Abstract
Hepatitis B and hepatitis C viruses (HCV) are frequently propagating blood borne pathogens in global community. Viral hepatitis is primarily associated with severe health complications, such as liver cirrhosis, hepatocellular carcinoma, hepatic fibrosis and steatosis. A literature review was conducted on hepatitis B virus (HBV), HBV genome, genotypic distribution and global epidemiology of HBV, HCV, HCV genome, HCV and host immune responses, HCV genotypic distribution and global epidemiology. The valued information was subjected for review. HBV has strict tissue tropism to liver. The virus infecting hepatocytes produces large amount of hepatitis B surface antigen particles which lack the DNA. It has capability to integrate into host genome. It has been found that genotype C is most emerging genotype associated with more severe liver diseases (cirrhosis). The approximate prevalence rate of genotype C is 27.7% which represents a major threat to future generations. Approximately 8% of population is chronic carrier of HBV in developing countries. The chronic carrier rate of HBV is 2%-7% in Middle East, Eastern and Southern Europe, South America and Japan. Among HCV infected individuals, 15% usually have natural tendency to overcome acute viral infection, where as 85% of individuals were unable to control HCV infection. The internal ribosomal entry site contains highly conserved structures important for binding and appropriate positioning of viral genome inside the host cell. HCV infects only in 1%-10% of hepatocytes, but production of tumor necrosis factor alpha (from CD8+ cells) and interferon-gamma cause destruction of both infected cells and non-infected surrounding cells. Almost 11 genotypes and above 100 subtypes of HCV exists worldwide with different geographical distribution. Many efforts are still needed to minimize global burden of these infections. For the complete eradication of HBV (just like small pox and polio) via vaccination strategies, sincere efforts would be required from government and nongovernmental organizations.
Keywords: Hepatitis B virus, HCV, Epidemiology of HBV and HCV, Genotypic distribution of HBV and HCV, Genome analysis of HBV and HCV
1. Introduction
Hepatitis B is life threatening liver disease caused by highly contagious blood borne viral pathogen known as hepatitis B virus (HBV). The HBV infection is one of the principle causes of severe liver disorders, including hepatocellular carcinoma, cirrhosis and end stage liver disease. In 1963, HBV was accidently discovered by Baruch Blumberg during his research on Australia antigen[1]. HBV is an enveloped virus which belongs to hepadnaviridae: with circular partially double stranded DNA representing highly compact organization. The HBV is smallest known DNA virus, having spherical shape with diameter of about 42 nm and genomic length of approximately 3.2 Kb[2],[3]. The infectious virus particle, also referred as Dane particle, is responsible for causing infection in approximately five percent of world's population with 2 billion people infected with the virus and 350 million as carrier of chronic infection. The virus is responsible for 600 000 deaths each year[4]–[6]. HBV has been recognized as an important global health problem. Tremendous efforts are being put forward by many scientists, from whole world, for prevention and control of viral infection. To date, successful vaccination strategies have been developed to arrest the viral spread among various populations. The need of time is to put major emphasis on awareness about risk factors associated with transmission of hepatitis viral infection and to equip with adequate strategies for prevention of disease at national and international level.
Hepatitis C virus (HCV) is blood borne pathogen which causes severe liver disorders, including hepatocellular carcinoma, hepatic steatosis, liver cirrhosis, end stage liver disease and various metabolic disorders. In 1989, HCV was identified by Choo et al. as a positive stranded RNA molecule related to Togaviridae or Flaviviridae[7]. HCV has been classified into the genus hepacivirus of the family Flaviviridae. This virus is responsible for causing infection in three percent of world's population with approximately 170 million persons at risk of developing chronic hepatitis[8]. Due to continuous increase in number of viral infected hepatitis patients, World Health Organization (WHO) has recognized HCV as a major global health problem. Various epidemiological patterns and worldwide surveillance strategies are being performed for prevention and control of this disease. The HCV is small spherical enveloped virion with icosahedral capsid. The structure consists of an icosahedral lipid membrane with 2 glycoproteins (termed E1 and E2) that form heterodimers. An icosahedral nucleocapsid is thought to be present inside the viral membrane[9]. The buoyant density of HCV in sucrose is 1.06 g/cm3, whereas in chronically infected individuals the density is approximately 1.17 g/cm3 which might be due to viral association with antibody[10]. HCV can live on various environmental surfaces for more than sixteen hours and possibly up to four days.
2. Literature search
Articles were searched from Google Scholar and Pubmed with key words of HBV and analysis of HBV DNA expression, genotypic distribution of HBV and global epidemiological patterns of HBV. HCV and analysis of HCV genome, genotypic distribution of HCV and global epidemiological patterns of HCV. The valued information was subjected for review.
2.1. HBV and genome
The nucleocapsid of Dane particle is about 28 nm in size and constitutes hepatitis B core antigen. It is involved in packaging of viral genome. The hepatitis B surface antigen (HBsAg) present on the surface of HBV particle and 22 nm particles and tubular form, act as complex antigenic determinant. The infectious Dane particle acquires its membrane by budding or through secretory transport mechanisms via Golgi apparatus and endoplasmic reticulum. Membrane at outer envelope forms HBsAg which contains three viral surface proteins named according to their size of small, middle and large as HBsAg, HBmAg and HBlAg respectively. These proteins are encoded on same open reading frame (ORF) that encodes 3 start codons and get overlaps with polymerase ORF[11].
The pregenomic RNA is the largest transcript which serves as template for viral replication. The only enzyme encoded by the viral genome, which reverse transcribes the pregenomic RNA, is viral polymerase and this enzyme is also located inside the nucleocapsid[12]. The alternative translation products of core gene include hepatitis B core antigen and hepatitis B envelope antigen (HBeAg). For the translation of HBeAg, an upstream pre-core region with ATG codon is required[13]. The HBeAg undergoes posttranslational modification. It plays very prominent role in molecular diagnosis of HBV infection by acting as an active marker for viral replication. The HBeAg (acting as antigenic determinant) also circulates in serum as a soluble protein[14]. There are two DNA strands known as long [L (negative) strand] and short [S (positive) strand]. The L strand contains fixed length of 3.2 Kb, but the length of S strand is variable at its 3′ end. The S strand usually spans 50% to 100% of the length of the L strand (Figure 1).
Figure1. Transcription of HBV.
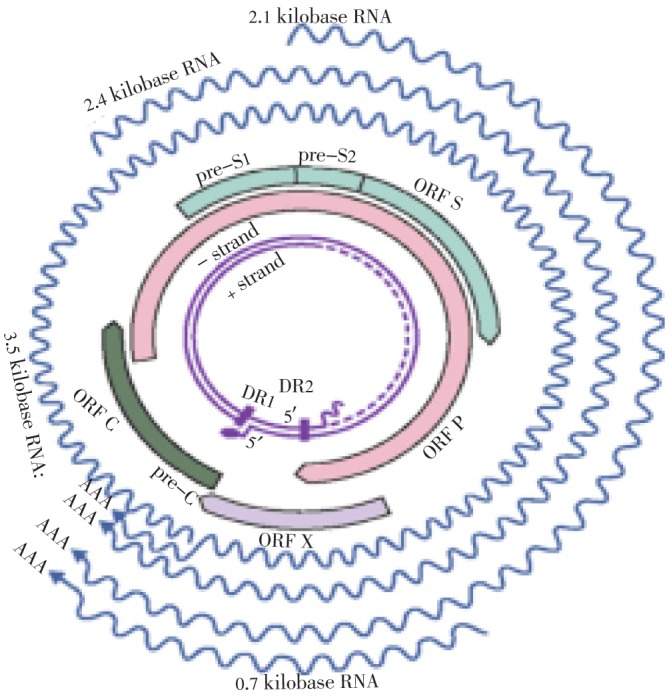
Basic source of information is Engleberg et al., Schaechter's Mechanisms of Microbial Disease.
There exists four conserved partially overlapping ORF in L strand, but in case of S strand, no partially overlapping ORF exist[15],[16]. Along with many DNA replication signals, the viral genome also contains two enhancer elements, six start codons, four promoters and a poly adenylation signal motif. The viral genome encodes for only seven proteins, that is pre-core/core proteins, the polymerase, three surface proteins (small, medium and large) and HBX protein as well. Most of RNA transcripts are capped and polyadenylated, like pre-C/C (3.5 Kb in length), pre S (of 2.4 Kb), smRNA (of 2.1 Kb) and an occasional xmRNA (of approximately 0.7 Kb). All of these HBV transcripts have 3′ end common which has been created by the polyadenylation signal of the core's gene[17],[18]. The level of HBsAg and IgG anti-HBc remains persistently detectable during chronic HBV infection. The level of HBeAg remains variable. Presence of HBsAg for more than six months indicates occurrence of chronic infection. A test negative for IgM anti-HBc together and positive for the HBsAg in same serum sample indicates onset of chronic infection in HBV infected patient[19].
Some of the possible causes of persistent viral infection includes high viral load, high replication rate, viral inhibition of antigen presentation, viral mutations that antagonize antigen recognition, immunosuppressive effects of virus, immunologic tolerance, exhausted T cell response, insufficient co-stimulation of virus specific T-cells, inefficient viral presenting cells and alteration of T Helper Type 1 and T Helper Type 2 balance[20]. On average the incubation period of HBV is approximately 60-90 d. Among patients of clinical illness (jaundice), the prevalence of HBV is approximately less than 10% for the patients of less than 5 years but the prevalence rate ranges from 30%-50% for patients of 5 years or above. For HBV infection, the acute case-fatality rate is approximately in the range of 0.5%-1%. Generally the rate of chronic infection, among patients of less than 5 years of age, is 30%-90%. But the rate of chronic infection lies between 2%-10% for the patients of 5 years of age or more. The premature mortality rate from chronic liver disease is approximately 15%-25%[21]. The HBV has strict tissue tropism to the liver. The virus infected hepatocytes produces large amount of HBsAg particles which lack the DNA. The viral DNA is capable of integrating into host chromosome. Normally HBV is not cytopathic itself, instead in case of chronic hepatitis B disease, the liver damage takes place because of immune clearance phase of host against HBV infected hepatocytes[22].
The primary liver carcinoma is considered as 5th most frequent cancer of world and hepatocellular carcinoma is the major type of primary liver carcinoma. In many areas of the world, more than 85% of hepatocellular carcinoma retains markers against hepatitis B and hepatitis C[23]. Two treatment options are available for the prophylaxis against HBV, which includes HBV and hepatitis B immune globin. For pre-exposure and post-exposure, hepatitis B vaccine is recommended. The vaccine is capable for long term protection against HBV infection. The hepatitis B immune globin can only provide temporary protection for approximately 3-6 months. This treatment option is usually recommended for post-exposure settings[24]. Hepatitis B viral form mutants are developed in some patients. These viral mutants are resistant against one or more antiviral drugs. Researchers tend to develop novel antiviral therapeutic options in order to prevent resistance and minimize viral load.
2.2. HCV and genome
The size of HCV genome is 9.5 Kb. On both 5′ and 3′ termini of the genome there exists highly conserved untranslated regions, which flanks a large translational ORF (of approximately 9 000 nucleotides) capable of encoding polyprotein of 3 000 amino acids. The uncapped 5′ noncoding region, of approximately 340 nucleotides, contains internal ribosomal entry site (IRES) which is essential for cap-independent translation of viral RNA. The 3′ region of the genome contains noncoding regions. The polyprotein encoded by the genome is initially translated as one single peptide which is later on co- and post transcriptionally processed by various cellular and viral enzymes[9],[25] (Figure 2).
Figure 2. HCV genome and information regarding HCV proteins.
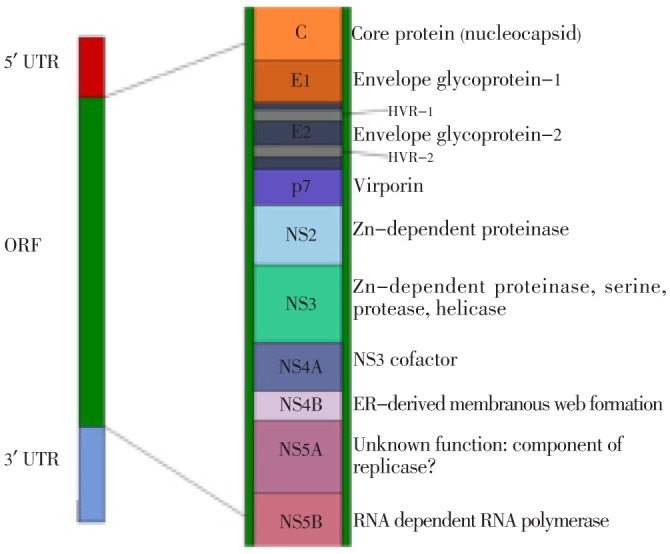
At the 5′ region of ORF there exists structural proteins including core, highly variable glycoproteins (E1 and E2) and small protein P7. The non structural proteins are encoded by rest of the genome (the proteins include NS2, NS3, NS4a, NS4b, NS5a, NS5b and untranslated regions). The E2 region contains two hypervariable regions and is considered as the most genetically variable region of the HCV genome. This region allows for evasion of host immune responses. The P7 region belongs to viroporin family and it is thought to be helpful for viral maturation and release. The expression of NS4B region leads to the formation of a membranous web, which is derived from endoplasmic reticulum of the host cell. This is thought to act as a scaffold with which viral RNA and proteins can associate during viral replication. All of these proteins are possible drug targets; however, the envelope glycoproteins may be less effective targets as they are extremely variable. The NS5B protein (RNA polymerase) is considered as target site of polymerase inhibitors and NS3 protein is considered as the target of protease inhibitors[9],[10],[26]. A newly discovered protein which belongs to HCV is protein F, which is produced by ribosomal frame shift mutation around codon 11 of core protein. HCV infected individuals contains antibodies against this protein, so it is hypothesized that this protein usually expresses in those persons who are HCV infected. The information regarding function of protein F and how often it is expressed in infected individuals is unknown[27]. A number of cellular pathways are affected by core protein of HCV. Example includes signaling of nuclear factor B (NF-B). NF-B is a transcription factor which is involved in anti-apoptotic genes and pro-inflammatory genes regulation. Activation of NF-B usually leads to HCV mediated pathogenesis[28]. The IRES contains highly conserved structures which are important for binding and appropriate positioning of viral genome inside the host cell[29]. At the 3′ end there exist a non coding region which contains 50 nucleotides polypyrimidine track and a highly conserved terminal sequence of 100 nucleotides.
A lot of information regarding HCV virion structure, binding mechanisms, functioning of viral proteins, release mechanisms and various host immune responses, is lacking due to the fact that HCV only infects humans and chimpanzees, and there is no reliable cell culture system or small animal model to facilitate complicated HCV research. Scientists are trying to develop subgenomic replicon systems and novel mice models with chimeric human livers. This approach would open new doors of scientific progress towards identification of most potent drug targets against HCV[30],[31]. In subgenomic replicon system, the structural genes of HCV are replaced by neomycine phosphotransferase II gene[30]. The internal ribosomal entry site acquired from encephalomyocarditis virus cause translation of non-structural proteins of HCV. Later on, for replication, the replicons are transferred to human hepatoma cells, for example Huh-7 cells. The structural genes were removed as a result of which virions cannot be assembled. The amplified RNA is either reused (for translation) or later on degraded by cellular ribonucleases[30]. There is an urgent demand for small animal models to study HCV replication and drug testing. In 2001, Mercer et al., successfully observed HCV replication (at two months post infection) in mice having chimeric liver with transplanted human hepatocytes[32].
2.3. HCV induced host immune response
HCV infection causes activation of host innate immune responses. Such responses usually take place after two days post infection, with increased involvement of interferon regulatory factors, protein kinase R and antiviral gene products, like interferon-inducible genes and immune transcription factors[27]. The innate immune response was observed in all individuals, regardless of whether they developed chronic infection or controlled the virus. Development of chronic infection suggests viral resistance to the innate immune responses, which probably exist due to interference of HCV proteins with pathways associated with innate immune system. It has been reported from in vitro studies that NS5A and E2 interfere with protein kinase R. The viral core protein causes inhibition of JAK-STAT pathway which is involved in interferon (IFN) signaling. The viral NS3/4A blocks the accumulation of phosphorylated interferon regulatory factor-3 (IRF-3), which inhibits expression of interferon stimulated genes and type I IFN[27],[33].
Among HCV infected individuals 15% had natural tendency to overcome acute viral infection, whereas 85% of individuals were unable to control HCV infection. The mystery behind this natural phenomenon is still unknown. However, among those who successfully control HCV infection, IFN-γ is preferentially expressed in liver (through T lymphocytes) several weeks post-infection. It results into expression of various chemokines which attract T cells and various proteins involved in antigen processing and presentation[27]. Among those individuals who control viral infection, there is increase production of CD4+ and CD8+ T cells. The chronic viral infection occurs when individuals are unable to mount HCV-specific T cell responses. Another possible reason is influence of strong immune responses which initially cause RNA clearance, followed by contraction in CD8+/CD4+ level and ultimately results rebound in viremia[25],[27],[33]. According to Sun et al.[34], there exist significant anomalies in immune response patterns among patients who exhibited viral clearance and those who were chronically infected with HCV. It includes, reduced frequency and limited capacity of HCV specific CD8+ cells, CD4+ cells with less production of interleukin-2 and impaired dendritic cells[34]. It has been reported that the T cell identifies dendritic cell via molecular signature at immunological synapse (a contact point). If the synapse signals for presence of foreign body, it would trigger T cell attack[34].
Chronically infected HCV patients possess decreased frequency of natural killer (NK) T cells and impaired NK cell activity, this response get reversed in patients undergoing alpha-interferon therapy. These cells (NK and NKT cells) tend to alleviate pathogenic attack before activation of adaptive immune system. It is further supported by literature that these cells are responsible for activation of adaptive immune responses and regulation of autoimmune responses[33]. HCV cause infection only in 1-10% of hepatocytes, but production of IFN-γ and tumor necrosis factor alpha from CD8+ cells cause destruction of both infected cells and non-infected surrounding cells. Such high level of cell death and regeneration rate leads to onset of hepatocellular carcinoma[33]. The common treatment available for chronic HCV infected patients is combination therapy including pegylated alpha-interferon and the ribavirin which is a nucleoside analog. Usually it requires single injection per week[35]. Although exact mechanism of action of these drugs is difficult to understand, yet it has been implicated that degradation of positive strand RNA and suppression of viral protein synthesis are principle mechanisms involved to arrest viral replication. It has been reported that this treatment option is only 50% effective against HCV genotype 1. And the efficacy for genotypes 2 and 3 was reported as 80%[30],[36]. Unfortunately there is no alternative treatment for non-responders. Severe adverse reactions in response to interferon/ribavirin therapy usually drag doctors towards prescription of lower dose. One of the most sadden side effect of the aforementioned therapy is that combination therapy can worsen liver disease (with major symptom of viral infection) due to which only a subset of patients could get relief from HCV infection. There is an urgent need for identification of novel antiviral strategies and designing new drugs to combat lethal viral infection[36],[37].
2.4. Genotypic distribution of hepatitis B virus
Hepatitis B virus infection is distributed worldwide in the form of eight different genotypes (A-H)[38]. A newly described genotype I is also a pivot of interest for future researchers[39]. There exists at least 8% nucleotide sequence dissimilarity among eight known HBV genotypes. In Pakistan genotype D is most prevalent with estimated prevalence rate of 63.71%. This genotype is usually less responsive towards interferon therapy. The prevalence of genotype A, C, B in Pakistan is 10.036%, 7.550% and 5.335% respectively. The reported prevalence of mixed genotypes and untypable genotypes is 9.93% and 2.37% respectively[40]. According to the most recent study conducted by Awan et al. it has been reported that genotype C is most emerging genotype associated with more severe liver diseases (cirrhosis). The approximate prevalence rate of this genotype is 27.7% which represents a major threat to future generations[41].
The genotype A is most prevalent in Africa, South East Asia including Philippines and Europe. It has two subtypes named as A1/Aa (most prevalent in Asia and Africa) and A2/Ae (most prevalent in Europe and United States). The genotypes B and C are most prevalent in Asia. The genotype B exist in two geographical locations named as B1/Bj (most prevalent in Japan) and B2/Ba (most prevalent in Asia) which is further classified into four distinct clades (B2-B4). The genotype C exit in two geographical locations named as C1/Cs (most prevalent in South-East Asia) and C2/Ce (most prevalent in East Asia) which is further classified into five distinct clades (C1 present in Myanmar, Thailand and Vietnam, C2 present in China, Korea and Japan, C3 present in New Caledonia and Polynesia, C4 present in Australia and C5 present in Philippines). The genotype D is most prevalent in India, Mediterranean and Middle East. It has been further classified into seven subtypes (D1-D7). The genotype F or H are mostly prevalent in Central and South America. The genotype F is further classified into four subtypes (F1-F4). The genotype G is most prevalent in France and Germany. It has been reported that genotypes A, D and F are most occurring in Brazil. In United States all genotypes exist with difference in frequencies based on ethnicity[42]–[45].
2.5. Genotypic distribution of HCV
HCV is increasingly recognized as major health care problem in the whole word. Despite of strenuous efforts from scientists, antiviral approaches could not completely eradicate it due to the fact that HCV is extremely heterogeneous. HCV is an RNA virus and lacks effective proofreading ability after its replication, that's why it introduces several mutations and keeps on evolution with respect to time. Mutations are not randomly distributed in the whole genome, instead these exist at hyper-variable regions of the genome which encode for envelop proteins; hence enable the virus to escape from host immune surveillance[46]. HCV has been categorized into various genotypes, having 67% nucleotide sequence identity of members with each other[47]. Up to date 11 genotypes and above 100 subtypes of HCV exists worldwide with different geographical distribution[48]. In Pakistan the most prevalent genotype is 3 followed by genotype 1 and 2[49]. Similarly genotype 3 and 1 are also foremost in Indian population[50]. According to P Simmonds et al., in United States of America and Canada, most prevalent genotypes are 1a, 1b, 2a, 2b and 3a. Similarly in South America, genotypes 1a, 1b, 2 and 3a are more common. In Northern Europe, genotypes 1a, 1b, 2b and 3a are more prevalent, where as in Western Europe genotypes 1a, 1b, 2a, 2b and 3a are majorly distributed. In Southern Europe there is higher prevalence of genotypes 1b and 2c, whereas in Eastern Europe, genotype 1b is significantly distributed. In Africa, prevalence of genotype 4 is higher in parts Northern Central Africa; prevalence of genotype 4a is greater in Egypt; and prevalence of genotypes 1, 2, 3 and 5a is foremost in South Africa. In Pacific region, there is increased prevalence of genotypes 1a, 1b, 2a, 2b and 3a in Australia; in Taiwan genotypes 1b, 2a and 2b are more common; in Japan, there is an increased prevalence of genotypes 1a, 2a and 2b; in Hong Kong, genotypes 6a, 1b, 2a and 2b are highly distributed; in Thailand, genotypes 1b, 2, 3 and 6 are most prevalent; in Malaysia, genotypes 1b, 2 and 3 are mostly present; and in Vietnam, there is an increased prevalence of genotypes 1b, 2 and 6. In Asia, prevalence of genotype 1b is higher in Turkey; prevalence of genotype 4 is greater in Middle East; and prevalence of genotypes 1b, 2a and 2b is higher in China[51],[52].
2.6. Global epidemiology of HBV
The epidemiology of chronic HBV infection is distinct and diverse worldwide (Figure 3). Various seroprevalence studies conducted in different areas of world can easily be categorized into three distinct groups of higher, intermediate and lower endemicity[53]. In developing countries with larger population (South East Asia, Sub Sahara Africa, China, Indonesia, Nigeria and Amazon Basin), there is higher prevalence of endemicity with approximately 8% of population as chronic carrier of HBV. In aforementioned areas of world, 70% to 95% of population represents present or past serological markers against HBV. In another study, it has been reported that 60% of world population exist in high endemic zone of HBV infection[54]–[56]. The intermediate endemic zone of HBV infection, Middle East, Eastern and Southern Europe, South America and Japan exist. Among these populations the estimated infection is approximately 10-60% and the chronic carrier rate is 2-7%. In the region of intermediate endemicity, majority of infection develop in adults but rate of chronic infection are higher in infants due to early childhood exposure to viral infection[57]. The seroprevalence of HBV infection has been reported 5% in India, while in Italy, Russia and Turkey the prevalence rate ranges from 3%-10%[58]–[61].
Figure 3. Global epidemiology of HBV and prevalence of HBV carriers. Basic source of information is Murray et al., Medical Microbiology.
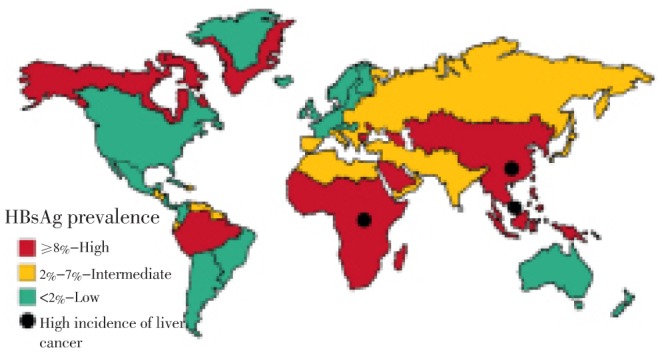
The HBV zone of lowest endemicity includes most developed countries such as Australia, North America and Northern and Western Europe. In aforementioned regions of world approximately 5-7% of population gets HBV infected with nearly 0.5% to 2.0% rate of chronic carriers. The most probable reasons of HBV infection in young adolescents could be exposure to high risk population groups, injection drug users, health care professionals, sex workers and unhealthy blood transfusion setups[62].
2.7. Global epidemiology of HCV
Geographic distribution of HCV is not uniform (Figure 4). Several seroprevalence studies on different populations tend to describe the epidemiological patterns of disease. Although seroprevalence studies conducted on general population best describes the actual status of disease in that particular region, however majorities of such studies are conducted on high risk populations. Among various seroprevalence studies, majorities are based on cross sectional design; however population based studies are better representative of entire community. Limited prevalence studies have been documented in various regions of world due to expenses and practical difficulties involved in detection of viral RNA in serum. Available data suggests that HCV is prevalent in 3% of world population[63]. The prevalence of HCV is considered highest in African and Asian regions, but countries with limited prevalence include Northern and Western Europe, Australia and North America[64]. China is world's most populous country with approximately 1 347 million population with estimated 38 million people infected with HCV[65],[66]. India is second world's most populous country with estimated population of 1 210 million population which covers approximately 17.25% of world population. According to a community-based study conducted in West Bengal, the estimated prevalence of HCV was reported to be of 0.9% (Populous countries wiki; prevalence of HCV was reported to be of 0.9%)[65],[67].
Figure 4. Global estimated prevalence of HCV.
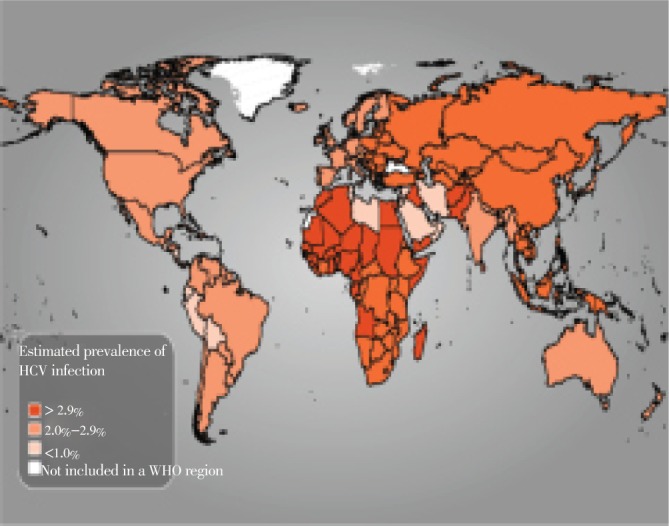
United States of America is ranked as third world's most populous country with population of 313 million, which covers approximately 4.47% of world population. According to a study conducted on 21 214 nationally representative samples, the seroprevalence of HCV was reported as 1.8%[55],[65]. Indonesia is ranked as fourth world most populous country with approximately 237 million people, which covers approximately 3.39% of world population. According to study conducted on 7 572 voluntary blood donors, the seroprevalence of HCV was reported as 2.1%[65],[68]. Brazil is ranked as fifth world most populous country having population of 192 million people, covering 2.74% of world population. According to a study conducted on 66 414 voluntary blood donors, the estimated HCV prevalence was approximately 1.1%[65],[69]. Pakistan is the sixth populous country of the world with approximate population of 179 million individuals covering 2.56% of world population. Khattak et al. conducted a study on 103 858 voluntary blood donors, it was reported that the seroprevalence of HCV was 4%[65],[70]. Therefore on the basis of HCV infection prevalence in the six most populous nations in the world, Pakistan can be inferred as country with highest HCV prevalence among other most populous nations of the world.
Viral hepatitis has been increasingly recognized as sever healthcare problem in world. This review provides comprehensive information regarding important HBV and HCV regions for conducting future valuable research projects. A lot of efforts are required to completely eradicate HBV and HCV from this world like previous small pox viral pathogen which had been eradicated from world (decades ago) and polio virus which would be eradicated from world in near future. It is a well known fact that one in twelve people worldwide is infected with viral hepatitis. Due to its rampant proliferation, virus quasispecies and increased viral is associated mortalities. It is anticipated that hepatitis virus would soon emerge as most dangerous viral pathogen. HCV prevalence is unfortunately increasing day by day in developing countries due to limited awareness among general population. Up to date no HCV vaccine has been successfully prepared therefore a lot of efforts are still needed to reduce global burden of this disease. The most important way to prevent future burden of HCV is through knowledge and awareness via prevention strategies designed by public health specialists.
Comments
Background
HBV and HCV are the most important cause of chronic viral hepatitis. They are responsible for thousand of deaths worldwide because of end stage liver diseases related to viral infection. So, chronic hepatitis B and C is a major health problem and preventive strategies need to be implemented as well as therapies to eradicate chronic infection in the case of HCV or to keep viral replication suppressed in the case of HBV.
Research frontiers
This paper summarizes some epidemiological information on HBV and HCV infection in the world. However, misleading information is reported on the classification of HCV genotypes such as classification into 11 genotypes. Further, the authors don't even mention the new antiviral drugs Boceprevir and Telaprevir approved for treatment of HCV genotype 1 chronic infection. I understand that antiviral therapy is not the focus of this review, but since the authors mention IFN-ribavarin treatment, they cannot overlook the new therapies.
Related reports
Several reviews and specific papers are published every year on these topics. For instance, the paper by Stroffolini in Digestive and Liver Disease, 2005, reports updated information on the prevalence of HBV in Italy.
Applications
The knowledge of the HBV and HCV epidemiology helps the health authorities to implement preventive measures to fight these two important causes of chronic viral hepatitis.
Peer review
Overall, the paper is informative and provides some valuable information on the epidemiology of HBV and HCV and their genomic organization. Details of the importance of a correct genotype determination should be further addressed since HCV genotypes can respond differently to antiviral therapy.
Footnotes
Conflict of interest statement: We declare that we have no conflict of interest.
References
- 1.Scarborough A, Hepatitus B. 2010. [Online] Available from: http://www.austincc.edu/microbio/2704z/hbv.htm. [Accessed on 27 July, 2013].
- 2.Yates S, Penning M, Goudsmit J, Frantzen I, Weijer BV, Strijp DV, Gemen BV. Quantitative detection of hepatitis B Virus DNA by real-time nucleic acid sequence-based amplification with molecular beacon detection. J Clin Microbiol. 2011;39(10):36–56. doi: 10.1128/JCM.39.10.3656-3665.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yen TSB. Hepadnaviral X protein: Review of recent progress. J Biomed Sci. 1996;3:20–30. doi: 10.1007/BF02253575. [DOI] [PubMed] [Google Scholar]
- 4.Hruska JF, Robinson WS. The proteins of hepatitis B Dane particle cores. J Med Virol. 1977;1(2):119–131. doi: 10.1002/jmv.1890010205. [DOI] [PubMed] [Google Scholar]
- 5.World Health Organization Hepatitis B. 2008. [Online] Available from: http://www.who.int/mediacentre/factsheets/fs204/en/ [Accessed on 27 July, 2013].
- 6.Everson GT, Weinberg H. Living with hepatitis B: A survivor's guide. US: Hatherleigh Press; 2002. [Google Scholar]
- 7.Choo QL, Kuo G, Weiner AJ, Overby LR, Bradley DW, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244(4902):359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 8.Conca P, Tarantino G. Hepatitis C virus lymphotropism and peculiar immunological phenotype: Effects on natural history and antiviral therapy. World J Gastroenterol. 2009;15(19):2305–2308. doi: 10.3748/wjg.15.2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Penin F, Dubuisson J. Structural biology of hepatitis C virus. Hepatology. 2004;39(1):5–19. doi: 10.1002/hep.20032. [DOI] [PubMed] [Google Scholar]
- 10.Appel N, Pietschmann T, Bartenschlager R. Mutational analysis of hepatitis C virus nonstructural protein 5A: Potential role of differential phosphorylation in RNA replication and identification of a genetically flexible domain. J Virol. 2005;79(5):3187–3194. doi: 10.1128/JVI.79.5.3187-3194.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seeger C, Mason WS. Hepatitis B virus biology. Microbiol Mol Biol Rev. 2000;64:51–68. doi: 10.1128/mmbr.64.1.51-68.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Summers J, Mason WS. Replication of the genome of a hepatitis B like virus by reverse transcription of an RNA intermediate. Cell. 1982;29:403–415. doi: 10.1016/0092-8674(82)90157-x. [DOI] [PubMed] [Google Scholar]
- 13.Tong S, Kim KH, Chante C, Wands J, Jisu L. Hepatitis B virus e antigen variants. Int J Med Sci. 2005;2(1):2–7. doi: 10.7150/ijms.2.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen MT, Billaud JN, Sallberg M, Guidotti LG, Chisari FV, Jones J, et al. A function of HBV precore protein is to regulate immune response to core antigen. Proc Natl Acad Sci USA. 2004;101(41):14913–14918. doi: 10.1073/pnas.0406282101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Summers J, Connel A. Genome of Hepatitis B virus restriction enzyme cleavage abd structure of DNA extracted from Dane particles. Proc Natl Acad Sci USA. 1975;72:4597–4601. doi: 10.1073/pnas.72.11.4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tiollais P, Pourcel C, Dejean A. The hepatitis B virus. Nature. 1985;317:489–495. doi: 10.1038/317489a0. [DOI] [PubMed] [Google Scholar]
- 17.Cattaneo R, Will H, Schaller H. Hepatitis B virus transcription in the infected liver. EMBO J. 1984;3(9):2191–2196. doi: 10.1002/j.1460-2075.1984.tb02113.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Enders GH, Ganem D, Varmus H. Mapping the major transcripts of ground squirrel hepatitis virus: the presumptive template for reverse transcriptase is terminally redundant. Cell. 1985;42(1):297–308. doi: 10.1016/s0092-8674(85)80125-2. [DOI] [PubMed] [Google Scholar]
- 19.Center for Disease Control and Prevention Hepatitis. 2010. [Online] Available from: http://www.cdc.gov/ncidod/diseases/hepatitis/slideset/hep_b/slide_4.htm. [Accessed on 27 July, 2013].
- 20.Sprengers D. Immune response and immunomodulation in chronic hepatitis B virus infection. [Online] Available from: http://repub.eur.nl/res/pub/8063/061101_Sprengers,%20Dave.pdf [Accessed on 27 July, 2013].
- 21.Butel JS. Papovaviruses. In: Baron S, Albrecht T, Castro G, Couch RB, Davis CP, et al., editors. Medical microbiology. 5th ed. Philadelphia: Mosby; 2005. Chapter 66. [Google Scholar]
- 22.Lok AS, McMahon BJ. Chronic hepatitis B: Update 2009. AASLD practice guideline update. [Online] Available from: http://www.aasld.org/practiceguidelines/documents/bookmarked%20practice%20guidelines/chronic_hep_b_update_2009%208_24_2009.pdf [Accessed on 27 July, 2013].
- 23.Kim JW, Ye Q, Forgues M, Chen Y, Budhu A, Sime J, et al. Cancer associated molecular signature in the tissue samples of patients with cirrhosis. Hepatology. 2004;39:518–527. doi: 10.1002/hep.20053. [DOI] [PubMed] [Google Scholar]
- 24.Samuel LK, Ronald C, Stanley E, James D, Caroline B, Cleveland, et al. Hepatitis B virus: A comprehensive strategy for eliminating transmission in the United States through universal childhood vaccination: Recommendations of the Immunization Practices Advisory Committee (ACIP) 1991. [Online] Available from: http://www.cdc.gov/mmwr/preview/mmwrhtml/00033405.htm [Accessed on 27 July, 2013]. [PubMed]
- 25.Moradpour D, Blum HE. A primer on the molecular virology of hepatitis C. Liver Int. 2004;24(6):519–525. doi: 10.1111/j.1478-3231.2004.0965.x. [DOI] [PubMed] [Google Scholar]
- 26.Ahmad A, Alvarez F. Role of NK and NKT cells in the immunopathogenesis of HCV-induced hepatitis. J Leukoc Biol. 2004;76(4):743–759. doi: 10.1189/jlb.0304197. [DOI] [PubMed] [Google Scholar]
- 27.Shoukry NH, Cawthon AG, Walker CM. Cell-mediated immunity and the outcome of hepatitis C virus infection. Annu Rev Microbiol. 2004;58:391–424. doi: 10.1146/annurev.micro.58.030603.123836. [DOI] [PubMed] [Google Scholar]
- 28.Mann EA, Stanford S, Sherman KE. Prevalence of mutations in hepatitis C virus core protein associated with alteration of NF-kB activation. Vir Res. 2006;121:51–57. doi: 10.1016/j.virusres.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 29.Kieft JS, Zhou K, Grech A, Jubin R, Doudna JA. Crystal structure of an RNA tertiary domain essential to HCV IRES-mediated translation initiation. Nat Struct Mol Biol. 2002;5:370–374. doi: 10.1038/nsb781. [DOI] [PubMed] [Google Scholar]
- 30.Guo JT, Sohn JA, Zhu Q, Seeger C. Mechanism of the interferon alpha response against hepatitis C virus replicons. Virology. 2004;325:71–81. doi: 10.1016/j.virol.2004.04.031. [DOI] [PubMed] [Google Scholar]
- 31.Trujillo-Murillo KC, Garza-Rodeiguex ML, Martinez-Rodiguex HG, Barrera-Saladana HA, Bosques-Padilla F, Ramos-Jimenez J, et al. Experimental models for hepatitis C virus (HCV) new opportunities for combating hepatitis C. Ann Hepatol. 2004;3(2):54–62. [PubMed] [Google Scholar]
- 32.Mercer DF, Schiller DE, Elliott JF, Douglas DN, Hao C, Rinfret A, et al. Hepatitis C virus replication in mice with chimeric human livers. Nat Med. 2001;7(8):927–933. doi: 10.1038/90968. [DOI] [PubMed] [Google Scholar]
- 33.Sun DX, Liu L, Heinz B, Kolykhalov A, Lamar J, Johnson RB, et al. P4 cap modified tetrapeptidyl alpha-ketoamides as potent HCV NS3 protease inhibitors. Bioorg Med Chem Lett. 2004;14:4333–4338. doi: 10.1016/j.bmcl.2004.05.078. [DOI] [PubMed] [Google Scholar]
- 34.American Liver Foundation . Liver awareness month. New York: American Liver Foundation; [Online] Available from: http://www.liverfoundation.org [Accessed on 27 July, 2013]. [Google Scholar]
- 35.Carroll SS, Ludmerer S, Handt L, Koeplinger K, Zhang NR, Graham D, et al. Robust antiviral efficacy upon administration of a nucleoside analog to hepatitis C virus-infected chimpanzees. Antimicrob Agents Chemother. 2009;53(3):926–934. doi: 10.1128/AAC.01032-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Center for Disease Control and Prevention Hepatitis C FAQs for the public. 2012. [Online] Available from: http://www.cdc.gov/hepatitis/c/cfaq.htm [Accessed on 27 July, 2013].
- 37.Kowdley KV. Hematologic side effects of interferon and ribavirin therapy. J Clin Gastroenterol. 2005;39:3–8. doi: 10.1097/01.mcg.0000145494.76305.11. [DOI] [PubMed] [Google Scholar]
- 38.Kramvis A, Kew M, François G. Hepatitis B virus genotypes. Vaccine. 2005;23(19):2409–2423. doi: 10.1016/j.vaccine.2004.10.045. [DOI] [PubMed] [Google Scholar]
- 39.Olinger CM, Jutavijittum P, Hubschen JM, Yousukh A, Sumountry B, Thammavong T, et al. Possible new hepatitis B virus genotype, southeast Asia. Emerg Infect Dis. 2008;14(11):1777–1780. doi: 10.3201/eid1411.080437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ali M, Idrees M, Ali L, Hussain A, Rehman I, Saleem S, et al. Hepatitis B virus in Pakistan: A systematic review of prevalence, risk factors, awareness status and genotypes. Virol J. 2011;8:102. doi: 10.1186/1743-422X-8-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Awan Z, Idrees M, Amin I, Butt S, Afzal S, Akbar H, et al. Pattern and molecular epidemiology of hepatitis B virus genotypes circulating in Pakistan. Infect Genet Evol. 2010;10:1242–1246. doi: 10.1016/j.meegid.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 42.Cavinta L, Cao G, Schaefer S. A new isolate of hepatitis B virus from the Philippines possibly representing a new subgenotype C6. J Med Virol. 2009;81(6):983–987. doi: 10.1002/jmv.21475. [DOI] [PubMed] [Google Scholar]
- 43.Mahtab MA, Rahman S, Khan M, Karim F. Hepatitis B virus genotypes: an overview. Hepatobiliary Pancreat Dis Int. 2008;7(5):457–464. [PubMed] [Google Scholar]
- 44.Palumbo E. Hepatitis B genotypes and response to antiviral therapy: a review. Am J Ther. 2007;14(3):306–309. doi: 10.1097/01.pap.0000249927.67907.eb. [DOI] [PubMed] [Google Scholar]
- 45.Kurbanov F, Tanaka Y, Kramvis A, Simmonds P, Mizokami M. When should “I” consider a new hepatitis B virus genotype. J Virol. 2008;82(16):8241–8242. doi: 10.1128/JVI.00793-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.World Health Organization . Hepatitis C. 2003. Geneva: World Health Organization; 2003. [Online] Available from: http://www.who.int/csr/disease/hepatitis/whocdscsrlyo2003/en/index2.html. [Assessed on 16 July, 2013] [Google Scholar]
- 47.Walewski JL, Gutierrez JA, Branch-Elliman W, Stump DD, Keller TR, Rodriguez A, et al. Mutation master: Profiles of substitutions in hepatitis C virus RNA of the core, alternate reading frame, and NS2 coding regions. RNA. 2002;8:557–571. doi: 10.1017/s1355838202029023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alestig E. Geographic and genetic diversity of hepatitis B. [Online] Available from: https://gupea.ub.gu.se/handle/2077/22932. [Accessed on 27 July, 2013].
- 49.Idrees M, Riazuddin S. Frequency distribution of hepatitis C virus genotypes in different gepgraphical regions of Pakistan and their possible routes of transmission. BMC Infect Dis. 2008;8:69. doi: 10.1186/1471-2334-8-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Narahari S, Juwle A, Basak S, Saranath D. Prevalence and geographic distribution of Hepatitis C virus genotypes in Indian patient cohort. Infect Genet Evol. 2009;9(4):636–645. doi: 10.1016/j.meegid.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 51.Simmonds P, Bukh J, Combet C, Deleage G, Enomoto N, Feinstone S, et al. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology. 2005;42(4):962–973. doi: 10.1002/hep.20819. [DOI] [PubMed] [Google Scholar]
- 52.Viral Hepatitis Prevention Report The hepatitis C virus. 2002. [Online] Available from: http://www.vhpb.org/files/html/Meetings_and_publications/VHPB_Meetings/geneva2002.htm [Accessed on 27 July, 2013].
- 53.Hou JL, Liu ZH, Gu F. Epidemiology and prevention of hepatitis B virus infection. Int J Med Sci. 2005;2(1):50–57. doi: 10.7150/ijms.2.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Colin WS, Simard EP, Finelli L, Fiore AE, Bell BP. Hepatitis B virus infection. epidemiology and vaccination. Oxford J Epidemol Rev. 2006;28:112–125. doi: 10.1093/epirev/mxj009. [DOI] [PubMed] [Google Scholar]
- 55.Alter MJ. Epidemiology and prevention of hepatitis B. Semin Liver Dis. 2003;23(1):39–46. doi: 10.1055/s-2003-37583. [DOI] [PubMed] [Google Scholar]
- 56.Margolis HS, Alter MJ, Hadler SC. Hepatitis B: Evolving epidemiology and implications for control. Semin Liver Dis. 1991;11:84–92. doi: 10.1055/s-2008-1040427. [DOI] [PubMed] [Google Scholar]
- 57.Toukan A. Strategy for the control of hepatitis B virus infection in the Middle East and North Africa. Vaccine. 1990;8:S117–S121. [PubMed] [Google Scholar]
- 58.Kurien T, Thyagarajan SP, Jeyaseelan L. Community prevalence of hepatitis B infection and modes of transmission in Tamil Nadu, India. Indian J Med Res. 2005;121:670–675. [PubMed] [Google Scholar]
- 59.Iashina T L, Favorov MO, Shakhgil'dian IV. The prevalence of the markers of viral hepatitis B and delta among the population in regions differing in the level of morbidity. Vopr Virusol. 1992;37:194–196. [PubMed] [Google Scholar]
- 60.Erden S, Buyukozturk S, Calangu S. A study of serological markers of hepatitis B and C viruses in Istanbul, Turkey. Med Princ Pract. 2003;12:184–188. doi: 10.1159/000070757. [DOI] [PubMed] [Google Scholar]
- 61.Da-Villa G, Sepe A. Immunization programme against hepatitis B virus infection in Italy: cost-effectiveness. Vaccine. 1999;17:1734–1738. doi: 10.1016/s0264-410x(98)00414-9. [DOI] [PubMed] [Google Scholar]
- 62.McQuillan GM, Townsend TR, Fields HA, Carroll M, Leahy M, Polk BF. Seroepidemiology of hepatitis B virus infection in the United States. Am J Med. 1989;87(3):S5–S10. doi: 10.1016/0002-9343(89)90523-8. [DOI] [PubMed] [Google Scholar]
- 63.World Health Organization . The world health report 2006-working together for health. Geneva: World Health Organization; 2006. [Online] Available from: http://www.who.int/whr/2006/en/ [Accessed on 27 July, 2013]. [Google Scholar]
- 64.Shepard CW, Finelli L, Alter MJ. Global epidemiology of hepatitis C virus infection. Lancet Infect Dis. 2005;5:558–567. doi: 10.1016/S1473-3099(05)70216-4. [DOI] [PubMed] [Google Scholar]
- 65.Wikipedia List of countries by population. 2011. [Online] Available from: http://en.wikipedia.org/wiki/List_of_countries _by_population . [Accessed on 27 July, 2013].
- 66.Hepatitis C articles (HCV) [Online] Available from: http://www.natap.org/2011/HCV/080211_01.htm [Accessed on 27 July, 2013].
- 67.Chowdhury A, Santra A, Chaudhuri S, Dhali GK, Chaudhuri S, Maity SG, et al. Hepatitis C virus infection in the general population: a community-based study in West Bengal, India. Hepatology. 2003;37:802–809. doi: 10.1053/jhep.2003.50157. [DOI] [PubMed] [Google Scholar]
- 68.Sulaiman HA, Julitasari, Sie A, Rustam M, Melani W, Corwin A, et al. Prevalence of hepatitis B and C viruses in healthy Indonesian blood donors. Trans R Soc Trop Med Hyg. 1995;89:167–170. doi: 10.1016/0035-9203(95)90480-8. [DOI] [PubMed] [Google Scholar]
- 69.Brandao AB, Costa FS. Risk factors for hepatitis C virus infection among blood donors in southern Brazil: a case-control study. BMC Gastroenterol. 2002;2:18. doi: 10.1186/1471-230X-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Khattak MF, Salamat N, Bhatti FA, Qureshi TZ. Seroprevalence of hepatitis B, C and HIV in blood donors in northern Pakistan. J Pak Med Assoc. 2002;52:398–402. [PubMed] [Google Scholar]


