Abstract
Steric stabilization of cationic liposome–DNA (CL–DNA) complexes is required for in vivo applications such as gene therapy. PEGylation (PEG: poly(ethylene glycol)) of CL–DNA complexes by addition of PEG2000-lipids yields sterically stabilized nanoparticles but strongly reduces their gene delivery efficacy. PEGylation-induced weakening of the electrostatic binding of CL–DNA nanoparticles to cells (leading to reduced uptake) has been considered as a possible cause, but experimental results have been ambiguous. Using quantitative live-cell imaging in vitro, we have investigated cell attachment and uptake of PEGylated CL–DNA nanoparticles with and without a custom synthesized RGD-peptide grafted to the distal ends of PEG2000-lipids. The RGD-tagged nanoparticles exhibit strongly increased cellular attachment as well as uptake compared to nanoparticles without grafted peptide. Transfection efficiency of RGD-tagged PEGylated CL-DNA NPs increases by about an order of magnitude between NPs with low and high membrane charge density (σM; the average charge per unit area of the membrane; controlled by the molar ratio of cationic to neutral lipid), even though uptake of RGD-tagged particles is only slightly enhanced by high σM. This suggests that endosomal escape and subsequent transfection efficiency of RGD-tagged NPs is facilitated by high σM. We present a model describing the interactions between PEGylated CL–DNA nanoparticles and the anionic cell membrane which shows how the PEG grafting density and membrane charge density affect adhesion of nanoparticles to the cell surface.
Introduction
Driven by the promises of gene therapy, the field of research centered on delivery of exogenous nucleic acids into cells remains extremely active [1–11]. Of the over 1900 clinical trials in gene therapy, 5.5% employ strategies classified as lipofection [12,13]. The current preference for viral vectors over synthetic vectors such as cationic lipids and polymers exists because viral vectors are more efficient for gene delivery in vivo. However, they suffer from a variety of issues, the most important of which are safety concerns [14–18]. Furthermore, the capacity of viral vectors is small and limited by their capsid size, while synthetic vectors are able to deliver very large pieces of DNA [19]. Another advantage of synthetic vectors is that they are less immunogenic because they lack viral capsid proteins. Commercial synthetic vectors with high transfection efficiencies are available for in vitro applications but the same success has not been achieved in vivo. The low transfection efficiency (TE; a measure of expression of the transfected gene) of synthetic vectors in vivo is due to the numerous biological and physico-chemical barriers that must be overcome in the process: protecting genetic material during circulation in the bloodstream, targeting it to the desired tissue, and then transferring it from the extracellular environment into the target cell, through the cytoplasm, and finally into the nucleus [20–22].
Surface functionalization of synthetic vectors with an inert polymer such as poly(ethylene glycol) (PEG) sterically stabilizes them and can help them avoid macrophage removal and thus remain in circulation [23,24]. This is essential to enable tissue targeting. However, PEGylation also reduces TE [25]. A possible cause of this is reduced electrostatic attraction between the PEGylated CL–DNA complex and the cell plasma membrane, resulting in inefficient uptake. Previous work investigating the uptake of PEGylated vectors has yielded ambiguous results [26– 29], possibly because no systematic study of the impact of important compositional parameters was performed. A variety of ligands such as transferrin, epidermal growth factor or cell penetrating peptides has been used to target CL–DNA complexes to specific cells or increase their uptake by cells [30–33]. However, many of these approaches, such as noncovalent complexation, do not lend themselves well to systematic studies. CL–DNA NPs, on the other hand, allow a high degree of control over NP charge, membrane charge density, and PEG grafting density. We thus created a model system to investigate specific and nonspecific attachment and uptake of CL–DNA NPs by covalently grafting a linear RGD (arginine-glycineaspartic acid) peptide to their surface. To this end, we used a custom synthesized lipid with a GRGDSP-OH peptide tethered to dioleyl lipid tails via PEG2000 (see Fig. S2 in the Supplementary Material). RGD-peptides bind to integrin receptors on the cell surface and have found broad applications in drug delivery and bioengineering [34–36]. While the linear RGD-peptide employed in this work is a good model system, future applications will make use of cyclic RGD peptides which exhibit higher specificity and affinity. For example, certain cyclic RGD peptides are very effective tumor targeting ligands by virtue of their ability to selectively target αvβ3 and αvβ5 integrins [37].
To quantify the efficiency of RGD-mediated uptake of CL–DNA complexes, we investigated the biophysical properties, in vitro transfection efficiency and biological activity of PEGylated CL–DNA NPs with and without RGD-tagging as well as of CL–DNA complexes without PEGylation. We also studied the effect of complex composition on electrostatic interactions between NPs and cells, by preparing complexes and NPs at both high and low membrane charge density (σM) (by varying the ratio of neutral and cationic lipid) and varied lipid/DNA charge ratio (ρ). Membrane charge density is a key parameter governing the TE of lamellar CL–DNA complexes [20,38]. We used quantitative live-cell imaging with particle tracking to assess the effect of RGD-tagging on the attachment and cellular uptake of CL–DNA NPs and measured TE to determine whether RGD-tagging can recover TE to the level of complexes without PEGylation and how this depends on σM.
Materials and methods
Materials
DOTAP, DOPC and DOPE-PEG2000 (referred to here as PEG2K-lipid) were purchased as solutions in chloroform from Avanti Polar Lipids (Alabaster, AL). The RGD-PEG2K-lipid contains a GRGDSP peptide (Gly-Arg-Gly-Asp-Ser-Pro-OH) covalently attached to the distal end of the PEG-chain of a custom PEG2000-lipid. It was synthesized via Fmoc solid phase synthesis, employing a lipid-PEG-acid building block in the final coupling step. The chemical structures of the lipids are shown in the Supplementary Material (Fig. S2). TRITC-DHPE (N-(6-tetramethylrhodaminethiocarbamoyl)-1,2-dihexadecanoyl-sn-glycero-3-phosphatidylethanolamine) was purchased from Invitrogen and has an excitation and emission maximum of 555 nm and 580 nm, respectively. The luciferase plasmid (pGL3) used in transfection experiments was purchased from Promega. The GFP-tubulin (Clontech) and pGL3 plasmids were propagated in E. coli and purified using a Qiagen Plasmid Mega Prep Kit. For live-cell imaging studies, the pGL3 vector was labeled using the Mirus Bio Label IT Nucleic Acid Labeling Kit with Cy5 (excitation/emission maximum: 649 nm/670 nm) according to the manufacturer’s protocol.
Liposome preparation
Lipid solutions in chloroform/methanol (3:1, v:v; for the RGD-PEG2K-lipid) or chloroform were combined at the desired molar ratio of lipids in glass vials. Lipid molar ratios of DOTAP/DOPC/PEG2K-lipid were 80/20–x/x for high-σM complexes and 30/70–x/x for low-σM complexes. To liposomes for live-cell imaging, 0.5 wt% (of total lipid) TRITC-DHPE lipid label were added. After mixing, the lipid solutions in organic solvent were dried, first by a stream of nitrogen and then in a vacuum for 12 h. The appropriate amount of sterile, high resistivity (18.2 MΩ cm) water to achieve a final concentration of 1 mM lipid was then added to the dried lipid films, and the resulting mixtures were incubated at 37°C for 16 h to form liposomes. Following this incubation, the liposome solutions were sonicated using a tip sonicator to form small unilamellar vesicles.
Cell culture and transfection
Mouse L-cells (ATCC number: CCL-1) were cultured in DMEM (Invitrogen) supplemented with 5% fetal bovine serum (HyClone) and 1% Penicillin/Streptomyocin (Invitrogen). Cells were kept at 37 °C in a humidified atmosphere containing 5% CO2 and were reseeded every 72 h to maintain subconfluency. For transfection studies, cells were seeded in 24 well-plates such that confluency at transfection was 60–80%. CL–DNA complexes were formed by diluting 1 μg of DNA and the appropriate amount of liposome solution to 250 μL each with Optimem (Invitrogen) and mixing. Complexes were incubated for 20 minutes at room temperature before addition to cells. Cells were washed once with PBS and then incubated with 200 μL of complex suspension (0.4 μg of DNA per well) for 6 h. After 6 h, the transfection medium was removed, and cells were rinsed once with PBS and then incubated in supplemented DMEM for 18 h. Cells were harvested in 150 μL of Passive Lysis Buffer (Promega) and subjected to one freeze-thaw cycle. Luciferase expression was measured using a Perkin-Elmer 1420 Victor3 V multilabel counter following the assay manufacturer’s (Promega) instructions. TE results are normalized to total cellular protein as measured by a Bradford Assay (BioRad). Data points represent an average of two measurements with error bars showing the standard deviation. All experiments were performed at least two times to ensure reproducibility.
Dynamic light scattering and zeta potential
The size and effective charge measurement of CL–DNA complexes and nanoparticles (NPs) was measured using a Malvern Nanosizer ZS (Fig. 1D, E) or Brookhaven Goniometer Light Scattering system (Fig. 2A). CL-DNA particles were prepared in light-scattering vials at the same concentration used in the transfection assay. A total of 2 μg of DNA and the appropriate amount of liposome (to achieve the desired lipid/DNA charge ratio) were mixed in 1 mL of the appropriate buffer and incubated at room temperature for 20 minutes. Dynamic light scattering was performed in both DMEM and high resistivity water as indicated below. Plots show the z-average diameter. All zeta potential measurements were performed in high resistivity water. All data points for dynamic light scattering and zeta potential are the average of two measurements performed on the same sample. Error bars show the standard deviation.
Fig. 1.
Biophysical characterization of cationic liposome (CL)–DNA complexes with and without PEGylation. (A-C) Schematic drawings of CL–DNA complexes prepared without PEG-lipid (A), with PEG2K-lipid (B), and with RGD-PEG2K-lipid (C). The drawings depict complexes in the prevalent lamellar phase (with an onion-like internal structure). (D, E) Hydrodynamic diameter of DOTAP-based CL–DNA complexes as a function of ρ (the lipid/DNA charge ratio), determined by dynamic light scattering. Measurements were performed 20 min and 24 h after complexes were formed in cell culture medium (DMEM). (D) Data for complexes at low σM (membrane charge density) containing varied amounts of PEG2K-lipid or RGD-PEG2K-lipid (i.e., with a lipid composition of 30/70–x/x, mol/mol/mol, DOTAP/DOPC/PEG-lipid; x=0, 5, and 10). (E) Data for complexes at high σM (i.e., at 80/20–x/x, mol/mol/mol, DOTAP/DOPC/PEG-lipid; x=0, 5, and 10). (F) Zeta potential of complexes prepared with 0 or 10 mol% PEG-lipid (see above) as a function of ρ. (G) Cryo-EM micrograph of a CL–DNA complex without PEG-lipid (DOTAP/DOPC=80/20, mol/mol) at ρ=10 in 50 mM aqueous NaCl. The multilamellar internal structure of the complex is clearly visible. (H) Cryo-EM micrograph showing PEGylated CL– DNA complexes (DOTAP/DOPC/PEG2K-lipid=80/15/5, mol/mol/mol) at ρ=10 in 50 mM NaCl. Although the sample underwent extensive centrifugation, the complex particles maintain well defined shapes and sub-100 nm sizes. Oligolamellar complexes (solid arrow) and unilamellar liposomes (dashed arrow) coexist. All scale bars represent 100 nm.
Fig. 2.
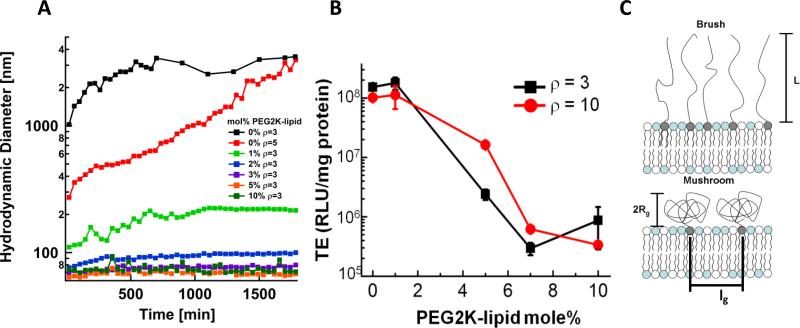
The effect of PEG2K-lipid content on complex size, transfection efficiency , and the conformation of grafted PEG chains. (A) High-σM CL–DNA nanoparticles (80 mol% DOTAP) form stable nanoparticles at as little as 1 mol% PEG2K-lipid. CL–DNA complexes without PEG2K-lipid (black curve) form μm-sized aggregates in cell culture medium. (B) Transfection efficiency of CL–DNA nanoparticles at high membrane charge density (DOTAP/DOPC/PEG2K-lipid=80/20–x/x, mol/mol/mol) as a function of PEG2K-lipid content, for two values of the lipid/DNA charge ratio (g=r). The significant drop in TE at 5 mol% PEG2K-lipid coincides with the mushroom to brush transition. (C) Schematic depiction of the chain conformation regimes and the relevant length scales of membrane-grafted polymers. Here, Rg is the radius of gyration of the polymer chain, L is the brush length, and lg characterizes the grafting density. At a critical grafting density (≈5 mol% PEG2K-lipid), the polymer chains transition from a coiled conformation (the “mushroom” conformation) to a more extended conformation (the “brush” conformation).
Cyro-electron microscopy
CL–DNA complexes (lipid molar ratios of DOTAP//DOPC=80//20 and DOTAP//DOPC//PEG2K-lipid=80//15//5) were formed in 50 mM NaCl at a lipid/DNA charge ratio of ρ=10 and a final DNA concentration of 3 mg/mL from a 30 mM liposome stock solution. The sample containing 5 mol% PEG2K-lipid was centrifuged at 5000 rpm for 15 minutes after mixing. Samples were preserved in vitrified ice supported by holey carbon films on 400-mesh copper grids. Each sample was prepared by applying a 3 μL drop of sample suspension to a freshly plasma cleaned grid, blotting with filter paper, and immediately proceeding with vitrification in liquid ethane. Grids were stored under liquid nitrogen until transferred to the electron microscope for imaging. Electron microscopy was performed using an FEI Tecnai F20 electron microscope, operating at 120 keV equipped with a Gatan 4kχ4k CCD camera. Vitreous ice grids were transferred into the electron microscope using a cryo-stage that maintains the grids at a temperature below –170 °C. Images were acquired using the automated acquisition software Leginon [39] using settings of ~2 μm defocus, electron dose of ~20 e-/Å2, and a pixel size at the image of 0.22 nm.
Live cell imaging
Live-cell imaging was performed using dually labeled (see Materials) complexes at ρ=10. The concentration of CL–DNA NPs was the same as that used in the transfection assay. Cells were grown to 60% confluency on poly(L-lysine)-coated coverslips (22 mm) and maintained at 37 °C using a Harvard Warner flow chamber (Harvard Apparatus Model #P2 and RC21-B). Images were taken on a Nikon Diaphot 300 using a Nikon 1.4 NA 60χ Plan Apo DIC Objective and Sensicam QE CCD. Brightfield images were captured at a magnification of 60χ in differential interference contract (DIC) mode. Fluorescent images are composed of two merged channels where one channel shows lipid (TRITC-DHPE) and the other DNA (Cy5). Images were analyzed using a Matlab routine that measured the intracellular spatial distribution of fluorescently labeled DNA. Data points and error bars represent the average and standard deviation of 10 to 20 representative cells. The Matlab routine first locates the cell boundary and nuclear membrane using the DIC image. Next, all intracellular fluorescent particles are located by fitting a 2D Gaussian to all fluorescent spots contained within the cell boundary. Finally, the closest distance to the nuclear membrane is measured and recorded for each intracellular particle.
Results
We measured the physico-chemical characteristics, transfection efficiency and intracellular trafficking for CL–DNA complexes without PEG2K-lipid (schematically shown in Fig. 1A), with PEG2K-lipid (Fig. 1B), or with RGD-PEG2K-lipid (Fig. 1C). The chemical structures of the employed lipids (monovalent cationic DOTAP and neutral DOPC) and PEG2K-lipids are shown in the Supplementary Material. In addition to the surface functionalization, we varied the membrane charge density (σM) and the lipid/DNA charge ratio (ρ) of the complexes. The membrane charge density is controlled by the molar ratio of cationic to neutral lipid. We chose 30 and 80 mol% DOTAP for low and high σM, respectively; less than 30 mol% DOTAP results in phase separation, while the TE of DOTAP/DOPC–DNA complexes is highest at the molar ratio of 80/20 (DOTAP/DOPC). The lipid/DNA charge ratio is an important parameter governing TE, and high TE requires ρ>1, typically 3≤ρ≤15. (The onset of lipid toxicity places an upper limit on the usable range of ρ.) We varied ρ by adjusting the amount of lipid, keeping the amount of DNA that the cells were exposed to constant. For some experiments, we varied the mole fraction of PEG2K-lipid in the lipid mixture. Phase separation typically occurs at PEG2K-lipid contents above 10 mol%, which is why we did not investigate higher contents. To minimize nonspecific interactions, we used 10 mol% PEG2K-lipid for most of our experiments.
Biophysical characterization
We used dynamic light scattering to determine the particle size and zeta potential of CL– DNA complexes. Complexes prepared without PEG2K-lipid and at low charge ratios near the isoelectric point (ρ=2 and ρ=3) in cell culture medium are comparatively large (>300 nm) as soon as 20 min after mixing of cationic liposomes and DNA (Fig. 1D, E). Their size increases over time: 24 h after mixing, these complexes have fused into μm-size aggregates. At higher ρ, the size of complexes without PEG2K-lipid is smaller and essentially stable over time.
The incorporation of 5 and 10 mol% PEG2K-lipid or RGD-PEG2K-lipid into the complexes yields stable nanoparticles (Fig. 1 D,E). At high σM (Fig. 1E), the size and stability of all complexes containing PEG2K-lipids is very similar. At low σM, small differences are visible.
Comparing NPs with 10% PEG2K-lipid and 10% RGD-PEG2K-lipid (Fig. 1D), RGD-functionalization of the distal end of the PEG chain results in slightly larger particles, especially at low ρ.
Fig. 2A shows the time dependence of the size of high-σM CL–DNA complexes (at ρ=3) in more detail. Complexes without PEG2K-lipid rapidly form large (>2 μm) aggregates, but as little as 1 mol% PEG2K-lipid is sufficient for steric stabilization into time-stable nanoparticles of a little over 200 nm diameter. Complexes at higher PEG2K-lipid content form stable NPs with a diameter below 100 nm. The sizes of NPs containing 3, 5, and 10 mol% PEG2K-lipid are identical, while the size of those containing 2 mol% are slightly larger.
Zeta potential measurements for representative examples of the three classes of CL–DNA complexes investigated in this work (Fig. 1 A–C) are shown in Fig. 1F. The zeta potential increases with ρ for almost all complexes (the only exception being low-σM, higher-ρcomplexes without PEG2K-lipid). This increase is much steeper for complexes at high ρM, independent of surface functionalization. Complexes without PEG2K-lipid exhibit the highest zeta potentials. For low-σM complexes, PEGylation by incorporation of PEG2K-lipid or RGD-PEG2K-lipid reduces their zeta potential strongly (from ≈41 mV to ≈23 mV) and also strongly reduces (to nearly zero) the increase of the zeta potential with ρ (filled black and red lines). PEGylation also strongly reduces the zeta potential of high-σM complexes, with PEG2K-lipid effecting a larger reduction than RGD-PEG2K-lipid. However, these complexes still show a strong increase of the zeta potential with ρ.
Cryo-TEM demonstrates that complexes formed without PEG2K-lipid form large aggregates in the presence of as little as 50 mM salt (Fig. 1G; the salt concentration (≈150 mM) in cell culture medium is much larger). The lamellar structure of these complexes [40] is readily apparent. In contrast, PEGylation with only 5 mol% PEG2K-lipid allows CL–DNA complexes to remain as sub-200 nm, well-defined nanoparticles with an internal oligolamellar structure at the same salt concentration, even after extensive centrifugation (Fig. 1H). These CL–DNA NPs coexist with unilamellar vesicles (Fig. 1H).
Transfection efficiency
The dependence of the TE of high-σM CL–DNA complexes on the amount of incorporated PEG2K-lipid at two lipid/DNA charge ratios, ρ=3 and ρ=10 is shown in Fig. 2B. The high TE of complexes without PEGylation remains unchanged at 1 mol% PEG2K-lipid. TE then drops steeply for 5 mol% (which is near the transition from mushroom regime to brush regime) and further for 7 mol% to a very low level where it remains for 10 mol%. The schematic in Fig. 2C shows how surface grafted PEG2K in the mushroom regime has a globular conformation where the spacing between grafting points (lg) is greater than the radius of gyration (Rg), while the brush regime occurs when lg < RG, which forces the PEG chains into an elongated conformation.
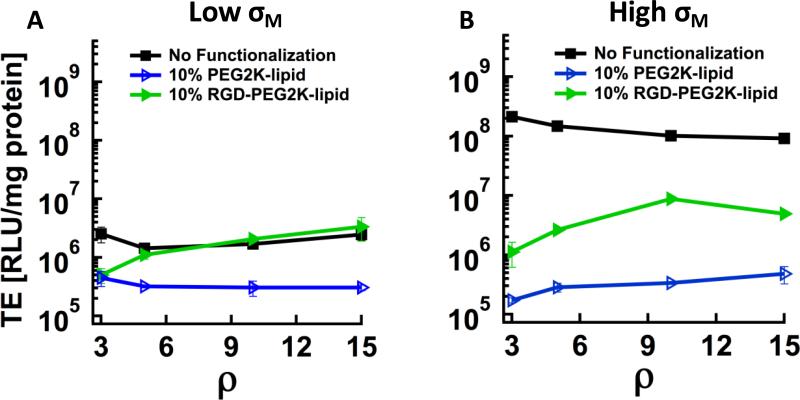
The transfection efficiencies (TE) of the three classes of CL–DNA complexes investigated in this work (Fig. 1A–C) as a function of ρ, for low and high σM are shown in Fig. 3. PEGylation with PEG2K-lipid reduces the (low) TE of low-σM CL–DNA complexes by about an order of magnitude, independent of ρ (Fig. 3A, black squares and blue open triangles). The TE of RGD-tagged CL–DNA NPs at low σM increases with ρ, and for ρ≥5, their TE matches that of the corresponding complexes without PEG2K-lipid (Fig. 3A, green triangles).
Fig. 3.
Transfection efficiency (TE) of CL–DNA complexes with and without PEGylation (10 mol% PEG-lipid) as a function of lipid/DNA charge ratio (g=r).(A) Complexes at low membrane charge density (30 mol% DOTAP) show low TE (black squares), which is further reduced by an order of magnitude upon incorporation of PEG2K-lipid (blue hollow triangles). RGD-tagging (incorporation of RGDPEG2K-lipid instead of PEG2K-lipid) fully recovers TE (green triangles). (B) The high TE of complexes at high membrane charge density (80 mol% DOTAP) without PEG-lipid (black squares) drops strongly upon incorporation of PEG2K-lipid (blue hollow triangles); RGD-tagging recovers TE, but only partially (green triangles).
High-σM DOTAP/DOPC–DNA complexes without PEG2K-lipid exhibit high TE which is comparable to that of commercial reagents. PEGylation reduces their TE by nearly three orders of magnitude (Fig. 3B), with the reduction in TE decreasing slightly as ρ increases. RGD-tagging of high-σM CL–DNA NPs partially recovers TE, with the biggest increase occurring at ρ=10. For all investigated values of ρ, the TE of RGD-tagged CL–DNA NPs at high σM is larger than at low σM.
The TE of CL–DNA complexes without PEG2K-lipid decreases slightly with increasing ρ. In contrast, TE increases with ρ for high-σM CL–DNA NPs both with and without RGD-tagging (the only exception being RGD-tagged NPs at ρ=15) as well as for RGD-tagged low-σM CL–DNA NPs. The TE of low-σM CL–DNA NPs without RGD-tagging decreases slightly from ρ=3 to ρ=5 and then remains constant (Fig. 4A, blue open triangles). PEGylated CL–DNA complexes at low σM and high σM show near-identical TE without RGD-tagging, but the high-σM RGD-tagged NPs transfect more efficiently (up to an order of magnitude) than their low-σM counterparts.
Fig. 4.
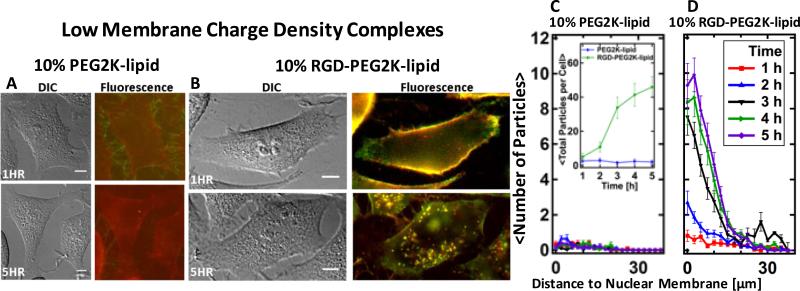
Live-cell imaging results for CL–DNA nanoparticles at low membrane charge density (DOTAP/DOPC/PEG-lipid=30/60/10, mol/mol/mol). (A,B) Representative differential interference contrast and merged fluorescence micrographs (DNA label: green; lipid label: red). (A) CL–DNA nanoparticles formed using PEG2K-lipid show some particles rich in DNA attached to filopodia and slight staining of the plasma membrane with lipid label 1 h after incubation of the cells. A single CL– DNA particle is visible within the perimeter of the cell 5 h after incubation. (B) RGD-tagged nanoparticles (formed using RGD-PEG2K-lipid) coat the plasma membrane 1 h after incubation, and numerous fluorescently labeled particles are visible inside the cell 5 h after incubation. (C, D) Spatial distribution of intracellular particles at various time points after incubation, determined using exogenous DNA fluorescence. Each curve represents an average over ≈20 cells. The inset in (C) shows the average total particle count per cell as a function of time. The difference in total uptake between NPs with and without RGD was statistically significant (P < 0.05). RGD-tagged particles are taken up efficiently and accumulate in the perinuclear region. In contrast, uptake of nanoparticles without RGD-tagging is negligible. All scale bars represent 10 μm.
Live-cell imaging of the uptake and trafficking of CL–DNA nanoparticles
We transfected cells with dually-labeled fluorescent CL–DNA NPs to directly observe particle uptake and intracellular motion. To quantitate the results from these experiments, we measured the spatial distribution of condensed fluorescent particles (Cy5 channel, i.e., the DNA label) using an automated Matlab routine [41]. Suh et al. proposed a similar analysis to determine the dominant types of particle motion at different spatial locations within the cytoplasm [42]. By measuring the distance between each NP and the nuclear membrane, we obtained the intracellular distribution of CL–DNA NPs as an ensemble average over many cells.
Fig. 4 shows imaging results for low-σM CL–DNA NPs with and without RGD-tagging. Typical DIC and fluorescent images taken at the beginning (1 h after addition of complexes) and end (5 h after addition) of the incubation period of cells with CL–DNA complexes in our TE assay are shown in Fig. 4A and B. At the end of this incubation period, cells are washed with PBS and incubated in serum-containing medium for 20-24 h before they are harvested and protein expression is measured (see Materials and Methods). At 1 h after incubation, particles rich in DNA are seen attached to filopodia, while the plasma membrane shows some staining by the fluorescent lipid label. This label (a fluorescent lipid) is simply mixed with the lipids of the complex and thus can label neighboring membranes via lipid exchange. A single intact PEGylated CL–DNA particle is observed inside the cell 5 h after incubation. Quantitative particle localization (Fig. 4C,D) confirms that uptake of low-σM CL–DNA NPs containing 10 mol% PEG2K-lipid is nearly zero (Fig. 4C, inset). In contrast, cells incubated with RGD-tagged CL–DNA NPs (10 mol% RGD-PEG2K-lipid) show bright fluorescence along the outline of the cell (as well as some particles within it) 1 h after incubation and numerous fluorescently labeled particles within the cell perimeter 5 h after incubation. Quantitative particle localization shows that the RGD-tagged CL–DNA NPs accumulate in the perinuclear region of the cells (Fig. 4D). The inset in Fig. 4C shows the average total number of NPs per cell: 5 h after complex addition, approximately 45 spots exhibiting bright fluorescence from the DNA label are observed in a single focal plane.
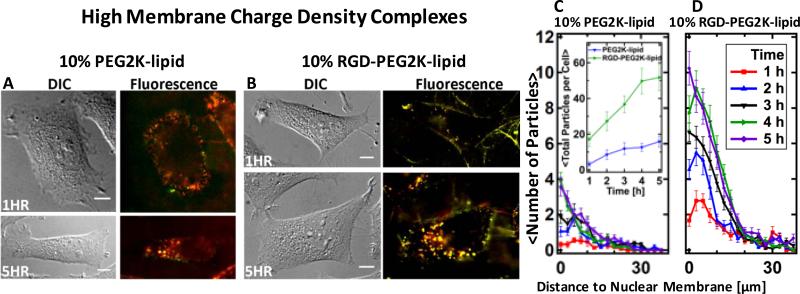
Live-cell imaging results for CL–DNA NPs at high σM are displayed in Fig. 5. In contrast to their low-σM counterparts, high-σM CL–DNA NPs without RGD-tagging adhere to cells at early time points (t<2 h), and a number of them are seen inside the cells 5 h after incubation (Fig. 5A, C). Nonetheless, the total uptake of RGD-tagged CL–DNA NPs is much higher (Fig. 5C, inset), as observed for low-σM NPs. Comparing RGD-tagged NPs of high and low σM, the intracellular distributions are similar (both showing perinuclear accumulation), but uptake occurs faster and is slightly larger for high-σM NPs (Fig. 5B,D).
Fig. 5.
Live-cell imaging results for CL–DNA nanoparticles at high membrane charge density (DOTAP/DOPC/PEG-lipid=80/10/10, mol/mol/mol). (A,B) Representative differential interference contrast and merged fluorescence micrographs (DNA label: green; lipid label: red). At this membrane charge density, CL–DNA nanoparticles formed using PEG2K-lipid (A) attach to cells and are internalized. For RGD-tagged CL–DNA nanoparticles (B), the extent of cell attachment and uptake is even higher. (C, D) Spatial distributions of intracellular particles at various time points after incubation of cells with CL–DNA nanoparticles, determined using exogenous DNA fluorescence. Each curve represents an average over ≈20 cells. The inset in (C) shows the average total particle count per cell as a function of time. Both types of particles are taken up and accumulate in the perinuclear region of the cell, but many more RGD-tagged particles are taken up per cell and their uptake is faster. The difference in total uptake between NPs with and without RGD was statistically significant (P < 0.05). All scale bars represent 10 μm.
Discussion
In this work, we aimed to investigate the importance of cell adhesion and uptake as a barrier to transfection by PEGylated CL–DNA complexes. In addition, we sought to investigate the effect of the important parameters σM and ρ (which are controlled by complex composition) on the uptake and in vitro TE of CL–DNA NPs. These parameters govern the strength of electrostatic, nonspecific interactions between CL–DNA NPs and cells. Thus, we not only compared PEGylated complexes with and without RGD-peptide functionalization, but also varied the molar ratio of cationic and neutral lipid and the lipid/DNA charge ratio. PEGylated complexes at low σM and high content of PEG2K-lipid, in particular, show negligible nonspecific interaction with cells and thus are an ideal system to investigate RGD-peptide-based specific interactions.
CL–DNA complexes prepared without PEG2K-lipid aggregate in salt-containing cell culture medium (Fig. 1D,E,G) because the salt screens the electrostatic repulsion of the particles. The lower the zeta potential of these complexes, i.e., the lower their ρ and σM (Fig. 1F), the smaller the electrostatic repulsion and thus the faster this aggregation occurs. Increasing ρ increases the zeta potential of complexes because at least part of the additional cationic lipid is incorporated into the complexes (a phenomenon called “overcharging” [43]). The extent of this overcharging (and thus the zeta potential) increases with σM [43]. While complex aggregation is not necessarily detrimental for in vitro transfection, it is undesirable for in vivo applications, because large aggregates will not remain in circulation but accumulate in “first pass” organs such as the lungs and the liver. Preventing aggregation by increasing electrostatic repulsion (i.e., increasing ρ and σM) is not a viable strategy to yield complexes for in vivo application because cationic particles are readily recognized by the complement system and removed from circulation [23].
Incorporation of PEG2K-lipid or RGD-PEG2K-lipid into CL–DNA complexes strongly reduces their zeta potential, which is a measure of effective surface charge (Fig. 1F). The zeta potential is defined as the electrostatic potential at the particles slipping plane, a hypothetical radius where enclosed counterions are weakly bound to the particle. Exchanging neutral DOPC for the negatively charged PEG2K-lipids reduces the overall charge of the complexes and thus the zeta potential, but this effect is not large, especially at high σM. The main reason for the measured decrease in zeta potential is an outward shift of the slipping plane due to the PEG/peptide-PEG shell surrounding each CL–DNA nanoparticle.
CL–DNA complexes prepared with PEG2K-lipid form nanoparticles that do not aggregate, regardless of charge ratio (Fig. 1D,E,G,H; Fig. 2A), because the PEG2K-lipid confers a sterically stabilizing PEG-shell to the CL–DNA complex particles [25]. Adding an RGD-peptide to the distal end of a PEG2K-lipid does not impair its ability to sterically stabilize complexes (Fig. 1 D,E). However, this stabilization against aggregation by attractive van der Waals interactions between particles does not necessarily imply that the attractive electrostatic interaction between nanoparticles and the plasma membrane is also screened. In fact, the uptake of PEGylated CL–DNA NPs increases with σM, whether or not they are tagged with RGD, and PEGylation even at 10 mol% is insufficient to fully inhibit the attachment and uptake of high-σM
CL–DNA NPs (Fig. 5A,C).
The drop of the TE of CL–DNA complexes with increasing content of PEG2K-lipid does not correlate with the extent of steric stabilization. Inclusion of 1 mol% PEG2K-lipid yields stable nanoparticles but leaves TE unaffected, and particle size and stabilization are the same for 5 and 7 mol% PEG2K-lipid, but their TE differs by about an order of magnitude. The drop in TE with PEG2K-lipid content does correlate, however, with a conformational transition of the PEG chains. At around 5 mol% PEG2K-lipid, full coverage of the complex with PEG chains in the mushroom conformation is achieved, corresponding to a drop in TE. However, the electrostatic interactions between the positively charged complexes and the negatively charged cell membrane are not fully suppressed, as evident from the relatively large increase of TE with ρ, i.e., with increasing zeta potential (effective surface charge) and therefore electrostatic interaction energy. At higher content of PEG2K-lipid, the PEG chains transition from the mushroom to the brush conformation (Fig. 2C). At this transition, the range and strength of repulsive forces due to the grafted PEG chains increases, strongly reducing the electrostatic interaction between complexes and cells. This reduces TE to about the level of uncomplexed DNA and all but eliminates the increase of TE with ρ (Fig. 2B, 7 and 10% PEG2K-lipid).
TE decreases to near the baseline level of transfection (the level obtained with uncomplexed DNA) at the inclusion of 10 mol% PEG2K-lipid for both low and high σM CL– DNA NPs (Fig. 3). This drop is most pronounced for complexes at high σM, which are highly efficient before PEGylation. Considering the main barriers to transfection, the observation that PEGylation sterically stabilizes CL–DNA complexes and reduces their zeta potential suggests two mechanisms that could cause the decrease in TE. One is a reduction in cell attachment and subsequent uptake, which is electrostatically mediated for CL–DNA complexes without PEGylation. The other is interference with endosomal escape, a step in the transfection process that requires fusion of the membranes of the complex and the endosome (also electrostatically mediated) and that has been shown to be the main barrier for lamellar complexes at low σM [20,38]. Prior work quantifying the uptake of PEGylated CL–DNA complexes has yielded ambiguous results; according to some reports, uptake was inhibited [26,27], but it was found to be unchanged or even enhanced in others [28,29]. Our data and model calculations (see below) point to an understanding that resolves this apparent contradiction: whether or not PEGylated complexes adhere to cells (which correlates with subsequent uptake in our experiments) depends on parameters such as PEG2K coverage, ρ, and σM, which are controlled by lipid choice and complex composition. In particular, σM is a key parameter affecting uptake and transfection efficiency of PEGylated CL–DNA complexes, as observed for complexes without PEGylation [20,38]. At sufficiently high σM, electrostatic interactions “leak through” even at high PEG coverage. This promotes attachment to cells and endosomal escape (if less efficiently than for complexes without PEG-lipids), even as the PEGylation prevents aggregation of the CL–DNA NPs.
Hypothesizing that the reduced TE of PEGylated CL–DNA complexes is at least in part due to reduced attachment to cells and subsequent uptake, we prepared a PEG2K-lipid with an RGD-peptide at the distal end of the PEG chain (RGD-PEG2K-lipid; see the Supplementary Material for the chemical structure). The linear GRGDSP-peptide employed in the RGDPEG2K-lipid binds to the ubiquitous integrin receptors on the cell surface. Thus, we sought to increase cell attachment, and potentially subsequent uptake, by substituting the RGD-PEG2K-lipid for the simple PEG2K-lipid (in other words, by RGD-tagging CL–DNA nanoparticles). (RGD-tagging has been shown to improve the uptake—not only the cell attachment—of metal nanoparticles and drugs [34].)
Low-σM CL–DNA NPs are an ideal system for studying the effect of RGD-tagging because their low surface charge prevents non-specific adhesion to the cell surface (Fig. 4A and C). Live-cell imaging revealed that RGD-tagging indeed leads to efficient attachment of CL– DNA NPs to cells and their subsequent uptake (Fig. 4B and D). Low-σM CL–DNA NPs with and without RGD-tagging have nearly identical zeta potentials (the zeta potential of the RGD-tagged NPs is in fact lower), strongly suggesting that the efficient attachment and subsequent uptake of the RGD-tagged NPs particles is solely attributable to ligand-receptor binding.
The fluorescence intensity of internalized particles 5 h after incubation is larger than, e.g., 1 h after incubation. This is likely due to fusion of endosomes containing fluorescently labeled nanoparticles, which would result in each fluorescent spot at 5 h corresponding to multiple CL– DNA particles within a single endosomal compartment.
At high σM, PEGylated CL–DNA NPs bind to the plasma membrane and are successfully uptaken, albeit both to a much smaller extent than their RGD-tagged counterparts (Fig. 5). Increasing σM slightly improves the uptake of RGD-tagged CL–DNA NPs (≈51 vs. ≈45 particles/cell at 5 h) but significantly improves the TE or RGD-tagged CL–DNA NPs (TE≈106 vs. 107 RLU/mg protein). This suggests that high σM can help RGD-tagged CL–DNA NPs overcome the barrier of endosomal entrapment. (We have previously shown that for lamellar CL–DNA complexes, σM is a critical parameter determining the propensity of internalized particles to escape endosomes [20,38].) Thus, both RGD-tagging and increased membrane charge density contribute to successful transfection, where RGD-tagged CL–DNA nanoparticles at high σM exhibit the highest TE of the PEGylated complexes investigated.
For most complex compositions (i.e., ρ and σM), RGD-tagging of CL–DNA NPs recovers about an order of magnitude in TE (Fig. 3). For low-σM NPs, this corresponds to full recovery of TE to the level of CL–DNA complexes without PEGylation. However, this level of TE is insufficient for applications. For high-σM NPs, the recovery of TE is only partial, and the difference in TE between RGD-tagged CL–DNA NPs at high and low σM is much smaller than for the corresponding complexes without PEGylation. An important question is whether the recovery of TE is partial because cell attachment and uptake of RGD-tagged NPs are lower than for complexes without PEGylation or whether another barrier (that is not addressed by RGD-tagging) is responsible. Various literature reports have suggested that PEGylation of CL–DNA complexes hinders endosomal escape [41,44,45]. The broad size distribution of non-PEGylated complexes prevents a direct comparison with NPs in live-cell imaging, but several lines of evidence suggest that the incomplete recovery of TE is indeed due to inefficient endosomal escape. Imaging results supporting this hypothesis are the increased intensity of fluorescent particles at later time points (see above), the observation of perinuclear accumulation of particles (observed for all particles taken up by the cells, likely as a result of endosomal trafficking), and the observation of modes of particle movement consistent with endosomal entrapment (see below). The fact that RGD-tagging fully recovers the TE of low-σM CL–DNA NPs is also consistent with the hypothesis. Previous work has shown that endosomal entrapment is the main barrier limiting TE for lamellar complexes at low σM (without PEGylation) [20,38] and thus their level of TE can be achieved without efficient endosomal escape. Finally, we recently showed that the TE of PEGylated complexes is enhanced by employing an acid-labile PEG2K-lipid, which enhances fusion with endosomal membranes by shedding its PEG-chains at low pH of late endosomes [41].
We analyzed the intracellular movement of RGD-tagged CL–DNA nanoparticles at high σM in live cells using particle tracking (see Supplementary Material for details). Analysis of the mean square displacement with time and labeling of microtubules with GFP-tubulin revealed both active transport of particles along microtubules and confined diffusion within a membrane-bounded compartment (Fig. S1). Both of these observations are consistent with endosomal entrapment of CL–DNA NPs. Particles entrapped in endosomes will show confined motion, and active transport via motor proteins is facilitated by specific interactions, suggesting that CL– DNA NPs undergoing such transport are enclosed within intact endosomes. Furthermore, another literature report has associated active transport of synthetic vectors with inefficient endosomal escape and low TE [46].
Modeling of the Interaction between PEGylated CL–DNA NPs and the Cell Membrane
We developed a model of the interaction between PEGylated CL–DNA NPs and the cell membrane to get a more detailed understanding of the parameters affecting this interaction and our experimental observations. For this model, we consider a charged, polymer-grafted sphere (as a model of the NP) approaching an oppositely-charged flat membrane (as a model of the plasma membrane). By calculating the free energy as a function of the distance between NP and plasma membrane, we gain insight into how parameters such as σM and PEG grafting density affect the adhesion of CL–DNA NPs to cells. The total free energy (ET) of interaction as a function of the distance D between the flat membrane and the outer edge of the polymer-grafted sphere comprises four separate terms of energy per unit area (Ex) and can be written as:
| (1) |
EE, EV, EH, and ES are the terms for electrostatic attraction, van der Waals attraction, hydration repulsion, and steric repulsion due to the grafted polymer shell, respectively. To compute the total free energy per particle, each of these terms is multiplied by a corresponding interaction area (Ax). The functional forms of the terms Ex are:
| (2) |
| (3) |
| (4) |
| (5) |
The first term (Eq. 2), which represents the electrostatic attraction, is the result of Debye– Hückel theory. Here, 1/κ is the Debye screening length, which we calculated to be 0.65 nm at the salt concentration in DMEM (150 mM of 1:1 electrolytes and 2.5 mM of 2:1 electrolytes); σPM is the membrane charge density of the plasma membrane. The value of ρPM used in the calculation shown in Fig. 6 is –2χ10-3e/Å2, which corresponds to a membrane containing 10 mol% anionic lipids. This is consistent with experimentally measured plasma membrane lipid compositions [47]. For the high values of ρ in our experiments, we assume that the surface charge density of the NP, σNP, is the same as that of the constituting lipid membrane, σM. The observation that the zeta potential approaches a saturation value as ρ increases (Fig. 1F) validates this assumption. For NPs at lower ρ (ρ<5), the calculated ρM is an upper limit for σNP. The surface charge density of the lipid membrane (σM=total charge/total area) can be calculated as described in [48], while also taking into account the negative charge of the PEG2K-lipids (see Fig. S2 for lipid chemical structures): σNP=(eZ+N + eZ–NPL)/(NCLACL+NNLANL +NPLAPL). Here, eZ+ and eZ– are the total charge of the cationic lipid and the PEG2K-lipid, respectively. The variables NCL, NNL, and NPL are the mole fraction of cationic lipid, neutral lipid, and PEG2K-lipid, respectively. The respective lipid head group areas are ACL, ANL, and APL. In our system both the cationic and PEGylated lipid are monovalent in charge thus eZ+=+1e and eZ–=–1e. For high-σM NPs, NCL=0.8, NNL=0.1, and NPL=0.1, while for low-σM NPs NCL=0.3, NNL=0.6, and NPL=0.1. We assumed the headgroup areas of the lipids to be equal at ACL=ANL=A =72 Å2 [49–51]. Using these parameters, σNP computes to 2.78χ10-3 e/Å2 and 9.72χ10-3 e/Å2 for low- and high-σM NPs, respectively. The van der Waals term (Eq. 3) contains only one constant, A=7χ10-21 J [52]. For the hydration repulsion term (Eq. 4) the amplitude and range of interaction were taken to be a =2χ10-8J/mm2 and λH=2 Å [52,53].
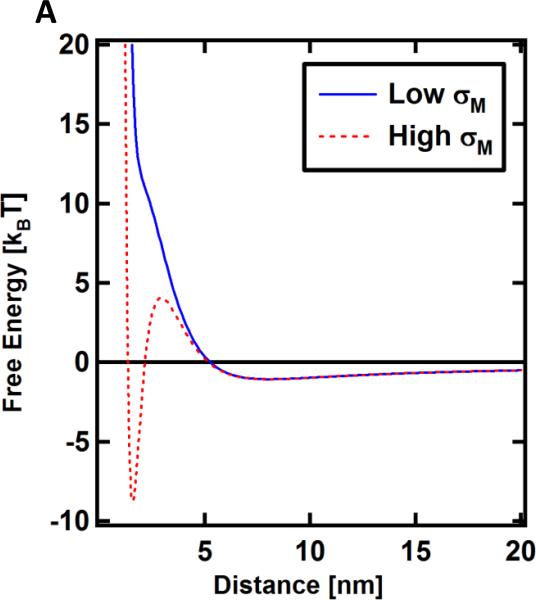
Fig. 6.
Calculated free energy of interaction of PEGylated CL–DNA complexes with the cell’s plasma membrane as a function of distance. Steric repulsion gives rise to a repulsive barrier, but a global minimum representative of adhesion appears at sufficiently high membrane charge density. Low and high σM refers to DOTAP/DOPC/PEG2K-lipid–DNA NPs with lipid molar ratios of 30/60/10 and 80/10/10, respectively.
The free energy per area of steric repulsion (Eq. 5) in the brush regime (>5 mol% PEG2K-lipid) is strongly affected by two parameters: lg, the distance between grafting points and L, the brush length [52] (Fig. 2C). Eq. 5 is a modified exponential approximation to a power law first proposed in Alexander de Gennes’ theory [54]. The two modifications made to that power law are replacing L with L/2 and E with E/2. The rationale for these modifications is that the original power law and exponential approximation describe a scenario where both surfaces are grafted with polymer, whereas in our case only one is (the PEGylated CL–DNA NP). These modifications have been successfully used to model various systems where only one surface contains grafted polymer [55,56]. When fit to experimental data, de Gennes’ theory typically overestimates the repulsion between polymer grafted surfaces [58,57]. In addition, accurate values for the headgroup size of the PEG2K-lipid are unavailable. Thus, we allowed lg to vary, within narrow boundaries. The value used in Fig. 6 is lg=4 nm, in reasonable agreement with the value (lg=3.02 nm) calculated by assuming equal headgroup size for all lipids as above (Aheadgroup=72 Å2). The brush height L, which has been measured via X-ray scattering and surface force apparatus, was set to 6.6 nm [58,59].
All four effective area terms Ax were calculated using the Langbein approximation [52]. For a potential of the form w(r)=Ce-βr, the effective area calculates to Aeff=2πR(1/β) [52], where R is the radius of the polymer-grafted sphere and the 1/β is the characteristic length scale of the interaction (β=κ for the electrostatic term; 1/β=λH=0.2 nm for the hydration term; 1/β=L/π for the steric term). For the van der Waals term, the Langbein approximation yields AV=2πRD.
Fig. 6 shows the free energy profile in units of kBT as a function of the distance D. Three notable features arise in the case of high σM: a minimum located at ≈3 nm, the depth and location of which are determined by the Debye screening length, σM and hydration layer thickness; a local maximum located at ≈4.5 nm (reflecting the steric barrier arising from the grafted polymer), the height and location of with are determined by grafting density and brush length, respectively; and a second minimum at ≈6 nm. The first minimum corresponds to binding events where complexes stick to the cell surface and remain immobilized until endocytosis. It is only present for high σM. The second, shallow minimum, indicates weak binding of the particle, which we interpret to correspond to “bump and run” events. It is present for both high and low σM. These features are in excellent agreement with the live-cell imaging results which showed that high-σM CL–DNA NPs bind to and are taken up by cells (Fig. 5) while low-σM NPs do not attach (Fig. 4).
Conclusions
PEGylation of CL–DNA complexes is mandatory for in vivo gene delivery applications but reduces transfection efficiency. Interestingly, steric stabilization of CL–DNA complexes against aggregation does not imply that cell attachment via electrostatic interaction between complexes and cells is inhibited, despite the fact that PEGylation severely reduces the effective surface potential. Uptake of PEGylated complexes (without RGD-tagging) is in fact significantly affected by σM: uptake of low-σM complexes is negligible, whereas high-σM complexes show some attachment and uptake. This explains seemingly inconsistent literature reports on the extent of uptake of PEGylated CL–DNA complexes. PEGylated CL–DNA complexes at low σM are an ideal model system to study targeting ligands and receptor-mediated uptake because for them the effects of specific interactions are not convoluted with those of nonspecific electrostatic attraction. RGD-tagging of PEGylated CL–DNA complexes strongly improves both attachment to cells and uptake, independent of σM. This fully recovers TE at low σM, where the TE of complexes without PEGylation is low and limited by endosomal escape. For high-σM complexes, TE recovery is only partial because of endosomal entrapment. Thus, the two main barriers increased by PEGylation are cellular uptake and endosomal escape. Future work to improve the TE of PEGylated CL–DNA complexes will need to combine tagging with RGD or similar targeting ligands with strategies to improve their endosomal escape, such as using acid-labile PEG-lipids.
Supplementary Material
Acknowledgment
This work was supported by NIH GM-59288 and NSF DMR-1101900. CLC was supported by NSRRC and Academia Sinica, Taiwan. NMR characterization was performed using the Central Facilities of the Materials Research Laboratory at UCSB which are supported by the MRSEC Program of the NSF under award no. DMR-1121053; a member of the NSF-funded Materials Research Facilities Network (www.mrfn.org). BFBS was supported by a Marie Curie International Outgoing Fellowship within the EU Seventh Framework Programme for Research and Technological Development (2007–2013), under grant agreement no. PIOF-GA-2009-252701. The EM work was conducted at the National Resource for Automated Molecular Microscopy at The Scripps Research Institute, which is supported by the U. S. NIH NIGMS Biomedical Technology Research Center program (GM103310) of the National Institute of General Medical Sciences (BC, CSP).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Guo X, Huang L. Recent advances in nonviral vectors for gene delivery. Acc Chem Res. 2012;45:971–9. doi: 10.1021/ar200151m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Salcher E, Wagner E. Chemically programmed polymers for targeted DNA and siRNA transfection. Top Curr Chem. 2010;296:227–49. doi: 10.1007/128_2010_69. [DOI] [PubMed] [Google Scholar]
- 3.Sioud M. Therapeutic siRNAs. Trends Pharmacol Sci. 2004;25:22–8. doi: 10.1016/j.tips.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 4.Caplen NJ. RNAi as a gene therapy approach. Expert Opin Biol Ther. 2003;3:575–86. doi: 10.1517/14712598.3.4.575. [DOI] [PubMed] [Google Scholar]
- 5.Yin JY, Ma ZY, Selliah N, Shivers DK, Cron RQ, Finkel TH. Effective gene suppression using small interfering RNA in hard-to-transfect human T cells. J Immunol Methods. 2006;312:1–11. doi: 10.1016/j.jim.2006.01.023. [DOI] [PubMed] [Google Scholar]
- 6.Bielke W, Erbacher C, editors. Topics in current chemistry, Vol. 296: Nucleic Acid Transfection. Heidelberg; Springer: 2010. [PubMed] [Google Scholar]
- 7.Ewert KK, Zidovska A, Ahmad A, Bouxsein NF, Evans HM, McAllister CS, et al. Cationic lipid-nucleic acid complexes for gene delivery and silencing: pathways and mechanisms for plasmid DNA and siRNA. Top Curr Chem. 2010;296:191–226. doi: 10.1007/128_2010_70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang L, Hung MC, Wagner E, editors. Advances in genetics, Vol 53: Non-viral vectors for gene therapy, part I. 2nd ed. Elsevier Academic Press; San Diego: 2005. [Google Scholar]
- 9.Li SD, Huang L. Gene therapy progress and prospects: non-viral gene therapy by systemic delivery. Gene Ther. 2006;13:1313–9. doi: 10.1038/sj.gt.3302838. [DOI] [PubMed] [Google Scholar]
- 10.Davis ME. The first targeted delivery of siRNA in humans via a self-assembling, cyclodextrin polymer-based nanoparticle: from concept to clinic. Mol Pharm. 2009;6:659–68. doi: 10.1021/mp900015y. [DOI] [PubMed] [Google Scholar]
- 11.Ewert K, Ahmad A, Evans HM, Safinya CR. Cationic lipid–DNA complexes for non-viral gene therapy: relating supramolecular structures to cellular pathways. Expert Opin Biol Ther. 2005:5;33–53. doi: 10.1517/14712598.5.1.33. [DOI] [PubMed] [Google Scholar]
- 12.Ginn SL, Alexander IE, Edelstein ML, Abedi MR, Wixon J. Gene therapy clinical trials worldwide to 2012 – an update. J Gene Med. 2013;15:65–77. doi: 10.1002/jgm.2698. [DOI] [PubMed] [Google Scholar]
- 13.Gene therapy clinical trials worldwide. Provided by the Journal of Gene Medicine. Online. 2013. Available from URL: http://www.wiley.com/legacy/wileychi/genmed/clinical/
- 14.Yi Y, Hahm SH, Lee KH. Retroviral gene therapy: safety issues and possible solutions. Curr Gene Ther. 2005;5:25–35. doi: 10.2174/1566523052997514. [DOI] [PubMed] [Google Scholar]
- 15.Young L, Mautner V. The promise and potential hazards of adenovirus gene therapy. Gut. 2001;48:733–6. doi: 10.1136/gut.48.5.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hacein-Bey-Abina S, von Kalle C, Schmidt M, Le Deist F, Wulffraat N, McIntyre E, et al. A serious adverse event after successful gene therapy for X-linked severe combined immunodeficiency. N Engl J Med. 2003;348:255–6. doi: 10.1056/NEJM200301163480314. [DOI] [PubMed] [Google Scholar]
- 17.Hacein-Bey-Abina S, von Kalle C, Schmidt M, McCormack M, Wulffraat N, Leboulch P, et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–9. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- 18.Raper S, Chirmule N, Lee F, Wivel N, Bagg A, Gao G, et al. Fatal systemic inflammatory response syndrome in a ornithine transcarbamylase deficient patient following adenoviral gene transfer. Mol Genet Metab. 2003;80:148–58. doi: 10.1016/j.ymgme.2003.08.016. [DOI] [PubMed] [Google Scholar]
- 19.Willard HF. Artifical chromosomes coming to life. Science. 2000;290:1308. doi: 10.1126/science.290.5495.1308. [DOI] [PubMed] [Google Scholar]
- 20.Lin AJ, Slack NL, Ahmad A, George CX, Samuel CE, Safinya CR. Three-dimensional imaging of lipid gene-carriers: membrane charge density controls universal transfection behavior in lamellar cationic liposome–DNA complexes. Biophys J. 2003;84:3307–16. doi: 10.1016/S0006-3495(03)70055-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suh J, Choy KL, Lai SK, Suk JS, Tang B, Prabhu S, et al. PEGylation of nanoparticles improves their cytoplasmic transport. Int J Nanomedicine. 2007;2:1–7. [PMC free article] [PubMed] [Google Scholar]
- 22.Schwake G, Youssef S, Kuhr JT, Gude S, David MP, Mendoza E, et al. Predictive modeling of non-viral gene transfer. Biotechnol Bioeng. 2010;105:805–13. doi: 10.1002/bit.22604. [DOI] [PubMed] [Google Scholar]
- 23.Plank C, Mechtler K, Szoka Jr FC, Wagner E. Activation of the complement system by synthetic DNA complexes: a potential barrier for intravenous gene delivery. Hum Gene Ther. 1996;7:1437–46. doi: 10.1089/hum.1996.7.12-1437. [DOI] [PubMed] [Google Scholar]
- 24.Fenske DB, MacLachlan I, Cullis PR. Stabilized plasmid-lipid particles: a systemic gene therapy vector. Methods Enzymol. 2002;346:36–71. doi: 10.1016/s0076-6879(02)46048-x. [DOI] [PubMed] [Google Scholar]
- 25.Martin-Herranz A, Ahmad A, Evans HM, Ewert KK, Schulze U, Safinya CR. Surface functionalized cationic lipid–DNA complexes for gene delivery: PEGylated lamellar complexes exhibit distinct DNA–DNA interaction regimes. Biophys J. 2004;86:1160–8. doi: 10.1016/S0006-3495(04)74190-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gjetting T, Arildsen NS, Christensen CL, Poulsen TT, Roth JA, Handlos VN, et al. In vitro and in vivo effects of polyethylene glycol (PEG)-modified lipid in DOTAP/cholesterol-mediated gene transfection. Int J Nanomedicine. 2010;9:371–83. doi: 10.2147/ijn.s10462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Immordino ML, Dosio F, Cattel L. Stealth liposomes: review of the basic science, rationale, and clinical applications, existing and potential. Int J Nanomedicine. 2006;1:297–315. [PMC free article] [PubMed] [Google Scholar]
- 28.Sadzuka Y, Kishi K, Hirota S, Sonobe T. Effect of polytheyleneglycol (PEG) chain on cell uptake of PEG-modified liposomes. J Liposome Res. 2003;13:157–72. doi: 10.1081/lpr-120020318. [DOI] [PubMed] [Google Scholar]
- 29.Kibria G, Hatakeyama H, Ohga N, Hida K, Harashima H. Dual-ligand modification of PEGylated liposomes shows better cell selectivity and efficient gene delivery. J Control Release. 2011;153:141–8. doi: 10.1016/j.jconrel.2011.03.012. [DOI] [PubMed] [Google Scholar]
- 30.Sakaguchi N, Kojima C, Harada A, Koiwai K, Shimizu K, Emi N, et al. Enhancement of transfection activity of lipoplexes by complexation with transferrin-bearing fusogenic polymer-modified liposomes. Int J Pharm. 2006;325:186–90. doi: 10.1016/j.ijpharm.2006.06.015. [DOI] [PubMed] [Google Scholar]
- 31.Bunuales M, Duzgunes N, Zalba S, Garrido MJ, Larduya I. Efficient gene delivery by EGF lipoplexes in vitro and in vivo. Nanomedicine (Lond) 2011;6:89–98. doi: 10.2217/nnm.10.100. [DOI] [PubMed] [Google Scholar]
- 32.Yu W, Pirollo KF, Rait A, Yu B, Xiang LM, Huang WQ, et al. A sterically stabilized immunolipoplex for systemic administration of a therapeutic gene. Gene Ther. 2004;11:1434–40. doi: 10.1038/sj.gt.3302304. [DOI] [PubMed] [Google Scholar]
- 33.Torchilin VP, Levchenko TS, Rammohan R, Volodina N, Papahadjopoulos-Sternberg B, D'Souza GGM. Cell transfection in vitro and in vivo with nontoxic TAT peptide-liposome–DNA complexes. Proc Natl Acad Sci U S A. 2003;100:1972–7. doi: 10.1073/pnas.0435906100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ruoslahti E, Bhatia SN, Sailor MJ. Targeting of drugs and nanoparticles to tumors. J Cell Biol. 2010;188:759–68. doi: 10.1083/jcb.200910104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Temming K, Schiffelers RM, Molema G, Kok RJ. RGD-based strategies for selective delivery of therapeutics and imaging agents to the tumour vasculature. Drug Resist Updat. 2005;8:381–402. doi: 10.1016/j.drup.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 36.Ruoslati E. RGD and other recognition sequences for integrins. Annu Rev Cell Dev Biol. 1996;12:697–715. doi: 10.1146/annurev.cellbio.12.1.697. [DOI] [PubMed] [Google Scholar]
- 37.Mas-Moruno C, Rechenmacher F, Kessler H. Cilengitide: the first anti-angiogenic small molecule drug candidate; design, synthesis and clinical evaluation. Anticancer Agents Med Chem. 2010;10:753–68. doi: 10.2174/187152010794728639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ahmad A, Evans HM, Ewert KK, George CX, Samuel CE, Safinya CR. New multivalent cationic lipids reveal bell curve for transfection efficiency versus membrane charge density: lipid–DNA complexes for gene delivery. J Gene Med. 2005;7:739–48. doi: 10.1002/jgm.717. [DOI] [PubMed] [Google Scholar]
- 39.Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, et al. Automated molecular microscopy: the new Leginon system. J Struct Biol. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 40.Rädler JO, Koltover I, Salditt T, Safinya CR. Structure of DNA–cationic liposomes: DNA intercalation in multilamellar membranes in distinct interhelical packing regimes. Science. 1997;275:810–4. doi: 10.1126/science.275.5301.810. [DOI] [PubMed] [Google Scholar]
- 41.Chan CL, Majzoub RN, Shirazi RS, Ewert KK, Chen YJ, Liang KS, et al. Endosomal escape and transfection efficiency of PEGylated cationic liposome–DNA complexes prepared with an acid-labile PEG-Lipid. Biomaterials. 2012;33:4928–35. doi: 10.1016/j.biomaterials.2012.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suh J, Wirtz J, Hanes J. Efficient active transport of gene nanocarriers to the cell nucleus. Proc Natl Acad Sci U S A. 2004;100:598–602. doi: 10.1073/pnas.0636277100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koltover I, Salditt T, Safinya CR. Phase diagram, stability, and overcharging of lamellar cationic lipid–DNA self-assembled complexes. Biophys J. 1999;77:915–24. doi: 10.1016/S0006-3495(99)76942-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sanders NN, Peeters L, Lentacker I, Demeester J, De Smedt SC. Wanted and unwanted properties of surface PEGylated nucleic acid nanoparticles in ocular gene transfer. J Control Release. 2007;122:226–35. doi: 10.1016/j.jconrel.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 45.Remaut K, Lucas B, Braeckmans K, Demeester J, De Smedt SC. PEGylation of liposomes favors the endosomal degradation of the delivered phosphodiester oligonucleotides. J Control Release. 2007;117:256–66. doi: 10.1016/j.jconrel.2006.10.029. [DOI] [PubMed] [Google Scholar]
- 46.Hasegawa S, Hirashima N, Nakanishi M. Microtubule involvement in the intracellular dynamics for gene transfection mediated by cationic liposomes. Gene Ther. 2001;8:1669–73. doi: 10.1038/sj.gt.3301573. [DOI] [PubMed] [Google Scholar]
- 47.Pankov R, Markovska T, Antonov P, Ivanova L, Momchilova A. The plasma membrane composition affects fusion between cells and model membranes. Chem Biol Interact. 2006;164:167–73. doi: 10.1016/j.cbi.2006.09.010. [DOI] [PubMed] [Google Scholar]
- 48.Ewert KK, Evans H, Ahmad A, Slack L, Lin A, Martin-Herranz A, et al. Lipoplex structures and their distinct cellular pathways. In: Huang L, Hung MC, Wagner E, editors. Advances in genetics, Vol 53: Non-Viral Vectors for Gene Therapy, part I. 2nd ed. Elsevier Academic Press; San Diego: 2005. pp. 119–55. [PubMed] [Google Scholar]
- 49.Tristram-Nagle S, Petrache HI, Nagle JF. Structure and interactions of fully hydrated dioleoylphosphatidylcholine bilayers. Biophys J. 1998;5:917–25. doi: 10.1016/S0006-3495(98)77580-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gruner SM, Tate MW, Kirk GL, So PTC, Turner DC, Keane DT, et al. X-ray diffraction study of the polymorphic behavior of N-methylated dioleoylphosphatidylethanolamine. Biochemistry. 1988;27:2853–66. doi: 10.1021/bi00408a029. [DOI] [PubMed] [Google Scholar]
- 51.Zhang HY, Hill RJ. Concentration dependence of lipopolymer self diffusion in supported bilayer membranes. J R Soc Interface. 2011;8:127–43. doi: 10.1098/rsif.2010.0200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Israelachvili J. Intermolecular and surface forces. Academic Press; London: 1992. [Google Scholar]
- 53.Roux D, Safinya CR. A synchrotron x-ray study of competing undulation and electrostatic interlayer interactions in fluid multimembrane lyotropic phases. J Phys (Paris) 1988;49:307–18. [Google Scholar]
- 54.deGennes PG. Polymers at an interface; a simplified view. Adv Colloid Interface Sci. 198(27):189–209. [Google Scholar]
- 55.Butt HJ, Kappl M, Mueller H, Raiteri R. Steric forces measured with the atomic force microscope at various temperatures. Langmuir. 1999;15:2559–65. [Google Scholar]
- 56.Lim RYH, Huang NP, Koser J, Deng J, Lau KHA, Schwarz-Herion K, et al. Flexible phenylalanine-glycine nucleoporins as entropic barriers to nucleocytoplasmic transport. Proc Natl Acad Sci U S A. 2006;103:9512–7. doi: 10.1073/pnas.0603521103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hansen PL, Cohen JA, Podgornik R, Parsegian VA. Osmotic properties of poly(ethylene glycols): quantitative features of brush and bulk scaling laws. Biophys J. 2003;84:350–5. doi: 10.1016/S0006-3495(03)74855-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kenworthy AK, Hristova K, Needham D, Mcintosh TJ. Range and magnitude of steric pressure between bilayers containing phospholipids with covalently attached poly(ethylene glycol). Biophys J. 1995;68:1921–36. doi: 10.1016/S0006-3495(95)80369-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kuhl TL, Leckband DE, Lasic DD, Israelachvili JN. Modulation of interaction forces between bilayers exposing short-chained ethylene oxide headgroups. Biophys J. 1995;66:1479–88. doi: 10.1016/S0006-3495(94)80938-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.