Abstract
The MYC proto-oncogene has been implicated in the pathogenesis of most types of human tumors. MYC activation alone in many normal cells is restrained from causing tumorigenesis through multiple genetic and epigenetically controlled checkpoint mechanisms, including proliferative arrest, apoptosis, and cellular senescence. When pathologically activated in a permissive epigenetic and/or genetic context, MYC bypasses these mechanisms, enforcing many of the “hallmark” features of cancer, including relentless tumor growth associated with DNA replication and transcription, cellular proliferation and growth, protein synthesis, and altered cellular metabolism. MYC mandates tumor cell fate, by inducing stemness and blocking cellular senescence and differentiation. Additionally, MYC orchestrates changes in the tumor microenvironment, including the activation of angiogenesis and suppression of the host immune response. Provocatively, brief or even partial suppression of MYC back to its physiological levels of activation can result in the restoration of intrinsic checkpoint mechanisms, resulting in acute and sustained tumor regression, associated with tumor cells undergoing proliferative arrest, differentiation, senescence, and apoptosis, as well as remodeling of the tumor microenvironment, recruitment of an immune response, and shutdown of angiogenesis. Hence, tumors appear to be “addicted” to MYC because of both tumor cell–intrinsic, cell-autonomous and host-dependent, immune cell–dependent mechanisms. Both the trajectory and persistence of many human cancers require sustained MYC activation. Multiscale mathematical modeling may be useful to predict when tumors will be addicted to MYC. MYC is a hallmark molecular feature of both the initiation and maintenance of tumorigenesis.
Many cancers require sustained MYC activation. MYC induces tumorigenesis by evading multiple tumor-suppressing mechanisms (e.g., apoptosis). But on MYC suppression, these mechanisms are restored.
The MYC proto-oncogene was first discovered as an etiologic agent of retrovirally mediated tumorigenesis. Later, MYC was illustrated to be activated through chromosomal translocation in Burkitt lymphoma (see Conacci-Sorrell et al. 2014). MYC is also commonly activated in tumorigenesis as a consequence of both oncogenic and epigenetic events (Boxer and Dang 2001; Eilers and Eisenman 2008; Dang 2012). Indeed, MYC is overexpressed and/or activated in more than half of human cancers (Escot et al. 1986; Ladanyi et al. 1993; Gamberi et al. 1998; Kawate et al. 1999; Stock et al. 2000; Boxer and Dang 2001).
MYC largely functions as a transcription factor that coordinates many biological processes (Dang et al. 1999). MYC activation can usurp these programs, resulting in the cardinal hallmark features of cancer. Thus, MYC activation contributes to autonomous proliferation and growth, relentless DNA replication, increased protein biogenesis, global changes in cellular metabolism, activation of the angiogenic switch, suppression of the response to autocrine and paracrine regulatory programs, and a restraint of host immune responses (Felsher 2003; Shachaf and Felsher 2005; van Riggelen et al. 2010b; Bachireddy et al. 2012; Dang 2012). Hence, MYC activation appears to be a molecular hallmark of cancer.
In this review, we examine the notion that MYC activation is one of the necessary events for the initiation of tumorigenesis and frequently results in tumor survival being dependent on high levels of MYC, herein referred to as MYC addiction.
MYC AS AN INITIATOR OF TUMORIGENESIS
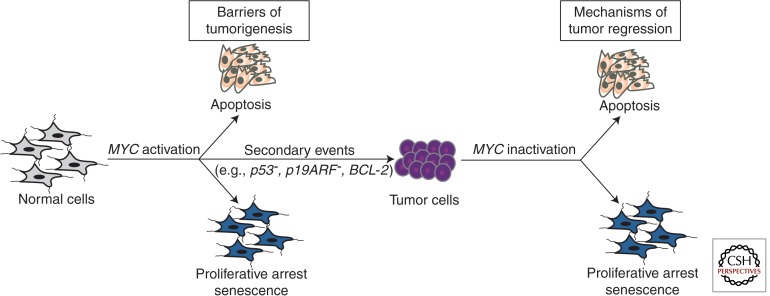
MYC is one of the most potent oncogenes as measured by many in vitro and in vivo assays for cell transformation phenotypes (Sheiness et al. 1978; Alitalo et al. 1983). However, MYC activation alone generally cannot induce tumorigenesis. Indeed, when MYC was first observed to induce neoplastic “transformation,” this was detected only in specific cell lines that were presumed to have acquired other genetic events that rendered them permissive (Spencer and Groudine 1991; Dang 1999). Thus, although it is one of the most commonly activated oncogenes implicated in the pathogenesis of human cancers, MYC overexpression alone is surprisingly incapable of inducing cellular proliferation or neoplastic transformation of most normal human cells. Instead, MYC overexpression in normal cells can have no effects or can be highly destructive, culminating in proliferative arrest, senescence, and/or apoptosis (Fig. 1) (Evan et al. 1992; Gibson et al. 1995; Felsher and Bishop 1999b; Grandori et al. 2003; Nilsson and Cleveland 2003; Hoffman and Liebermann 2008).
Figure 1.
MYC-induced cancer initiation and maintenance. MYC induces tumorigenesis by evading multiple tumor-suppressing checkpoint mechanisms, including proliferative arrest, apoptosis, and/or senescence. On MYC suppression these barriers are restored, enabling sustained tumor regression.
MYC overexpression has been observed to enforce DNA replication and entry into S phase (Cerni et al. 1986; Mai et al. 1996; Felsher et al. 2000; Dominguez-Sola et al. 2007). Indeed, MYC is part of the replication complex (Dominguez-Sola et al. 2007; Srinivasan et al. 2013; see Dominguez-Sola and Gautier 2014 and Kuzyk and Mai 2014). However, MYC alone cannot cause mitotic cellular division (Felsher et al. 2000). In some cases MYC causes normal cells to grow and replicate DNA, but they cannot divide; rather, these cells become polyploid (Mai et al. 1996; Dang 1999; Johnston et al. 1999; Felsher et al. 2000; Neto-Silva et al. 2010). Moreover, MYC overexpression can enforce replication in a manner that results in DNA breaks (Karlsson et al. 2003a). This appears to be the consequence of several mechanisms. MYC may directly block double-stranded DNA repair and/or increase oxidative damage, causing DNA damage (Vafa et al. 2002; Karlsson et al. 2003a; Ray et al. 2006). MYC overexpression can cause normal cells to undergo proliferative arrest under some circumstances (Felsher et al. 2000) and cellular senescence in other cases (Grandori et al. 2003). Thus, MYC deregulation alone cannot force complete transit through the cell division cycle.
The consequences of MYC overexpression in a normal cell are also dependent on epigenetic and genetic contexts. MYC overexpression in the embryonic liver induces cellular proliferation, whereas in adult liver it promotes cellular growth without mitotic division that is associated with polyploidy (Beer et al. 2004). Circumstances in an adult host that promote hepatocytes to proliferate, such as a partial hepatectomy or treatment with a liver toxin, can enable MYC to induce cellular proliferation in adult hepatocytes more readily (Makino et al. 1984; Beer et al. 2004). Similarly, the loss of the tumor suppressor p53 cooperates with MYC to induce cellular proliferation and tumorigenesis in adult hepatocytes (Beer et al. 2004). Thus, cellular context and specific genetic defects can enable MYC to more readily induce proliferation and tumorigenesis.
The gene dosage of MYC strongly influences the consequences of its activation. Highly robust activation of MYC is more commonly associated with DNA damage and apoptosis; conversely, less robust MYC activation appears to be associated with proliferative arrest and cellular senescence (Felsher et al. 2000; Grandori et al. 2003). Similarly, the gene dosage of MYC appears to strongly influence the consequences on cellular proliferation versus apoptosis (Murphy et al. 2008). Thus, the level and context of MYC dictate the consequences of its activation.
MYC COOPERATES TO INDUCE TUMORIGENESIS
MYC cooperates with many other oncogenic events to initiate tumorigenesis (Murray et al. 1983; Compere et al. 1989; DeoCampo et al. 2000; Welm et al. 2005; Clegg et al. 2011). Many oncogenes were first discovered as cooperating events in screens of MYC-induced tumor formation (Jonkers and Berns 1996; Mikkers et al. 2002; Mikkers and Berns 2003; Kool and Berns 2009; Mendrysa et al. 2010). Genetic events that abrogate cell-cycle checkpoints critical to the regulation of proliferative arrest, apoptosis, and/or senescence frequently synergize with MYC to induce proliferation as well as malignant transformation. Examples include the overexpression of BCL-2, loss of p53, or loss of p19ARF (Green 1997; Zindy et al. 1998; Jacobs et al. 1999; Schmitt et al. 1999; Schmitt and Lowe 2001). It is now axiomatic that normal cells possess multiple “intrinsic” mechanisms of tumor suppression that prevent malignant transformation by individual oncogenes such as MYC (Lowe et al. 2004).
Examination of the consequences of MYC activation using in vivo models has identified, in addition to cell-autonomous mechanisms, host-dependent mechanisms that influence MYC’s ability to initiate tumorigenesis. Thus, toxins or carcinogens associated with the activation of cellular proliferation can cooperate with MYC to induce tumorigenesis (Beer et al. 2008). Similarly, autocrine mechanisms involving the expression of transforming growth factor-α as well as other cytokines are critical in tumor initiation and maintenance, as described below (Calvisi and Thorgeirsson 2005; Cavin et al. 2005). Both innate immunity (Reimann et al. 2010) and direct autocrine effects (van Riggelen et al. 2010a) contribute to the suppression of tumorigenesis.
Finally, the precise physiological state of the cell is likely to influence the consequences of MYC activation. The particular stage of differentiation of a cellular lineage may determine the consequences of MYC activation. For example, MYC activation in embryonic hepatocytes induces robust cellular proliferation, but in adult cells MYC activation induces DNA replication associated with mitotic arrest and results in hyperdiploid cells that cannot become malignantly transformed (Beer et al. 2004). Similarly, MYC expression in embryonic heart induces hyperplasia, whereas in the adult heart MYC induces hypertrophy (DW Felsher, unpubl.). The mechanistic basis for these differences may relate to the recent findings that MYC amplifies the output of previously activated gene expression programs (Lin et al. 2012; Nie et al. 2012; see Levens 2013 and Rahl and Young 2014). Such expression programs would be expected to differ depending on cell type and stage of development. Thus, the phenotypic consequences of MYC expression may be influenced not just by cooperating genetic events but also perhaps by nongenetic or even epigenetic mechanisms.
Generally, MYC initiates tumorigenesis in a permissive epigenetic context that results from tumor-intrinsic mechanisms that regulate proliferation, apoptosis, and innate and adaptive immunity. Host programs may include modulation of the tumor microenvironment and the regulation of angiogenesis. Other genetic events may be required to perturb the normal regulation of the microenvironment, thus accelerating tumorigenesis. Changes in the microenvironment may create a circumstance that is more generally permissive for tumorigenesis. Thus, the local microenvironment may also contribute to susceptibility to MYC-induced tumorigenesis.
MYC-INDUCED TUMORS ARE ONCOGENE ADDICTED
The notion that cancer is a genetic disease associated with discrete and definable events that activate oncogenes and/or inactivate tumor suppressor genes suggested a rationale for the treatment of cancer. The repair of these mutant gene products could, in theory, reverse cancer (Bowden et al. 1994; Luo et al. 2009). Yet several questions arise: How does one decide which and how many events must be targeted? Will cancers exhibit genetic instability that enables the ready acquisition of compensatory mutations? Does an oncogene need to be mutated to be essential for maintenance of a neoplastic state? Could targeting oncogenes be toxic because most proto-oncogenes are essential?
To some investigators, the question of the potential efficacy of targeting oncogenes was moot, because the use of antisense oligonucleotides has established the potential utility of oncogene addiction (Tamm et al. 2001; Dias and Stein 2002; Pastorino et al. 2004; Maksimenko and Malvy 2005; Stein et al. 2005). However, many critics have been dismissive and have postulated that these results were nonspecific, obtained largely through in vitro study and unlikely to be relevant to more complex human tumors.
To experimentally address whether cancer is reversible, many research groups examined whether the conditional activation of an oncogene could induce reversible tumorigenesis. The tactic has been to use transgenic mouse models with a conditional oncogene. Mouse models using the Tet system and/or chimeric gene products that could be activated in an on/off fashion are the two most common approaches (Giuriato et al. 2004; Arvanitis and Felsher 2005; Felsher 2006). Many investigators have focused on MYC.
The suppression of MYC was shown to reverse tumorigenesis. Similar results were seen in a wide variety of tumors, including hematopoietic tumors (T- and B-cell lymphoma and leukemia), epithelial tumors (hepatocellular, breast, squamous carcinoma), and mesenchymal tumors (osteogenic sarcoma) (Felsher and Bishop 1999a; Jain et al. 2002; Pelengaris et al. 2002; Marinkovic et al. 2004). Identical results have been observed for several other oncogenes (Table 1) (Chin et al. 1999; Felsher and Bishop 1999a; Huettner et al. 2000; D’Cruz et al. 2001; Fisher et al. 2001; Jain et al. 2002; Marinkovic et al. 2004; Shachaf et al. 2004; Lawlor et al. 2006; Ji et al. 2007; Li et al. 2007; Chakravarty et al. 2011). Importantly, in at least some cases, it was confirmed that these tumors were clonal and genetically complex (Karlsson et al. 2003b). In the experimental mouse model, MYC-induced tumorigenesis is reversible. Importantly, endogenous MYC was not suppressed in these experiments.
Table 1.
Models illustrating MYC-associated oncogene addiction
| Oncogene | Conditional mouse model | Reference |
|---|---|---|
| MYC | T-cell acute lymphoblastic leukemia | Felsher and Bishop 1999a |
| Hepatocellular carcinoma | Shachaf et al. 2004 | |
| Osteosarcoma | Jain et al. 2002 | |
| T- and B-cell acute lymphoblastic leukemia | Marinkovic et al. 2004 | |
| Mammary adenocarcinoma | D’Cruz et al. 2001 | |
| Islet tumors | Lawlor et al. 2006 | |
| RAS | Melanoma (HRAS) | Chin et al. 1999 |
| Lung adenocarcinoma (KRAS) | Fisher et al. 2001 | |
| BRAF | Thyroid cancer | Chakravarty et al. 2011 |
| Lung adenocarcinoma | Ji et al. 2007 | |
| EGFR | Lung adenocarcinoma | Li et al. 2007 |
| BCR-ABL | B-cell acute lymphoblastic leukemia | Huettner et al. 2000 |
Yet it is important to note that MYC-induced tumorigenesis is not always reversible. In some genetic contexts MYC suppression resulted in the initial regression of a tumor that subsequently recurred. The introduction of additional genetic events, such as a mutant RAS, can abrogate the reversibility of MYC-induced breast adenocarcinoma (D’Cruz et al. 2001). Similarly, loss of p53 expression also prevents MYC-induced lymphoma from being reversible (Giuriato et al. 2006). However, when examined, all tumors that recurred after MYC suppression had reactivated MYC expression (Choi et al. 2011). Thus, tumors may not be able to completely escape MYC addiction.
Importantly, MYC-induced tumorigenesis may be reversible even when MYC is not the initiating oncogenic lesion. Using a dominant negative MYC, designated “omoMYC,” investigators illustrated that conditional suppression of MYC appears to reverse RAS-induced tumorigenesis (Soucek et al. 2008, 2013). However, it is also possible that RAS is activating endogenous MYC, an event that could possibly explain why these tumors exhibit MYC dependence. Indeed, the mechanism of action of omoMYC is not entirely clear. Recent observations suggest that omoMYC blocks some of the interactions between MYC and some of its partners (Savino et al. 2011). Less clear is whether it mediates effects independently of MYC. Further insight into the mechanism of omoMYC is likely to provide compelling clues to how best to target MYC therapeutically. Thus, addiction to MYC appears to be a feature of cancers driven by oncogenes other than MYC.
MECHANISMS OF MYC-ASSOCIATED ONCOGENE ADDICTION
Given that many experimental models demonstrated a dependency on continuous MYC expression for tumor progression, the key challenge to understanding MYC oncogene addiction is the delineation of the mechanisms underlying this dependency (Weinstein 2002; Sharma and Settleman 2007; Felsher 2008; Weinstein and Joe 2008). The inactivation of MYC is associated with sustained reversal of many of the hallmark features of tumorigenesis. On MYC inactivation, tumor cells appear to restore normal checkpoint mechanisms and undergo proliferative arrest, differentiation, apoptosis, and/or cellular senescence. In addition to cell-intrinsic mechanisms of MYC addiction, the tumor microenvironment is also remodeled on MYC inactivation, which results in the restoration of normal tissue architecture (Felsher and Bishop 1999a). Tumor angiogenesis is diminished with MYC withdrawal (Giuriato et al. 2006). Thus, there appears to be a restoration of normal physiologic programs both within the tumor cell and in the tumor microenvironment.
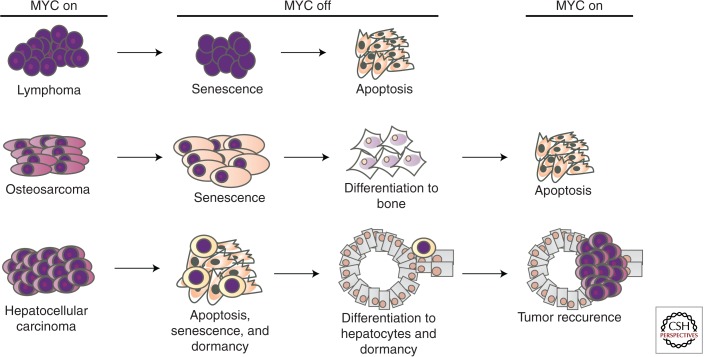
The specific mechanisms of MYC addiction appear to be tumor type specific (Fig. 2). Hematopoietic tumors appear to undergo proliferative arrest, differentiation, and senescence, followed by robust apoptosis on MYC withdrawal (Felsher and Bishop 1999a). In contrast, sarcomas undergo proliferative arrest, differentiation, and senescence with minimal if any evidence for apoptosis (Jain et al. 2002). Epithelial tumors appear to have two major fates; most tumor cells undergo proliferative arrest, senescence, and apoptosis, but a subpopulation of tumor cells undergoes differentiation into seemingly normal cells but actually exhibit evidence of tumor dormancy (Boxer et al. 2004; Shachaf et al. 2004). Whether these variations reflect programs unique to specific cell types or are a consequence of differences in the genetic events associated with each type of cancer is not known.
Figure 2.
Consequences of MYC inactivation in multiple types of cancer. MYC inactivation elicits oncogene addiction by multiple mechanisms that differ depending on tumor type. MYC inactivation in lymphoma induces proliferative arrest, differentiation/senescence, and widespread apoptosis. MYC inactivation in osteosarcoma induces proliferative arrest and differentiation/senescence but not apoptosis. MYC reactivation does not restore tumorigenesis. MYC inactivation in liver adenocarcinoma induces proliferative arrest, differentiation/senescence, and apoptosis. MYC reactivation can result in restoration of the tumor.
BRIEF OR PARTIAL SUPPRESSION OF MYC CAN REVERSE TUMORIGENESIS
A brief 2-day or partial (twofold decrease) suppression of MYC can result in sustained tumor regression. Brief suppression of MYC is associated with an irreversible change in the cellular program; in some contexts the tumors cannot be restored on MYC reactivation (Jain et al. 2002). Similarly, a twofold decrease in oncogenic levels of MYC was sufficient to induce tumor regression (Shachaf and Felsher 2005). This is tumor type specific, because lymphoma and osteosarcoma exhibit this phenotype (Jain et al. 2002; Giuriato et al. 2006), whereas epithelial tumors such as hepatocellular or breast carcinoma do not (Boxer et al. 2004; Shachaf et al. 2004).
In osteogenic sarcoma MYC suppression results in terminal cellular differentiation from osteoblasts into differentiated osteocytes (which are associated with bone formation in vivo) (Jain et al. 2002). The reactivation of MYC not only fails to restore the cancer but either has no consequence or is associated with apoptosis. Microarray analysis revealed that MYC suppression is associated with irreversible changes in gene expression as a result of the inability of MYC to bind to the promoters of many of these genes. In particular, MYC suppression results in the permanent shutdown of genes related to ribosome biosynthesis and protein synthesis (Wu et al. 2008; van Riggelen et al. 2010b).
Partial suppression of MYC can also result in sustained tumor regression. Notably, in this case the levels of MYC were below that of human tumor-derived cell lines and above that of proliferating normal human cells or Epstein–Barr virus–transformed lymphocytes (Shachaf et al. 2008). Thus, there appears to be a threshold level of MYC that is required to sustain a malignant phenotype (Shachaf et al. 2008). Protein and gene expression analysis identified many specific changes, but notably, ribosomal gene products were suppressed. Collectively, these results suggest that a global shift in protein biogenesis is an important feature of how MYC suppression results in tumor regression (Ruggero and Pandolfi 2003).
MYC activation is also associated with global changes in the energy metabolism of cancer cells. These changes may make tumors particularly susceptible to the inhibition of enzymes that are essential for energy metabolism (Dang 1999, 2013; Dang et al. 2009; O’Shea and Ayer 2013). Hence, the addiction to MYC observed in many cancer cells could at least in part relate to acute changes in metabolism. The suppression of MYC may induce tumor regression by acutely disrupting the ability of tumor cells to maintain sufficient metabolism to sustain survival and/or by directly regulating death signaling (see Dang 2013; O’Shea and Ayer 2013; Morrish and Hockenbery 2014).
An important implication of these results is that it may be sufficient to partially and/or briefly suppress MYC expression in at least some tumor types to induce a sustained clinical effect on human disease. That transient inactivation of MYC is effective may be due to the dependence of MYC-associated oncogene addiction on molecular features that are dictated shortly after oncogene inactivation (Tran et al. 2011). Whether tumor inhibition is entirely cell autonomous or results from a delayed host-dependent mechanism remains to be determined. For example, the host immune system seems to be critical for tumor regression on withdrawal of MYC.
MYC ONCOGENE ADDICTION AND THE IMMUNE SYSTEM
In addition to cell-autonomous effects of ectopic MYC expression, alterations in immune surveillance mechanisms by MYC may also contribute to tumorigenesis (Rooney et al. 1985; Soucek et al. 2007; Grivennikov et al. 2010). Indeed, we found that MYC inactivation induces tumor regression through both tumor cell–intrinsic as well as host-dependent mechanisms (Rakhra et al. 2010). MYC inactivation in an immunocompromised RAG1−/− host (deficient for B and T cells) or a CD4−/− host (deficient for CD4+ T-helper cells) exhibited reduced kinetics of tumor regression, increased minimal residual disease, and inevitable tumor recurrence. An immunocompetent host is hence essential for tumor regression on MYC withdrawal.
In situ analysis following MYC inactivation showed that the absence of host immune effectors had little effect on the proliferative arrest or apoptosis in the tumor, but the absence of immune effectors largely abrogated cellular senescence and diminished angiogenesis. Thrombospondins were implicated as one of the critical effectors. Similarly, suppression of MYC through omoMYC induces changes in the tumor microenvironment associated with tumor regression (Eilers and Eisenman 2008; Sodir et al. 2011). These observations illustrate that the suppression of MYC mediates tumorigenesis both through direct effects on tumor cells as well as through alteration of immunity (Restifo 2010).
Thus, oncogene addiction may occur via mechanisms that operate on multiple levels. First, there is a tumor cell–intrinsic induction of proliferative arrest, senescence, and apoptosis. Second, there is a recruitment of immune effectors that is probably mediated by a noncanonical CD4+ T-cell-specific mechanism. Third, there is a remodeling of the tumor microenvironment. The initial regression of a tumor is cell autonomous, but complete regression requires host-dependent mechanisms (Fig. 3).
Figure 3.
MYC inactivation elicits tumor regression through both cell-autonomous and non-cell-autonomous mechanisms of tumor regression. MYC activation leads to tumorigenesis through suppression of critical safeguards such as apoptosis, proliferative arrest, differentiation, and senescence. Activation of MYC also facilitates engagement of hallmarks of tumor growth, as well as cell-extrinsic phenomena such as host immunity. TGF, transforming growth factor.
An understanding of the role of the host immune system in the mechanism by which oncogene inactivation induces tumor regression could be important for development of cancer therapeutics. Therapies identified only through in vitro screens of isolated tumor cells are unlikely to accurately predict the consequences of oncogene suppression. Therapies that concomitantly target oncogenes but suppress the immune system may be counterproductive. A combination approach that targets oncogenes but also activates specific immune mechanisms may be complementary and most efficacious.
MODELING AND PREDICTING MYC-ASSOCIATED ONCOGENE ADDICTION
To understand the factors contributing to MYC oncogene addiction, differential changes in survival and death signals were mathematically modeled (Tran et al. 2011). Oncogene suppression could increase death signals, could suppress survival signals, or could activate or suppress both survival and death signals but with death outpacing survival. What was found through modeling was that most tumors regress because both survival and death signals dissipate on oncogene suppression, with death signals decaying at a slower rate. The specific mechanistic basis for differential decay of survival versus death signaling on MYC withdrawal is not known but may be related to differential regulation of the effectors of survival and death (Sharma et al. 2006; Tran et al. 2011), differential levels of metabolites that regulate or are required for survival or death (Wise et al. 2008; Gao et al. 2009), and/or non-cell-autonomous mechanisms such as changes in autocrine or paracrine signaling from the host (Eilers and Eisenman 2008).
The mathematical modeling described could at least in part reconcile how the activation of MYC during tumor initiation in normal cells and the inactivation of MYC in an established tumor both can induce apoptosis. MYC activation can induce apoptosis in many cells that can serve as a barrier to tumorigenesis, unless there is at least a partial defect in programs, for example, through mutation of p53 function, accelerating tumor progression (Lowe et al. 2004). However, MYC inactivation in a tumor seemingly paradoxically results in robust apoptosis because the reduction in proliferation and survival signals occurs much more rapidly and completely than the associated attenuation of apoptotic signals. Thus, although MYC inactivation in a tumor may suppress some mechanisms that contribute to apoptotic signals, the overall effect is overwhelming tumor regression because of the concurrent complete and rapid suppression of proliferation, combined with the suppression of prosurvival signals. Even in the circumstance of attenuated apoptosis signals, in summation, the dominant effect is the overall marked increase in cell death.
CONCLUDING REMARKS
Experimental evidence supporting the occurrence of MYC addiction in animal models suggests that some human cancers could also be addicted to MYC and hence could be treated by targeting MYC. However, MYC has yet to be successfully therapeutically targeted for the treatment of cancer, as most transcription factors are generally considered “undruggable” (Verdine and Walensky 2007; Filippakopoulos et al. 2010; Delmore et al. 2011; Yan and Higgins 2013). Nevertheless, several reports suggest that small interfering RNA, short hairpin RNA, and antisense oligonucleotides could be potential strategies to target MYC (Hermeking 2003; Pastorino et al. 2004; Vita and Henriksson 2006). Therapies that briefly or even partially suppress MYC directly or indirectly may be highly efficacious in the reversal of neoplasia, for example, as illustrated by inhibition via statins or BRD4 inhibition (Shachaf et al. 2007; Cao et al. 2011; Delmore et al. 2011; McKeown and Bradner 2014). Alternatively, synthetic lethal screens may identify therapeutic strategies to target MYC-addicted tumors (Yang et al. 2010; Kessler et al. 2012; Toyoshima et al. 2012; Cermelli et al. 2014).
ACKNOWLEDGMENTS
The authors acknowledge all current and past members of the Felsher lab for their contributions in characterizing various models of oncogene addiction. Our research has been generously funded by grants from the following government agencies and private foundations: National Cancer Institute (R01CA17037801, R01CA89305, R01CA105102, P50CA114747, P01CA034233, and U56CA112973), the Leukemia and Lymphoma Society (R6223-07), and Burroughs Welcome Fund Career Award.
Footnotes
Editors: Chi V. Dang and Robert N. Eisenman
Additional Perspectives on MYC and the Pathway to Cancer available at www.perspectivesinmedicine.org
REFERENCES
*Reference is also in this collection.
- Alitalo K, Bishop JM, Smith DH, Chen EY, Colby WW, Levinson AD 1983. Nucleotide sequence to the v-myc oncogene of avian retrovirus MC29. Proc Natl Acad Sci 80: 100–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvanitis C, Felsher DW 2005. Conditionally MYC: Insights from novel transgenic models. Cancer Lett 226: 95–99 [DOI] [PubMed] [Google Scholar]
- Bachireddy P, Rakhra K, Felsher DW 2012. Immunology in the clinic review series; focus on cancer: Multiple roles for the immune system in oncogene addiction. Clin Exp Immunol 167: 188–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beer S, Zetterberg A, Ihrie RA, McTaggart RA, Yang Q, Bradon N, Arvanitis C, Attardi LD, Feng S, Ruebner B, et al. 2004. Developmental context determines latency of MYC-induced tumorigenesis. PLoS Biol 2: e332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beer S, Komatsubara K, Bellovin DI, Kurobe M, Sylvester K, Felsher DW 2008. Hepatotoxin-induced changes in the adult murine liver promote MYC-induced tumorigenesis. PloS ONE 3: e2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowden GT, Schneider B, Domann R, Kulesz-Martin M 1994. Oncogene activation and tumor suppressor gene inactivation during multistage mouse skin carcinogenesis. Cancer Res 54: 1882s–1885s [PubMed] [Google Scholar]
- Boxer LM, Dang CV 2001. Translocations involving c-myc and c-myc function. Oncogene 20: 5595–5610 [DOI] [PubMed] [Google Scholar]
- Boxer RB, Jang JW, Sintasath L, Chodosh LA 2004. Lack of sustained regression of c-MYC-induced mammary adenocarcinomas following brief or prolonged MYC inactivation. Cancer Cell 6: 577–586 [DOI] [PubMed] [Google Scholar]
- Calvisi DF, Thorgeirsson SS 2005. Molecular mechanisms of hepatocarcinogenesis in transgenic mouse models of liver cancer. Toxicologic Pathol 33: 181–184 [DOI] [PubMed] [Google Scholar]
- Cao Z, Fan-Minogue H, Bellovin DI, Yevtodiyenko A, Arzeno J, Yang Q, Gambhir SS, Felsher DW 2011. MYC phosphorylation, activation, and tumorigenic potential in hepatocellular carcinoma are regulated by HMG-CoA reductase. Cancer Res 71: 286–2297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavin LG, Wang F, Factor VM, Kaur S, Venkatraman M, Thorgeirsson SS, Arsura M 2005. Transforming growth factor-α inhibits the intrinsic pathway of c-Myc-induced apoptosis through activation of nuclear factor-κB in murine hepatocellular carcinomas. Mol Cancer Res 3: 403–412 [DOI] [PubMed] [Google Scholar]
- *.Cermelli S, Jang IS, Bernard B, Grandori C 2014. Synthetic lethal screens as a means to understand and treat MYC-driven cancers. Cold Spring Harb Perspect Med 4: a014209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerni C, Mougneau E, Zerlin M, Julius M, Marcu KB, Cuzin F 1986. c-myc and functionally related oncogenes induce both high rates of sister chromatid exchange and abnormal karyotypes in rat fibroblasts. Curr Top Microbiol Immunol 132: 193–201 [DOI] [PubMed] [Google Scholar]
- Chakravarty D, Santos E, Ryder M, Knauf JA, Liao XH, West BL, Bollag G, Kolesnick R, Thin TH, Rosen N, et al. 2011. Small-molecule MAPK inhibitors restore radioiodine incorporation in mouse thyroid cancers with conditional BRAF activation. J Clin Invest 121: 4700–4711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin L, Tam A, Pomerantz J, Wong M, Holash J, Bardeesy N, Shen Q, O’Hagan R, Pantginis J, Zhou H, et al. 1999. Essential role for oncogenic Ras in tumour maintenance. Nature 400: 468–472 [DOI] [PubMed] [Google Scholar]
- Choi PS, van Riggelen J, Gentles AJ, Bachireddy P, Rakhra K, Adam SJ, Plevritis SK, Felsher DW 2011. Lymphomas that recur after MYC suppression continue to exhibit oncogene addiction. Proc Natl Acad Sci 108: 17432–17437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clegg NJ, Couto SS, Wongvipat J, Hieronymus H, Carver BS, Taylor BS, Ellwood-Yen K, Gerald WL, Sander C, Sawyers CL 2011. MYC cooperates with AKT in prostate tumorigenesis and alters sensitivity to mTOR inhibitors. PloS ONE 6: e17449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compere SJ, Baldacci P, Sharpe AH, Thompson T, Land H, Jaenisch R 1989. The ras and myc oncogenes cooperate in tumor induction in many tissues when introduced into midgestation mouse embryos by retroviral vectors. Proc Natl Acad Sci 86: 2224–2228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Conacci-Sorrell M, McFerrin L, Eisenman RN 2014. An overview of MYC and its interactome. Cold Spring Harb Perspect Med 4: a014357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV 1999. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol Cell Biol 19: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV 2012. MYC on the path to cancer. Cell 149: 22–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Dang CV 2013. MYC, metabolism, cell growth, and tumorigenesis. Cold Spring Harb Perspect Med 3: a014217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV, Resar LM, Emison E, Kim S, Li Q, Prescott JE, Wonsey D, Zeller K 1999. Function of the c-Myc oncogenic transcription factor. Exp Cell Res 253: 63–77 [DOI] [PubMed] [Google Scholar]
- Dang CV, Le A, Gao P 2009. MYC-induced cancer cell energy metabolism and therapeutic opportunities. Clin Cancer Res 15: 6479–6483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Cruz CM, Gunther EJ, Boxer RB, Hartman JL, Sintasath L, Moody SE, Cox JD, Ha SI, Belka GK, Golant A, et al. 2001. c-MYC induces mammary tumorigenesis by means of a preferred pathway involving spontaneous Kras2 mutations. Nat Med 7: 235–239 [DOI] [PubMed] [Google Scholar]
- Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, Kastritis E, Gilpatrick T, Paranal RM, Qi J, et al. 2011. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 146: 904–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeoCampo ND, Wilson MR, Trosko JE 2000. Cooperation of bcl-2 and myc in the neoplastic transformation of normal rat liver epithelial cells is related to the down-regulation of gap junction-mediated intercellular communication. Carcinogenesis 21: 1501–1506 [PubMed] [Google Scholar]
- Dias N, Stein CA 2002. Antisense oligonucleotides: Basic concepts and mechanisms. Mol Cancer Ther 1: 347–355 [PubMed] [Google Scholar]
- *.Dominguez-Sola D, Gautier J 2014. MYC and the control of DNA replication. Cold Spring Harb Perspect Med 10.1101/cshperspect.a014423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Sola D, Ying CY, Grandori C, Ruggiero L, Chen B, Li M, Galloway DA, Gu W, Gautier J, Dalla-Favera R 2007. Non-transcriptional control of DNA replication by c-Myc. Nature 448: 445–451 [DOI] [PubMed] [Google Scholar]
- Eilers M, Eisenman RN 2008. Myc’s broad reach. Genes Dev 22: 2755–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escot C, Theillet C, Lidereau R, Spyratos F, Champeme MH, Gest J, Callahan R 1986. Genetic alteration of the c-myc protooncogene (MYC) in human primary breast carcinomas. Proc Natl Acad Sci 83: 4834–4838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evan GI, Wyllie AH, Gilbert CS, Littlewood TD, Land H, Brooks M, Waters CM, Penn LZ, Hancock DC 1992. Induction of apoptosis in fibroblasts by c-myc protein. Cell 69: 119–128 [DOI] [PubMed] [Google Scholar]
- Felsher DW 2003. Cancer revoked: Oncogenes as therapeutic targets. Nat Rev Cancer 3: 375–380 [DOI] [PubMed] [Google Scholar]
- Felsher DW 2006. Tumor dormancy: Death and resurrection of cancer as seen through transgenic mouse models. Cell Cycle 5: 1808–1811 [DOI] [PubMed] [Google Scholar]
- Felsher DW 2008. Oncogene addiction versus oncogene amnesia: Perhaps more than just a bad habit? Cancer Res 68: 3081–3086; discussion 3086 [DOI] [PubMed] [Google Scholar]
- Felsher DW, Bishop JM 1999a. Reversible tumorigenesis by MYC in hematopoietic lineages. Mol Cell 4: 199–207 [DOI] [PubMed] [Google Scholar]
- Felsher DW, Bishop JM 1999b. Transient excess of MYC activity can elicit genomic instability and tumorigenesis. Proc Natl Acad Sci 96: 3940–3944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsher DW, Zetterberg A, Zhu J, Tlsty T, Bishop JM 2000. Overexpression of MYC causes p53-dependent G2 arrest of normal fibroblasts. Proc Natl Acad Sci 97: 10544–10548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, et al. 2010. Selective inhibition of BET bromodomains. Nature 468: 1067–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher GH, Wellen SL, Klimstra D, Lenczowski JM, Tichelaar JW, Lizak MJ, Whitsett JA, Koretsky A, Varmus HE 2001. Induction and apoptotic regression of lung adenocarcinomas by regulation of a K-Ras transgene in the presence and absence of tumor suppressor genes. Genes Dev 15: 3249–3262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamberi G, Benassi MS, Bohling T, Ragazzini P, Molendini L, Sollazzo MR, Pompetti F, Merli M, Magagnoli G, Balladelli A, et al. 1998. C-myc and c-fos in human osteosarcoma: Prognostic value of mRNA and protein expression. Oncology 55: 556–563 [DOI] [PubMed] [Google Scholar]
- Gao P, Tchernyshyov I, Chang TC, Lee YS, Kita K, Ochi T, Zeller KI, De Marzo AM, Van Eyk JE, Mendell JT, et al. 2009. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature 458: 762–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson AW, Cheng T, Johnston RN 1995. Apoptosis induced by c-myc overexpression is dependent on growth conditions. Exp Cell Res 218: 351–358 [DOI] [PubMed] [Google Scholar]
- Giuriato S, Rabin K, Fan AC, Shachaf CM, Felsher DW 2004. Conditional animal models: A strategy to define when oncogenes will be effective targets to treat cancer. Semin Cancer Biol 14: 3–11 [DOI] [PubMed] [Google Scholar]
- Giuriato S, Ryeom S, Fan AC, Bachireddy P, Lynch RC, Rioth MJ, van Riggelen J, Kopelman AM, Passegue E, Tang F, et al. 2006. Sustained regression of tumors upon MYC inactivation requires p53 or thrombospondin-1 to reverse the angiogenic switch. Proc Natl Acad Sci 103: 16266–16271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandori C, Wu KJ, Fernandez P, Ngouenet C, Grim J, Clurman BE, Moser MJ, Oshima J, Russell DW, Swisshelm K, et al. 2003. Werner syndrome protein limits MYC-induced cellular senescence. Genes Dev 17: 1569–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green DR 1997. A Myc-induced apoptosis pathway surfaces. Science 278: 1246–1247 [DOI] [PubMed] [Google Scholar]
- Grivennikov SI, Greten FR, Karin M 2010. Immunity, inflammation, and cancer. Cell 140: 883–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermeking H 2003. The MYC oncogene as a cancer drug target. Curr Cancer Drug Targets 3: 163–175 [DOI] [PubMed] [Google Scholar]
- Hoffman B, Liebermann DA 2008. Apoptotic signaling by c-MYC. Oncogene 27: 6462–6472 [DOI] [PubMed] [Google Scholar]
- Huettner CS, Zhang P, Van Etten RA, Tenen DG 2000. Reversibility of acute B-cell leukaemia induced by BCR-ABL1. Nat Genet 24: 57–60 [DOI] [PubMed] [Google Scholar]
- Jacobs JJ, Scheijen B, Voncken JW, Kieboom K, Berns A, van Lohuizen M 1999. Bmi-1 collaborates with c-Myc in tumorigenesis by inhibiting c-Myc-induced apoptosis via INK4a/ARF. Genes Dev 13: 2678–2690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M, Arvanitis C, Chu K, Dewey W, Leonhardt E, Trinh M, Sundberg CD, Bishop JM, Felsher DW 2002. Sustained loss of a neoplastic phenotype by brief inactivation of MYC. Science 297: 102–104 [DOI] [PubMed] [Google Scholar]
- Ji H, Wang Z, Perera SA, Li D, Liang MC, Zaghlul S, McNamara K, Chen L, Albert M, Sun Y, et al. 2007. Mutations in BRAF and KRAS converge on activation of the mitogen-activated protein kinase pathway in lung cancer mouse models. Cancer Res 67: 4933–4939 [DOI] [PubMed] [Google Scholar]
- Johnston LA, Prober DA, Edgar BA, Eisenman RN, Gallant P 1999. Drosophila myc regulates cellular growth during development. Cell 98: 779–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonkers J, Berns A 1996. Retroviral insertional mutagenesis as a strategy to identify cancer genes. Biochim Biophys Acta 1287: 29–57 [DOI] [PubMed] [Google Scholar]
- Karlsson A, Deb-Basu D, Cherry A, Turner S, Ford J, Felsher DW 2003a. Defective double-strand DNA break repair and chromosomal translocations by MYC overexpression. Proc Natl Acad Sci 100: 9974–9979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson A, Giuriato S, Tang F, Fung-Weier J, Levan G, Felsher DW 2003b. Genomically complex lymphomas undergo sustained tumor regression upon MYC inactivation unless they acquire novel chromosomal translocations. Blood 101: 2797–2803 [DOI] [PubMed] [Google Scholar]
- Kawate S, Fukusato T, Ohwada S, Watanuki A, Morishita Y 1999. Amplification of c-myc in hepatocellular carcinoma: Correlation with clinicopathologic features, proliferative activity and p53 overexpression. Oncology 57: 157–163 [DOI] [PubMed] [Google Scholar]
- Kessler JD, Kahle KT, Sun T, Meerbrey KL, Schlabach MR, Schmitt EM, Skinner SO, Xu Q, Li MZ, Hartman ZC, et al. 2012. A SUMOylation-dependent transcriptional subprogram is required for Myc-driven tumorigenesis. Science 335: 348–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kool J, Berns A 2009. High-throughput insertional mutagenesis screens in mice to identify oncogenic networks. Nat Rev Cancer 9: 389–399 [DOI] [PubMed] [Google Scholar]
- *.Kuzyk A, Mai S 2014. c-MYC induced genomic instability. Cold Spring Harb Perspect Med 4: a014373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladanyi M, Park CK, Lewis R, Jhanwar SC, Healey JH, Huvos AG 1993. Sporadic amplification of the MYC gene in human osteosarcomas. Diagn Mol Pathol 2: 163–167 [PubMed] [Google Scholar]
- Lawlor ER, Soucek L, Brown-Swigart L, Shchors K, Bialucha CU, Evan GI 2006. Reversible kinetic analysis of Myc targets in vivo provides novel insights into Myc-mediated tumorigenesis. Cancer Res 66: 4591–4601 [DOI] [PubMed] [Google Scholar]
- *.Levens D 2013. Cellular MYCRo economics: Balancing MYC function with MYC expression. Cold Spring Harb Perspect Med 3: a014233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Shimamura T, Ji H, Chen L, Haringsma HJ, McNamara K, Liang MC, Perera SA, Zaghlul S, Borgman CL, et al. 2007. Bronchial and peripheral murine lung carcinomas induced by T790M-L858R mutant EGFR respond to HKI-272 and rapamycin combination therapy. Cancer Cell 12: 81–93 [DOI] [PubMed] [Google Scholar]
- Lin CY, Loven J, Rahl PB, Paranal RM, Burge CB, Bradner JE, Lee TI, Young RA 2012. Transcriptional amplification in tumor cells with elevated c-Myc. Cell 151: 56–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe SW, Cepero E, Evan G 2004. Intrinsic tumour suppression. Nature 432: 307–315 [DOI] [PubMed] [Google Scholar]
- Luo J, Solimini NL, Elledge SJ 2009. Principles of cancer therapy: Oncogene and non-oncogene addiction. Cell 136: 823–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mai S, Hanley-Hyde J, Fluri M 1996. c-Myc overexpression associated DHFR gene amplification in hamster, rat, mouse and human cell lines. Oncogene 12: 277–288 [PubMed] [Google Scholar]
- Makino R, Hayashi K, Sugimura T 1984. C-myc transcript is induced in rat liver at a very early stage of regeneration or by cycloheximide treatment. Nature 310: 697–698 [DOI] [PubMed] [Google Scholar]
- Maksimenko A, Malvy C 2005. Oncogene-targeted antisense oligonucleotides for the treatment of Ewing sarcoma. Expert Opin Ther Targets 9: 825–830 [DOI] [PubMed] [Google Scholar]
- Marinkovic D, Marinkovic T, Mahr B, Hess J, Wirth T 2004. Reversible lymphomagenesis in conditionally c-MYC expressing mice. Int J Cancer J 110: 336–342 [DOI] [PubMed] [Google Scholar]
- *.McKeown MR, Bradner JE 2014. Therapeutic strategies to inhibit MYC. Cold Spring Harb Perspect Med 10.1101/cshperspect.a014266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendrysa SM, Akagi K, Roayaei J, Lien WH, Copeland NG, Jenkins NA, Eisenman RN 2010. An integrated genetic-genomic approach for the identification of novel cancer loci in mice sensitized to c-Myc-induced apoptosis. Genes Cancer 1: 465–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkers H, Berns A 2003. Retroviral insertional mutagenesis: Tagging cancer pathways. Adv Cancer Res 88: 53–99 [DOI] [PubMed] [Google Scholar]
- Mikkers H, Allen J, Knipscheer P, Romeijn L, Hart A, Vink E, Berns A 2002. High-throughput retroviral tagging to identify components of specific signaling pathways in cancer. Nat Genet 32: 153–159 [DOI] [PubMed] [Google Scholar]
- *.Morrish F, Hockenbery D 2014. MYC and mitochondrial biogenesis. Cold Spring Harb Perspect Med 4: a014225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy DJ, Junttila MR, Pouyet L, Karnezis A, Shchors K, Bui DA, Brown-Swigart L, Johnson L, Evan GI 2008. Distinct thresholds govern Myc’s biological output in vivo. Cancer Cell 14: 447–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray MJ, Cunningham JM, Parada LF, Dautry F, Lebowitz P, Weinberg RA 1983. The HL-60 transforming sequence: A ras oncogene coexisting with altered myc genes in hematopoietic tumors. Cell 33: 749–757 [DOI] [PubMed] [Google Scholar]
- Neto-Silva RM, de Beco S, Johnston LA 2010. Evidence for a growth-stabilizing regulatory feedback mechanism between Myc and Yorkie, the Drosophila homolog of Yap. Dev Cell 19: 507–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie Z, Hu G, Wei G, Cui K, Yamane A, Resch W, Wang R, Green DR, Tessarollo L, Casellas R, et al. 2012. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell 151: 68–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson JA, Cleveland JL 2003. Myc pathways provoking cell suicide and cancer. Oncogene 22: 9007–9021 [DOI] [PubMed] [Google Scholar]
- *.O’Shea JM, Ayer DE 2013. Coordination of nutrient availability and utilization by MAX- and MLX-centered transcription networks. Cold Spring Harb Perspect Med 3: a014258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastorino F, Brignole C, Marimpietri D, Di Paolo D, Zancolli M, Pagnan G, Ponzoni M 2004. Targeted delivery of oncogene-selective antisense oligonucleotides in neuroectodermal tumors: Therapeutic implications. Ann N Y Acad Sci 1028: 90–103 [DOI] [PubMed] [Google Scholar]
- Pelengaris S, Khan M, Evan GI 2002. Suppression of Myc-induced apoptosis in β cells exposes multiple oncogenic properties of Myc and triggers carcinogenic progression. Cell 109: 321–334 [DOI] [PubMed] [Google Scholar]
- *.Rahl PB, Young RA 2014. MYC and transcription elongation. Cold Spring Harb Perspect Med 4: a014340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakhra K, Bachireddy P, Zabuawala T, Zeiser R, Xu L, Kopelman A, Fan AC, Yang Q, Braunstein L, Crosby E, et al. 2010. CD4+ T cells contribute to the remodeling of the microenvironment required for sustained tumor regression upon oncogene inactivation. Cancer Cell 18: 485–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray S, Atkuri KR, Deb-Basu D, Adler AS, Chang HY, Herzenberg LA, Felsher DW 2006. MYC can induce DNA breaks in vivo and in vitro independent of reactive oxygen species. Cancer Res 66: 6598–6605 [DOI] [PubMed] [Google Scholar]
- Reimann M, Lee S, Loddenkemper C, Dorr JR, Tabor V, Aichele P, Stein H, Dorken B, Jenuwein T, Schmitt CA 2010. Tumor stroma-derived TGF-β limits Myc-driven lymphomagenesis via Suv39h1-dependent senescence. Cancer Cell 17: 262–272 [DOI] [PubMed] [Google Scholar]
- Restifo NP 2010. Can antitumor immunity help to explain “oncogene addiction?” Cancer Cell 18: 403–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooney CM, Rowe M, Wallace LE, Rickinson AB 1985. Epstein–Barr virus-positive Burkitt’s lymphoma cells not recognized by virus-specific T-cell surveillance. Nature 317: 629–631 [DOI] [PubMed] [Google Scholar]
- Ruggero D, Pandolfi PP 2003. Does the ribosome translate cancer? Nat Rev Cancer 3: 179–192 [DOI] [PubMed] [Google Scholar]
- Savino M, Annibali D, Carucci N, Favuzzi E, Cole MD, Evan GI, Soucek L, Nasi S 2011. The action mechanism of the Myc inhibitor termed Omomyc may give clues on how to target Myc for cancer therapy. PloS ONE 6: e22284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt CA, Lowe SW 2001. Bcl-2 mediates chemoresistance in matched pairs of primary Eμ-myc lymphomas in vivo. Blood Cells Mol Dis 27: 206–216 [DOI] [PubMed] [Google Scholar]
- Schmitt CA, McCurrach ME, de Stanchina E, Wallace-Brodeur RR, Lowe SW 1999. INK4α/ARF mutations accelerate lymphomagenesis and promote chemoresistance by disabling p53. Genes Dev 13: 2670–2677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shachaf CM, Felsher DW 2005. Tumor dormancy and MYC inactivation: Pushing cancer to the brink of normalcy. Cancer Res 65: 4471–4474 [DOI] [PubMed] [Google Scholar]
- Shachaf CM, Kopelman AM, Arvanitis C, Karlsson A, Beer S, Mandl S, Bachmann MH, Borowsky AD, Ruebner B, Cardiff RD, et al. 2004. MYC inactivation uncovers pluripotent differentiation and tumour dormancy in hepatocellular cancer. Nature 431: 1112–1117 [DOI] [PubMed] [Google Scholar]
- Shachaf CM, Perez OD, Youssef S, Fan AC, Elchuri S, Goldstein MJ, Shirer AE, Sharpe O, Chen J, Mitchell DJ, et al. 2007. Inhibition of HMGcoA reductase by atorvastatin prevents and reverses MYC-induced lymphomagenesis. Blood 110: 2674–2684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shachaf CM, Gentles AJ, Elchuri S, Sahoo D, Soen Y, Sharpe O, Perez OD, Chang M, Mitchel D, Robinson WH, et al. 2008. Genomic and proteomic analysis reveals a threshold level of MYC required for tumor maintenance. Cancer Res 68: 5132–5142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma SV, Settleman J 2007. Oncogene addiction: Setting the stage for molecularly targeted cancer therapy. Genes Dev 21: 3214–3231 [DOI] [PubMed] [Google Scholar]
- Sharma SV, Gajowniczek P, Way IP, Lee DY, Jiang J, Yuza Y, Classon M, Haber DA, Settleman J 2006. A common signaling cascade may underlie “addiction” to the Src, BCR-ABL, and EGF receptor oncogenes. Cancer Cell 10: 425–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheiness D, Fanshier L, Bishop JM 1978. Identification of nucleotide sequences which may encode the oncogenic capacity of avian retrovirus MC29. J Virol 28: 600–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodir NM, Swigart LB, Karnezis AN, Hanahan D, Evan GI, Soucek L 2011. Endogenous Myc maintains the tumor microenvironment. Genes Dev 25: 907–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soucek L, Lawlor ER, Soto D, Shchors K, Swigart LB, Evan GI 2007. Mast cells are required for angiogenesis and macroscopic expansion of Myc-induced pancreatic islet tumors. Nat Med 13: 1211–1218 [DOI] [PubMed] [Google Scholar]
- Soucek L, Whitfield J, Martins CP, Finch AJ, Murphy DJ, Sodir NM, Karnezis AN, Swigart LB, Nasi S, Evan GI 2008. Modelling Myc inhibition as a cancer therapy. Nature 455: 679–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soucek L, Whitfield JR, Sodir NM, Masso-Valles D, Serrano E, Karnezis AN, Swigart LB, Evan GI 2013. Inhibition of Myc family proteins eradicates KRas-driven lung cancer in mice. Genes Dev 27: 504–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer CA, Groudine M 1991. Control of c-myc regulation in normal and neoplastic cells. Adv Cancer Res 56: 1–48 [DOI] [PubMed] [Google Scholar]
- Srinivasan SV, Dominguez-Sola D, Wang LC, Hyrien O, Gautier J 2013. Cdc45 is a critical effector of myc-dependent DNA replication stress. Cell Reps 3: 1629–1639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein CA, Benimetskaya L, Mani S 2005. Antisense strategies for oncogene inactivation. Semin Oncol 32: 563–572 [DOI] [PubMed] [Google Scholar]
- Stock C, Kager L, Fink FM, Gadner H, Ambros PF 2000. Chromosomal regions involved in the pathogenesis of osteosarcomas. Genes Chromosomes Cancer 28: 329–336 [DOI] [PubMed] [Google Scholar]
- Tamm I, Dorken B, Hartmann G 2001. Antisense therapy in oncology: New hope for an old idea? Lancet 358: 489–497 [DOI] [PubMed] [Google Scholar]
- Toyoshima M, Howie HL, Imakura M, Walsh RM, Annis JE, Chang AN, Frazier J, Chau BN, Loboda A, Linsley PS, et al. 2012. Functional genomics identifies therapeutic targets for MYC-driven cancer. Proc Natl Acad Sci 109: 9545–9550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran PT, Bendapudi PK, Lin HJ, Choi P, Koh S, Chen J, Horng G, Hughes NP, Schwartz LH, Miller VA, et al. 2011. Survival and death signals can predict tumor response to therapy after oncogene inactivation. Sci Transl Med 3: 103–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vafa O, Wade M, Kern S, Beeche M, Pandita TK, Hampton GM, Wahl GM 2002. c-Myc can induce DNA damage, increase reactive oxygen species, and mitigate p53 function: A mechanism for oncogene-induced genetic instability. Mol Cell 9: 1031–1044 [DOI] [PubMed] [Google Scholar]
- van Riggelen J, Muller J, Otto T, Beuger V, Yetil A, Choi PS, Kosan C, Moroy T, Felsher DW, Eilers M 2010a. The interaction between Myc and Miz1 is required to antagonize TGFβ-dependent autocrine signaling during lymphoma formation and maintenance. Genes Dev 24: 1281–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Riggelen J, Yetil A, Felsher DW 2010b. MYC as a regulator of ribosome biogenesis and protein synthesis. Nat Rev Cancer 10: 301–309 [DOI] [PubMed] [Google Scholar]
- Verdine GL, Walensky LD 2007. The challenge of drugging undruggable targets in cancer: Lessons learned from targeting BCL-2 family members. Clin Cancer Res 13: 7264–7270 [DOI] [PubMed] [Google Scholar]
- Vita M, Henriksson M 2006. The Myc oncoprotein as a therapeutic target for human cancer. Semin Cancer Biol 16: 318–330 [DOI] [PubMed] [Google Scholar]
- Weinstein IB 2002. Cancer. Addiction to oncogenes—The Achilles heal of cancer. Science 297: 63–64 [DOI] [PubMed] [Google Scholar]
- Weinstein IB, Joe A 2008. Oncogene addiction. Cancer Res 68: 3077–3080; discussion 3080 [DOI] [PubMed] [Google Scholar]
- Welm AL, Kim S, Welm BE, Bishop JM 2005. MET and MYC cooperate in mammary tumorigenesis. Proc Natl Acad Sci 102: 4324–4329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise DR, DeBerardinis RJ, Mancuso A, Sayed N, Zhang XY, Pfeiffer HK, Nissim I, Daikhin E, Yudkoff M, McMahon SB, et al. 2008. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci 105: 18782–18787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CH, Sahoo D, Arvanitis C, Bradon N, Dill DL, Felsher DW 2008. Combined analysis of murine and human microarrays and ChIP analysis reveals genes associated with the ability of MYC to maintain tumorigenesis. PLoS Genet 4: e1000090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C, Higgins PJ 2013. Drugging the undruggable: Transcription therapy for cancer. Biochim Biophys Acta 1835: 76–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D, Liu H, Goga A, Kim S, Yuneva M, Bishop JM 2010. Therapeutic potential of a synthetic lethal interaction between the MYC proto-oncogene and inhibition of aurora-B kinase. Proc Natl Acad Sci 107: 13836–13841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zindy F, Eischen CM, Randle DH, Kamijo T, Cleveland JL, Sherr CJ, Roussel MF 1998. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev 12: 2424–2433 [DOI] [PMC free article] [PubMed] [Google Scholar]