Abstract
Development of the cerebellum involves the coordinated proliferation, differentiation, maturation, and integration of cells from multiple neuronal and glial lineages. In rodent models, much of this occurs in the early postnatal period. However, our understanding of the molecular mechanisms that regulate this phase of cerebellar development remains incomplete. Here, we address the role of the transcription factor nuclear factor one X (NFIX), in postnatal development of the cerebellum. NFIX is expressed by progenitor cells within the external granular layer and by cerebellar granule neurons within the internal granule layer. Using NFIX−/− mice, we demonstrate that the development of cerebellar granule neurons and Purkinje cells within the postnatal cerebellum is delayed in the absence of this transcription factor. Furthermore, the differentiation of mature glia within the cerebellum, such as Bergmann glia, is also significantly delayed in the absence of NFIX. Collectively, the expression pattern of NFIX, coupled with the delays in the differentiation of multiple cell populations of the developing cerebellum in NFIX−/− mice, suggest a central role for NFIX in the regulation of cerebellar development, highlighting the importance of this gene for the maturation of this key structure.
INDEXING TERMS: cerebellum, transcription factor, NFIX, cerebellar granule neurons, Bergmann glia
The cerebellum plays a pivotal role in many brain functions, including balance control, sensorimotor function, and the vestibular ocular reflex, as well as contributing to cognitive processes such as feed-forward sensory-motor learning and spatial memory (Hatten and Roussel, 2011; Sotelo, 2004). The prevalence of human congenital disorders associated with malformation of the cerebellum, such as Dandy-Walker malformation, Joubert syndrome, and cerebellar vermis hypoplasia, highlights the importance of understanding the critical developmental events regulating cerebellar formation (Chizhikov and Millen, 2003). The fundamental anatomical organization of the cerebellum was elucidated over a century ago (for review see Sotelo, 2004), demonstrating that the afferent inputs from the climbing and mossy fibers ultimately converge on the efferent output from the cerebellum, the Purkinje cells. However, despite our understanding of the morphological development of the laminar structure of the cerebellum, our understanding of the genes that control the development of this structure remains limited.
In mice, cerebellar development is initiated during midembryogenesis through the induction of the cerebellar territory from rhombomere 1 (Millen and Gleeson, 2008; Sajan et al., 2010). However, the majority of cerebellar development occurs postnatally, culminating in the formation of the mature cerebellum by approximately postnatal day 20 (P20) in mice (Miale and Sidman, 1961). Recent research has identified some of the molecules involved in the development of distinct cellular populations within the cerebellum. For instance, the transcription factors Zic1 and Zic2 regulate the proliferation of progenitor cells within the external granular layer (EGL; Aruga et al., 2002). Furthermore, both sonic hedgehog (Shh), which is expressed by Purkinje cells (Spassky et al., 2008; Wechsler-Reya and Scott, 1999), and the Notch signalling pathway (Solecki et al., 2001) have also been implicated in regulating the proliferation of EGL progenitors. However, the molecular circuitry regulating the differentiation of other cerebellar cells, such as the Bergmann glia, is less well understood.
One family of transcription factors, the nuclear factor one (NFI) genes, has recently been shown to be central to the process of gliogenesis within different areas of the nervous system (Mason et al., 2009; Piper et al., 2007). For instance, NFIA has been demonstrated to modulate gliogenesis within the developing spinal cord (Deneen et al., 2006), hippocampus (Piper et al., 2010), and neocortex (Namihira et al., 2009). NFIB has also been implicated in glial development within the brain, with NFIB−/− mice exhibiting delayed glial development within the developing neocortex and hippocampus (Barry et al., 2008; Piper et al., 2009a). At a mechanistic level, NFIA has been proposed to act downstream of the Notch signalling pathway to induce gliogenesis (Namihira et al., 2009) but has also been shown to regulate Notch signalling negatively, promoting glial differentiation via the repression of progenitor pathways and the activation of glial gene transcription (Piper et al., 2010). Both NFIA and NFIB have also been implicated in cerebellar development.
These genes are broadly expressed in the postnatal cerebellum, and NFIB−/− mice have been reported to have foliation defects within the cerebellum (Steele-Perkins et al., 2005). NFIA−/− mice also display cerebellar defects, in particular a decrease in cerebellar size at P17, coupled with aberrant foliation and a delay in the postmitotic maturation of cerebellar granule neurons (Wang et al., 2007). More recent research has identified potential targets of NFI genes in the regulation of cerebellar granule neurons, including Tag1, N-cadherin, and GABRA6 (Wang et al., 2004, 2007, 2010), demonstrating that these transcription factors orchestrate a complex series of developmental events during cerebellar granule neuron development. These lines of evidence indicate that NFI genes are central for cerebellar granule neuron development. However, given the role of other NFI family members in regulating gliogenesis within the cortex and spinal cord, we postulated that these transcription factors might also contribute to this process within the developing cerebellum. To test this, we used mice lacking the NFIX gene, which survive postnatally (Campbell et al., 2008), as a model in which to investigate the molecular regulation of neurogenesis and gliogenesis within the cerebellum. Within the postnatal and adult cerebellum, NFIX was expressed in the granule neurons and was also expressed by progenitors within the EGL during development. Analysis of postnatal NFIX−/− mice demonstrated that, in the absence of this transcription factor, the size of the cerebellum was significantly reduced, and the development of cerebellar granule neurons, Purkinje cells, and Bergmann glia was markedly delayed. These data implicate NFIX in the regulation of multiple aspects of cerebellar development and in particular demonstrate that this transcription factor is critical for glial differentiation within the cerebellum.
MATERIALS AND METHODS
Animals
Male and female wild-type C57Bl/6J and NFIX−/− mice were used in this study. These animals were bred at The University of Queensland under approval from the institutional Animal Ethics Committee. No cerebellar defects were detected in wild-type or heterozygote animals. Heterozygous NFIX mice were bred to obtain wild-type, heterozygous, and homozygous progeny. Embryos were genotyped by PCR as previously described (Campbell et al., 2008).
Haematoxylin staining
The cerebella from wild-type C57Bl/6J or NFIX−/− embryos were dissected from the skull, blocked in 3% noble agar (Difco, Sparks, MS), and sectioned sagittally at 50 µm on a vibratome (Leica, Nussloch, Germany). Sections were then mounted and stained with Mayer’s hematoxylin using standard protocols.
Antibody characterization
Antibodies, sources, and the concentrations at which they were used are listed in Table 1.
TABLE 1.
Summary of Primary Antibodies Used in This Study
| Antibody | Host | Source, catalog No. | Antigen | Dilution |
|---|---|---|---|---|
| NFIX | Rabbit polyclonal | Active motif, 39348 | Amino acid residues 277–291 of human NFIX | 1/10,000 |
| GFAP | Rabbit polyclonal | Dako, Z0334 | Bovine spinal cord isolate | 1/20,000 |
| GLAST | Rabbit polyclonal | Gift | Glast full-length protein | 1/50,000 |
| Nestin | Mouse monoclonal | Developmental Studies Hybridoma Bank, Rat-401 | Full-length nestin | 1/1,500 |
| Pax6 | Rabbit polyclonal | Millipore, AB2237 | The 17 C-terminal residues of mouse Pax6 | 1/25,000 |
| NeuN | Mouse monoclonal | Millipore, MAB377 | Purified cell nuclei from mouse brain | 1/2,000 |
| Calbindin | Rabbit polyclonal | Swant, CB38 | Full length rat calbindin D-28K | 1/20,000 |
| Cleaved caspase 3 | Rabbit polyclonal | Cell Signaling Technology, 9661 | RGTELDCGIETD | 1/5,000 |
| Phosphohistone H3 | Rabbit polyclonal | Millipore, 06-570 | Human histone H3 peptide (amino acids 7–20) | 1/5,000 |
NFIX
The anti-NFIX antibody specifically detects a single band at 46.3 kDa in Western blots of JEG-3 cells expressing HA-tagged NFIX (manufacturer’s information), corresponding to the predicted molecular weight for NFIX. The specificity of this antibody has previously been confirmed using immunohistochemistry with a blocking peptide and via immunohistochemistry on NFIX−/− cortical tissue (Campbell et al., 2008). The staining pattern observed here matches previous descriptions (Wang et al., 2007).
GFAP
The pattern of glial fibrillary acidic protein (GFAP) immunoreactivity observed here matches previous descriptions of GFAP expression within the cerebellum (Wiencken-Barger et al., 2007). The specificity of the anti-GFAP antibody has previously been confirmed through a lack of immunoreactivity within samples from Gfap−/− mice (Hanbury et al., 2003).
Nestin
As described previously, the nestin antiserum recognizes a doublet of 200 kDa in Western blots performed on homogenates from embryonic day (E) 15 rat spinal cord (Hockfield and McKay, 1985; Petkau et al., 2010). The pattern of nestin immunoreactivity observed within the developing cerebellum matches previous descriptions (Sergaki et al., 2010).
GLAST
The glutamate/aspartate transporter (GLAST) antibody recognizes a single band on Western blots at ~59 kDa, corresponding to the predicted size for GLAST (Plachez et al., 2000). The anti-GLAST antibody has been previously shown to label radial glial cells in both cultured cells (Plachez et al., 2000) and embryonic brains (Piper et al., 2009a). The staining pattern observed here in the cerebellum matches previous descriptions (Fukaya et al., 1999; Furuta et al., 1997).
Pax6
The anti-Pax6 antibody specifically detects a single band in Western blots at ~48 kDa on lysates of fetal mouse brain (manufacturer’s information) and has previously been shown to label specifically cerebellar neurons in vivo (Janmaat et al., 2009). Furthermore, the global staining pattern observed in the cerebellum here matches previous descriptions (Engelkamp et al., 1999).
Calbindin
The anticalbindin antibody specifically detects a single band in Western blots at ~28 kDa on lysates of mouse brain (manufacturer’s information) and has previously been demonstrated to be specific for this antigen (Haverkamp et al., 2008). The staining pattern described here for the cerebellum matches previous descriptions (Fukaya et al., 1999).
Cleaved caspase 3
The anticleaved caspase 3 antibody specifically detects a single band in Western blots at ~17 kDa on lysates of HeLa cells (manufacturer’s information). This antibody does not recognize full-length caspase 3 or other cleaved caspases, instead recognizing only the large fragment of activated caspase 3. This antibody has previously been demonstrated to label apoptotic cells within the brain (Schwartz et al., 2010).
Phosphohistone H3
The antiphosphohistone H3 antibody specifically detects a single band in Western blots at ~17 kDa on lysates of HeLa cells (manufacturer’s information) and has previously been shown to label mitotic cells within the developing rhombic lip (Volkmann et al., 2010) and within the P6 cerebellum in a manner matching that described here (Flora et al., 2009).
NeuN
The anti-NeuN antibody recognizes postmitotic neuronal nuclei, and has been used extensively to label neurons specifically via immunohistochemistry (Ge et al., 2010). The pattern of NeuN expression described here matches previous descriptions of NeuN expression within the postnatal cerebellum (Flora et al., 2009).
Immunohistochemistry
Postnatal pups and adults were deeply anesthetised with sodium pentobarbital, before being transcardially perfused with 0.9% saline, followed by 4% paraformaldehyde (PFA), with postfixation in 4% PFA at 4°C. Cerebella were removed and sectioned sagittally at 50 µm on a vibratome. Immunohistochemistry of floating sections was performed using the chromogen 3,3′-diaminobenzidine as described previously (Piper et al., 2009b; Plachez et al., 2008). Biotin-conjugated goat anti-rabbit IgG (BA-1000; Vector Laboratories, Burlingame, CA) and donkey anti-mouse IgG (715-065-150; Jackson Immunoresearch, West Grove, PA) secondary antibodies were used for chromogenic immunohistochemistry. For all immunohistochemical analyses, at least three wild-type and three NFIX−/− brains were analyzed. Sections from the vermis of the cerebellum were imaged using an upright microscope (Zeiss upright Axio-Imager Z1) fitted with an Axio-Cam HRc camera. Figures were cropped for presentation in Adobe Photoshop (San Jose, CA).
Reverse transcription and quantitative real-time PCR
Cerebella were dissected and immediately snap frozen. Total RNA was extracted using an RNeasy Micro Kit (Qiagen, Valencia, CA). Reverse transcription was performed using Superscript III (Invitrogen, Carlsbad, CA), and qPCR was performed as described previously (Piper et al., 2010). Briefly, 0.5 µg total RNA was reverse transcribed with random hexamers. qPCRs were carried out in a Rotor-Gene 3000 (Corbett Life Science, Valencia, CA) with SYBR Green PCR Master Mix (Invitrogen). All the samples were diluted 1/100 with RNase/DNase free water and 5 µl of these dilutions were used for each SYBR Green PCR reaction containing 10 µl SYBR Green PCR Master Mix, 10 µM of each primer, and deionized water. The reactions were incubated for 10 minutes at 95°C, followed by 40 cycles with 15 seconds denaturation at 95°C, 20 seconds annealing at 60°C, and 30 seconds extension at 72°C. Primer sequences used are listed in Table 2.
TABLE 2.
QPCR Primers Used in This Study
| Gene | Forward primer | Reverse primer |
|---|---|---|
| Pax6 | ACGTATATCCCAGTTCTCAGA | ATTCACTCCGCTGTGACTGT |
| Calbindin | CGAAAGAAGGCTGGATTGGA | AAGACGTGAGCCAACTCTAC |
| Gfap | AGTGGTATCGGTCTAAGTTTG | CGATAGTCGTTAGCTTCGTG |
| Glast | CACTGCTGTCATTGTGGGTA | AGCATCCTCATGAGAAGCTC |
| Nestin | GAAGTGGCTACATACAGGAC | AGCTTCAGCTTGGGGTCAG |
| Astrotactin 1 | TGCACATCTCAGTCATGGGT | CTCTGCACTGGCACTCTTC |
| NeuN | GCCCAAACGACTACATGTCT | CAGCATCTGAGCTAGTTTCAA |
| Gabra6 | TCCTCTATACCATGAGGCTC | TAGGATAAGCATAGCTCCCAA |
| N-cadherin | CTGAGGACCCTATGAAGGAA | GATGACCCAGTCTCTCTTCT |
| Ephrin B1 | TTACTACATTACATCAACGTCC | TGTCACAGCATTTGGATCTTG |
| Tag1 | GCTTCGGCTTTCTACAGGAA | CGGTAGGACAAACCTGGGT |
| Parvalbumin | AGGATGAGCTGGGGTCCAT | TTCAACCCCAATCTTGCCGT |
qPCR data expression and analysis
After completion of the PCR amplification, the data were analyzed with the Rotor-Gene software as described previously (Piper et al., 2009b). When quantifying the mRNA expression levels, the housekeeping gene hypoxanthine ribosyltransferase (HPRT) was used as a relative standard. All the samples were tested in triplicate. By means of this strategy, we achieved a relative PCR kinetic of standard and sample. For all qPCR analyses, RNA from three biological replicates for both wild-type and NFIX−/− mice were interrogated. Statistical analyses were performed using a two-tailed unpaired t-test. Error bars represent the standard error of the mean.
Quantification of cerebellar size and cell counts
For quantification of sagittal sections at all ages, measurements were made at the vermis of the cerebellum. For quantification of cerebellar size, matched sections from the vermis of wild-type and NFIX−/− cerebella were imaged, and size was measured in Zeiss Axiovision software. To measure the width of the EGL, cerebella were imaged, and then 20 measurements from each replicate were taken from folia three in both the wild-type and the NFIX knockouts. For analysis of apoptosis, cleaved caspase 3-positive cells across the entire sagittal section were counted and then normalized to area by dividing the total number of immunopositive cells by the cross-sectional area. For analysis of phosphohistone H3-positive cells, the total number of immunopositive cells in the EGL of each section was divided by the cross-sectional area. To calculate the number of Pax6-positive cells in the premigratory and molecular zones, the number of immunopositive cells within these zones was counted and divided by the area of the section. For Pax6 and phosphohistone H3 cell counts, we also measured the size of the nucleus to determine whether there was a difference between genotypes (Guillery, 2002). No size differences were noted, so we did not apply the Abercrombie correction factor. Quantification was performed blind to the genotype of the sample, and statistical analyses were performed using a two-tailed unpaired t-test. Error bars represent the standard error of the mean.
Luciferase reporter assay
The constructs used in the luciferase assay were a full-length NFIX expression construct driven by the chick β-actin promoter (NFIX pCAGIG), and a construct containing 2.1 kb of the mouse Gfap promoter (Zhou et al., 2004). DNA was transfected into HEK 293 cells using FuGene (Invitrogen). Renilla luciferase (pRL SV40; Promega, Madison, WI) was added to each transfection as a normalization control. After 48 hours, luciferase activity was assessed using a dual-luciferase system (Promega) per the manufacturer’s instructions. Statistical analyses were performed via ANOVA. Error bars represent the standard error of the mean.
RESULTS
NFIX is expressed in the postnatal and adult cerebellum
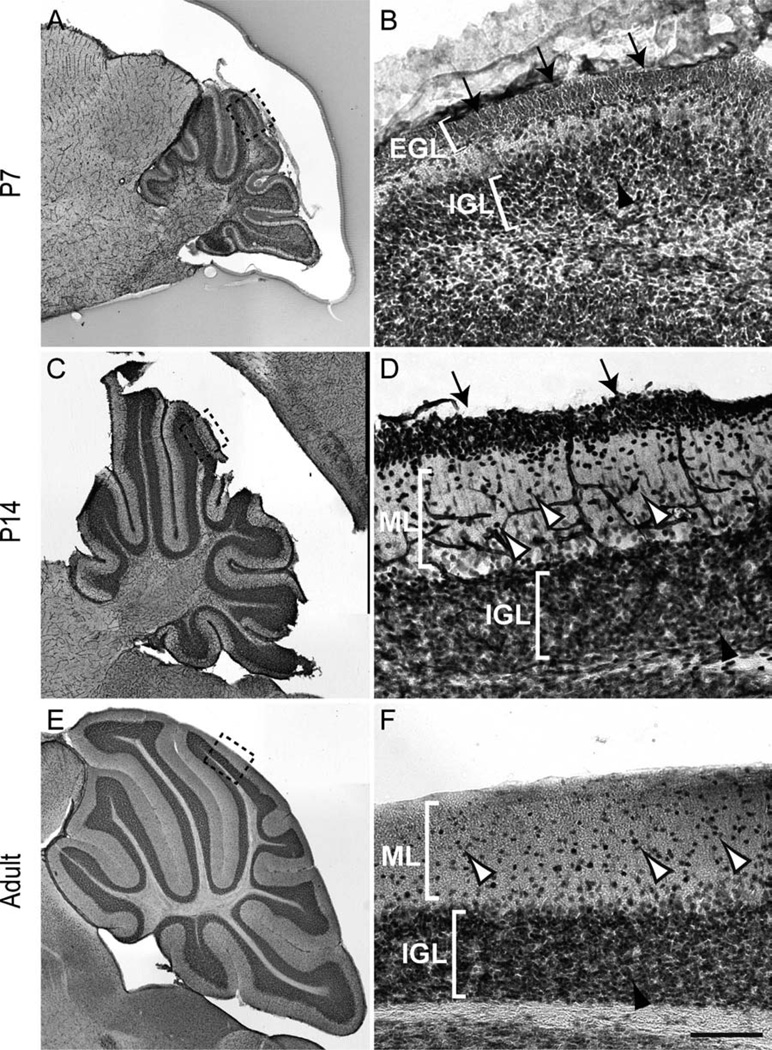
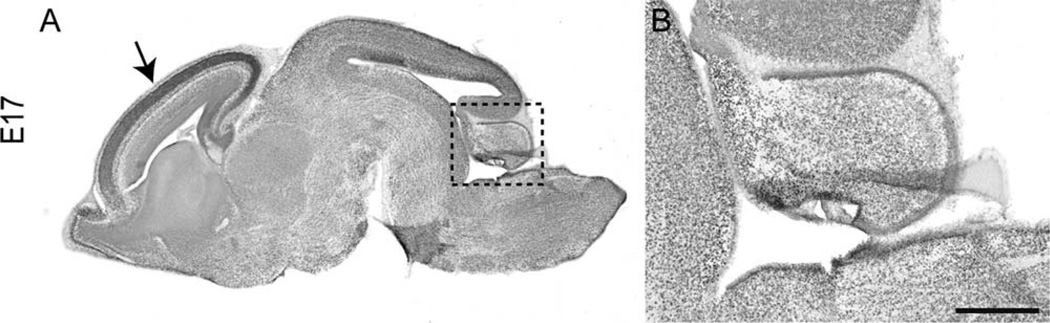
To address the role of NFIX in postnatal cerebellar development, we first analyzed the expression of this transcription factor in the cerebellum via immunohistochemistry. At P7, cells within both the EGL and the internal granule layer (IGL) expressed NFIX (Fig. 1A,B). At P14, cells within the EGL, which by this stage are predominantly premigratory, immature granule neurons, were still seen to express NFIX, as did the mature granule neurons within the IGL (Fig. 1C,D). Expression of NFIX was also observed in cells within the molecular layer at this age (Fig. 1D). In the adult cerebellum, cerebellar granule neurons within the IGL expressed high levels of NFIX, with isolated cells within the molecular layer also expressing NFIX (Fig. 1E,F). NFIX was also broadly expressed within the cerebellar anlage at E17, albeit at a low level (Fig. 2), indicating that progenitor cells within the embryonic cerebellum are likely to express this transcription factor.
Figure 1.
Expression of NFIX in the cerebellum. Expression of NFIX in sagittal sections of the postnatal (A–D) and adult (E,F) cerebellum. A: NFIX was broadly expressed in the cerebellum at P7. B: Higher magnification view of the boxed region (lobule VI) in A, showing expression of NFIX by cells within the external granular layer (EGL; arrows) and inner granule cell layer (IGL; arrowhead). C: Expression of NFIX at P14. D: Higher magnification view of the boxed region (lobule VI) in C, showing that immature granule neurons within the premigratory zone of the EGL (arrows) and cells within the molecular layer (ML; open arrowheads) and IGL (solid arrowhead) express NFIX. E: Expression of NFIX in the adult cerebellum. F: Higher magnification of the boxed region (lobule VI) in E. NFIX was expressed by cells within the IGL (solid arrowhead), as well as by scattered cells within the ML (open arrowheads). Scale bar = 25 µm in F (applies to D,F); 200 µm for A,C,E; 15 µm for B.
Figure 2.
Expression of NFIX in the embryonic cerebellum. A,B: Sagittal section of an E17 brain, showing expression of NFIX. A: NFIX was strongly expressed within the dorsal telencephalon at E17 (arrow in A). B: Higher magnification view of the boxed region in A, showing that NFIX was expressed widely, but at a low level, throughout the developing cerebellar anlage. Scale bar = 200 µm in B; 1,000 µm for A.
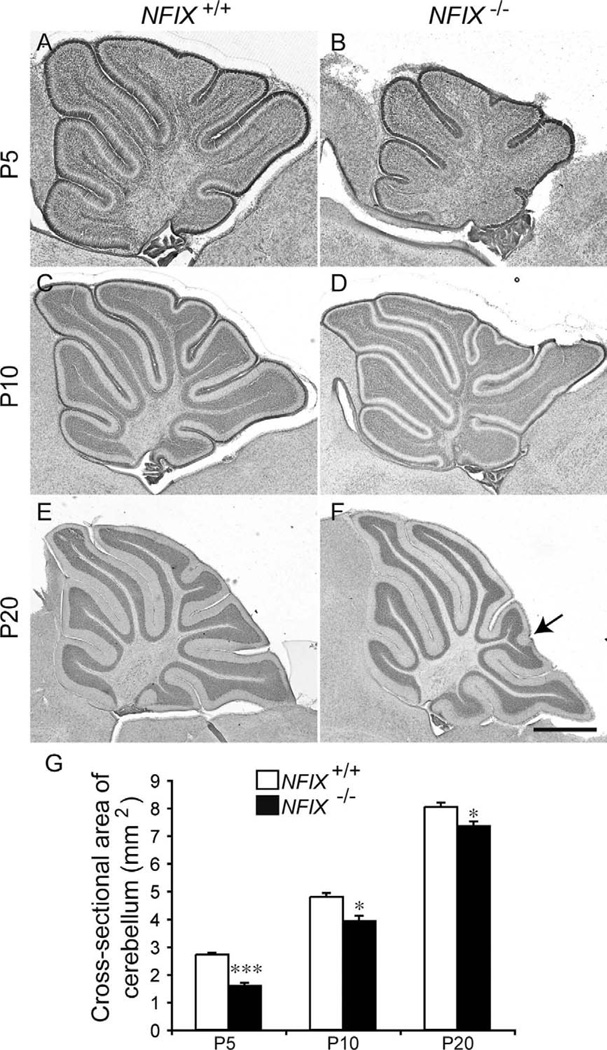
Cerebellar size is diminished in NFIX null mutants
Unlike mice lacking NFIA or NFIB, NFIX−/− mice are viable postnatally and exhibit a variety of forebrain deficits, including an increase in size of the cingulate cortex and malformation of the hippocampal dentate gyrus (Campbell et al., 2008). Analysis of sagittal sections of the cerebellum at the level of the vermis revealed that NFIX−/− mice also display abnormal development of this brain region. At P5, hematoxylin staining revealed that the pattern of foliation in the NFIX−/− cerebellum was abnormal compared with that in wild-type controls, with the mutant having folia of markedly reduced size (Fig. 3A,B). Analysis of the cross-sectional area of the cerebellum revealed that the cerebellum was significantly smaller in NFIX−/− mice than in controls at P5 (Fig. 3G). At P10 and P20, this difference was less pronounced. However, mutants still exhibited a significantly smaller cerebellum at these ages (Fig. 3C–G). Furthermore, there were subtle foliation defects in the mutant cerebellum, including a reduction in the size of lobule VII in the mutant and a bifurcation within the distal end of lobule VIII (arrow in Fig. 3F). Finally, the area of the IGL appeared to be reduced in the NFIX−/− mutant cerebellum.
Figure 3.
Reduction in cerebellum size in NFIX−/− mice. Hematoxylin-stained sagittal sections of the cerebellum of wild-type (A,C,E) and NFIX−/− (B,D,F) mice at P5 (A,B), P10 (C,D), and P20 (E,F). G: Quantification of the area of the cerebellum at the level of the vermis. At P5, the cross-sectional area of the NFIX−/− cerebellum (B) was markedly smaller than that of the wild-type (A). At P10 and P20 the size of the NFIX−/− cerebellum (D,F) had partially recovered but was still significantly reduced in comparison with that of wild-type controls (C,E). The folia of NFIX−/− mice appeared subtly different at P20; for instance, the tip of lobule VIII appears bifurcated in the mutant (arrow in F). *P < 0.05; ***P < 0.001, t-test. Scale bar = 200 µm in F (applies to E,F); 100 µm for A,B; 150 µm for C,D.
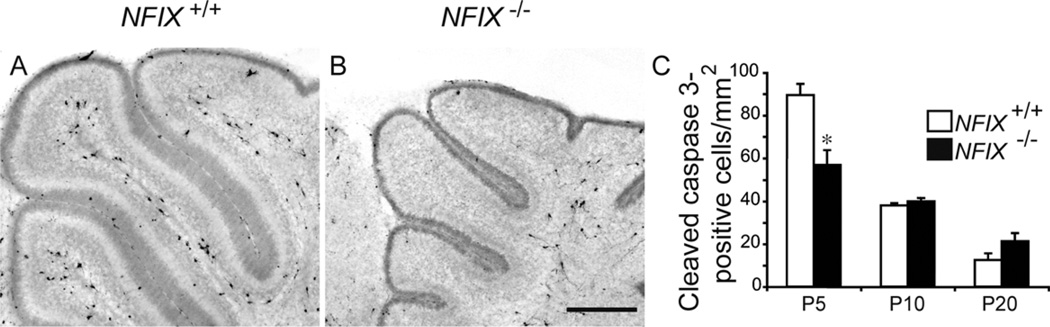
Increased apoptotic cell death does not underlie the reduction in the size of the NFIX−/− cerebellum
The reduction in cerebellar size in the NFIX−/− mice suggested that increased apoptosis may underlie the deficit observed in these mice. To investigate this, we analyzed the expression of a marker for apoptotic cells, cleaved caspase 3. In both wild-type and NFIX−/− mice, the majority of apoptotic cells were observed within the cerebellar white matter. Interestingly, there appeared to be fewer apoptotic cells in the mutant cerebellum at P5 in comparison with wild-type controls (Fig. 4A,B). Determination of the total number of cleaved caspase 3-positive cells normalized against the cross-sectional area of the cerebellum confirmed this finding, indicating that at P5 there were actually fewer apoptotic cells/mm2 in the mutant than in the control, whereas at P10 and P20 no significant difference was observed between wild-type and NFIX−/− mice (Fig. 4C).
Figure 4.
Reduction in cerebellum size in NFIX−/− mice is not due to apoptosis. Expression of the apoptotic marker cleaved caspase 3 in sagittal sections of wild-type (A) and NFIX−/− (B) mice at P5. C: Quantification of the number of cleaved caspase 3-positive cells/mm2 revealed that at P5 there were actually fewer apoptotic cells per unit area within the NFIX−/− cerebellum compared with that of wild-type controls. At P10 and P20 no significant differences were observed. *P < 0.05, t-test. Scale bar = 80 µm.
Development of the EGL is delayed in the NFIX−/− cerebellum
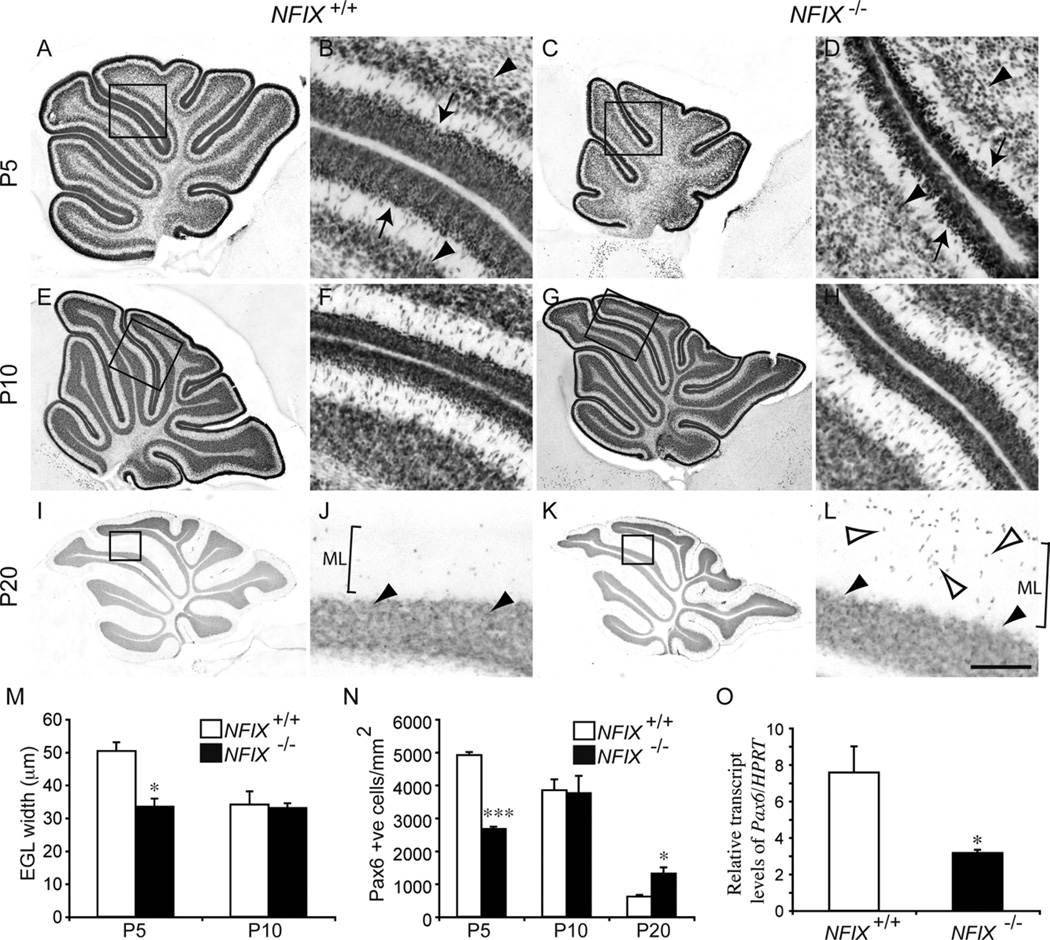
Because increased apoptosis was not the mechanism underlying the reduction in the size of the NFIX−/− cerebellum, we next examined the development of the EGL. The EGL is populated by progenitor cells originally derived from the rhombic lip. These progenitor cells proliferate and differentiate into cerebellar granule neurons, which migrate radially through the molecular layer of the cerebellum to reach their final destination, the IGL (Sotelo, 2004). The majority of cerebellar granule neurons are produced during the first 2 postnatal weeks in mice (Miale and Sidman, 1961), such that the EGL disappears by P15. We used the expression of Pax6, a marker for both EGL progenitors and cerebellar granule neurons (Engelkamp et al., 1999), to analyze EGL and IGL development in NFIX−/− mice. In wild-type P5 mice, the expression of Pax6 delineated the EGL and cerebellar granule neurons within the IGL (Fig. 5A,B). In the NFIX mutant, however, the width of the EGL was markedly reduced, and the IGL was poorly defined (Fig. 5C,D). Measurement of EGL width confirmed a significant reduction in the NFIX−/− cerebellum at P5 (Fig. 5M). Moreover, analysis of mRNA levels by quantitative real-time PCR (qPCR) revealed a significant reduction in Pax6 mRNA in the mutant at this age (Fig. 5O). By P10, expression of Pax6 within the EGL of NFIX−/− mice was comparable to that in controls, a finding confirmed by measurement of the EGL width, which revealed no significant differences between NFIX mutants and controls (Fig. 5E–H,M). By P20, the EGL had disappeared in both the wild-type and the NFIX mutant. However, there appeared to be more Pax6-positive cells in the molecular layer of the mutant than in the control (Fig. 5I–L). To investigate this, we counted the number of Pax6-positive cells within the molecular layer of wild-type and NFIX−/− mice at P5, P10, and P20 (Fig. 5N). At P5, there were significantly fewer Pax6-positive cells in the molecular layer of the mutant, a finding that is probably linked to the delayed development of the EGL in the mutant at this age (Fig. 5D). At P10 the number of migrating Pax6-positive cells was not significantly different between the two groups. However, at P20 a significantly greater number of cells within the molecular layer of the mutant expressed Pax6 (Fig. 5N), suggesting that migration or differentiation of EGL cells is delayed in the NFIX−/− cerebellum. Unfortunately, on a C57Bl/6J background, NFIX−/− mice die at weaning, precluding analysis of mutants at later stages of development. Collectively, these data indicate that the reduction in the size of the cerebellum of NFIX−/− mice is due, at least in part, to delayed development and differentiation of progenitor cells within the EGL.
Figure 5.
Delayed differentiation of the EGL layer in NFIX−/− mice. Expression of Pax6 at P5 (A–D), P10 (E–H), and P20 (I–L) in wild-type and NFIX−/− mice. A: Expression of Pax6 in the wild-type cerebellum at P5. B: Higher magnification of the boxed region in A (lobules IV– VI), revealing that cells within the EGL (arrows) and the nascent IGL (arrowheads) express Pax6. C: Expression of Pax6 in the NFIX−/− cerebellum at P5. D: Higher magnification of the boxed region in C (lobules IV–VI), revealing that the width of the EGL was markedly reduced (arrows) and that the IGL (arrowheads) was less well defined. By P10, expression of Pax6 in the cerebellum of wild-type (E,F) and NFIX−/− mice was comparable. By P20, the EGL was no longer evident in either the wild-type (I,J) or the NFIX−/− (K,L) cerebella. Cells within the IGL maintained Pax6 expression (solid arrowheads in J,L). However, in the mutant cerebellum, there were markedly more Pax6-positive cells in the molecular layer (open arrowheads in L) than were observed in the control. F,H,J,L are higher magnification views of the boxed regions in E,G,I,K, respectively (lobules IV–VI in F,H; lobules IV/V in J,L). M: The width of the EGL was significantly reduced in the cerebellum of NFIX−/− mice at P5 compared with controls. N: Counts of Pax6-positive cells in the molecular layer of wild-type and mutant cerebella. At P5 there were significantly fewer Pax6-positive cells in the molecular layer of NFIX−/− mice. However, at P20 there were significantly more Pax6-positive cells in the molecular layer of the mutant compared with wild-type controls. O: qPCR on P5 cerebellar tissue revealed a significant reduction in Pax6 mRNA in the NFIX−/− mutant compared with controls. ML, molecular layer. *P < 0.05; ***P < 0.001, t-test. Scale bar = 40 µm in L (applies to B,D,F,H,J,L); 100 µm for A,C; 150 µm for E,G; 200 µm for I,K.
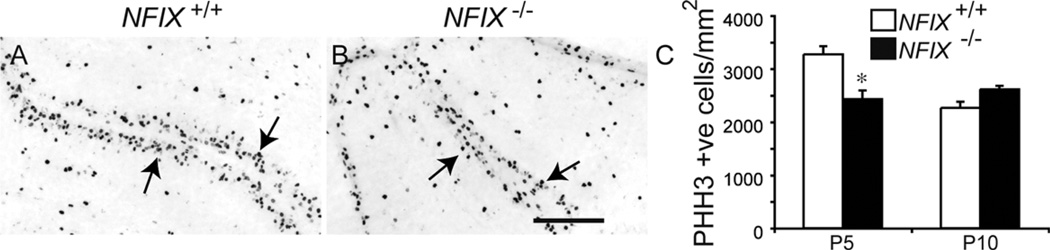
Proliferation in the EGL of NFIX mutants is reduced in the early postnatal period
Pax6-expressing cells within the EGL proliferate during the early postnatal period. The EGL was reduced at P5 in the NFIX mutant, so we hypothesized that a reduction in proliferation could underlie this defect. To test this hypothesis, we analyzed the expression of the mitotic marker phosphohistone H3 in the EGL of wild-type and NFIX−/− mice at P5 and P10. At P5 there appeared to be fewer phosphohistone H3-positive cells in the EGL of the mutant (Fig. 6A,B). Counts of the total number of mitotic cells within the EGL (normalized against the cross-sectional area of the cerebellum) confirmed this conclusion, revealing a significant reduction in phosphohistone H3-positive cells in the EGL of the NFIX mutant (Fig. 6C). At P10 the number of mitotic cells in the EGL in NFIX−/− mice did not differ significantly from that in wild-type controls (Fig. 6C), a finding in line with the congruence of EGL width in NFIX+/+ and NFIX−/− mice at this age (Fig. 5M).
Figure 6.
Proliferation in the EGL of early postnatal NFIX−/− mice is reduced. Expression of the mitotic marker phosphohistone H3 (PHH3) in lobules IV–VI of the cerebellum of wild-type (A) and NFIX−/− (B) mice at P5. Phosphohistone H3-positive cells were observed in the EGL of both wild-type and NFIX−/− mice (arrows). C: Counts of phosphohistone H3-positive cells in the EGL revealed significantly fewer mitotic cells within the EGL of P5 NFIX−/− mice compared with controls. No significant difference was observed at P10. *P < 0.05, t-test. Scale bar = 50 µm.
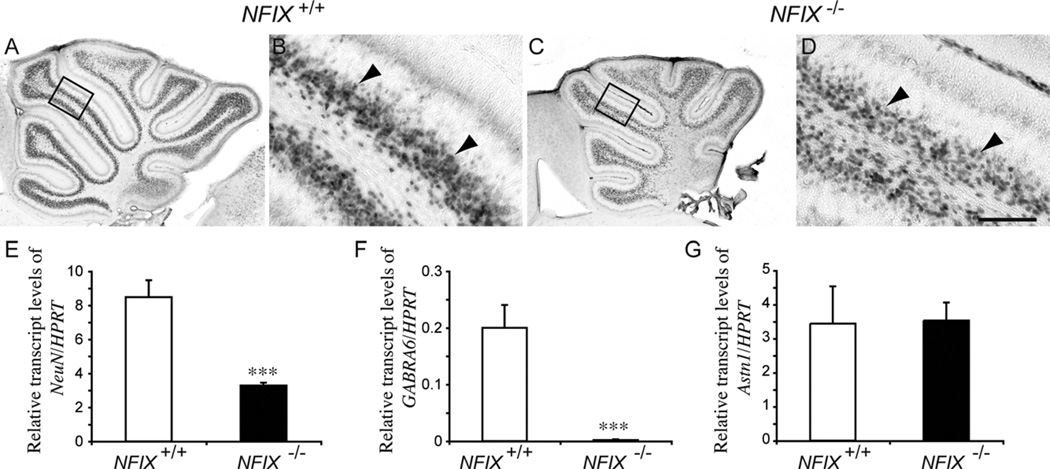
Development of cerebellar granule neurons is delayed in NFIX−/− mice
As EGL progenitors differentiate, they produce cerebellar granule neurons, which migrate radially into the IGL. The presence of a less well-defined IGL in P5 NFIX−/− cerebella based on Pax6 immunohistochemistry (Fig. 5C), coupled with reduced EGL proliferation at this age, suggested delays in cerebellar granule cell differentiation in the absence of NFIX. To investigate this further, we analyzed the expression of the neuron-specific nuclear protein NeuN at P5. In the wild-type, a clearly demarcated band of NeuN-expressing cells was observed within the IGL (Fig. 7A,B). In the NFIX mutant, however, NeuN expression was reduced, and the IGL was less well developed (Fig. 7C,D). This finding was supported by qPCR analysis of NeuN mRNA levels, which were significantly reduced in NFIX−/− mice compared with controls at P5 (Fig. 7E). Within the IGL, postmigratory cerebellar granule neurons express cell type-specific genes, such as the GABAA α6 receptor subunit (GABRA6; Kilpatrick et al., 2010). Expression of this gene was also downregulated in the cerebellum of P5 NFIX−/− mice (Fig. 7F), indicative of delayed development of the cerebellar granule neurons in the absence of NFIX. Finally, we analyzed the expression of astrotactin, a glycoprotein expressed on migratory cerebellar granule neurons that is involved in mediating their migration along the fibers of Bergmann glia (Edmondson et al., 1988; Stitt and Hatten, 1990). Levels of astrotactin were not significantly different in the NFIX−/− cerebellum (Fig. 7G), suggesting that it is the differentiation of neurons that is deficient in these mutants, not their subsequent astrotactin-mediated migration into the IGL.
Figure 7.
Neuronal differentiation is delayed in the cerebellum of NFIX−/− mice. Expression of the neuronal marker NeuN in the cerebellum of wild-type (A,B) and NFIX−/− (C,D) mice at P5. NeuN-expressing cells in the IGL of the wild-type and mutant were apparent (arrowheads), but there appeared to be more NeuN-positive cells in the IGL of the wild-type compared with the NFIX knockout. B,D are higher magnification views of the boxed regions (lobules IV/V) in A,C, respectively. E–G: qPCR on wild-type and NFIX−/− P5 cerebellar tissue demonstrated significantly reduced levels of NeuN (E) and GABRA6 (F) mRNA in the mutant. However, levels of astrotactin 1 mRNA (G) were not significantly different compared with controls. ***P < 0.001, t-test. Scale bar = 20 µm in D (applies to B,D); 100 µm for A,C.
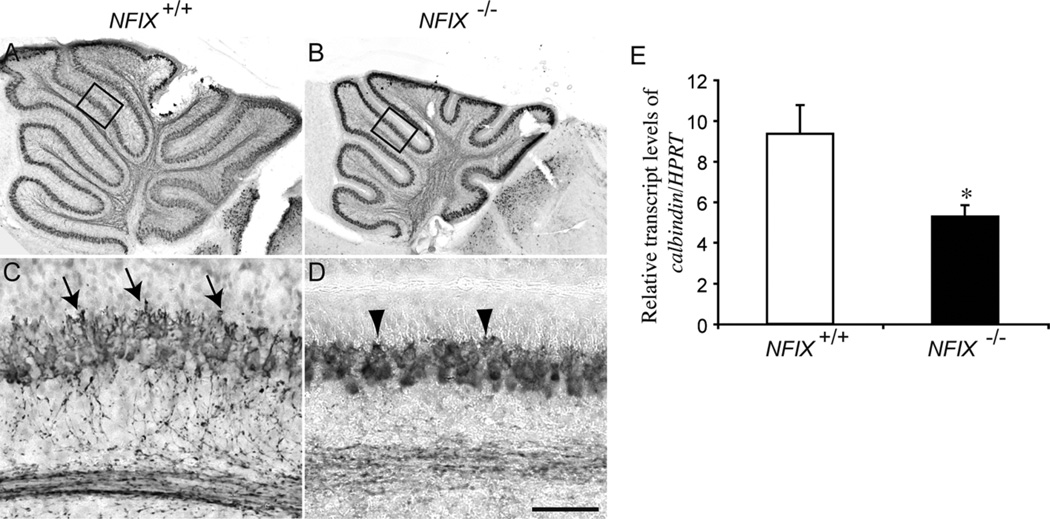
Purkinje cell development is delayed in NFIX−/− mice
The Purkinje cells form the sole efferent output from the cerebellum. Unlike cerebellar granule neurons, which are developmentally derived from EGL progenitors that have their origins in the rhombic lip, Purkinje cells are derived from progenitor cells in the dorsomedial ventricular zone along the fourth ventricle adjacent to the rhombic lip (Sotelo, 2004). We analyzed development of the Purkinje cell layer based on the expression of calbindin, a specific marker for this population of neurons in the cerebellum (Lordkipanidze and Dunaevsky, 2005). In P5 wild-type mice, the Purkinje cell layer was clearly defined. Furthermore, at this age the dendritic processes of the Purkinje cells were developing, and branched processes were observed extending toward the EGL (Fig. 8A,C). In the mutant, however, although calbindin-expressing Purkinje cells were evident in the cerebellum, they were not as morphologically mature as in wild-type mice. Specifically, expression of calbindin in the Purkinje cell axonal extensions was reduced, and the dendritic arborizations of the cells were markedly shorter and less branched than in the wild-type controls (Fig. 8B,D). As with the developing cerebellar granule neurons, the development of Purkinje cells in the mutant had caught up with the wild-type by P10, and by P20 Purkinje cell development in the mutant appeared relatively normal (data not shown). These data indicate that the early development of Purkinje cells is delayed in NFIX−/− mice. Analysis of calbindin mRNA levels supported these findings, revealing a significant reduction in calbindin mRNA in the cerebellum of NFIX−/− mice at P5 (Fig. 8E).
Figure 8.
Expression of calbindin in the cerebellum of NFIX−/− mice. Expression of the Purkinje cell marker calbindin in the cerebellum of wild-type (A,C) and NFIX−/− (B,D) mice at P5. In the wild type, Purkinje cells were beginning to extend dendritic processes radially into the molecular layer (arrows in C). In the knockout, elaboration of dendritic processes was markedly delayed (arrowheads in D). C,D are higher magnification views of the boxed regions (lobules IV/V) in A,B, respectively. E: qPCR on wild-type and NFIX−/− P5 cerebellar tissue demonstrated significantly reduced levels of calbindin mRNA in the mutant. *P < 0.05, t-test. Scale bar = 20 µm in D (applies to B,D); 100 µm for A,C.
Absence of NFIX culminates in delayed glial development within the cerebellum
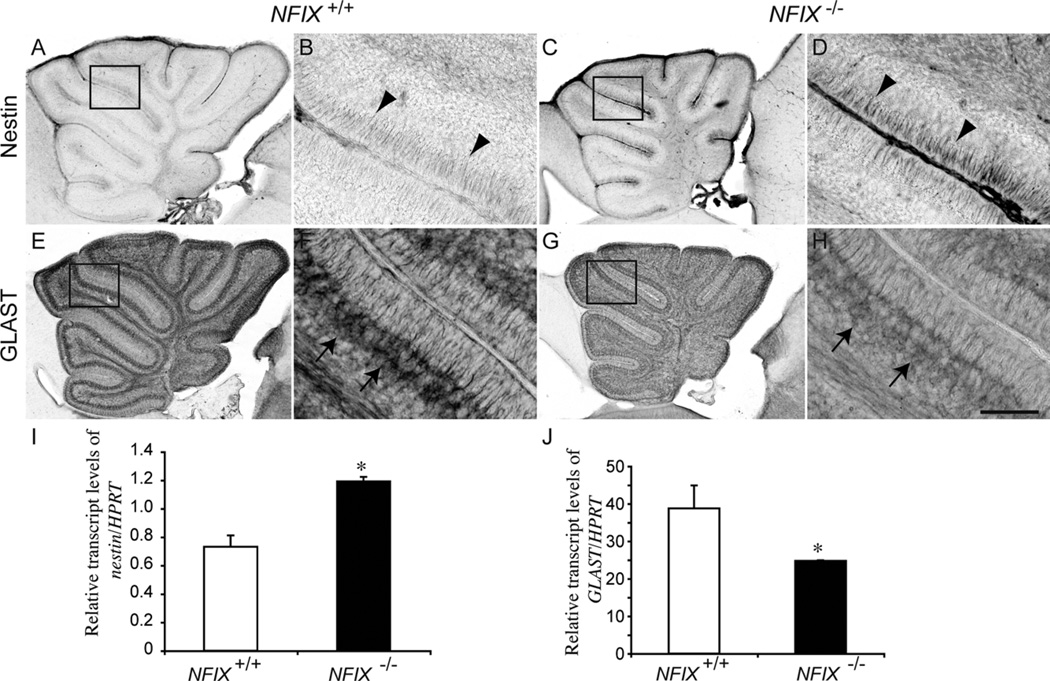
Surprisingly little is known about the genetic determinants of glial development in the cerebellum. Developmentally, the dorsomedial ventricular zone along the fourth ventricle gives rise to the progenitors of the astrocytic populations of the cerebellum (Bergmann glia and velate protoplasmic astrocytes), as well as Purkinje cells (Hoogland and Kuhn, 2009). Given the role of NFI genes in regulating glial differentiation within the brain (Barry et at., 2008; Piper et al., 2010; Shu et al., 2003) and spinal cord (Deneen et al., 2006), we next investigated whether NFIX contributes to the formation of astrocytes within the cerebellum. Ventricular zone-derived radial progenitors that give rise to cerebellar cells, including Bergmann glia, express the intermediate filament protein nestin (Zhang et al., 2010). In normal animals, nestin expression declines rapidly in the early postnatal period (Zhang et al., 2010). In P5 wild-type mice, nestin expression was detected in the radial fibers of the progenitor cells (Fig. 9A,B). In the NFIX mutant, however, nestin expression was markedly higher (Fig. 9C,D), a finding supported by an increase in nestin mRNA in the mutant as assayed by qPCR (Fig. 9I), indicative of delayed progenitor cell differentiation. Upon differentiation, astroglial cells express cell type-specific markers such as the glutamate/aspartate transporter (GLAST; Hartfuss et al., 2001). In the cerebellum, GLAST is highly localized to the cell bodies and processes of Bergmann glia (Storck et al., 1992), which was clearly seen in the wild-type cerebellum at P5 (Fig. 9E,F). In the mutant, however, GLAST expression was reduced within the Bergmann glia, with markedly lower GLAST immunoreactivity observed within the cell bodies and radial fibers of these cells (Fig. 9G,H). A reduction in Glast expression in NFIX−/− mice was also seen at the mRNA level at this age (Fig. 9J). Collectively, the expression of nestin and GLAST indicate that the differentiation of Bergmann glia is also delayed in NFIX−/− mice.
Figure 9.
Delayed differentiation of progenitors in the NFIX−/− cerebellum. Expression of the neural progenitor cell marker nestin (A–D) and the astroglial marker GLAST (E–H) in the cerebellum of wild-type and NFIX−/− mice at P5. In the wild-type at P5, expression of nestin was low, with expression observed on radial fibers of progenitors at the pial surface of the cerebellum (arrowheads in B). In the mutant, however, expression of nestin was markedly higher (arrowheads in D). Conversely, expression of GLAST by Bergmann glia was markedly higher in the wild type (arrows in F) compared with the NFIX knockout cerebellum (arrows in H). B,D,F,H are higher magnification views of the boxed regions (lobules IV–VI) in A,C,E,G, respectively. I,J: qPCR on wild-type and NFIX−/− P5 cerebellar tissue demonstrated significantly increased levels of nestin (I) and significantly reduced levels of Glast (J) mRNA in the mutant. *P < 0.05, t-test. Scale bar = 20 µm in H (applies to B,D,F,H); 100 µm for A,C,E,G.
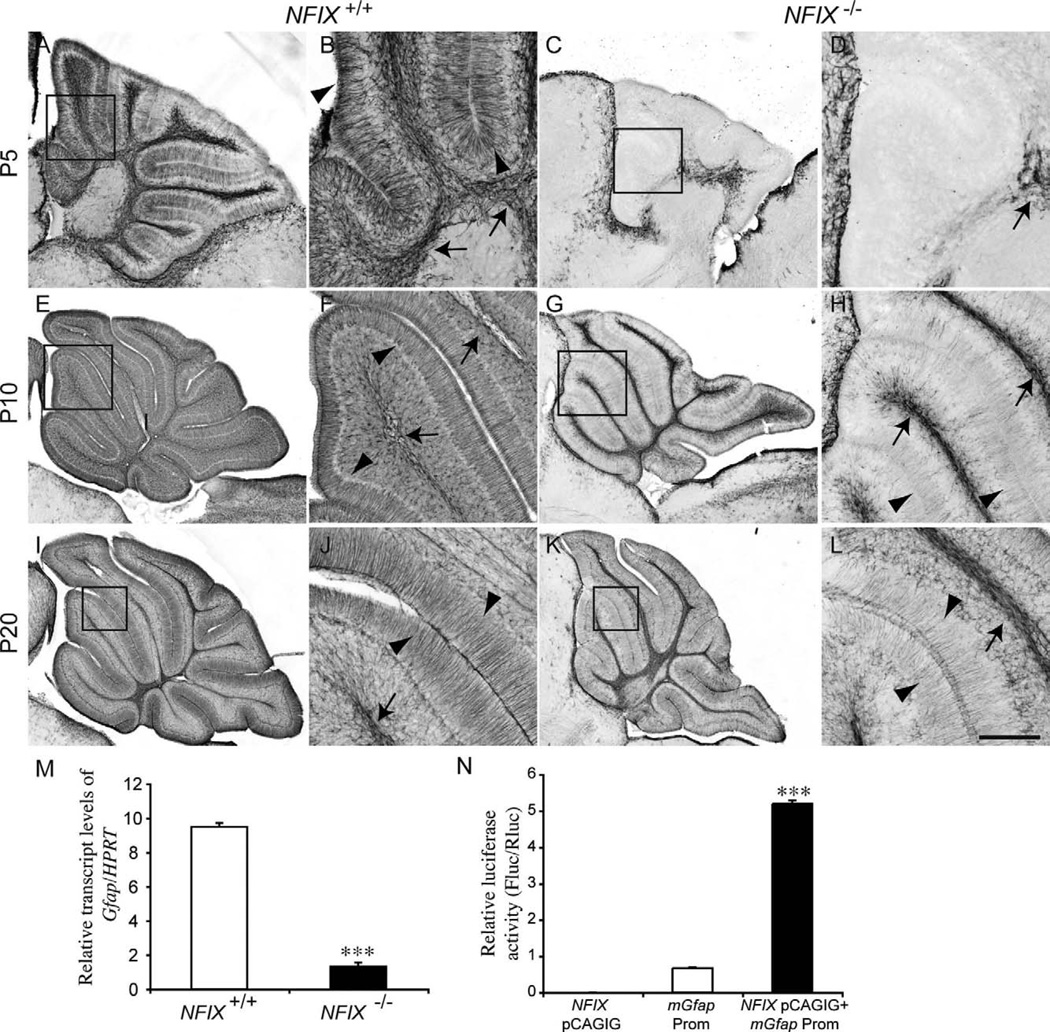
As they mature, cerebellar astrocytes begin expressing glial fibrillary acidic protein (Gfap; Gimenez et al., 2000). GFAP is strongly expressed within the radial fibers of mature Bergmann glia. In P5 wild-type mice, GFAP immunoreactivity was observed in the velate protoplasmic astrocytes within the white matter and within the radial fibers of Bergmann glia (Fig. 10A,B). In the NFIX mutant, however, there was no GFAP immunoreactivity within the molecular layer of the cerebellum, and very little expression within the cerebellar white matter (Fig. 10C,D), further suggesting a delay in the differentiation of cerebellar astrocytes. Together with the reduced detection of apoptosis within the cerebellar white matter at P5 (Fig. 4), these data indicate that the differentiation or survival of glial progenitors is abnormal in NFIX−/− mice. The reduction in GFAP expression at P5 was also mirrored at the mRNA level, with significantly reduced Gfap expression in the mutant at this age (Fig. 10M). Between P10 (Fig. 10E–H) and P20 (Fig. 10I–L), expression of GFAP in the mutant did become evident within the white matter and in the Bergmann glia, but expression levels were reduced compared with those of age-matched controls. NFI genes, including NFIX, have previously been implicated in the direct regulation of Gfap expression (Cebolla and Vallejo, 2006). By using a transcriptional reporter assay, we confirmed these findings, demonstrating that NFIX directly activates gene transcription under control of the Gfap promoter (Fig. 10N). Taken together, these data reveal that astrocytic development is delayed in the cerebellum of NFIX−/− mice and that NFIX may drive the differentiation of cerebellar astrocytes through the transcriptional activation of glial-specific genes such as Gfap.
Figure 10.
Delayed glial differentiation in the cerebellum of NFIX−/− mice. Expression of the mature astrocyte marker GFAP at P5 (A–D), P10 (E–H), and P20 (I–L). In the wild type at P5, GFAP expression was evident in both velate protoplasmic astrocytes (arrows in B) and Bergmann glia (arrowheads in B). In the mutant, however, no GFAP expression was detected in the Bergmann glia, and there appeared to be far fewer velate protoplasmic astrocytes in the white matter (arrow in D). At P10 and P20 expression of GFAP within the velate protoplasmic astrocytes (arrows in F,J) and Bergmann glia (arrowheads in F,J) of the wild-type became more pronounced. In the mutant, however, although GFAP expression became more apparent in velate protoplasmic astrocytes (arrows H,L) and Bergmann glia (arrowheads H,L), the level of expression was reduced in comparison with the controls. B,D,F,H,J,L are higher magnification views of the boxed regions (lobules III–V) in A,C,E,G,I,K, respectively. M: qPCR on wild-type and NFIX−/− P5 cerebellar tissue demonstrated significantly reduced levels of Gfap mRNA in the NFIX mutant. N: Transcription reporter assay, demonstrating that expression of NFIX (NFIX pCAGIG) elicits robust transcriptional activation of the reporter gene (firefly luciferase) under the control of the Gfap promoter (Gfap Prom). ***P < 0.001, t-test (M), ANOVA (N). Scale bar = 40 µm in L (applies to J,L); 100 µm for A,C; 25 µm for B,D,F,H; 150 µm for E,G; 200 µm for I,K.
DISCUSSION
In the embryonic murine nervous system, development of the cerebellum is initiated at approximately midgestation, with the specification of cerebellar progenitors within rhombomere 1, and concludes approximately 3 weeks into the postnatal period (Millen and Gleeson, 2008; Sotelo, 2004). The cerebellum is distinct in that the majority of its development occurs postnatally (Miale and Sidman, 1961). Indeed, only Purkinje cells and neurons within the deep cerebellar nuclei exit the cell cycle completely during embryogenesis, with the progenitors of the remaining neuronal and glial lineages continuing to proliferate postnatally. The transcription factors NFIA and NFIB have previously been shown to regulate the differentiation and maturation of cerebellar granule neurons (Steele-Perkins et al., 2005; Wang et al., 2007). Here, we extend these findings by demonstrating that NFIX regulates both neuronal and glial development in the postnatal cerebellum, providing insight into the molecular mechanisms controlling the morphogenesis of this critical brain structure.
The NFI genes have been implicated in regulating the development of many regions within the central nervous system, including the cerebellum (Steele-Perkins et al., 2005; Wang et al., 2007), spinal cord (Deneen et al., 2006), neocortex (Piper et al., 2009a), pons (Kumbasar et al., 2009), and hippocampus (Barry et al., 2008). Much of this work has focussed on two family members, NFIA and NFIB, demonstrating that NFI genes orchestrate multiple developmental processes in vivo via transcriptional activation and/or repression (Mason et al., 2009). NFIA, for instance, has been implicated in the formation of cortical axon tracts (Shu et al., 2003) and controlling granule cell development within the nascent cerebellum (Wang et al., 2007), promoting astrocytogenesis within the neocortex and hippocampus (Shu et al., 2003), and driving the generation of astrocyte and oligodendrocyte precursors within the spinal cord (Deneen et al., 2006). Such multiple effects indicate that NFI gene function is likely to be dependent on the developmental context in which each gene is expressed. Analysis of NFIX−/− mice also suggests that this gene is required for correct morphological development of the neocortex and hippocampus (Campbell et al., 2008). The role of NFIX in cerebellar development was previously unclear, despite the broad pattern of NFIX expression in this structure during postnatal development (Wang et al., 2007). Our data show that the cerebellum is significantly smaller in NFIX−/− mice and that neuronal populations, such as cerebellar granule neurons and Purkinje cells, display delayed developmental profiles. Moreover, development of astrocytes is markedly delayed in the absence of NFIX, highlighting the fact that this gene controls the development of multiple cell lineages during cerebellar development.
At a mechanistic level, our understanding of the molecular mechanisms regulating granule cell development are well advanced, with a suite of genes known to contribute to the specification, migration and maturation of these neurons (Sotelo, 2004). The genes regulating glial development within the cerebellum, however, have received far less attention, so our understanding of the mechanisms regulating cerebellar gliogenesis is less complete. Two distinct types of astrocytes are found within the mature cerebellar cortex: Bergmann glia and velate protoplasmic astrocytes, both of which are initially derived from progenitor cells found within the dorsomedial ventricular zone along the fourth ventricle (Hoogland and Kuhn, 2009; Milosevic and Goldman, 2002). Mature Bergmann glia, like Müller glia within the retina, maintain radial fibers that project through the molecular layer to the pial surface. The cell bodies of Bergmann glia migrate from the ventricular zone through the mantle to form a monolayer within the Purkinje cell layer. This process is regulated by Notch signalling (Komine et al., 2007). Our data show that the development of Bergmann glia is delayed in the absence of NFIX, insofar as expression of GLAST and GFAP is markedly reduced in NFIX mutants, whereas the expression of nestin, a progenitor marker, is upregulated. However, both GLAST and GFAP label glial processes, so it is impossible to determine via their expression whether the migration of Bergmann glia into the Purkinje cell layer is delayed or whether monolayer formation is disrupted in NFIX−/− mice. However, in light of recent evidence linking NFI genes to the induction of gliogenesis (Namihira et al., 2009), coupled with our data demonstrating that NFIX activates transcription under control of the Gfap promoter, it seems likely that NFIX plays a key role in driving the differentiation of Bergmann glia during development. The development of velate protoplasmic astrocytes is even less well understood. Our data also indicate that NFIX is required for the development of these astrocytes, because their appearance is markedly delayed in NFIX−/− mice. Collectively, these findings highlight a role for NFIX in driving astrocytic differentiation within the cerebellar cortex, providing a significant advance in our understanding of how these critical cells form during development.
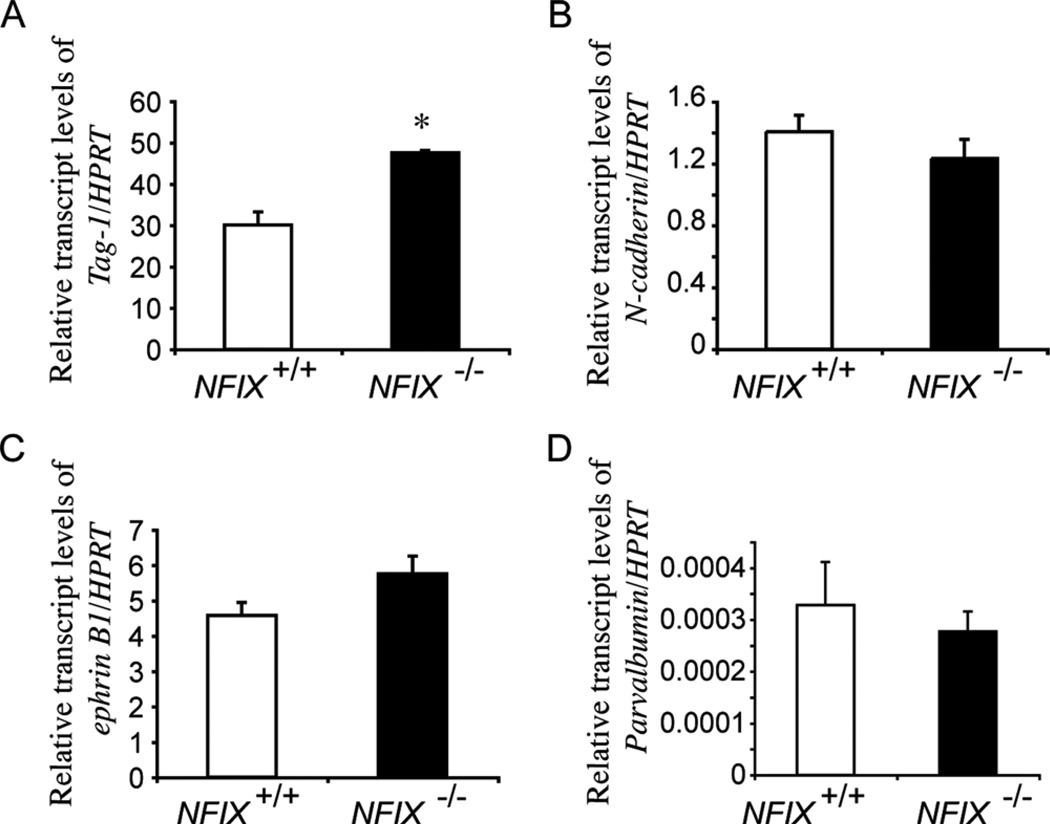
Cerebellar granule cells are the most abundant population of neurons within the brain, and their developmental ontogeny is well defined. At a molecular level, many genes have been implicated in granule cell development, including NFIA and NFIB (Kilpatrick et al., 2010). In an elegant series of studies, NFI genes were shown to contribute to multiple stages of granule cell development, including maturation, migration, axon guidance, dendrite formation, and expression of genes including Gabra6, Tag-1, N-cadherin, and ephrin B1 (Wang et al., 2004, 2007, 2010). Our data demonstrate that granule cell development also requires NFIX, as the development of these neurons was delayed in the absence of this gene. Exactly how NFIX contributes to granule cell development is unclear, although previous studies suggest possible mechanisms. For example, studies linking NFI gene function to granule cell migration, dendrite development, and axon outgrowth employed an NFI dominant repressor construct, which should repress all members of this transcription factor family (Wang et al., 2007). Given the expression of NFIX in EGL progenitors, premigratory granule cells, and IGL granule cells (Fig. 1), it is possible that the effects observed with the dominant repressor might have occurred in part via repression of NFIX function. In support of this, qPCR on P5 tissue demonstrated that the expression of the cell-adhesion molecule Tag-1 was significantly higher in NFIX−/− mice, whereas levels of N-cadherin and ephrin B1 were not significantly different (Fig. 11), thereby providing further insight into the mechanism by which NFIX regulates granule cell development. Furthermore, the delayed migration of granule cells through the molecular layer (Fig. 5L,N) at P20 could also result from cell-extrinsic mechanisms. Granule cells migrate into the IGL in close association with radially oriented Bergmann glial fibers (Rakic, 1971). Thus, the delay in the differentiation of the Bergmann glia might also have contributed to the delayed migration of postmitotic granule cells.
Figure 11.
qPCR analysis of P5 wild-type and NFIX−/− cerebellar tissue. qPCR on P5 cerebellar tissue revealed a significant increase in Tag-1 mRNA in the NFIX−/− mutant compared with controls (A). Levels of N-cadherin (B), ephrin B1 (C), and parvalbumin (D) were not significantly different between controls and NFIX−/− mice. *P < 0.05, t-test.
Recently, NFIA was shown to act through at least two distinct mechanisms during development of the cerebral cortex, driving progenitor cell differentiation via the activation of glial-specific genes, and also repressing genes responsible for progenitor cell maintenance such as the Notch effector gene Hes1 (Piper et al., 2010). Hes1 has also been shown to promote progenitor cell maintenance within the cerebellum (Solecki et al., 2001). These findings led us to hypothesize that Notch pathway activity may be misregulated in NFIX−/− mice. However, qPCR at P5 revealed no significant alterations in Hes1 or Hes5 mRNA in the mutant (data not shown), suggesting that NFIX does not regulate the Notch pathway in this developmental context during the early postnatal period. Another molecule implicated in the proliferation of EGL progenitor cells is Shh, which is expressed by Purkinje cells (Spassky et al., 2008; Wechsler-Reya and Scott, 1999). The levels of Shh mRNA were also unchanged in the cerebellum of NFIX−/− mice at P5 (data not shown). From these findings, we infer that NFIX may regulate the differentiation of progenitor cells directly, rather that regulating the expression of genes controlling EGL progenitor proliferation.
Purkinje cells provide the efferent output of the cerebellum. The development of these neurons is well characterized at both a morphological and a molecular level. Our data show that the development of Purkinje cells is also reliant on NFIX, insofar as development of the dendritic processes of these neurons is delayed at P5 in NFIX−/− mice. The levels of calbindin mRNA were also significantly reduced in the NFIX−/− mutants at P5, providing a further indication of a developmental delay in Purkinje cell development. Parvalbumin expression, on the other hand, was not significantly different at P5, suggesting that parvalbumin-expressing stellate and basket cells might not be delayed in their development at this time (Fig. 11). The delay in the elaboration of Purkinje cell dendrites might also be cell extrinsic, because the Purkinje cell dendritic tree elaborates along the scaffold of the Bergmann glial fibers (Lordkipanidze and Dunaevsky, 2005). Thus, the delays in Purkinje cell development in NFIX−/− mice are likely to be the result of multiple deficits.
In conclusion, our data provide a comprehensive insight into the developmental abnormalities within the cerebellum of mice lacking NFIX. The results show that NFIX contributes to multiple aspects of cerebellar development and that the differentiation of both neuronal and glial lineages is delayed in the absence of this transcription factor. Future studies will focus on the specific mechanisms through which NFIX mediates these important developmental events and whether they are conserved in other regions affected by the loss of this gene, including the cortex and hippocampus.
ACKNOWLEDGMENTS
We thank John Baisden, Erica Little, and Oressia Zalucki for technical assistance; Rowan Tweedale for critical analysis of the manuscript; and Niels Danbolt (University of Oslo) for providing the anti-GLAST antibody.
Grant sponsor: National Health and Medical Research Council (NHMRC; to M.P., L.J.R.); Grant number: MP-1003462; Grant number: LJR-569504; Grant sponsor: National Institute of Health; Grant number: RMGHL080624.
LITERATURE CITED
- Aruga J, Inoue T, Hoshino J, Mikoshiba K. Zic2 controls cerebellar development in cooperation with Zic1. J Neurosci. 2002;22:218–225. doi: 10.1523/JNEUROSCI.22-01-00218.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry G, Piper M, Lindwall C, Moldrich R, Mason S, Little E, Sarkar A, Tole S, Gronostajski RM, Richards LJ. Specific glial populations regulate hippocampal morphogenesis. J Neurosci. 2008;28:12328–12340. doi: 10.1523/JNEUROSCI.4000-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell CE, Piper M, Plachez C, Yeh YT, Baizer JS, Osinski JM, Litwack ED, Richards LJ, Gronostajski RM. The transcription factor Nfix is essential for normal brain development. BMC Dev Biol. 2008;8:52. doi: 10.1186/1471-213X-8-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebolla B, Vallejo M. Nuclear factor-I regulates glial fibrillary acidic protein gene expression in astrocytes differentiated from cortical precursor cells. J Neurochem. 2006;97:1057–1070. doi: 10.1111/j.1471-4159.2006.03804.x. [DOI] [PubMed] [Google Scholar]
- Chizhikov V, Millen KJ. Development and malformations of the cerebellum in mice. Mol Genet Metab. 2003;80:54–65. doi: 10.1016/j.ymgme.2003.08.019. [DOI] [PubMed] [Google Scholar]
- Deneen B, Ho R, Lukaszewicz A, Hochstim CJ, Gronostajski RM, Anderson DJ. The transcription factor NFIA controls the onset of gliogenesis in the developing spinal cord. Neuron. 2006;52:953–968. doi: 10.1016/j.neuron.2006.11.019. [DOI] [PubMed] [Google Scholar]
- Edmondson JC, Liem RK, Kuster JE, Hatten ME. Astrotactin: a novel neuronal cell surface antigen that mediates neuron-astroglial interactions in cerebellar microcultures. J Cell Biol. 1988;106:505–517. doi: 10.1083/jcb.106.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelkamp D, Rashbass P, Seawright A, van Heyningen V. Role of Pax6 in development of the cerebellar system. Development. 1999;126:3585–3596. doi: 10.1242/dev.126.16.3585. [DOI] [PubMed] [Google Scholar]
- Flora A, Klisch TJ, Schuster G, Zoghbi HY. Deletion of Atoh1 disrupts Sonic Hedgehog signaling in the developing cerebellum and prevents medulloblastoma. Science. 2009;326:1424–1427. doi: 10.1126/science.1181453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukaya M, Yamada K, Nagashima M, Tanaka K, Watanabe M. Down-regulated expression of glutamate transporter GLAST in Purkinje cell-associated astrocytes of reeler and weaver mutant cerebella. Neurosci Res. 1999;34:165–175. doi: 10.1016/s0168-0102(99)00052-8. [DOI] [PubMed] [Google Scholar]
- Furuta A, Rothstein JD, Martin LJ. Glutamate transporter protein subtypes are expressed differentially during rat CNS development. J Neurosci. 1997;17:8363–8375. doi: 10.1523/JNEUROSCI.17-21-08363.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge SN, Ma YF, Hioki H, Wei YY, Kaneko T, Mizuno N, Gao GD, Li JL. Coexpression of VGLUT1 and VGLUT2 in trigeminothalamic projection neurons in the principal sensory trigeminal nucleus of the rat. J Comp Neurol. 2010;518:3149–3168. doi: 10.1002/cne.22389. [DOI] [PubMed] [Google Scholar]
- Gimenez YRM, Langa F, Menet V, Privat A. Comparative anatomy of the cerebellar cortex in mice lacking vimentin, GFAP, and both vimentin and GFAP. Glia. 2000;31:69–83. doi: 10.1002/(sici)1098-1136(200007)31:1<69::aid-glia70>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- Guillery RW. On counting and counting errors. J Comp Neurol. 2002;447:1–7. doi: 10.1002/cne.10221. [DOI] [PubMed] [Google Scholar]
- Hanbury R, Ling ZD, Wuu J, Kordower JH. GFAP knockout mice have increased levels of GDNF that protect striatal neurons from metabolic and excitotoxic insults. J Comp Neurol. 2003;461:307–316. doi: 10.1002/cne.10667. [DOI] [PubMed] [Google Scholar]
- Hartfuss E, Galli R, Heins N, Gotz M. Characterization of CNS precursor subtypes and radial glia. Dev Biol. 2001;229:15–30. doi: 10.1006/dbio.2000.9962. [DOI] [PubMed] [Google Scholar]
- Hatten ME, Roussel MF. Development and cancer of the cerebellum. Trends Neurosci. 2011;34:134–142. doi: 10.1016/j.tins.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haverkamp S, Specht D, Majumdar S, Zaidi NF, Brandstatter JH, Wasco W, Wassle H, Tom Dieck S. Type 4 OFF cone bipolar cells of the mouse retina express calsenilin and contact cones as well as rods. J Comp Neurol. 2008;507:1087–1101. doi: 10.1002/cne.21612. [DOI] [PubMed] [Google Scholar]
- Hockfield S, McKay RD. Identification of major cell classes in the developing mammalian nervous system. J Neurosci. 1985;5:3310–3328. doi: 10.1523/JNEUROSCI.05-12-03310.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogland TM, Kuhn B. Recent developments in the understanding of astrocyte function in the cerebellum in vivo. Cerebellum. 2009;9:264–271. doi: 10.1007/s12311-009-0139-z. [DOI] [PubMed] [Google Scholar]
- Janmaat S, Frederic F, Sjollema K, Luiten P, Mariani J, van der Want J. Formation and maturation of parallel fiber-Purkinje cell synapses in the Staggerer cerebellum ex vivo. J Comp Neurol. 2009;512:467–477. doi: 10.1002/cne.21910. [DOI] [PubMed] [Google Scholar]
- Kilpatrick DL, Wang W, Gronostajski R, Litwack ED. Nuclear factor I and cerebellar granule neuron development: an intrinsic-extrinsic interplay. Cerebellum. 2010 doi: 10.1007/s12311-010-0227-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komine O, Nagaoka M, Watase K, Gutmann DH, Tanigaki K, Honjo T, Radtke F, Saito T, Chiba S, Tanaka K. The monolayer formation of Bergmann glial cells is regulated by Notch/RBP-J signaling. Dev Biol. 2007;311:238–250. doi: 10.1016/j.ydbio.2007.08.042. [DOI] [PubMed] [Google Scholar]
- Kumbasar A, Plachez C, Gronostajski RM, Richards LJ, Litwack ED. Absence of the transcription factor Nfib delays the formation of the basilar pontine and other mossy fiber nuclei. J Comp Neurol. 2009;513:98–112. doi: 10.1002/cne.21943. [DOI] [PubMed] [Google Scholar]
- Lordkipanidze T, Dunaevsky A. Purkinje cell dendrites grow in alignment with Bergmann glia. Glia. 2005;51:229–234. doi: 10.1002/glia.20200. [DOI] [PubMed] [Google Scholar]
- Mason S, Piper M, Gronostajski RM, Richards LJ. Nuclear factor one transcription factors in CNS development. Mol Neurobiol. 2009;39:10–23. doi: 10.1007/s12035-008-8048-6. [DOI] [PubMed] [Google Scholar]
- Miale IL, Sidman RL. An autoradiographic analysis of histogenesis in the mouse cerebellum. Exp Neurol. 1961;4:277–296. doi: 10.1016/0014-4886(61)90055-3. [DOI] [PubMed] [Google Scholar]
- Millen KJ, Gleeson JG. Cerebellar development and disease. Curr Opin Neurobiol. 2008;18:12–19. doi: 10.1016/j.conb.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milosevic A, Goldman JE. Progenitors in the postnatal cerebellar white matter are antigenically heterogeneous. J Comp Neurol. 2002;452:192–203. doi: 10.1002/cne.10384. [DOI] [PubMed] [Google Scholar]
- Namihira M, Kohyama J, Semi K, Sanosaka T, Deneen B, Taga T, Nakashima K. Committed neuronal precursors confer astrocytic potential on residual neural precursor cells. Dev Cell. 2009;16:245–255. doi: 10.1016/j.devcel.2008.12.014. [DOI] [PubMed] [Google Scholar]
- Petkau TL, Neal SJ, Orban PC, MacDonald JL, Hill AM, Lu G, Feldman HH, Mackenzie IR, Leavitt BR. Progranulin expression in the developing and adult murine brain. J Comp Neurol. 2010;518:3931–3947. doi: 10.1002/cne.22430. [DOI] [PubMed] [Google Scholar]
- Piper M, Dawson AL, Lindwall C, Barry G, Plachez C, Richards LJ. Emx and Nfi genes regulate cortical development and axon guidance in the telencephalon. Novartis Found Symp. 2007;288:230–242. discussion 242-235, 276–281. [PubMed] [Google Scholar]
- Piper M, Moldrich RX, Lindwall C, Little E, Barry G, Mason S, Sunn N, Kurniawan ND, Gronostajski RM, Richards LJ. Multiple non-cell-autonomous defects underlie neocortical callosal dysgenesis in Nfib-deficient mice. Neural Dev. 2009a;4:43. doi: 10.1186/1749-8104-4-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper M, Plachez C, Zalucki O, Fothergill T, Goudreau G, Erzurumlu R, Gu C, Richards LJ. Neuropilin 1-Sema signaling regulates crossing of cingulate pioneering axons during development of the corpus callosum. Cereb Cortex. 2009b;19(Suppl 1):i11–i21. doi: 10.1093/cercor/bhp027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper M, Barry G, Hawkins J, Mason S, Lindwall C, Little E, Sarkar A, Smith AG, Moldrich RX, Boyle GM, Tole S, Gronostajski RM, Bailey TL, Richards LJ. NFIA controls telencephalic progenitor cell differentiation through repression of the Notch effector Hes1. J Neurosci. 2010;30:9127–9139. doi: 10.1523/JNEUROSCI.6167-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plachez C, Danbolt NC, Recasens M. Transient expression of the glial glutamate transporters GLAST and GLT in hippocampal neurons in primary culture. J Neurosci Res. 2000;59:587–593. doi: 10.1002/(SICI)1097-4547(20000301)59:5<587::AID-JNR1>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Plachez C, Lindwall C, Sunn N, Piper M, Moldrich RX, Campbell CE, Osinski JM, Gronostajski RM, Richards LJ. Nuclear factor I gene expression in the developing forebrain. J Comp Neurol. 2008;508:385–401. doi: 10.1002/cne.21645. [DOI] [PubMed] [Google Scholar]
- Rakic P. Neuron-glia relationship during granule cell migration in developing cerebellar cortex. A Golgi and electron microscopic study in macacus rhesus. J Comp Neurol. 1971;141:283–312. doi: 10.1002/cne.901410303. [DOI] [PubMed] [Google Scholar]
- Sajan SA, Waimey KE, Millen KJ. Novel approaches to studying the genetic basis of cerebellar development. Cerebellum. 2010;9:272–283. doi: 10.1007/s12311-010-0169-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz CM, Cheng A, Mughal MR, Mattson MP, Yao PJ. Clathrin assembly proteins AP180 and CALM in the embryonic rat brain. J Comp Neurol. 2010;518:3803–3818. doi: 10.1002/cne.22425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergaki MC, Guillemot F, Matsas R. Impaired cerebellar development and deficits in motor coordination in mice lacking the neuronal protein BM88/Cend1. Mol Cell Neurosci. 2010;44:15–29. doi: 10.1016/j.mcn.2010.01.011. [DOI] [PubMed] [Google Scholar]
- Shu T, Butz KG, Plachez C, Gronostajski RM, Richards LJ. Abnormal development of forebrain midline glia and commissural projections in Nfia knock-out mice. J Neurosci. 2003;23:203–212. doi: 10.1523/JNEUROSCI.23-01-00203.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solecki DJ, Liu XL, Tomoda T, Fang Y, Hatten ME. Activated Notch2 signaling inhibits differentiation of cerebellar granule neuron precursors by maintaining proliferation. Neuron. 2001;31:557–568. doi: 10.1016/s0896-6273(01)00395-6. [DOI] [PubMed] [Google Scholar]
- Sotelo C. Cellular and genetic regulation of the development of the cerebellar system. Prog Neurobiol. 2004;72:295–339. doi: 10.1016/j.pneurobio.2004.03.004. [DOI] [PubMed] [Google Scholar]
- Spassky N, Han YG, Aguilar A, Strehl L, Besse L, Laclef C, Ros MR, Garcia-Verdugo JM, Alvarez-Buylla A. Primary cilia are required for cerebellar development and Shh-dependent expansion of progenitor pool. Dev Biol. 2008;317:246–259. doi: 10.1016/j.ydbio.2008.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele-Perkins G, Plachez C, Butz KG, Yang G, Bachurski CJ, Kinsman SL, Litwack ED, Richards LJ, Gronostajski RM. The transcription factor gene Nfib is essential for both lung maturation and brain development. Mol Cell Biol. 2005;25:685–698. doi: 10.1128/MCB.25.2.685-698.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitt TN, Hatten ME. Antibodies that recognize astrotactin block granule neuron binding to astroglia. Neuron. 1990;5:639–649. doi: 10.1016/0896-6273(90)90218-5. [DOI] [PubMed] [Google Scholar]
- Storck T, Schulte S, Hofmann K, Stoffel W. Structure, expression, and functional analysis of a Na+-dependent glutamate/aspartate transporter from rat brain. Proc Natl Acad Sci U S A. 1992;89:10955–10959. doi: 10.1073/pnas.89.22.10955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkmann K, Chen YY, Harris MP, Wullimann MF, Koster RW. The zebrafish cerebellar upper rhombic lip generates tegmental hindbrain nuclei by long-distance migration in an evolutionary conserved manner. J Comp Neurol. 2010;518:2794–2817. doi: 10.1002/cne.22364. [DOI] [PubMed] [Google Scholar]
- Wang W, Stock RE, Gronostajski RM, Wong YW, Schachner M, Kilpatrick DL. A role for nuclear factor I in the intrinsic control of cerebellar granule neuron gene expression. J Biol Chem. 2004;279:53491–53497. doi: 10.1074/jbc.M410370200. [DOI] [PubMed] [Google Scholar]
- Wang W, Mullikin-Kilpatrick D, Crandall JE, Gronostajski RM, Litwack ED, Kilpatrick DL. Nuclear factor I coordinates multiple phases of cerebellar granule cell development via regulation of cell adhesion molecules. J Neurosci. 2007;27:6115–6127. doi: 10.1523/JNEUROSCI.0180-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Crandall JE, Litwack ED, Gronostajski RM, Kilpatrick DL. Targets of the nuclear factor I regulon involved in early and late development of postmitotic cerebellar granule neurons. J Neurosci Res. 2010;88:258–265. doi: 10.1002/jnr.22199. [DOI] [PubMed] [Google Scholar]
- Wechsler-Reya RJ, Scott MP. Control of neuronal precursor proliferation in the cerebellum by Sonic Hedgehog. Neuron. 1999;22:103–114. doi: 10.1016/s0896-6273(00)80682-0. [DOI] [PubMed] [Google Scholar]
- Wiencken-Barger AE, Djukic B, Casper KB, McCarthy KD. A role for Connexin43 during neurodevelopment. Glia. 2007;55:675–686. doi: 10.1002/glia.20484. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Niu B, Yu D, Cheng X, Liu B, Deng J. Radial glial cells and the lamination of the cerebellar cortex. Brain Struct Funct. 2010;215:115–122. doi: 10.1007/s00429-010-0278-5. [DOI] [PubMed] [Google Scholar]
- Zhou BY, Liu Y, Kim B, Xiao Y, He JJ. Astrocyte activation and dysfunction and neuron death by HIV-1 Tat expression in astrocytes. Mol Cell Neurosci. 2004;27:296–305. doi: 10.1016/j.mcn.2004.07.003. [DOI] [PubMed] [Google Scholar]