Abstract
Objective
To evaluate the ability of Epithelial-to-mesenchymal transition (EMT)-related microRNAs (miRNAs) as serum biomarkers for prognosis and prediction of metastasis in colorectal cancer (CRC) patients.
Background
EMT-related miRNAs drive CRC progression and metastasis. However, their potential as serum biomarkers in CRC has not been studied.
Methods
This was a three-phase study using 446 colorectal specimens. In the first phase, we selected candidate miRNAs associated with metastasis by analyzing the expression of four miR-200 family members (miR-200b, -200c, -141 and -429) in serum samples from 12 stage I and IV CRC patients. The second phase involved independent validation of candidate miRNAs in serum from 182 CRC patients and 24 controls. Lastly, we analyzed expression in matched 156 tumor tissues from 182 CRC patients, as well as an independent set of 20 matched primary CRC and corresponding liver metastases to identify source of circulating miRNAs.
RESULTS
Following initial screening, miR-200c was selected as the candidate serum miRNA best associated with metastasis. Validation analysis revealed that serum miR-200c levels were significantly higher in stage IV compared with stage I–III CRCs. High serum miR-200c demonstrated a significant positive correlation with lymph node, distant metastasis and prognosis (P=0.0026, P=0.0023 and P=0.0064, respectively). More importantly, serum miR-200c was an independent predictor for lymph node metastasis (OR=4.81, 95% CI=1.98–11.7 P=0.0005), tumor recurrence (HR=4.51, 95% CI=1.56–13.01 P=0.005) and emerged as an independent prognostic marker for CRC (HR: 2.67, 95%CI: 1.28–5.67, P=0.01).
CONCLUSION
Serum miR-200c has a strong potential to serve as a noninvasive biomarker for CRC prognosis and predicting metastasis.
Keywords: MiR-200c, EMT, colorectal cancer, prognostic marker, noninvasive biomarker
Introduction
Colorectal cancer (CRC) is one of the most common malignancies worldwide, and is a major cause of cancer-related deaths1. Survival rates of patients with CRC have increased in the past few years, possibly as a result of earlier diagnosis and improved treatment regimens, nonetheless, approximately 30–50% of patients who undergo curative resection subsequently experience local tumor recurrence or metastasis2. This subgroup of patients usually receive chemotherapy often in combination with monoclonal antibody therapy, with a median overall survival duration of ~20 months, and the response rates at best around 50%3. However, the substantial financial costs associated with CRC treatment not only present an economic burden, but treatment of all patients with chemotherapy without a priori selection leads to overtreatment of patients with toxic agents that produce severe adverse effects4. In order to overcome this clinical challenge, there is a clear need to identify biomarkers that will facilitate the identification of patients with a poor prognosis, and permit personalized treatment strategies for patients with high risk of CRC recurrence.
Blood-based tumor markers are gaining acceptance as a potential alternative for noninvasive detection of cancer. Serum carcinoembryonic antigen (CEA) is one marker that is frequently used for predicting prognosis in patients with CRC5, 6. Unfortunately, CEA levels do not always correlate with the presence of metastasis, and the incidence of false-positive and false-negative results are very high7, 8. Consequently, there is a dire need to identify highly robust biomarkers that can clinically determine cancer prognosis, and are better indicators of patient outcome than the existing TNM staging system or other conventional tumor markers of CRC9.
MicroRNAs (miRNAs) are non-coding RNA molecules of approximately 21–23 nucleotides in length that regulate target gene expression by interfering with their transcription or by inhibiting translation10. miRNAs play crucial roles in diverse cellular biological processes, including differentiation, proliferation, growth, migration and survival. The discovery that miRNA expression is frequently dysregulated in malignant tumors underpins their critical role, which is a matter of active investigation, both from a basic science perspective and for its clinical usefulness11. Recently, several studies have highlighted the diagnostic and prognostic utility of plasma and serum-based miRNA levels, because tumor-derived miRNAs are present in human circulation in remarkably stable forms that are protected from endogenous ribonuclease activity12. These reports suggest that plasma/serum miRNA-based assays may constitute accurate methods for diagnosis and prognosis of human cancer, although to date only a few studies have specifically addressed the clinical significance of circulating miRNAs in patients with CRC13–17.
Epithelial-to-mesenchymal transition (EMT) manifests through downregulation of E-cadherin and successive loss of cell-cell adhesion, leading to a mesenchymal phenotype18. This contributes to accelerated invasiveness, dissemination and metastasis of epithelial tumor cells in several carcinomas, including CRC19–21. The miR-200 family (miR-200a, miR-200b, miR-200c, miR-141, and miR-429) inhibits the E-cadherin-suppressor targets such as zinc finger E-box binding homeobox-1 (ZEB1) and 02 (ZEB2), which are important initiators of EMT in CRC22, 23. In addition, we have recently reported that dysregulated expression of miR-200b, -200c, -141 and -429 is responsible for EMT-MET switch in colorectal metastasis. In this study, we for the first time demonstrated that miR-200c/429 cluster was significantly over-expressed in liver metastasis compared to primary colorectal cancer, and the expression of these miRNAs was specifically regulated by aberrant methylation of their promoter regions.24. In spite of their involvement in metastasis, none of the previous studies has explored the clinical significance of miR-200 family expression in serum of patients with CRC.
In view of these limitations in the current literature, we focused this study on the expression analysis of miR-200 family (miR-200b, miR-200c, miR-141 and miR-429) in the serum of CRC patients, by adopting a three-step approach. First, in screening a subset of samples, we selected candidate miRNAs that were associated with metastasis by comparing expression levels in the serum from stage I and stage IV CRC patients. In the second phase, using a larger and independent cohort of serum specimens from CRC patients and healthy controls, we validated the clinical significance of selected miRNAs as potential noninvasive biomarkers for predicting metastasis, tumor recurrence or the prognosis of CRC patients. Finally, we investigated the expression of selected miRNAs in primary CRC and distant metastasis tissues in an effort to identify the origin of these miRNAs in serum. Using this systematic approach, we demonstrate that serum levels of miR-200c, which is a bona fide EMT-related miRNA, are not only significantly associated with a metastatic phenotype in the colon, but also serve as a potential biomarker for predicting lymph node metastasis, tumor recurrence and prognosis in CRC patients.
Methods
Study design and clinical specimens
This study included analysis of 446 colorectal specimens that which were obtained at Mie University Medical Hospital, Mie prefecture in Japan between 2005 and 2011. This was a three phase study, which aimed to screen, validate, and determine the potential contribution of serum miRNAs in CRC patients.
During the initial screening phase, we analyzed serum levels of several candidate miR-200 family in a subset of 24 serum samples from stage I (n=12) and stage IV (n=12) CRC patients. In the second phase, candidate miRNAs that were overexpressed in serum of stage IV vs. stage I patients in the initial screening step were further validated in a larger, independent cohort, which included serum samples from 182 CRC patients and 24 normal controls. The final phase aimed to evaluate the potential source of miRNAs in the serum in CRC patients by comparing expression of selected miRNAs in matched surgical FFPE tissues (n=156) from 182 CRC patients and 20 adjacent normal mucosa. In addition, we analyzed an independent set of matched primary CRC specimens (n=20) and their corresponding liver metastasis tissues (n=20) during this step of the study.
Patients treated with radiotherapy or chemotherapy prior to surgery were not included in this study. Patients with stage III and IV disease received 5-fluorouracil-based chemotherapy, whereas no adjuvant therapy was given to stage I and II CRC patients. CEA levels in serum samples were measured by standard enzyme immunoassay as a routine clinical test. Both serum- and tissue-based studies were approved by the Institutional Review Broad (IRB) of the Mie University Hospital, Japan and Baylor University Medical Center, Dallas, USA. All participants gave written consent for their information to be stored in the hospital database and used for research.
RNA isolation from serum and qRT-PCR
Small RNAs were enriched from all serum samples using the Qiagen miRNAeasy Kit (Qiagen, Valencia, CA). Briefly, 250 µL of serum was thawed on ice and centrifuged at 10,000 rpm for 5 minutes to remove cellular debris. Next, 200 µL of supernatant was lysed with five volumes of Qiazol solution. For normalization of sample-to-sample variation during the RNA isolation procedures, 25 fmol of synthetic C. elegans miRNA (cel-miR-39) was added to each denatured sample. Small RNAs were then enriched and purified according to the manufacturer’s protocol, with the exception that the enriched small RNAs were eluted in 40 µL of preheated nuclease-free water. For miRNA-based RT-PCR assays, 1.67 µL of enriched small RNAs from serum samples were reverse-transcribed using the TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems, San Diego, CA) in a total reaction volume of 5.0 µL, according to the manufacturer’s instructions. RT products were diluted 1:15 and used as PCR template. PCR reactions for quantification of miR-200b, miR-200c, miR-141, miR-429 and cel-miR-39 were performed in duplicate using TaqMan 2× Universal PCR Master Mix using conditions described previously25. The qRT-PCR reactions were performed using an Applied Biosystems 7000 Sequence Detection System with the following cycling conditions: 95°C for 10 min, followed by 45 cycles of 95°C for 15s and 60°C for 1 min. The cycle threshold (Ct) values were calculated with SDS 1.4 software (Applied Biosystems, Foster City, CA).
RNA isolation from FFPE tissues and qRT-PCR
Total RNA was isolated from FFPE samples using the RecoverAll Total Nucleic Acid Isolation Kit (Ambion Inc., Austin, Texas, USA). Briefly, tissue sections were microdissected to enrich for neoplastic cells, followed by deparaffinization and RNA extraction using the manufacturer’s protocol. Total RNA was eluted in appropriate buffer, and quantified using a NanoDrop spectrophotometer (NanoDrop Technologies, Wilmington, DE). Reverse transcription reactions were carried out using the TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems, San Diego, CA) in a total reaction volume of 15 µL. MiR-200c and miR-16 were quantified in duplicate by qRT-PCR, using TaqMan MicroRNA Assay Kits (Applied Biosystems, Foster City, CA). qRT-PCR was performed on an Applied Biosystems 7000 Sequence Detection System using the following cycling conditions: 95°C for 10 min, followed by 45 cycles of 95°C for 15s and 60°C for 1 min. Cycle threshold (Ct) values were calculated with SDS 1.4 software (Applied Biosystems, Foster City, CA).
Calculation of miRNA expression
The average expression levels of serum and tissue miRNAs were normalized against cel-miR-3912, 25 and miR-1626, 27 using the 2−ΔCt method. Differences between the groups are presented as ΔCt, indicating the difference between the Ct value of the miRNA of interest and the Ct value of the normalizer miRNA. To ensure consistent measurements throughout all assays, for each PCR amplification reaction, three independent RNA samples were loaded as internal controls to account for any plate to plate variation, and the results from each plate were normalized against internal normalization controls.
In situ hybridization
Five micrometer-thick FFPE tissue sections were hybridized with the miR-200c probe (LNA-modified and 5`- and 3`-DIG-labeled oligonucleotide; Exiqon, Woburn, Massachusetts, USA), followed by incubation with anti–DIG-AP Fab fragments conjugated to alkaline phosphatase, and the hybridization signal was detected by applying nitroblue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate color substrate (Roche Applied Science, Mannheim, Germany). Positive controls (U6 snRNA, LNA-modified and 5`- and 3`-DIG-labeledOligonucleotide; Exiqon) and negative controls (scrambled microRNA control, LNA-modified and 5`- and 3`-DIG-labeled oligonucleotide; Exiqon) were included in each hybridization procedure.
Statistical analysis
The significance of serum and tissue miRNA levels was determined by the Mann–Whitney test, Kruskal-Wallis test or the χ2 test where appropriate. Logistic regression analysis was used to predict the factors influencing lymph node metastasis. Overall and disease free survival curves were analyzed using the Kaplan-Meier method, and differences were examined using Log-rank tests. Cox’s proportional hazard regression test was used to estimate univariate and multivariate hazard ratios for recurrence and prognosis. Receiver operating characteristic (ROC) curves with Youden’s Index correction28 were established for determining optimal miRNA expression cut-off thresholds for analyzing lymph node metastasis prediction, disease free survival and overall survival. All P values were two-sided, and those less than 0.05 were considered statistically significant. All statistical analyses were carried out using Medcalc 7.2 for Windows (Broekstraat 52, 9030, Mariakerke, Belgium).
Results
Serum miR-200c is a candidate miRNA that is associated with CRC metastasis
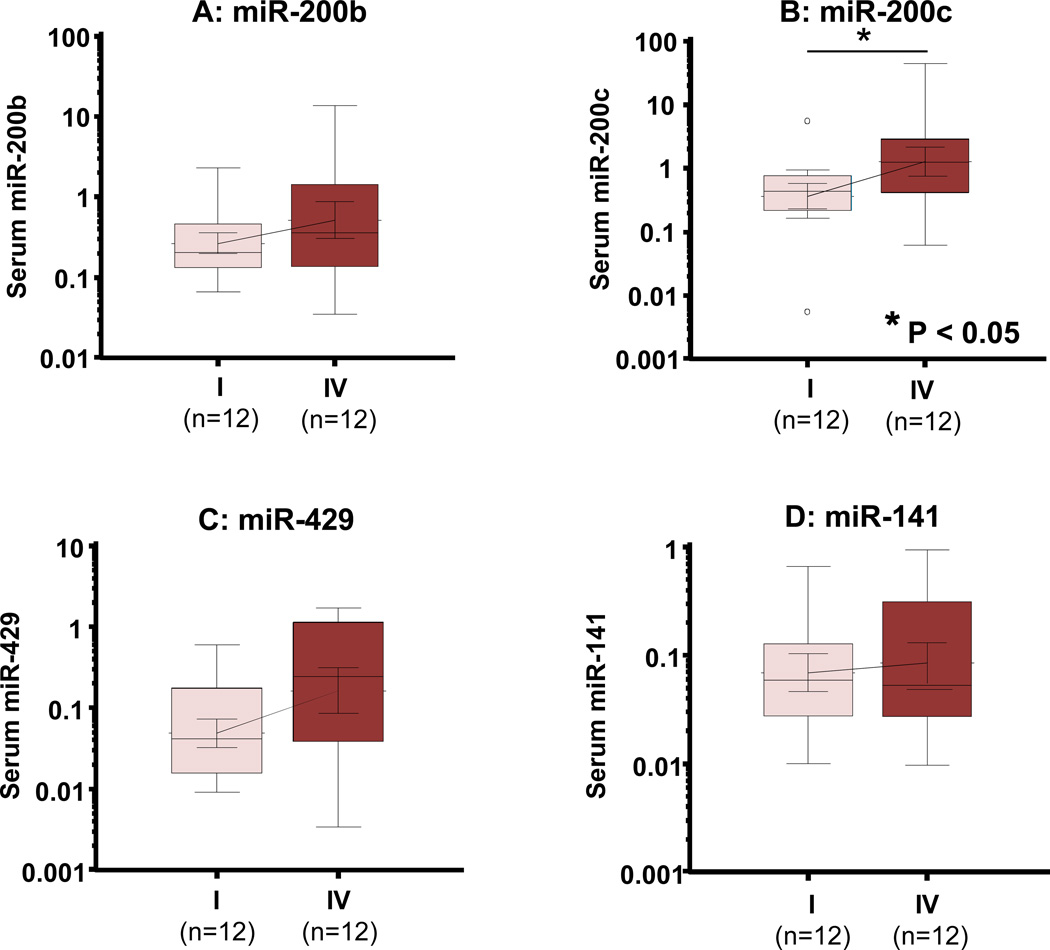
In the initial screening step aimed at identifying metastasis-associated serum miRNA biomarkers as noninvasive prognostic markers, we investigated the relative expression levels of miR-200 family (miR-200b, miR-200c, miR-141 and miR-429) in a subset of serum specimens from 12 stage IV and 12 stage I CRC patients (Table S1). Among all miRNA analyzed (Fig. 1), miR-200c was significantly elevated in the serum of stage IV patients compared to stage I CRC patients (P<0.05; Fig. 1B). In contrast, no significant differences were observed in miR-141, miR-200b and miR-429 expression between stage I and stage IV CRC patients (Fig. 1A, 1C and 1D). Based upon these observations, we subsequently focused on validating and further exploring the clinical significance of miR-200c in an independent set of serum samples from 182 CRC patients. In addition, we also attempted to investigate the potential origin of miR-200c in serum by analyzing matched serum and tumor tissues samples from patients who had -matched primary tissues from 182 CRC patients and an independent set of tissues from 20 pairs of primary CRCs and matched liver metastases.
Figure 1. Expression analysis of miR-200 family members in the serum of stage I and stage IV CRC patients.
Box plots of serum levels of miR-200b (A), miR-200c (B), miR-429 (C) and miR-141 (D) in stage I (n=12) and stage IV (n=12) CRC patients. MiR-200c levels in serum from stage IV patients were significantly higher than that of stage I patients. The boxes represent the interquartile range, and the lines across the boxes indicate the median values. Expression levels of these miRNAs (log10 scale on the y-axis) were normalized to cel-miR-39. Statistical analysis was performed using Mann-Whitney test.
Serum miR-200c expression levels serve as a predictive and prognostic biomarker in CRC patients
Patient characteristics and distribution of tumor stages are summarized in Table S2. There were no significant differences in the mean age between CRC patients (67 ± 10.4 years) and healthy controls (64 ± 12.9 years; P>0.05; ANOVA). The gender distribution in the CRC group was 105:77 (males:females), and in the control group it was 13:11 (P>0.05; Chi-square test). For miR-200c expression analysis in matched primary CRCs tissues and serum, 156 of 182 samples were available from the CRC patient cohort.
Serum miR-200c levels predict lymph node metastasis in patients with CRC
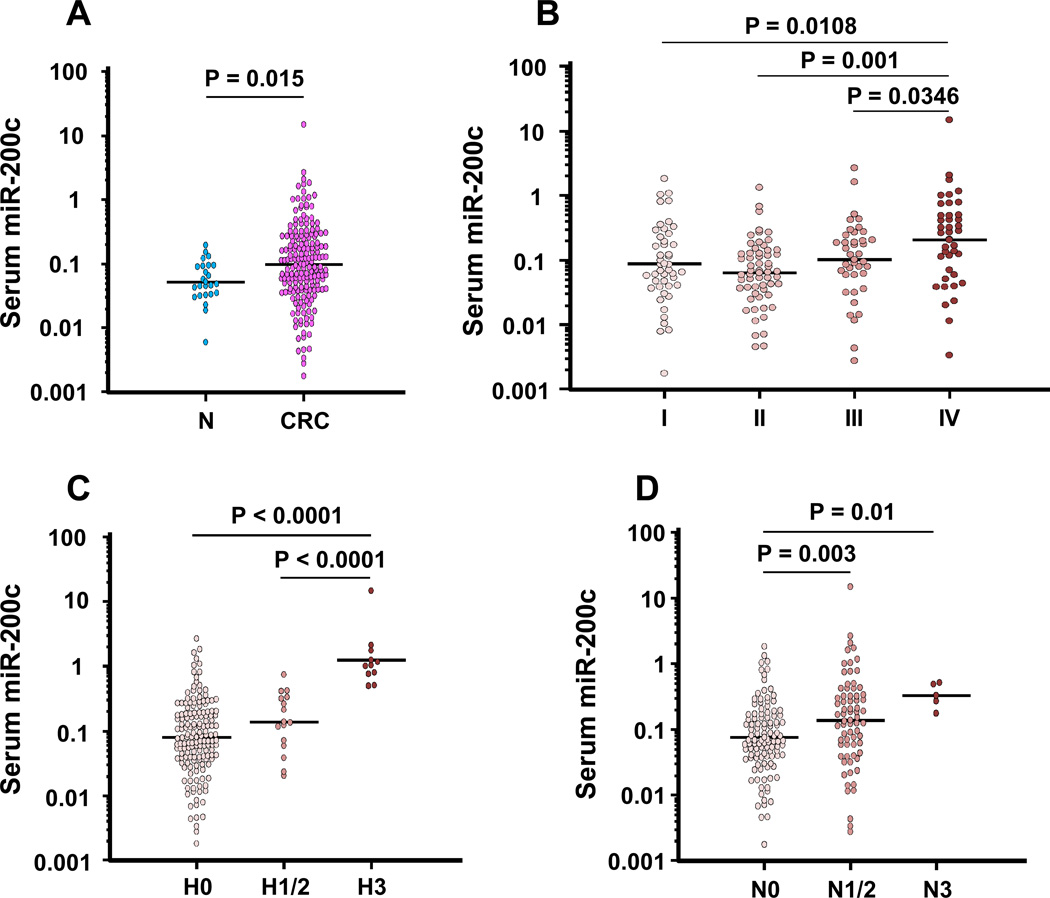
The expression levels of serum miR-200c in CRC were significantly higher compared to that in normal controls (P=0.015; Fig. 2A). Serum miR-200c levels were significantly higher in stage IV patients than in normal controls, and stage I, II and III CRC patients (Fig. 2B). The potential clinical significance of serum miR-200c expression is presented in Table 1. As shown, high expression of serum miR-200c was associated with a metastatic phenotype, including lymph node metastasis (P=0.0026), liver metastasis (P=0.0015) and the development of distant metastases (P=0.0023) in CRC patients.
Figure 2. Validation of miR-200c expression in a validation cohort of CRC patients.
(A) Dot plots of serum miR-200c levels in healthy normal controls (NC) (n=24) and patients with CRC (n=182). (B) Dot plots of serum miR-200c levels across various stages of CRCs. MiR-200c levels in serum from CRC patients were significantly elevated compared with those of normal controls, and the expression levels in stage IV CRC patients were significantly higher than those in stage I–III patients. (C) Dot plots of serum miR-200c levels subdivided by H Stage. (D) Dot plots of serum miR-200c levels subdivided by N Stage. Serum miR-200c increased depending on the higher N and H stages. The lines indicate the mean values. Expression levels of miR-200c (log10 scale on the y-axis) were normalized to cel-miR-39. Statistically significant differences were determined using Mann-Whitney tests and Kruskal-Wallis test.
Table 1.
Association between miR-200c expression in serum and primary CRC and various clinicopathological characteristics
| Serum miR-200c (n =182) | Tissue miR-200c (n=156) | ||||||
|---|---|---|---|---|---|---|---|
| Factors | high (n = 91) |
low (n = 91) |
p value | high (n = 78) |
low (n = 78) |
p value | |
| Age | ≦68 | 43 | 43 | 0.88 | 31 | 43 | 0.078 |
| > 68 | 48 | 48 | 47 | 35 | |||
| Gender | Male | 49 | 56 | 0.36 | 41 | 48 | 0.29 |
| Female | 42 | 35 | 37 | 30 | |||
| Histological grade | well/mod | 83 | 82 | 0.99 | 69 | 71 | 0.79 |
| poor/ mucinous | 8 | 9 | 9 | 7 | |||
| Tumor size | ≦40(small) | 47 | 42 | 0.50 | 38 | 40 | 0.87 |
| > 40(large) | 44 | 49 | 40 | 38 | |||
| Serosal invasion | Absent | 26 | 29 | 0.78 | 27 | 18 | 0.12 |
| Present | 65 | 62 | 51 | 60 | |||
| Lymph node met. | Absent | 43 | 64 | 0.0026 | 49 | 38 | 0.10 |
| Present | 48 | 27 | 29 | 40 | |||
| Venous invasion | Absent | 52 | 53 | 0.99 | 49 | 38 | 0.10 |
| Present | 39 | 38 | 29 | 40 | |||
| Lymphatic invasion | Absent | 20 | 26 | 0.39 | 26 | 10 | 0.0044 |
| Present | 71 | 65 | 52 | 68 | |||
| Liver metastasis | Absent | 70 | 86 | 0.0015 | 71 | 67 | 0.45 |
| Present | 21 | 5 | 7 | 11 | |||
| Peritoneal metastasis | Absent | 83 | 88 | 0.21 | 76 | 70 | 0.10 |
| Present | 8 | 3 | 2 | 8 | |||
| Distant metastasis | Absent | 62 | 80 | 0.0023 | 68 | 59 | 0.099 |
| Present | 29 | 11 | 10 | 19 | |||
well, well differentiated; mod, moderately differentiated; poor, poorly differentiated
We next analyzed miR-200c levels in serum based upon pathological extension of colorectal neoplasia to hepatic or lymph node metastasis. We first determined associations between miR-200c expression and the H-classification of CRCs; with H0 indicating no liver metastasis; H1 representing liver metastasis with less than five nodules smaller than 5 cm; H2 indicative of metastasis that does not involve H1 and H3; and H3 indicating liver metastasis with more than five metastasis larger than 5 cm. The miR-200c levels were significantly higher in H3 CRC patients than those in H0 or H1–2 CRC patients (P<0.0001; Fig. 2C). Likewise, serum expression of miR-200c was significantly higher in patients with lymph node metastasis to the aorta (n3) compared with regional (n1–2: P=0.003; Fig. 2D) or absent lymph node metastasis (n0: P=0.01; Fig. 2D) in CRC patients. Additionally in stage I–III patients, serum miR-200c expression increased in accordance with progression of lymph node metastasis, and miR-200c expression in N3 patients was significantly higher than that in N0 patients (P=0.036; Fig. S1A). Furthermore in stage II–III CRCs, serum miR-200c expression of lymph node-positive patients (stage III) was significantly higher than that of node-negative patients (stage II) (P=0.037; Fig. S1B). Taken together, these results indicate that serum miR-200c levels in CRC could be influenced by tumor volume or its dissemination to lymph node or hepatic metastatic sites.
To further evaluate whether serum miR-200c levels can serve as a predictor of lymph node metastasis, we performed logistic regression analysis. Univariate analysis demonstrated that highly invasive tumors (T3/4; P=0.0024), with lymphatic (P<0.0001) and venous invasion (P=0.0001), high CEA levels (P=0.0001) and high levels of serum miR-200c (P=0.0001) were all significantly associated with lymph node metastasis (Table 2). Furthermore, serum miR-200c expression was an independent predictor of lymph node metastasis in CRC based upon multivariate logistic regression analysis (HR=4.81, 95% CI=1.98–11.7 P=0.0005), suggesting that serum miR-200c as a predictor of lymph node metastasis is superior to pathological findings that are known to be risk factors.
Table 2.
Uni- and multi-variate analyses for predicting lymph node metastasis in CRC patients
| Univariate | Multivariate | |||||
|---|---|---|---|---|---|---|
| Variables | HR$ | 95%CI | p value | HR$ | 95%CI | p value |
| Pathological T stage (T3/4 vs. 1/2) | 6.46 | 2.61–15.96 | 0.0024 | 2.38 | 0.81–7.03 | 0.11 |
| Pathology (poor vs. mod/well differentiated) | 2 | 0.57–6.98 | 0.2715 | 1.39 | 0.38–5.07 | 0.62 |
| Venous Invasion (positive vs. negative) | 4.59 | 2.13–9.89 | 0.0001 | 1.24 | 0.52–2.96 | 0.62 |
| Lymphatic Invasion (positive vs. negative) | 18.26 | 5.43–61.38 | <0.0001 | 6.56 | 1.55–27.8 | 0.010 |
| CEA (≥ 5 vs. <5) | 6.25 | 2.45–15.92 | 0.0001 | 2.44 | 1.02–5.84 | 0.044 |
| miR-200c in serum (high vs. low) | 3.61 | 1.85–7.09 | 0.0001 | 4.81 | 1.98–11.7 | 0.0005 |
HR, Hazard Ratio; CI, Confidence Interval; CEA, Carcinoembryonic antigen; mod, moderately; TNM, tumor-node-metastasis staging
HR, hazard ratio for presence of lymph node metastasis in CRC patients
Serum miR-200c is a prognostic and tumor recurrence predictive biomarker in CRC
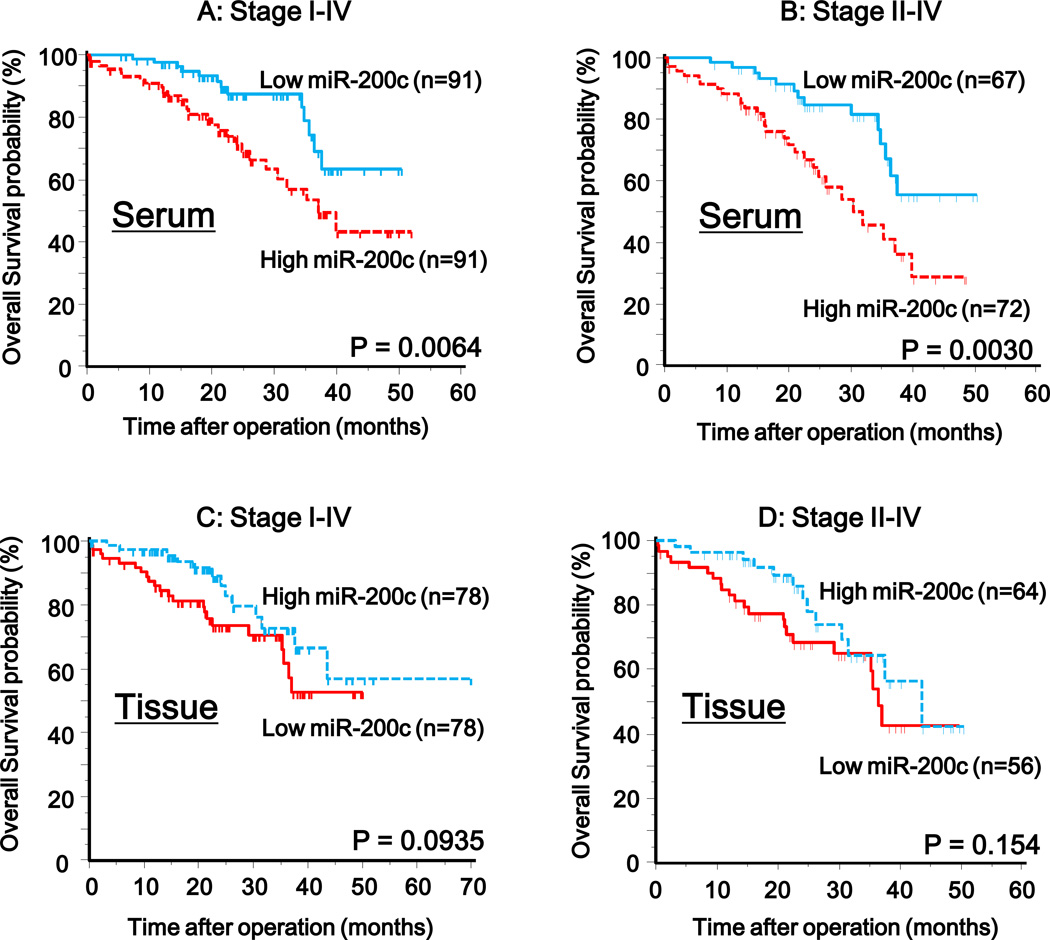
To further evaluate whether serum miR-200c levels can predict CRC prognosis, we next performed survival analysis. Kaplan-Meier analysis showed that patients with higher levels of serum miR-200c had significantly poorer survival than those with lower expression of this miRNA in stage I–IV and II–IV CRC patients, respectively (P=0.0064 and P=0.0030; log-rank test; Fig. 3A, B). To determine whether serum miR-200c expression was an independent risk factor for prognosis, the Cox proportional hazard regression model was employed (Table 3). In univariate analysis, high levels of miR-200c in serum (P=0.006), high levels of CEA (P=0.0001), high pathological T stage (T3/4; P=0.0024), lymph node metastasis (P<0.0001), poor differentiation (P=0.036) and high TNM stage (stage III/IV; P<0.0001) were significantly associated with poor prognosis. On the other hand, multivariate analysis showed that high serum miR-200c expression was an independent prognostic marker for predicting poorer overall survival in CRC patients (HR=2.67, 95% CI=1.28–5.67 P=0.01; Table 3). In addition, patients with high serum miR-200c in stage II but not stage III had shorter disease free survival than those with low serum miR-200c, respectively (P=0.025, log-rank test; Fig. S2A, P=0.11, log-rank test; Fig. S2B). To determine whether serum miR-200c can serve as a predictor for tumor recurrence in curative patients (stage II–III), Cox’s proportional hazard regression model was utilized (Table 4). Univariate analysis showed that venous invasion (positive; P=0.038), lymph node metastasis (P=0.0015) and high serum miR-200c levels (P=0.024) were significantly associated with disease free survival. In contrast, multivariate analysis revealed that high serum miR-200c was an independent predictor for tumor recurrence in stage II–III CRC patients (HR=4.51, 95% CI=1.56–13.01 P=0.005). In addition, high serum miR-200c expression was the only factor that allowed prediction of stage II CRC patients that experienced tumor recurrence (HR=8.43, 95% CI=1.00–72.96 P=0.01; Table 5). Therefore, serum miR-200c levels may not only serve as predictive marker of lymph node metastasis, but also predict poor prognosis and early recurrence in patients with higher accuracy than serum CEA levels or pathological staging.
Figure 3. Kaplan-Meier survival curves of CRC patients subdivided by miR-200c levels in serum and matched primary tumors from CRC patients.
Overall survival rates of stage I–IV (A) and stage II–IV (B) CRC patients with high serum miR-200c levels were significantly lower than for those with low miR-200c expression (stage I–IV, P=0.0064; stage II–IV, P=0.003; Log-rank test). Overall survival rates of stage I–IV (C) and stage II–IV (D) CRC patients with low miR-200c expression in primary CRC were lower than for those with high miR-200c expression (stage I–IV, P=0.0935; stage II–IV, P=0.154; Log-rank test). The miRNA expression cut-off thresholds for miR-200c expression in serum were deduced from the ROC curves with Youden’s Index.
Table 3.
Uni- and multi-variate analyses for prognostic factors in CRC patients
| Univariate | Multivariate | |||||
|---|---|---|---|---|---|---|
| Variables | HR$ | 95%CI | p value | HR$ | 95%CI | p value |
| Age (≥ 67 vs. <67) | 0.76 | 0.42–1.35 | 0.35 | - | - | - |
| Gender (Female vs. Male) | 1.02 | 0.56–1.86 | 0.92 | - | - | - |
| Pathological T stage (T3/4 vs.1/2) | 8.97 | 2.19–36.7 | 0.0024 | 3.63 | 0.82–16.1 | 0.091 |
| Pathology (poor vs. mod/well differentiated) | 2.26 | 1.05–4.84 | 0.036 | 2.07 | 0.86–4.96 | 0.105 |
| Lymph node metastasis (positive vs. negative) | 17.1 | 6.18–47.8 | < 0.0001 | 1.24 | 0.28–5.46 | 0.78 |
| TNM stage (III/IV vs. I/II) | 33.4 | 8.12–136.9 | < 0.0001 | 10.2 | 1.27–81.7 | 0.03 |
| CEA (≥ 5 vs. <5) | 4.84 | 2.15–10.89 | 0.0001 | 1.46 | 0.57–3.74 | 0.43 |
| miR-200c in serum (high vs. low) | 2.43 | 1.26–4.68 | 0.006 | 2.67 | 1.28–5.67 | 0.01 |
| miR-200c in primary tumor (high vs. low) | 0.56 | 0.28–1.10 | 0.092 | - | - | - |
HR, Hazard ratio; CI, Confidence interval; CEA, Carcinoembryonic antigen; mod, moderately; TNM, tumor-node-metastasis staging
HR, hazard ratio for survival outcome in CRC patients
Table 4.
Uni- and multi-variate analyses for predictive factors of recurrence in stage II–III CRC patients
| Univariate | Multivariate | |||||
|---|---|---|---|---|---|---|
| Variables | HR$ | 95%CI | p value | HR$ | 95%CI | p value |
| Age (≥ 67 vs. <67) | 0.89 | 0.40–1.99 | 0.78 | - | - | - |
| Gender (Female vs. Male) | 0.65 | 0.29–1.44 | 0.29 | - | - | - |
| Pathological T stage (T3/4 vs.1/2) | 0.78 | 0.18–3.33 | 0.75 | - | - | - |
| Pathology (poor vs. mod/well differentiated) | 1.29 | 0.39–4.34 | 0.67 | - | - | - |
| Venous invasion (positive vs. negative) | 2.47 | 1.05–5.78 | 0.038 | 3.76 | 1.43–9.93 | 0.008 |
| Lymphatic invasion (positive vs. negative) | 1.92 | 0.45–8.11 | 0.34 | - | - | - |
| TNM stage (III vs. II) | 3.98 | 1.71–9.30 | 0.0015 | 3.24 | 1.32–7.96 | 0.01 |
| CEA (≥ 5 vs. <5) | 1.89 | 0.77–4.62 | 0.1646 | - | - | - |
| miR-200c in serum (high vs. low) | 3.01 | 1.08–8.39 | 0.024 | 4.51 | 1.56–13.01 | 0.005 |
CI, Confidence interval; CEA, Carcinoembryonic antigen; mod, moderately; TNM, tumor-node-metastasis staging
HR, hazard ratio for recurrence in CRC patients
Table 5.
Univariate analyses for predictive factors of recurrence in stage II CRC patients
| Variables | HR$ | 95%CI | p value |
|---|---|---|---|
| Age (≥ 67 vs. <67) | 0.91 | 0.20–4.05 | 0.9 |
| Gender (Female vs. Male) | 1.33 | 0.26–6.81 | 0.73 |
| Pathological T stage (T3/4 vs.1/2) | 0.21 | 0.02–1.85 | 0.16 |
| Pathology (poor vs. mod/well differentiated) | 4.55 | 0.87–23.93 | 0.07 |
| Venous invasion (positive vs. negative) | 2.72 | 0.52–14.21 | 0.24 |
| Lymphatic invasion (positive vs. negative) | 0.38 | 0.09–2.46 | 0.47 |
| CEA (≥ 5 vs. <5) | 1.51 | 0.25–9.00 | 0.65 |
| miR-200c in serum (high vs. low) | 8.43 | 1.00–72.96 | 0.01 |
CI, Confidence interval; CEA, Carcinoembryonic antigen; mod, moderately;
HR, hazard ratio for recurrence in CRC patients
Investigation of miR-200c source in serum of CRC patients
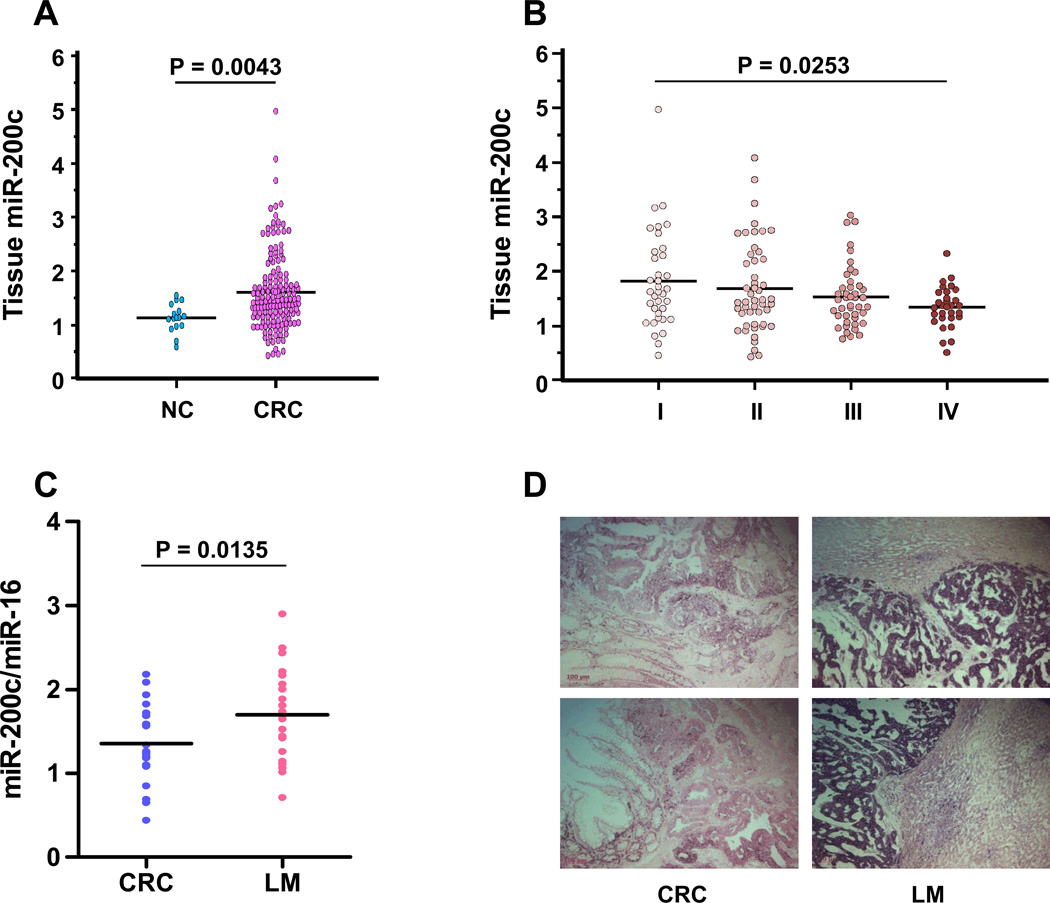
In an effort to determine whether miR-200c levels in serum are of tumor origin, we quantified the expression levels of miR-200c in matched CRC tissues. As expected, miR-200c expression in CRC tissues was significantly higher than in normal colonic mucosa (P=0.0043; Fig. 4A). Surprisingly however, miR-200c levels in CRC tissues gradually decreased with increasing tumor stage and the levels in stage IV CRCs were significantly lower than in stage I CRC (P=0.0253; Fig. 4B), suggesting a lack of direct correlation between serum and matched tissue miR-200c expression. In addition, there were no associations between miR-200c expression in CRC and clinicopathological findings, except for lymphatic invasion (Table 1, Fig. 3C, D). We next analyzed miR-200c expression in both primary CRC and matched liver metastases from 20 independent CRC patients. Of interest, the levels of miR-200c in liver metastases were significantly higher than in primary CRC (P=0.0135; Fig. 4C). Furthermore, we also confirmed miR-200c expression in both primary CRC tumors and matched liver metastases by in situ hybridization, illustrating that miR-200c was highly expressed in liver metastases compared with primary CRC (Fig. 4D). Taken together, we hypothesize that a potential source of miR-200c in serum of CRC patients might be the metastatic sites; hence, serum miR-200c levels may serve as a superior metastasis, recurrence-predictive and prognostic marker of CRC.
Figure 4. Expression of miR-200c in tissues from CRC patients.
(A) Dot plots of miR-200c levels in adjacent normal mucosae (NC) (n=20) and CRC tissues (n=156). (B) Dot plots of miR-200c tissue levels across various CRC stages. (C) Dot plots of miR-200c tissue levels for comparisons between matched primary CRCs (CRC) (n=20) and their corresponding liver metastasis (LM) (n=20). (D) In situ hybridization analysis of miR-200c in matched primary CRCs and their corresponding LM. Representative photomicrographs are shown from 2 primary CRCs (left panels) and matched LM (right panels). MiR-200c expressions in LMs were higher than that in primary CRCs expressed. In contrast, miR-200c expression in normal liver tissues was either very low or absent. Line indicates the mean value. Expression levels of miR-200c were normalized to has-miR-16. Statistically significant differences were determined using Mann-Whitney tests and Kruskal-Wallis test.
Discussion
This study investigates the potential clinical utility of four critical miR-200 family members to serve as noninvasive prognostic and metastasis-predictive biomarkers in CRC patients. To this end, in a small screening subset, we found that serum miR-200c levels in stage IV CRCs were significantly higher than in stage I CRC. Accordingly, we selected miR-200c and subsequently validated it as a candidate serum miRNA with a potential promise as a predictive biomarker for metastasis in CRC. We performed independent validation experiments using a large cohort of samples from 182 CRC patients and 24 control subjects. These data provided strong evidence that serum miR-200c levels in CRC patients were significantly higher than in healthy controls. Furthermore, serum miR-200c levels were significantly higher in stage IV compared to stage I–III CRC patients. Additionally, high serum miR-200c levels were associated with shorter overall survival, and served as an independent prognostic biomarker in CRC patients. Interestingly, instead of lymph node metastasis, serum miR-200c was the best independent predictor of tumor recurrence in stage II–III patients. From a clinical standpoint, our study showed that the preoperative serum miR-200c expression was the only factor for detecting early tumor recurrence in stage II CRC patients; a target population that can significantly benefit from timely clinical intervention and result in an improvement of cancer survival in patients with this malignancy. Collectively, these results indicate that evaluation of miR-200c in CRC patients presents a clinically promising biomarker that can facilitate disease risk assessment and severity in patients with CRC.
Our data are of particular interest because these highlight that over-expression of miR-200c in serum provided an independent predictor of lymph node metastasis. Currently, there are several options for curative treatment of early CRC without lymph node metastasis, including endoscopic mucosal resection, endoscopic submucosal dissection and laparoscopic-assisted partial colectomy. However, the routine choice of these less invasive treatments is essentially determined by the clinicopathological findings that include macroscopic type, tumor size, presence of an ulcer, and the histology of biopsy specimens29, 30. Considering the clinical challenge that a significant proportion of these patients are erroneously misdiagnosed using these conventional pathological criteria, incorporation of more robust molecular biomarkers such as quantification of miR-200c expression in serum, followed by preoperative selection of patients without lymph node metastasis may be possible, and could promote minimally invasive treatments for early CRC.
The miR-200 family is known to inhibit EMT by maintaining the epithelial phenotype through directly targeting the E-cadherin transcriptional repressors ZEB1 and ZEB2, thereby inducing upregulation of E-cadherin31. Conversely, inhibition of the miR-200 family induces mesenchymal-like spindle morphology, accompanied by an increase in cell invasion and migration, which is considered to be an important initial step in the metastatic spread of cancer31. Once metastasized, cancer cells undergo mesenchymal-to-epithelial transition (MET), a process that facilitates the subsequent settlement and proliferation of metastasized cells in distant organs32. Furthermore, we recently demonstrated that miR-200c is overexpressed in metastatic CRC compared with matched primary CRC, and its expression is epigenetically regulated24. These data collectively indicate that the miR-200 family, in particular miR-200c, play a critical role in regulating the EMT-MET switch to assist cancer cells moving from local to distant sites and establishing metastatic tumors.
Considering the role of the EMT-MET processes in mediating and facilitating tumor metastasis, the source of miR-200c in serum is intriguing. In the current study, we observed a gradual decrease in miR-200c expression in matched primary CRC tissues in a stage-dependent manner, with significantly lower expression in stage IV CRCs. Consequently, the significant elevation of miR-200c expression in the serum of stage IV CRC patients was not reflected in primary CRC tissues. In contrast, miR-200c was significantly overexpressed in liver metastases compared to matched primary CRCs, which was consistent with markedly high intensity in situ expression of miR-200c in the liver metastasis. Furthermore, high levels of miR-200c in serum were significantly associated with lymph node, liver and metastases to other organs. Interestingly, the increase in serum miR-200c levels occurred in accordance with the N category in the TNM classification (were involvement of number of nodes) and the H-system of the Japanese Society for Cancer of the Colon and Rectum33, in which metastases are classified according to the size of maximum metastasis diameter and the number of metastases. Although speculative, our results suggest that the origin of serum miR-200c from patients with metastasis might be from the metastatic site, from which the cancer cells are secreted abundantly into the systemic circulation in CRC patients. Alternatively, the fall in tissue miR-200c might reflect changes taking place in the primary tumor associated with EMT that facilitate exit from the primary tumor, a process that is later reversed in the metastatic deposit.
In conclusion, this study demonstrates several novel pieces of evidence favoring serum levels of miR-200c as a robust prognostic and metastasis-predictive biomarker in patients with CRC. First, miR-200c in serum was significantly associated with a metastatic phenotype in CRC; particularly, serum miR-200c expression is a good predictive marker for lymph node metastasis in CRC. Second, miR-200c in serum is an independent prognostic marker and predictive biomarker of tumor recurrence in curative patients, especially stage II CRCs. and appears to be superior compared to CEA levels or pathological TNM staging in CRC patients. Finally, our data suggest that the source of miR-200c in serum might be contributed by foci of tumor metastasis within lymph nodes, liver or other distant organs. Therefore, we propose that evaluation of preoperative serum miR-200c is a promising clinical tool for determining tumor recurrence in curative patients and overall prognosis, and in particular might aid in the identification of patients with micrometastasis that require intensive monitoring following surgery.
Supplementary Material
(A) Dot plots of serum miR-200c levels based upon N Stage in stage I–III CRC patients. (B) Dot plots of serum miR-200c levels subdivided by status of lymph node metastasis in stage II–III CRC patients. The lines indicate the mean values. Expression levels of miR-200c (log10 scale on the y-axis) were normalized to cel-miR-39. Statistically significant differences were determined using Mann-Whitney tests and Kruskal-Wallis test.
(A) Kaplan- Meier curves for disease free survival (DFS) in stage II CRC patients according to serum miR-200c expression (P=0.025; Log-rank test). (B) Kaplan-Meyer curves for DFS in stage III CRC patients according to serum miR-200c expression (P=0.11; Log-rank test). The cut-off values for miR-200c expression in serum were deduced from the ROC curves with Youden’s Index.
Acknowledgments
Grants Funding: The present work was supported by grants R01 CA72851 and CA129286 from the National Cancer Institute, National Institutes of Health, and funds from the Baylor Research Institute to CRB and AG. This study was also supported by a pilot project grant from the Charles A Sammons Cancer Center, Baylor University Medical Center to AG.
Footnotes
Author Contributions: Study concept and design (Y.T, A.G); Provision of samples (K.T, Y.I, M.K); Acquisition of data (Y.T, A.G); Analysis and interpretation of data (Y.T, K.H, A.G); Statistical analysis (Y.T, K.H, A.G); Drafting of the manuscript (Y.T, C.R.B, A.G)
Conflict of interest: The authors have no conflict of interests to disclose
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Lieberman DA, Rex DK, Winawer SJ, et al. Guidelines for Colonoscopy Surveillance After Screening and Polypectomy: A Consensus Update by the US Multi-Society Task Force on Colorectal Cancer. Gastroenterology. 2012;143:844–857. doi: 10.1053/j.gastro.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 3.Halama N, Herrmann C, Jaeger D, Herrmann T. Treatment with cetuximab, bevacizumab and irinotecan in heavily pretreated patients with metastasized colorectal cancer. Anticancer Res. 2008;28:4111–4115. [PubMed] [Google Scholar]
- 4.Meropol NJ, Schulman KA. Cost of cancer care: issues and implications. J Clin Oncol. 2007;25:180–186. doi: 10.1200/JCO.2006.09.6081. [DOI] [PubMed] [Google Scholar]
- 5.Duffy MJ, van Dalen A, Haglund C, et al. Tumour markers in colorectal cancer: European Group on Tumour Markers (EGTM) guidelines for clinical use. Eur J Cancer. 2007;43:1348–1360. doi: 10.1016/j.ejca.2007.03.021. [DOI] [PubMed] [Google Scholar]
- 6.Reiter W, Stieber P, Reuter C, et al. Multivariate analysis of the prognostic value of CEA and CA 19-9 serum levels in colorectal cancer. Anticancer Res. 2000;20:5195–5198. [PubMed] [Google Scholar]
- 7.Fakih MG, Padmanabhan A. CEA monitoring in colorectal cancer. What you should know. Oncology. 2006;20:579–587. [PubMed] [Google Scholar]
- 8.Tan E, Gouvas N, Nicholls RJ, et al. Diagnostic precision of carcinoembryonic antigen in the detection of recurrence of colorectal cancer. Surg Oncol. 2009;18:15–24. doi: 10.1016/j.suronc.2008.05.008. [DOI] [PubMed] [Google Scholar]
- 9.Duffy MJ. Carcinoembryonic antigen as a marker for colorectal cancer: is it clinically useful? Clin Chem. 2001;47:624–630. [PubMed] [Google Scholar]
- 10.Cortez MA, Calin GA. MicroRNA identification in plasma and serum: a new tool to diagnose and monitor diseases. Expert Opin Biol Ther. 2009;9:703–711. doi: 10.1517/14712590902932889. [DOI] [PubMed] [Google Scholar]
- 11.van Kouwenhove M, Kedde M, Agami R. MicroRNA regulation by RNA-binding proteins and its implications for cancer. Nat Rev Cancer. 2011;11:644–656. doi: 10.1038/nrc3107. [DOI] [PubMed] [Google Scholar]
- 12.Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ng EK, Chong WW, Jin H, et al. Differential expression of microRNAs in plasma of patients with colorectal cancer: a potential marker for colorectal cancer screening. Gut. 2009;58:1375–1381. doi: 10.1136/gut.2008.167817. [DOI] [PubMed] [Google Scholar]
- 14.Huang Z, Huang D, Ni S, et al. Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int J Cancer. 2010;127:118–126. doi: 10.1002/ijc.25007. [DOI] [PubMed] [Google Scholar]
- 15.Wang LG, Gu J. Serum microRNA-29a is a promising novel marker for early detection of colorectal liver metastasis. Cancer Epidemiol. 2012;36:e61–e67. doi: 10.1016/j.canep.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 16.Pu XX, Huang GL, Guo HQ, et al. Circulating miR-221 directly amplified from plasma is a potential diagnostic and prognostic marker of colorectal cancer and is correlated with p53 expression. J Gastroenterol Hepatol. 2010;25:1674–1680. doi: 10.1111/j.1440-1746.2010.06417.x. [DOI] [PubMed] [Google Scholar]
- 17.Cheng H, Zhang L, Cogdell DE, et al. Circulating plasma MiR-141 is a novel biomarker for metastatic colon cancer and predicts poor prognosis. PLoS One. 2011;6:e17745. doi: 10.1371/journal.pone.0017745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 19.Spaderna S, Schmalhofer O, Hlubek F, et al. A transient, EMT-linked loss of basement membranes indicates metastasis and poor survival in colorectal cancer. Gastroenterology. 2006;131:830–840. doi: 10.1053/j.gastro.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 20.Hugo H, Ackland ML, Blick T, et al. Epithelial--mesenchymal and mesenchymal--epithelial transitions in carcinoma progression. J Cell Physiol. 2007;213:374–383. doi: 10.1002/jcp.21223. [DOI] [PubMed] [Google Scholar]
- 21.Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2:442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 22.Burk U, Schubert J, Wellner U, et al. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008;9:582–589. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wellner U, Schubert J, Burk UC, et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol. 2009;11:1487–1495. doi: 10.1038/ncb1998. [DOI] [PubMed] [Google Scholar]
- 24.Hur K, Toiyama Y, Takahashi M, et al. MicroRNA-200c modulates epithelial-to-mesenchymal transition (EMT) in human colorectal cancer metastasis. Gut. 2012 Jul 10; doi: 10.1136/gutjnl-2011-301846. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kroh EM, Parkin RK, Mitchell PS, Tewari M. Analysis of circulating microRNA biomarkers in plasma and serum using quantitative reverse transcription-PCR (qRT-PCR) Methods. 2010;50:298–301. doi: 10.1016/j.ymeth.2010.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Link A, Balaguer F, Shen Y, et al. Fecal MicroRNAs as novel biomarkers for colon cancer screening. Cancer Epidemiol Biomarkers Prev. 2010;19:1766–1774. doi: 10.1158/1055-9965.EPI-10-0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chang KH, Mestdagh P, Vandesompele J, et al. MicroRNA expression profiling to identify and validate reference genes for relative quantification in colorectal cancer. BMC Cancer. 2010;10:173. doi: 10.1186/1471-2407-10-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ruopp MD, Perkins NJ, Whitcomb BW, Schisterman EF. Youden Index and optimal cut-point estimated from observations affected by a lower limit of detection. Biom J. 2008;50:419–430. doi: 10.1002/bimj.200710415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tominaga K, Nakanishi Y, Nimura S, et al. Predictive histopathologic factors for lymph node metastasis in patients with nonpedunculated submucosal invasive colorectal carcinoma. Dis Colon Rectum. 2005;48:92–100. doi: 10.1007/s10350-004-0751-4. [DOI] [PubMed] [Google Scholar]
- 30.Egashira Y, Yoshida T, Hirata I, et al. Analysis of pathological risk factors for lymph node metastasis of submucosal invasive colon cancer. Mod Pathol. 2004;17:503–511. doi: 10.1038/modpathol.3800030. [DOI] [PubMed] [Google Scholar]
- 31.Gregory PA, Bert AG, Paterson EL, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 32.Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9:265–273. doi: 10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- 33.Japanease Society of Colon of the Colon and Rectum. Japanese Classification of Colorectal Carcinoma. Second English Edition. Tokyo: Kanehara&Co., Ltd.; 2009. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Dot plots of serum miR-200c levels based upon N Stage in stage I–III CRC patients. (B) Dot plots of serum miR-200c levels subdivided by status of lymph node metastasis in stage II–III CRC patients. The lines indicate the mean values. Expression levels of miR-200c (log10 scale on the y-axis) were normalized to cel-miR-39. Statistically significant differences were determined using Mann-Whitney tests and Kruskal-Wallis test.
(A) Kaplan- Meier curves for disease free survival (DFS) in stage II CRC patients according to serum miR-200c expression (P=0.025; Log-rank test). (B) Kaplan-Meyer curves for DFS in stage III CRC patients according to serum miR-200c expression (P=0.11; Log-rank test). The cut-off values for miR-200c expression in serum were deduced from the ROC curves with Youden’s Index.