Abstract
The MYC oncogene is a multifunctional protein that is aberrantly expressed in a significant fraction of tumors from diverse tissue origins. Because of its multifunctional nature, it has been difficult to delineate the exact contributions of MYC’s diverse roles to tumorigenesis. Here, we review the normal role of MYC in regulating DNA replication as well as its ability to generate DNA replication stress when overexpressed. Finally, we discuss the possible mechanisms by which replication stress induced by aberrant MYC expression could contribute to genomic instability and cancer.
MYC influences several steps during the initiation of DNA replication (e.g., it promotes Cdc45 recruitment at replication origins). Replication stress induced by aberrant MYC expression and subsequent misregulation of this activity may contribute to genomic instability and cancer.
NORMAL ROLE OF MYC IN CYCLING CELLS AND CONTROL OF THE G1/S TRANSITION
C-MYC (MYC) and its cellular homologs are essential for the initiation and maintenance of normal cell cycles, both in vivo and in vitro (Shichiri et al. 1993; de Alboran et al. 2001; Prathapam et al. 2006), and only in rare cases is MYC dispensable for cell growth (Shichiri et al. 1993; Mateyak et al. 1997; Pierce et al. 2008; Steiger et al. 2008). MYC proteins are strongly induced during cell-cycle entry from quiescence, and are critical in the decision of cells to enter or exit cell cycle (Holzel et al. 2001). For example, resting lymphocytes require c-MYC expression to initiate and sustain the proliferative burst triggered by immune activation signals (de Alboran et al. 2001). During neurogenesis, the cerebellar primordium relies on MYCN to sustain the rapid proliferation of neural progenitors (Knoepfler et al. 2002). This is also the case for MYC in other progenitor and transiently amplifying tissue compartments (Wilson et al. 2004; Muncan et al. 2006; Sansom et al. 2007; Laurenti et al. 2008). In addition, constitutive MYC expression is sufficient to promote cell-cycle entry (G0 to S transition) and sustain replicative cycles in specific cellular settings, like mouse and rat fibroblasts, or postmitotic neurons (Kaczmarek et al. 1985; Eilers et al. 1991; Steiner et al. 1995; Mateyak et al. 1997). Although the mechanism by which MYC drives cell-cycle progression is not fully understood (Amati et al. 1998; Obaya et al. 1999), it is becoming increasingly clear that transcriptional and nontranscriptional mechanisms mediate the ability of MYC to initiate and sustain proliferative cycles.
Transcriptional Control of Cell-Cycle Entry
As discussed by Dang (2013), Campbell and White (2014), and Morrish and Hockenbery (2014), a major biological end point of MYC activity is the increase in cell mass, achieved through its global transcriptional effects on cellular and mitochondrial metabolism and ribosomal biogenesis (Gomez-Roman et al. 2003; O’Connell et al. 2003; Grandori et al. 2005; Liu et al. 2008). Coupling of cell growth and cell-cycle progression could explain, at least in part, the ability of MYC to induce cell-cycle entry (Obaya et al. 1999; Schorl and Sedivy 2003). This is supported by the observations that inhibition of specific metabolic pathways activated in response to MYC activity (e.g., oxidative phosphorylation or nucleotide biosynthesis) can prevent MYC-dependent cell-cycle entry and progression in MYC−/− cells (Liu et al. 2008; Morrish et al. 2008).
In cell culture, MYC activation also correlates with changes in expression levels of cyclins, cyclin-dependent kinases (CDKs), and cyclin-dependent kinase inhibitors (CKIs) (Obaya et al. 1999; Hermeking et al. 2000). By favoring the relative abundance of activating (cyclin D1, Cdk-4, and Cdk-6) versus inhibitory (p15 and p21) complexes, MYC can promote cell-cycle entry and progression. Of note, some of these studies have been performed in engineered Rat1A MYC null cells, which have the unusual ability to slowly proliferate in absence of MYC (Shichiri et al. 1993; Holzel et al. 2001). Therefore, it is unclear whether these observations extend beyond this cell type.
D-type cyclins (D1, D2, and D3) associate with Cdk-4/Cdk-6 and drive exit from quiescence and commitment to cell cycle. In B lymphocytes, cyclin D2 is required for MYC-induced cell-cycle entry and proliferation in response to immune activating cues (de Alboran et al. 2001; Calado et al. 2012; Dominguez-Sola et al. 2012; Poe et al. 2012). Cyclin D2 transcriptional activation by MYC requires the PI3K pathway (Bouchard et al. 2001), and activation of the PI3K pathway is also required for cells to exit quiescence and commit to the initiation of DNA replication (Kumar et al. 2006; Marques et al. 2008). The relationship between cyclin D2 and PI3K is an important example of the connection between external cues (e.g., growth factors) and the downstream effects of MYC activity.
Control of the G1/S Transition by Cyclin E/Cdk-2 Activity
Although D-type cyclins and Cdk-4/6 are critical for promoting cell-cycle entry, they are dispensable during DNA replication, when cyclin E/Cdk-2 complexes are instead required (Sherr and Roberts 2004). Cyclin E/Cdk-2 complexes target several substrates directly involved in DNA replication, including proteins necessary for replication origin licensing (TopBP1, MCM) and origin firing in mammals and other organisms (Yu and Sicinski 2004). Once cyclin E/Cdk-2 reach critical levels, they are sufficient to drive cell-cycle completion even in the absence of active mitogenic signals (Sherr and Roberts 2004).
The relationship between MYC and cyclinE/Cdk-2 is complex and is only partially understood. For example, acute MYC depletion in cycling cells via genetic manipulation leads to cell-cycle arrest and subsequent quiescence, effects that can be rescued by constitutive expression of specific growth factor-like signals like those provided by cellular Src (c-Src). In this context, c-Src activates early G1 regulators but cannot drive G1/S transition and DNA synthesis unless MYC is reintroduced or cyclin E/Cdk-2 complex is expressed (Prathapam et al. 2006). These observations point to a rate-limiting role for MYC at the G1/S transition and further suggest that MYC alters the activity of G1/S-specific CDK complexes, consistent with several reports indicating that MYC regulates cyclin E/Cdk-2 activity.
The activity of cyclin E/Cdk-2 complexes is markedly decreased in myc−/− cells (Obaya et al. 2002), and MYC appears to be required for cyclin E/Cdk-2 activation by either displacing or inactivating p27Kip1 via poorly defined mechanisms (Steiner et al. 1995; Vlach et al. 1996; Berns et al. 1997; Muller et al. 1997; Perez-Roger et al. 1997; Pusch et al. 1997; O’Hagan et al. 2000). p27Kip1 is a CDK inhibitor that is loaded with cyclin E/Cdk-2 at origins of replication at each cell cycle and specifically blocks its activity. Degradation—or functional inactivation—of p27 at these sites is coupled to the initiation of DNA replication (Furstenthal et al. 2001; You et al. 2002). Therefore, a potential mechanism by which MYC could induce origin firing and trigger DNA synthesis would involve the modulation of cyclin E/Cdk-2 complex activity at origins of replication (see below) (Srinivasan et al. 2013).
Nontranscriptional Control of DNA Replication Initiation
An additional way by which MYC contributes to cell-cycle progression is by directly controlling DNA replication initiation and S-phase entry. The ability of MYC to directly promote DNA synthesis has been experimentally documented for a long time. Some early reports suggested that this biological function would involve direct interactions between MYC and the DNA replication machinery (Kaczmarek et al. 1985; Studzinski et al. 1986; Iguchi-Ariga et al. 1987, 1988; Hermeking et al. 1994; Lemaitre et al. 1995). However, the idea was later challenged (Gutierrez et al. 1987, 1988). Subsequently, the prevalent idea was that MYC effect on DNA synthesis was a consequence of its transcriptional activity on cell-cycle regulators, as described above. Nevertheless, different groups had reported the existence of protein–protein interactions between MYC and components of the prereplication protein complex (Table 1, and references therein). These findings, and the apparent overlap between specific origins of replication and MYC-binding sites (Iguchi-Ariga et al. 1988) pointed to a direct role for MYC in the initiation of DNA replication, which was characterized two decades later (Dominguez-Sola et al. 2007, and see below).
Table 1.
Known functional interactions between MYC and the DNA replication and replication surveillance machinery
| Factor | Functional interaction | Notes | References |
|---|---|---|---|
| Cdc6 | Abrogation of E-box-dependent transcriptional activity Physical interaction and colocalization at assembled replication origins |
Takayama et al. 2000a; Dominguez-Sola et al. 2007 |
|
| Orc1 | Abrogation of E-box-dependent transcriptional activity | Takayama et al. 2000b | |
| Orc2 | Colocalization at chromatin enhancers throughout cell cycle | Drosophila | Yang et al. 2013 |
| ORC | Physical interaction and colocalization at assembled replication origins | Dominguez-Sola et al. 2007 | |
| Cdt1 | Physical interaction and colocalization at assembled replication origins | Dominguez-Sola et al. 2007 | |
| Mcm2-7 | Physical interaction and colocalization at assembled replication origins | Dominguez-Sola et al. 2007; Koch et al. 2007 | |
| Cdc45 | Chromatin recruitment facilitated by MYC | Dominguez-Sola et al. 2007; Sankar et al. 2009; Srinivasan et al. 2013 | |
| Pol-α | Cosegregation in common functional protein complexes | Results challenged in a publication by Gutierrez et al. (1987) | Studzinski et al. 1986 |
| BRCA1 | Abrogation of MYC-dependent transcriptional activation, or active transcriptional repression of target genes | BRCA1 defects and MYC overexpression seem to coincide in some breast cancer subtypes (Ren et al. 2013) | Wang et al. 1998; Li et al. 2002; Kennedy et al. 2005 |
| TopBP1 | Competition for Miz-1 binding | Possible effects in DNA damage response (modulation of ATR checkpoint?) | Herold et al. 2008 |
| WRN | Genetic interaction (see text) | Grandori et al. 2003 | |
| ATR | Genetic interaction (see text) | Murga et al. 2011 |
ORC, origin recognition complex.
MYC CONTROL OF INITIATION OF DNA REPLICATION
DNA replication, the process by which the entire genome is accurately duplicated during S phase, requires the unwinding, copying, and reassembling of three billions base pairs in human cells. Replication contributes significantly to genomic instability and chromosome alterations owing to the fragile replication intermediates generated at replication forks, the presence of obstacles on the DNA template, and potential replication errors.
Origin Assembly and Mechanism of Activation
Chromosomal replication in eukaryotes starts at multiple discrete loci, the origins of replication. Origins of replication are assembled and activated in a stepwise manner from proteins and protein complexes (Blow and Dutta 2005; DePamphilis et al. 2006). In addition to regulation of origin activity, some of these factors also participate in genome surveillance to activate checkpoints and DNA damage responses when the DNA replication machinery encounters problems. This concept is important to understand the consequences of tampering with normal DNA replication, and the significance of specific genetic interactions between MYC and some of these factors.
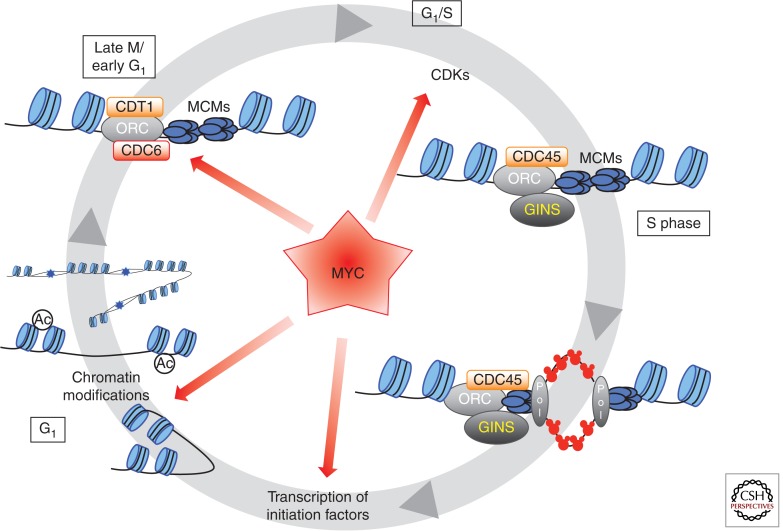
The mechanisms that control the timing, distribution, and efficiency of replication origins vary considerably among eukaryotes and are still not fully understood, especially in mammalian cells (Mechali 2010). In contrast, the mechanisms that regulate the initiation of a single origin of replication are conserved throughout evolution and well characterized. The functional unit that assembles on chromatin, the prereplicative complex (pre-RC) is a multiprotein complex conserved from yeast to human (Bell and Dutta 2002; Errico and Costanzo 2010; Costa et al. 2013). Pre-RC assembly starts with the binding of the origin recognition complex (ORC). In budding yeast, this six-subunit complex recognizes specific DNA sequences. In other eukaryotes, ORC proteins are thought to recognize specific DNA structures rather than specific sequences. ORC is loaded in late M phase or early G1 phase of the cell cycle (Fig. 1). ORC, together with Cdc6 and Cdt1 recruit the minichromosome maintenance (MCM) hexameric helicase complex at origins. MCMs are the catalytic components of the replicative DNA helicase responsible for unwinding genomic DNA bidirectionally. MCM assembly completes the formation of an inactive pre-RC.
Figure 1.
Regulation of origin assembly and origin firing by c-MYC. The stepwise assembly of the prereplicative complex takes place in late mitosis (M phase) or early G1 with the binding of ORC (origin recognition complex), followed by the Cdt1- and Cdc6-dependent loading of the MCM (minichromosome maintenance) helicase. At the G1/S transition, CDK (cyclin-dependent kinase) activity is required for the activation of the pre-RC into an initiation complex, coinciding with the assembly of the helicase cofactors CDC45 and GINS. Subsequently, DNA polymerases are recruited and DNA synthesis occurs on unwound DNA. Origins of replication are assembled within permissive regions of chromatin, which are nucleosome-free and open. MYC can influence several steps in these processes, as indicated by the red arrows. MYC physically interacts with the pre-RC proteins CDC6 and MCMs. MYC also regulates the activity of CDKs at origins and moreover, influences the generation of open chromatin, which in turn is thought to regulate the spatiotemporal pattern of origin positioning and firing. Finally, MYC also stimulates the transcription of many factors required for origin assembly and activation (see text for details).
Although the MCM complex harbors the catalytic activity that unwinds DNA, the formation of an active helicase requires the presence of Cdc45 and the GINS complex. These three components assemble into the CMG (Cdc45/MCM/GINS) complex, which is the active enzyme (Fig. 1). Productive activation of the helicase requires additional factors Sld2, Sld3, and Dbp11 in budding yeast, which are regulated by phosphorylation by Dbf4-dependent kinase (DDK) and by CDKs (Yabuuchi et al. 2006; Tanaka et al. 2007). In yeast, DDK phosphorylates MCM2-7 complex and promotes the loading of Sld3 and Cdc45. CDK promotes the loading of GINS, and phosphorylates Sld2 and Sld3 triggering the binding of these proteins to the BRCT tandem repeats of Dbp11. The combined actions of these kinases convert the pre-RC into an “initiation” complex. RecQ4, Treslin/Ticrr, and TopBP1 are the vertebrate orthologs of Sld2, Sld3, and Dbp11, respectively (Mueller et al. 2011). They are thought to play similar roles as their yeast counterparts, yet the mechanistic details of their mode of action are not fully understood. On activation, the CMG complex undergoes significant structural changes that allow the hexameric MCM ring (the helicase engine) to switch from encircling dsDNA (MCM is loaded in G1) to encircling ssDNA and unwinding DNA by strand exclusion (Pacek et al. 2006; Costa et al. 2011). As mentioned above, MYC physically interacts with several components of the pre-RC providing a possible mechanism for MYC’s role in the initiation of DNA synthesis (Fig. 1; Table 1).
To ensure that DNA replication is taking place once and only once at each cell cycle (i.e., that no part of the genome is rereplicated), loading of the pre-RC is temporally uncoupled from its activation. Pre-RCs are assembled in late mitosis and G1 when CDK activity is low and permissive for the stepwise assembly of its components. Activation of pre-RCs into functional initiation complexes requires CDK activity, which normally rises at the onset of S phase when DNA synthesis is initiated. Because elevated CDK activity also inhibits further pre-RC assembly, CDKs play a dual role during DNA replication. They activate initiation of DNA replication and prevent pre-RC assembly once DNA synthesis has started. As discussed above, MYC is known to influence the activity of S-phase CDK, thus providing another possible mechanism by which MYC could regulate origin assembly or activation (Fig. 1).
Replication Timing and Origin Selection
In budding yeast, in which ORC complexes bind specific DNA sequences, the position and numbers of origins are primarily dictated by the genome’s nucleotide sequence. In contrast, the positioning of origins of replication in other eukaryotes is largely independent of specific sequences. It is estimated that there are approximately 30,000 origins in human cells. The structural determinants of the positioning of mammalian cell origins are starting to be unraveled, thanks to complementary genome-wide origin mapping studies (Evertts and Coller 2012). Large numbers of origins have been predicted by computational approaches using strategies such as the genome-wide identification of discontinuities in nucleotide composition strand asymmetry (Huvet et al. 2007), or have been mapped experimentally by RNA–DNA hybrid analysis, nascent strand, or origin trapping methods (Mesner et al. 2006; Cadoret et al. 2008; Karnani et al. 2010). A consensus emerges from these diverse approaches as well as some differences. Origins are found in nucleosome-free regions, they are enriched at DNase-hypersensitive sites, and correlate with acetylated histone H4 and other chromatin marks (di- and trimethyl H3K4). Origins are also enriched near CpG islands. Notably, origins of replication are significantly enriched at transcription start sites including the c-MYC locus itself (Tao et al. 2000), although transcription per se is not required for origin activity (Cohen et al. 2003).
Chromatin modifications, and more specifically histone acetylation, can influence origin assembly and activity (Aggarwal and Calvi 2004; Danis et al. 2004; Miotto and Struhl 2010) and could potentially regulate DNA replication dynamics. Notably, MYC influences histone and chromatin acetylation and associates with several chromatin modifying enzymes (Frank et al. 2001; Knoepfler et al. 2006; Martinato et al. 2008) and could therefore influence the position and the activity of origins through this mechanism (Fig. 1). For example, MYC binds to an E box within the Lamin B origin to recruit the MLL1 (mixed lineage leukemia 1) methyltransferase, which modifies surrounding nucleosomes by methylating H3K4. These marks subsequently recruit the HBO1 histone acetylase, resulting in the generation of a nucleosome-free open chromatin region and the loading of MCM at the origin (Swarnalatha et al. 2012).
Studies, mostly performed in yeast, are thus consistent with the idea that modification of the chromatin landscape can directly alter the spatiotemporal program of DNA replication. In agreement with a role for histone acetylation in origin activity, deletion of the Rdp3 histone deacetylase in yeast advances the firing of late origins (Knott et al. 2009). Conversely, deletion of the forkhead box transcription factor Fkh1 and Fkh2 delays the firing of early origins (Knott et al. 2012). Finally, Rif1 deletion results in delayed firing of early origins and advanced firing of late origins, thus compressing S phase (Hayano et al. 2012). Notably deletion of Rif1 suppresses the lethality of DDK deletion. Rif1 also controls replication timing in human and mouse cells (Cornacchia et al. 2012; Yamazaki et al. 2012) in a way that is similar to yeast. Rif1-depleted cells show increased chromatin-bound Cdc45 in G1 suggesting that Rif1, a chromatin remodeling enzyme, normally prevents Cdc45 loading.
Loading onto chromatin of Cdc45, along with Treslin and the GINS protein complex, is required for the activation of assembled origins of replication. Cdc45, GINS, and Treslin are low abundance proteins and are loaded only at selected origins. Thus, the regulation of their association to replication origins is essential in determining the pattern and timing of origin firing in different organisms, from yeast to humans (Mantiero et al. 2011; Tanaka et al. 2011). As described above, chromatin remodeling enzymes, such as Rif1, regulate Cdc45 binding to chromatin, which is a rate-limiting step of origin activation (Pryde et al. 2009; Wong et al. 2011; Cornacchia et al. 2012; Knott et al. 2012; Yamazaki et al. 2012).
MYC and Initiation of DNA Replication
Work in Xenopus cell-free extracts and mammalian cells recently showed that MYC directly influences DNA replication initiation by promoting recruitment of Cdc45 to chromatin, in a manner independent of MYC transcriptional activities (Dominguez-Sola et al. 2007; Sankar et al. 2009; Srinivasan et al. 2013). Through this mechanism, MYC regulates the number of replication origins activated early in S phase, which is proportional to the number of MYC molecules available in the cell. MYC cellular protein concentration is low suggesting that MYC is limiting under physiologic conditions (Waters et al. 1991; Rosales et al. 2013). The precise molecular events by which MYC controls Cdc45 recruitment to chromatin are not yet fully understood, although they could involve the modulation of cyclin E/Cdk-2 activity at origins (Srinivasan et al. 2013). Notably, Treslin mutants insensitive to cyclin E/Cdk-2-dependent phosphorylation are severely deficient in DNA replication (Kumagai et al. 2010, 2011). Because GINS/Treslin is critical for proper recruitment of Cdc45 to origins (Kumagai et al. 2010), GINS/Treslin could regulate the ability of MYC to recruit Cdc45 to replication origins.
The interaction between Treslin and TopBP1 is essential for the recruitment of Cdc45 and GINS to replication origins and the initiation of DNA replication (Boos et al. 2011; Kumagai et al. 2011; Mueller et al. 2011). TopBP1 is a BRCT domain-containing protein with essential functions in DNA replication and for the activation of ATR during checkpoint responses (Kumagai et al. 2006). In addition, TopBP1 physically interacts with Miz-1 (ZBTB17), a POZ-ZF protein required for MYC-dependent transcriptional repression of specific gene targets like p21 (CDKN1A) (Herold et al. 2002; Seoane et al. 2002; van de Wetering et al. 2002; Wu et al. 2003; further detailed by Eilers and colleagues in Wiese et al. 2013). The interaction between Miz-1 and TopBP1 is disrupted following UV irradiation, or by MYC (Herold et al. 2002, 2008). These observations could reflect the physiological engagement of TopBP1 at origins and its essential role in regulating initiation of DNA replication.
MYC, REPLICATION STRESS, AND TUMORIGENESIS
Challenges to DNA Replication: Stress
DNA replication stress is used widely to refer to conditions that challenge the normal process of DNA replication. For example, stress can be generated experimentally by decreasing the nucleotide pool or by inhibiting/slowing down the replicative polymerases. However, DNA replication stress is also intrinsic to this physiological process and can be exacerbated by the deregulation of oncogenes. The resulting “oncogene-induced” stress often has a replication component (see below).
When faced with replicative stress, cells stall replication forks to avoid their collapse or damage. Broken forks are indeed a source of double-strand breaks and are prone to illegitimate recombination. For example, hydroxyurea (an inhibitor of ribonucleotide reductase) and aphidicolin (an inhibitor of replicative DNA polymerases) induce gene rearrangements with microhomologies (Arlt et al. 2009, 2011). Furthermore, deep sequencing studies reveal complex chromosome rearrangements thought to arise from replication fork failure associated with genetic disorders (Lee et al. 2007; Carr et al. 2011). Therefore, checkpoint pathways that ensure replication fork stability are most critical to cell viability (Segurado and Tercero 2009).
Major challenges to DNA replication progression are physical barriers and obstacles along the DNA molecule. These include problematic genomic sequences (palindromes, G-quartet, telomeric repeats, tRNA genes), proteins bound to DNA and DNA adducts, including RNA polymerases, which can cause replication fork stalling. One of the most frequent problems is thought to be collision between a replication fork and transcribing RNA polymerases. MYC deregulation has the unique potential to trigger such collisions because it acutely stimulates replication and is a general activator of transcription (reviewed in Bermejo et al. 2012). Fork stalling is more pronounced following head-on collisions with an RNA polymerase than following codirectional encounters (Pomerantz and O’Donnell 2008). As mentioned above, open, nucleosome-free zones within acetylated chromatin favor the assembly of origins, but these chromatin features are also characteristic of transcription start sites. Initiating transcription and replication from the same open chromatin regions has the potential advantage of allowing codirectionality in these processes. Nonetheless, the most recent genome-wide analysis did not find evidence for transcription and replication codirectionality, suggesting that head-on collisions might not be the major problem for replication forks (Necsulea et al. 2009). Instead, the presence of a transcription bubble could be the most problematic encounter (Azvolinsky et al. 2009). It has been proposed that replicating through nuclear membrane tethered genes might also be a significant obstacle to faithful replication (Evertts and Coller 2012).
Replication Stress and Preneoplasia
Replication stress, as seen by robust staining for activated (phosphorylated) forms of the DNA damage response (DDR) proteins ATM, Chk1, Chk2, γH2AX, and p53, as well as 53BP1 foci, is a common phenomenon in early, preneoplastic lesions, which are the earliest detectable morphological evidence of tumor initiation and are associated with increased risk of cancer (Bartkova et al. 2005; Gorgoulis et al. 2005). Replication stress has been primarily documented in epithelial and melanocytic preneoplasias but not in other tissues (Bartkova et al. 2005; Gorgoulis et al. 2005; Di Micco et al. 2006; Kuilman et al. 2010). This could either reflect tissue specificity or the fact that routine histopathological analysis of superficial, epithelial lesions is generally more thorough. The constitutive DDR elicited by replication stress is thought to be responsible for the elevated level of genomic instability observed in tumors, especially solid tumors (Negrini et al. 2010). In turn, this potentially leads to the accumulation of point mutations, deletions, and chromosome rearrangements (Hartwell and Kastan 1994).
These observations strongly support the concept that several (but not all) oncogenes can trigger DNA replication stress followed by activation of persistent DDR and subsequent genomic instability in tumors (what has been conceptualized as “oncogenic stress”). Furthermore, forced expression of certain oncogenes recapitulates the DDR activation seen in preneoplastic lesions, in both cell culture and xenograft tumor models (see Table 2). Importantly, replication stress and subsequent DDR, both in experimental settings and in preneoplastic lesions, are intimately associated with the activation of cell-intrinsic tumor-suppressor mechanisms, checkpoints, senescence, and apoptosis, which have been clearly shown in different preneoplastic lesions (Bartkova et al. 2005, 2006; Gorgoulis et al. 2005; Di Micco et al. 2006). These tumor suppressor mechanisms are believed to be responsible for the delayed progression of preneoplastic lesions to overt cancers, and not surprisingly, are frequently and selectively lost during tumor progression (Bartek et al. 2007; Halazonetis et al. 2008). However, genetic inactivation of tumor suppressor pathways such as the ATM-p53 axis occurs subsequent to the activation of the DDR in preneoplastic lesions (Bartek et al. 2007; Halazonetis et al. 2008).
Table 2.
Factors known to induce replication stress in experimental settings
| Factor | Function | Associated features | Original references |
|---|---|---|---|
| Ras | Membrane-associated small GTPase | Cellular senescence Coincides with Cdc6 up-regulation |
Di Micco et al. 2006 |
| Cyclin E | S-phase cyclin | Bartkova et al. 2005 | |
| Cdc25A | Dual specificity phosphatase, required for G1 to S transition | Bartkova et al. 2005 | |
| E2F1 | Transcriptional activator of DNA replication and cell-cycle genes | Bartkova et al. 2005 | |
| MYC | Transcriptional amplifier DNA replication initiation factor |
Cellular senescence in some cell types | Grandori et al. 2003; Dominguez-Sola et al. 2007 |
| Mos | Serine/threonin kinase, activated MAP kinase cascade | Cellular senescence | Bartkova et al. 2005 |
| Cdc6 | Prereplication complex component | Cellular senescence | Bartkova et al. 2005; Di Micco et al. 2006 |
| Cdc45 | Preinitiation complex, required for GINS loading and replication origin firing | Epistatic to MYC | Srinivasan et al. 2013 |
The mechanistic connection between replication stress and senescence or apoptosis is, however, poorly understood. Senescence and apoptosis appear to be context dependent and are mutually exclusive responses to persistent replication stress and DDR triggered by oncogenes (d’Adda di Fagagna 2008). One possibility is that factors involved in the control of DNA replication or DDR might also be involved in the control of senescence or apoptosis responses. For example, Cdk-2 differentially modulates the ability of MYC to trigger cellular senescence and replication stress. Specifically, loss of Cdk-2 in cells and tissues with deregulated MYC expression triggers a senescence response that is not associated with increased replication stress (Campaner et al. 2010), perhaps owing to the requirement of Cdk-2 activity in MYC-dependent replication (Srinivasan et al. 2013). Similarly, MYC and Cdk-2 synergize to suppress senescence downstream from Ras hyperactivation (Hydbring et al. 2010), and MYC is also required to suppress BRAF-induced senescence in melanoma (Zhuang et al. 2008). Synergy between MYC and Cdk-2 relies in part on Cdk-2-dependent MYC phosphorylation at Ser62 by Cdk-2, a critical MYC residue normally phosphorylated in response to Ras activation (Sears et al. 2000; Hydbring et al. 2010). The ability of Cdk-2 to dictate the cellular responses to MYC deregulation seems to also be supported by the observation that this kinase is also essential for MYC-dependent apoptosis (Deb-Basu et al. 2006).
Limiting MYC-Dependent Replication Stress
As outlined above, replication stress triggers diverse cellular responses, including DDR and senescence, which normally determine the fate of cells with deregulated oncogenes in a cell context-dependent manner. Indeed, the outcomes of oncogene- and MYC-induced replication stress are determined by the status and the activity of specific factors and signaling pathways (Fig. 2; see also Table 2).
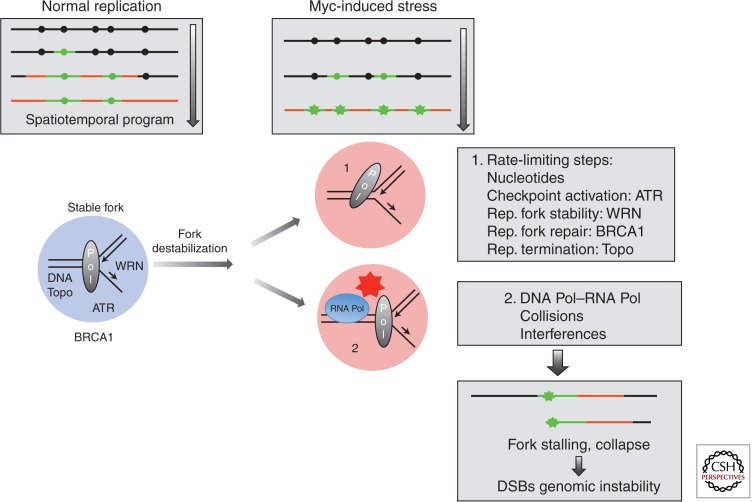
Figure 2.
Possible sources of MYC-dependent replication stress. Top panels depict the distribution of inactive (black dots) and active (green dots) replication origins, and regions with newly synthesized DNA (green—early tracks, and red—late tracks). A green star marks a stalled replication fork. The temporal sequence of events is depicted from top to bottom. Work in Xenopus egg cell-free extracts and in mammalian cells indicates that MYC overexpression perturbs the spatiotemporal program of DNA replication, as an excess number of replication origins are activated in response to MYC deregulation (Dominguez-Sola et al. 2007; Srinivasan et al. 2013). An excess number of active replication origins creates important challenges to normal DNA replication progression (“replication stress”): (1) It imposes an important demand on the replication machinery and the availability of factors essential for normal replication which importantly, are in limiting amounts in all cells. Under conditions of replication stress induced by MYC, cells are extremely dependent on the availability and activity of these factors to sustain normal replication progression (see “rate-limiting steps” box) (Table 2). (2) On the other hand, an increased number of active replication forks may lead to complex spatial and structural interactions with ongoing RNA polymerase transcriptional complexes. This situation can potentially lead to collisions or interferences between replication and transcription machineries. Both scenarios, (1) and (2), can result in the destabilization of the replisome and the collapse of active replication forks, which in turn lead to the generation of DNA double-strand breaks (DSBs). DSBs can only be resolved by the DNA damage repair machinery (through ATM activation), and if unresolved, can lead to the acquisition of gene rearrangements and genomic instability. Rep., replication; RNA pol, RNA polymerase II; Pol, DNA polymerase.
WRN
WRN is a member of the RecQ family of DNA helicases, commonly mutated in Werner syndrome, a genetic disease characterized by premature aging and cancer predisposition. WRN can resolve aberrant DNA intermediates arising at DNA replication forks and thus plays a crucial role in the response to replication stress (Fig. 2) (Sidorova 2008). Notably, loss of WRN activity synergizes with MYC deregulation to induce replication stress and cellular senescence (Grandori et al. 2003; Robinson et al. 2009; Moser et al. 2012). WRN is required to allow MYC overexpressing cells to grow or to form tumors (Grandori et al. 2003; Moser et al. 2012). MYC deregulation in absence of WRN leads to activation of cellular senescence similar to that found in preneoplastic lesions on oncogene deregulation (Grandori et al. 2003, 2004). Furthermore, Emu-MYC mice with deficient WRN helicase activity show a significant delay in lymphoma onset, coinciding with signs of increased DNA damage and senescence in the tumor cells (Moser et al. 2012). The role of WRN is not restricted to limiting MYC-induced replication stress, as deregulation of other oncogenes capable of inducing replication stress (i.e., cyclin E and E2F1) leads to cell death in absence of WRN, or in absence of MUS81, an endonuclease required to process stalled forks and certain replication intermediates in response to replication stress (Franchitto et al. 2008; Murfuni et al. 2013; Neelsen et al. 2013).
ATM and ATR
The timing of origin firing during DNA replication is regulated by the activity of ATM and ATR, which control the density of active replication origins allowing replication to proceed normally and be completed at the end of S phase (Shechter et al. 2004; Shechter and Gautier 2005). Interestingly, ATM and ATR show strong genetic interactions with MYC deregulation during tumorigenesis.
Loss of ATM accelerates the development and progression of MYC-driven tumors, coinciding with impaired p53 activation and reduced apoptosis (Pusapati et al. 2006; Shreeram et al. 2006; Maclean et al. 2007; Reimann et al. 2007). Similarly, addition of caffeine—a known ATM inhibitor—to Xenopus cell-free extracts with MYC overexpression results in bypass of MYC-dependent checkpoints, and replication proceeding despite active replication stress (Dominguez-Sola et al. 2007). ATM activation in response to DNA breaks requires its acetylation by Tip60 (KAT5) (Sun et al. 2005), a histone acetyltransferase recruited to chromatin by different transcription factors including MYC. Tip60 is also involved in chromatin transactions critical to the DDR (Ikura et al. 2000; Frank et al. 2003; Squatrito et al. 2006), and is required to mount an effective DDR on MYC deregulation. Tip60 haploinsufficiency synergizes with MYC deregulation in the development of lymphomas in Emu-MYC mice, coinciding with the abrogation of the DDR in tumor cells, despite the integrity of p53 function (Gorrini et al. 2007). The genetic interaction between MYC, ATM, and Tip60 suggests that the inability to elicit a functional DDR on MYC deregulation facilitates MYC-dependent tumorigenesis.
The role of ATR has been studied in patients of Seckel syndrome, a rare human autosomal recessive disorder caused by homozygous or heterozygous compound mutations in the ATR gene (O’Driscoll et al. 2003). ATR is a crucial factor in the cellular response to replication stress, and its acute inhibition is deleterious to proliferating cells (Cimprich and Cortez 2008; Branzei and Foiani 2010; Couch et al. 2013). Notably, MYC-induced lymphomagenesis driven by the Emu-MYC transgene is inhibited in a faithful mouse model of Seckle syndrome, coinciding with an exacerbation of MYC-induced replicative stress, massive DNA damage, robust checkpoint activation, and cell-cycle arrest in tumor cells (Murga et al. 2011).
Overall, the distinct contributions of ATM and Tip60, or WRN and ATR during MYC-dependent tumorigenesis reflect the specific roles of these factors in the response to MYC deregulation. ATR is essential for the viability of cycling cells, and plays a crucial role during DNA replication by acting locally at active replication forks to monitor fork progression, maintain replisome stability and engage the checkpoint signaling machinery (Cimprich and Cortez 2008). Comparably, WRN is thought to resolve specific replication intermediates during normal DNA replication (Enomoto 2001; Pichierri et al. 2011). In contrast, ATM is not required for cell-cycle progression, and mostly responds to DNA double-strand breaks, which can be a consequence of collapsed replication forks or defects in ATR activity (Cimprich and Cortez 2008; Branzei and Foiani 2010). Thus, both ATR and WRN seem to sit upstream of ATM, which in turn would promote checkpoint activation and DNA damage repair in response to DNA DSBs arising from unresolved replication intermediates.
BRCA1
MYC has been shown to interact with BRCA1 tumor suppressor (see Table 1) (Kennedy et al. 2005). The functional outcome of this interaction is thought to be transcriptional, but it is conceivable that BRCA1 limits MYC-induced DNA damage, especially double-strand breaks (DSBs). BRCA1 tumor suppressor activity is associated with its ability to facilitate homology-dependent repair of DSBs. Of note, MYC deregulation correlates best with the basal-like breast cancer type (Alles et al. 2009), the breast tumor class that is associated with BRCA1 loss.
Nucleotides
Alterations of the cellular nucleotide pools can promote mutagenesis, genome instability, and tumorigenesis, mainly because physiologic nucleotide levels are limiting for normal DNA replication progression (Poli et al. 2012). Increased DNA replication can severely reduce cellular nucleotide pools, which can limit normal replication progression and can trigger replication stress. This phenomenon is caused by an imbalance of factors required for replication progression. In agreement with this idea, experiments in cell culture show that addition of nucleosides to cell culture reduces oncogene-induced replication stress and subsequent DNA damage (Bester et al. 2011). This is consistent with other studies showing how cells may adapt to chronic replication stress by up-regulating their nucleotide pools, thus avoiding activation of checkpoints (Poli et al. 2012). Such adaptive response might be of special significance in cells under replication stress induced by MYC deregulation, as MYC is known to regulate nucleotide pools and stimulate nucleotide biosynthesis (Liu et al. 2008; Mannava et al. 2008). This particular trait could explain why MYC-dependent replication stress fails to activate robust checkpoints in mammalian cells (Dominguez-Sola et al. 2007).
Replication Stress as a Source of Cancer-Related Gene Rearrangements and MYC-Dependent Genomic Instability
As detailed by Kuzyk and Mai (2014) and Gabay et al. (2014), most malignancies triggered by MYC deregulation show clear signs of genomic instability. In vitro experiments, and in vivo experiments in MYC transgenic mice originally showed that transient MYC expression results in activation of the DDR, genomic instability, and chromosomal aberrations typically seen in MYC-dependent human tumors (Felsher and Bishop 1999; Fest et al. 2002; Kuttler and Mai 2006; Prochownik 2008). Induction of karyotypic abnormalities in cells with MYC deregulation occurs via pathways that are in part independent of its transcriptional activity (Menssen et al. 2007).
Genetic rearrangements associated with oncogenic stress/activation can occur as a result of telomere shortening and deprotection when telomerase becomes rate limiting during replication. This phenomenon yields DSBs, which favor aberrant telomere fusions and breakage-fusion cycles, resulting in additional damage and subsequent chromosome rearrangements (Artandi and DePinho 2010). In addition, DSBs may arise following alteration of the normal process of DNA replication and the generation of DNA replication stress by oncogenes such as MYC. Replication stress has been recently linked experimentally to the appearance of genomic instability in cancer (Barlow et al. 2013; Burrell et al. 2013). Abnormal DNA replication events could promote the acquisition of tandem duplications in basal-like breast cancers and analogous rearrangements in different tumor types (Stephens et al. 2009; Dereli-Oz et al. 2011). In this scenario, replication stress would leave behind a DNA “footprint,” in a similar fashion to what has been proposed for UV radiation, alkylating agents, or tobacco exposure (Pleasance et al. 2010; Alexandrov et al. 2013a,b).
One of the mechanisms by which replication stress can impact on genome stability is by promoting the appearance of DSBs during S phase (Branzei and Foiani 2010). S-phase DSBs are then carried over into mitosis and can cause cytokinesis failure and abnormal chromosome segregation (Ichijima et al. 2010; Burrell et al. 2013; Neelsen et al. 2013). Defects in chromosome segregation result in chromosomal instability (CIN), which is a common feature in solid cancers of epithelial origin (Schvartzman et al. 2010). It has been proposed that CIN could have a main impact during preneoplastic stages, by facilitating the loss of tumor suppressor genes (Nowak et al. 2002; Michor et al. 2005; Negrini et al. 2010). In agreement with this notion, the contribution of replication stress to chromosomal instability in colon cancer, a solid cancer commonly studied as a model of CIN, appears to be most significant at the progression point between adenoma (the preneoplastic stage) and carcinoma (Burrell et al. 2013).
DNA replication stress has also been proposed to arise from overreplication. Loss of the once-per-cell-cycle regulation of DNA synthesis results in rereplication of DNA segments owing to persistent origin activation. This phenomenon, which can result in the generation of tandem duplications and gene amplifications (Green et al. 2010; Black et al. 2013), can be detected on activation of the K-Ras oncogene (Di Micco et al. 2006). However, rereplication events are not observed on MYC deregulation in Xenopus cell-free extracts or in mammalian cells (Dominguez-Sola et al. 2007), suggesting the existence of specific differences in how oncogene deregulation disturbs the normal control of DNA replication.
Specific features at the chromosomal region under stress seem to also be important in determining the outcome of the cellular responses to replication stress (Branzei and Foiani 2010). In most mammalian cell types, induction of replication stress alone by oncogene deregulation fails to activate effective checkpoints. In contrast, oncogene-induced replication stress is able to trigger checkpoint responses in ATR- or WRN-deficient cells (see above). Hence, it is possible that the type of lesions induced by oncogene-induced replication stress remain “cryptic” and undetected as the cell cycle proceeds in cells with functional responses (Ichijima et al. 2010). But importantly, not all regions in the genome are equally sensitive to the challenges imposed by replication stress. Replication stress preferentially targets DNA regions located in so-called fragile sites (Bartkova et al. 2005; Gorgoulis et al. 2005; Bartek et al. 2007; Halazonetis et al. 2008), which are defined as genomic regions that selectively accumulate structural abnormalities in response to replication defects (Casper et al. 2002; Durkin and Glover 2007). A fraction of these fragile sites, known as “common fragile sites,” correspond to DNA segments enriched in A-T rich sequences and located in late replicating regions of the genome (Glover 2006; Durkin and Glover 2007), which are characterized by a paucity of replication origins. This architecture makes them extremely vulnerable to defects in replication progression, as they tend to be left unreplicated (Letessier et al. 2011) and can serve as sites of genetic rearrangements at mitosis. A second group of fragile sites is associated instead to early replicating regions, which accumulate damage under replication stress and on MYC deregulation (Barlow et al. 2013). “Early replicating fragile sites” constitute a challenge in situations in which DNA replication initiation is exaggerated, as it occurs on MYC deregulation (Dominguez-Sola et al. 2007; Srinivasan et al. 2013). Notably, a significant fraction of fragile sites overlaps with regions recurrently involved in gene rearrangements in cancer mouse models and in human tumor samples, suggesting that genomic structural abnormalities in cancer can also be the result of replication stress downstream from oncogene activation (Barlow et al. 2013).
CONCLUDING REMARKS AND FUTURE DIRECTIONS
As detailed in this article, the ability of MYC to control DNA replication initiation can provide an alternative explanation to some critical biological effects of MYC in normal and in cancer cells with deregulated MYC alleles. Nonetheless, the relative contribution of this activity to the spectrum of MYC biological effects remains to be explored.
A critical outcome of MYC activity at the onset of DNA replication is to enforce the recruitment of Cdc45 to chromatin. It will be important to assess whether MYC modulates the activity of protein complexes (e.g., Treslin/TopBP1/RecQL4) that influence Cdc45 loading and/or dictates the chromatin context (acetylation or other histone modifications). In turn, these studies could shed light on the significance of known interactions between MYC and some of these factors (e.g., TopBP1), and help understand the functional significance of certain cancer-associated mutations, like those found in RecQL4 (Fang et al. 2013) (refer to the COSMIC database, cancer.sanger.ac.uk/cancergenome/projects/cosmic). Similarly, understanding the molecular and functional relationship between MYC and Cdk-2 should provide important insights into how MYC regulates replication initiation, but most notably, on the possible connection between replication stress and senescence, which as of now, remains a mystery.
A second important question is to understand the genome-wide consequences of MYC on DNA replication initiation, as it would condition our understanding on the global control of DNA replication and the extent of the genomic abnormalities caused by MYC-dependent replication stress. MYC localizes at validated origins of replication in mammalian cells (Dominguez-Sola et al. 2007; Sankar et al. 2009; Swarnalatha et al. 2012), and in Drosophila, MYC colocalizes with Orc2, a key element of the pre-RC, throughout the genome (Yang et al. 2013). However, no genome-wide analysis correlating the location of active origins of replication and MYC-bound sites has been reported to date. Given the unparalleled extent of MYC binding across the genome (Fernandez et al. 2003; Orian et al. 2003; Lin et al. 2012; Nie et al. 2012) and our limited knowledge on how origins of replication are selected in every cell cycle to initiate productive DNA synthesis, the implications for such studies would be far-reaching. The close relationship between MYC chromatin binding and global transcription could provide important clues to understand how the long appreciated connection between transcription and replication origin selection is established (MacAlpine et al. 2004; Kohzaki and Murakami 2005; Sasaki et al. 2006; Sequeira-Mendes et al. 2009; Karnani et al. 2010; Dellino et al. 2013). Moreover, these studies would also help us understand how MYC influences cell differentiation and facilitates cellular reprogramming (Wilson et al. 2004; Cartwright et al. 2005; Leon et al. 2009; Smith et al. 2010), because it is plausible that changes in the distribution, choice, and timing of active replication origins throughout the genome would underlie cell fate transitions (Gilbert 2010; Ryba et al. 2011).
Given the prevalence of oncogene-dependent replication stress in early neoplasms, and the causal relationship between replication stress and genomic instability in cancer cells, it will be critical to understand the contribution of MYC-dependent replication stress to tumorigenesis. Indeed, and as detailed by Gabay et al. (2014), the molecular mechanisms that underlie MYC tumorigenic potential, and those that explain its addictive nature in tumor cells remain poorly understood. Altogether, we anticipate that these studies will bring us closer to defining a unifying theory on the role of MYC in normal and cancer cells.
ACKNOWLEDGMENTS
The authors thank the members of the Gautier and Dalla-Favera laboratory for helpful comments during the preparation of this manuscript. D.D-S. is supported by a Howard Temin Award of the National Cancer Institute (U.S. National Institutes of Health (4R00CA151827). J.G. is supported by NCI awards CACA92245 and CA167826.
Footnotes
Editors: Chi V. Dang and Robert N. Eisenman
Additional Perspectives on MYC and the Pathway to Cancer available at www.perspectivesinmedicine.org
REFERENCES
*Reference is also in this collection.
- Aggarwal BD, Calvi BR 2004. Chromatin regulates origin activity in Drosophila follicle cells. Nature 430: 372–376 [DOI] [PubMed] [Google Scholar]
- Alexandrov LB, Nik-Zainal S, Wedge DC, Campbell PJ, Stratton MR 2013a. Deciphering signatures of mutational processes operative in human cancer. Cell Rep 3: 246–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Borresen-Dale AL, et al. 2013b. Signatures of mutational processes in human cancer. Nature 500: 415–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alles MC, Gardiner-Garden M, Nott DJ, Wang Y, Foekens JA, Sutherland RL, Musgrove EA, Ormandy CJ 2009. Meta-analysis and gene set enrichment relative to ER status reveal elevated activity of MYC and E2F in the “basal” breast cancer subgroup. PloS ONE 4: e4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amati B, Alevizopoulos K, Vlach J 1998. Myc and the cell cycle. Front Biosci 3: d250–d268 [DOI] [PubMed] [Google Scholar]
- Arlt MF, Mulle JG, Schaibley VM, Ragland RL, Durkin SG, Warren ST, Glover TW 2009. Replication stress induces genome-wide copy number changes in human cells that resemble polymorphic and pathogenic variants. Am J Hum Genet 84: 339–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlt MF, Ozdemir AC, Birkeland SR, Wilson TE, Glover TW 2011. Hydroxyurea induces de novo copy number variants in human cells. Proc Natl Acad Sci 108: 17360–17365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artandi SE, DePinho RA 2010. Telomeres and telomerase in cancer. Carcinogenesis 31: 9–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azvolinsky A, Giresi PG, Lieb JD, Zakian VA 2009. Highly transcribed RNA polymerase II genes are impediments to replication fork progression in Saccharomyces cerevisiae. Mol Cell 34: 722–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow JH, Faryabi RB, Callen E, Wong N, Malhowski A, Chen HT, Gutierrez-Cruz G, Sun HW, McKinnon P, Wright G, et al. 2013. Identification of early replicating fragile sites that contribute to genome instability. Cell 152: 620–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartek J, Bartkova J, Lukas J 2007. DNA damage signalling guards against activated oncogenes and tumour progression. Oncogene 26: 7773–7779 [DOI] [PubMed] [Google Scholar]
- Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, Zieger K, Guldberg P, Sehested M, Nesland JM, Lukas C, et al. 2005. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature 434: 864–870 [DOI] [PubMed] [Google Scholar]
- Bartkova J, Rezaei N, Liontos M, Karakaidos P, Kletsas D, Issaeva N, Vassiliou LV, Kolettas E, Niforou K, Zoumpourlis VC, et al. 2006. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature 444: 633–637 [DOI] [PubMed] [Google Scholar]
- Bell SP, Dutta A 2002. DNA replication in eukaryotic cells. Ann Rev Biochem 71: 333–374 [DOI] [PubMed] [Google Scholar]
- Bermejo R, Lai MS, Foiani M 2012. Preventing replication stress to maintain genome stability: Resolving conflicts between replication and transcription. Mol Cell 45: 710–718 [DOI] [PubMed] [Google Scholar]
- Berns K, Hijmans EM, Bernards R 1997. Repression of c-Myc responsive genes in cycling cells causes G1 arrest through reduction of cyclin E/CDK2 kinase activity. Oncogene 15: 1347–1356 [DOI] [PubMed] [Google Scholar]
- Bester AC, Roniger M, Oren YS, Im MM, Sarni D, Chaoat M, Bensimon A, Zamir G, Shewach DS, Kerem B 2011. Nucleotide deficiency promotes genomic instability in early stages of cancer development. Cell 145: 435–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black JC, Manning AL, Van Rechem C, Kim J, Ladd B, Cho J, Pineda CM, Murphy N, Daniels DL, Montagna C, et al. 2013. KDM4A lysine demethylase induces site-specific copy gain and rereplication of regions amplified in tumors. Cell 154: 541–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow JJ, Dutta A 2005. Preventing re-replication of chromosomal DNA. Nat Rev Mol Cell Biol 6: 476–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boos D, Sanchez-Pulido L, Rappas M, Pearl LH, Oliver AW, Ponting CP, Diffley JF 2011. Regulation of DNA replication through Sld3-Dpb11 interaction is conserved from yeast to humans. Curr Biol 21: 1152–1157 [DOI] [PubMed] [Google Scholar]
- Bouchard C, Dittrich O, Kiermaier A, Dohmann K, Menkel A, Eilers M, Luscher B 2001. Regulation of cyclin D2 gene expression by the Myc/Max/Mad network: Myc-dependent TRRAP recruitment and histone acetylation at the cyclin D2 promoter. Genes Dev 15: 2042–2047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branzei D, Foiani M 2010. Maintaining genome stability at the replication fork. Nat Rev Mol Cell Biol 11: 208–219 [DOI] [PubMed] [Google Scholar]
- Burrell RA, McClelland SE, Endesfelder D, Groth P, Weller MC, Shaikh N, Domingo E, Kanu N, Dewhurst SM, Gronroos E, et al. 2013. Replication stress links structural and numerical cancer chromosomal instability. Nature 494: 492–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadoret JC, Meisch F, Hassan-Zadeh V, Luyten I, Guillet C, Duret L, Quesneville H, Prioleau MN 2008. Genome-wide studies highlight indirect links between human replication origins and gene regulation. Proc Natl Acad Sci 105: 15837–15842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calado DP, Sasaki Y, Godinho SA, Pellerin A, Kochert K, Sleckman BP, de Alboran IM, Janz M, Rodig S, Rajewsky K 2012. The cell-cycle regulator c-Myc is essential for the formation and maintenance of germinal centers. Nat Immunol 13: 1092–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campaner S, Doni M, Hydbring P, Verrecchia A, Bianchi L, Sardella D, Schleker T, Perna D, Tronnersjo S, Murga M, et al. 2010. Cdk2 suppresses cellular senescence induced by the c-myc oncogene. Nat Cell Biol 12: 54–59 [DOI] [PubMed] [Google Scholar]
- *.Campbell KJ, White RJ 2014. MYC regulation of cell growth through control of transcription by RNA polymerases I and III. Cold Spring Harb Perspect Med 4: a018408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr AM, Paek AL, Weinert T 2011. DNA replication: Failures and inverted fusions. Semin Cell Dev Biol 22: 866–874 [DOI] [PubMed] [Google Scholar]
- Cartwright P, McLean C, Sheppard A, Rivett D, Jones K, Dalton S 2005. LIF/STAT3 controls ES cell self-renewal and pluripotency by a Myc-dependent mechanism. Development 132: 885–896 [DOI] [PubMed] [Google Scholar]
- Casper AM, Nghiem P, Arlt MF, Glover TW 2002. ATR regulates fragile site stability. Cell 111: 779–789 [DOI] [PubMed] [Google Scholar]
- Cimprich KA, Cortez D 2008. ATR: An essential regulator of genome integrity. Nat Rev Mol Cell Biol 9: 616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen SM, Brylawski BP, Cordeiro-Stone M, Kaufman DG 2003. Same origins of DNA replication function on the active and inactive human X chromosomes. J Cell Biochem 88: 923–931 [DOI] [PubMed] [Google Scholar]
- Cornacchia D, Dileep V, Quivy JP, Foti R, Tili F, Santarella-Mellwig R, Antony C, Almouzni G, Gilbert DM, Buonomo SB 2012. Mouse Rif1 is a key regulator of the replication-timing programme in mammalian cells. EMBO J 31: 3678–3690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa A, Ilves I, Tamberg N, Petojevic T, Nogales E, Botchan MR, Berger JM 2011. The structural basis for MCM2-7 helicase activation by GINS and Cdc45. Nat Struct Mol Biol 18: 471–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa A, Hood IV, Berger JM 2013. Mechanisms for initiating cellular DNA replication. Annu Rev Biochem 82: 25–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couch FB, Bansbach CE, Driscoll R, Luzwick JW, Glick GG, Betous R, Carroll CM, Jung SY, Qin J, Cimprich KA, et al. 2013. ATR phosphorylates SMARCAL1 to prevent replication fork collapse. Genes Dev 27: 1610–1623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- d’Adda di Fagagna F 2008. Living on a break: Cellular senescence as a DNA-damage response. Nat Rev Cancer 8: 512–522 [DOI] [PubMed] [Google Scholar]
- *.Dang CV 2013. MYC, metabolism, cell growth, and tumorigenesis. Cold Spring Harb Perspect Med 3: a014217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danis E, Brodolin K, Menut S, Maiorano D, Girard-Reydet C, Mechali M 2004. Specification of a DNA replication origin by a transcription complex. Nat Cell Biol 6: 721–730 [DOI] [PubMed] [Google Scholar]
- de Alboran IM, O’Hagan RC, Gartner F, Malynn B, Davidson L, Rickert R, Rajewsky K, DePinho RA, Alt FW 2001. Analysis of C-MYC function in normal cells via conditional gene-targeted mutation. Immunity 14: 45–55 [DOI] [PubMed] [Google Scholar]
- Deb-Basu D, Aleem E, Kaldis P, Felsher DW 2006. CDK2 is required by MYC to induce apoptosis. Cell Cycle 5: 1342–1347 [DOI] [PubMed] [Google Scholar]
- Dellino GI, Cittaro D, Piccioni R, Luzi L, Banfi S, Segalla S, Cesaroni M, Mendoza-Maldonado R, Giacca M, Pelicci PG 2013. Genome-wide mapping of human DNA-replication origins: Levels of transcription at ORC1 sites regulate origin selection and replication timing. Genome Res 23: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePamphilis ML, Blow JJ, Ghosh S, Saha T, Noguchi K, Vassilev A 2006. Regulating the licensing of DNA replication origins in metazoa. Curr Opin Cell Biol 18: 231–239 [DOI] [PubMed] [Google Scholar]
- Dereli-Oz A, Versini G, Halazonetis TD 2011. Studies of genomic copy number changes in human cancers reveal signatures of DNA replication stress. Mol Oncol 5: 308–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Micco R, Fumagalli M, Cicalese A, Piccinin S, Gasparini P, Luise C, Schurra C, Garre M, Nuciforo PG, Bensimon A, et al. 2006. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature 444: 638–642 [DOI] [PubMed] [Google Scholar]
- Dominguez-Sola D, Ying CY, Grandori C, Ruggiero L, Chen B, Li M, Galloway DA, Gu W, Gautier J, Dalla-Favera R 2007. Non-transcriptional control of DNA replication by c-Myc. Nature 448: 445–451 [DOI] [PubMed] [Google Scholar]
- Dominguez-Sola D, Victora GD, Ying CY, Phan RT, Saito M, Nussenzweig MC, Dalla-Favera R 2012. The proto-oncogene MYC is required for selection in the germinal center and cyclic reentry. Nat Immunol 13: 1083–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durkin SG, Glover TW 2007. Chromosome fragile sites. Ann Rev Genet 41: 169–192 [DOI] [PubMed] [Google Scholar]
- Eilers M, Schirm S, Bishop JM 1991. The MYC protein activates transcription of the α-prothymosin gene. EMBO J 10: 133–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enomoto T 2001. Functions of RecQ family helicases: Possible involvement of Bloom’s and Werner’s syndrome gene products in guarding genome integrity during DNA replication. J Biochem 129: 501–507 [DOI] [PubMed] [Google Scholar]
- Errico A, Costanzo V 2010. Differences in the DNA replication of unicellular eukaryotes and metazoans: Known unknowns. EMBO Rep 11: 270–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evertts AG, Coller HA 2012. Back to the origin: Reconsidering replication, transcription, epigenetics, and cell cycle control. Genes Cancer 3: 678–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang H, Nie L, Chi Z, Liu J, Guo D, Lu X, Hei TK, Balajee AS, Zhao Y 2013. RecQL4 helicase amplification is involved in human breast tumorigenesis. PloS ONE 8: e69600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsher DW, Bishop JM 1999. Transient excess of MYC activity can elicit genomic instability and tumorigenesis. Proc Natl Acad Sci 96: 3940–3944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez PC, Frank SR, Wang L, Schroeder M, Liu S, Greene J, Cocito A, Amati B 2003. Genomic targets of the human c-Myc protein. Genes Dev 17: 1115–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fest T, Mougey V, Dalstein V, Hagerty M, Milette D, Silva S, Mai S 2002. c-MYC overexpression in Ba/F3 cells simultaneously elicits genomic instability and apoptosis. Oncogene 21: 2981–2990 [DOI] [PubMed] [Google Scholar]
- Franchitto A, Pirzio LM, Prosperi E, Sapora O, Bignami M, Pichierri P 2008. Replication fork stalling in WRN-deficient cells is overcome by prompt activation of a MUS81-dependent pathway. J Cell Biol 183: 241–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank SR, Schroeder M, Fernandez P, Taubert S, Amati B 2001. Binding of c-Myc to chromatin mediates mitogen-induced acetylation of histone H4 and gene activation. Genes Dev 15: 2069–2082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank SR, Parisi T, Taubert S, Fernandez P, Fuchs M, Chan HM, Livingston DM, Amati B 2003. MYC recruits the TIP60 histone acetyltransferase complex to chromatin. EMBO Rep 4: 575–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furstenthal L, Swanson C, Kaiser BK, Eldridge AG, Jackson PK 2001. Triggering ubiquitination of a CDK inhibitor at origins of DNA replication. Nat Cell Biol 3: 715–722 [DOI] [PubMed] [Google Scholar]
- *.Gabay M, Li Y, Felsher DW 2014. MYC activation is a hallmark of cancer initiation and maintenance. Cold Spring Harb Perspect Med 10.1101/cshperspect.a014241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert DM 2010. Cell fate transitions and the replication timing decision point. J Cell Biol 191: 899–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover TW 2006. Common fragile sites. Cancer Lett 232: 4–12 [DOI] [PubMed] [Google Scholar]
- Gomez-Roman N, Grandori C, Eisenman RN, White RJ 2003. Direct activation of RNA polymerase III transcription by c-Myc. Nature 421: 290–294 [DOI] [PubMed] [Google Scholar]
- Gorgoulis VG, Vassiliou LV, Karakaidos P, Zacharatos P, Kotsinas A, Liloglou T, Venere M, Ditullio RA Jr, Kastrinakis NG, Levy B, et al. 2005. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature 434: 907–913 [DOI] [PubMed] [Google Scholar]
- Gorrini C, Squatrito M, Luise C, Syed N, Perna D, Wark L, Martinato F, Sardella D, Verrecchia A, Bennett S, et al. 2007. Tip60 is a haplo-insufficient tumour suppressor required for an oncogene-induced DNA damage response. Nature 448: 1063–1067 [DOI] [PubMed] [Google Scholar]
- Grandori C, Wu KJ, Fernandez P, Ngouenet C, Grim J, Clurman BE, Moser MJ, Oshima J, Russell DW, Swisshelm K, et al. 2003. Werner syndrome protein limits MYC-induced cellular senescence. Genes Dev 17: 1569–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandori C, Robinson KL, Galloway DA, Swisshelm K 2004. Functional link between Myc and the Werner gene in tumorigenesis. Cell Cycle 3: 22–25 [PubMed] [Google Scholar]
- Grandori C, Gomez-Roman N, Felton-Edkins ZA, Ngouenet C, Galloway DA, Eisenman RN, White RJ 2005. c-Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nat Cell Biol 7: 311–318 [DOI] [PubMed] [Google Scholar]
- Green BM, Finn KJ, Li JJ 2010. Loss of DNA replication control is a potent inducer of gene amplification. Science 329: 943–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez C, Guo ZS, Farrell-Towt J, Ju G, DePamphilis ML 1987. c-myc protein and DNA replication: Separation of c-myc antibodies from an inhibitor of DNA synthesis. Mol Cell Biol 7: 4594–4598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez C, Guo ZS, Burhans W, DePamphilis ML, Farrell-Towt J, Ju G 1988. Is c-myc protein directly involved in DNA replication? Science 240: 1202–1203 [DOI] [PubMed] [Google Scholar]
- Halazonetis TD, Gorgoulis VG, Bartek J 2008. An oncogene-induced DNA damage model for cancer development. Science 319: 1352–1355 [DOI] [PubMed] [Google Scholar]
- Hartwell LH, Kastan MB 1994. Cell cycle control and cancer. Science 266: 1821–1828 [DOI] [PubMed] [Google Scholar]
- Hayano M, Kanoh Y, Matsumoto S, Renard-Guillet C, Shirahige K, Masai H 2012. Rif1 is a global regulator of timing of replication origin firing in fission yeast. Genes Dev 26: 137–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermeking H, Wolf DA, Kohlhuber F, Dickmanns A, Billaud M, Fanning E, Eick D 1994. Role of c-myc in simian virus 40 large tumor antigen-induced DNA synthesis in quiescent 3T3-L1 mouse fibroblasts. Proc Natl Acad Sci 91: 10412–10416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermeking H, Rago C, Schuhmacher M, Li Q, Barrett JF, Obaya AJ, O’Connell BC, Mateyak MK, Tam W, Kohlhuber F, et al. 2000. Identification of CDK4 as a target of c-MYC. Proc Natl Acad Sci 97: 2229–2234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold S, Wanzel M, Beuger V, Frohme C, Beul D, Hillukkala T, Syvaoja J, Saluz HP, Haenel F, Eilers M 2002. Negative regulation of the mammalian UV response by Myc through association with Miz-1. Mol Cell 10: 509–521 [DOI] [PubMed] [Google Scholar]
- Herold S, Hock A, Herkert B, Berns K, Mullenders J, Beijersbergen R, Bernards R, Eilers M 2008. Miz1 and HectH9 regulate the stability of the checkpoint protein, TopBP1. EMBO J 27: 2851–2861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzel M, Kohlhuber F, Schlosser I, Holzel D, Luscher B, Eick D 2001. Myc/Max/Mad regulate the frequency but not the duration of productive cell cycles. EMBO Rep 2: 1125–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huvet M, Nicolay S, Touchon M, Audit B, d’Aubenton-Carafa Y, Arneodo A, Thermes C 2007. Human gene organization driven by the coordination of replication and transcription. Genome Res 17: 1278–1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hydbring P, Bahram F, Su Y, Tronnersjo S, Hogstrand K, von der Lehr N, Sharifi HR, Lilischkis R, Hein N, Wu S, et al. 2010. Phosphorylation by Cdk2 is required for Myc to repress Ras-induced senescence in cotransformation. Proc Natl Acad Sci 107: 58–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichijima Y, Yoshioka K, Yoshioka Y, Shinohe K, Fujimori H, Unno J, Takagi M, Goto H, Inagaki M, Mizutani S, et al. 2010. DNA lesions induced by replication stress trigger mitotic aberration and tetraploidy development. PloS ONE 5: e8821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iguchi-Ariga SM, Itani T, Kiji Y, Ariga H 1987. Possible function of the c-myc product: Promotion of cellular DNA replication. EMBO J 6: 2365–2371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iguchi-Ariga SM, Okazaki T, Itani T, Ogata M, Sato Y, Ariga H 1988. An initiation site of DNA replication with transcriptional enhancer activity present upstream of the c-myc gene. EMBO J 7: 3135–3142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikura T, Ogryzko VV, Grigoriev M, Groisman R, Wang J, Horikoshi M, Scully R, Qin J, Nakatani Y 2000. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell 102: 463–473 [DOI] [PubMed] [Google Scholar]
- Kaczmarek L, Hyland JK, Watt R, Rosenberg M, Baserga R 1985. Microinjected c-myc as a competence factor. Science 228: 1313–1315 [DOI] [PubMed] [Google Scholar]
- Karnani N, Taylor CM, Malhotra A, Dutta A 2010. Genomic study of replication initiation in human chromosomes reveals the influence of transcription regulation and chromatin structure on origin selection. Mol Biol Cell 21: 393–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy RD, Gorski JJ, Quinn JE, Stewart GE, James CR, Moore S, Mulligan K, Emberley ED, Lioe TF, Morrison PJ, et al. 2005. BRCA1 and c-Myc associate to transcriptionally repress psoriasin, a DNA damage-inducible gene. Cancer Res 65: 10265–10272 [DOI] [PubMed] [Google Scholar]
- Knoepfler PS, Cheng PF, Eisenman RN 2002. N-myc is essential during neurogenesis for the rapid expansion of progenitor cell populations and the inhibition of neuronal differentiation. Genes Dev 16: 2699–2712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoepfler PS, Zhang XY, Cheng PF, Gafken PR, McMahon SB, Eisenman RN 2006. Myc influences global chromatin structure. EMBO J 25: 2723–2734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knott SR, Viggiani CJ, Tavare S, Aparicio OM 2009. Genome-wide replication profiles indicate an expansive role for Rpd3L in regulating replication initiation timing or efficiency, and reveal genomic loci of Rpd3 function in Saccharomyces cerevisiae. Genes Dev 23: 1077–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knott SR, Peace JM, Ostrow AZ, Gan Y, Rex AE, Viggiani CJ, Tavare S, Aparicio OM 2012. Forkhead transcription factors establish origin timing and long-range clustering in S. cerevisiae. Cell 148: 99–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch HB, Zhang R, Verdoodt B, Bailey A, Zhang CD, Yates JR 3rd, Menssen A, Hermeking H 2007. Large-scale identification of c-MYC-associated proteins using a combined TAP/MudPIT approach. Cell Cycle 6: 205–217 [DOI] [PubMed] [Google Scholar]
- Kohzaki H, Murakami Y 2005. Transcription factors and DNA replication origin selection. BioEssays 27: 1107–1116 [DOI] [PubMed] [Google Scholar]
- Kuilman T, Michaloglou C, Mooi WJ, Peeper DS 2010. The essence of senescence. Genes Dev 24: 2463–2479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai A, Lee J, Yoo HY, Dunphy WG 2006. TopBP1 activates the ATR-ATRIP complex. Cell 124: 943–955 [DOI] [PubMed] [Google Scholar]
- Kumagai A, Shevchenko A, Shevchenko A, Dunphy WG 2010. Treslin collaborates with TopBP1 in triggering the initiation of DNA replication. Cell 140: 349–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai A, Shevchenko A, Shevchenko A, DUnphy WG 2011. Direct regulation of Treslin by cyclin-dependent kinase is essential for the onset of DNA replication. J Cell Biol 193: 995–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Marques M, Carrera AC 2006. Phosphoinositide 3-kinase activation in late G1 is required for c-Myc stabilization and S-phase entry. Mol Cell Biol 26: 9116–9125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuttler F, Mai S 2006. c-Myc, genomic instability and disease. Genome Dyn 1: 171–190 [DOI] [PubMed] [Google Scholar]
- *.Kuzyk A, Mai S 2014. c-MYC-induced genomic instability. Cold Spring Harb Perspect Med 4: a014373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurenti E, Varnum-Finney B, Wilson A, Ferrero I, Blanco-Bose WE, Ehninger A, Knoepfler PS, Cheng PF, MacDonald HR, Eisenman RN, et al. 2008. Hematopoietic stem cell function and survival depend on c-Myc and N-Myc activity. Cell Stem Cell 3: 611–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JA, Carvalho CM, Lupski JR 2007. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell 131: 1235–1247 [DOI] [PubMed] [Google Scholar]
- Lemaitre JM, Bocquet S, Buckle R, Mechali M 1995. Selective and rapid nuclear translocation of a c-Myc-containing complex after fertilization of Xenopus laevis eggs. Mol Cell Biol 15: 5054–5062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leon J, Ferrandiz N, Acosta JC, Delgado MD 2009. Inhibition of cell differentiation: A critical mechanism for MYC-mediated carcinogenesis? Cell Cycle 8: 1148–1157 [DOI] [PubMed] [Google Scholar]
- Letessier A, Millot GA, Koundrioukoff S, Lachages AM, Vogt N, Hansen RS, Malfoy B, Brison O, Debatisse M 2011. Cell-type-specific replication initiation programs set fragility of the FRA3B fragile site. Nature 470: 120–123 [DOI] [PubMed] [Google Scholar]
- Li H, Lee TH, Avraham H 2002. A novel tricomplex of BRCA1, Nmi, and c-Myc inhibits c-Myc-induced human telomerase reverse transcriptase gene (hTERT) promoter activity in breast cancer. J Biol Chem 277: 20965–20973 [DOI] [PubMed] [Google Scholar]
- Lin CY, Loven J, Rahl PB, Paranal RM, Burge CB, Bradner JE, Lee TI, Young RA 2012. Transcriptional amplification in tumor cells with elevated c-Myc. Cell 151: 56–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YC, Li F, Handler J, Huang CR, Xiang Y, Neretti N, Sedivy JM, Zeller KI, Dang CV 2008. Global regulation of nucleotide biosynthetic genes by c-Myc. PloS ONE 3: e2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAlpine DM, Rodriguez HK, Bell SP 2004. Coordination of replication and transcription along a Drosophila chromosome. Genes Dev 18: 3094–3105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maclean KH, Kastan MB, Cleveland JL 2007. Atm deficiency affects both apoptosis and proliferation to augment Myc-induced lymphomagenesis. Mol Cancer Res 5: 705–711 [DOI] [PubMed] [Google Scholar]
- Mannava S, Grachtchouk V, Wheeler LJ, Im M, Zhuang D, Slavina EG, Mathews CK, Shewach DS, Nikiforov MA 2008. Direct role of nucleotide metabolism in C-MYC-dependent proliferation of melanoma cells. Cell Cycle 7: 2392–2400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantiero D, Mackenzie A, Donaldson A, Zegerman P 2011. Limiting replication initiation factors execute the temporal programme of origin firing in budding yeast. EMBO J 30: 4805–4814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques M, Kumar A, Cortes I, Gonzalez-Garcia A, Hernandez C, Moreno-Ortiz MC, Carrera AC 2008. Phosphoinositide 3-kinases p110α and p110β regulate cell cycle entry, exhibiting distinct activation kinetics in G1 phase. Mol Cell Biol 28: 2803–2814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinato F, Cesaroni M, Amati B, Guccione E 2008. Analysis of Myc-induced histone modifications on target chromatin. PloS ONE 3: e3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateyak MK, Obaya AJ, Adachi S, Sedivy JM 1997. Phenotypes of c-Myc-deficient rat fibroblasts isolated by targeted homologous recombination. Cell Growth Differ 8: 1039–1048 [PubMed] [Google Scholar]
- Mechali M 2010. Eukaryotic DNA replication origins: Many choices for appropriate answers. Nat Rev Mol Cell Biol 11: 728–738 [DOI] [PubMed] [Google Scholar]
- Menssen A, Epanchintsev A, Lodygin D, Rezaei N, Jung P, Verdoodt B, Diebold J, Hermeking H 2007. c-MYC delays prometaphase by direct transactivation of MAD2 and BubR1: Identification of mechanisms underlying c-MYC-induced DNA damage and chromosomal instability. Cell Cycle 6: 339–352 [DOI] [PubMed] [Google Scholar]
- Mesner LD, Crawford EL, Hamlin JL 2006. Isolating apparently pure libraries of replication origins from complex genomes. Mol cell 21: 719–726 [DOI] [PubMed] [Google Scholar]
- Michor F, Iwasa Y, Vogelstein B, Lengauer C, Nowak MA 2005. Can chromosomal instability initiate tumorigenesis? Semin Cancer Biol 15: 43–49 [DOI] [PubMed] [Google Scholar]
- Miotto B, Struhl K 2010. HBO1 histone acetylase activity is essential for DNA replication licensing and inhibited by Geminin. Mol Cell 37: 57–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Morrish F, Hockenbery D 2014. MYC and mitochondrial biogenesis. Cold Spring Harb Perspect Med 4: a014225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrish F, Neretti N, Sedivy JM, Hockenbery DM 2008. The oncogene c-Myc coordinates regulation of metabolic networks to enable rapid cell cycle entry. Cell Cycle 7: 1054–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser R, Toyoshima M, Robinson K, Gurley KE, Howie HL, Davison J, Morgan M, Kemp CJ, Grandori C 2012. MYC-driven tumorigenesis is inhibited by WRN syndrome gene deficiency. Mol Cancer Res 10: 535–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller AC, Keaton MA, Dutta A 2011. DNA replication: Mammalian Treslin-TopBP1 interaction mirrors yeast Sld3-Dpb11. Curr Biol 21: R638–R640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller D, Bouchard C, Rudolph B, Steiner P, Stuckmann I, Saffrich R, Ansorge W, Huttner W, Eilers M 1997. Cdk2-dependent phosphorylation of p27 facilitates its Myc-induced release from cyclin E/cdk2 complexes. Oncogene 15: 2561–2576 [DOI] [PubMed] [Google Scholar]
- Muncan V, Sansom OJ, Tertoolen L, Phesse TJ, Begthel H, Sancho E, Cole AM, Gregorieff A, de Alboran IM, Clevers H, et al. 2006. Rapid loss of intestinal crypts upon conditional deletion of the Wnt/Tcf-4 target gene c-Myc. Mol Cell Biol 26: 8418–8426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murfuni I, Nicolai S, Baldari S, Crescenzi M, Bignami M, Franchitto A, Pichierri P 2013. The WRN and MUS81 proteins limit cell death and genome instability following oncogene activation. Oncogene 32: 610–620 [DOI] [PubMed] [Google Scholar]
- Murga M, Campaner S, Lopez-Contreras AJ, Toledo LI, Soria R, Montana MF, D’Artista L, Schleker T, Guerra C, Garcia E, et al. 2011. Exploiting oncogene-induced replicative stress for the selective killing of Myc-driven tumors. Nat Struct Mol Biol 18: 1331–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Necsulea A, Guillet C, Cadoret JC, Prioleau MN, Duret L 2009. The relationship between DNA replication and human genome organization. Mol Biol Evol 26: 729–741 [DOI] [PubMed] [Google Scholar]
- Neelsen KJ, Zanini IM, Herrador R, Lopes M 2013. Oncogenes induce genotoxic stress by mitotic processing of unusual replication intermediates. J Cell Biol 200: 699–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrini S, Gorgoulis VG, Halazonetis TD 2010. Genomic instability—An evolving hallmark of cancer. Nat Rev Mol Cell Biol 11: 220–228 [DOI] [PubMed] [Google Scholar]
- Nie Z, Hu G, Wei G, Cui K, Yamane A, Resch W, Wang R, Green DR, Tessarollo L, Casellas R, et al. 2012. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell 151: 68–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak MA, Komarova NL, Sengupta A, Jallepalli PV, Shih Ie M, Vogelstein B, Lengauer C 2002. The role of chromosomal instability in tumor initiation. Proc Natl Acad Sci 99: 16226–16231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obaya AJ, Mateyak MK, Sedivy JM 1999. Mysterious liaisons: The relationship between c-Myc and the cell cycle. Oncogene 18: 2934–2941 [DOI] [PubMed] [Google Scholar]
- Obaya AJ, Kotenko I, Cole MD, Sedivy JM 2002. The proto-oncogene c-myc acts through the cyclin-dependent kinase (Cdk) inhibitor p27Kip1 to facilitate the activation of Cdk4/6 and early G1 phase progression. J Biol Chem 277: 31263–31269 [DOI] [PubMed] [Google Scholar]
- O’Connell BC, Cheung AF, Simkevich CP, Tam W, Ren X, Mateyak MK, Sedivy JM 2003. A large scale genetic analysis of c-Myc-regulated gene expression patterns. J Biol Chem 278: 12563–12573 [DOI] [PubMed] [Google Scholar]
- O’Driscoll M, Ruiz-Perez VL, Woods CG, Jeggo PA, Goodship JA 2003. A splicing mutation affecting expression of ataxia-telangiectasia and Rad3-related protein (ATR) results in Seckel syndrome. Nat Genet 33: 497–501 [DOI] [PubMed] [Google Scholar]
- O’Hagan RC, Ohh M, David G, de Alboran IM, Alt FW, Kaelin WG Jr, DePinho RA 2000. Myc-enhanced expression of Cul1 promotes ubiquitin-dependent proteolysis and cell cycle progression. Genes Dev 14: 2185–2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orian A, van Steensel B, Delrow J, Bussemaker HJ, Li L, Sawado T, Williams E, Loo LW, Cowley SM, Yost C, et al. 2003. Genomic binding by the Drosophila Myc, Max, Mad/Mnt transcription factor network. Genes Dev 17: 1101–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacek M, Tutter AV, Kubota Y, Takisawa H, Walter JC 2006. Localization of MCM2-7, Cdc45, and GINS to the site of DNA unwinding during eukaryotic DNA replication. Mol Cell 21: 581–587 [DOI] [PubMed] [Google Scholar]
- Perez-Roger I, Solomon DL, Sewing A, Land H 1997. Myc activation of cyclin E/Cdk2 kinase involves induction of cyclin E gene transcription and inhibition of p27Kip1 binding to newly formed complexes. Oncogene 14: 2373–2381 [DOI] [PubMed] [Google Scholar]
- Pichierri P, Ammazzalorso F, Bignami M, Franchitto A 2011. The Werner syndrome protein: Linking the replication checkpoint response to genome stability. Aging 3: 311–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce SB, Yost C, Anderson SA, Flynn EM, Delrow J, Eisenman RN 2008. Drosophila growth and development in the absence of dMyc and dMnt. Dev Biol 315: 303–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleasance ED, Stephens PJ, O’Meara S, McBride DJ, Meynert A, Jones D, Lin ML, Beare D, Lau KW, Greenman C, et al. 2010. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature 463: 184–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poe JC, Minard-Colin V, Kountikov EI, Haas KM, Tedder TF 2012. A c-Myc and surface CD19 signaling amplification loop promotes B cell lymphoma development and progression in mice. J Immunol 189: 2318–2325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poli J, Tsaponina O, Crabbe L, Keszthelyi A, Pantesco V, Chabes A, Lengronne A, Pasero P 2012. dNTP pools determine fork progression and origin usage under replication stress. EMBO J 31: 883–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomerantz RT, O’Donnell M 2008. The replisome uses mRNA as a primer after colliding with RNA polymerase. Nature 456: 762–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prathapam T, Tegen S, Oskarsson T, Trumpp A, Martin GS 2006. Activated Src abrogates the Myc requirement for the G0/G1 transition but not for the G1/S transition. Proc Natl Acad Sci 103: 2695–2700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prochownik EV 2008. c-Myc: Linking transformation and genomic instability. Curr Mol Med 8: 446–458 [DOI] [PubMed] [Google Scholar]
- Pryde F, Jain D, Kerr A, Curley R, Mariotti FR, Vogelauer M 2009. H3 k36 methylation helps determine the timing of cdc45 association with replication origins. PloS ONE 4: e5882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pusapati RV, Rounbehler RJ, Hong S, Powers JT, Yan M, Kiguchi K, McArthur MJ, Wong PK, Johnson DG 2006. ATM promotes apoptosis and suppresses tumorigenesis in response to Myc. Proc Natl Acad Sci 103: 1446–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pusch O, Bernaschek G, Eilers M, Hengstschlager M 1997. Activation of c-Myc uncouples DNA replication from activation of G1-cyclin-dependent kinases. Oncogene 15: 649–656 [DOI] [PubMed] [Google Scholar]
- Reimann M, Loddenkemper C, Rudolph C, Schildhauer I, Teichmann B, Stein H, Schlegelberger B, Dorken B, Schmitt CA 2007. The Myc-evoked DNA damage response accounts for treatment resistance in primary lymphomas in vivo. Blood 110: 2996–3004 [DOI] [PubMed] [Google Scholar]
- Ren J, Jin F, Yu Z, Zhao L, Wang L, Bai X, Zhao H, Yao W, Mi X, Wang E, et al. 2013. MYC overexpression and poor prognosis in sporadic breast cancer with BRCA1 deficiency. Tumour Biol 34: 3945–3958 [DOI] [PubMed] [Google Scholar]
- Robinson K, Asawachaicharn N, Galloway DA, Grandori C 2009. c-Myc accelerates S phase and requires WRN to avoid replication stress. PloS ONE 4: e5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosales T, Nie Z, Kapoor V, Casellas R Jr, Knutson JR, Levens D 2013. Partition of Myc into immobile vs. mobile complexes within nuclei. Sci Rep 3: 1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryba T, Hiratani I, Sasaki T, Battaglia D, Kulik M, Zhang J, Dalton S, Gilbert DM 2011. Replication timing: A fingerprint for cell identity and pluripotency. PLoS Comput Biol 7: e1002225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankar N, Kadeppagari RK, Thimmapaya B 2009. c-Myc-induced aberrant DNA synthesis and activation of DNA damage response in p300 knockdown cells. J Biol Chem 284: 15193–15205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansom OJ, Meniel VS, Muncan V, Phesse TJ, Wilkins JA, Reed KR, Vass JK, Athineos D, Clevers H, Clarke AR 2007. Myc deletion rescues Apc deficiency in the small intestine. Nature 446: 676–679 [DOI] [PubMed] [Google Scholar]
- Sasaki T, Ramanathan S, Okuno Y, Kumagai C, Shaikh SS, Gilbert DM 2006. The Chinese hamster dihydrofolate reductase replication origin decision point follows activation of transcription and suppresses initiation of replication within transcription units. Mol Cell Biol 26: 1051–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorl C, Sedivy JM 2003. Loss of protooncogene c-Myc function impedes G1 phase progression both before and after the restriction point. Mol Biol Cell 14: 823–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schvartzman JM, Sotillo R, Benezra R 2010. Mitotic chromosomal instability and cancer: Mouse modelling of the human disease. Nat Rev Cancer 10: 102–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears R, Nuckolls F, Haura E, Taya Y, Tamai K, Nevins JR 2000. Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev 14: 2501–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segurado M, Tercero JA 2009. The S-phase checkpoint: Targeting the replication fork. Biol Cell 101: 617–627 [DOI] [PubMed] [Google Scholar]
- Seoane J, Le HV, Massague J 2002. Myc suppression of the p21Cip1 Cdk inhibitor influences the outcome of the p53 response to DNA damage. Nature 419: 729–734 [DOI] [PubMed] [Google Scholar]
- Sequeira-Mendes J, Diaz-Uriarte R, Apedaile A, Huntley D, Brockdorff N, Gomez M 2009. Transcription initiation activity sets replication origin efficiency in mammalian cells. PLoS Genet 5: e1000446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shechter D, Gautier J 2005. ATM and ATR check in on origins: A dynamic model for origin selection and activation. Cell Cycle 4: 235–238 [PubMed] [Google Scholar]
- Shechter D, Costanzo V, Gautier J 2004. ATR and ATM regulate the timing of DNA replication origin firing. Nat Cell Biol 6: 648–655 [DOI] [PubMed] [Google Scholar]
- Sherr CJ, Roberts JM 2004. Living with or without cyclins and cyclin-dependent kinases. Genes Dev 18: 2699–2711 [DOI] [PubMed] [Google Scholar]
- Shichiri M, Hanson KD, Sedivy JM 1993. Effects of c-myc expression on proliferation, quiescence, and the G0 to G1 transition in nontransformed cells. Cell Growth Differ 4: 93–104 [PubMed] [Google Scholar]
- Shreeram S, Hee WK, Demidov ON, Kek C, Yamaguchi H, Fornace AJ Jr, Anderson CW, Appella E, Bulavin DV 2006. Regulation of ATM/p53-dependent suppression of myc-induced lymphomas by Wip1 phosphatase. J Exp Med 203: 2793–2799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidorova JM 2008. Roles of the Werner syndrome RecQ helicase in DNA replication. DNA Repair 7: 1776–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KN, Singh AM, Dalton S 2010. Myc represses primitive endoderm differentiation in pluripotent stem cells. Cell Stem Cell 7: 343–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squatrito M, Gorrini C, Amati B 2006. Tip60 in DNA damage response and growth control: Many tricks in one HAT. Trend Cell Biol 16: 433–442 [DOI] [PubMed] [Google Scholar]
- Srinivasan SV, Dominguez-Sola D, Wang LC, Hyrien O, Gautier J 2013. Cdc45 is a critical effector of myc-dependent DNA replication stress. Cell Rep 3: 1629–1639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiger D, Furrer M, Schwinkendorf D, Gallant P 2008. Max-independent functions of Myc in Drosophila melanogaster. Nat Genet 40: 1084–1091 [DOI] [PubMed] [Google Scholar]
- Steiner P, Philipp A, Lukas J, Godden-Kent D, Pagano M, Mittnacht S, Bartek J, Eilers M 1995. Identification of a Myc-dependent step during the formation of active G1 cyclin-cdk complexes. EMBO J 14: 4814–4826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens PJ, McBride DJ, Lin ML, Varela I, Pleasance ED, Simpson JT, Stebbings LA, Leroy C, Edkins S, Mudie LJ, et al. 2009. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature 462: 1005–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studzinski GP, Brelvi ZS, Feldman SC, Watt RA 1986. Participation of c-myc protein in DNA synthesis of human cells. Science 234: 467–470 [DOI] [PubMed] [Google Scholar]
- Sun Y, Jiang X, Chen S, Fernandes N, Price BD 2005. A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proc Natl Acad Sci 102: 13182–13187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarnalatha M, Singh AK, Kumar V 2012. The epigenetic control of E-box and Myc-dependent chromatin modifications regulate the licensing of lamin B2 origin during cell cycle. Nucleic Acids Res 40: 9021–9035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama M, Taira T, Iguchi-Ariga SM, Ariga H 2000a. CDC6 interacts with c-Myc to inhibit E-box-dependent transcription by abrogating c-Myc/Max complex. FEBS Lett 477: 43–48 [DOI] [PubMed] [Google Scholar]
- Takayama MA, Taira T, Tamai K, Iguchi-Ariga SM, Ariga H 2000b. ORC1 interacts with c-Myc to inhibit E-box-dependent transcription by abrogating c-Myc-SNF5/INI1 interaction. Genes Cells 5: 481–490 [DOI] [PubMed] [Google Scholar]
- Tanaka S, Umemori T, Hirai K, Muramatsu S, Kamimura Y, Araki H 2007. CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature 445: 328–332 [DOI] [PubMed] [Google Scholar]
- Tanaka S, Nakato R, Katou Y, Shirahige K, Araki H 2011. Origin association of Sld3, Sld7, and Cdc45 proteins is a key step for determination of origin-firing timing. Curr Biol 21: 2055–2063 [DOI] [PubMed] [Google Scholar]
- Tao L, Dong Z, Leffak M, Zannis-Hadjopoulos M, Price G 2000. Major DNA replication initiation sites in the c-myc locus in human cells. J Cell Biochem 78: 442–457 [DOI] [PubMed] [Google Scholar]
- van de Wetering M, Sancho E, Verweij C, de Lau W, Oving I, Hurlstone A, van der Horn K, Batlle E, Coudreuse D, Haramis AP, et al. 2002. The β-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell 111: 241–250 [DOI] [PubMed] [Google Scholar]
- Vlach J, Hennecke S, Alevizopoulos K, Conti D, Amati B 1996. Growth arrest by the cyclin-dependent kinase inhibitor p27Kip1 is abrogated by c-Myc. EMBO J 15: 6595–6604 [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Zhang H, Kajino K, Greene MI 1998. BRCA1 binds c-Myc and inhibits its transcriptional and transforming activity in cells. Oncogene 17: 1939–1948 [DOI] [PubMed] [Google Scholar]
- Waters CM, Littlewood TD, Hancock DC, Moore JP, Evan GI 1991. c-myc protein expression in untransformed fibroblasts. Oncogene 6: 797–805 [PubMed] [Google Scholar]
- *.Wiese KE, Walz S, von Eyss B, Wolf E, Athineos D, Sansom O, Eilers M 2013. The role of MIZ-1 in Myc-dependent tumorigenesis. Cold Spring Harb Perspect Med 3: a014290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson A, Murphy MJ, Oskarsson T, Kaloulis K, Bettess MD, Oser GM, Pasche AC, Knabenhans C, Macdonald HR, Trumpp A 2004. c-Myc controls the balance between hematopoietic stem cell self-renewal and differentiation. Genes Dev 18: 2747–2763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong PG, Winter SL, Zaika E, Cao TV, Oguz U, Koomen JM, Hamlin JL, Alexandrow MG 2011. Cdc45 limits replicon usage from a low density of preRCs in mammalian cells. PloS ONE 6: e17533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Cetinkaya C, Munoz-Alonso MJ, von der Lehr N, Bahram F, Beuger V, Eilers M, Leon J, Larsson LG 2003. Myc represses differentiation-induced p21CIP1 expression via Miz-1-dependent interaction with the p21 core promoter. Oncogene 22: 351–360 [DOI] [PubMed] [Google Scholar]
- Yabuuchi H, Yamada Y, Uchida T, Sunathvanichkul T, Nakagawa T, Masukata H 2006. Ordered assembly of Sld3, GINS and Cdc45 is distinctly regulated by DDK and CDK for activation of replication origins. EMBO J 25: 4663–4674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki S, Ishii A, Kanoh Y, Oda M, Nishito Y, Masai H 2012. Rif1 regulates the replication timing domains on the human genome. EMBO J 31: 3667–3677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Sung E, Donlin-Asp PG, Corces VG 2013. A subset of Drosophila Myc sites remain associated with mitotic chromosomes colocalized with insulator proteins. Nat Commun 4: 1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You Z, Harvey K, Kong L, Newport J 2002. Xic1 degradation in Xenopus egg extracts is coupled to initiation of DNA replication. Genes Dev 16: 1182–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q, Sicinski P 2004. Mammalian cell cycles without cyclin E-CDK2. Cell Cycle 3: 292–295 [PubMed] [Google Scholar]
- Zhuang D, Mannava S, Grachtchouk V, Tang WH, Patil S, Wawrzyniak JA, Berman AE, Giordano TJ, Prochownik EV, Soengas MS, et al. 2008. C-MYC overexpression is required for continuous suppression of oncogene-induced senescence in melanoma cells. Oncogene 27: 6623–6634 [DOI] [PMC free article] [PubMed] [Google Scholar]