Abstract
In this work we review the current knowledge on the prehistory, origins, and evolution of spliceosomal introns. First, we briefly outline the major features of the different types of introns, with particular emphasis on the nonspliceosomal self-splicing group II introns, which are widely thought to be the ancestors of spliceosomal introns. Next, we discuss the main scenarios proposed for the origin and proliferation of spliceosomal introns, an event intimately linked to eukaryogenesis. We then summarize the evidence that suggests that the last eukaryotic common ancestor (LECA) had remarkably high intron densities and many associated characteristics resembling modern intron-rich genomes. From this intron-rich LECA, the different eukaryotic lineages have taken very distinct evolutionary paths leading to profoundly diverged modern genome structures. Finally, we discuss the origins of alternative splicing and the qualitative differences in alternative splicing forms and functions across lineages.
Spliceosomal introns are ubiquitous in nearly every eukaryotic genome. The α-proteobacterial ancestor of mitochondria may have possessed group II self-splicing introns that gave rise to these spliceosomal introns.
SURPRISES AND MYSTERIES OF INTRONS AND INTRON EVOLUTION
With mid-20th century breakthroughs, molecular cell biology finally seemed to obey a relatively simple logic. Genetic information was encoded in DNA genes (Avery et al. 1944; Watson and Crick 1953), which were transcribed into RNA and subsequently translated into functional proteins (Crick 1958). However, a most unexpected finding “interrupted” this logic. The coding information of DNA genes was sometimes broken into pieces separated by sequences whose sole apparent purpose was to generate an extra RNA sequence that then had to be removed to generate intact protein-coding messenger RNAs. The initial findings in viruses (Berget et al. 1977; Chow et al. 1977) were soon extended to many cellular genes. With the advent of large-scale sequencing projects, it became clear that one kind of intron (the spliceosomal introns), as well as the cellular machinery that removes them (the spliceosome), are ubiquitous in eukaryotic genomes. For example, the average human transcript contains ∼9 introns, totaling several hundred thousand introns across the genome and comprising a quarter of the DNA content of each cell (Lander et al. 2001; Venter et al. 2001). Moreover, functions for some introns began to emerge. In particular, by regulating the removal of introns and subsequent rejoining of exons (so-called intron splicing), eukaryotic genes can generate multiple transcripts, vastly expanding molecular diversity. First hypothesized by Gilbert (1978), this process, known as alternative splicing (AS), appears to be widespread in eukaryotes, seemingly reaching its apex in mammals (Barbosa-Morais et al. 2012), in which 95% of multiexon genes undergo AS (Pan et al. 2008; Wang et al. 2008).
Nearly four decades after the discovery of introns, many questions remain unanswered. The most fascinating questions are still the most fundamental ones: Why do introns exist? When and how did they arise? These questions were first formulated as part of the exciting and contentious “introns early/late” debate. Introns early held that introns were very ancient structures that predated cellular life, and that modern organisms with few or no introns had lost them secondarily (in particular, all prokaryotic genomes contain either no introns or only a few nonspliceosomal introns) (see below) (Darnell 1978; Gilbert 1987; Long et al. 1995; de Souza et al. 1996). The related “introns first” hypothesis holds that exons emerged from noncoding regions between RNA genes in the RNA world (Poole et al. 1998; Penny et al. 2009). On the other hand, introns late counters that spliceosomal introns arose later, at some point during eukaryotic evolution (Cavalier-Smith 1985, 1991; Dibb and Newman 1989; Stoltzfus 1994; Logsdon et al. 1995; Logsdon 1998). This debate spanned over 20 years despite (or perhaps owing to) the scarcity of directly relevant data, finding resolution only with the availability of many whole genome sequences. Although adherents to both perspectives remain, the introns early perspective has been weakened by the finding of low or zero intron density in all prokaryotic lineages, and the gradual weakening of (and emergence of potential alternative explanations for) statistical signals suggestive of early introns. However, although, formally, the data tipped the scales in favor of introns late, the current consensus may be seen as a mixture of the two perspectives (Koonin 2006): spliceosomal introns appeared abruptly at the time of the origin of eukaryotes (and thus are quite ancient even if not primordial), and originated from preexisting self-splicing introns, which were likely present at very early stages of life.
TYPES OF LARIAT INTRONS: SELF-SPLICING INTRONS AND THE PREHISTORY OF THE SPLICEOSOMAL SYSTEM
Spliceosomal introns are just one of the four major classes of introns found in nature, together with group I and group II self-splicing introns, and tRNA introns. Intron types are defined based on various structural and mechanistic features, and they have distinct phylogenetic distributions. In this section we discuss different intron types and compare crucial aspects of the evolutionarily related spliceosomal and group II introns, collectively known as lariat introns.
Spliceosomal Introns
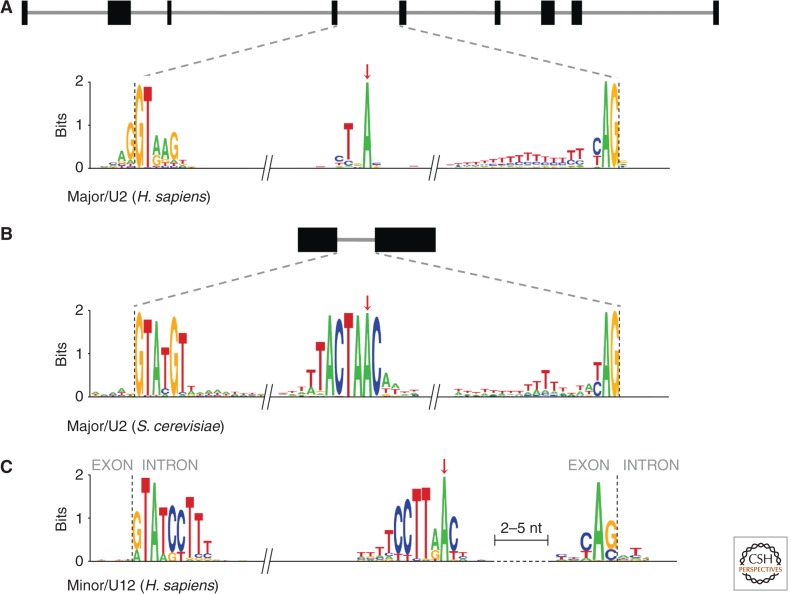
Spliceosomal introns are found in all studied eukaryotic nuclear genomes (with two possible exceptions) (Andersson et al. 2007; Lane et al. 2007) and only in eukaryotic nuclear genomes, although the total number of introns in each species varies by orders of magnitude. They are characterized by their mechanism of splicing, which is catalyzed by the spliceosome, a complex ribonucleoprotein machinery formed by five small RNAs (the U1, U2, U4, U5, and U6 snRNAs) and more than 200 proteins (Wahl et al. 2009). Although there are also marked lineage-specific differences, the sequence structure of most spliceosomal introns consists of a short 5′ splice site (5′ss) boundary, a minimal AG dinucleotide 3′ splice site (3′ss) boundary, a catalytic adenosine (the branch point [BP]), and a polypyrimidine tract between the BP and the 3′ss (Fig. 1). These sequences are recognized and bound by the core components of the spliceosome and are crucial for the splicing reaction (Ruskin and Green 1985; Chiara and Reed 1995; Umen and Guthrie 1995; Chiara et al. 1997; Du and Rosbash 2002). The rest of the intronic sequence, which in some instances in vertebrates may reach up to a million nucleotides, is generally evolutionarily unconstrained and highly variable (Irimia and Roy 2008b). The splicing process consists of two consecutive transesterification reactions, catalyzed by the snRNAs of the spliceosome (Domdey et al. 1984; Padgett et al. 1984; Ruskin et al. 1984; Lin et al. 1985). First, the BP adenosine performs a nucleophilic attack on the 5′ end of the intron, resulting in the cleavage of the 5′ nucleotide of the intron (generally the “G” in the GT) from the upstream exon. Second, the intron is fully excised from the mRNA by another nucleophilic attack by the upstream exon, which is ligated to the 5′ of the downstream exon, generating a precise exon–exon junction and liberating the intronic sequence in a characteristic lariat structure.
Figure 1.
Consensus sequences of spliceosomal intron core splicing signals for (A) human major/U2 introns, (B) yeast major/U2 introns, and (C) human minor/U12 introns. The branch point (BP) adenosine is indicated by a red arrow.
Interestingly, there is another subtype of spliceosomal introns in eukaryotes, the so-called minor or U12 introns (in contrast to the major or U2 subtype described above). The mechanism of splicing is nearly identical, but the minor spliceosome is assembled around four distinct snRNAs—U11, U12, U4atac, and U6atac—and a few specific accessory proteins (Hall and Padgett 1996; Tarn and Steitz 1996; Will et al. 1999); the rest of the protein machinery and the U5 snRNA are shared with the U2 spliceosome. Although the sequence structure of both subtypes is similar overall, U12 introns have a different and stricter consensus sequence at the 5′ss and near the BP, and the distance between the BP and the 3′ss is highly constrained (typically 10–15 nucleotides) (Fig. 1C) (Jackson 1991; Hall and Padgett 1994). Although the much more limited phylogenetic distribution of U12 introns initially suggested they might have arisen more recently in evolution, U12 introns and spliceosomal components have since been discovered in diverse eukaryotic lineages, implying the existence of the U12 system in the last eukaryotic common ancestor (LECA) (Russell et al. 2006; Dávila López et al. 2008).
Group II Self-Splicing Introns
Found in bacterial genomes and in chloroplast and mitochondrial genomes of widely diverged eukaryotes (Ferat and Michel 1993; Lambowitz and Zimmerly 2011; Candales et al. 2012), group II self-splicing introns are believed to predate the origin of eukaryotes, perhaps even preceding the origin of cellular life (Koonin 2006). They have been identified, always in low numbers, in around a quarter of the sequenced bacterial genomes (Lambowitz and Zimmerly 2011). On the other hand, they are rare in archaea (only described, so far, in two related species) (Dai and Zimmerly 2003; Rest and Mindell 2003; Doose et al. 2013). The mechanism of splicing of group II introns is very similar to that of spliceosomal introns—and likely evolutionarily related—involving two transesterification reactions and the release of an excised intron lariat. However, in this case, these reactions are catalyzed by the intronic RNA itself, which has ribozymatic activity (Peebles et al. 1986; Schmelzer and Schweyen 1986; van der Veen et al. 1986). Accordingly, group II intronic RNA sequences show very complex and conserved secondary structures that span 400–800 nucleotides (Lambowitz and Zimmerly 2011). Group II intron sequences are organized in six main domains (DI–DVI) consisting of various functional “loops” and “bulges,” which are the basis of the ribozymatic activity (Michel and Ferat 1995; Qin and Pyle 1998; Lambowitz and Zimmerly 2011). Although group II introns are capable of self-splicing in vitro, efficient splicing in vivo requires specific proteins, which are usually encoded by the intron, but can also be recruited from the host (Solem et al. 2009). The protein encoded by the intron (the “intron-encoded protein,” IEP) usually acts in cis, assisting the splicing only of its own intron. However, in some cases, an IEP can evolve the ability to aid splicing of multiple related introns that proliferated from a single intron copy, providing a common splicing apparatus, and allowing some IEP copies to degenerate (Dai and Zimmerly 2003; Meng et al. 2005). In addition to the role in assisting intron splicing, IEPs are multifunctional proteins that also have reverse transcriptase activity, which allows group II introns to reverse transcribe into DNA, conferring them their nature of mobile genetic elements (Cousineau et al. 2000; Lambowitz and Zimmerly 2011).
There are at least three major subgroups of group II introns, with further subdivisions, distributed among eight phylogenetically supported lineages (Zimmerly et al. 2001; Simon et al. 2008). Each group is characterized by specific RNA secondary structures, and by peculiarities of splicing and mobility (Lambowitz and Zimmerly 2011). In particular, although bacterial introns are usually highly active and functionally complete, introns in eukaryotic organelles often lack important secondary domains and/or encode degenerate IEPs (Copertino and Hallick 1993; Michel and Ferat 1995; Bonen 2008; Barkan 2009). Importantly, the splicing of these degenerate introns likely relies on the action of trans-acting RNAs from other introns and/or host-encoded proteins (Jarrell et al. 1988; Goldschmidt-Clermont et al. 1991; Suchy and Schmelzer 1991; Lambowitz and Zimmerly 2011), potentially mirroring intermediate steps hypothesized for the origin of spliceosomal introns.
Other Types of Introns
(i) Group I introns: present in representatives of most eukaryotic supergroups, either in nuclear ribosomal RNA genes or in mitochondrial and/or plastid genes, as well as in some bacteria and viruses (Haugen et al. 2005). They also catalyze their own splicing reaction by a very different mechanism, utilizing an exogenous guanosine (exoG) as a cofactor that does not release a lariat (Cech et al. 1981; Bass and Cech 1984; van der Horst and Tabak 1985).
(ii) Introns in nuclear and archaeal transfer RNA genes: present in tRNA genes of eukaryotic nuclear or archaeal genomes (Marck and Grosjean 2003; Randau and Söll 2008), and also in some coding genes in archaea (Yokobori et al. 2009; Doose et al. 2013). They do not share functional or structural similarities with the other types of introns, as they are typically very short and their splicing is fully catalyzed by protein enzymes (rather than ribozymes) (Randau and Söll 2008).
ORIGIN AND ESTABLISHMENT OF THE SPLICEOSOMAL SYSTEM DURING EUKARYOGENESIS
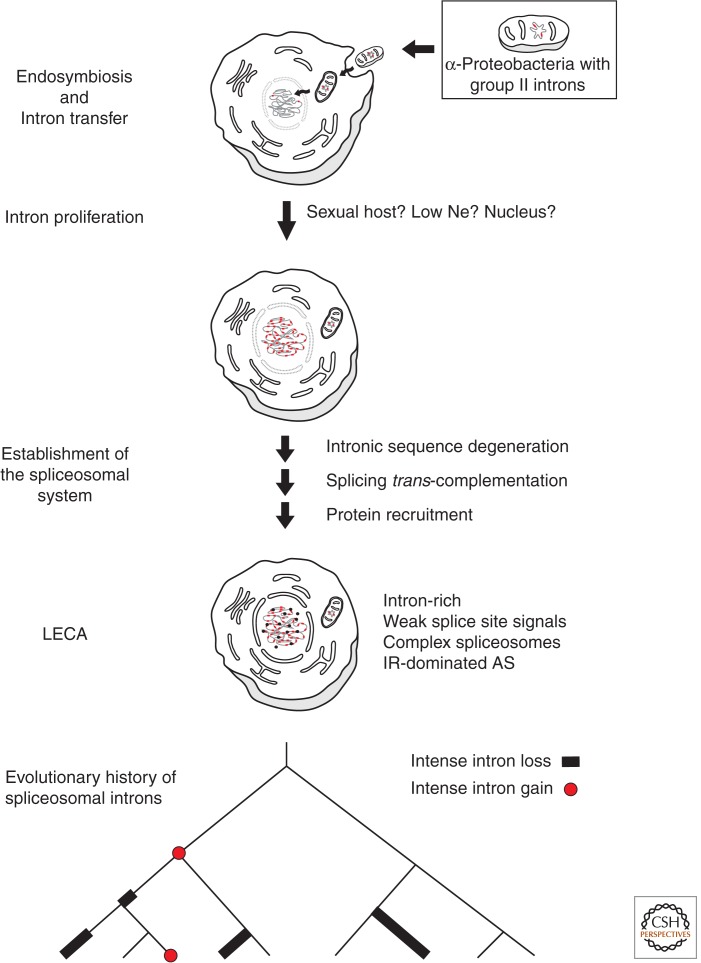
In this section, we discuss the major steps leading to the origin and establishment of the spliceosomal system in eukaryotes. Despite the persistence of disagreement on some important aspects, there is a general consensus about how this process may have unfolded (Fig. 2). According to this general model, spliceosomal introns evolved from invading group II introns, perhaps derived from the early mitochondrion (thought to be descended from an engulfed member of the α-proteobacteria, whose modern members contain group II introns). For some reason(s), these introns then proliferated to an unprecedented level in the host genome. Over time, the self-splicing activities of these many intron copies degenerated, which was associated with the increase of trans-encoded RNAs and proteins that promoted efficient intron splicing, setting the basis of the protospliceosomal machinery, and further releasing selective pressure on cis-intronic splicing elements. As this protospliceosomal machinery recruited more proteins and became more efficient, introns became increasingly reliant on the emerging spliceosome for proper splicing.
Figure 2.
Steps leading to the origin and establishment of the complex spliceosomal system of LECA during eukaryogenesis (see text).
Transfer of Group II Introns to the Host Genome
Structural and functional evidence suggests that spliceosomal and group II self-splicing introns are evolutionarily related. Both types of introns are spliced through a similar two-step catalytic reaction that relies on an endogenous adenosine (the BP), and releases the excised intron as a lariat structure. The two intron types have similar boundary sequences (GT-AY in group II introns and usually GT-AG in spliceosomal introns, although some U12 introns are AT-AC) (Lambowitz and Zimmerly 2011), and there are striking structural similarities between key regions of group II intron domains and spliceosomal snRNAs (Lambowitz and Zimmerly 2011). These include at least (i) domain DV and U6 snRNA, with divalent metal-ion binding sites involved in catalysis and similar base-pairing interactions (Jarrell et al. 1988; Peebles et al. 1995; Yu et al. 1995; Abramovitz et al. 1996; Konforti et al. 1998; Yean et al. 2000; Shukla and Padgett 2002), further supported by crystal structure (Toor et al. 2008; Keating et al. 2010); (ii) ID3 subdomain and d–d′ motifs and U5 snRNA stem loop, involved in the recognition of 5′ and 3′ exons (Hetzer et al. 1997); and (iii) DVI and the U2-intron pairing that include the BP adenosine (Schmelzer and Schweyen 1986; Parker et al. 1987; Li et al. 2011). In addition, it has also been shown that extracts of snRNAs can catalyze both splicing reactions without proteins in vitro (Valadkhan et al. 2007, 2009), in a similar manner to complete group II introns.
Whereas it is theoretically possible that group II introns evolved from spliceosomal introns (or that both share a distinct common ancestor [Vesteg et al. 2012]), it seems much more likely that group II introns gave rise to spliceosomal introns (Cech 1986). The presence of spliceosomal introns in all extant eukaryotic supergroups indicates that this transformation occurred before LECA. The most favored hypothesis is that spliceosomal introns originated from α-proteobacterial group II introns transferred from the protomitochondria to the host genome soon after the endosymbiotic event (Cavalier-Smith 1991), along with other parts of the protomitochondrial genome (Thorsness and Weber 1996; Adams and Palmer 2003). Supporting this view, complete and mobile group II introns are relatively common in modern α-proteobacteria, but (nearly) absent in the archaeal domain (Lambowitz and Zimmerly 2011), to which the preendosymbiotic protoeukaryote is thought to be more closely related (see Guy et al. 2014). In addition, few intron positions are shared between ancient paralogs (i.e., gene duplicates originated before eukaryogenesis), suggesting that at least most introns arose during or after the advent of eukaryotes (Sverdlov et al. 2007).
Unprecedented Group II Intron Proliferation in the Host Genome
It is also widely accepted that, following the transfer of (presumably few) group II introns from the endosymbiont to the host genome, at least one event of massive intron proliferation took place (Fig. 2). From a number perhaps similar to modern α-proteobacteria—usually <30 introns per genome (Lambowitz and Zimmerly 2004)—the ancestors of LECA experienced an expansion of introns that populated their genomes with thousands of elements, estimated at ≥2 introns per kbp of coding sequence, and hypothesized to account for >70% of the ancestral protoeukaryotic genome (Koonin 2009). This proliferation is orders of magnitude larger than any known expansion in prokaryotes, suggesting exceptional circumstances. Although most researchers agree on the evolutionary singularity of this event, the nature of these circumstances is strongly debated. Three (nonmutually exclusive) major explanations have been proposed: (i) the archaeal host lacked unknown defense mechanism against retroelement multiplication (Koonin 2006); (ii) an extremely low effective population size (Ne) after an evolutionary bottleneck allowed the protoeukaryote to fix thousands of intron copies by simple genetic drift, despite the selective disadvantage (Koonin 2006); and (iii) the occurrence of frequent meiosis or meiosislike sexual reproduction in the eukaryotic ancestor allowed large-scale spreading of introns at the expense of the host's fitness (Poole 2006), as previously proposed by Hickey more generally for any class of mobile genetic element (Hickey 1982).
The discovery of two archaea species with a few (4 and 21) group II introns, likely laterally transferred from bacteria (Dai and Zimmerly 2003; Rest and Mindell 2003), for which no dramatic proliferation has occurred, argues that the first explanation is insufficient (the lack of an unknown defense system against group II intron expansion in this lineage). The second proposed explanation, which can be integrated within a more general hypothesis of the origin and evolution of eukaryotes and their genomes (Lynch and Conery 2003; Lynch 2006), currently has considerable support. This hypothesis posits that many eukaryotic genomic features initially proliferated owing to selection being too inefficient to purge them from the population, which situation might arise if the effective population size of these ancestral organisms was very small (a conjecture for which direct tests are very difficult). The presence of these slightly deleterious elements is nonetheless argued to have driven the emergence of profound and defining eukaryotic characteristics, such as the nucleus, as a defense mechanism (Koonin 2006; López-García and Moreira 2006; Martin and Koonin 2006). The third hypothesis (Poole 2006) invokes a very different causality, drawing on the classic arguments on transposable elements by Hickey (1982). Hickey showed that frequent sexual reproduction is crucial to the success of transposable elements: whereas with no or infrequent sex, the fitness costs of aggressively propagating elements will doom them to extinction, in the presence of frequent sex, even highly deleterious elements can spread through the genome. This suggests the possibility that the advent of frequent sexual reproduction could have brought an unprecedented spread of type II introns. Consistent with this notion, meiosis and sexual reproduction appear to have been well established by LECA (Ramesh et al. 2005), although, crucially, the order of appearance in the evolution of frequent sexual reproduction and high intron density remains unknown.
Need and Benefits of Trans-Complementation: Emergence of the Spliceosome
Whatever the cause, the effect of this intron propagation was a genome newly riddled with thousands of elements interrupting most coding genes that required removal from pre-mRNAs. Although intact group II introns are capable of self-splicing and thus may be (nearly) functionally neutral, the need to maintain a high number of constrained functional sequences and tertiary structures in every intron implies strong mutational pressure. Although the number of intronic and exonic sites required for splicing of spliceosomal introns has been estimated to be around 20–40 per intron (Lynch 2002, 2006), the number of potentially constrained sites in a group II intron seems to be at least one order of magnitude higher (Lambowitz and Zimmerly 2011). Therefore, a spliceosomal system that made use of only a few trans-acting factors to replace hundreds of thousands of constrained sequences in cis may have significantly relieved much of the mutational pressure associated with an extremely high number of introns.
There is also compelling mechanistic evidence that the transition from a system of self-spliced introns to one based on trans-complementation may have evolved gradually. The first step would involve the partial degeneration of a number of introns, which would lose important functional structural elements and/or their IEPs. As in many of modern organelle's group II introns (Jarrell et al. 1988; Goldschmidt-Clermont et al. 1991; Suchy and Schmelzer 1991; Lambowitz and Zimmerly 2011), the splicing of these partially degenerated introns would have been assisted in trans by (partial) RNA sequences of other introns. Presumably, some of these sequences were more efficient than others at assisting exogenous splicing, and thus conferred a benefit and became fixed in the populations, eventually giving rise to the snRNAs of the two types of modern spliceosomes (Sharp 1991). Various pieces of evidence support this idea. In addition to their structural similarities, some domains of group II introns can directly replace snRNAs in spliceosomal splicing in vitro (e.g., group II domain DV RNA can act instead of U6atac snRNA [Shukla and Padgett 2002]). Conversely, U5 snRNA can complement group II introns missing the ID3 region (Hetzer et al. 1997).
These (probably quite inefficient) initial protospliceosomes could then have evolved increased efficiency by recruiting auxiliary proteins. Some of these factors could have been derived from the multiple available IEP copies, which can also act in trans in the splicing of related introns (Dai and Zimmerly 2003; Meng et al. 2005). Others could have been coopted from various other processes of RNA metabolism (e.g., Lsm/Sm proteins, whose homologs play diverse roles in prokaryotes) (Wilusz and Wilusz 2005; Mura et al. 2013), or recruited from sequences of retrotransposons or other mobile elements (e.g., Prp8, the largest protein of the spliceosome and an integral part of its catalytic center, is thought to have evolved from a retroelement-encoded reverse transcriptase) (Dlakić and Mushegian 2011). Gene duplication and functional specialization are likely to have also played a role. For example, the aforementioned Sm/Lsm proteins experienced several waves of early gene duplication (Veretnik et al. 2009), which, based on their phylogenetic distribution (present in all eukaryotic groups, and thus in LECA) and the presence of shared intron positions among paralogs, can be traced to the period between the initial intron proliferation and the completion of the fully modern spliceosome (Veretnik et al. 2009). By the time of LECA, a very complex, modern-looking spliceosome, composed of at least 78 proteins and all the snRNAs, had already been established (Collins and Penny 2005).
The Origins of the Minor and Major Spliceosomal Systems
In fact, LECA had not only one complex spliceosome, but two. Comparative genomics have revealed that both U2/major and U12/minor spliceosomal introns, as well as RNA and protein components associated with their distinct splicing machineries, were present in LECA. These two subtypes are quite similar, and they share most of the protein machinery involved in their splicing; however, each subtype uses a specific subset of snRNAs (with the exception of U5), which are likely the most ancestral part of the spliceosome. These subsets of snRNAs are nonetheless evolutionarily related to one another and probably originated by gene duplication some time before LECA (Russell et al. 2006). Thus, debate has emerged as to which subtype originated first in evolution and which one is derived. Several nonconclusive arguments can be put forward for each case (Roy and Irimia 2009; Rogozin et al. 2012). Supporting U12 introns’ ancestrality is their higher structural similarity with group II introns. The BP in both types typically consist of an A within a polypyrimidine tract located at a constant, short distance from the 3′ss (domain DIV in group II introns) (Bonen and Vogel 2001; Lambowitz and Zimmerly 2011); on the other hand, U2 introns’ BPs are usually located further away from the 3′ss, and at a much wider range of distances. In fact, U2 introns ancestrally have a polypyrimidine tract between the BP and the 3′ss (Irimia and Roy 2008a), which has been hypothesized to derive from the ancestral BP sequences of U12 introns (Burge et al. 1998). Along these lines, U12 introns can be viewed in some sense as a transitional form between the highly constrained structures of group II introns (Bonen and Vogel 2001; Lambowitz and Zimmerly 2011) and the diffuse splicing signals of most U2 spliceosomal introns (Irimia et al. 2007b; Irimia and Roy 2008a; Schwartz et al. 2008). Consistent with such a directional process, U12 introns are known to often convert into U2 introns, whereas the reverse process has never been described (e.g., Burge et al. 1998; Alioto 2007). Second, positions of U12 introns may be more conserved between Arabidopsis and humans than those of U2 introns, and they are also more often localized at the 5′ of the gene—a signature of ancestral introns that have resisted intron loss (Basu et al. 2008a).
On the other hand, phylogenetic reconstruction of the insertion sites of ancient (pre-LECA) introns revealed a U2-like insertion bias (reminiscent of the protosplice site model of intron insertion) (Dibb and Newman 1989). This result suggests that U2 introns dominated at least by the time of LECA (Basu et al. 2008c). However, this leaves open the possibility that this proliferation postdates the secondary advent of U2 introns, because group II introns may not have an insertion bias similar to U2 introns. Finally, neither type may be truly ancestral: The two types may have emerged semi-independently from a larger pool of primitive snRNA duplicates with a shared protein machinery.
HISTORY OF THE SPLICEOSOMAL SYSTEM
In this section we will describe the evolutionary history of introns since LECA. First, we discuss attempts to reconstruct LECA's intron–exon structures using comparisons of modern genomes. Then, we summarize the evolutionary trajectories taken by different eukaryotic lineages, ranging from near complete loss and transformation of their ancestral introns, to the intronic stasis observed in many modern lineages.
Reconstruction of Intron–Exon Structures in LECA
The evolutionary steps described in the previous section were largely complete by the time of LECA. The time point corresponding to LECA represents a quite clear divide in our confidence about evolutionary inferences. Reconstruction of steps leading up to LECA (origins of spliceosomal-like introns, reasons for the unprecedented spread of introns, and transfer of splicing from cis- to trans-encoded elements, etc.) remains uncertain because of a lack of close protoeukaryotic-like relatives. In contrast, inferences of features of LECA are relatively much more straightforward because of the possibility of comparing lineages that diverged from LECA. That is, whereas the former by necessity relies on largely indirect evidence, the latter can draw on direct (not to say incontrovertible) evidence.
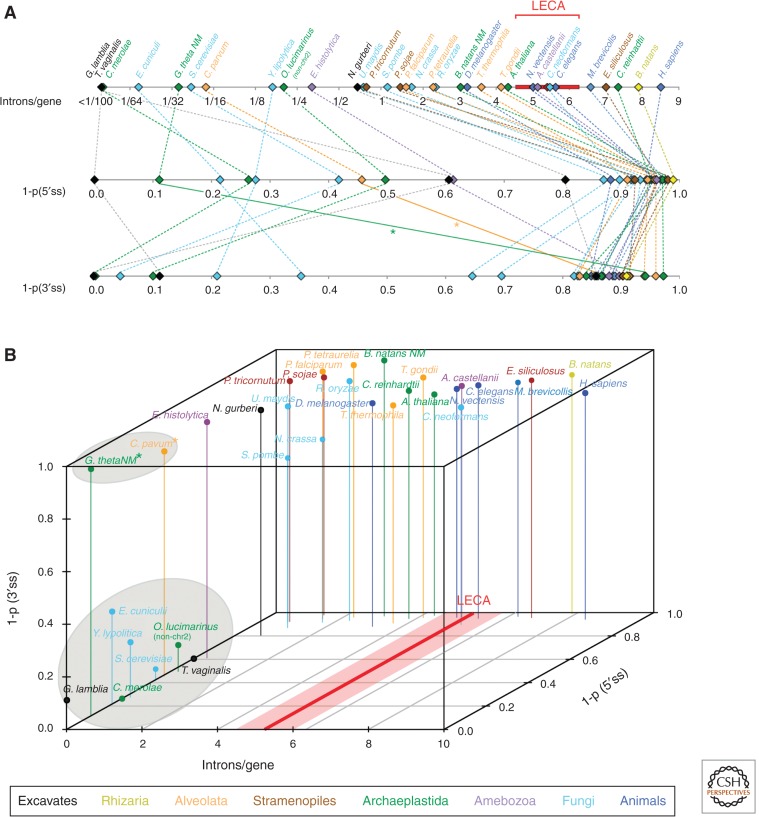
Comparative studies of modern genomes have drawn a surprising picture of the intron–exon structures of genes in the genome of LECA (Roy and Irimia 2009; Rogozin et al. 2012; Koonin et al. 2013), all of them pointing to a complex spliceosomal system. First, comparisons of intron positions across orthologous genes from dozens of eukaryotic genomes have shown that LECA was remarkably intron rich. At its simplest, the logic is that if modern introns were created after the various species diverged, the positions of these introns in homologous genes might be expected to be quite different across species, whereas if ancestral genes already contained many introns and those introns have been retained in modern species, intron positions in homologous genes might largely coincide across species (Fig. 3). This general strategy can be modified to account for the possibility that general preferences in intron insertion (e.g., into protosplice sites) and/or intron phase biases (reviewed in Rogozin et al. 2012) could lead to independent intron insertions occurring at homologous positions in different lineages (Sverdlov et al. 2005; Yoshihama et al. 2006). A variety of studies have shown that genes from intron-rich species from any major eukaryotic supergroup share a significant number of intron positions with other groups (Fedorov et al. 2002; Rogozin et al. 2003; Csurös et al. 2008, 2011). Different investigators have used different methods of evolutionary inference, ranging from parsimony to maximum likelihood, to attempt to reconstruct ancestral intron densities (Rogozin et al. 2003; Qiu et al. 2004; Csurös 2005, 2008; Nguyen et al. 2005; Roy and Gilbert 2005; Carmel et al. 2007, 2009). With increasing taxonomic sampling usually yielding increasingly higher estimates for LECA's intron density, the latest reconstructions (Csurös et al. 2011) used a Markov chain Monte Carlo approach to investigate 99 genomes from five eukaryotic supergroups, and estimated that LECA had 53%–74% of modern human intron density, or 4.5–6.3 introns per gene (assuming similar average protein-coding lengths). That is, available evidence suggests that the intron–exon structures of ancestral eukaryotes were exceedingly complex, with intron densities comparable to the most intron-dense modern densities, and more intron dense than, for instance, modern plants and insects (Fig. 4).
Figure 3.
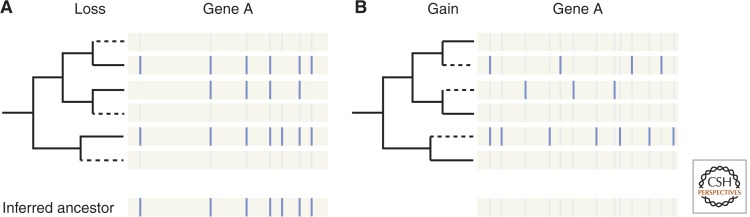
Scenarios for intron evolution: shared ancestral introns versus independent gain. Hypothetical comparisons of intron positions across orthologous genes of distantly related species are shown. (A) Many introns are in homologous positions, suggesting that these introns represent ancestral introns that have been retained. (B) Lack of correspondence between intron positions in different lineages suggest that most introns have been independently gained within each lineage.
Figure 4.
Diversity of intron densities and homogeneity of splicing signals across eukaryotes. (A) Number of introns per gene (top), 1-probability (1-p) that two introns from a species share the same 5′ss (middle) or the same BP motif (bottom) for 33 species from all major eukaryotic supergroups (see color key). (B) Association between these three features: all very intron-poor species (<0.2 introns per gene) show high levels of across-intron homogeneity for 5′ss; nearly all very intron-poor species show high levels of homogeneity for BP (clusters of species inside gray ellipses). Exceptions for BP motifs (Cryptosporidium parvum and Guillardia theta NM) are indicated by asterisks. Inferred intron densities in LECA (Csurös et al. 2011) are indicated in red.
The second unexpected characteristics of LECA's intron–exon structures involve the intron sequences themselves. Nearly all studied eukaryotic species show similar consensus sequences for core splicing sequence motifs—the 5′ splice site (ss), BP, and 3′ss (Fig. 1)—reflecting largely similar sequence preferences across species determined by a shared spliceosomal machinery. However, the strength of these preferences—the extent to which individual introns’ specific motifs adhere to this consensus—varies dramatically (Fig. 4). For instance, whereas human introns use a vast array of 5′ss, 75% of all introns in the model yeast S. cerevisiae have the same 5′ss, GTATGT. Put another way, the probability that two randomly chosen Saccharomyces cerevisiae introns have the same hexamer splice site is 58%, whereas two human 5′ss's will correspond only 6% of the time. Following the intuitive sense that (i) genome complexity will reflect organismal complexity; and (ii) more “highly evolved” organisms have more transformed genomes, it was initially thought that ancestral unicellular eukaryotes would have had “simple” yeastlike intron sequences, with regular and predictable splice signals, and that “complex” diverse signals arose by sequence divergence in multicellular lineages (perhaps related to AS, which makes extensive use of splice site heterogeneity) (Ast 2004). However, comparisons across species revealed that in fact yeast is the exception, with the majority of unicellular and multicellular species alike (and thus likely LECA) utilizing heterogeneous splicing signals, a finding that holds both for the 5′ss and the BP (Irimia et al. 2007a; Irimia and Roy 2008a; Schwartz et al. 2008; Keren et al. 2010). Moreover, LECA's introns also likely harbored a polypyrimidine tract between the BP and the 3′ss—similar to modern animals and plants, but not yeasts—as inferred by the general presence of this signal in representatives of most eukaryotic supergroups (Irimia and Roy 2008a). This sequence is bound in the first steps of spliceosome assembly by the U2AF65 (Zamore et al. 1992), a factor already present in the ancestral spliceosome (Collins and Penny 2005). In addition to modern-looking canonical splicing signals, LECA likely also made use of some kind of auxiliary splicing signals called exonic splicing enhancers (ESE) (Warnecke et al. 2008); again consistent with this inference, most families of SR proteins, which are responsible for binding ESEs, were present in the last common ancestor of eukaryotes (Collins and Penny 2005; Plass et al. 2008).
Beyond the basic splicing machinery, LECA has also been hypothesized to possess other cellular processes often associated with splicing. For example, nonsense-mediated decay (NMD), a mechanism to eliminate spurious or unwanted transcripts, is found in diverse eukaryotes (Amrani et al. 2004; Chen et al. 2008; Jaillon et al. 2008; Kerényi et al. 2008; Roy and Irimia 2009). Indeed NMD and intron spread are likely closely linked, because frequent splicing errors would necessitate NMD, whereas the presence of NMD would decrease the costs of intron spread (Koonin 2006; Lynch et al. 2006). Other “policing” processes, such as the ubiquitin signaling at the proteomic level, could also have evolved as a defense mechanism against massive intron presence (Koonin 2006). The timing of emergence of other secondary aspects of the spliceosomal system, most notably the functional associations between chromatin and splicing seen in animals (Braunschweig et al. 2013), still remain to be elucidated.
Diverging from LECA: Different Modes of Spliceosomal Intron Evolution across Eukaryotes
From these intron-rich ancestors with heterogeneous splicing signals and complex spliceosomes, the full diversity of modern eukaryotic intron–exon structures has emerged over the past 1.5 billion years of evolution. The results of this evolutionary process have been dramatic. Most noticeably, intron densities vary by orders of magnitude, from several per gene and thousands or tens of thousands per genome in many unicellular and multicellular lineages (Merchant et al. 2007; Roy and Irimia 2009; Curtis et al. 2012), to a handful per genome in the genomes of some other species (Fig. 4) (Matsuzaki et al. 2004; Vanacova et al. 2005; Morrison et al. 2007). Notably, these densities do not respect our intuitive sense of biological simplicity and complexity. Many parasitic lineages have intron densities comparable to those of most intron-rich multicellular lineages, and at least one multicellular lineage has intron densities comparable to some of the most intron-poor known unicellulars (Collén et al. 2013). Although the origins of nearly intronless lineages—massive intron loss—are clear, the origins of the highly intron-dense lineages are less so. For some lineages, phylogenetic reconstruction of intron loss and gain suggest that modern high intron density is almost completely owing to retention of ancestral introns (e.g., in the case of vertebrates, intron number appears to have decreased somewhat since the ancestor of animals 1 billion years ago) (Sullivan et al. 2006; Srivastava et al. 2008; Csurös et al. 2011). In other cases, many modern introns appear to have been gained through a variety of processes, most dramatically creation of hundreds or thousands of introns through propagation of intron-creating elements (Worden et al. 2009; Roy and Irimia 2012; van der Burgt et al. 2012). Similarly, organelle-derived or horizontally transferred genes sometimes acquire similar densities to those of more ancestral nuclear host genes (Basu et al. 2008b; Ahmadinejad et al. 2010; Clarke et al. 2013, although see Roy et al. 2006 and Flot et al. 2013). Interestingly, both nearly complete loss of introns and dramatic intron proliferation have occurred several times independently across the eukaryotic tree. For instance, nearly complete loss has occurred in groups as diverged as fungi (at least twice), green and red algae, apicomplexa, and excavates (three times) (Irimia and Roy 2008a). The reasons for this are still unclear, and the proposed explanations range from selection for genome streamlining to runaway intron loss mutation (reviewed in Maeso et al. 2012b). In lineages that have experienced many new intron gains, new introns appear not to have been created at constant rates over millions of years of evolution, but instead to be concentrated in episodes of gain (Roy 2006; Roy and Penny 2006; Csurös et al. 2011). Over recent times, many studied modern lineages appear to be under a sort of intronic evolutionary stasis, experiencing low (or very low) numbers of intron gain and loss (Roy et al. 2003, 2006; Nielsen et al. 2004; Roy and Hartl 2006; Coulombe-Huntington and Majewski 2007a,b; Roy and Penny 2007a; Stajich et al. 2007; Loh et al. 2008; Zhang et al. 2010). On the contrary, only a few known exceptions scattered across the eukaryotic tree show significant ongoing intron gain-dominated evolution (Carmel et al. 2007; Roy and Penny 2007b; Worden et al. 2009; Denoeud et al. 2010; van der Burgt et al. 2012).
In addition to intron densities, splicing signals also show marked differences across genomes (Fig. 4). In particular, 5′ss consensus in a given species may be either very strict, with nearly all introns in the genome having an identical or similar sequence, or highly heterogeneous, as in the case of the human genome (Irimia et al. 2007b). Heterogeneous 5′ss signals are observed in most lineages, whereas the few lineages with strict signals are scattered across the phylogenetic tree, representing exceptions to the general pattern in nearly all major eukaryotic supergroups (Irimia et al. 2007b). This distribution strongly suggests that these exceptions have evolved independently within each lineage, echoing the case for lineages that have undergone nearly complete intron loss. Unexpectedly, these two processes were found to have a very clear correspondence: In nearly intronless species, the few remaining introns have highly similar and constrained 5′ss signals (Fig. 4) (Irimia et al. 2007b). A similar pattern of strict signals has been observed for the BP (Irimia and Roy 2008a; Schwartz et al. 2008), although with a few exceptions involving species with few introns but heterogeneous BPs (Fig. 4). Finally, in a few instances, a further constraint on splicing signals has evolved, in which the BP is “anchored” to the 3′ss, always present at exactly or almost the same number of nucleotides away (Irimia and Roy 2008a). Thus, in these extreme cases, the remaining U2 introns show similarities to U12 introns, which are also found in low numbers per genome: extended constrained 5′ss and BP signals and BP position. Although several explanations have been proposed to explain the coevolution between splicing signals and intron numbers (Irimia et al. 2007b, 2009), the cause remains obscure. It seems likely that an important component is changes in the spliceosome: Massive introns loss is often accompanied by loss of some auxiliary spliceosomal factors largely involved in recognition of auxiliary splicing sequence signals such as ESEs (Plass et al. 2008), presumably leading to a greater emphasis on core splicing signals, and thus requiring increased information content in those signals.
Intron lengths have also experienced considerable divergence across eukaryotes. Although it is difficult to confidently estimate average and median intron length in LECA, they may have been similar to those in most eukaryotic genomes studied so far, perhaps around ∼150 nucleotides on average and with a median centered around 70–90 nucleotides. It should be noted, however, that introns were probably significantly longer at least in earlier ancestors, as group II introns from which they derived are at least 400–800 nucleotides long (Lambowitz and Zimmerly 2011), and they may reach 2.5 kbp when they encode their own IEP (Lambowitz and Zimmerly 2004; Koonin 2009). Surveys of intron lengths across eukaryotes reveal striking deviations in intron length distributions in both directions. First, tiny introns (18–36 nucleotides) have independently evolved in a few species scattered across the eukaryotic tree, in groups as diverged as animals (Ogino et al. 2010; Tsai et al. 2013), green algae (Gilson et al. 2006), and ciliates (Russell et al. 1994). Moreover, in all of these species the distributions of lengths are very sharp—suggesting strong pressure to maintain a seemingly optimal small length—and their splicing signals are highly diffuse (with the noted exception of microsporidians) (Irimia and Roy 2008a; Lee et al. 2010). On the opposite side, introns from certain lineages are unusually long. These include some plants (Jiang and Goertzen 2011), brown algae (Cock et al. 2010), and, especially, vertebrates, with averages one order of magnitude higher than most species, and with some introns reaching up to one megabase. These expansions seem likely to be associated with insertion of transposable elements into intronic sequences (Jiang and Goertzen 2011), although, in some cases, exceptionally long introns may be associated with the presence of large numbers of cis-regulatory elements (Irimia et al. 2011).
FUNCTIONS OF INTRONS: ORIGINS AND EVOLUTION OF ALTERNATIVE SPLICING
Given the fascinating and complex evolutionary paths that intron–exon structures have taken from LECA to each extant eukaryotic lineage, perhaps the major paradox is that, for most cases, introns are not known to play a general functional role in the biology of the cell: They are simply intervening sequences that must be removed for proper gene expression to occur. Several evolutionary functions have been proposed for introns, including enhancing within-gene homologous recombination (Comeron and Kreitman 2000), and facilitation of exon shuffling and thus the evolution of new genes (Gilbert 1987; Liu and Grigoriev 2004). In addition, certain intron sequences are known to play regulatory roles in transcription, either by acting as a limiting factor in cotranscriptional splicing (Patel et al. 2002), or indirectly by harboring DNA regulatory elements (e.g., Epstein et al. 1999; Irimia et al. 2012; Maeso et al. 2012). Similarly, other introns harbor nested bona fide protein-coding or noncoding RNA genes (e.g., Assis et al. 2008; Kumar 2009; Chorev and Carmel 2013). However, the most widespread function of introns is probably through AS, which has been estimated to occur in nearly all human multiexonic genes (Pan et al. 2008; Wang et al. 2008). By differentially processing introns (and exons), eukaryotic cells can generate multiple mRNA and protein isoforms from a single gene. This differential processing may consist of nonsplicing (or retention) of introns, in the simplest case, but also of exclusion (or skipping) of an entire exon, alternative choice of multiple 5′ss, and/or 3′ss within an intron (which would produce exon truncations/expansions), or more complex splicing patterns (Irimia and Blencowe 2012).
AS may itself perform different functions. First, AS can generate protein isoforms with highly distinct sequences that may have dramatically different activities and biological roles (e.g., Boise et al. 1993; Gabut et al. 2011); but also proteins with only slight variations that contribute to fine-tuning of cellular processes (Lopez 1998). Second, AS can produce nonfunctional transcript variants, serving as an extra layer of down-regulation of gene expression. This may be achieved by on/off protein switches (Bingham et al. 1988), or by coupling missense AS to NMD (Lewis et al. 2003; Lareau et al. 2007; Yap et al. 2012), leading to transcript degradation before translation. Third, inclusion of alternative sequences may lead to differences in intracellular transport, either at the mRNA (Buckley et al. 2011) or protein level (Freitag et al. 2012; Kabran et al. 2012). Nevertheless, despite a plethora of described examples of each kind, a major unanswered question is still to what extent AS is functional or simply splicing “noise” (Sorek et al. 2004; Irimia et al. 2008; Roy and Irimia 2008).
AS was initially believed a relatively recent innovation, largely associated with the emergence of multicellularity (Ast 2004). However, the wealth of genomes and transcriptomes accumulated over the past decade has turned around this view. First, significant amounts of AS (usually intron retention) have now been described in nearly all well-studied species. These include representatives of all eukaryotic supergroups (McGuire et al. 2008; Labadorf et al. 2010; Otto et al. 2010; Rhind et al. 2011; Shen et al. 2011; Curtis et al. 2012; Sebé-Pedrós et al. 2013), and even some intron-poor species (Pleiss et al. 2007; Kabran et al. 2012). Second, LECA's intron–exon structures seem to have met all classic requirements for AS to occur (Roy and Irimia 2009; Koonin et al. 2013): It was intron-rich (Irimia et al. 2007b) and had heterogeneous splice signals, which are associated with splicing variation within and across genomes (Stamm et al. 2000; Ast 2004; Baek and Green 2005). Moreover, functional and evolutionary analyses of alternatively spliced genes showed that ancient eukaryotic genes (i.e., those present in LECA) show high levels of AS in different eukaryotic lineages, suggesting that they could have been amenable to AS also in LECA (Irimia et al. 2007b). Thus, all these lines of evidence strongly suggest that LECA could have had at least a simple program of AS, perhaps comparable to those observed in most modern unicellular eukaryotes, dominated by intron retention and perhaps acting mainly as an extra layer of gene regulation. However, as for any modern eukaryote, it is not clear what fraction of that transcriptional diversity played any relevant biological role.
Despite the similar patterns of AS in most eukaryotes, AS has also undergone remarkable diversification in some specific lineages. For instance, and most unexpectedly (Ast 2004), the intron-poor yeast S. cerevisiae may have evolved regulated, functional AS for the majority, if not all, of its intron-containing genes. S. cerevisiae has ∼260 introns, with a strong bias toward ribosomal genes (Juneau et al. 2006). In-depth transcriptomic analyses have shown that splicing of specific sets of introns can be down-regulated in response to different growth conditions (Pleiss et al. 2007; Parenteau et al. 2011). For example, only a few minutes after amino acid starvation, splicing of most ribosomal introns is down-regulated, thereby reducing production of ribosomal proteins, thus presumably reducing global translation (Pleiss et al. 2007).
Nonetheless, the most famous example of extreme AS diversification is found in animals, particularly in primates (Barbosa-Morais et al. 2012). Not only is AS much more common in metazoans, but the mode of AS is also qualitatively different from that of other eukaryotic lineages. Unlike most eukaryotes, in which intron retention dominates, animals show frequent usage of exon skipping (McGuire et al. 2008). In exon skipping, AS leads to the inclusion/exclusion of full exons that behave as “cassettes” of sequence and that can be deployed in a tissue-specific manner. Consistent with the cassette idea, most regulated alternative exons are multiple of three nucleotides (Xing and Lee 2005), thereby preserving the reading frame when included/excluded. Thus, cassette exons can have a much broader impact on proteome diversity, by introducing or disrupting functional protein domains, or by regulating disordered regions often involved in protein–protein interactions (Buljan et al. 2012; Ellis et al. 2012; Colak et al. 2013). Intriguingly, the reasons for this transition from intron retention (mostly impacting gene regulation) to exon skipping (increasing proteomic diversity) during the evolution of early animals are still unknown. Given the potential centrality of AS for the origin of animal complexity, understanding this crucial transition is a central priority for future studies.
Strikingly, another eukaryotic lineage has recently been reported to have mammalian-like levels of exon skipping, the chlorarachniophyte Bigelowiella natans (Curtis et al. 2012). This species has intron densities comparable to humans (eight to nine introns per gene), and displays extremely high levels of splicing variation, including both intron retention and exon skipping. RNAseq quantification of exon skipping shows levels comparable only to the human cortex (Curtis et al. 2012), the highest AS levels described so far in any tissue and organism (Barbosa-Morais et al. 2012). Whereas much of B. natans AS may represent noisy splicing, a few potential cases in which AS drives alternative subcellular targeting were identified (Curtis et al. 2012), suggesting that AS may play important and diverse roles in the biology of B. natans. Surveys of diverse eukaryotes to identify additional cases of independent evolution of widespread functional exon skipping (or other forms of protein-diversifying AS) are crucial to a full understanding of the functional impact of the spliceosomal system in eukaryotes.
CONCLUDING REMARKS
Evidence from biochemistry and comparative genomics strongly suggests that spliceosomal introns originated and proliferated during the origin of eukaryotes, in a process intimately linked to other evolutionary events of eukaryogenesis. The resulting portrait of the LECA spliceosomal system was comparable to that of intron-rich modern eukaryotic organisms, with myriad introns, complex splicing machinery, and coupling of splicing to other cellular processes. From that complex ancestor, intron–exon structures have been highly plastic throughout eukaryotic evolution, with cases of extreme divergence from their ancestors that shaped the genomic landscapes of eukaryotes probably like no other. Particularly remarkable are the multiple instances of recurrent evolution of multiple features, including massive intron loss, constraint of splicing signals, restriction of BP-3′ss distance, loss of polypyrimidine tract, and complete loss of U12 introns.
ACKNOWLEDGMENTS
We thank Eesha Sharma for critical reading of the manuscript. M.I. holds a PDF from the Human Frontiers Science Program Organization.
Footnotes
Editors: Patrick J. Keeling and Eugene V. Koonin
Additional Perspectives on The Origin and Evolution of Eukaryotes available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Abramovitz DL, Friedman RA, Pyle AM 1996. Catalytic role of 2′-hydroxyl groups within a group II intron active site. Science 271: 1410–1413 [DOI] [PubMed] [Google Scholar]
- Adams KL, Palmer JD 2003. Evolution of mitochondrial gene content: Gene loss and transfer to the nucleus. Mol Phylogenet Evol 29: 380–395 [DOI] [PubMed] [Google Scholar]
- Ahmadinejad N, Dagan T, Gruenheit N, Martin W, Gabaldón T 2010. Evolution of spliceosomal introns following endosymbiotic gene transfer. BMC Evol Biol 10: 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alioto TS 2007. U12DB: A database of orthologous U12-type spliceosomal introns. Nucl Acids Res 35: D110–D115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amrani N, Ganesan R, Kervestin S, Mangus DA, Ghosh S, Jacobson A 2004. A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature 432: 112–118 [DOI] [PubMed] [Google Scholar]
- Andersson JO, Sjögren AM, Horner DS, Murphy CA, Dyal PL, Svärd SG, Logsdon JMJ, Ragan MA, Hirt RP, Roger AJ 2007. A genomic survey of the fish parasite Spironucleus salmonicida indicates genomic plasticity among diplomonads and significant lateral gene transfer in eukaryote genome evolution. BMC Genomics 8: 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assis R, Kondrashov AS, Koonin EV, Kondrashov FA 2008. Nested genes and increasing organizational complexity of metazoan genomes. Trends Genet 24: 475–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ast G 2004. How did alternative splicing evolve? Nat Rev Genet 5: 773–782 [DOI] [PubMed] [Google Scholar]
- Avery OT, Macleod CM, McCarty M 1944. Studies on the chemical nature of the substance inducing transformation of Pneumococcal types: Induction of transformation by a desoxyribonucleic acid fraction isolated from Pneumococcus type III. J Exp Med 79: 137–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek D, Green P 2005. Sequence conservation, relative isoform frequencies, and nonsense-mediated decay in evolutionarily conserved alternative splicing. Proc Natl Acad Sci 102: 12813–12818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa-Morais NL, Irimia M, Pan Q, Xiong HY, Gueroussov S, Lee LJ, Slobodeniuc V, Kutter C, Watt S, Colak R, et al. 2012. The evolutionary landscape of alternative splicing in vertebrate species. Science 338: 1587–1593 [DOI] [PubMed] [Google Scholar]
- Barkan A 2009. Genome-wide analysis of RNA-protein interactions in plants. Methods Mol Biol 553: 13–37 [DOI] [PubMed] [Google Scholar]
- Bass BL, Cech TR 1984. Specific interaction between the self-splicing RNA of Tetrahymena and its guanosine substrate: Implications for biological catalysis by RNA. Nature 308: 820–826 [DOI] [PubMed] [Google Scholar]
- Basu MK, Makalowski W, Rogozin IB, Koonin EV 2008a. U12 intron positions are more strongly conserved between animals and plants than U2 intron positions. Biol Direct 3: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu MK, Rogozin IB, Deusch O, Dagan T, Martin W, Koonin EV 2008b. Evolutionary dynamics of introns in plastid-derived genes in plants: Saturation nearly reached but slow intron gain continues. Mol Biol Evol 25: 111–119 [DOI] [PubMed] [Google Scholar]
- Basu MK, Rogozin IB, Koonin EV 2008c. Primordial spliceosomal introns were probably U2-type. Trends Genet 24: 525–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berget SM, Moore C, Sharp PA 1977. Spliced segments at the 5′ terminus of adenovirus 2 late mRNA. Proc Natl Acad Sci 74: 3171–3175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingham PM, Chou TB, Mims I, Zachar Z 1988. On/off regulation of gene expression at the level of splicing. Trends Genet 4: 134–138 [DOI] [PubMed] [Google Scholar]
- Boise LH, González-García M, Postema CE, Ding L, Lindsten T, Turka LA, Mao X, Nuñez G, Thompson CB 1993. bcl-x, a bcl-2-related gene that functions as a dominant regulator of apoptotic cell death. Cell 74: 597–608 [DOI] [PubMed] [Google Scholar]
- Bonen L 2008. Cis- and trans-splicing of group II introns in plant mitochondria. Mitochondrion 8: 26–34 [DOI] [PubMed] [Google Scholar]
- Bonen L, Vogel J 2001. The ins and outs of group II introns. Trends Genet 17: 322–331 [DOI] [PubMed] [Google Scholar]
- Braunschweig U, Gueroussov S, Plocik AM, Graveley BR, Blencowe BJ 2013. Dynamic integration of splicing within gene regulatory pathways. Cell 152: 1252–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley PT, Lee MT, Sul JY, Miyashiro JY, Bell TJ, Fisher SA, Kim J, Eberwine J 2011. Cytoplasmic intron sequence-retaining transcripts can be dendritically targeted via ID element retrotransposons. Neuron 69: 877–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buljan M, Chalancon G, Eustermann S, Wagner GP, Fuxreiter M, Bateman A, Babu MM 2012. Tissue-specific splicing of disordered segments that embed binding motifs rewires protein interaction networks. Mol Cell 46: 871–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge CB, Padgett RA, Sharp PA 1998. Evolutionary fates and origins of U12-type introns. Mol Cell 2: 773–785 [DOI] [PubMed] [Google Scholar]
- Candales MA, Duong A, Hood KS, Li T, Neufeld RA, Sun R, McNeil BA, Wu L, Jarding AM, Zimmerly S 2012. Database for bacterial group II introns. Nucleic Acids Res 40: D187–D190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmel L, Wolf YI, Rogozin IB, Koonin EV 2007. Three distinct modes of intron dynamics in the evolution of eukaryotes. Genome Res 17: 1034–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmel L, Rogozin IB, Wolf YI, Koonin EV 2009. A maximum likelihood method for reconstruction of the evolution of eukaryotic gene structure. Methods Mol Biol 541: 357–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T 1985. Selfish DNA and the origin of introns. Nature 315: 283–284 [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T 1991. Intron phylogeny: A new hypothesis. Trends Genet 7: 145–148 [PubMed] [Google Scholar]
- Cech TR 1986. The generality of self-splicing RNA: Relationship to nuclear mRNA splicing. Cell 44: 207–210 [DOI] [PubMed] [Google Scholar]
- Cech TR, Zaug AJ, Grabowski PJ 1981. In vitro splicing of the ribosomal RNA precursor of Tetrahymena: Involvement of a guanosine nucleotide in the excision of the intervening sequence. Cell 27: 487–496 [DOI] [PubMed] [Google Scholar]
- Chen YH, Su LH, Sun CH 2008. Incomplete nonsense-mediated mRNA decay in Giardia lamblia. Int J Parasitol 38: 1305–1317 [DOI] [PubMed] [Google Scholar]
- Chiara MD, Reed R 1995. A two-step mechanism for 5′ and 3′ splice-site pairing. Nature 375: 510–513 [DOI] [PubMed] [Google Scholar]
- Chiara MD, Palandjian L, Feld Kramer R, Reed R 1997. Evidence that U5 snRNP recognizes the 3′ splice site for catalytic step II in mammals. EMBO J 16: 4746–4759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chorev M, Carmel L 2013. Computational identification of functional introns: High positional conservation of introns that harbor RNA genes. Nucleic Acids Res 41: 5604–5613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow LT, Gelinas RE, Broker TR, Roberts RJ 1977. An amazing sequence arrangement at the 5′ ends of adenovirus 2 messenger RNA. Cell 12: 1–8 [DOI] [PubMed] [Google Scholar]
- Clarke M, Lohan AJ, Liu B, Lagkouvardos I, Roy S, Zafar N, Bertelli C, Schilde C, Kianianmomeni A, Bürglin TR, et al. 2013. Genome of Acanthamoeba castellanii highlights extensive lateral gene transfer and early evolution of tyrosine kinase signaling. Genome Biol 14: R11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cock JM, Sterck L, Rouzé P, Scornet D, Allen, Amoutzias G, Anthouard V, Artiguenave F, Aury JM, Badger JH, et al. 2010. The Ectocarpus genome and the independent evolution of multicellularity in brown algae. Nature 465: 617–621 [DOI] [PubMed] [Google Scholar]
- Colak R, Kim T, Michaut M, Sun M, Irimia M, Bellay J, Myers CL, Blencowe BJ, Kim PM 2013. Distinct types of disorder in the human proteome: Functional implications for alternative splicing. PLoS Comput Biol 9: e1003030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collén J, Porcel B, Carré W, Ball SG, Chaparro C, Tonon T, Barbeyron T, Michel G, Noel B, Valentin K, et al. 2013. Genome structure and metabolic features in the red seaweed Chondrus crispus shed light on evolution of the Archaeplastida. Proc Natl Acad Sci 110: 5247–5252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins L, Penny D 2005. Complex spliceosomal organization ancestral to extant eukaryotes. Mol Biol Evol 22: 1053–1066 [DOI] [PubMed] [Google Scholar]
- Comeron JM, Kreitman M 2000. The correlation between intron length and recombination in Drosophila. Dynamic equilibrium between mutational and selective forces. Genetics 156: 1175–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copertino DW, Hallick RM 1993. Group II and group III introns of twintrons: Potential relationships with nuclear pre-mRNA introns. Trends Biochem Sci 18: 467–471 [DOI] [PubMed] [Google Scholar]
- Coulombe-Huntington J, Majewski J 2007a. Characterization of intron loss events in mammals. Genome Res 17: 23–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulombe-Huntington J, Majewski J 2007b. Intron loss and gain in Drosophila. Mol Biol Evol 24: 2842–2850 [DOI] [PubMed] [Google Scholar]
- Cousineau B, Lawrence S, Smith D, Belfort M 2000. Retrotransposition of a bacterial group II intron. Nature 404: 1018–1021 [DOI] [PubMed] [Google Scholar]
- Crick FH 1958. On protein synthesis. Symp Soc Exp Biol 12: 138–163 [PubMed] [Google Scholar]
- Csurös M 2005. Likely scenarios of intron evolution. Third RECOMB Satellite Workshop on Comparative Genomics, pp. 47–60 Springer, New York [Google Scholar]
- Csurös M 2008. Malin: Maximum likelihood analysis of intron evolution in eukaryotes. Bioinformatics 24: 1538–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csurös M, Rogozin IB, Koonin EV 2008. Extremely intron-rich genes in the alveolate ancestors inferred with a flexible maximum-likelihood approach. Mol Biol Evol 25: 903–911 [DOI] [PubMed] [Google Scholar]
- Csurös M, Rogozin IB, Koonin EV 2011. A detailed history of intron-rich eukaryotic ancestors inferred from a global survey of 100 complete genomes. PLoS Comput Biol 7: e1002150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis BA, Tanifuji G, Burki F, Gruber A, Irimia M, Maruyama S, Arias MC, Ball SG, Gile GH, Hirakawa Y, et al. 2012. Algal genomes reveal evolutionary mosaicism and the fate of nucleomorphs. Nature 492: 59–65 [DOI] [PubMed] [Google Scholar]
- Dai L, Zimmerly S 2003. ORF-less and reverse-transcriptase-encoding group II introns in archaebacteria, with a pattern of homing into related group II intron ORFs. RNA 9: 14–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell J Jr 1978. Implications of RNA-RNA splicing in evolution of eukaryotic cells. Science 202: 1257–1260 [DOI] [PubMed] [Google Scholar]
- Dávila López M, Rosenblad MA, Samuelsson T 2008. Computational screen for spliceosomal RNA genes aids in defining the phylogenetic distribution of major and minor spliceosomal components. Nucl Acids Res 36: 3001–3010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denoeud F, Henriet S, Mungpakdee S, Aury JM, Da Silva C, Brinkmann H, Mikhaleva J, Olsen LC, Jubin C, Cañestro C, et al. 2010. Plasticity of animal genome architecture unmasked by rapid evolution of a pelagic tunicate. Science 330: 381–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza SJ, Long M, Schoenbach L, Roy SW, Gilbert W 1996. Intron positions correlate with module boundaries in ancient proteins. Proc Natl Acad Sci 93: 14632–14636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibb NJ, Newman AJ 1989. Evidence that introns arose at proto-splice sites. EMBO J 8: 2015–2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dlakić M, Mushegian A 2011. Prp8, the pivotal protein of the spliceosomal catalytic center, evolved from a retroelement-encoded reverse transcriptase. RNA 17: 799–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domdey H, Apostol B, Lin RJ, Newman A, Brody E, Abelson J 1984. Lariat structures are in vivo intermediates in yeast pre-mRNA splicing. Cell 39: 611–621 [DOI] [PubMed] [Google Scholar]
- Doose G, Alexis M, Kirsch R, Findeiß S, Langenberger D, Machné R, Mörl M, Hoffmann S, Stadler PF 2013. Mapping the RNA-Seq trash bin: Unusual transcripts in prokaryotic transcriptome sequencing data. RNA Biol 10: 1204–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H, Rosbash M 2002. The U1 snRNP protein U1C recognizes the 5′ splice site in the absence of base pairing. Nature 419: 86–90 [DOI] [PubMed] [Google Scholar]
- Ellis JD, Barrios-Rodiles M, Colak R, Irimia M, Kim T, Calarco JA, Wang X, Pan Q, O'Hanlon D, Kim PM, et al. 2012. Tissue-specific alternative splicing remodels protein-protein interaction networks. Mol Cell 46: 884–892 [DOI] [PubMed] [Google Scholar]
- Epstein DJ, McMahon AP, Joyner AL 1999. Regionalization of Sonic hedgehog transcription along the anteroposterior axis of the mouse central nervous system is regulated by Hnf3-dependent and -independent mechanisms. Development 126: 281–292 [DOI] [PubMed] [Google Scholar]
- Fedorov A, Merican A, Gilbert W 2002. Large-scale comparison of intron positions among animal, plant, and fungal genes. Proc Natl Acad Sci 99: 16128–16133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferat JL, Michel F 1993. Group II self-splicing introns in bacteria. Nature 364: 358–361 [DOI] [PubMed] [Google Scholar]
- Flot JF, Hespeels B, Li X, Noel B, Arkhipova I, Danchin EG, Hejnol A, Henrissat B, Koszul R, Aury JM, et al. 2013. Genomic evidence for ameiotic evolution in the bdelloid rotifer Adineta vaga. Nature 22: 453–457 [DOI] [PubMed] [Google Scholar]
- Freitag J, Ast J, Bölker M 2012. Cryptic peroxisomal targeting via alternative splicing and stop codon read-through in fungi. Nature 485: 522–525 [DOI] [PubMed] [Google Scholar]
- Gabut M, Samavarchi-Tehrani P, Wang X, Slobodeniuc V, O’Hanlon D, Sung HK, Alvarez M, Talukder S, Pan Q, Mazzoni EO, et al. 2011. An alternative splicing switch regulates embryonic stem cell pluripotency and reprogramming. Cell 147: 132–146 [DOI] [PubMed] [Google Scholar]
- Gilbert W 1978. Why genes in pieces? Nature 271: 501. [DOI] [PubMed] [Google Scholar]
- Gilbert W 1987. The exon theory of genes. Cold Spring Harb Symp Quant Biol 52: 901–905 [DOI] [PubMed] [Google Scholar]
- Gilson PR, Su V, Slamovits CH, Reith ME, Keeling PJ, McFadden GI 2006. Complete nucleotide sequence of the chlorarachniophyte nucleomorph: Nature's smallest nucleus. Proc Natl Acad Sci 103: 9566–9571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldschmidt-Clermont M, Choquet Y, Girard-Bascou J, Michel F, Schirmer-Rahire M, Rochaix JD 1991. A small chloroplast RNA may be required for trans-splicing in Chlamydomonas reinhardtii. Cell 65: 135–143 [DOI] [PubMed] [Google Scholar]
- *.Guy L, Saw JH, Ettema TJG 2014. The archaeal legacy of eukaryotes: A phylogenomic perspective. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a016022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall SL, Padgett RA 1994. Conserved sequences in a class of rare eukaryotic nuclear introns with non-consensus splice sites. J Mol Biol 239: 357–365 [DOI] [PubMed] [Google Scholar]
- Hall SL, Padgett RA 1996. Requirement of U12 snRNA for in vivo splicing of a minor class of eukaryotic nuclear pre-mRNA introns. Science 271: 1716–1718 [DOI] [PubMed] [Google Scholar]
- Haugen P, Simon DM, Bhattacharya D 2005. The natural history of group I introns. Trends Genet 21: 111–119 [DOI] [PubMed] [Google Scholar]
- Hetzer M, Wurzer G, Schweyen RJ, Muelle MW 1997. Trans-activation of group II intron splicing by nuclear U5 snRNA. Nature 386: 417–420 [DOI] [PubMed] [Google Scholar]
- Hickey DA 1982. Selfish DNA: A sexually-transmitted nuclear parasite. Genetics 101: 519–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irimia M, Blencowe BJ 2012. Alternative splicing: Decoding an expansive regulatory layer. Curr Opin Cell Biol 24: 323–332 [DOI] [PubMed] [Google Scholar]
- Irimia M, Roy SW 2008a. Evolutionary convergence on highly-conserved 3′ intron structures in intron-poor eukaryotes and insights into the ancestral eukaryotic genome. PLoS Genet 4: e1000148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irimia M, Roy SW 2008b. Spliceosomal introns as tools for genomic and evolutionary analysis. Nucleic Acids Res 36: 1703–1712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irimia M, Penny D, Roy SW 2007a. Co-evolution of genomic intron number and splice sites. Trends Genet 23: 321–325 [DOI] [PubMed] [Google Scholar]
- Irimia M, Rukov JL, Penny D, Roy SW 2007b. Functional and evolutionary analysis of alternatively spliced genes is consistent with an early eukaryotic origin of alternative splicing. BMC Evol Biol 7: 188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irimia M, Rukov JL, Penny D, Garcia-Fernandez J, Vinther J, Roy SW 2008. Widespread evolutionary conservation of alternatively spliced exons in Caenorhabditis. Mol Biol Evol 25: 375–382 [DOI] [PubMed] [Google Scholar]
- Irimia M, Roy SW, Neafsey DE, Abril JF, Garcia-Fernandez J, Koonin EV 2009. Complex selection on 5′ splice sites in intron-rich organisms. Genome Res 19: 2021–2027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irimia M, Maeso I, Burguera D, Hidalgo-Sánchez M, Puelles L, Garcia-Fernàndez J, Roy SW, Ferran JL 2011. Contrasting 5′ and 3′ evolutionary histories and frequent evolutionary convergence in Meis/hth gene structures. Genome Biol Evol 3: 551–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irimia M, Tena JJ, Alexis MS, Fernandez-Miñan A, Maeso I, Bogdanovic O, de la Calle-Mustienes E, Roy SW, Gómez-Skarmeta JL, Fraser HB 2012. Extensive conservation of ancient microsynteny across metazoans due to cis-regulatory constraints. Genome Res 22: 2356–2367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson LJ 1991. A reappraisal of non-consensus mRNA splice sites. Nucl Acids Res 19: 3795–3798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillon O, Bouhouche K, Gout JF, Aury JM, Noel B, Nowacki M, Serrano V, Porcel BM, Ségurens B, Mouël AL, et al. 2008. Translational control of intron splicing in eukaryotes. Nature 451: 359–362 [DOI] [PubMed] [Google Scholar]
- Jarrell KA, Dietrich RC, Perlman PS 1988. Group II intron domain 5 facilitates a trans-splicing reaction. Mol Cell Biol 8: 2361–2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang K, Goertzen LR 2011. Spliceosomal intron size expansion in domesticated grapevine (Vitis vinifera). BMC Res Notes 4: 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juneau K, Miranda M, Hillenmeyer ME, Nislow C, Davis RW 2006. Introns regulate RNA and protein abundance in yeast. Genetics 174: 511–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabran P, Rossignol T, Gaillardin C, Nicaud JM, Neuvéglise C 2012. Alternative splicing regulates targeting of malate dehydrogenase in Yarrowia lipolytica. DNA Res 19: 231–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keating KS, Toor N, Perlman PS, Pyle AM 2010. A structural analysis of the group II intron active site and implications for the spliceosome. RNA 16: 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren H, Lev-Maor G, Ast G 2010. Alternative splicing and evolution: Diversification, exon definition and function. Nat Rev Genet 11: 345–355 [DOI] [PubMed] [Google Scholar]
- Kerényi Z, Mérai Z, Hiripi L, Benkovics A, Gyula P, Lacomme C, Barta E, Nagy F, Silhavy D 2008. Inter-kingdom conservation of mechanism of nonsense-mediated mRNA decay. EMBO J 27: 1585–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konforti BB, Abramovitz DL, Duarte CM, Karpeisky A, Beigelman L, Pyle AM 1998. Ribozyme catalysis from the major groove of group II intron domain 5. Mol Cell 1: 433–441 [DOI] [PubMed] [Google Scholar]
- Koonin EV 2006. The origin of introns and their role in eukaryogenesis: A compromise solution to the introns-early versus introns-late debate? Biol Direct 1: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV 2009. Intron-dominated genomes of early ancestors of eukaryotes. J Hered 100: 618–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Csurös M, Rogozin IB 2013. Whence genes in pieces: Reconstruction of the exon-intron gene structures of the last eukaryotic common ancestor and other ancestral eukaryotes. Wiley Interdiscip Rev RNA 4: 93–105 [DOI] [PubMed] [Google Scholar]
- Kumar A 2009. An overview of nested genes in eukaryotic genomes. Eukaryot Cell 8: 1321–1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labadorf A, Link A, Rogers MF, Thomas J, Reddy AS, Ben-Hur A 2010. Genome-wide analysis of alternative splicing in Chlamydomonas reinhardtii. BMC Genomics 11: 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambowitz AM, Zimmerly S 2004. Mobile group II introns. Annu Rev Genet 38: 1–35 [DOI] [PubMed] [Google Scholar]
- Lambowitz AM, Zimmerly S 2011. Group II introns: Mobile ribozymes that invade DNA. Cold Spring Harb Perspect Biol 3: a003616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921 [DOI] [PubMed] [Google Scholar]
- Lane CE, van den Heuvel K, Kozera C, Curtis BA, Parsons BJ, Bowman S, Archibald JM 2007. Nucleomorph genome of Hemiselmis andersenii reveals complete intron loss and compaction as a driver of protein structure and function. Proc Natl Acad Sci 104: 19908–19913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lareau LF, Brooks AN, Soergel DAW, Meng Q, Brenner SE 2007. The coupling of alternative splicing and nonsense mediated mRNA Decay. In Alternative splicing in the postgenomic era (ed. Blencowe BJ, Graveley BR), pp. 190–211 Landes Bioscience, Austin, TX: [DOI] [PubMed] [Google Scholar]
- Lee RC, Gill EE, Roy SW, Fast NM 2010. Constrained intron structures in a microsporidian. Mol Biol Evol 27: 1979–1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Green RE, Brenner SE 2003. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc Natl Acad Sci 100: 189–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CF, Costa M, Michel F 2011. Linking the branchpoint helix to a newly found receptor allows lariat formation by a group II intron. EMBO J 30: 3040–3051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin RJ, Newman AJ, Cheng SC, Abelson J 1985. Yeast mRNA splicing in vitro. J Biol Chem 260: 14780–14792 [PubMed] [Google Scholar]
- Liu M, Grigoriev A 2004. Protein domains correlate strongly with exons in multiple eukaryotic genomes—Evidence of exon shuffling? Trends Genet 20: 399–403 [DOI] [PubMed] [Google Scholar]
- Logsdon J 1998. The recent origins of spliceosomal introns revisited. Curr Opin Genet Dev 8: 637–648 [DOI] [PubMed] [Google Scholar]
- Logsdon J Jr, Tyshenko M, Dixon C, Jafari JD, Walker V, Palmer J 1995. Seven newly discovered intron positions in the triose-phosphate isomerase gene: Evidence for the introns-late theory. Proc Natl Acad Sci 92: 8507–8511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh YH, Brenner S, Venkatesh B 2008. Investigation of loss and gain of introns in the compact genomes of pufferfishes (Fugu and Tetraodon). Mol Biol Evol 25: 526–535 [DOI] [PubMed] [Google Scholar]
- Long M, de Souza S, Gilbert W 1995. Evolution of the intron-exon structure of eukaryotic genes. Curr Opin Genet Dev 5: 774–778 [DOI] [PubMed] [Google Scholar]
- Lopez AJ 1998. Alternative splicing of pre-mRNA: Developmental consequences and mechanisms of regulation. Annu Rev Genet 32: 279–305 [DOI] [PubMed] [Google Scholar]
- López-García P, Moreira D 2006. Selective forces for the origin of the eukaryotic nucleus. Bioessays 28: 525–533 [DOI] [PubMed] [Google Scholar]
- Lynch M 2002. Intron evolution as a population-genetic process. Proc Natl Acad Sci 99: 6118–6123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M 2006. The origins of eukaryotic gene structure. Mol Biol Evol 23: 450–468 [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery JS 2003. The origins of genome complexity. Science 302: 1401–1404 [DOI] [PubMed] [Google Scholar]
- Lynch M, Hong X, Scofield DG 2006. NMD and the evolution of eukaryotyic gene structure. In Nonsense-mediated mRNA decay (ed. Maquat LE), pp. 197–211 Landes Bioscience, Georgetown [Google Scholar]
- Maeso I, Irimia M, Tena JJ, González-Pérez E, Tran D, Ravi V, Venkatesh B, Campuzano S, Gómez-Skarmeta JL, Garcia-Fernàndez J 2012a. An ancient genomic regulatory block conserved across bilaterians and its dismantling in tetrapods by retrogene replacement. Genome Res 22: 642–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeso I, Roy SW, Irimia M 2012b. Widespread recurrent evolution of genomic features. Genome Biol Evol 4: 486–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marck C, Grosjean H 2003. Identification of BHB splicing motifs in intron-containing tRNAs from 18 archaea: Evolutionary implications. RNA 9: 1516–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin W, Koonin EV 2006. Introns and the origin of nucleus-cytosol compartmentalization. Nature 440: 41–45 [DOI] [PubMed] [Google Scholar]
- Matsuzaki M, Misumi O, Shin-i T, Maruyama S, Takahara M, Miyagishima SY, Mori T, Nishida K, Yagisawa F, Nishida K, et al. 2004. Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature 428: 653–657 [DOI] [PubMed] [Google Scholar]
- McGuire A, Pearson M, Neafsey D, Galagan J 2008. Cross-kingdom patterns of alternative splicing and splice recognition. Genome Biol 9: R50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Q, Wang Y, Liu XQ 2005. An intron-encoded protein assists RNA splicing of multiple similar introns of different bacterial genes. J Biol Chem 280: 35085–35088 [DOI] [PubMed] [Google Scholar]
- Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Marechal-Drouard L, et al. 2007. The Chlamydomonasgenome reveals the evolution of key animal and plant functions. Science 318: 245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel F, Ferat JL 1995. Structure and activities of group II introns. Ann Rev Biochem 64: 435–461 [DOI] [PubMed] [Google Scholar]
- Morrison HG, McArthur AG, Gillin FD, Aley SB, Adam RD, Olsen GJ, Best AA, Cande WZ, Chen F, Cipriano MJ, et al. 2007. Genomic minimalism in the early diverging intestinal parasite Giardia lamblia. Science 317: 1921–1926 [DOI] [PubMed] [Google Scholar]
- Mura C, Randolph PS, Patterson J, Cozen AE 2013. Archaeal and eukaryotic homologs of Hfq: A structural and evolutionary perspective on Sm function. RNA Biol 10: 608–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen H, Yoshihama M, Kenmochi N 2005. New maximum likelihood estimators for eukaryotic intron evolution. PLoS Comput Biol 1: e79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen C, Friedman B, Birren B, Burge C, Galagan J 2004. Patterns of intron gain and loss in fungi. PLoS Biol 2: e422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino K, Tsuneki K, Furuya H 2010. Unique genome of dicyemid mesozoan: Highly shortened spliceosomal introns in conservative exon/intron structure. Gene 449: 70–76 [DOI] [PubMed] [Google Scholar]
- Otto TD, Wilinski D, Assefa S, TM Keane, Sarry LR, Böhme U, Lemieux J, Barrell B, Pain A, Berriman M, et al. 2010. New insights into the blood-stage transcriptome of Plasmodium falciparum using RNA-Seq. Mol Microbiol 76: 12–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padgett RA, Konarska MM, Grabowski PJ, Hardy SF, Sharp PA 1984. Lariat RNA's as intermediates and products in the splicing of messenger RNA precursors. Science 225: 898–903 [DOI] [PubMed] [Google Scholar]
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ 2008. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet 40: 1413–1415 [DOI] [PubMed] [Google Scholar]
- Parenteau J, Durand M, Morin G, Gagnon J, Lucier JF, Wellinger RJ, Chabot B, Elela SA 2011. Introns within ribosomal protein genes regulate the production and function of yeast ribosomes. Cell 147: 320–331 [DOI] [PubMed] [Google Scholar]
- Parker R, Siliciano PG, Guthrie C 1987. Recognition of the TACTAAC box during mRNA splicing in yeast involves base pairing to the U2-like snRNA. Cell 49: 229–239 [DOI] [PubMed] [Google Scholar]
- Patel AA, McCarthy M, Steitz JA 2002. The splicing of U12-type introns can be a rate-limiting step in gene expression. EMBO J 21: 3804–3815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peebles CL, Perlman PS, Mecklenburg KL, Petrillo ML, Tabor JH, Jarrell KA, Cheng HL 1986. A self-splicing RNA excises an intron lariat. Cell 44: 213–223 [DOI] [PubMed] [Google Scholar]
- Peebles CL, Zhang M, Perlman PS, Franzen JS 1995. Catalytically critical nucleotide in domain 5 of a group II intron. Proc Natl Acad Sci 92: 4422–4426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penny D, Hoeppner MP, Poole AM, Jeffares DC 2009. An overview of the introns-first theory. J Mol Evol 69: 527–540 [DOI] [PubMed] [Google Scholar]
- Plass M, Agirre E, Reyes D, Camara EE 2008. Co-evolution of the branch site and SR proteins in eukaryotes. Trends Genet 24: 590–594 [DOI] [PubMed] [Google Scholar]
- Pleiss JA, Whitworth GB, Bergkessel M, Guthrie C 2007. Rapid, transcript-specific changes in splicing in response to environmental stress. Mol Cell 27: 928–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole AM 2006. Did group II intron proliferation in an endosymbiont-bearing archaeon create eukaryotes? Biol Direct 1: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole AM, Jeffares DC, Penny D 1998. The path from the RNA world. J Mol Evol 46: 1–17 [DOI] [PubMed] [Google Scholar]
- Qin PZ, Pyle AM 1998. The architectural organization and mechanistic function of group II intron structural elements. Curr Opin Struct Biol 8: 301–308 [DOI] [PubMed] [Google Scholar]
- Qiu W, Schisler N, Stoltzfus A 2004. The evolutionary gain of spliceosomal introns: Sequence and phase preferences. Mol Biol Evol 21: 1252–1263 [DOI] [PubMed] [Google Scholar]
- Ramesh MA, Malik SB, Logsdon JMJ 2005. A phylogenomic inventory of meiotic genes; evidence for sex in Giardia and an early eukaryotic origin of meiosis. Curr Biol 15: 185–191 [DOI] [PubMed] [Google Scholar]
- Randau L, Söll D 2008. Transfer RNA genes in pieces. EMBO Rep 9: 623–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rest JS, Mindell DP 2003. Retroids in archaea: Phylogeny and lateral origins. Mol Biol Evol 20: 1134–1142 [DOI] [PubMed] [Google Scholar]
- Rhind N, Chen Z, Yassour M, Thompson DA, Haas BJ, Habib N, Wapinski I, Roy S, Lin MF, Heiman DI, et al. 2011. Comparative functional genomics of the fission yeasts. Science 332: 930–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogozin IB, Wolf YI, Sorokin AV, Mirkin BG, Koonin EV 2003. Remarkable interkingdom conservation of intron positions and massive, lineage-specific intron loss and gain in eukaryotic evolution. Curr Biol 13: 1512–1517 [DOI] [PubMed] [Google Scholar]
- Rogozin IB, Carmel L, Csurös M, Koonin EV 2012. Origin and evolution of spliceosomal introns. Biol Direct 7: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy SW 2006. Intron-rich ancestors. Trends Genet 22: 468–471 [DOI] [PubMed] [Google Scholar]
- Roy SW, Gilbert W 2005. Complex early genes. Proc Natl Acad Sci 102: 1986–1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy SW, Hartl DL 2006. Very little intron loss/gain in Plasmodium: Intron loss/gain mutation rates and intron number. Genome Res 16: 750–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy SW, Irimia M 2008. Intron mis-splicing: No alternative? Genome Biol 9: 208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy SW, Irimia M 2009. Splicing in the eukaryotic ancestor: Form, function and dysfunction. Trends Ecol Evol 24: 447–455 [DOI] [PubMed] [Google Scholar]
- Roy SW, Irimia M 2012. Genome evolution: Where do new introns come from? Curr Biol 22: R529–R531 [DOI] [PubMed] [Google Scholar]
- Roy SW, Penny D 2006. Large-scale intron conservation and order-of-magnitude variation in intron loss/gain rates in apicomplexan evolution. Genome Res 16: 1270–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy SW, Penny D 2007a. Patterns of intron loss and gain in plants: Intron loss-dominated evolution and genome-wide comparison of O. sativa and A. thaliana. Mol Biol Evol 24: 171–181 [DOI] [PubMed] [Google Scholar]
- Roy SW, Penny D 2007b. A very high fraction of unique intron positions in the intron-rich diatom Thalassiosira pseudonana indicates widespread intron gain. Mol Biol Evol 24: 1447–1457 [DOI] [PubMed] [Google Scholar]
- Roy SW, Fedorov A, Gilbert W 2003. Large-scale comparison of intron positions in mammalian genes shows intron loss but no gain. Proc Natl Acad Sci 100: 7158–7162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy SW, Irimia M, Penny D 2006. Very little intron gain in Entamoeba histolytica genes laterally transferred from prokaryotes. Mol Biol Evol 23: 1824–1827 [DOI] [PubMed] [Google Scholar]
- Ruskin B, Green MR 1985. Role of the 3′ splice site consensus sequence in mammalian pre-mRNA splicing. Nature 317: 732–734 [DOI] [PubMed] [Google Scholar]
- Ruskin B, Krainer AR, Maniatis T, Green MR 1984. Excision of an intact intron as a novel lariat structure during pre-mRNA splicing in vitro. Cell 38: 317–331 [DOI] [PubMed] [Google Scholar]
- Russell CB, Fraga D, Hinrichsen RD 1994. Extremely short 20–33 nucleotide introns are the standard length in Paramecium tetraurelia. Nucleic Acids Res 22: 1221–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell AG, Charette JM, Spencer DF, Gray MW 2006. An early evolutionary origin for the minor spliceosome. Nature 443: 863–866 [DOI] [PubMed] [Google Scholar]
- Schmelzer C, Schweyen RJ 1986. Self-splicing of group II introns in vitro: Mapping of the branch point and mutational inhibition of lariat formation. Cell 46: 557–565 [DOI] [PubMed] [Google Scholar]
- Schwartz S, Silva J, Burstein D, Pupko T, Eyras E, Ast G 2008. Large-scale comparative analysis of splicing signals and their corresponding splicing factors in eukaryotes. Genome Res 18: 88–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebé-Pedrós A, Irimia M, Del Campo J, Parra-Acero H, Russ C, Nusbaum C, Blencowe BJ, Ruiz-Trillo I 2013. Regulated aggregative multicellularity in a close unicellular relative of metazoa. Elife 2: e01287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PA 1991. Five easy pieces. Science 254: 663. [DOI] [PubMed] [Google Scholar]
- Shen D, Ye W, Dong S, Wang Y, Dou D 2011. Characterization of intronic structures and alternative splicing in Phytophthora sojae by comparative analysis of expressed sequence tags and genomic sequences. Can J Microbiol 57: 84–90 [DOI] [PubMed] [Google Scholar]
- Shukla GC, Padgett RA 2002. A catalytically active group II intron domain 5 can function in the U12-dependent spliceosome. Mol Cell 9: 1145–1150 [DOI] [PubMed] [Google Scholar]
- Simon DM, Clarke NA, McNeil BA, Johnson I, Pantuso D, Dai L, Chai D, Zimmerly S 2008. Group II introns in eubacteria and archaea: ORF-less introns and new varieties. RNA 14: 1704–1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solem A, Zingler N, Pyle AM, Li-Pook-Than J 2009. Group II introns and their protein collaborators. In Non-protein coding RNAs (ed. Walter NG). Springer, Berlin [Google Scholar]
- Sorek R, Shamir R, Ast G 2004. How prevalent is functional alternative splicing in the human genome? Trends Genet 20: 68–71 [DOI] [PubMed] [Google Scholar]
- Srivastava M, Begovic E, Chapman J, Putnam NH, Hellsten U, Kawashima T, Kuo A, Mitros T, Salamov A, Carpenter ML, et al. 2008. The Trichoplax genome and the nature of placozoans. Nature 454: 955–960 [DOI] [PubMed] [Google Scholar]
- Stajich JE, Dietrich FS, Roy SW 2007. Comparative genomic analysis of fungal genomes reveals intron-rich ancestors. Genome Biol 8: R223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamm S, Zhu J, Nakai K, Stoilov P, Stoss O, Zhang MQ 2000. An alternative-exon database and its statistical analysis. DNA Cell Biol 19: 739–756 [DOI] [PubMed] [Google Scholar]
- Stoltzfus A 1994. Origin of introns-early or late? Nature 369: 526–527 [DOI] [PubMed] [Google Scholar]
- Suchy M, Schmelzer C 1991. Restoration of the self-splicing activity of a defective group II intron by a small trans-acting RNA. J Mol Biol 222: 179–187 [DOI] [PubMed] [Google Scholar]
- Sullivan JC, Reitzel AM, Finnerty JR 2006. A high percentage of introns in human genes were present early in animal evolution: Evidence from the basal metazoan Nematostella vectensis. Genome Inform 17: 219–229 [PubMed] [Google Scholar]
- Sverdlov A, Rogozin I, Babenko V, Koonin E 2005. Conservation versus parallel gains in intron evolution. Nucl Acids Res 33: 1741–1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sverdlov AV, Csurös M, Rogozin IB, Koonin EV 2007. A glimpse of a putative pre-intron phase of eukaryotic evolution. Trends Genet 23: 105–108 [DOI] [PubMed] [Google Scholar]
- Tarn WY, JA Steitz 1996. A novel spliceosome containing U11, U12, and U5 snRNPs excises a minor class (AT-AC) intron in vitro. Cell 84: 801–811 [DOI] [PubMed] [Google Scholar]
- Thorsness PE, Weber ER 1996. Escape and migration of nucleic acids between chloroplasts, mitochondria, and the nucleus. Int Rev Cytol 165: 207–234 [DOI] [PubMed] [Google Scholar]
- Toor N, Keating KS, Taylor SD, Pyle AM 2008. Crystal structure of a self-spliced group II intron. Science 320: 77–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai IJ, Zarowiecki M, Holroyd N, Garciarrubio A, Sanchez-Flores A, Brooks KL, Tracey A, Bobes RJ, Fragoso G, Sciutto E, et al. 2013. The genomes of four tapeworm species reveal adaptations to parasitism. Nature 496: 57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umen JG, Guthrie C 1995. A novel role for a U5 snRNP protein in 3′ splice site selection. Genes Dev 9: 855–868 [DOI] [PubMed] [Google Scholar]
- Valadkhan S, Mohammadi A, Wachtel C, Manley JL 2007. Protein-free spliceosomal snRNAs catalyze a reaction that resembles the first step of splicing. RNA 13: 2300–2311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valadkhan S, Mohammadi A, Jaladat Y, Geisler S 2009. Protein-free small nuclear RNAs catalyze a two-step splicing reaction. Proc Natl Acad Sci 106: 11901–11906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanacova S, Yan W, Carlton JM, Johnson PJ 2005. Spliceosomal introns in the deep-branching eukaryote Trichomonas vaginalis. Proc Natl Acad Sci 102: 4430–4435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Burgt A, Severing E, de Wit PJGM, Collemare J 2012. Birth of new spliceosomal introns in fungi by multiplication of introner-like elements. Curr Biol 22: 1260–1265 [DOI] [PubMed] [Google Scholar]
- van der Horst G, Tabak HF 1985. Self-splicing of yeast mitochondrial ribosomal and messenger RNA precursors. Cell 40: 759–766 [DOI] [PubMed] [Google Scholar]
- van der Veen R, Arnberg AC, van der Horst G, Bonen L, Tabak HF, Grivell LA 1986. Excised group II introns in yeast mitochondria are lariats and can be formed by self-splicing in vitro. Cell 44: 225–234 [DOI] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. 2001. The sequence of the human genome. Science 291: 1304–1351 [DOI] [PubMed] [Google Scholar]
- Veretnik S, Wills C, Youkharibache P, Valas RE, Bourne PE 2009. Sm/Lsm genes provide a glimpse into the early evolution of the spliceosome. PLoS Comput Biol 5: e1000315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vesteg M, Sándorová Z, Krajčovič J 2012. Selective forces for the origin of spliceosomes. J Mol Evol 74: 226–231 [DOI] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Lührmann R 2009. The spliceosome: Design principles of a dynamic RNP machine. Cell 136: 701–718 [DOI] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB 2008. Alternative isoform regulation in human tissue transcriptomes. Nature 456: 470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnecke T, Parmley JL, Hurst LD 2008. Finding exonic islands in a sea of non-coding sequence: Splicing related constraints on protein composition and evolution are common in intron-rich genomes. Genome Biol 9: R29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson JD, Crick FH 1953. Molecular structure of nucleic acids: A structure for deoxyribose nucleic acid. Nature 171: 737–738 [DOI] [PubMed] [Google Scholar]
- Will CL, Schneider C, Reed R, Luhrmann R 1999. Identification of both shared and distinct proteins in the major and minor spliceosomes. Science 284: 2003–2005 [DOI] [PubMed] [Google Scholar]
- Wilusz CJ, Wilusz J 2005. Eukaryotic Lsm proteins: Lessons from bacteria. Nat Struct Mol Biol 12: 1031–1036 [DOI] [PubMed] [Google Scholar]
- Worden AZ, Lee JH, Mock T, Rouzé P, Simmons MP, Aerts AL, Allen AE, Cuvelier ML, Derelle E, Everett MV, et al. 2009. Green evolution and dynamic adaptations revealed by genomes of the marine picoeukaryotes Micromonas. Science 324: 268–272 [DOI] [PubMed] [Google Scholar]
- Xing Y, Lee CJ 2005. Protein modularity of alternatively spliced exons is associated with tissue-specific regulation of alternative splicing. PLoS Genet 1: e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap K, Lim ZQ, Khandelia P, Friedman B, Makeyev EV 2012. Coordinated regulation of neuronal mRNA steady-state levels through developmentally controlled intron retention. Genes Dev 28: 1209–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yean SL, Wuenschell G, Termini J, Lin RJ 2000. Metal-ion coordination by U6 small nuclear RNA contributes to catalysis in the spliceosome. Nature 408: 881–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokobori S, Itoh T, Yoshinari S, Nomura N, Sako Y, Yamagishi A, Oshima T, Kita K, Watanabe Y 2009. Gain and loss of an intron in a protein-coding gene in Archaea: The case of an archaeal RNA pseudouridine synthase gene. BMC Evol Biol 9: 198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihama M, Nakao A, Nguyen HD, Kenmochi N 2006. Analysis of ribosomal protein gene structures: Implications for intron evolution. PLoS Genet 2: e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu YT, Maroney PA, Darzynkiwicz E, Nilsen TW 1995. U6 snRNA function in nuclear pre-mRNA splicing: A phosphorothioate interference analysis of the U6 phosphate backbone. RNA 1: 46–54 [PMC free article] [PubMed] [Google Scholar]
- Zamore PD, Patton JG, Green MR 1992. Cloning and domain structure of the mammalian splicing factor U2AF. Nature 355: 609–614 [DOI] [PubMed] [Google Scholar]
- Zhang LY, Yang YF, Niu DK 2010. Evaluation of models of the mechanisms underlying intron loss and gain in Aspergillus fungi. J Mol Evol 71: 364–373 [DOI] [PubMed] [Google Scholar]
- Zimmerly S, Hausner G, Wu X 2001. Phylogenetic relationships among group II intron ORFs. Nucleic Acids Res 29: 1238–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]