are disease specific.
To develop an approach common for most LSDs, it is necessary to understand how common cellular pathways are affected by lipid storage. Several recent studies reveal that lysosomal exocytosis, one of the two most important lysosomal trafficking pathways in the “outward” or “anterograde” direction (Fig. 1), are defective in multiple LSDs (27–29). Most intriguingly, induction of lysosomal exocytosis was found to facilitate the clearance of stored materials regardless of the nature and cause of the storage (14, 27, 29, 30). In this review we will first describe the core machinery of lysosomal exocytosis, and then discuss how it can be regulated for the purpose of cellular clearance as a therapeutic target for LSDs.
LYSOSOMAL EXOCYTOSIS
Unlike the “conventional secretion” mediated by the ER-Golgi-secretory vesicle pathway, lysosomal exocytosis belongs to the poorly understood “unconventional secretion”, which also includes the fusion of MVBs (exosome release), autophagosomes, autolysosomes, and early endosomes to the plasma membrane (31). Exocytosis of secretory lysosomes (lysosome-related organelles) has been long-known to be present in specialized cell types such as hematopoietic cells (32). The regulated exocytosis of conventional lysosomes was first observed in CHO and NRK fibroblasts, but is now known to be present in all cell types (33). Upon stimulation, lysosomes translocate from the perinuclear and cytosolic regions to the plasma membrane along the microtubules (33–36). After docking, lysosomes fuse directly with the plasma membrane to release the lysosomal content into the extracellular space (Fig. 3A) (33). Lysosomal exocytosis represents the major output pathway of the lysosome and is important for a variety of cellular processes, including plasma membrane repair (33), secretion and transmitter release (37), neurite outgrowth (38), and particle uptake in macrophages (28, 39).
Fig. 3.
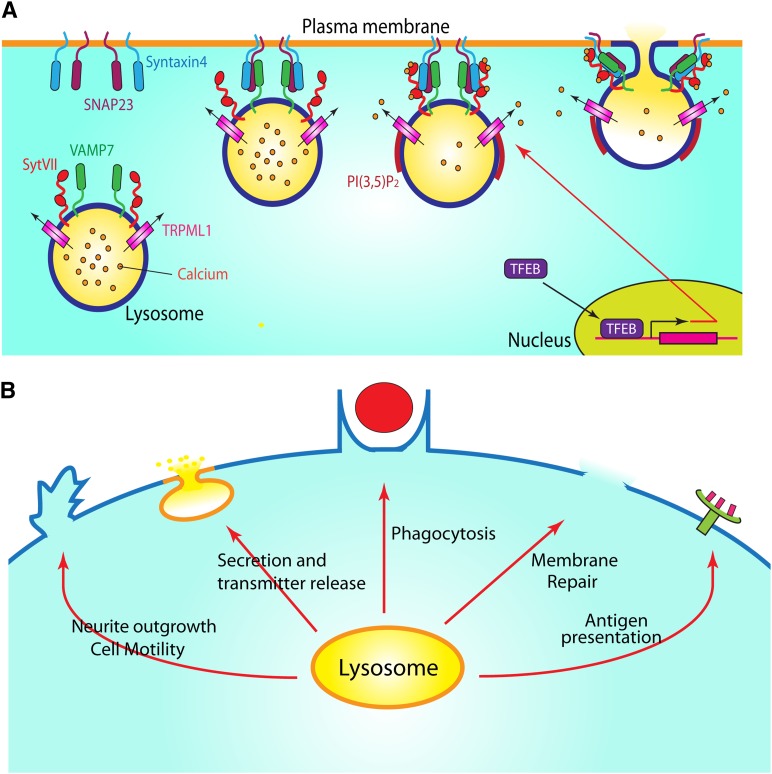
Measuring Ca2+-dependent lysosomal exocytosis. A: Lysosomal exocytosis is a Ca2+-dependent process. Early endosomes are generated from nascent endocytic vesicles via endocytosis. Autophagosomes are generated via autophagy. Upon a series of sorting and maturation processes, autophagosomes and endosomes fuse with lysosomes to become endolysosomes and autolysosomes. Lysosomes are high Ca2+ (∼0.5 mM) compartments. Lysosomes fuse with the plasma membrane in response to an increase in extralysosomal [Ca2+]i (>1 μM). B: Methods for the detection of lysosomal exocytosis. Method 1 (lysosomal enzyme release) is used to measure the release of lysosomal contents, such as hydrolytic enzymes, into the cell culture medium. Method 2 (Lamp1 surface staining) is to monitor the insertion of lysosomal membrane proteins into the plasma membrane by using a monoclonal antibody against a luminal epitope of lysosomal membrane proteins, such as Lamp1, in nonpermeabilized cells. Confocal images in the right panel show that ML-SA1 treatment resulted in the localization of Lamp1 (red) on the plasma membrane in nonpermeabilized WT, but not TRPML1 KO macrophages. The total amount of Lamp1 (green) proteins is detected using the same antibody in permeabilized cells [modified from (28) with permission]. Method 3 (electrophysiology-based exocytosis assay) is used to detect the plasma membrane insertion of lysosomal ion channels as a result of lysosomal exocytosis.
Measuring lysosomal exocytosis
Several assays have been developed to detect lysosomal exocytosis in vitro. The most commonly used method is to measure the activity of lysosomal hydrolytic enzymes, such as β-hexosaminidase and acid phosphatase, which are released into the extracellular medium during exocytosis (40). This lysosomal enzyme release assay measures accumulatively the extent of lysosomal exocytosis over a period of time (Fig. 3B). By comparing with the activity of the cell-associated enzymes, this assay can be used to estimate the size of the releasable pool for specific stimuli and conditions in a given time.
Another way to measure lysosomal exocytosis is to monitor the insertion of lysosomal membrane proteins into the surface of the plasma membrane (28, 33, 40). After exocytosis, lysosomal membrane proteins, such as the most abundant Lamp1 proteins, are present on the plasma membrane. Hence, measuring the surface expression of Lamp1 using a monoclonal antibody against a luminal epitope of Lamp1 can provide readout for lysosomal exocytosis (Fig. 3B). To avoid the contamination from the lysosomal pool of Lamp1 proteins, the experiment should be performed in nonpermeabilized cells (28, 33, 40). This Lamp1 surface staining assay detects lysosomal exocytosis, either acutely or accumulatively. The approach is also applicable to other lysosome membrane proteins. Even if antibodies against luminal epitopes are not available, one can engineer a FLAG or HA tag to the luminal side of the protein for detection. Given the heterogeneity in the pools of lysosomes, this assay may help determine whether specific lysosomal resident proteins are sorted “in” or “out” before or during lysosomal docking.
An electrophysiology-based “exocytosis assay” has been recently developed to provide a temporal resolution higher than the above-mentioned exocytosis/secretion methods (Fig. 3B). In this assay, whole-cell currents of lysosomal ion channels, for example, transient receptor potential mucolipin 1 (TRPML1), are used to “detect” the plasma membrane insertion of lysosomal channels during exocytosis (28). Most lysosomal channels, unlike the plasma membrane ones, are presumed to have short half-lives at the plasma membrane. They may undergo rapid endocytosis to retrieve from a nonnative environment, i.e., the plasma membrane. Hence, to maximize detection, it might be necessary to block endocytosis (28). Capacitance measurement has been another well-established electrophysiology-based exocytosis assay (41). However, such an assay requires a large amount of exocytosis, and does not distinguish lysosomal exocytosis from other forms of exocytosis in conventional and unconventional secretion.
Exocytosis of lysosomes can also be measured using total internal reflection fluorescent microscopy (TIRF), if a fluorescence tag is engineered to a lysosomal membrane protein. TIRF only detects sub-plasma membrane events, thus providing a very precise and quantitative approach to capture single exocytosis events (42). As lysosomes are highly acidified compartments, lysosome-targeted pH-sensitive probes, such as pHluorin, could be combined with TIRF to monitor lysosomal exocytosis based on the fluorescence changes (42).
The molecular machinery and regulators of lysosomal exocytosis
Compared with synaptic vesicle exocytosis, a process sharing many similarities with lysosomal exocytosis (43), the molecular mechanisms underlying lysosomal exocytosis are much less understood (40, 43). Both processes involve several prefusion steps in tethering/docking and several postdocking steps in membrane fusion. However, distinct sets of exocytosis machinery are employed, which include soluble N-ethylmaleimide-sensitive factor-attachment protein receptors (SNAREs) that bring the two membranes closer to each other. Importantly, both fusion events are regulated by Ca2+ and the same family of Ca2+ sensors. As common mechanisms and regulation may exist for different forms of unconventional secretion, the molecular mechanisms described below may also apply to the fusion of endocytic or autophagic vesicles with the plasma membrane. In many LSDs, the lysosomes undergoing exocytosis are indeed mostly autolysosomes (29).
Cytoskeleton and motor proteins.
Lysosomes undergo both short-range actin-based and long-range microtubule-based movements (3, 5). Under resting conditions, lysosomes are mostly localized in cytosol and the perinuclear microtubule-organizing center (29). Upon stimulation, lysosome mobility and the numbers of lysosomes that are localized sub-plasma membrane (lysosomal docking to plasma membrane) are increased (29). While lysosome mobility is increased when the microtubule-dependent kinesin motor proteins are upregulated (44), lysosomal docking to the plasma membrane is also dependent on the microtubule (27). In MCF-7 cells, KIF5B is the only microtubule-dependent kinesin motor protein associated with lysosomes (45). Increasing lysosome mobility may increase lysosomal docking and exocytosis. For example, in NRK cells, both lysosomal distribution and lysosomal exocytosis are regulated by cAMP (46), which may affect the cytosolic pH to influence microtubule-dependent lysosomal movement in cells (47).
Rabs and tethering factors.
The small GTPase Rab7 protein and its effectors, such as HOPS complex proteins, are known to tether late endosomes and lysosomes for homotypic and heterotypic membrane fusion (3). Whether these proteins perform a similar task for lysosome-plasma membrane fusion remains to be investigated. Rab7 effectors may also affect lysosomal exocytosis by regulating motor proteins in lysosome mobility (44).
SNAREs.
Similar to the exocytosis of synaptic vesicles and endosome-endosome membrane fusion, the fusion of lysosomes with the plasma membrane is initiated by the formation of the SNARE complexes (48). SNARE proteins are characterized by the existence of coiled-coil homology domains (48). During lysosomal exocytosis, vesicle-associated membrane protein 7 (VAMP7), on the surface of lysosomes, forms a trans-SNARE complex with syntaxin-4 and synaptosome-associated protein of 23 kDa (SNAP23) on the plasma membrane (49) (Fig. 4A). The formation of the trans-SNARE complex pulls the lysosomes and plasma membrane toward each other and docks the lysosomes on the cytoplasmic side of the plasma membrane (Fig. 4A). Lysosomal exocytosis, similar to late endosome-lysosome fusion (3), is blocked by the expression of dominant-negative VAMP7 constructs in cells (39).
Fig. 4.
The molecular machinery and biological functions of lysosomal exocytosis. A: The core machinery of lysosomal exocytosis. Upon stimulation, lysosomes are tethered/docked to the plasma membrane. The subsequent formation of SNARE complexes requires the interaction of lysosomal SNARE protein, VAMP7, with SNAP23, and syntaxin 4 on the cytoplasmic side of the plasma membrane. Upon preassembly of the SNARE complex, lysosomes are pulled closer to the plasma membrane at the exocytosis site. Localized lysosomal Ca2+ release, which may be mediated by PI(3,5)P2 activation of TRPML1, activates Ca2+ sensor Syt VII in the lysosome to promote the fusion of lysosomes with the plasma membrane. Lysosomal docking and fusion is regulated by TFEB, a transcription factor for lysosome biogenesis and autophagy. B: Lysosomal exocytosis is involved in various cellular processes: neurite outgrowth, secretion, transmitter release, phagocytosis, and membrane repair.
Ca2+.
After lysosomes are docked to the plasma membrane by the formation of the VAMP7- syntaxin-4-SNAP23 trans-SNARE complex, final fusion is triggered by a localized rise in intracellular Ca2+ ([Ca2+]i) levels (33, 40). In response to [Ca2+]i increase, artificially induced by Ca2+ ionophores or membrane disruptions, lysosomal enzyme release is increased and lysosomes are observed to fuse directly with the plasma membrane (33, 40). The [Ca2+]i required for lysosomal exocytosis is predicted to be >1 μM (40), which is higher than the 0.5 μM required for late endosome-lysosome fusion in the cell-free vesicle fusion assays (50) (Fig. 3A). During synaptic vesicle exocytosis, a very rapid increase in [Ca2+]i is required to mediate the fast (<1 ms) fusion of synaptic vesicles with the presynaptic membrane (43). The increase in Ca2+ occurs locally near the site of exocytosis, and is mediated by the depolarization-triggered Ca2+ flux through the voltage-gated Ca2+ channels on the presynaptic membrane (43). For lysosomal exocytosis, however, the source of Ca2+ and the identity of the Ca2+ channel(s) have remained elusive until recently.
The Ca2+ required for late endosome-lysosome and lysosome-plasma membrane fusion has been suggested to come from the lysosomal lumen. Like the ER, the main intracellular Ca2+ store with an estimated [Ca2+]lumen of 0.5–2 mM (51), lysosomes are also Ca2+ stores with a free [Ca2+]lumen of 400–600 μM (52, 53). In the cell-free vesicle fusion assays, late endosome-lysosome fusion is inhibited by the Ca2+ chelator 1,2-bis(o-aminophenoxy)ethane-N,N,N’,N’-tetraacetic acid (BAPTA), but not EGTA (3, 50). Although both BAPTA and EGTA have high binding affinity for Ca2+, BAPTA binds to Ca2+ approximately 100 times faster than EGTA does. Therefore, such distinct BAPTA versus EGTA sensitivity suggests that the source of Ca2+ must be very close to the fusion spot (54). In other words, lysosomes themselves most likely provide the Ca2+ required for lysosomal fusion (50, 54). Interestingly, preloading cells with the fast membrane-permeant Ca2+ chelator BAPTA-acetoxymethyl ester (BAPTA-AM) (1–10 μM) dramatically reduces lysosomal exocytosis and particle uptake in macrophages, while the slow Ca2+ chelator EGTA-AM was less effective (28). It is worth noting that high (millimolar range) concentrations of EGTA-AM were known to chelate luminal Ca2+ to block in vitro late-endosome-lysosome fusion (50, 54). Collectively, lysosomes may provide Ca2+ for membrane fusion between lysosomes and other compartments, including late endosomes and plasma membrane.
Ca2+ sensors: synaptotagmin-VII and others.
The synaptotagmin (Syt) family of Ca2+ sensor proteins has 16 members, each containing two Ca2+-binding C2 domains in the cytosolic side (55). During synaptic vesicle-plasma membrane fusion, a localized increase in [Ca2+]i activates vesicle-localized Syt-I to trigger final fusion (43). For lysosomal exocytosis, Syt-VII is likely to be the lysosomal Ca2+ sensor (Fig. 4A). First, Syt-VII is ubiquitously expressed and localized on the lysosome (33, 39). Second, ionomycin-induced lysosomal exocytosis is reduced in cells isolated from Syt-VII KO mice (39), or cells transfected with dominant-negative Syt-VII constructs (28, 33). Because lysosomal exocytosis is not completely abolished in the Syt-VII KO mice, other Syt proteins may also participate in lysosomal exocytosis. In addition, other candidate lysosomal Ca2+ sensors include calmodulin, which has been previously implicated in the late endosome-lysosome fusion (50), and ALG-2, a lysosome-targeted EF hand Ca2+ binding protein (56).
The mechanisms by which Syt-VII promotes lysosomal exocytosis are not clear. Extrapolated from synaptic vesicle studies (43, 55), the Ca2+-bound C2 domain in Syt-VII may increase phosphoinositide binding and association with the preassembled SNARE complexes to further pull the lysosomes to the proximity of the plasma membrane (55). The resulting force may increase membrane curvature to facilitate lipid bilayer mixing (55).
TRPML1 Ca2+ channels.
Although Ca2+ and Ca2+ sensors have been implicated in lysosomal exocytosis, more definitive evidence requires the molecular identification of Ca2+ channels in the lysosome. Transient receptor potentials are a family of Ca2+-permeable nonselective cation channels (57). Among them, members of TRPML subfamily, TRPML1–3 channels, are primarily localized in the endosome and lysosome (58). Human mutations of TRPML1 cause mucolipidosis type IV (ML-IV), a neurodegenerative LSD characterized by retinal degeneration and mental retardation (59–61). At the cellular level, lysosomal trafficking defects are observed in every single cell type (58, 62).
ML-IV or TRPML1-deficient cells are filled with a spectrum of undigested lipids, such as sphingoglycolipids, phospholipids, and cholesterol (63). However, the activities of most hydrolases are relatively normal in TRPML1-deficient cells (58, 62). As the accumulation of lipids in the ML-IV lysosomes could be explained by defective lysosomal trafficking associated with these cells (27), it was hypothesized that TRPML1 is involved in lysosomal membrane fusion/fission by acting as a lysosomal Ca2+ release channel (Fig. 4A) (64).
Several pieces of evidence suggest a role of TRPML1 in Ca2+-dependent lysosomal exocytosis. First, fibroblasts obtained from ML-IV patients exhibited reduced ionomycin-induced lysosomal exocytosis (65). However, as ionomycin induces Ca2+ flux non-physiologically independent of any specific ion channels, the effect of ionomycin on lysosomal exocytosis could be a secondary observation, because ionomycin presumably acts directly on the Ca2+ sensor Syt-VII, thereby bypassing upstream TRPML1 activation. Second, enhanced lysosomal exocytosis was seen in HEK293 cells overexpressing gain-of-function mutant TRPML1 channels (66). Third, TRPML1 knockdown in a macrophage cell line reduced transport of the major histocompatibility complex II (MHC-II) from lysosomal compartments to the plasma membrane (67). Fourth, transcription factor EB (TFEB)-induced lysosomal exocytosis (see below) was almost abolished in ML-IV cells (27). Taken together, these studies suggest that TRPML1 is required for lysosomal exocytosis.
The direct evidence to support a role of TRPML1 in lysosomal exocytosis was recently obtained using TRPML1-specific small molecule activator (ML-SA1) (28). In the primary macrophages, a 15–30 min ML-SA1 treatment induced the secretion of up to 20% of the total cell-associated lysosomal acid phosphatase enzymes (28). In addition, ML-SA1 also induced a significant amount of Lamp1 surface staining in WT macrophages, but not in ML1 KO macrophages (28) (Fig. 3B). Moreover, ML-SA1-induced lysosomal exocytosis was abolished by BAPTA-AM, but not EGTA (28). Collectively, these results suggest that activation of TRPML1 is sufficient to induce lysosomal exocytosis in a Ca2+-dependent manner.
TFEB transcription factor.
bHLH-leucine zipper TFEB is a master regulator of both lysosomal biogenesis and autophagy (14, 30, 68). When autophagy is triggered or when lysosomes are under stress conditions, TFEB proteins translocate from the cytoplasm to the nucleus, inducing the expression of its target genes (14). Bioinformatics analysis suggested that TFEB regulates the expression of hundreds of autophagy-related and lysosome-related genes by binding to the coordinated lysosomal expression and regulation motif (30, 68). Indeed, overexpression of TFEB in HeLa cells is sufficient to increase the mRNA expression of 291 genes that are mostly involved in autophagy and lysosomal biogenesis (30, 68). As a TFEB-regulated lysosomal gene, TRPML1 contains a coordinated lysosomal expression and regulation motif upstream from its transcription start site, and its mRNA expression is upregulated by more than several fold by TFEB overexpression (27, 68). Putting these results together, lysosomal stress, such as lipid storage, may increase the expression/activity of TFEB and its target genes. Although still speculative, it is possible that TFEB may mediate the bulk of the compensatory changes associated with LSDs.
TFEB overexpression significantly increases both docking and fusion of lysosomes with the plasma membrane (27). Consistent with the TFEB-induced upregulation of TRPML1 (Fig. 4A), the TFEB-induced increase in lysosomal exocytosis is abolished in ML-IV fibroblasts (27). Therefore, TRPML1 appears to be an essential target gene of TFEB. However, in ML-IV cells, TFEB overexpression still increases lysosomal docking to the plasma membrane. Therefore, in addition to TRPML1 upregulation, TFEB may also increase the expression/activity of tethering factors or proteins involved in lysosome mobility (14). For instance, TFEB may upregulate kinesin motor proteins that are responsible for the trafficking of lysosomes to the plasma membrane along microtubules (27, 69).
Neuraminidase 1 as the negative regulator of lysosomal exocytosis.
Lysosomal exocytosis is also regulated by posttranslation modification of lysosomal proteins. Neuraminidase 1 (Neu1) is a lysosomal enzyme responsible for the cleavage of the terminal sialic acid residues from glycoproteins and glycolipids (70). Mutations in the Neu1 gene cause sialidosis (71). The lysosomal membrane protein Lamp1, the most abundant lysosomal membrane protein that is heavily glycosylated, is a substrate of Neu1 (70). Interestingly, lysosomal docking and exocytosis are increased in Neu1 KO cells and this effect is dependent on Lamp1. However, how Lamp1 and its sialylation regulates lysosomal docking is still not very clear.
Releasable lysosome pool.
Although lysosomes can undergo Ca2+-dependent exocytosis, it is not clear what percentage of the total lysosome population can be docked and undergo exocytosis upon stimulation in different cell types (35). Lysosomes are heterogeneous in size, but the molecular markers to label different pools of lysosomes have remained to be identified. In LSDs, it is primarily the autolysosomes, but not the endolysosomes, that undergo exocytosis (29). Lysosomal mobility and subcellular location may be the key determinants of lysosomal docking, and hence, the size of the releasable lysosome pool. Cell types and the nature of the stimulus may determine the size of the releasable pool. While activation of TRPML1 promotes postdocking fusion, TFEB regulates both lysosomal docking and fusion (27). In NRK and MEF cells, under normal conditions, only about 10–15% of total lysosomal β-hexosaminidase can be released by ionomycin within minutes (40). In macrophages, however, up to 40–50% of total lysosomal acid phosphatases were releasable by ionomycin within 15 min (28). As lysosomal enzyme release is an accumulative assay, a high level of lysosome mobility that is known to be associated with macrophages (44) may account for the observed differences in the releasable pool. An outstanding question is whether an increase in lysosome-plasma membrane fusion causes a feedback to facilitate lysosomal mobility and docking. Even more generally, how do cells determine the size, location, and number of lysosomes under different environmental conditions? The nutrient-sensing machinery in the lysosome (LYNUS), including mTOR and TFEB, may play a critical role in regulating these basic parameters for lysosomes (14).
Directionality of lysosomal exocytosis
In nonpolarized cells, lysosomal exocytosis may occur at the sites of action, so called “focal exocytosis”. For example, in macrophages, upon large particle binding, lysosomal exocytosis occurs specifically at the site of particle uptake (28, 39, 72). Likewise, in muscle cells and other cell types, lysosomal exocytosis occurs at the sites of membrane disruptions (33, 73). Localized Ca2+ increase is likely to be the underlying mechanism for focal exocytosis. In addition, the microtubule-dependent transport of lysosomes may be regulated by large particle binding or membrane injury (35, 44).
In polarized epithelial cells, lysosomal exocytosis is directed toward the basolateral membranes, but not the apical membranes (74). The directionality of lysosomal exocytosis in these cells could be caused by the localization of the plasma membrane SNARE, syntaxin4, exclusively at the basolateral membrane, resulting in a “polarized” formation of the SNARE complex (74). Whether other proteins of the lysosomal exocytosis machinery, such as Syt-VII and VAMP7, are also polarized in their localization remains unknown.
The biological implications of lysosomal exocytosis
There are two primary purposes for lysosomal exocytosis: membrane remodeling and secretion. The former may contribute to neurite outgrowth (38), membrane repair (33), and phagocytosis (28, 39). The latter may contribute to antigen presentation (75), bone absorption in osteoclasts (76), axonal myelination (77), and transmitter release (37).
Secretion.
In hematopoietic cells, including neutrophils and cytotoxic T cells (32), secretory lysosomes (lysosome-related organelles) were observed to undergo exocytosis in a Ca2+-dependent manner (75). The release of lysosomal hydrolytic enzymes into the extracellular space may help digest and defend against pathogens (75) (Fig. 4B). Lysosomal exocytosis is dramatically upregulated in osteoclasts during bone reabsorption (76). Additionally, lysosomal exocytosis is crucial for antigen presentation mediated by major histocompatibility complex II (MHC-II) in macrophages (78) (Fig. 4B). Because exocytosis of conventional lysosomes is now known to be present in all cell types, it is possible that lysosomes play a general role in secretion. Secreted lysosomal contents may lead to various cellular responses, including altered cell mobility and cellular transformation (79). Secretion of lysosomal acid (a)SMase is essential for membrane repair (80).
Transmitter release.
In astrocytes, ATP release is thought to be dependent on lysosomal exocytosis (37). Lysosomes contain high levels of ATP and undergo exocytosis in a Ca2+-dependent manner upon various stimulations (37). In contrast, glutamate release in the astrocytes requires nonlysosome small vesicles, and is independent of lysosomal exocytosis (36).
Membrane repair.
The integrity of the plasma membrane is maintained through an active repair process (81). The repair of plasma membrane damage in skeletal muscle and other cell types requires a rapid increase in Ca2+ to trigger the recruitment of intracellular vesicles that fuse with the plasma membrane to replace the disrupted membranes (33, 81) (Fig. 4B). As membrane damage triggers lysosomal exocytosis, the major candidate for these vesicles is the lysosomes. Indeed, in Syt-VII KO muscle, membrane repair is impaired (82). Ca2+ influx has been proposed to be the source of Ca2+ for lysosomal exocytosis, as removal of the extracellular Ca2+ impaired the resealing process in most in vitro damage/repair assays (82, 83). However, it remains unknown whether such manipulation secondarily affects other intracellular Ca2+-dependent processes. It also remains to be tested whether lysosomal Ca2+ release can serve as a more physiological trigger of membrane repair in vivo. Importantly, in addition to provide intracellular membranes, lysosomal exocytosis is also crucial for the release of lysosomal aSMases to mediate rapid endocytosis of damaged membranes, which in turn is essential for membrane repair (80).
Phagocytosis.
Lysosomal exocytosis is required for the uptake of large particles in macrophages (39, 72, 84) (Fig. 4B). Professional phagocytes, such as macrophages, engulf large cellular particles, such as apoptotic cells, by forming filopodia-like structures called pseudopods to surround the particles (85). A large amount of membranes are required to form pseudopods (84). Because the overall surface area remains relatively constant during pseudopod formation (41), intracellular membranes, especially those from endosomes and lysosomes, are delivered to the sites of particle ingestion (so-called focal exocytosis) (39, 72). When lysosomal exocytosis is inhibited by dominant negative forms of VAMP7 or Syt-VII, large particle uptake is significantly reduced (39, 72). Interestingly, TRPML1 KO macrophages exhibit defects in large particle uptake, a phenotype that closely resembles the phagocytic defects observed in Syt-VII KO macrophages (28). By using the lysosome-targeted genetically encoded Ca2+ sensor GCaMP3 (86), which is much more sensitive in detecting lysosomal Ca2+ release than conventional dye-based methods (87), it was found that particles binding to macrophages induced TRPML1-mediated lysosomal Ca2+ release specifically at the site of particle uptake (28). As defective clearance of late apoptotic neurons may contribute to neurodegeneration (88), a loss of TRPML1 channel activity may result in the pathogenic accumulation of extracellular particles and brain inflammation seen in ML-IV or other LSDs.
Neurite outgrowth.
Lysosomal exocytosis may also play a role in neurite outgrowth (38). Neurite outgrowth is mediated by the exocytosis of intracellular vesicles at the leading edges of developing neurites. While the Ca2+ ionophore induces Lamp1 surface staining in the neuronal processes, neurite outgrowth is significantly reduced in Syt-VII KO neurons (38).
THE INTERACTION LOOP OF LYSOSOMAL LIPIDS AND LYSOSOMAL EXOCYTOSIS
The interaction between lysosomal lipids and lysosomal exocytosis is two-fold. First, lysosomal exocytosis and lysosomal trafficking are regulated by both “lumen-facing” and “cytoplasm-facing” lysosomal lipids. Second, exocytosis of lysosomes may “dump” undigested lipids into the extracellular space, reducing lipid accumulation in the lysosome.
Regulation of lysosomal exocytosis by lysosomal lipids
Lysosomal lipids are important regulators of lysosomal membrane trafficking. Cytoplasm-facing lysosomal lipids, such as endolysosome-localized phosphoinositides, may function as signaling molecules to regulate lysosomal trafficking. On the other hand, lumen-facing lysosomal lipids on the perimeter membrane and in intraluminal vesicles, such as cholesterol and sphingolipids, may affect membrane fusion machinery in the lysosome. First, lumen-facing lysosomal lipid accumulation may affect the composition of the lipid bilayer, which in turn influences the activity of membrane proteins involved in fusion and fission. For example, lipids such as cholesterol may affect the formation of SNARE complexes (89, 90). Second, accumulation of lumen-facing lysosomal lipids, such as SMs, may influence exocytosis by affecting the channel activity of TRPML1 (86). Furthermore, lipid precipitation reduces the activity of lysosomal hydrolases, which causes the secondary accumulation to inhibit lysosomal trafficking (4).
Luminal lipids: cholesterol and SM.
Cholesterol is an important lipid in mammalian cells participating in cell signaling and maintenance of membrane integrity (91). Unlike other complex lipids in the intraluminal vesicles, cholesterol is not degraded by lysosomal hydrolases. Instead, this lipid is transported out of the lysosome via the cholesterol exporter NPC1 (16, 92). Mutations in NPC1 lead to the accumulation of cholesterol and other lipids in the endolysosomal compartments, resulting in a LSD called NPC (93, 94). Soluble NPC2 protein is a small glycoprotein in the lysosome lumen that transfers cholesterol to NPC1. Mutations in NPC2 also result in cholesterol accumulation and NPC disease (94). Interestingly, in addition to these primary cholesterol storage disorders, most other LSDs exhibit secondary accumulation of cholesterol in their lysosomes as well (17).
SNAREs are partially associated with the cholesterol-rich domains on the membrane (90). Hence, the cholesterol level may be a critical determinant of membrane fusion. In fibroblasts isolated from two different LSDs, multiple sulfatase deficiency and mucopolysaccharidosis type III, the secondary cholesterol accumulation was shown to affect the localization and function of lysosomal SNARE VAMP7, but not plasma membrane SNARE SNAP23 (90). Subsequently, the SNARE complex formation was impaired, and the late endosome-lysosome fusion was inhibited (90). Reducing membrane cholesterol levels with methyl-β-cyclodextrin consistently increases ionomycin-induced lysosomal exocytosis in MCDK cells (74). In contrast, treating cells with U18666A, a hydrophobic amine that causes cholesterol accumulation in the late endosomes and lysosomes, almost completely inhibited ionomycin-induced lysosomal exocytosis (74).
SM is a plasma membrane lipid that accumulates in the lysosome when aSMases are defective, as is the case for NPA and NPB cells (4). In NPC cells, cholesterol accumulation causes reduced activity in aSMases and subsequent SM accumulation in the lysosome (4). Cholesterol and SM accumulation may have a synergistic effect on lysosomal trafficking defects observed in NPC cells (86). Lysosomal accumulation of SM in NPC cells directly inhibits TRPML1 channel activity and TRPML1-mediated lysosomal Ca2+ release (95). Intriguingly, TRPML-specific activator ML-SA1 together with TRPML1 overexpression was able to reduce cholesterol accumulation in NPC cells (86). Hence, luminal lipids could potentially regulate lysosomal trafficking by influencing lysosomal Ca2+ release. The mechanisms by which TRPML1 activation promotes cholesterol clearance from the lysosome are still not clear. Because induction of lysosomal exocytosis by cyclodextrin was sufficient to promote cholesterol clearance from NPC cells (96), increased lysosomal exocytosis could be the primary underlying mechanism.
Cytoplasmic lipids: phosphoinositides.
Cytoplasmic lipids are known to be important regulators of membrane trafficking. Ptdlns, a group of membrane phospholipids localized on the cytosolic side of cellular compartments, are reversibly phosphorylated on their inositol ring at positions 3, 4, and 5, resulting in the generation of seven phosphoinositides [PI3P, PI4P, PI5P, PI(3,4)P2, PI(3,5)P2, PI(4,5)P2, and PI(3,4,5)P3] (97). Among them, PI(3)P and PI(3,5)P2 are localized on the endosomes and lysosomes (97). Meanwhile, PI(4)P and PI(4,5)P2 may also be transiently produced in the lysosome under certain conditions (98). Lysosomal trafficking and lysosomal exocytosis are regulated by endolysosome-localized PI(3,5)P2, whose synthesis requires the kinase PIKfyve in association with the phosphatase Fig4 and the scaffolding protein Vac14 (99, 100). Genetic disruption or pharmacological inactivation of any of these components results in a decrease in the cellular PI(3,5)P2 level and defects in lysosomal trafficking (99, 100).
Lysosomal exocytosis is significantly reduced in macrophages isolated from Fig4 KO mice (28), suggesting that PI(3,5)P2 is a crucial regulator of lysosomal exocytosis. PI(3,5)P2 may regulate lysosomal trafficking via three potential mechanisms. First, PI(3,5)P2 may increase the fusogenic potential of lysosomes by affecting the properties of the lipid bilayer, especially at the fusion spot (101). At the plasma membrane, PI(4,5)P2 helps generate the membrane curvature necessary for membrane fusion (97). Second, PI(3,5)P2 may recruit a variety of cytoplasmic effector proteins to facilitate membrane trafficking (102). For synaptic vesicle exocytosis, PI(4,5)P2 recruits the priming factors to the site of membrane fusion (43, 55). It is conceivable that PI(3,5)P2 recruits motor proteins to increase lysosomal mobility and docking. Third, PI(3,5)P2 may regulate the activity of lysosomal membrane proteins that are parts of the trafficking machinery. For example, Syt-VII may contain a PI(3,5)P2 binding site, analogous to the PI(4,5)P2 binding site in Syt-I (43, 55). However, direct evidences to support the above-mentioned mechanisms are still lacking.
Direct evidence, however, does exist to support that PI(3,5)P2 may directly regulate the activity of lysosomal channels and transporters. Many plasma membrane ion channels and transporters require PI(4,5)P2 as a positive cofactor (103). It is likely that PI(3,5)P2 plays a similar role for lysosomal channels/transporters. Indeed, whole-lysosome patch-clamp recordings have demonstrated that PI(3,5)P2 binds directly to activate TRPML1 in a physiologically-relevant low nanomolar range (64). TRPML1 contains a cluster of positively charged amino acid residues on the N terminus, which was shown to bind directly to PI(3,5)P2 in in vitro lipid-protein binding assays (64). Charge removal mutations abolished the PI(3,5)P2 activation and, importantly, the effect of TRPML1 on lysosomal trafficking (64). Collectively, PI(3,5)P2 may regulate lysosomal trafficking events, such as exocytosis, through the activation of TRPML1 and lysosomal Ca2+ release.
PI(3,5)P2 may also regulate lysosomal exocytosis via modulation of other lysosomal membrane proteins. PI(3,5)P2 may regulate two-pore TPC proteins to alter the membrane potential of lysosomes, which might play a role in membrane fusion (104). In addition, lysosomal acidification is impaired in PI(3,5)P2-deficient cells. Lysosomal pH is known to be an important regulator of membrane trafficking (102). It remains to be established whether PI(3,5)P2 may directly regulate V-ATPase and other proton transporters (102).
Whether PI(3,5)P2 plays a permissive or instructive role in lysosomal exocytosis is not clear. To distinguish these two possibilities, it is necessary to visualize PI(3,5)P2 dynamics during lysosomal exocytosis (105). Using a genetically-encoded PI(3,5)P2 indicator based on the PI(3,5)P2-binding domain in TRPML1, it was recently shown that PI(3,5)P2 levels increased transiently prior to fusion of two Lamp1-positive vesicles (105), and during particle uptake (28). However, in another set of Lamp1-positive vesicles, PI(3,5)P2 levels remained constant (105). Therefore, PI(3,5)P2 may play multiple roles in regulating lysosomal trafficking and exocytosis.
Stimulating exocytosis to clear lysosomal lipid storage
Because lipid storage is the primary cause of lysosomal dysfunction, pharmacological and genetic manipulations that could clear the accumulated materials can potentially serve as novel therapeutic approaches for LSDs. Because TFEB overexpression and TRPML1 activation increase lysosomal exocytosis, manipulating the expression and activity of TFEB and TRPML1 may provide exciting opportunities to clear lysosomal lipid storage.
TFEB approach.
In the neuronal stem cells isolated from the mouse models of multiple sulfatase deficiency and mucopolysaccharidosis type IIIA, two neurodegenerative LSDs caused by lysosomal accumulation of glycosaminoglycans (GAGs), TFEB overexpression was sufficient to reduce the storage of GAGs in the lysosomes (27). Additionally, TFEB overexpression strongly reduced the lysosomal accumulation of lipofuscin in cells from the murine model of the juvenile form of neuronal ceroid lipofuscinoses (NCL) known as Batten disease (27), as well as fibroblast cells obtained from patients with glycogen-storage Pompe’s disease (29). Viral-mediated gene transfer of TFEB leads to the clearance of stored materials, a decrease in lysosome size, and alleviation of the pathological symptoms in multiple LSDs (14). Additionally, TFEB overexpression was also able to induce clearance in the mouse models of Parkinson’s and Huntington’s disease (106, 107).
The beneficial effect of TFEB on various LSDs is dependent on TRPML1 and lysosomal exocytosis (27). TFEB-mediated clearance is largely abolished when TRPML1 channel activity is compromised or when lysosomal exocytosis is inhibited (27). Therefore, it is possible that TFEB promotes the clearance of lysosomal storage through lysosomal exocytosis, in which the storage materials are secreted upon induction of lysosomal exocytosis by TFEB activation. Considering that TRPML1 is upregulated by TFEB overexpression (108), it is likely TFEB may directly upregulate TRPML1 to promote lysosomal exocytosis. However, TFEB may also mediate the expression of Vac14 (108) that is required for PI(3,5)P2 production (109). Therefore, TFEB can also regulate lysosomal exocytosis by acting on other regulators in the pathway, including the machinery for lysosome mobility.
Unexpectedly, the beneficial effect TFEB has on various LSDs is also dependent on autophagy (29). When autophagy was genetically inhibited, the rescue effect of TFEB in Pompe cells was abolished (29). The lysosomes undergoing exocytosis were enlarged and contained markers for both lysosomes and autophagosomes (29). Hence, it is likely that the autolysosomes are the primary compartments undergoing exocytosis in LSD cells. Consistently, the majority of the undigested lipids are of autophagic origin. Indeed, most LSDs exhibit a slightly increased autophagy initiation but impaired completion resulting in autophagic arrest and accumulation of autolysosomes (8). Collectively, in LSD cells, TFEB regulates cellular clearance via exocytosis of arrested, enlarged autolysosomes.
Given that TFEB induces cellular clearance in LSD cells by accelerating lysosomal trafficking and lysosomal exocytosis, modulating TFEB activity represents a very attractive therapeutic strategy for LSDs. However, genetic approaches can only provide proof-of-principle studies in mouse models of LSDs. To extend the observations to human LSD patients, it is desirable to develop reagents that can be used to “activate” TFEB. TFEB activity and its translocation to the nucleus are shown to be regulated by protein phosphorylation (30). Thus, TFEB phosphorylation and dephosphorylation could be an attractive target for large-scale drug screens.
TRPML1 approach.
Unlike other LSDs, the overexpression of TFEB failed to induce the clearance of stored materials in ML-IV cells, indicating that TFEB induced cellular clearance most likely through TRPML1 (27). This places TRPML1 in a unique situation for LSDs. However, is activation of TRPML1 sufficient for cellular clearance?
Impaired lysosomal Ca2+ release through TRPML1 could contribute to secondary lipid accumulation, acting as a common pathogenic mechanism for many LSDs (86). Most LSDs are caused by impaired lysosomal degradation due to the lack of a specific hydrolytic enzyme. However, the accumulation of specific substrates may also cause trafficking defects, which in turn cause secondary lysosome storage (Fig. 2). Lysosome storage can in turn affect lysosomal degradation and membrane trafficking, resulting in a vicious cycle. Indeed, TRPML1 could be directly inhibited by accumulated lipids in the case of SM for NPA and NPC cells (86). Whether TRPML1 is similarly inhibited in other LSDs by their accumulated lipids is not clear. For NPA/B and NPC cells, deinhibition of TRPML1 could break the vicious cycle and reduce lipid accumulation, which may serve as an effective approach for these LSDs. Indeed, TRPML1 overexpression and small molecule TRPML1 agonists were shown to promote cholesterol clearance in NPC cells (86). As cholesterol accumulation is commonly seen in many LSDs, activating TRPML1 may represent a novel therapeutic approach for many other LSDs. TRPML-specific agonists such as SF compounds (SF-51) and ML-SA1 have been successfully used for in vitro studies (86, 110). They, together with the next generation TRPML-specific compounds, may also facilitate the in vivo investigation of TRPML1 as a therapeutic strategy for lipid storage disorders.
Although enhancing lysosomal exocytosis seems to be an attractive approach for LSDs, there are several limitations that should be considered. The major concern is that cellular clearance through lysosomal exocytosis may lead to the secretion of a large amount of biomaterials into the extracellular space. The secreted lysosomal enzymes and undigested lipids could potentially trigger immune responses. It is worth noting that the clearance of large extracellular particles by macrophages is defective in ML-IV and some LSDs (28). In addition, microglia activation is evident in the brain of TRPML1 KO mice (28) as well as in mouse models of other LSDs (M. A. Samie and H. Xu, unpublished observations). As TRPML1 activation may be able to boost phagocytic clearance of apoptotic debris in the brain, the TRPML1 approach may increase both cellular clearance and extracellular clearance. However, unlike the TFEB approach, the clearance effect of TRPML1 approach has not been established in vivo.
CONCLUSION AND FUTURE DIRECTIONS
Recent studies in lysosome cell biology and LSDs have clearly demonstrated that lysosomes are not the dead-end of degradation pathways. Instead, lysosomes are very dynamic organelles that play active roles in numerous cellular processes. Lysosomes receive inputs through endocytic and autophagic pathways and send outputs through retrograde trafficking and lysosomal exocytosis. Cells must maintain a balance between endocytosis and exocytosis. Misregulation of these two cellular processes could result in accumulation of undigested materials and dysfunctional lysosomes. Now that the basic mechanisms of lysosomal exocytosis are known, the new focus is to investigate how primary storage causes defective lysosomal trafficking and exocytosis to cause secondary lipid accumulation.
The central concept that enhancing lysosomal trafficking, and particularly lysosomal exocytosis, may be able to alleviate the pathological symptoms in most LSDs regardless of the primary deficiency is appealing. Indeed, the TFEB approach has been shown to be effective in various LSDs, but not in ML-IV. However, a small molecule drug that specifically stimulates TFEB activity, ideally downstream of mTOR, is still lacking. The TRPML1 approach is effective in some in vitro cell culture models, but in vivo studies are still lacking. On the other hand, TRPML1-specific small molecule activators such as ML-SA1 have already been developed and proven helpful for studying the basic regulatory mechanisms for lysosomal exocytosis. Future years may see more exciting work on the role of lysosomal exocytosis in LSDs and other diseases.
Acknowledgments
The authors appreciate the encouragement and helpful comments of other members of the Xu laboratory and apologize to colleagues whose works are not cited due to space limitations.
Footnotes
Abbreviations:
- aSMase
- acid SMase
- BAPTA
- 1,2-bis(o-aminophenoxy)ethane-N,N,N’,N’-tetraacetic acid
- BAPTA-AM
- BAPTA-acetoxymethyl ester
- ER
- endoplasmic reticulum
- ERT
- enzyme replacement therapy
- LSD
- lysosomal storage disease
- LYNUS
- lysosomal nutrient sensing system
- ML-IV
- mucolipidosis type IV
- ML-SA1
- transient receptor potential mucolipin 1-specific small molecule activator
- mTOR
- mammalian target of rapamycin
- MVB
- multi-vesicular body
- Neu1
- neuraminidase 1
- NPC
- Niemann-Pick type C
- SNAP23
- synaptosome-associated protein of 23 kDa
- SNARE
- soluble N-ethylmaleimide-sensitive factor-attachment protein receptor
- Syt
- synaptotagmin
- TFEB
- transcription factor EB
- TIRF
- total internal reflection fluorescent microscopy
- TRPML1
- transient receptor potential mucolipin 1
- VAMP7
- vesicle-associated membrane protein 7
- vacuolar-ATPase
- V-ATPase
The work in the authors’ laboratory is supported by National Institutes of Health Grants NS062792, MH096595, and AR060837 (to H.X.).
REFERENCES
- 1.Kolter T., Sandhoff K. 2005. Principles of lysosomal membrane digestion: stimulation of sphingolipid degradation by sphingolipid activator proteins and anionic lysosomal lipids. Annu. Rev. Cell Dev. Biol. 21: 81–103. [DOI] [PubMed] [Google Scholar]
- 2.Kornfeld S., Mellman I. 1989. The biogenesis of lysosomes. Annu. Rev. Cell Biol. 5: 483–525. [DOI] [PubMed] [Google Scholar]
- 3.Luzio J. P., Pryor P. R., Bright N. A. 2007. Lysosomes: fusion and function. Nat. Rev. Mol. Cell Biol. 8: 622–632. [DOI] [PubMed] [Google Scholar]
- 4.Schulze H., Sandhoff K. 2011. Lysosomal lipid storage diseases. Cold Spring Harb. Perspect. Biol. 3: a004804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huotari J., Helenius A. 2011. Endosome maturation. EMBO J. 30: 3481–3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mellman I. 1989. Organelles observed: lysosomes. Science. 244: 853–854. [DOI] [PubMed] [Google Scholar]
- 7.Neiss W. F. 1984. A coat of glycoconjugates on the inner surface of the lysosomal membrane in the rat kidney. Histochemistry. 80: 603–608. [PubMed] [Google Scholar]
- 8.Lieberman A. P., Puertollano R., Raben N., Slaugenhaupt S., Walkley S. U., Ballabio A. 2012. Autophagy in lysosomal storage disorders. Autophagy. 8: 719–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mizushima N., Levine B., Cuervo A. M., Klionsky D. J. 2008. Autophagy fights disease through cellular self-digestion. Nature. 451: 1069–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berg T. O., Fengsrud M., Strømhaug P. E., Berg T., Seglen P. O. 1998. Isolation and characterization of rat liver amphisomes. Evidence for fusion of autophagosomes with both early and late endosomes. J. Biol. Chem. 273: 21883–21892. [DOI] [PubMed] [Google Scholar]
- 11.Ruivo R., Anne C., Sagné C., Gasnier B. 2009. Molecular and cellular basis of lysosomal transmembrane protein dysfunction. Biochim. Biophys. Acta. 1793: 636–649. [DOI] [PubMed] [Google Scholar]
- 12.Sagné C., Gasnier B. 2008. Molecular physiology and pathophysiology of lysosomal membrane transporters. J. Inherit. Metab. Dis. 31: 258–266. [DOI] [PubMed] [Google Scholar]
- 13.Gillard B. K., Clement R. G., Marcus D. M. 1998. Variations among cell lines in the synthesis of sphingolipids in de novo and recycling pathways. Glycobiology. 8: 885–890. [DOI] [PubMed] [Google Scholar]
- 14.Settembre C., Fraldi A., Medina D. L., Ballabio A. 2013. Signals from the lysosome: a control centre for cellular clearance and energy metabolism. Nat. Rev. Mol. Cell Biol. 14: 283–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu L., McPhee C. K., Zheng L., Mardones G. A., Rong Y., Peng J., Mi N., Zhao Y., Liu Z., Wan F., et al. 2010. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature. 465: 942–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Infante R. E., Wang M. L., Radhakrishnan A., Kwon H. J., Brown M. S., Goldstein J. L. 2008. NPC2 facilitates bidirectional transfer of cholesterol between NPC1 and lipid bilayers, a step in cholesterol egress from lysosomes. Proc. Natl. Acad. Sci. USA. 105: 15287–15292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parkinson-Lawrence E. J., Shandala T., Prodoehl M., Plew R., Borlace G. N., Brooks D. A. 2010. Lysosomal storage disease: revealing lysosomal function and physiology. Physiology (Bethesda). 25: 102–115. [DOI] [PubMed] [Google Scholar]
- 18.Neufeld E. F. 1991. Lysosomal storage diseases. Annu. Rev. Biochem. 60: 257–280. [DOI] [PubMed] [Google Scholar]
- 19.Deleted in proof.
- 20.Walkley S. U., Vanier M. T. 2009. Secondary lipid accumulation in lysosomal disease. Biochim. Biophys. Acta. 1793: 726–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dong X. P., Cheng X., Mills E., Delling M., Wang F., Kurz T., Xu H. 2008. The type IV mucolipidosis-associated protein TRPML1 is an endolysosomal iron release channel. Nature. 455: 992–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ballabio A., Gieselmann V. 2009. Lysosomal disorders: from storage to cellular damage. Biochim. Biophys. Acta. 1793: 684–696. [DOI] [PubMed] [Google Scholar]
- 23.Boya P., Kroemer G. 2008. Lysosomal membrane permeabilization in cell death. Oncogene. 27: 6434–6451. [DOI] [PubMed] [Google Scholar]
- 24.Cox T. M., Cachon-Gonzalez M. B. 2012. The cellular pathology of lysosomal diseases. J. Pathol. 226: 241–254. [DOI] [PubMed] [Google Scholar]
- 25.Eng C. M., Banikazemi M., Gordon R. E., Goldman M., Phelps R., Kim L., Gass A., Winston J., Dikman S., Fallon J. T., et al. 2001. A phase 1/2 clinical trial of enzyme replacement in fabry disease: pharmacokinetic, substrate clearance, and safety studies. Am. J. Hum. Genet. 68: 711–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sawkar A. R., Adamski-Werner S. L., Cheng W. C., Wong C. H., Beutler E., Zimmer K. P., Kelly J. W. 2005. Gaucher disease-associated glucocerebrosidases show mutation-dependent chemical chaperoning profiles. Chem. Biol. 12: 1235–1244. [DOI] [PubMed] [Google Scholar]
- 27.Medina D. L., Fraldi A., Bouche V., Annunziata F., Mansueto G., Spampanato C., Puri C., Pignata A., Martina J. A., Sardiello M., et al. 2011. Transcriptional activation of lysosomal exocytosis promotes cellular clearance. Dev. Cell. 21: 421–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Samie M., Wang X., Zhang X., Goschka A., Li X., Cheng X., Gregg E., Azar M., Zhuo Y., Garrity A. G., et al. 2013. A TRP channel in the lysosome regulates large particle phagocytosis via focal exocytosis. Dev. Cell. 26: 511–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spampanato C., Feeney E., Li L., Cardone M., Lim J. A., Annunziata F., Zare H., Polishchuk R., Puertollano R., Parenti G., et al. 2013. Transcription factor EB (TFEB) is a new therapeutic target for Pompe disease. EMBO Mol. Med. 5: 691–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Settembre C., Di Malta C., Polito V. A., Garcia Arencibia M., Vetrini F., Erdin S., Erdin S. U., Huynh T., Medina D., Colella P., et al. 2011. TFEB links autophagy to lysosomal biogenesis. Science. 332: 1429–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nickel W., Rabouille C. 2009. Mechanisms of regulated unconventional protein secretion. Nat. Rev. Mol. Cell Biol. 10: 148–155. [DOI] [PubMed] [Google Scholar]
- 32.Stinchcombe J. C., Griffiths G. M. 1999. Regulated secretion from hemopoietic cells. J. Cell Biol. 147: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reddy A., Caler E. V., Andrews N. W. 2001. Plasma membrane repair is mediated by Ca(2+)-regulated exocytosis of lysosomes. Cell. 106: 157–169. [DOI] [PubMed] [Google Scholar]
- 34.Coorssen J. R., Schmitt H., Almers W. 1996. Ca2+ triggers massive exocytosis in Chinese hamster ovary cells. EMBO J. 15: 3787–3791. [PMC free article] [PubMed] [Google Scholar]
- 35.Jaiswal J. K., Andrews N. W., Simon S. M. 2002. Membrane proximal lysosomes are the major vesicles responsible for calcium-dependent exocytosis in nonsecretory cells. J. Cell Biol. 159: 625–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu T., Sun L., Xiong Y., Shang S., Guo N., Teng S., Wang Y., Liu B., Wang C., Wang L., et al. 2011. Calcium triggers exocytosis from two types of organelles in a single astrocyte. J. Neurosci. 31: 10593–10601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dou Y., Wu H. J., Li H. Q., Qin S., Wang Y. E., Li J., Lou H. F., Chen Z., Li X. M., Luo Q. M., et al. 2012. Microglial migration mediated by ATP-induced ATP release from lysosomes. Cell Res. 22: 1022–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arantes R. M., Andrews N. W. 2006. A role for synaptotagmin VII-regulated exocytosis of lysosomes in neurite outgrowth from primary sympathetic neurons. J. Neurosci. 26: 4630–4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Czibener C., Sherer N. M., Becker S. M., Pypaert M., Hui E., Chapman E. R., Mothes W., Andrews N. W. 2006. Ca2+ and synaptotagmin VII-dependent delivery of lysosomal membrane to nascent phagosomes. J. Cell Biol. 174: 997–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rodríguez A., Webster P., Ortego J., Andrews N. W. 1997. Lysosomes behave as Ca2+-regulated exocytic vesicles in fibroblasts and epithelial cells. J. Cell Biol. 137: 93–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holevinsky K. O., Nelson D. J. 1998. Membrane capacitance changes associated with particle uptake during phagocytosis in macrophages. Biophys. J. 75: 2577–2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jaiswal J. K., Chakrabarti S., Andrews N. W., Simon S. M. 2004. Synaptotagmin VII restricts fusion pore expansion during lysosomal exocytosis. PLoS Biol. 2: E233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Südhof T. C. 2013. Neurotransmitter release: the last millisecond in the life of a synaptic vesicle. Neuron. 80: 675–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mrakovic A., Kay J. G., Furuya W., Brumell J. H., Botelho R. J. 2012. Rab7 and Arl8 GTPases are necessary for lysosome tubulation in macrophages. Traffic. 13: 1667–1679. [DOI] [PubMed] [Google Scholar]
- 45.Cardoso C. M. P., Groth-Pedersen L., Høyer-Hansen M., Kirkegaard T., Corcelle E., Andersen J. S., Jäättelä M., Mylandsted J. 2009. Depletion of kinesin 5B affects lysosomal distribution and stability and induces peri-nuclear accumulation of autophagosomes in cancer cells. PLoS ONE. 4: e4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rodríguez A., Martinez I., Chung A., Berlot C. H., Andrews N. W. 1999. cAMP regulates Ca2+-dependent exocytosis of lysosomes and lysosome-mediated cell invasion by trypanosomes. J. Biol. Chem. 274: 16754–16759. [DOI] [PubMed] [Google Scholar]
- 47.Heuser J. 1989. Changes in lysosome shape and distribution correlated with changes in cytoplasmic pH. J. Cell Biol. 108: 855–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jahn R., Scheller R. H. 2006. SNAREs–engines for membrane fusion. Nat. Rev. Mol. Cell Biol. 7: 631–643. [DOI] [PubMed] [Google Scholar]
- 49.Rao S. K., Huynh C., Proux-Gillardeaux V., Galli T., Andrews N. W. 2004. Identification of SNAREs involved in synaptotagmin VII-regulated lysosomal exocytosis. J. Biol. Chem. 279: 20471–20479. [DOI] [PubMed] [Google Scholar]
- 50.Pryor P. R., Mullock B. M., Bright N. A., Gray S. R., Luzio J. P. 2000. The role of intraorganellar Ca(2+) in late endosome-lysosome heterotypic fusion and in the reformation of lysosomes from hybrid organelles. J. Cell Biol. 149: 1053–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Burdakov D., Petersen O. H., Verkhratsky A. 2005. Intraluminal calcium as a primary regulator of endoplasmic reticulum function. Cell Calcium. 38: 303–310. [DOI] [PubMed] [Google Scholar]
- 52.Christensen K. A., Myers J. T., Swanson J. A. 2002. pH-dependent regulation of lysosomal calcium in macrophages. J. Cell Sci. 115: 599–607. [DOI] [PubMed] [Google Scholar]
- 53.Lloyd-Evans E., Morgan A. J., He X., Smith D. A., Elliot-Smith E., Sillence D. J., Churchill G. C., Schuchman E. H., Galione A., Platt F. M. 2008. Niemann-Pick disease type C1 is a sphingosine storage disease that causes deregulation of lysosomal calcium. Nat. Med. 14: 1247–1255. [DOI] [PubMed] [Google Scholar]
- 54.Luzio J. P., Bright N. A., Pryor P. R. 2007. The role of calcium and other ions in sorting and delivery in the late endocytic pathway. Biochem. Soc. Trans. 35: 1088–1091. [DOI] [PubMed] [Google Scholar]
- 55.Chapman E. R. 2008. How does synaptotagmin trigger neurotransmitter release? Annu. Rev. Biochem. 77: 615–641. [DOI] [PubMed] [Google Scholar]
- 56.Vergarajauregui S., Martina J. A., Puertollano R. 2009. Identification of the penta-EF-hand protein ALG-2 as a Ca2+-dependent interactor of mucolipin-1. J. Biol. Chem. 284: 36357–36366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu L. J., Sweet T. B., Clapham D. E. 2010. International Union of Basic and Clinical Pharmacology. LXXVI. Current progress in the mammalian TRP ion channel family. Pharmacol. Rev. 62: 381–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cheng X., Shen D., Samie M., Xu H. 2010. Mucolipins: Intracellular TRPML1–3 channels. FEBS Lett. 584: 2013–2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bargal R., Avidan N., Ben-Asher E., Olender Z., Zeigler M., Frumkin A., Raas-Rothschild A., Glusman G., Lancet D., Bach G. 2000. Identification of the gene causing mucolipidosis type IV. Nat. Genet. 26: 118–123. [DOI] [PubMed] [Google Scholar]
- 60.Bassi M. T., Manzoni M., Monti E., Pizzo M. T., Ballabio A., Borsani G. 2000. Cloning of the gene encoding a novel integral membrane protein, mucolipidin, and identification of the two major founder mutations causing mucolipidosis type IV. Am. J. Hum. Genet. 67: 1110–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sun M., Goldin E., Stahl S., Falardeau J. L., Kennedy J. C., Acierno J. S., Jr, Bove C., Kaneski C. R., Nagle J., Bromley M. C., et al. 2000. Mucolipidosis type IV is caused by mutations in a gene encoding a novel transient receptor potential channel. Hum. Mol. Genet. 9: 2471–2478. [DOI] [PubMed] [Google Scholar]
- 62.Puertollano R., Kiselyov K. 2009. TRPMLs: in sickness and in health. Am. J. Physiol. Renal Physiol. 296: F1245–F1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Micsenyi M. C., Dobrenis K., Stephney G., Pickel J., Vanier M. T., Slaugenhaupt S. A., Walkley S. U. 2009. Neuropathology of the Mcoln1(-/-) knockout mouse model of mucolipidosis type IV. J. Neuropathol. Exp. Neurol. 68: 125–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dong X. P., Shen D., Wang X., Dawson T., Li X., Zhang Q., Cheng X., Zhang Y., Weisman L. S., Delling M., et al. 2010. PI(3,5)P(2) controls membrane trafficking by direct activation of mucolipin Ca(2+) release channels in the endolysosome. Nat. Commun. 1: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.LaPlante J. M., Sun M., Falardeau J., Dai D., Brown E. M., Slaugenhaupt S. A., Vassilev P. M. 2006. Lysosomal exocytosis is impaired in mucolipidosis type IV. Mol. Genet. Metab. 89: 339–348. [DOI] [PubMed] [Google Scholar]
- 66.Dong X. P., Wang X., Shen D., Chen S., Liu M., Wang Y., Mills E., Cheng X., Delling M., Xu H. 2009. Activating mutations of the TRPML1 channel revealed by proline-scanning mutagenesis. J. Biol. Chem. 284: 32040–32052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thompson E. G., Schaheen L., Dang H., Fares H. 2007. Lysosomal trafficking functions of mucolipin-1 in murine macrophages. BMC Cell Biol. 8: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sardiello M., Palmieri M., di Ronza A., Medina D. L., Valenza M., Gennarino V. A., Di Malta C., Donaudy F., Embrione V., Polishchuk R. S., et al. 2009. A gene network regulating lysosomal biogenesis and function. Science. 325: 473–477. [DOI] [PubMed] [Google Scholar]
- 69.Matteoni R., Kreis T. E. 1987. Translocation and clustering of endosomes and lysosomes depends on microtubules. J. Cell Biol. 105: 1253–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yogalingam G., Bonten E. J., van de Vlekkert D., Hu H., Moshiach S., Connell S. A., d’Azzo A. 2008. Neuraminidase 1 is a negative regulator of lysosomal exocytosis. Dev. Cell. 15: 74–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pshezhetsky A. V., Richard C., Michaud L., Igdouran S., Wang S., Elsliger M. A., Qu J., Leclerc D., Gravel R., Dallaire L., et al. 1997. Cloning, expression and chromosomal mapping of human lysosomal sialidase and characterization of mutations in sialidosis. Nat. Genet. 15: 316–320. [DOI] [PubMed] [Google Scholar]
- 72.Braun V., Fraisier V., Raposo G., Hurbain I., Sibarita J. B., Chavrier P., Galli T., Niedergang F. 2004. TI-VAMP/VAMP7 is required for optimal phagocytosis of opsonised particles in macrophages. EMBO J. 23: 4166–4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Piper R. C., Luzio J. P. 2004. CUPpling calcium to lysosomal biogenesis. Trends Cell Biol. 14: 471–473. [DOI] [PubMed] [Google Scholar]
- 74.Xu J., Toops K. A., Diaz F., Carvajal-Gonzalez J. M., Gravotta D., Mazzoni F., Schreiner R., Rodriguez-Boulan E., Lakkaraju A. 2012. Mechanism of polarized lysosome exocytosis in epithelial cells. J. Cell Sci. 125: 5937–5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Blott E. J., Griffiths G. M. 2002. Secretory lysosomes. Nat. Rev. Mol. Cell Biol. 3: 122–131. [DOI] [PubMed] [Google Scholar]
- 76.Mostov K., Werb Z. 1997. Journey across the osteoclast. Science. 276: 219–220. [DOI] [PubMed] [Google Scholar]
- 77.Chen G., Zhang Z., Wei Z., Cheng Q., Li X., Li W., Duan S., Gu X. 2012. Lysosomal exocytosis in Schwann cells contributes to axon remyelination. Glia. 60: 295–305. [DOI] [PubMed] [Google Scholar]
- 78.Cresswell P. 1994. Assembly, transport, and function of MHC class II molecules. Annu. Rev. Immunol. 12: 259–293. [DOI] [PubMed] [Google Scholar]
- 79.Kirkegaard T., Jäättelä M. 2009. Lysosomal involvement in cell death and cancer. Biochim. Biophys. Acta. 1793: 746–754. [DOI] [PubMed] [Google Scholar]
- 80.Corrotte M., Almeida P. E., Tam C., Castro-Gomes T., Fernandes M. C., Millis B. A., Cortez M., Miller H., Song W., Maugel T. K., et al. 2013. Caveolae internalization repairs wounded cells and muscle fibers. Elife. 2: e00926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.McNeil P. L., Kirchhausen T. 2005. An emergency response team for membrane repair. Nat. Rev. Mol. Cell Biol. 6: 499–505. [DOI] [PubMed] [Google Scholar]
- 82.Chakrabarti S., Kobayashi K. S., Flavell R. A., Marks C. B., Miyake K., Liston D. R., Fowler K. T., Gorelick F. S., Andrews N. W. 2003. Impaired membrane resealing and autoimmune myositis in synaptotagmin VII-deficient mice. J. Cell Biol. 162: 543–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cai C., Masumiya H., Weisleder N., Matsuda N., Nishi M., Hwang M., Ko J. K, Lin P., Thornton A., Zhao X., et al. 2009. MG53 nucleates assembly of cell membrane repair machinery. Nat. Cell Biol. 11: 56–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Huynh K. K., Kay J. G., Stow J. L., Grinstein S. 2007. Fusion, fission, and secretion during phagocytosis. Physiology (Bethesda). 22: 366–372. [DOI] [PubMed] [Google Scholar]
- 85.Aderem A., Underhill D. M. 1999. Mechanisms of phagocytosis in macrophages. Annu. Rev. Immunol. 17: 593–623. [DOI] [PubMed] [Google Scholar]
- 86.Shen D., Wang X., Li X., Zhang X., Yao Z., Dibble S., Dong X. P., Yu T., Lieberman A. P., Showalter H. D., et al. 2012. Lipid storage disorders block lysosomal trafficking by inhibiting a TRP channel and lysosomal calcium release. Nat. Commun. 3: 731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li X., Garrity A. G., Xu H. 2013. Regulation of membrane trafficking by signalling on endosomal and lysosomal membranes. J. Physiol. 591: 4389–4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Venkatachalam K., Long A. A., Elsaesser R., Nikolaeva D., Broadie K., Montell C. 2008. Motor deficit in a Drosophila model of mucolipidosis type IV due to defective clearance of apoptotic cells. Cell. 135: 838–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chamberlain L. H., Burgoyne R. D., Gould G. W. 2001. SNARE proteins are highly enriched in lipid rafts in PC12 cells: implications for the spatial control of exocytosis. Proc. Natl. Acad. Sci. USA. 98: 5619–5624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fraldi A., Annunziata F., Lombardi A., Kaiser H. J., Medina D. L., Spampanato C., Fedele A. O., Polishchuk R., Sorrentino N. C., Simons K., et al. 2010. Lysosomal fusion and SNARE function are impaired by cholesterol accumulation in lysosomal storage disorders. EMBO J. 29: 3607–3620. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 91.Maxfield F. R., Tabas I. 2005. Role of cholesterol and lipid organization in disease. Nature. 438: 612–621. [DOI] [PubMed] [Google Scholar]
- 92.Cheruku S. R., Xu Z., Dutia R., Lobel P., Storch J. 2006. Mechanism of cholesterol transfer from the Niemann-Pick type C2 protein to model membranes supports a role in lysosomal cholesterol transport. J. Biol. Chem. 281: 31594–31604. [DOI] [PubMed] [Google Scholar]
- 93.Ory D. S. 2004. The Niemann-Pick disease genes: regulators of cellular cholesterol homeostasis. Trends Cardiovasc. Med. 14: 66–72. [DOI] [PubMed] [Google Scholar]
- 94.Pentchev P. G. 2004. Niemann-Pick C research from mouse to gene. Biochim. Biophys. Acta. 1685: 3–7. [DOI] [PubMed] [Google Scholar]
- 95.Shen D., Wang X., Li X., Zhang X., Yao Z., Dibble S., Dong X. P., Yu T., Lieberman A. P., Showalter H. D., et al. Lipid storage disorders block lysosomal trafficking by inhibiting a TRP channel and lysosomal calcium release. Nat. Commun. 3: 731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chen F. W., Li C., Ioannou Y. A. 2010. Cyclodextrin induces calcium-dependent lysosomal exocytosis. PLoS ONE. 5: e15054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Di Paolo G., De Camilli P. 2006. Phosphoinositides in cell regulation and membrane dynamics. Nature. 443: 651–657. [DOI] [PubMed] [Google Scholar]
- 98.Rong Y., Liu M., Ma L., Du W., Zhang H., Tian Y., Cao Z., Li Y., Ren H., Zhang C., et al. 2012. Clathrin and phosphatidylinositol-4,5-bisphosphate regulate autophagic lysosome reformation. Nat. Cell Biol. 14: 924–934. [DOI] [PubMed] [Google Scholar]
- 99.Duex J. E., Nau J. J., Kauffman E. J., Weisman L. S. 2006. Phosphoinositide 5-phosphatase Fig 4p is required for both acute rise and subsequent fall in stress-induced phosphatidylinositol 3,5-bisphosphate levels. Eukaryot. Cell. 5: 723–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zolov S. N., Bridges D., Zhang Y., Lee W. W., Riehle E., Verma R., Lenk G. M., Converso-Baran K., Weide T., Albin R. L., et al. 2012. In vivo, Pikfyve generates PI(3,5)P2, which serves as both a signaling lipid and the major precursor for PI5P. Proc. Natl. Acad. Sci. USA. 109: 17472–17477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Shen D., Wang X., Xu H. 2011. Pairing phosphoinositides with calcium ions in endolysosomal dynamics: phosphoinositides control the direction and specificity of membrane trafficking by regulating the activity of calcium channels in the endolysosomes. Bioessays. 33: 448–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.McCartney A. J., Zhang Y., Weisman L. S. 2014. Phosphatidylinositol 3,5-bisphosphate: Low abundance, high significance. Bioessays. 36: 52–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Falkenburger B. H., Jensen J. B., Dickson E. J., Suh B. C., Hille B. 2010. Phosphoinositides: lipid regulators of membrane proteins. J. Physiol. 588: 3179–3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wang X., Zhang X., Dong X. P., Samie M., Li X., Cheng X., Goschka A., Shen D., Zhou Y., Harlow J., et al. 2012. TPC proteins are phosphoinositide-activated sodium-selective ion channels in endosomes and lysosomes. Cell. 151: 372–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Li X., Wang X., Zhang X., Zhao M., Tsang W. L., Zhang Y., Yau R. G., Weisman L. S., Xu H. 2013. Genetically encoded fluorescent probe to visualize intracellular phosphatidylinositol 3,5-bisphosphate localization and dynamics. Proc. Natl. Acad. Sci. USA. 110: 21165–21170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Dehay B., Bové J., Rodríguez-Muela N., Perier C., Recasens A., Boya P., Vila M. 2010. Pathogenic lysosomal depletion in Parkinson’s disease. J. Neurosci. 30: 12535–12544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tsunemi T., Ashe T. D., Morrison B. E., Soriano K. R., Au J., Roque R. A., Lazarowski E. R., Damian V. A., Masliah E., Spada A. R. 2012. PGC-1α rescues Huntington’s disease proteotoxicity by preventing oxidative stress and promoting TFEB function. Sci. Transl. Med. 4: 142ra97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sardiello M., Ballabio A. 2009. Lysosomal enhancement: a CLEAR answer to cellular degradative needs. Cell Cycle. 8: 4021–4022. [DOI] [PubMed] [Google Scholar]
- 109.Duex J. E., Tang F., Weisman L. S. 2006. The Vac14p-Fig4p complex acts independently of Vac7p and couples PI3,5P2 synthesis and turnover. J. Cell Biol. 172: 693–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Grimm C., Hassan S., Wahl-Schott C., Biel M. 2012. Role of TRPML and two-pore channels in endolysosomal cation homeostasis. J. Pharmacol. Exp. Ther. 342: 236–244. [DOI] [PubMed] [Google Scholar]