Abstract
Inflammation is associated with the activation of genes that contribute to immune defense and tissue repair. The bromodomain-containing proteins of the BET family, which recognize histone lysine acetylation, play a key role in the transcriptional control of inflammatory genes. Inhibition of BET proteins by the small-molecule inhibitor I-BET affects the expression of a particular subset of inflammatory genes—namely, ones that follow an “analog-like,” but not “digital-like” activation pattern. This ability of I-BET to target genes based on the dynamic pattern of their activation may facilitate the further development of anti-inflammatory treatment protocols that are tuned to the individual or to disease-specific patterns of gene expression.
The small-molecule inhibitor I-BET affects the expression of a subset of inflammatory genes. It may facilitate the development of anti-inflammatory treatments that are tuned to individual- or disease-specific gene expression patterns.
In humans as well as other organisms, tissue damage or interaction with pathogens leads to an inflammatory response associated with the production of numerous biologically active proteins and metabolites (see Nathan 2002). These mediators of the inflammatory response contribute to the elimination of pathogens as well as to the repair of tissue damage caused either by infection or other factors. The persistence of pathogens and environmental stresses makes inflammation an unavoidable part of life, yet the scope and timing of inflammation are crucial for the health of the individual. Recent research shows that chromatin plays an important role in the regulation of inflammation by activating cohorts of inflammatory genes (see Medzhitov and Horng 2009; Smale 2010). The Nicodeme et al. (2010) study describes a novel approach for the therapeutic intervention of inflammation, using a synthetic compound that targets bromodomain and extraterminal domain (BET) family epigenetic regulators of the transcriptional response. BET proteins are “readers” of the acetylated lysines found on the amino-terminal tails of histone proteins in chromatin. Acetylated histones are typically associated with transcriptionally active chromatin regions, and BET proteins play a role in achieving transcriptional competence as effector molecules, via the recruitment and association with other activating factors. Therefore, it follows that inhibitors of BET proteins may have a transcriptionally repressive effect by blocking the capacity of these epigenetic readers to recognize their acetylated lysine targets.
Excessive inflammatory responses are associated with numerous acute and chronic disorders that range from acute bacteria-induced sepsis to lingering chronic inflammatory conditions such as rheumatoid arthritis or Crohn's colitis (see Nathan 2002). Chronic inflammation has also been associated with cancer, as persistent repair of inflamed tissues may increase the likelihood of a cancerous mutation in dividing cells.
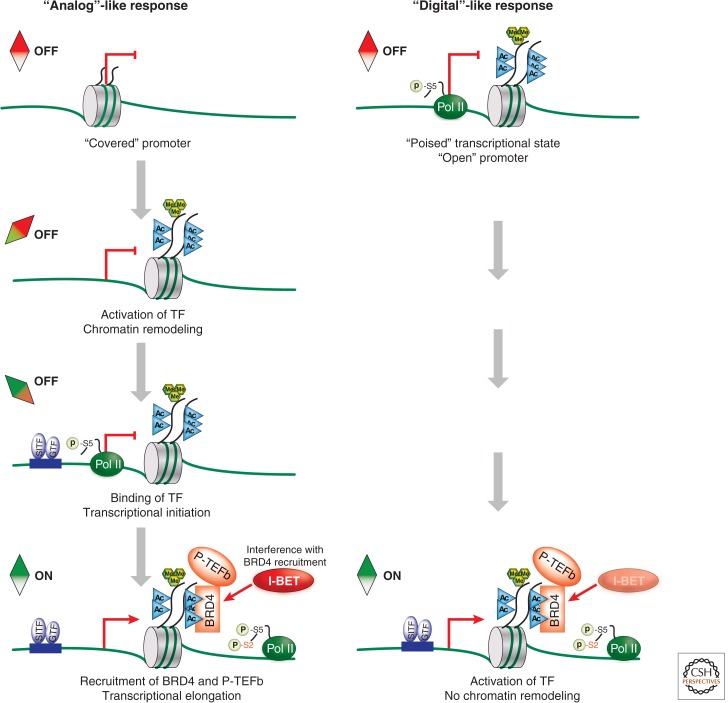
The magnitude and timing of inflammatory responses reflect tightly controlled and cell-type-specific gene expression patterns. Tissue-localized macrophages are the most immediate and perhaps the most potent cellular carriers of inflammatory responses on interaction with pathogens or tissue-derived proinflammatory triggers (see Medzhitov and Horng 2009). The patterns of gene expression associated with inflammation are likely to be trigger-specific. However, much of the general principles that govern inflammation can be discerned from the analysis of the macrophage response to Gram-negative bacteria-derived lipopolysaccharide (LPS), one of the best-studied triggers of inflammation in mice and man. Exposure of macrophages to LPS results in the activation of Toll-like receptor 4 followed by initiation of the signaling program that triggers the inflammatory response (see Medzhitov and Horng 2009; Smale 2010). At the gene level, this response has a well-defined pattern. Shortly after LPS triggering, macrophages up-regulate early response genes that can be divided into generic (e.g., c-Fos) and more inflammation-specific genes (e.g., TNF and Cxcl1/2). As the inflammatory response progresses, the expression of primary response genes is followed by the activation of secondary response genes that encode various regulators of the local and systemic inflammatory response. The stepwise recruitment of primary and secondary response genes into the inflammatory response reflects certain distinct features of these genes. These features include, but are not limited to, differences in AT content of gene promoters, dependence on chromatin-remodeling factors, and abundance of RNA polymerase II at gene promoters before cell activation by LPS (see Smale 2010; Busslinger and Tarakhovsky 2014). To summarize, primary response genes appear to be activated according to a “digital”-like on/off response pattern, whereas secondary response genes follow an “analog”-like activation pattern that requires progression through discrete activation events to enable the initiation of transcription and elongation of mature RNA (see Fig. 1).
Figure 1.
I-BET selectively inhibits genes that follow an “analog”-like activation pattern while not affecting the “digital”-like response. In response to an inflammatory stimulus, secondary response genes follow an “analog”-like activation pattern. This activation involves chromatin remodeling and exposure of the nucleosome-covered gene promoter. Next, transcriptional initiation commences with the binding of stimulus-induced transcription factors (SITF) and general transcription factors (GTF) to the accessible DNA. Induced acetylation of histones H3 and H4 (H3/H4Kac, illustrated as cyan triangles) recruits BRD4 and P-TEFb to chromatin. P-TEFb phosphorylates RNA polymerase II (Pol II) on serine 2 (S2) and allows pause-release of Pol II, resulting in elongation of mature RNA. Conversely, primary response genes follow a “digital”-like activation pattern in response to a stimulus. These genes already have relatively high levels of Pol II and the permissive histone marks, H3K4me3 (green hexagons) and H3/H4Kac, before stimulation, indicating a “poised” state that does not require chromatin remodeling. Stimulation results in TF binding and a H3/H4Kac-dependent recruitment of BRD4 and P-TEFb, allowing for the productive transcription of mature RNA. I-BET selectively prevents the transcription of genes that follow the “analog”-like, but not the “digital”-like activation pattern. This specificity suggests that “analog”-like secondary response genes are more dependent on BET protein function than “digital”-like primary response genes.
The apparent differences in activation of inflammatory genes have suggested that pharmacological regulation of inflammation could be based on a gene's dependence on common regulators of transcriptional responses rather than protein target specificity. Indeed, studies from 2009 showed that a deficiency in the BRD4 epigenetic transcriptional regulator (a BET family protein) resulted in greatly reduced LPS-driven expression of key inflammatory genes (Hargreaves et al. 2009). The natural ligands of BET proteins are the acetyl moieties at the amino-terminal portions of histones H3 and H4. Thus, our pharmacologically driven studies focused on determining the effects of inhibiting the BET family of proteins (BRD2, -3, and -4) implicated in the epigenetic regulation of gene expression during inflammation (Nicodeme et al. 2010). To our advantage was the availability of highly selective BET protein–specific ligands that affect the interaction between BET proteins and their natural ligands. The studies centered on the bromodomain inhibitor I-BET (initially called compound GSK525762A), which binds to BET proteins. We argued that I-BET would affect the ability of BET proteins to bind to their histone ligands, leading to a reduction in gene expression. Moreover, we speculated that the consequence of pharmacological intervention of BET protein–histone association is likely to be gene-specific and reflects the dependence of individual inflammatory genes on BET proteins.
BET proteins are associated with both primary and secondary response genes at relatively low but similar levels before macrophages are triggered with LPS. However, primary response genes have relatively higher levels of RNA polymerase II, and the “permissive” H3K4me3 and H3/H4Kac marks (see Nicodeme et al. 2010). These features suggested that primary genes are poised for immediate “digital” type transcriptional activation. Consequently, they may not require BET proteins as much as secondary response genes, which respond in a more “analog”-like way (see Fig. 1). In contrast, the incremental changes of secondary response genes during their activation require subsequent steps of chromatin remodeling, transcriptional initiation, and transcriptional elongation, making them more susceptible to I-BET. Indeed, we found that treatment of macrophages in vitro with I-BET resulted in strong and selective attenuation of secondary response gene expression while leaving the expression of primary response genes largely unaffected. Significantly, the selective effect of I-BET on secondary response genes holds true for both macrophage and fibroblast responses, not only to LPS, but to secondary mediators of the inflammatory responses such as TNF or IFN-β. We conclude that I-BET appears to “recognize” and target genes based on their epigenetic state.
This selective action of I-BET on secondary response gene activation has potential therapeutic advantages. First, most of the housekeeping genes do not rely on BET proteins for their expression. Second, the gene-state-specific effect of I-BET may enable accurate dosing of the drug within ranges that affect only a particular set of inflammatory genes. Thus, combined with the high rate of I-BET clearance in the organism (see Nicodeme et al. 2010), the duration and dose of I-BET could be computed for a highly selective treatment protocol.
The therapeutic potential of I-BET in in vivo inflammation has been validated by the successful treatment of bacteria- or LPS-induced sepsis in mice. The future of BET inhibitors, including I-BET and related compounds such as JQ1 (Filippakopoulos et al. 2010; Qi 2014), for the treatment of inflammatory disorders will require the disease-tailored administration of I-BET alone or in combination with other drugs that control inflammatory responses. At this time, some of these questions are being addressed in the Phase I clinical trials of I-BET for the treatment of NUT midline carcinoma and also other cancers that are dependent on the continuous expression of MYC.
I-BET's ability to interfere with particular patterns of gene expression has set a strong precedent, validating further searches for synthetic ligands that can interfere with the binding of epigenetic regulators of gene expression to histone or DNA motifs or posttranslational modifications. Such synthetic histone or DNA mimics could represent a novel generation of drugs that target epigenetically defined gene states during inflammation and conceivably other diseases such as cancer (as discussed in Qi 2014).
Footnotes
Editors: C. David Allis, Marie-Laure Caparros, Thomas Jenuwein, and Danny Reinberg
Additional Perspectives on Epigenetics available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- *.Busslinger M, Tarakhovsky A 2014. Epigenetic control of immunity. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, et al. 2010. Selective inhibition of BET bromodomains. Nature 468: 1067–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargreaves DC, Horng T, Medzhitov R 2009. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell 138: 129–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov R, Horng T 2009. Transcriptional control of the inflammatory response. Nat Rev Immunol 9: 692–703 [DOI] [PubMed] [Google Scholar]
- Nathan C 2002. Points of control in inflammation. Nature 420: 846–852 [DOI] [PubMed] [Google Scholar]
- Nicodeme E, Jeffrey KL, Schaefer U, Beinke S, Dewell S, Chung C-W, Chandwani R, Marazzi I, Wilson P, Coste H, et al. 2010. Suppression of inflammation by a synthetic histone mimic. Nature 468: 1119–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Qi J 2014. Bromodomain and extraterminal domain inhibitors (BETi) for cancer therapy: Chemical modulation of chromatin structure. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a018663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smale ST 2010. Selective transcription in response to an inflammatory stimulus. Cell 140: 833–844 [DOI] [PMC free article] [PubMed] [Google Scholar]