Abstract
Recent progress in DNA manipulation and gene circuit engineering has greatly improved our ability to programme and probe mammalian cell behaviour. These advances have led to a new generation of synthetic biology research tools and potential therapeutic applications. Programmable DNA-binding domains and RNA regulators are leading to unprecedented control of gene expression and elucidation of gene function. Rebuilding complex biological circuits such as T cell receptor signalling in isolation from their natural context has deepened our understanding of network motifs and signalling pathways. Synthetic biology is also leading to innovative therapeutic interventions based on cell-based therapies, protein drugs, vaccines and gene therapies.
The field of synthetic biology seeks to make biology more efficient, reliable and predictable to engineer and in doing so to increase the scope of possible biological functions for therapeutic and research applications. Synthetic biologists rewire biological systems by modifying and recombining existing genetic elements and creating entirely new genetic parts. This approach has become possible owing to the increasing number of available genetic building blocks and a greater under-standing of biomolecular modules ranging from DNA regulatory sequences to protein interaction networks and how to recombine them. The synthetic biology approach also benefits from advances in mathematical modelling and principles that have been developed in engineering disciplines1. Such principles include the standardization of genetic modules, such as promoters or transcriptional terminator regions, and the concept of abstraction; that is, breaking down biological systems into component parts and collections of parts into devices2, 3.
Early synthetic biology studies focused on engineering circuits in bacterial hosts. The first systems built were inspired by electronics and included the construction of genetic switches4, oscillators5, and digital logic gates6. These synthetic networks showed that engineering-based methods could be used to programme computational behaviour into cells. They also helped to elucidate how naturally occurring gene networks can generate dynamic output behaviours such as oscillations or memory of transient stimuli. Engineering of unicellular organisms has led to interesting practical applications in biosensing, therapeutics and the production of biofuels and pharmaceuticals7. Although in the beginning the field of mammalian synthetic biology merely mimicked and lagged behind this early bacterial work, it is now rapidly advancing owing to major developments in manipulating mammalian genomes and in methods for cloning large DNA circuits (Box 1).
Box 1. Constructing large DNA circuits.
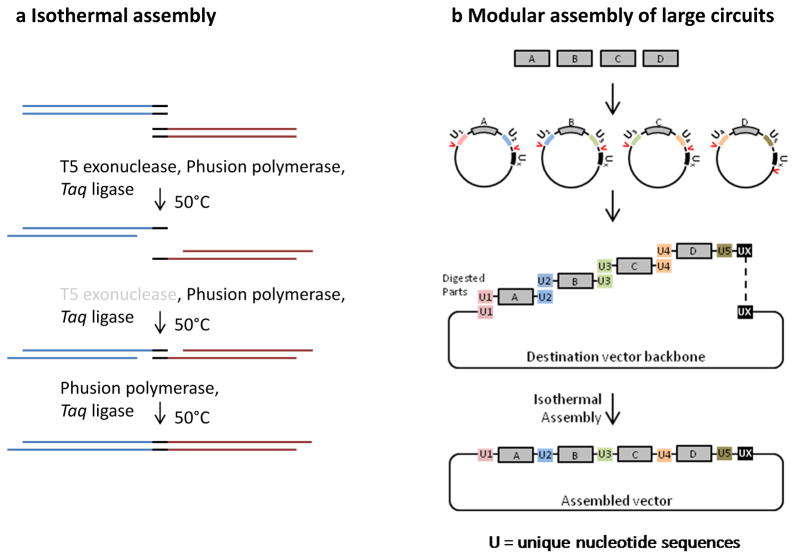
The efficiency and ease of construction of multipart DNA constructs, one of the core technologies that defines the success of synthetic biology, has improved immensely over the past decade. On one hand, this has been spurred by an exponential increase in the efficiency of DNA sequencing and short oligonucleotide synthesis142. On the other hand, newly developed methods have simplified the assembly of synthesized or pre-existing small DNA parts into large circuits. By circumventing the use of restriction endonucleases, a single-step isothermal in vitro recombination reaction has revolutionized the way researchers manipulate and join DNA molecules143 (see the figure part a). In an isothermal cloning reaction, pieces of DNA that share terminal sequence overlaps (shown in dark blue) of 20–40 base pairs in length are assembled at a constant temperature of 50 °C using three different enzymes. In the first step, T5 exonuclease removes nucleotides from the 5′ ends of double-stranded DNA and generates single-strand DNA overhangs that can anneal to DNA molecules with complementary terminal sequences. In the second step, Phusion DNA polymerase fills the gaps, and Taq DNA ligase seals the nicks. T5 exonuclease is heat-labile and gradually loses its activity during the incubation at 50° C, therefore only short overhangs of 15–20 bp are formed in the first step. Recently developed plasmid systems rely on the isothermal assembly method for the rapid and modular construction of large mammalian genetic circuits starting from a library of small sub-parts144, 145 (see the figure part b). These methods make use of unique nucleotide sequences (UNSes) that flank the library parts and facilitate isothermal assembly. Of note, one of these cloning methods has been used for the integration of large genetic circuits into single genomic sites43.
Box 1.
In this Review, we focus on advances in engineering synthetic circuits in mammalian cells and how they are both improving our understanding of cellular processes and stimulating the development of novel therapeutic approaches. We first describe the molecular tools and basic circuits that have been developed for engineering mammalian cells, highlighting major advances such as programmable transcription factors, as well as RNA and protein signalling devices. We then discuss how these tools are being used to study gene regulatory mechanisms, gene networks and signalling pathways. We also review applications of synthetic biology in the delivery of therapeutic agents and in the development of novel therapeutics such as chimeric antigen receptor (CAR)-modified T cells, and RNA- and cell-based vaccines. Throughout, we discuss key developments, but also highlight some genetic engineering advances that are not traditionally attributed to synthetic biology studies. Finally, we conclude by describing major technical hurdles and discuss what the future may hold for mammalian synthetic biology.
Tools and basic circuits
Synthetic biology is built around the concept of engineering biomolecular tools on the basis of genetic modules from different organisms and combining them into circuits to impart novel biological functionalities to host organisms. We provide an overview of these tools and discuss their applications in engineering synthetic networks.
Tools for transcriptional control
Transcription factor circuits make up the largest number of mammalian synthetic circuits to date, in part because they are intuitive to design and implement. Transcription factors consist of DNA-binding domains and transcriptional activation or repression domains for positive and negative regulation of target genes, respectively (FIG. 1a). Transcription factors bind to well-defined DNA sequences that can be used to engineer synthetic promoters8, 9. The first generation of synthetic gene circuits was based on naturally existing transcription factors, such as LacI, TetR and GAL4, and synthetic promoters that contain corresponding transcription factor-binding sequences. The bacterial transcription factors LacI and TetR offer the advantage that small molecules such as isopropyl-β-d-thiogalactopyranoside (IPTG) and doxycycline can regulate their DNA-binding activity, thereby providing the ability to induce target genes at a range of expression levels8, 9.
Figure 1. Tools used in mammalian synthetic biology.
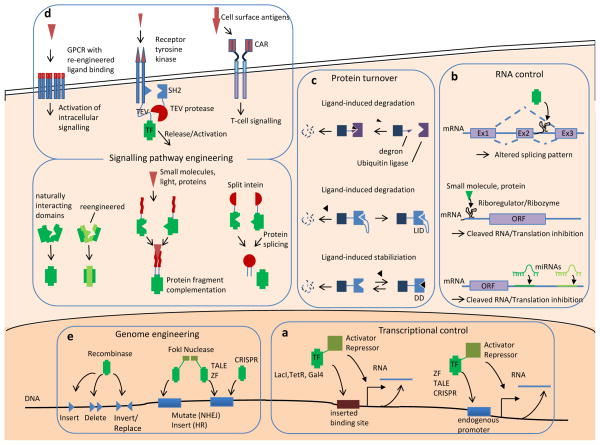
a | Tools for transcriptional control. DNA-binding domains from transcription factors (TFs) such as LacI, TetR or GAL4 are fused to protein domains that activate or repress transcription in mammalian cells8, 9. These artificial transcription factors regulate genes that contain their target-binding sites. Synthetic transcription factors based on zinc-fingers (ZFs)10, transcription activator-like effectors (TALEs)13, 14 and clustered regularly interspaced short palindromic repeats (CRISPR)23–27 can be used to target any endogenous genomic region. b | Tools for RNA control. Aptamers can bind to proteins or small molecules. Addition of a protein-binding aptamer to an intron has been used to control the exclusion of an alternatively spliced exon (Ex2)49. When combined with ribozymes, aptamers can degrade the RNAs in which they reside47. Similarly, aptamers placed in 5′ untranslated regions (UTRs) can regulate translation42. mRNA translation can also be controlled by placing combinations of sequences complementary to endogenous microRNAs (miRNAs) in the mRNA 3′ UTR29. c | Tools for protein turnover regulation. Ligand-induced protein degradation has been achieved by fusing a degradation recognition site (degron) to the target protein. In the presence of a ligand, the degron is bound by an E3 ubiquitin ligase complex54. Alternatively, a ligand-induced degradation (LID) domain can be fused to the protein of interest. The LID domain mediates ligand-dependent degradation55. This approach can also stabilize the target protein, by using a modified degradation domain (DD), the activity of which is blocked by a ligand56. d | Tools for signalling pathway engineering. Rerouting the signalling of cell surface receptors can be achieved by engineering their ligand specificity58. Intracellular receptor domains have been modified by fusing the intracellular receptor domains with a tobacco etch virus (TEV) protease cleavage peptide followed by an artificial transcription factor59, 60. In the shown example, the TEV peptide was fused to a SRC-homology 2 (SH2) protein signalling domain, which leads to its recruitment to the receptor upon activation and subsequent release of the transcription factor60. Chimeric antigen receptors (CARs) are synthetic T cell receptors that enable the retargeting of T cell activity towards cells with the targeted surface antigen107. Rerouting of intracellular signalling proteins has been achieved by reengineering their interaction domain such that it only recognizes an engineered binding partner but not the natural occurring counterpart63. Alternatively, proteins have been brought into close proximity to each other by fusing them to protein dimerization domains, which are either constitutively active or induced by small molecules or light37, 64, 91, 146. Inteins and split (trans-acting) inteins are proteins that can self-excise and ligate the peptides fused to them. e | Tools for genome engineering. Recombinases catalyse the recombination of a pair of short target sequences (triangles), which are pre-integrated into the genome. Depending on the target sequence configuration, DNA elements can be inserted, deleted, inverted or replaced70. Nucleases fused to DNA-binding factors such as zinc-finger nucleases (ZFNs)74, 75 or TALE nucleases (TALENs)76, as well as CRISPR-based systems19–21 have been used to induce a double-strand break at any given DNA locus. Upon double-strand break formation, the non-homologous end joining (NHEJ) or homologous recombination pathway induces a mutation or insertion of sequence fragments, respectively. GPCR, G protein-coupled receptor; ORF, open reading frame; RTK, receptor Tyr kinase.
Although these natural transcription factors continue to be used in specific applications, they are increasingly being replaced by programmable transcription factors such as zinc-finger-containing factors, transcription activator-like effectors (TALEs) and clustered regularly interspaced short palindromic repeats (CRISPR)-based regulators, each of which can be engineered to bind to desired DNA sequences. All of these factors are highly customizable; however, each class of transcription factor comes with advantages and trade-offs and is ideally suited to different applications (Supplementary information S1 (table)). The design of zinc-fingers does not follow a simple code, and in vitro selection assays are often part of their design pipeline10. In comparison, TALEs are more straightforward to design than zinc-fingers. The DNA-binding domains of TALEs are composed of repeated domains of 34–35 amino acids, each of which recognizes and binds to a single DNA base pair with high specificity, the binding code for which was recently discovered11, 12. By stringing together the DNA-binding repeat domains with known specificities, TALEs can be designed to bind 7–34 bp-long DNA sequences13, 14. Although TALEs often display off-target binding to DNA sequences with up to 3 mismatches in the target DNA sequence, computational algorithms have been designed to address this non-specificity12, 14. TALEs are large proteins with highly repetitive sequences in the DNA-binding domain, which poses challenges to cloning and delivery into host genomes. However, strategies for the efficient generation of large libraries of TALEs15–17 and for their viral delivery into host cells have been recently developed18.
The newest class of artificial transcription factors, CRISPR-based regulators, is based on Cas9, a protein from the bacterial CRISPR system that can bind to DNA by using a short guide RNA that is complementary to desired target DNA sequences. As a first application in mammalian cells, the endonuclease activity of Cas9 was used to generate double-strand breaks at genomic sites to induce gene mutations or homologous recombination19–22. It has also been shown recently that the catalytically inactive Cas9, lacking endonuclease activity, retains its DNA binding capability and can interfere with transcriptional elongation23. The efficiency of this repressor system can be improved by fusing the KRAB (Krüppel associated box) repressor domain to Cas924. Similarly, by fusing the VP64 transcriptional activation domain to Cas9, the CRISPR system can act as a transcriptional activator25–27. The ability to specify a target using an RNA molecule rather than a protein domain makes CRISPR transcription factors versatile and facilitates the generation of large libraries of these transcription factors.
Circuits based on synthetic transcription factors can be engineered to interact with endogenous signalling networks through native promoter elements, transcription factors, microRNAs (miRNAs), small molecules, proteases and cell surface receptor ligands28–30. To recapitulate the expression and regulation of an endogenous gene, the promoter sequence of the endogenous gene can be fused upstream of a synthetic transcription factor coding sequence to drive its expression. Alternatively, in applications in which the synthetic circuit should sense the expression of specific endogenous transcription factors as input signals, synthetic promoter elements can be designed to harbour multiple copies of the binding sites of the transcription factor31. Synthetic transcription factors have also been engineered to sense metabolites and small molecules, including IPTG, doxycycline, 4-hydroxy-tamoxifen (4HT), uric acid, rapamycin, macrolides and streptogramin30, 32–35. Recently, TALEs have been designed to respond to the activation of hypoxia-inducible factor 1α (HIF1α), an endogenous transcription factor that is expressed upon induction of hypoxia signalling30. Physical signals such as light can also be used as input signals for transcriptional circuits, whereby light triggers the assembly of transcription factor protein fragments into a functional transcription factor36. A recent study described a light-inducible TALE system, consisting of a TALE DNA-binding domain fused to the light-sensitive cryptochrome protein CRY2 and the transcriptional activator VP64 fused to CIB1 (Ca2+- and integrin-binding protein 1)37. Illumination with blue light triggers CIB1 recruitment to CRY2 and induces VP64-mediated transcription at the site of TALE binding. The authors demonstrated that this system can be used for optogenetic modulation of endogenous transcription in the mouse brain37. Similar modulation of gene expression in mammalian cells in the presence of visible light background can be achieved using ultraviolet B light-mediated protein–protein interaction systems38, 39.
Synthetic transcription factors and promoter elements have been used to engineer complex circuits such as networks that perform logic computation40–43, feedback loops to make devices that can confer a long lasting memory in response to a transient stimulus44 and genetic switches for tight control of gene expression14, 32, 40. Transcription factor-based circuits have also been built that specifically respond to intermediate levels of input, or generate time-delayed or oscillatory transcriptional output45, 46. Early examples of these gene circuits have used prokaryotic-based transcription factors, whereas newer synthetic transcription factors are aiming for higher scalability using programmable factors. Although many of these gene circuits were demonstrated as proof-of-concept systems, these circuits are now advancing to the stage at which they can be used as tools to modulate gene activities and cell behaviours as discussed in the applications sections.
Tools for RNA control
Although the earliest synthetic systems were largely transcription factor-based, RNA regulators are having an ever-increasing role in mammalian synthetic biology47. RNA-based parts and regulatory mechanisms for use in mammalian cells have been engineered to respond to several different types of molecular inputs, including small molecules, metabolites and proteins47 (FIG. 1b). These systems use aptamers, which are structured RNAs that can bind to small molecules and proteins and in response regulate the activity of other RNAs effectors on the same molecule or in trans. One commonly used type of RNA effector is a self-cleaving ribozyme that can degrade the RNA it resides in upon binding of a protein or small molecule to the aptamer48. In other applications, aptamers have also been placed within introns, where they can change the splicing pat-tern of the mRNA in response to protein binding49. In a more recent example, aptamers that sense the presence of specific proteins were placed in the 5′ untranslated region (UTR) of transcripts, resulting in translational inhibition of these transcripts in the presence of the protein inputs42. Endogenous miRNAs can also be sensed by placing complementary target sequences in the 3′ UTR of transcripts that code for output proteins or transcription factor regulators29. In the presence of the miRNA, the mRNA transcript is either degraded or translationally repressed by RNAi activity. Circuits composed of up to six different miRNA inputs have been demonstrated to classify cell types on the basis of miRNA expression patterns29. Furthermore, siRNA- and miRNA-based regulators have also been shown as viable options for Boolean logic computing frameworks50.
RNA-based parts and devices hold many advantages but also trade-offs compared with other types of regulators. RNA-based systems are relatively fast-acting, as they do not require translation. They are also well-suited for sensing certain types of molecules such as miRNAs and further benefit from directed molecular evolution methods for quick screening for RNA aptamers with new specified binding properties51. Furthermore, RNA-targeted systems are very effective when combined with transcriptional regulation. Inhibitory RNA combined with transcriptional repressors can lead to near complete repression of gene activity14, 32. However, although miRNA regulation is very robust, some of the other RNA-based sensors can be less sensitive as compared to transcription factor-based systems49.
Tools for protein turnover regulation
Altering protein stability is a powerful way to rapidly and post-translationally control protein activity. Protein stability depends on several factors such as the length of the peptide sequence and the occurrence of specific amino acids that can be phosphorylated52. Furthermore, proteins can be actively degraded through the ubiquitylation path-way, wherein an E3 ubiquitin ligase recognizes proteins that harbour a specific domain and catalyses the transfer of ubiquitin to this target protein, ultimately leading to its recognition and degradation by the proteasome53. In some instances, target recognition by an E3 ubiquitin ligase can be regulated by a small-molecule ligand. For example, the Arabidopsis thaliana TRANSPORT INHIBITOR RESPONSE 1 (TIR1) protein has recently been shown to form an active E3 ubiquitin ligase complex in mammalian cells, and it binds its recognition site upon addition of auxins, which are plant hormones that control gene expression54. This system has been used to reversibly induce degradation of a fluorescent protein that was engineered to contain a recognition site for TIR1 54 (FIG. 1c).
An alternative method for ligand-induced protein degradation has been developed by screening variants of the FK506- and rapamycin-binding protein (FKBP) that have additional amino acid residues appended to the carboxyl terminus55. This screen has revealed a ligand-induced degradation (LID) domain, which when fused to a protein confers stability in the absence of a ligand and causes rapid degradation in the presence of the high-affinity ligand Shield-155. Conversely, a different variant of FKBP has been engineered to confer protein stability specifically in the presence of the Shield-1 ligand56. Work in yeast has also demonstrated a novel light-reactive protein degradation domain, which opens the possibility of using light to regulate protein stability in mammalian cells57.
Tools for signalling pathway engineering
Several synthetic signalling networks have been engineered to introduce novel control schemes or reroute information flow (FIG. 1d). Modified membrane receptors that detect unnatural small molecules represent one class of tools that can offer orthogonal external control of cellular function. For instance, directed molecular evolution has been used to change the ligand specificity of G protein coupled receptors (GPCRs)58. Rerouting signalling output has also been accomplished by modifying the intracellular domain of receptors. In one such approach, artificial transcription factors are fused to the intracellular domain of membrane receptors to translate extracellular signals into transcriptional outputs. This mechanism has been implemented in the Notch receptor by replacing its intracellular domain with the transcriptional activator GAL–VP16, which gets cleaved upon binding to the Delta ligand59. Similarly, the output of GPCRs has been rerouted using tobacco etch virus (TEV) protease. In this system, the intracellular domain of the GPCR has been fused to a TEV protease recognition site followed by a synthetic transcription factor. Upon GPCR activation, the small signalling transduction protein arrestin fused to TEV protease is recruited to the GPCR, which leads to the release of the artificial transcription factor60. An analogous system has been developed for receptor Tyr kinases, in which the TEV protease is fused to a SRC-homology 2 (SH2) protein domain that is recruited to the activated receptor60. By fusing the death effector domain FADD to SH2, receptor activation has also been rerouted to induce cell death61. Another notable example of reengineered receptors are CARs, which are synthetic T cell receptors that enable the retargeting of T cell activity towards cells with the targeted surface antigen (see the following sections for more details).
Cytosolic proteins involved in intracellular signalling consist of multiple domains that define their catalytic activity, localization and binding to scaffold proteins. Proteins with novel functionalities can be engineered by recombining these domains62. The engineering of such signalling networks requires modifying protein–protein interactions, which for many applications are ideally orthogonal to the protein interaction network of the host cell. One recent example of designing orthogonal signalling proteins is the engineering of the GTPase CDC42 and its activator intersectin63. The engineered version of CDC42 is exclusively induced by its cognate partner, while maintaining its ability to interact with other GTPase signalling components63. Alternatively, specificity and orthogonality in synthetic signalling networks can be achieved by fusing interaction domains to dimerizing proteins such as synthetic coiled-coil peptides64.
A unique class of tools for post-translational circuit engineering are inteins, which are protein splicing domains that can self-excise and ligate attached peptides. Inteins and split (trans-acting) inteins that are activated in response to small molecules, light and protein inputs have been generated, enabling the post-translational control of circuits65–67. There are also several systems available that use light to induce protein-protein interactions in a reversible manner. For instance, it has been shown that fusing the kinase CRAF (also known as RAF1) to CRY2, which dimerizes in response to blue light, allows the regulation of the kinase activity using light68. Similarly, light-induced oligomerization of a WNT receptor domain has been used to activate the β-catenin pathway69.
Tools for genome engineering
Site-specific recombination was among the first technologies that enabled the precise manipulation of mammalian genomes. The method relies on bacterial or fungal recombinases that catalyse the recombination of a pair of short target sequences (for example, LoxP sites in the case of Cre recombinase) that are pre-integrated into the genome around loci of interest70 (FIG. 1e). Upon induction of the recombinase, the intervening DNA is either flipped or excised depending on the orientation of the recombinase-target sites. In mice, increased specificity of conditional gene knockouts has been achieved through an expression cassette that is controlled by the combined activity of two different recombinases, which in turn are induced by different tissue-specific promoters71. Such an in vivo ‘AND’ gate, which induces a knock-out in response to a combination of two signals, has also been created by expressing the two fragments of a split Cre recombinase from two different tissue-specific promoters72, 73.
The traditional method for site-specific gene targeting in mammalian cells involves transfection of DNA containing the desired alterations together with flanking long homologous DNA strands and selection of rare modified cells. However, this technique is highly inefficient. The development of programmable DNA nucleases to introduce site-specific DNA breaks, a potent inducer of localized homologous recombination, has greatly improved the process. In the case of zinc-finger nucleases74, 75 and TALE nucleases (TALENs)76 a pair of engineered DNA-binding proteins is fused to a FokI nuclease domain (FIG. 1d). The fusion proteins are engineered to bind in close proximity on DNA, leading to the dimerization of FokI and the induction of a double-strand break at the target site. The recently developed CRISPR system relies on a DNA endonuclease that is targeted to DNA by a short RNA rather than a protein domain, which makes this method very versatile19–21. Thus far, editing of the mammalian genome by CRISPR seems to be very efficient and enables multiple alleles to be mutated in parallel22. However, high levels of off-target cleavage by CRISPR-Cas nucleases have been observed and still need to be addressed77.
Multiplexed genome engineering (MAGE), which relies on oligonucleotide-mediated recombineering, is an alternative method for editing multiple genetic loci in parallel78. Demonstrated to be useful in bacteria78, the adaptation of this method to mammalian cells is still under way79.
The tools described in this section enable the manipulation of one or more genetic loci. Moreover, there have also been efforts towards synthesizing whole genomes de novo. Synthesis of a mitochondrial genome has been reported in 2010 80, and de novo synthesis of a yeast genome is currently being developed81.
Synthetic biology in basic research
The past decade has seen enormous advances in mapping the genome and proteome in different biological systems, including mammalian cells. Although this has led to an increasingly comprehensive picture of gene and protein networks, there is still a need for deeper mechanistic understanding of these systems. Synthetic biology approaches offer potential insight into the logic of cellular systems at different levels by rebuilding and studying them in a context isolated from their high degree of natural interconnectivity.
Studying chromatin and gene regulation
Gene activity in eukaryotes is regulated by a complex interplay between cis- and trans-acting DNA elements such as promoters and enhancers. Novel high-throughput methods for testing the activity of synthetic DNA sequences have helped to understand the architecture of these gene regulatory regions. For instance, a library of tandem repeats of all possible 10-mer DNA sequences has been cloned upstream of GFP under the control of a minimal promoter and tested for its activity after retroviral-mediated transfection into human cell lines82. This approach led to the identification of a novel strong synthetic promoter and revealed transcription factor-binding motifs that can induce high levels of transcription82. The efficiency of testing such libraries has been improved by ‘barcoding’ promoter variants with short DNA sequences and measuring their activity by high-throughput RNA sequencing83 (FIG. 2A). This method, termed massively parallel reporter assay, has recently been used to study a large number of variants of endogenous enhancers as well as combinations of conserved regulatory motifs found in enhancers84–87.
Figure 2. Studying chromatin and gene regulation.
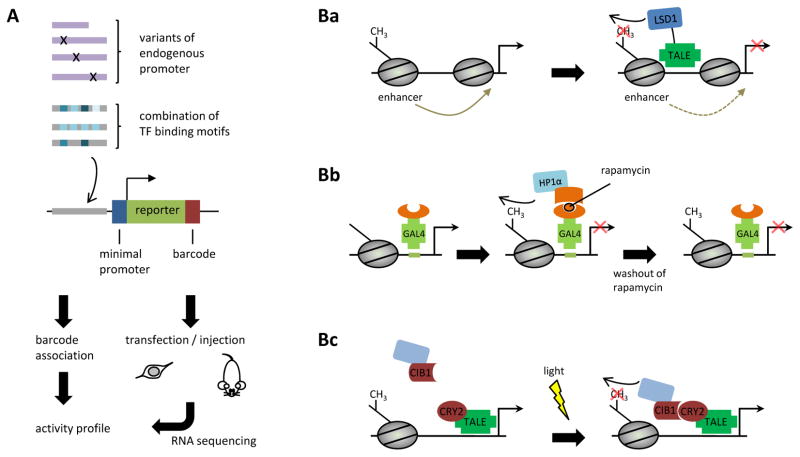
A | Testing libraries of gene regulatory regions. A large number of synthetic variants of endogenous promoters or sequences containing combinations of transcription factor (TF)-binding motifs are cloned upstream of a minimal promoter driving a reporter gene. The constructs also contain a short variable nucleotide sequence that serves as a ‘barcode’ and can be associated with the inserted promoter sequence by high-throughput sequences of the library. The library is injected into mice or transfected into cultured cells, and its activity is measured by RNA sequencing. B | Recruitment of chromatin-modifying enzymes by zinc-fingers (ZFs) or transcription activator-like effectors (TALEs). Site-specific targeting of the histone demethylase LSD1 has been used to study the interplay between histone marks found at enhancers and nearby genes90 (Ba). Reversible recruitment of chromatin-modifying enzymes. Rapamycin-inducible dimerization with the transcription factor GAL4 has been used to recruit heterochromatin-binding protein 1α (HP1α), a component of repressive chromatin, to GAL4-binding sites integrated at the Oct4 promoter91. The histone H3 Lys9 methylation mark, indirectly induced by HP1α, has been shown to be epigenetically transmitted after washout of rapamycin and loss of HP1α binding (Bb). Light-inducible transcriptional effectors (LITEs). Light stimulation induces dimerization of the cryptochrome protein CRY2 and CIB1 (Ca2+- and integrin-binding protein 1), leading to the recruitment of a chromatin-modifying enzyme (shown in light blue) to the target promoter37 (Bc).
In addition to transcription factor occupancy of regulatory DNA elements, eukaryotic gene expression is also influenced by changes to chromatin through DNA methylation, nucleosome positioning and post-translational modifications of histones. In order to change the chromatin environment at specific genomic locations, enzymes that catalyse the addition or removal of chromatin marks have been fused to programmable transcription factors that can be directed to any sequence of interest. This approach was first demonstrated by the fusion of a zinc-finger transcription factor with a DNA methyltransferase88. Recently, the same principle has been adapted to TALEs37, 89, 90. In one such application, directing the activity of ten-eleven translocation 1 (TET1), an enzyme involved in the demethylation of DNA, to gene promoters has revealed methylation sites that are crucially involved in gene repression89. In another similar application, site-specific targeting of the histone demethylase Lys-specific demethylase 1 (LSD1) has been used to study the interplay between histone marks found at enhancers and nearby genes90 (FIG. 2Ba). Small molecule-induced dimerization has also been used to recruit chromatin-modifying enzymes in a rapid and reversible manner91. It has been shown that transient induction of a repressive histone modification at the Oct4 (also known as Pou5f1) gene leads to its heritable transmission through multiple cell generations91 (FIG. 2Bb). Transient induction can also be triggered by light, as has been recently demonstrated by a new system termed light-inducible transcriptional effectors (LITEs) for reversible, TALE-guided targeting of chromatin-modifying enzymes37 (FIG. 2Bc).
Studying gene networks
Genes and their regulators form highly interconnected networks, the topology of which can be inferred (or reverse-engineered) from perturbation and gene expression data. In order to benchmark reverse-engineering algorithms, small synthetic gene networks have been engineered within the natural cellular environment of yeast and human cells92, 93 (FIG. 3a).
Figure 3. Studying gene and signalling networks.
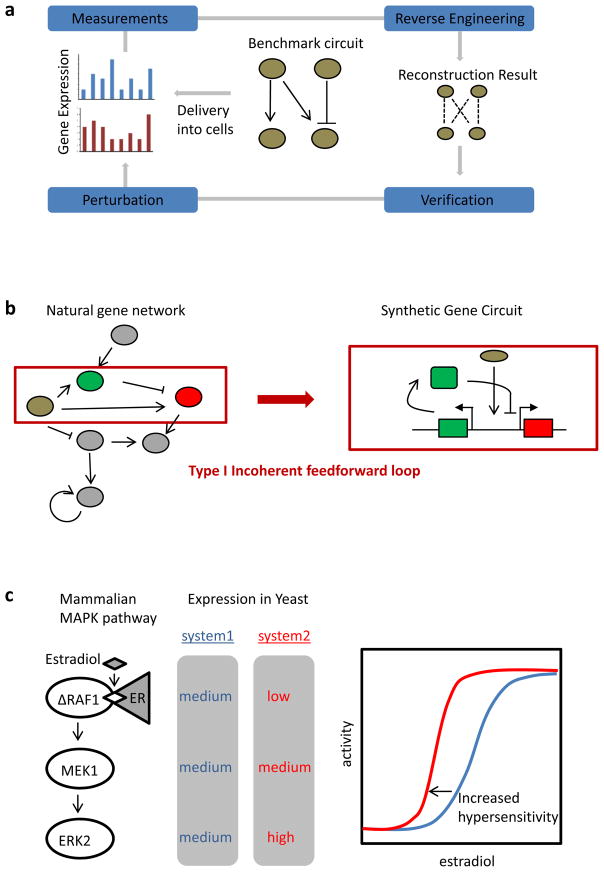
a | Schematic of reverse engineering with synthetic circuits. A benchmark gene circuit is integrated into a mammalian cell line. Different reverse engineering algorithms are used to generate a network model from perturbation and gene expression analysis. The quality of reverse engineering algorithms is assessed and improved by comparing the resulting models to the actual circuit architecture in an iterative cycle93. b | Studying gene network architectures. Synthetic gene circuits enable the study of network modules in isolation from their natural context in which they may be interconnected with other endogenous signalling pathways (shown in blue). The schematic shows the design of a type I incoherent feedforward loop circuit, consisting of an input (shown in brown) that activates both an output (shown in red) and an 97 heterologous reconstitution. A mammalian MAPK pathway, consisting of an estradiol-inducible RAF1 protein that activates MEK1, which in turn activates ERK2, has been expressed in yeast. Increased concentration of each protein at each step of the cascade has been shown to augment the degree of input ultrasensitivity101. ER, oestrogen receptor.
Gene regulatory networks from bacteria to humans are characterized by the occurrence of several over-represented network motifs94, 95. Synthetic gene circuits have enabled the study of these network motifs in isolation from their natural interconnections (FIG. 3b). The positive feedback loop, in which transcription of a gene is activated by its own output, is one common motif94, 95. By rebuilding this motif as a synthetic gene circuit, it has been shown that it generates hysteresis, meaning that the induction of the repressed state requires a reduction of the input beyond the amount that is necessary for activation96. Another common motif is a type I incoherent feedforward loop, which consists of an input that activates both an output and an auxiliary gene, which in turn represses the output (FIG. 3b). A circuit with this regulatory motif has been demonstrated to adapt in response to the amount of genetic template and was therefore suggested to have a role in gene dosage compensation97. Synthetic circuits have also been used to generate oscillatory gene expression as found for genes controlled by the circadian clock. A genetic circuit consisting of a positive feedback loop and a time-delayed negative feedback loop has produced robust oscillations and revealed system parameters for tuning the oscillatory behaviour in mammalian cells46. Of note, combinations of positive and negative feedback loops have also been found to underlie natural oscillatory networks, such as the cell cycle in Xenopus laevis embryos98. For more examples of how synthetic gene circuits have been used to study their natural counterparts, we refer the reader to a recent review99.
Studying cell signalling
Rebuilding artificial signalling networks and studying them in an exogenous context obviates the challenges of interconnectivity that involve endogenous competing signalling networks and enables the essential components of a signalling pathway to be defined. Following such an approach, the genes encoding proteins involved in the T cell receptor (TCR) signalling pathway, which are normally only active in T lymphocytes, were expressed in a non-immune cell100. With a set of more than 10 heterologously expressed proteins, the authors of this study recapitulated TCR signalling and tested competing biophysical models of TCR activation.
Mammalian signalling pathways have also been studied by heterologous reconstitution in different organisms. For example, a MAPK activation cascade based on the mammalian ERK1 and ERK2 pathway and including RAF1, MEK1 and ERK2 has been shown to be functional and well insulated (that is, not interfering with other endogenous pathways) when expressed in yeast101. Perturbation of the system in yeast has revealed insights into natural MAPK cascades, which, in certain contexts, show a graded response whereas, in others, show a switch-like (ultrasensitive) activation. Specifically, it has been demonstrated that increasing the concentration of each sequential protein at each step of the cascade increases the degree of input ultrasensitivity (an increase in the Hill coefficient) and reduces the activation threshold (FIG. 3c).
Synthetic biology in therapy
Synthetic biology aims to develop new approaches for therapeutic interventions through enhanced efficiency, predictability and safety of engineered systems, as well as by expanding the possibilities for regulating biological systems. We provide an overview of novel therapeutic strategies and outline where synthetic biology approaches are helping to improve existing treatment options.
Gene therapy
After suffering a number of setbacks, the field of gene therapy has recently regained traction by the approval of the first commercial gene therapy treatment, Glybera (Uniqure), for lipoprotein lipase deficiency102. For this and many other current therapies, genes are packaged within viruses, which can be locally administered to the tissue of interest. However, higher degrees of specificity are needed and targeting the right cell types can be critical. Gene therapies are beginning to benefit from synthetic biology approaches aimed at both improving the specificity of the gene delivery and the specificity of the expression of therapeutic genes. For instance, in a recent study, a large library of adeno-associated virus variants was generated and in vivo-directed evolution was used to identify a virus that can target the cells of the outer retina103. Cell type-specific activity could also be achieved by expressing therapeutic genes from promoter elements that are active only in certain cell types31.
Gene therapy has also been performed in combination with autologous cell transplantation, in which stem cells from a patient are removed, treated and later reintroduced in the same individual. Gene replacement therapy in autologous haematopoietic stem cells has recently shown promising outcomes for the treatment of inherited diseases104, 105. This method of delivery has also been explored for direct manipulation of endogenous genes to either correct or induce mutations. For instance, there are ongoing clinical trials for HIV therapeutics that are based on targeted gene disruption in T cells. In this case, a ZFN is used to mutate the chemokine receptor 5 (CCR5) gene in autologous CD4+ T cells, which then become resistant to HIV and outlive HIV-infected cells106. Such therapeutic approaches based on genome editing are expected to benefit from tools recently developed in the synthetic biology field, such as TALENs and CRISPR.
Chimeric Antigen Receptor-based T cell therapy
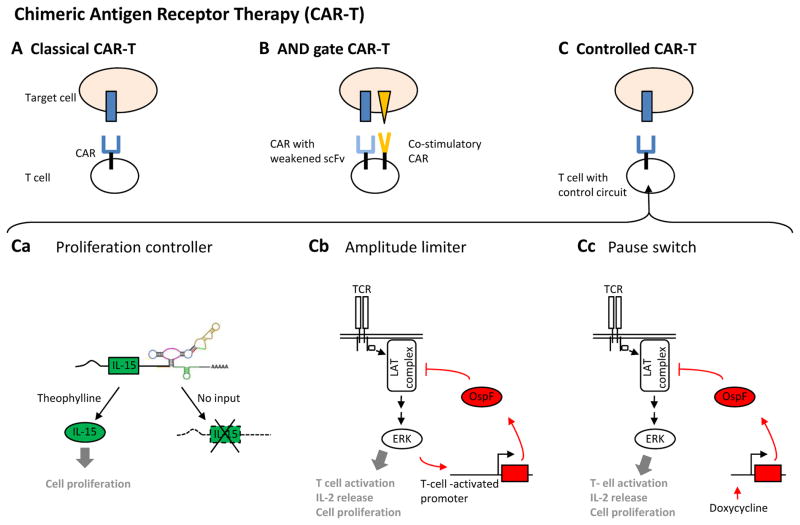
CARs are synthetic T cell receptors that enable the retargeting of T cell activity towards cells with the targeted surface antigen107. In this approach, T cells of a patient are engineered ex vivo to express the CAR and then adoptively transferred into the patient, thus killing targeted cells. Recently, CAR-based immunotherapies have been successful in targeting cancer types for which no other appropriate treatment was available108. CARs are modular fusion proteins that consist of cytoplasmic signalling domains from the T cell receptor and T cell co-receptor, a trans-membrane region, an extracellular linker and an antigen-targeting element, which is most often a single chain variable fragment (scFv) antibody. In all CAR-based therapies that are currently in the clinic, cancer cells are targeted on the basis of single cancer-specific antigens and thus can essentially kill every cell that contains the targeted antigen (FIG. 4A). Although showing very promising results, even in current therapies many ‘on-target off-tumour’ effects have been observed that can lead to lethal toxicity108. Thus, a current focus is engineering greater specificity to CAR-modified T cells.
Figure 4. Chimeric antigen receptor therapy.
A | The classic chimeric antigen receptor (CAR) therapy (CAR-T) approach. T cells are engineered ex vivo to express a CAR and then transferred into the original donor patient, where they destroy cells that display the target antigen. B | AND gate CAR therapy. T cells are engineered to express two CARs, one with a weakened single chain variable fragment (scFv) domain and one that contains co-stimulatory domains in its intracellular domain. These engineered T cells have been shown to preferentially target cells that display two antigens together109. C | Gene circuits for controlling CAR therapy activity. A RNA device enables the control of T cell proliferation. The device stabilizes the expression of secreted interleukin-15 (IL-15), a proliferation-inducing cytokine, in the presence of the small-molecule drug Theophylline111 (Ca). The amplitude limiter device uses a promoter that is activated upon T cell signalling and induces the expression of the bacterial virulence protein OspF, which in turn irreversibly inactivates T cell signalling. This negative feedback loop has been shown to dampen the amplitude of T cell activation112 (Cb). The pause switch device consists of a construct that induces OspF expression and thereby inhibits T cell activation in response to doxycycline112 (Cc). LAT, linker activator for T cells; TCR, T cell receptor; ZAP70, ζ-chain-associated protein kinase 70.
One recent approach to increase specificity involved the creation of a CAR-based AND logic gate that used novel CARs to target and kill cells that express two antigens but not the cells that displayed only one or none of the antigens109, 110 (FIG. 4B). A different system focused on controlling T cell proliferation and comprised an RNA control device that allowed stabilization of interleukin-15 (IL-15; a proliferation-inducing cytokine) only in the presence of a small-molecule drug111 (FIG. 4Ca). In yet another recent paper, kinase inhibitors from human pathogens have been used to rewire the TCR signalling pathway to produce novel behaviours in T cell signalling, including a delayed TCR signalling ‘pause switch’ and feedback modulators in order to tune the amplitude of the T cell signalling response112 (FIG. 4Cb, Cc).
Prosthetic networks
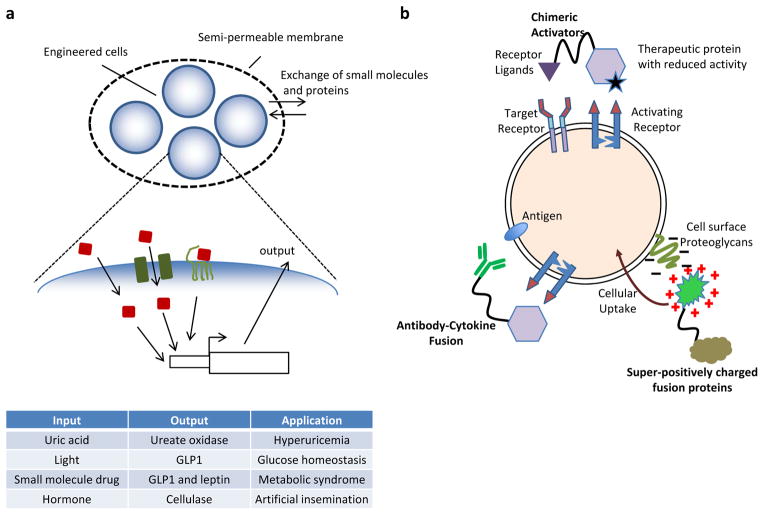
Implantation of genetically engineered encapsulated cells is currently being explored as a method for delivering complex gene control circuits113. Encapsulating cells in a biocompatible and semi-permeable material such as alginate enables diffusion of small molecules and proteins yet prevents immune reactivity and potential release of genetic information (FIG. 5a). In 1980, encapsulated pancreatic islet cells have been used to sense blood glucose levels and secrete insulin in a rat model of diabetes114. Recently, this encapsulation method has been extended to xenogeneic cells, which are engineered to harbour ‘prosthetic networks’, which are synthetic circuits that sense disease-relevant metabolites and can coordinate diagnostic, preventive and therapeutic responses35, 113. In one study, encapsulated mammalian cells were engineered to release uric acid eliminating urate oxidase in response to high levels of uric acid, thus controlling hyperuricemia33. In another instance, encapsulated cells contained a synthetic signalling cascade for light-inducible transgene expression115. Upon transdermal illumination, the implanted cells released glucagon-like peptide 1 (GLP1), which regulated glucose homeostasis115. Finally, a combined drug- and gene-based therapy has recently been described, consisting of encapsulated cells engineered to release the metabolically active peptides GLP1 and leptin in response to administration of the antihypertensive drug guanabenz35.
Figure 5. Prosthetic networks and protein-based therapies.
a | Prosthetic networks. Implantation of genetically engineered cells that are encapsulated in a semi-permeable membrane enables diffusion of small molecules and proteins and at the same time acts as a barrier to the immune system. Cells implemented in such systems have been engineered to release small effector proteins (the ‘output’) in response to specific molecular inputs (shown in blue and orange)33, 35, 113, 115. b | Protein-based therapies. When fused to antibodies or receptor ligands, therapeutic proteins such as cytokines can be targeted to cells that express the corresponding antigen or receptor, respectively116. In the case of chimeric activators, the therapeutic protein has been mutated to have low affinity for its receptor, resulting in a reduced response in cells that only bind the therapeutic protein but are not bound by the targeting element118, 119. Fusion to proteins that carry a high positive surface charge has been shown to facilitate intracellular delivery of proteins128–130. GLP1, glucagon-like peptide 1.
Protein-based therapies
Protein-based therapies have various advantages over nucleic acid-based or cell-based therapies as they can be administered locally and transiently and override the safety concerns associated with genomic manipulation strategies. Most protein-based drugs are naturally occurring proteins such as growth factors and antibodies that work through cell surface receptors to modulate the activity of signalling pathways. Synthetic biology aims at improving such therapeutics by testing combinations and variants of these proteins (FIG. 5b). For instance, direct fusion of anti-bodies with therapeutic proteins has led to enhanced targeting to cells116. In addition, bi-specific antibodies have enabled retargeting of T cell activities to targeted cells117. Another system, which is an extension of fusion proteins and is called chimeric activators enables even better cell targeting of therapeutic molecules118, 119. Chimeric activators comprise a targeting molecule such as an antibody or growth factor specific to target cells and an activity protein that is mutated to have low affinity for its receptor on the target cell surface. Cell surface binding of the targeting domain of chimeric activators increases the local concentration of the activity protein target cells. Chimeric activators have been generated to target mutated versions of the therapeutic proteins interferon-α2a (IFNα2a) and erythropoietin (EPO) to cells with epidermal growth factor receptor (EGFR) and glycophorin, respectively118, 119.
Another focus has been intracellular protein delivery to mammalian cells based on liposomes120–122, nanoparticles123–125, fusion with receptor ligands126, 127 and ‘supercharged’ tags, which are polypeptides with an unusually high net charge128, 129. In particular, fusing proteins with naturally occurring high net positively charged proteins and synthetic supercharged GFP, a GFP re-designed to have high positive surface charge, has been shown to be an effective mechanism to improve the cellular uptake of biologically active protein drugs in mouse tissues130. Supercharged fusion proteins have also been demonstrated to improve the stability and block the aggregation of proteins129.
Synthetic biology in vaccines
To prevent outbreaks of rapidly evolving pathogens, the swift development of a vaccine is of paramount importance. Recently, it was demonstrated that a vaccine for influenza could be completed in under a week’s time using a synthetic biology approach that involved bioinformatics, gene synthesis and a mammalian cell line production system131. Computer-aided design in combination with whole-genome synthesis has also been used to recode polio and influenza viruses to contain infrequently used codon pairs, representing a novel method for creating safe live attenuated viruses for vaccination132, 133. RNA-based vaccines represent another exciting area; synthetic RNA transfected into cells can produce vaccine antigens for rapid and efficient cell presentation and immunity. As the RNA does not mutate the genome, it does not have the safety concerns associated with DNA vaccines, and, although it is less stable than DNA, RNA can be produced at large scale in a timely fashion. Recently, the use of self-replicating RNA machinery has allowed long-term expression of antigens, which leads to a more stable response of RNA vaccines134.
Vaccines are also being developed to stimulate immunity against cancers. A cancer vaccine based on dendritic cells, Provenge (Dendreon), was recently approved for use in prostate cancer, and many other therapeutic cancer vaccines are currently in clinical trials. Provenge is formulated from autologous dendritic cells of a patient loaded with prostate cancer antigen, and granulocyte-macrophage colony-stimulating factor (GM-CSF), a cytokine that helps in improving dendritic cell maturation. The success rate of these cell vaccines could perhaps be further improved through genetic engineering of dendritic cells in combination with ways to overcome systemic immunosuppression135.
Perspective
The combination of genetically engineered cells with recent developments in mimicking complex tissue structures and living organs has great potential to advance tissue engineering. Driven by progress in microfluidics, novel microdevices are able to emulate tissue structures, their dynamic mechanical properties and their biochemical functions136. These devices are currently built using primary or immortalized cells, and the development of genetically engineered cells offers the opportunity to create even more sophisticated organ models. Cells that contain genetic circuits for synthetic pattern formation137 or induction of distinct cellular states49, 138 could be used to create complex patterns of cells with different functionality. Besides offering insights into normal and diseased organ function, organ-mimicking devices are useful for preclinical drug development and toxicity screening136. For example, engineered cells could be used to monitor cellular responses, specifically by genetic circuits that sense cellular states29, 139 and confer memory to transient stimuli44.
Engineered cells have also helped to advance therapeutic applications such as CAR therapies and cancer vaccines. However, mammalian system engineering still needs to overcome a number of technical hurdles, such as the scalability, orthogonality and predictability of synthetic circuit behaviours. Although programmable transcription factors and engineered protein–protein interaction domains are leading the way in addressing the problem of orthogonality, the predictability of genomically integrated DNA-based circuits still remains a cause of concern due to site-and cell type-specific effects. As one possible solution, the mammalian synthetic biology community could agree on characterizing circuits in a set of genomic loci such as safe harbour sites known to tolerate the integration of transgenes140. Alternatively, one could consider expressing circuits from a human artificial chromosome, which can hold large amounts of DNA and does not integrate into the host genome. For the challenge of scalability, the key might be to use different types of regulators that act on different classes of molecules, for instance an assortment of transcriptional and post-translational regulators of RNA and proteins, as recently demonstrated for circuits that performed multi-bit (that is, the integration of multiple digital inputs) processing in mammalian cells42.
For therapeutic applications that are based on genetically engineered cells, the field also needs to address the issue of immunogenicity of the circuit components and cells. In this case, insights can be gained from CAR therapy technologies. For example, it has recently been shown that T cells made from induced pluripotent stem (iPS) cells can be used for CAR therapy, which offers an off-the-shelf alternative to autologous T cell isolation141. The authors of this study proposed that the alloreactivity of iPS cell-derived T cells could be eliminated by disrupting the endogenous TCR, whereas allorejection could be minimized by generating iPS cells from common human leukocyte antigen (HLA) haplotypes.
We anticipate that continuing progress in mammalian synthetic biology will lead to new interesting applications in basic cell biology and the development of novel therapeutics.
Supplementary Material
Acknowledgments
The authors apologize to their colleagues whose work could not be cited owing to space limitations. They thank M. Inniss, J. Torella, T. Ford and J. Chen for helpful comments on the manuscript. The work in the authors laboratory was supported by: a European Molecular Biology Organization Fellowship and a Human Frontier Science Program Fellowship to F.L.; a Swiss National Science Foundation Fellowship to A.G. and funds from NIH and the Defense Advanced Research Projects Agency to P.A.S..
Glossary
- Digital logic gate
An idealized or physical device that implements Boolean logic (such as AND, OR or NOT) on one or more inputs to produce a single output
- Optogenetic
The combination of genetics and optics to control light-sensitive proteins within specific cells
- Orthogonal
A system is orthogonal when changes to one component do not influence the other components
- Directed molecular evolution
A method that mimics the process of natural selection to evolve proteins or nucleic acids towards a user defined goal
- Recombineering
Genetic engineering that is based on homologous recombination systems
- Gene dosage compensation
Mechanisms that dampen fluctuations in gene expression levels in response to changes in gene copy numbers
- Xenogeneic cells
Cells belonging to individuals of different species
- Alloreactivity
The immunologic reactions that occur when cells or tissues are transplanted between two individuals of the same species
References
- 1.Endy D. Foundations for engineering biology. Nature. 2005;438:449–53. doi: 10.1038/nature04342. [DOI] [PubMed] [Google Scholar]
- 2.Knight T. DARPA BioComp Plasmid Distribution 1.00 of Standard Biobrick Components. MIT Synthetic Biology Working Group Reports. 2002 [Google Scholar]
- 3.Mutalik VK, et al. Precise and reliable gene expression via standard transcription and translation initiation elements. Nat Methods. 2013;10:354–60. doi: 10.1038/nmeth.2404. [DOI] [PubMed] [Google Scholar]
- 4.Gardner TS, Cantor CR, Collins JJ. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403:339–42. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- 5.Elowitz MB, Leibler S. A synthetic oscillatory network of transcriptional regulators. Nature. 2000;403:335–8. doi: 10.1038/35002125. [DOI] [PubMed] [Google Scholar]
- 6.Guet CC, Elowitz MB, Hsing W, Leibler S. Combinatorial synthesis of genetic networks. Science. 2002;296:1466–70. doi: 10.1126/science.1067407. [DOI] [PubMed] [Google Scholar]
- 7.Khalil AS, Collins JJ. Synthetic biology: applications come of age. Nat Rev Genet. 2010;11:367–79. doi: 10.1038/nrg2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown M, et al. lac repressor can regulate expression from a hybrid SV40 early promoter containing a lac operator in animal cells. Cell. 1987;49:603–12. doi: 10.1016/0092-8674(87)90536-8. [DOI] [PubMed] [Google Scholar]
- 9.Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci U S A. 1992;89:5547–51. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maeder ML, et al. Rapid “open-source” engineering of customized zinc-finger nucleases for highly efficient gene modification. Mol Cell. 2008;31:294–301. doi: 10.1016/j.molcel.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boch J, et al. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326:1509–12. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- 12.Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science. 2009;326:1501. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- 13.Morbitzer R, Romer P, Boch J, Lahaye T. Regulation of selected genome loci using de novo-engineered transcription activator-like effector (TALE)-type transcription factors. Proc Natl Acad Sci U S A. 2010;107:21617–22. doi: 10.1073/pnas.1013133107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garg A, Lohmueller JJ, Silver PA, Armel TZ. Engineering synthetic TAL effectors with orthogonal target sites. Nucleic Acids Res. 2012;40:7584–95. doi: 10.1093/nar/gks404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reyon D, et al. FLASH assembly of TALENs for high-throughput genome editing. Nat Biotechnol. 2012;30:460–5. doi: 10.1038/nbt.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morbitzer R, Elsaesser J, Hausner J, Lahaye T. Assembly of custom TALE-type DNA binding domains by modular cloning. Nucleic Acids Res. 2011;39:5790–9. doi: 10.1093/nar/gkr151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmid-Burgk JL, et al. Rapid hierarchical assembly of medium-size DNA cassettes. Nucleic Acids Res. 2012;40:e92. doi: 10.1093/nar/gks236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holkers M, et al. Differential integrity of TALE nuclease genes following adenoviral and lentiviral vector gene transfer into human cells. Nucleic Acids Res. 2013;41:e63. doi: 10.1093/nar/gks1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–23. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jinek M, et al. RNA-programmed genome editing in human cells. Elife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–6. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang H, et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–8. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qi LS, et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–83. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gilbert LA, et al. CRISPR-Mediated Modular RNA-Guided Regulation of Transcription in Eukaryotes. Cell. 2013;154:442–51. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maeder ML, et al. CRISPR RNA-guided activation of endogenous human genes. Nat Methods. 2013 doi: 10.1038/nmeth.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mali P, et al. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol. 2013 doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perez-Pinera P, et al. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat Methods. 2013 doi: 10.1038/nmeth.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wehr MC, et al. Monitoring regulated protein-protein interactions using split TEV. Nat Methods. 2006;3:985–93. doi: 10.1038/nmeth967. [DOI] [PubMed] [Google Scholar]
- 29.Xie Z, Wroblewska L, Prochazka L, Weiss R, Benenson Y. Multi-input RNAi-based logic circuit for identification of specific cancer cells. Science. 2011;333:1307–11. doi: 10.1126/science.1205527. [DOI] [PubMed] [Google Scholar]
- 30.Li Y, Moore R, Guinn M, Bleris L. Transcription activator-like effector hybrids for conditional control and rewiring of chromosomal transgene expression. Sci Rep. 2012;2:897. doi: 10.1038/srep00897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Whitfield TW, et al. Functional analysis of transcription factor binding sites in human promoters. Genome Biol. 2012;13:R50. doi: 10.1186/gb-2012-13-9-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deans TL, Cantor CR, Collins JJ. A tunable genetic switch based on RNAi and repressor proteins for regulating gene expression in mammalian cells. Cell. 2007;130:363–72. doi: 10.1016/j.cell.2007.05.045. [DOI] [PubMed] [Google Scholar]
- 33.Kemmer C, et al. Self-sufficient control of urate homeostasis in mice by a synthetic circuit. Nat Biotechnol. 2010;28:355–60. doi: 10.1038/nbt.1617. [DOI] [PubMed] [Google Scholar]
- 34.Leisner M, Bleris L, Lohmueller J, Xie Z, Benenson Y. Rationally designed logic integration of regulatory signals in mammalian cells. Nat Nanotechnol. 2010;5:666–70. doi: 10.1038/nnano.2010.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ye H, et al. Pharmaceutically controlled designer circuit for the treatment of the metabolic syndrome. Proc Natl Acad Sci U S A. 2013;110:141–6. doi: 10.1073/pnas.1216801110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Polstein LR, Gersbach CA. Light-inducible spatiotemporal control of gene activation by customizable zinc finger transcription factors. J Am Chem Soc. 2012;134:16480–3. doi: 10.1021/ja3065667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Konermann S, et al. Optical control of mammalian endogenous transcription and epigenetic states. Nature. 2013;500:472–6. doi: 10.1038/nature12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Crefcoeur RP, Yin R, Ulm R, Halazonetis TD. Ultraviolet-B-mediated induction of protein-protein interactions in mammalian cells. Nat Commun. 2013;4:1779. doi: 10.1038/ncomms2800. [DOI] [PubMed] [Google Scholar]
- 39.Muller K, et al. Multi-chromatic control of mammalian gene expression and signaling. Nucleic Acids Res. 2013;41:e124. doi: 10.1093/nar/gkt340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kramer BP, Fischer C, Fussenegger M. BioLogic gates enable logical transcription control in mammalian cells. Biotechnol Bioeng. 2004;87:478–84. doi: 10.1002/bit.20142. [DOI] [PubMed] [Google Scholar]
- 41.Lohmueller JJ, Armel TZ, Silver PA. A tunable zinc finger-based framework for Boolean logic computation in mammalian cells. Nucleic Acids Res. 2012;40:5180–7. doi: 10.1093/nar/gks142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Auslander S, Auslander D, Muller M, Wieland M, Fussenegger M. Programmable single-cell mammalian biocomputers. Nature. 2012;487:123–7. doi: 10.1038/nature11149. [DOI] [PubMed] [Google Scholar]
- 43.Lienert F, et al. Two- and three-input TALE-based AND logic computation in embryonic stem cells. Nucleic Acids Res. 2013 doi: 10.1093/nar/gkt758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Burrill DR, Inniss MC, Boyle PM, Silver PA. Synthetic memory circuits for tracking human cell fate. Genes Dev. 2012;26:1486–97. doi: 10.1101/gad.189035.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weber W, et al. A synthetic time-delay circuit in mammalian cells and mice. Proc Natl Acad Sci U S A. 2007;104:2643–8. doi: 10.1073/pnas.0606398104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tigges M, Marquez-Lago TT, Stelling J, Fussenegger M. A tunable synthetic mammalian oscillator. Nature. 2009;457:309–12. doi: 10.1038/nature07616. [DOI] [PubMed] [Google Scholar]
- 47.Liang JC, Bloom RJ, Smolke CD. Engineering biological systems with synthetic RNA molecules. Mol Cell. 2011;43:915–26. doi: 10.1016/j.molcel.2011.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Win MN, Smolke CD. Higher-order cellular information processing with synthetic RNA devices. Science. 2008;322:456–60. doi: 10.1126/science.1160311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Culler SJ, Hoff KG, Smolke CD. Reprogramming cellular behavior with RNA controllers responsive to endogenous proteins. Science. 2010;330:1251–5. doi: 10.1126/science.1192128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rinaudo K, et al. A universal RNAi-based logic evaluator that operates in mammalian cells. Nat Biotechnol. 2007;25:795–801. doi: 10.1038/nbt1307. [DOI] [PubMed] [Google Scholar]
- 51.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–22. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 52.Yen HC, Xu Q, Chou DM, Zhao Z, Elledge SJ. Global protein stability profiling in mammalian cells. Science. 2008;322:918–23. doi: 10.1126/science.1160489. [DOI] [PubMed] [Google Scholar]
- 53.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–79. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 54.Nishimura K, Fukagawa T, Takisawa H, Kakimoto T, Kanemaki M. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat Methods. 2009;6:917–22. doi: 10.1038/nmeth.1401. [DOI] [PubMed] [Google Scholar]
- 55.Bonger KM, Chen LC, Liu CW, Wandless TJ. Small-molecule displacement of a cryptic degron causes conditional protein degradation. Nat Chem Biol. 2011;7:531–7. doi: 10.1038/nchembio.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Banaszynski LA, Chen LC, Maynard-Smith LA, Ooi AG, Wandless TJ. A rapid, reversible, and tunable method to regulate protein function in living cells using synthetic small molecules. Cell. 2006;126:995–1004. doi: 10.1016/j.cell.2006.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Renicke C, Schuster D, Usherenko S, Essen LO, Taxis C. A LOV2 domain-based optogenetic tool to control protein degradation and cellular function. Chem Biol. 2013;20:619–26. doi: 10.1016/j.chembiol.2013.03.005. [DOI] [PubMed] [Google Scholar]
- 58.Dong S, Rogan SC, Roth BL. Directed molecular evolution of DREADDs: a generic approach to creating next-generation RASSLs. Nat Protoc. 2010;5:561–73. doi: 10.1038/nprot.2009.239. [DOI] [PubMed] [Google Scholar]
- 59.Struhl G, Adachi A. Nuclear access and action of notch in vivo. Cell. 1998;93:649–60. doi: 10.1016/s0092-8674(00)81193-9. [DOI] [PubMed] [Google Scholar]
- 60.Barnea G, et al. The genetic design of signaling cascades to record receptor activation. Proc Natl Acad Sci U S A. 2008;105:64–9. doi: 10.1073/pnas.0710487105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Howard PL, Chia MC, Del Rizzo S, Liu FF, Pawson T. Redirecting tyrosine kinase signaling to an apoptotic caspase pathway through chimeric adaptor proteins. Proc Natl Acad Sci U S A. 2003;100:11267–72. doi: 10.1073/pnas.1934711100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lim WA. Designing customized cell signalling circuits. Nat Rev Mol Cell Biol. 2010;11:393–403. doi: 10.1038/nrm2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kapp GT, et al. Control of protein signaling using a computationally designed GTPase/GEF orthogonal pair. Proc Natl Acad Sci U S A. 2012;109:5277–82. doi: 10.1073/pnas.1114487109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Reinke AW, Grant RA, Keating AE. A synthetic coiled-coil interactome provides heterospecific modules for molecular engineering. J Am Chem Soc. 2010;132:6025–31. doi: 10.1021/ja907617a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mootz HD, Blum ES, Tyszkiewicz AB, Muir TW. Conditional protein splicing: a new tool to control protein structure and function in vitro and in vivo. J Am Chem Soc. 2003;125:10561–9. doi: 10.1021/ja0362813. [DOI] [PubMed] [Google Scholar]
- 66.Berrade L, Kwon Y, Camarero JA. Photomodulation of protein trans-splicing through backbone photocaging of the DnaE split intein. Chembiochem. 2010;11:1368–72. doi: 10.1002/cbic.201000157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Selgrade DF, Lohmueller JJ, Lienert F, Silver PA. Protein Scaffold-Activated Protein Trans-Splicing in Mammalian Cells. J Am Chem Soc. 2013 doi: 10.1021/ja401689b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wend S, et al. Optogenetic Control of Protein Kinase Activity in Mammalian Cells. ACS Synth Biol. 2013 doi: 10.1021/sb400090s. [DOI] [PubMed] [Google Scholar]
- 69.Bugaj LJ, Choksi AT, Mesuda CK, Kane RS, Schaffer DV. Optogenetic protein clustering and signaling activation in mammalian cells. Nat Methods. 2013;10:249–52. doi: 10.1038/nmeth.2360. [DOI] [PubMed] [Google Scholar]
- 70.Garcia-Otin AL, Guillou F. Mammalian genome targeting using site-specific recombinases. Front Biosci. 2006;11:1108–36. doi: 10.2741/1867. [DOI] [PubMed] [Google Scholar]
- 71.Taniguchi H, et al. A resource of Cre driver lines for genetic targeting of GABAergic neurons in cerebral cortex. Neuron. 2011;71:995–1013. doi: 10.1016/j.neuron.2011.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hirrlinger J, et al. Split-cre complementation indicates coincident activity of different genes in vivo. PLoS One. 2009;4:e4286. doi: 10.1371/journal.pone.0004286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang P, et al. Intersectional Cre driver lines generated using split-intein mediated split-Cre reconstitution. Sci Rep. 2012;2:497. doi: 10.1038/srep00497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bibikova M, Beumer K, Trautman JK, Carroll D. Enhancing gene targeting with designed zinc finger nucleases. Science. 2003;300:764. doi: 10.1126/science.1079512. [DOI] [PubMed] [Google Scholar]
- 75.Porteus MH, Baltimore D. Chimeric nucleases stimulate gene targeting in human cells. Science. 2003;300:763. doi: 10.1126/science.1078395. [DOI] [PubMed] [Google Scholar]
- 76.Christian M, et al. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 2010;186:757–61. doi: 10.1534/genetics.110.120717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fu Y, et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013;31:822–6. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang HH, et al. Programming cells by multiplex genome engineering and accelerated evolution. Nature. 2009;460:894–8. doi: 10.1038/nature08187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rios X, et al. Stable gene targeting in human cells using single-strand oligonucleotides with modified bases. PLoS One. 2012;7:e36697. doi: 10.1371/journal.pone.0036697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gibson DG, Smith HO, Hutchison CA, 3rd, Venter JC, Merryman C. Chemical synthesis of the mouse mitochondrial genome. Nat Methods. 2010;7:901–3. doi: 10.1038/nmeth.1515. [DOI] [PubMed] [Google Scholar]
- 81.Dymond JS, et al. Synthetic chromosome arms function in yeast and generate phenotypic diversity by design. Nature. 2011;477:471–6. doi: 10.1038/nature10403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schlabach MR, Hu JK, Li M, Elledge SJ. Synthetic design of strong promoters. Proc Natl Acad Sci U S A. 2010;107:2538–43. doi: 10.1073/pnas.0914803107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Patwardhan RP, et al. High-resolution analysis of DNA regulatory elements by synthetic saturation mutagenesis. Nat Biotechnol. 2009;27:1173–5. doi: 10.1038/nbt.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kheradpour P, et al. Systematic dissection of regulatory motifs in 2000 predicted human enhancers using a massively parallel reporter assay. Genome Res. 2013;23:800–11. doi: 10.1101/gr.144899.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Melnikov A, et al. Systematic dissection and optimization of inducible enhancers in human cells using a massively parallel reporter assay. Nat Biotechnol. 2012;30:271–7. doi: 10.1038/nbt.2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Patwardhan RP, et al. Massively parallel functional dissection of mammalian enhancers in vivo. Nat Biotechnol. 2012;30:265–70. doi: 10.1038/nbt.2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Smith RP, et al. Massively parallel decoding of mammalian regulatory sequences supports a flexible organizational model. Nat Genet. 2013;45:1021–8. doi: 10.1038/ng.2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Xu GL, Bestor TH. Cytosine methylation targetted to pre-determined sequences. Nat Genet. 1997;17:376–8. doi: 10.1038/ng1297-376. [DOI] [PubMed] [Google Scholar]
- 89.Maeder ML, et al. Targeted DNA demethylation and activation of endogenous genes using programmable TALE-TET1 fusion proteins. Nat Biotechnol. 2013 doi: 10.1038/nbt.2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mendenhall EM, et al. Locus-specific editing of histone modifications at endogenous enhancers. Nat Biotechnol. 2013 doi: 10.1038/nbt.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hathaway NA, et al. Dynamics and memory of heterochromatin in living cells. Cell. 2012;149:1447–60. doi: 10.1016/j.cell.2012.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cantone I, et al. A yeast synthetic network for in vivo assessment of reverse-engineering and modeling approaches. Cell. 2009;137:172–81. doi: 10.1016/j.cell.2009.01.055. [DOI] [PubMed] [Google Scholar]
- 93.Kang T, et al. Reverse engineering validation using a benchmark synthetic gene circuit in human cells. ACS Synth Biol. 2013;2:255–62. doi: 10.1021/sb300093y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shen-Orr SS, Milo R, Mangan S, Alon U. Network motifs in the transcriptional regulation network of Escherichia coli. Nat Genet. 2002;31:64–8. doi: 10.1038/ng881. [DOI] [PubMed] [Google Scholar]
- 95.Gerstein MB, et al. Architecture of the human regulatory network derived from ENCODE data. Nature. 2012;489:91–100. doi: 10.1038/nature11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kramer BP, Fussenegger M. Hysteresis in a synthetic mammalian gene network. Proc Natl Acad Sci U S A. 2005;102:9517–22. doi: 10.1073/pnas.0500345102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bleris L, et al. Synthetic incoherent feedforward circuits show adaptation to the amount of their genetic template. Mol Syst Biol. 2011;7:519. doi: 10.1038/msb.2011.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pomerening JR, Kim SY, Ferrell JE., Jr Systems-level dissection of the cell-cycle oscillator: bypassing positive feedback produces damped oscillations. Cell. 2005;122:565–78. doi: 10.1016/j.cell.2005.06.016. [DOI] [PubMed] [Google Scholar]
- 99.Riccione KA, Smith RP, Lee AJ, You L. A synthetic biology approach to understanding cellular information processing. ACS Synth Biol. 2012;1:389–402. doi: 10.1021/sb300044r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.James JR, Vale RD. Biophysical mechanism of T-cell receptor triggering in a reconstituted system. Nature. 2012;487:64–9. doi: 10.1038/nature11220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.O’Shaughnessy EC, Palani S, Collins JJ, Sarkar CA. Tunable signal processing in synthetic MAP kinase cascades. Cell. 2011;144:119–31. doi: 10.1016/j.cell.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gaudet D, et al. Efficacy and long-term safety of alipogene tiparvovec (AAV1-LPLS447X) gene therapy for lipoprotein lipase deficiency: an open-label trial. Gene Ther. 2013;20:361–9. doi: 10.1038/gt.2012.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dalkara D, et al. In vivo-directed evolution of a new adeno-associated virus for therapeutic outer retinal gene delivery from the vitreous. Sci Transl Med. 2013;5:189ra76. doi: 10.1126/scitranslmed.3005708. [DOI] [PubMed] [Google Scholar]
- 104.Aiuti A, et al. Lentiviral Hematopoietic Stem Cell Gene Therapy in Patients with Wiskott-Aldrich Syndrome. Science. 2013 doi: 10.1126/science.1233151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Biffi A, et al. Lentiviral Hematopoietic Stem Cell Gene Therapy Benefits Metachromatic Leukodystrophy. Science. 2013 doi: 10.1126/science.1233158. [DOI] [PubMed] [Google Scholar]
- 106.Perez EE, et al. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat Biotechnol. 2008;26:808–16. doi: 10.1038/nbt1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gross G, Waks T, Eshhar Z. Expression of immunoglobulin-T-cell receptor chimeric molecules as functional receptors with antibody-type specificity. Proc Natl Acad Sci U S A. 1989;86:10024–8. doi: 10.1073/pnas.86.24.10024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Brentjens RJ, et al. Safety and persistence of adoptively transferred autologous CD19-targeted T cells in patients with relapsed or chemotherapy refractory B-cell leukemias. Blood. 2011;118:4817–28. doi: 10.1182/blood-2011-04-348540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kloss CC, Condomines M, Cartellieri M, Bachmann M, Sadelain M. Combinatorial antigen recognition with balanced signaling promotes selective tumor eradication by engineered T cells. Nat Biotechnol. 2013;31:71–5. doi: 10.1038/nbt.2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lanitis E. Chimeric Antigen Receptor T Cells with Dissociated Signaling Domains Exhibit Focused Antitumor Activity with Reduced Potential for Toxicity In Vivo. Cancer Immunology Research. 2013 doi: 10.1158/2326-6066.CIR-13-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chen YY, Jensen MC, Smolke CD. Genetic control of mammalian T-cell proliferation with synthetic RNA regulatory systems. Proc Natl Acad Sci U S A. 2010;107:8531–6. doi: 10.1073/pnas.1001721107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wei P, et al. Bacterial virulence proteins as tools to rewire kinase pathways in yeast and immune cells. Nature. 2012;488:384–8. doi: 10.1038/nature11259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Folcher M, Fussenegger M. Synthetic biology advancing clinical applications. Curr Opin Chem Biol. 2012;16:345–54. doi: 10.1016/j.cbpa.2012.06.008. [DOI] [PubMed] [Google Scholar]
- 114.Lim F, Sun AM. Microencapsulated islets as bioartificial endocrine pancreas. Science. 1980;210:908–10. doi: 10.1126/science.6776628. [DOI] [PubMed] [Google Scholar]
- 115.Ye H, Daoud-El Baba M, Peng RW, Fussenegger M. A synthetic optogenetic transcription device enhances blood-glucose homeostasis in mice. Science. 2011;332:1565–8. doi: 10.1126/science.1203535. [DOI] [PubMed] [Google Scholar]
- 116.Ortiz-Sanchez E, Helguera G, Daniels TR, Penichet ML. Antibody-cytokine fusion proteins: applications in cancer therapy. Expert Opin Biol Ther. 2008;8:609–32. doi: 10.1517/14712598.8.5.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Byrne H, Conroy PJ, Whisstock JC, O’Kennedy RJ. A tale of two specificities: bispecific antibodies for therapeutic and diagnostic applications. Trends Biotechnol. 2013 doi: 10.1016/j.tibtech.2013.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Cironi P, Swinburne IA, Silver PA. Enhancement of cell type specificity by quantitative modulation of a chimeric ligand. J Biol Chem. 2008;283:8469–76. doi: 10.1074/jbc.M708502200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Taylor ND, Way JC, Silver PA, Cironi P. Anti-glycophorin single-chain Fv fusion to low-affinity mutant erythropoietin improves red blood cell-lineage specificity. Protein Eng Des Sel. 2010;23:251–60. doi: 10.1093/protein/gzp085. [DOI] [PubMed] [Google Scholar]
- 120.Zelphati O, et al. Intracellular delivery of proteins with a new lipid-mediated delivery system. J Biol Chem. 2001;276:35103–10. doi: 10.1074/jbc.M104920200. [DOI] [PubMed] [Google Scholar]
- 121.Akinc A, et al. Targeted delivery of RNAi therapeutics with endogenous and exogenous ligand-based mechanisms. Mol Ther. 2010;18:1357–64. doi: 10.1038/mt.2010.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Akinc A, et al. A combinatorial library of lipid-like materials for delivery of RNAi therapeutics. Nat Biotechnol. 2008;26:561–9. doi: 10.1038/nbt1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kam NW, Liu Z, Dai H. Functionalization of carbon nanotubes via cleavable disulfide bonds for efficient intracellular delivery of siRNA and potent gene silencing. J Am Chem Soc. 2005;127:12492–3. doi: 10.1021/ja053962k. [DOI] [PubMed] [Google Scholar]
- 124.Hasadsri L, Kreuter J, Hattori H, Iwasaki T, George JM. Functional protein delivery into neurons using polymeric nanoparticles. J Biol Chem. 2009;284:6972–81. doi: 10.1074/jbc.M805956200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lee SK, Han MS, Asokan S, Tung CH. Effective gene silencing by multilayered siRNA-coated gold nanoparticles. Small. 2011;7:364–70. doi: 10.1002/smll.201001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Nishina K, et al. Efficient in vivo delivery of siRNA to the liver by conjugation of alpha-tocopherol. Mol Ther. 2008;16:734–40. doi: 10.1038/mt.2008.14. [DOI] [PubMed] [Google Scholar]
- 127.Rizk SS, et al. An engineered substance P variant for receptor-mediated delivery of synthetic antibodies into tumor cells. Proc Natl Acad Sci U S A. 2009;106:11011–5. doi: 10.1073/pnas.0904907106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Bidwell GL, 3rd, Raucher D. Cell penetrating elastin-like polypeptides for therapeutic peptide delivery. Adv Drug Deliv Rev. 2010;62:1486–96. doi: 10.1016/j.addr.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Thompson DB, Cronican JJ, Liu DR. Engineering and identifying supercharged proteins for macromolecule delivery into mammalian cells. Methods Enzymol. 2012;503:293–319. doi: 10.1016/B978-0-12-396962-0.00012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Cronican JJ, et al. A class of human proteins that deliver functional proteins into mammalian cells in vitro and in vivo. Chem Biol. 2011;18:833–8. doi: 10.1016/j.chembiol.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Dormitzer PR, et al. Synthetic generation of influenza vaccine viruses for rapid response to pandemics. Sci Transl Med. 2013;5:185ra68. doi: 10.1126/scitranslmed.3006368. [DOI] [PubMed] [Google Scholar]
- 132.Coleman JR, et al. Virus attenuation by genome-scale changes in codon pair bias. Science. 2008;320:1784–7. doi: 10.1126/science.1155761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Mueller S, et al. Live attenuated influenza virus vaccines by computer-aided rational design. Nat Biotechnol. 2010;28:723–6. doi: 10.1038/nbt.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Geall AJ, Mandl CW, Ulmer JB. RNA: The new revolution in nucleic acid vaccines. Semin Immunol. 2013;25:152–9. doi: 10.1016/j.smim.2013.05.001. [DOI] [PubMed] [Google Scholar]
- 135.Boudreau JE, Bonehill A, Thielemans K, Wan Y. Engineering dendritic cells to enhance cancer immunotherapy. Mol Ther. 2011;19:841–53. doi: 10.1038/mt.2011.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Huh D, et al. A human disease model of drug toxicity-induced pulmonary edema in a lung-on-a-chip microdevice. Sci Transl Med. 2012;4:159ra147. doi: 10.1126/scitranslmed.3004249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Greber D, Fussenegger M. An engineered mammalian band-pass network. Nucleic Acids Res. 2010;38:e174. doi: 10.1093/nar/gkq671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Galloway KE, Franco E, Smolke CD. Dynamically Reshaping Signaling Networks to Program Cell Fate via Genetic Controllers. Science. 2013 doi: 10.1126/science.1235005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Nissim L, Bar-Ziv RH. A tunable dual-promoter integrator for targeting of cancer cells. Mol Syst Biol. 2010;6:444. doi: 10.1038/msb.2010.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Papapetrou EP, et al. Genomic safe harbors permit high beta-globin transgene expression in thalassemia induced pluripotent stem cells. Nat Biotechnol. 2011;29:73–8. doi: 10.1038/nbt.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Themeli M, et al. Generation of tumor-targeted human T lymphocytes from induced pluripotent stem cells for cancer therapy. Nat Biotechnol. 2013 doi: 10.1038/nbt.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Carr PA, Church GM. Genome engineering. Nat Biotechnol. 2009;27:1151–62. doi: 10.1038/nbt.1590. [DOI] [PubMed] [Google Scholar]
- 143.Gibson DG, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–5. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 144.Guye P, Li Y, Wroblewska L, Duportet X, Weiss R. Rapid, modular and reliable construction of complex mammalian gene circuits. Nucleic Acids Res. 2013 doi: 10.1093/nar/gkt605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Torella JP, et al. Rapid construction of insulated genetic circuits via synthetic sequence-guided isothermal assembly. Nucleic Acids Res. 2013 doi: 10.1093/nar/gkt860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Pathak GP, Vrana JD, Tucker CL. Optogenetic control of cell function using engineered photoreceptors. Biol Cell. 2013;105:59–72. doi: 10.1111/boc.201200056. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.