SUMMARY
We have developed a technique, called Ubiquitin Ligase Substrate Trapping, for the isolation of ubiquitinated substrates in complex with their ubiquitin ligase (E3). By fusing a ubiquitin associated (UBA) domain to an E3 ligase, we were able to selectively purify the polyubiquitinated forms of E3 substrates. Using Ligase Traps of eight different F-box proteins (SCF specificity factors) coupled with mass spectrometry, we identified known, as well as previously uncharacterized substrates. Polyubiquitinated forms of candidate substrates associated with their cognate F-box partner, but not other Ligase Traps. Interestingly, the four most abundant candidate substrates identified for the F-box protein Saf1 were all vacuolar/lysosomal proteins. Analysis of one of these substrates, Prb1, showed that Saf1 selectively promotes ubiquitination of the unprocessed form of the zymogen. This suggests that Saf1 is part of a pathway that targets protein precursors for proteasomal degradation.
INTRODUCTION
The ubiquitin-proteasome system regulates protein activities through degradation and performs quality control of misfolded, mistranslated or aggregated peptides (Deshaies and Joazeiro, 2009; Finley et al., 2012; Ravid and Hochstrasser, 2008). The Skp1-Cul1-F-box (SCF) complex is one of several families of ubiquitin ligases that mediate the timely proteolysis of regulatory proteins (Cardozo and Pagano, 2004; Petroski and Deshaies, 2005). This complex is assembled around a cullin scaffold (Cdc53) that bridges a small RING finger protein (Rbx1) and an adaptor protein (Skp1), which in turn recruits an F-box protein. The F-box subunit serves as the substrate recognition module (Bai et al., 1996; Patton et al., 1998). In general, F-box proteins recognize their substrates only after they are modified, typically by phosphorylation (Hsiung et al., 2001; Orlicky et al., 2003; Skowyra et al., 1997). Budding yeast encodes 20 putative F-box proteins (Willems et al., 2004), whereas the humans encode 68 (Jin et al., 2004). Although numerous substrates have been identified for the well-characterized Cdc4 and Grr1 F-box proteins (Jonkers and Rep, 2009; Reed, 2003), many of the remaining yeast F-box proteins have few, if any, known substrates.
Several strategies have previously been used to search for in vivo substrates of ubiquitin ligases. In most studies, substrates were identified as proteins that were no longer ubiquitinated or were selectively stabilized when a ligase was inactivated (Benanti et al., 2007; Emanuele et al., 2011; Kim et al., 2011; Yen and Elledge, 2008). While this approach has identified some substrates, it has significant drawbacks. First, loss of ligase activity may perturb cellular physiology, causing cell cycle alterations or DNA damage, indirectly affecting the ubiquitin proteome. In addition, some substrates are targeted by more than one ligase and the absence of a single ligase may not lead to a significant change in the level of the target protein (Landry et al., 2012). Furthermore, by using protein levels as the indicator of ubiquitination, polyubiquitination events that result in a non-proteolytic outcome or which target only a specific subpopulation will not be detected. Other approaches to identifying ligase targets exploit the physical interaction between a ligase and its substrate (Busino et al., 2007; Davis et al., 2013). In these studies, immunoaffinity purification techniques are used to isolate ligase-substrate complexes. The major challenge in using this strategy is that ligase-substrate interactions are often too weak for successful co-purification of the target protein.
In this paper, we describe a method called Ubiquitin Ligase Substrate Trapping (“Ligase Trapping”), which we used to identify substrates of ubiquitin ligases. In this approach, a polyubiquitin-binding domain (UBA) is fused to an E3 ligase. The UBA increases the affinity of the ligase for its polyubiquitinated substrate, thereby enhancing ligase-substrate stability and permitting the isolation of polyubiquitinated substrates by affinity purification. We used this approach to look for target substrates of eight different F-box proteins and identified 17 known substrates specific to the F-box proteins examined. In addition, 18 previously uncharacterized candidates were shown to bind specifically to their targeting F-box protein as a polyubiquitinated species and/or were enriched in mutants lacking that F-box protein. Interestingly, the most abundant group of candidates isolated from the poorly described F-box protein Saf1 was vacuolar/lysosomal proteases. Characterization of one of these proteases, the Prb1 zymogen, indicated that the Saf1 Ligase Trap specifically bound the polyubiquitinated form of the unprocessed protein. This suggests a model in which the SCFSaf1 ligase is part of a pathway that targets incorrectly, or incompletely, processed vacuolar/lysosomal proteins.
Results
Fusion of ubiquitin-associated domains to F-box proteins increases their binding affinity to ubiquitinated substrates
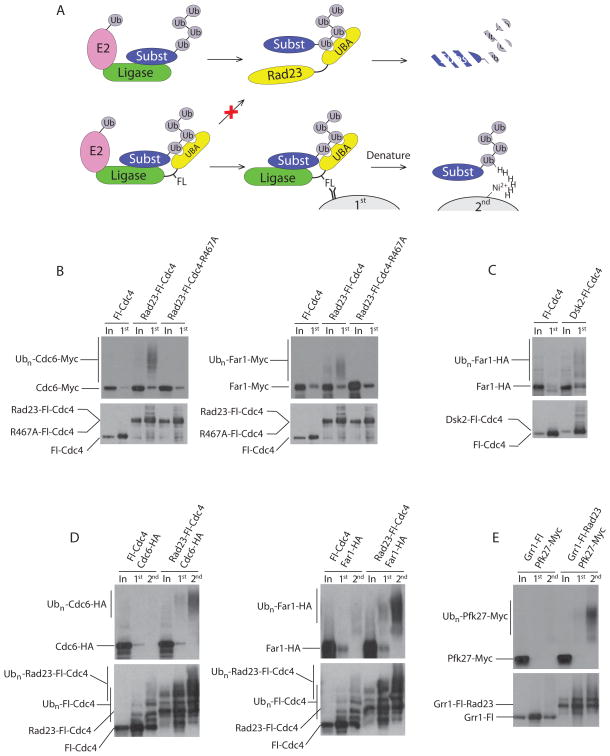
To increase the binding affinity between F-box proteins and their ubiquitinated substrates, we fused a ubiquitin-associated (UBA) domain from the soluble ubiquitin receptor Dsk2 or Rad23 to F-box proteins (Figure 1A). These UBAs have a strong preference for polyubiquitin, but bind both the K48- and K63-linked forms (Raasi et al., 2005; Sims et al., 2009). Using three tandem Flag epitopes as a linker, we fused the amino terminus of the F-box protein Cdc4 to the tandem UBAs of Rad23 (Rad23-Fl-Cdc4). Polyubiquitinated Cdc6 and Far1, known Cdc4 substrates, co-purified with Rad23-Fl-Cdc4 in a manner that required the UBA domain (Figure 1B). To determine whether association of the Ligase Trap with polyubiquitinated substrates required substrate recognition by Cdc4, we mutated arginine 467 of Cdc4’s WD40 domain in the fusion protein (Rad23-Fl-R467A). Since this mutation abolishes the ability of Cdc4 to bind substrates (Nash et al., 2001), it is lethal and thus the experiments in Figure 1B were performed with a galactose-inducible tagged allele in the presence of wild type CDC4. The R467A mutation strongly impaired the ability of Rad23-Fl-Cdc4 to interact with ubiquitinated Cdc6 and Far1 (Figure 1B), indicating that the binding of the fusion protein to ubiquitinated substrate depends upon both the binding of Cdc4 to the substrate and the UBA to ubiquitin chains. The single UBA domain of Dsk2 could also increase the binding affinity of Cdc4 for ubiquitinated substrates, as demonstrated using a Dsk2-Fl-Cdc4 fusion (Figure 1C). We named this approach ‘Ubiquitin Ligase Substrate Trapping’.
Figure 1. UBA fusions to ubiquitin ligases increase their affinity for ubiquitinated substrates.
(A) Schematic representation of Ligase Trapping and purification procedures. Top panel: During normal degradation, ubiquitin ligases like the SCF associate with both substrates and E2s. Ubiquitin-charged E2s then transfer their ubiquitin to the substrate, leading to the formation of a polyubiquitin chain. This is shuttled to the proteasome with the help of ubiquitin receptors, such as Rad23. Bottom panel: Ligase Trapping. A UBA domain is fused to a ubiquitin ligase via a Flag linker. The UBA binds the nascent ubiquitin chain while the linker allows a Flag immunoprecipitation of the ligase in complex with the substrate. The expression of 6xHis-tagged ubiquitin allows a second purification step that specifically isolates ubiquitinated species using Ni-NTA agarose beads under denaturing conditions. (B) Western blots of input whole cell extract (In) and the first purification step (1st). Flag immunoprecipitation of Fl-Cdc4, Rad23-Fl-Cdc4 and Rad23-Fl-Cdc4-R467A (containing a R467A mutation in CDC4) in cells expressing Cdc6-Myc or Far1-Myc. Ligase Traps were under the GAL1 promoter. (C) Flag immunoprecipitation was performed as in (B) using Dsk2-Fl-Cdc4 as bait and Far1 tagged with HA. (D) Two-step purification of Fl-Cdc4 and Rad23-Fl-Cdc4 expressed from the Cdc4 promoter. 6xHis-tagged ubiquitin was expressed under the GAL1 promoter in strains expressing Cdc6-Myc or Far1-Myc. Input (In), Flag immunoprecipitation (1st) followed by Ni-NTA purification (2nd) as illustrated in (A). (E) As in (D), using Grr1 and its known substrate Pfk27-Myc. Unlike Cdc4, Grr1 Traps are C-terminal fusions.
A proteomic screen for ubiquitinated substrates of the SCF using ‘Ligase Trapping’
To identify SCF substrates by Ligase Trapping, we analyzed ubiquitinated proteins bound to the UBA-F-box fusions using liquid chromatography and tandem mass spectrometry (LC-MS/MS). A two-step purification procedure was adopted using cells that express hexahistidine (6xHis) tagged ubiquitin. After an initial anti-Flag immunoprecipitation, a second purification was performed under denaturing conditions (6M urea) using Ni-NTA beads to enrich for ubiquitinated substrates (Figures 1A and 1D). This second step reduced nonspecific binding and eliminated proteins that were associated with the ubiquitinated species, but were not themselves substrates.
To examine whether Ligase Trapping worked for other F-box proteins, we produced UBA fusions with Grr1, an F-box protein involved in cell cycle regulation and nutrient response. A Grr1-Fl-Rad23 fusion protein bound ubiquitinated Pfk27, a known substrate of Grr1 (Benanti et al., 2007) (Figure 1E). Interestingly, since the Rad23 UBA domain was fused to the carboxy terminus of Grr1 (Grr1-Fl-Rad23), the successful isolation of tagged ubiquitinated substrates demonstrates that UBAs work at either terminus. Given these results, we performed a proteomic screen for yeast SCF substrates using UBA fusions of eight different F-box proteins (Figure S1). For these and all experiments, except those in Figure 1B, Ligase Traps were expressed under their endogenous F-box promoter and represent the only copy of the F-box protein in the cell.
A library of yeast strains, each expressing a galactose-inducible 6xHis-ubiquitin allele and a different UBA-F-box fusion protein (Figure 2), was used for our initial Ligase Trapping experiment. For the Cdc4 Ligase Trap, both N-terminal and C-terminal fusions were used. In addition, we generated Ligase Traps with a UBA from both Rad23 and Dsk2 for each F-box protein. In the case of Mdm30, Saf1, and Skp2, the addition of the tandem UBAs of Rad23 (but not the single Dsk2 UBA) resulted in reduced expression of that Ligase Trap, and therefore these constructs were not used. We performed a two-step purification of each fusion followed by LC-MS/MS analysis and identified 17 known SCF substrates (Figure 2 and Supplemental Table 1). Two replicates of each Grr1 Trap were performed identically, and the results of both are shown.
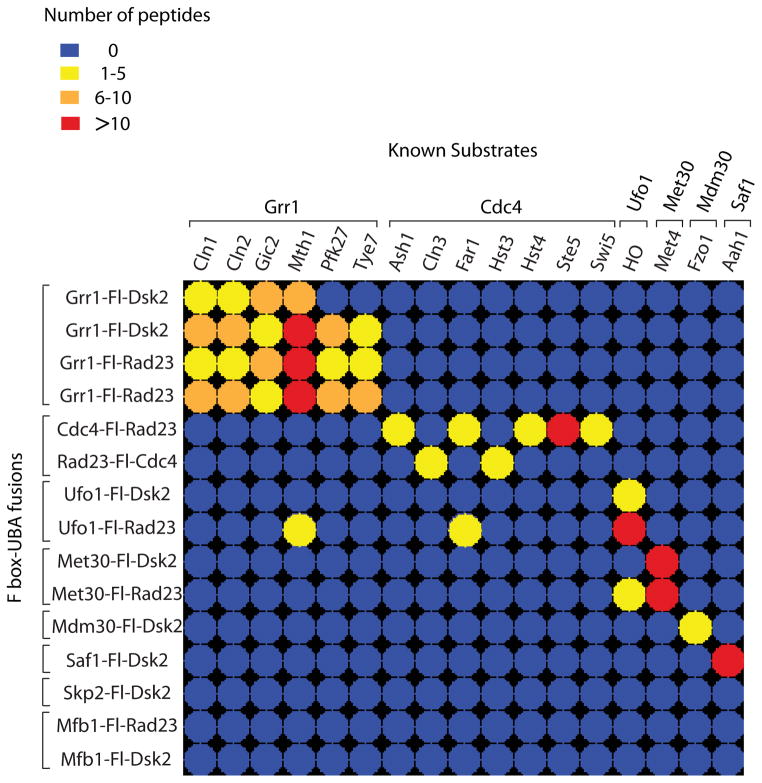
Figure 2. LC-MS/MS analysis of two-step purifications of Ligase Traps identifies known SCF substrates.
Color-coded matrix showing known SCF substrates identified by Ligase Trapping. Two-step purifications were performed from cell extracts expressing UBA fusions of eight F-box proteins. These represent data from initial purifications performed in parallel. In the case of Cdc4, both N- and C-terminal purifications are shown. Dsk2 and Rad23 fusions are shown when both fusions were well expressed. Repeats of two identical pairs of Grr1 traps are shown for comparison. Colors represent spectral counts for each protein in each purification. For full list of substrates and references, see Table S1.
Importantly, Ligase Trapping exhibited a high degree of specificity, as most known substrates co-purified uniquely with their expected F-box protein. Only three of the 17 substrates were found associated with an unrelated F-box protein: the Grr1 substrate, Mth1, and the Cdc4 substrate, Far1, were also detected in the Ufo1-Fl-Rad23 purification. While we can’t exclude a functional redundancy between these F-box proteins, the Ufo1-Fl-Rad23 Ligase Trap exhibited higher nonspecific binding to ubiquitinated targets than the other seven Traps. Ufo1 is unique among yeast F-box proteins for having a C-terminal ubiquitin-interacting motif (UIM), another class of ubiquitin binding domain, possibly contributing to this background. The Ufo1 substrate HO was captured not only by the Ufo1-Fl-Dsk2 and Ufo1-Fl-Rad23 traps, but also by Met30-Fl-Rad23.
The criteria used to select candidate SCF substrates is based on both an enrichment factor of >25-fold over other Ligase Traps and an average spectral count of ≥1.8. The exception was Ufo1, for which we instituted a threshold of ≥6 spectral counts and was not included in the fold enrichment calculation because of its high background. Hits that appear in more than one F-box protein may represent redundant targeting by F-box proteins, however these were put aside for our initial analysis. Thirty-eight candidate substrates met these criteria (Supplemental Table 1). Since our nonspecific binding was so low, the majority of these showed no binding to the other Ligase Traps analyzed. To determine whether proteins with fewer peptides also represent substrates, we examined several candidates that fell below our 1.8 spectral count cutoff for F-box proteins Grr1 and Cdc4.
Validation of Grr1 candidate substrates
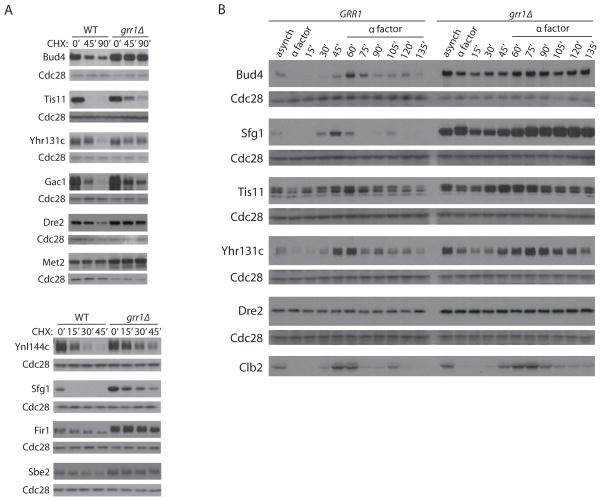
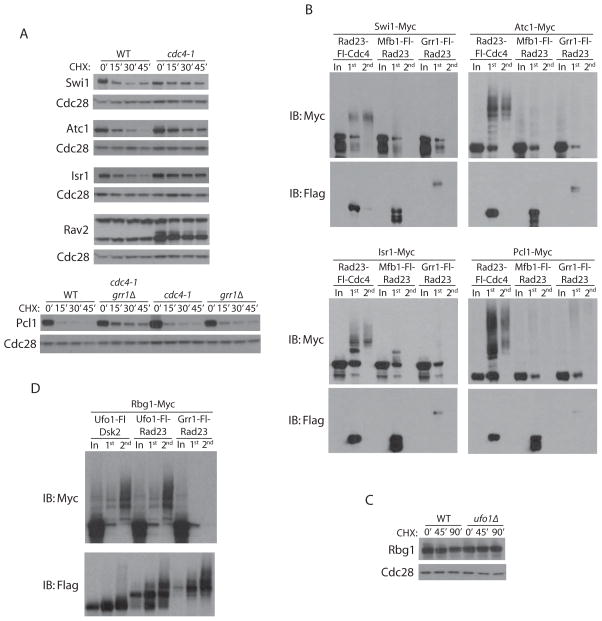
To assess the specificity of Ligase Trapping, we validated candidate substrates of Grr1 most extensively. First, we determined if the stability of Grr1 candidate substrates was increased in grr1Δ cells. Of 12 examined, six showed a significant increase in stability in asynchronous populations of grr1Δ cells (Bud4, Tis11, Gac1, Ynl144c, Sfg1, and Fir1), three others showed more modest changes (Yhr131c, Dre2, and Sbe2) and three appeared stable (Met2, Npl4, and Ykr045c) (Figure 3A and Table 1). While these nine candidate substrates were stabilized in grr1Δ cells relative to GRR1, some still exhibited significant turnover (Sfg1 and Tis11, Figure 3A). Destabilization that only occurs during a particular phase of the cell cycle might be less detectable in asynchronous cultures. Therefore, we analyzed five substrates in synchronized GRR1 and grr1Δ cells (Figure 3B). Grr1 was largely responsible for the cell cycle-dependent expression of Bud4 and Sfg1. Tis11 and Yhr131c also showed some cell cycle regulation, and this was modestly reduced in grr1Δ cells. In contrast, Dre2 was not regulated in a cell cycle-dependent fashion.
Figure 3. Stability of Grr1 candidate substrates.
(A) Epitope-tagged candidate Grr1 substrates were expressed in GRR1 or grr1Δ cells. To rescue slow growth, all grr1Δ cells are also rgt1Δ. Asynchronous cultures were treated with cycloheximide (CHX) for the indicated number of minutes and anti-Myc western blots were performed on whole cell extracts. Western blot of Cdc28 is shown as a loading control. (B) A subset of candidate substrates were analyzed during the cell cycle by Western blots of whole cell extracts. Cells were released from α-factor-mediated G1 arrest and collected at different time points. α-factor was again added after 60 minutes to re-arrest cells in the subsequent G1. Clb2 and Cdc28 are shown to monitor cell cycle progression and as a loading control, respectively. Asynchronous cells are shown for comparison.
Table 1. Substrates identified for Grr1.
A list of all proteins that were ≥25-fold enriched and had an average of ≥1.8 spectral counts are shown above the bar. Below the bar are three verified candidates that had an average of fewer than 1.8 spectral counts. Under t½ “+”, “+/−”, and “−” indicates a significant, slight or no change in grr1Δ cells. “*” indicates an increase in steady state levels but not in half-life. Under “Ubiquit”, “+”, “+/−”, and “−” indicate direct visualization of smears associated with a significant, slight, or no ubiquitination by either Ligase Trap or direct examination (for Ykr045c). Spectral counts for four independent experiments are shown, along with mean values. “BKGD” reflects the mean spectral counts observed from all other Ligase Traps. Known substrates identified in these four experiments are highlighted in gray.
| t1/2 | Ubiquit | Spectral count | Gene function | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Exp1 | Exp2 | Exp3 | Exp4 | mean | BKGD | ||||
| Bud4 | + | + | 47 | 53 | 24 | 23 | 36.8 | 0 | Anillin-like. Bud site selection. |
| Mth1 | KNOWN | 25 | 30 | 10 | 13 | 13.8 | 0 | Repression of transcription by Rgt1 | |
| Ynl144c | + | N.D. | 13 | 10 | 6 | 10 | 9.8 | 0 | Unknown function |
| Sfg1 | + | + | 14 | 5 | 6 | 13 | 9.5 | 0 | Transcription factor/pseudohyphal growth |
| Tis11 | + | + | 11 | 6 | 5 | 9 | 7.8 | 0 | 3′ UTR binding/mRNA turnover |
| Cln2 | KNOWN | 9 | 10 | 3 | 3 | 6.3 | 0.07 | G1 cyclin | |
| Las17 | N.D. | − | 6 | 3 | 5 | 9 | 5.8 | 0.11 | WASP-like, Actin assembly |
| Gic2 | KNOWN | 5 | 2 | 6 | 8 | 5.3 | 0 | Cdc42 effector | |
| Cln1 | KNOWN | 8 | 6 | 3 | 4 | 5.3 | 0 | G1 cyclin | |
| Pfk27 | KNOWN | 10 | 7 | 0 | 1 | 4.5 | 0 | 6-Phosphofructose 2 kinase | |
| Met2 | * | N.D. | 7 | 3 | 2 | 4 | 4 | 0 | Methionine Biosynthesis |
| Tye7 | KNOWN | 6 | 4 | 0 | 4 | 3.5 | 0 | bHLH Transcription factor | |
| Gac1 | + | N.D. | 11 | 0 | 0 | 2 | 3.3 | 0 | PP1 regulatory subunit |
| Npl4 | − | N.D. | 10 | 0 | 0 | 1 | 2.8 | 0 | Binds Cdc48 |
| Fir1 | + | N.D. | 4 | 1 | 0 | 2 | 1.8 | 0 | 3′ mRNA processing |
| Yhr131c | +/− | +/− | 4 | 0 | 0 | 3 | 1.8 | 0 | Unknown; binds phosphatidylinositols |
| Dre2 | +/− | + | 0 | 0 | 0 | 4 | 1 | 0 | Cytosolic Fe-S protein assembly |
| Ykr045c | − | + | 2 | 0 | 0 | 1 | 0.8 | 0 | Unknown function |
| Sbe2 | +/− | + | 2 | 0 | 0 | 0 | 0.5 | 0 | Bud growth/transport from Golgi to bud |
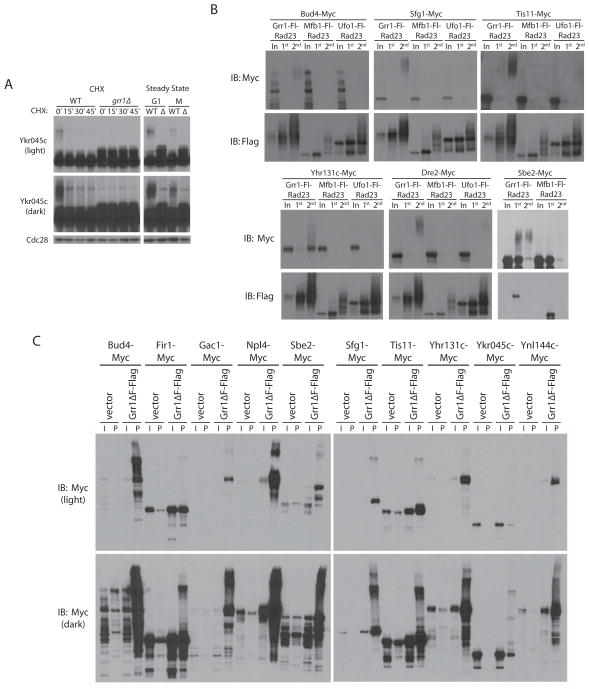
The SCF often targets phosphorylated substrates. Several stabilized substrates (Sfg1, Tis11, Fir1, Ynl144c and Sbe2) were enriched for an electrophoretically shifted, and likely phosphorylated, form in grr1Δ strains (Figure 3A). This is particularly evident for Ykr045c, which shows a species in grr1Δ cells with slightly reduced mobility in both G1- and nocodazole-arrested cells (Figure 4A). This form, which is most likely phosphorylated, cannot be detected in GRR1 cells, possibly due to its selective degradation upon ubiquitination. While bulk levels of Ykr045c do not decrease following addition of cycloheximide, a very high-molecular-weight (and likely ubiquitinated) smear, seen only in the GRR1 cells, disappears. This finding underscores the fact that a lack of bulk turnover of a candidate substrate does not necessarily indicate that the F-box protein in question is not targeting this substrate. Three other Grr1 candidate substrates (Met2, Npl4 and Sbe2) also showed no significant difference in stability between GRR1 and grr1Δ cells, although Met2 and Sbe2 showed a significant increase in steady state levels in grr1Δ strains. Npl4 is a component of the Cdc48 complex, which is thought to help disassemble the SCF (Yen et al., 2012), and is likely not a Grr1 substrate. We previously found Met2 to be strongly transcriptionally induced by deletion of GRR1, suggesting that this may not be a direct target (Benanti et al., 2007).
Figure 4. Ubiquitination of Grr1 candidate substrates.
(A) Short and long exposures of anti-Myc Western blots of a CHX chase assay of the uncharacterized ORF YKR045C in GRR1 and grr1Δ cells. Steady state levels of Ykr045c are shown in G1- and nocodazole-arrested cells. (B) Six candidate Grr1 substrates were expressed in cells containing Ligase Traps of Grr1 as well as Mfb1 and/or Ufo1 as negative controls. Western blots of two-step purifications, as in Figure 1D, are shown. (C) Western blots of whole cell extract (I) and anti-Flag pulldowns (P) from strains expressing Myc-tagged candidate Grr1 substrates and transformed with either empty vector (pRS426) or pYES2-Grr1ΔF-Fl (a galactose-inducible copy of Grr1-Flag lacking the F-box domain).
Our Ligase Trapping technology allows not only the initial identification of substrates, but also a means by which to examine the ubiquitination of that substrate directly. To validate that SCFGrr1 candidate substrates were ubiquitinated in vivo, we performed two-step purifications of Grr1-Fl-Rad23 Ligase Traps in cells expressing Myc-tagged alleles of four proteins that met our criteria for candidates (Sfg1, Bud4, Tis11, and Yhr131c) and four that did not (Sbe2, Dre2, Gin4 and Mps1) (Table 1). Purifications were performed in Grr1-Fl-Rad23 and two control Ligase Traps. We employed Mfb1-Fl-Rad23 because it used the same UBA and was expressed at similar levels as Grr1. We also chose Ufo1-Fl-Rad23, as this Ligase Trap showed the highest nonspecific binding, and therefore set an upper threshold for background. All four candidates that fell within the 1.8 peptide cutoff co-purified as ubiquitinated proteins specifically with Grr1-Fl-Rad23 (Table 1 and Figure 4B). Of these, Yhr131c reproducibly showed less extensive ubiquitination. For the remaining four, Dre2 and Sbe2 showed Grr1-specific ubiquitination (Figure 4B), whereas Mps1 and Gin4 did not (data not shown). Thus, of the ten Grr1 candidates that fell within our cutoff, at least seven appeared to be genuine substrates by at least one criterion. Moreover, these data suggest that several of the lower abundance hits are likely to be substrates as well. To show that our candidate substrates are direct targets of Grr1, we determined whether each could associate with a Grr1 construct lacking the F-box motif, Grr1ΔF, which fails to incorporate into functional SCF complexes (Bai et al., 1996). As shown in Figure 4C, all 10 substrates tested copurified strongly with the Flag-tagged Grr1ΔF construct (some in a shifted polyubiquitinated form). While copurification with Grr1ΔF is useful as a follow-up to confirm binding, this technique provides levels of background too high for initial substrate identification.
Examining substrates of other F-box proteins
We also examined candidates obtained for four additional F-box proteins: Cdc4, Ufo1, Skp2 and Saf1. Unlike the other Ligase Traps, we generated Traps of Cdc4 fused at either terminus. This was carried out because C-terminal tagging of Cdc4 appeared to compromise its function. Moreover, all Cdc4 fusions purified less polyubiquitinated product than other Ligase Traps (Figure S1), consistent with our finding that Cdc4 is particularly difficult to modify while retaining functionality. Since the N-terminally tagged CDC4 strains were healthier, we employed the RAD23-Fl-CDC4 in parallel. Despite its compromised function in vivo, the CDC4-Fl-RAD23 identified known substrates equally well, if not better than, the RAD23-Fl-CDC4 allele (Figure 2). We examined seven candidate Cdc4 substrates above the 1.8 peptide cutoff (Atc1, Isr1, Swi1, Amn1, Sac3, Osh3, Ipt1) and two that were below it (Pcl1 and Rav2). In the case of Atc1, Isr1, and Swi1, stabilization could be observed in the cdc4-1 strain (Figure 5A). Partial stabilization was also observed for Pcl1, a G1 cyclin. Cdc4 and Grr1 have previously been shown to target the G1 cyclin Cln3 redundantly (Landry et al., 2012), but we found that Grr1 contributed only slightly to Pcl1 turnover. Rav2 is a protein component of the RAVE complex, which functions in assembly of the vacuolar ATPase and binds the SCF core subunit Skp1 (Seol et al., 2001). Interestingly, levels of a smaller form of Rav2 are elevated in cdc4-1 strains and a shifted form is seen to accumulate. CDC4 did not affect the stability of Osh3, Ipt1, or Amn1. To better characterize some of these potential Cdc4 substrates, we determined whether their ubiquitinated forms associated with the Rad23-Fl-Cdc4 Ligase Trap (Figure 5B) and two similarly abundant control Ligase Traps. Four of the tested candidates (Swi, Atc1, Isr1, and Pcl1) showed specific association of a ubiquitin smear with the Cdc4 Trap. In contrast, Osh3 and Ipt1 did not (data not shown).
Figure 5. Ubiquitination of Cdc4 and Ufo1 candidate substrates.
(A) Myc-tagged Cdc4 candidate substrates were expressed in CDC4 cells or cdc4-1 temperature sensitive mutants. Cultures were simultaneously shifted from 23°C to 37°C and treated with CHX for the indicated number of minutes. Western blots were probed with anti-Myc (substrates) or anti-Cdc28 (loading control). A cdc4-1 grr1Δ double mutant was examined for Pcl1. (B) Western blots of two-step purifications performed on cell extracts expressing Rad23-Fl-Cdc4 and Myc-tagged Swi1, Atc1, Isr1 or Pcl1, as in Figure 1. (C) The Ufo1 candidate substrate Rbg1 was Myc-tagged in UFO1 and ufo1Δ cells, treated with CHX for the indicated times, and examined by Western blot. (D) Western blot of two-step purifications performed on cell extracts expressing Ufo1-Fl-Rad23, Ufo1-Fl-Dsk2 or Grr1-Fl-Rad23 together with Myc-tagged Rbg1.
For the F-box protein Ufo1, we identified HO as a substrate, as has previously been reported (Kaplun et al., 2006). Because the Ufo1 Traps showed unusually high background, we set the threshold for substrates for this Trap to be six spectral counts and 25-fold enrichment. Despite this, we were able to discern several strongly enriched candidates, many of which had roles in translation. The strongest of these, the polysome-associated Rbg1 protein, is a member of the Obg/CgtA GTP-binding proteins conserved from bacteria to humans (Wout et al., 2009). Although Rbg1 levels were not affected in ufo1Δ strains (Figure 5C), ubiquitinated Rbg1 purified with two forms of the Ufo1 Trap, but not the Grr1-Fl-Rad23 control (Figure 5D). This suggests that only a subset of Rbg1 in the cell is targeted by Ufo1 under these conditions. Alternatively, Rbg1 may be targeted in only a subset of cells.
Saf1 promotes ubiquitination of proteins of the secretory pathway
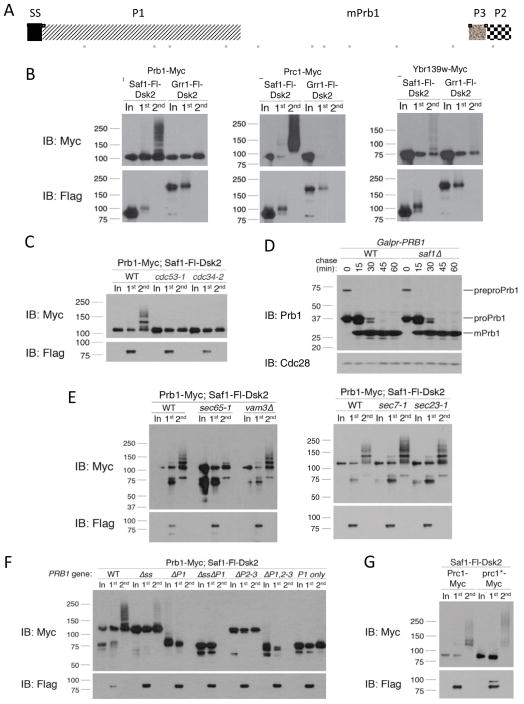
All four of the candidate substrates identified for Saf1 were vacuolar/lysosomal enzymes (Figure 6B and Supplemental Table 1): three proteases (Prb1, Prc1 and the putative Ybr139w) and an alkaline phosphatase (Pho8), two of which have previously been shown to associate with Saf1 in a large-scale study (Ho et al., 2002). The yeast vacuole is thought to be similar to the mammalian lysosome as it contains a large number of degradative enzymes. The uncharacterized protein Ybr139w has been suggested to be a serine carboxypeptidase (Baxter et al., 2004; Wunschmann et al., 2007) and shows vacuolar localization. Prb1, Prc1 and Pho8 are synthesized as the inactive zymogens preproPrb1, preproPrc1 and proPho8. (The processing of Ybr139w, if any, is uncharacterized.) During ER-to-vacuole progression, precursors are cleaved to generate the active enzymes that can be distinguished by their lower molecular weight. For Prb1, there are at least four proteolytic steps. The signal sequence (SS) is first removed upon translocation into the ER, followed by a second cleavage event to remove P1, generating “proPrb1” (Figure 6A). After exiting the ER, two C-terminal cleavages remove P2 and P3 to generate the mature form (mPrb1).
Figure 6. Saf1 targets vacuolar zymogens that fail to properly mature.
(A) Full length Prb1 (preproPrb1) is constituted by 635 amino acids. The N-terminal signal peptide (SP; 20 amino acids) is cleaved during translocation into the ER. In the ER, an intramolecular proteolytic cleavage removes P1 (260 amino acids) to yield proPrb1. In the late Golgi/vacuole, P2 (~30 amino acids) and P3 (~30 amino acids) are cleaved off. Mature Prb1 (mPrb1) is roughly 295 amino acids (~31 kDa). Positions of Prb1 peptides identified by LC-MS/MS analysis of the two-step purification of Saf1-Fl-Dsk2 are represented as solid gray bars below. (B) Two-step purification from extracts expressing Saf1-Fl-Dsk2 and Myc-tagged Prb1, Prc1 or Ykr139w. Grr1-Fl-Dsk2 was used as a negative control for binding specificity. (C) Two-step purification was performed as in (B) in wild type, cdc53-1 or cdc34-2 mutants. Cells were maintained at 23°C then shifted to 38°C for 45 minutes prior to collection. (D) Strains containing PRB1 under the inducible GAL1 promoter were maintained in 2% raffinose and induced with 2% galactose for 15 min, collected and resuspended in 2% glucose. Time points were collected, Western blotted and probed with anti-Prb1 antibody, which recognizes all forms of Prb1. (E) Two-step purification in wild type, sec65-1, vam3Δ, sec7-1 or sec23-1 mutants performed as in (B), except strains were grown/induced at 23°C (permissive temp) and shifted to the 38°C (restrictive temp) for 45 minutes prior to collection. (F) Prb1 constructs, under the control of the TEF1 promoter, were examined by two-step purification as in (B). (G) Two-step purification was performed as in (B) with strains expressing either PRC1 or prc1-G255R, which encodes the Prc1* (aka CPY*) allele.
To show that SCFSaf1 ubiquitinates these vacuolar/lysosomal substrates, we C-terminally Myc-tagged each in cells expressing Saf1-Fl-Dsk2 or the control Trap Grr1-Fl-Dsk2. Two-step purifications immunoprecipitated ubiquitinated Prb1-Myc, Prc1-Myc and Ybr139w-Myc, but not Pho8-Myc (Figure 6B and data not shown) specifically with the Saf1 Ligase Trap. The high-molecular-weight species indicative of ubiquitinated Prb1-Myc are detected almost exclusively above the ~110 kDa band corresponding to preproPrb1-Myc (Figures 6E, 6F and S3B). This suggests that SCFSaf1 ubiquitinates Prb1 prior to proteolytic activation, although C-terminal tagging of Prb1 did delay processing somewhat (Figure S3A). LC-MS/MS data from Saf1 Ligase Trap purifications confirmed that Saf1 interacts with preproPrb1. Of the 29% sequence coverage we obtained for Prb1, most of it (17%) was located in portions of Prb1 that are removed during proteolytic activation, whereas only 12% represented portions of mPrb1 (Figures 6A and S2). This is despite the fact that mPrb1 represents the vast majority of Prb1 in wild type cells, with preproPrb1 barely visible (Figure S3A). Unfortunately, only two distinct peptides, representing 4% coverage, were obtained for Prc1. Given that only a small portion (about 20%) of Prc1 is removed during processing, this data set does not allow us to determine if the Prc1 precursor was targeted. We could not detect Pho8-Myc in Saf1-Fl-Dsk2 purifications by Western blot. While we cannot exclude the possibility that Pho8 is a false-positive hit of the LC-MS/MS analysis, it is also possible that the epitope tag on Pho8 interferes with its ubiquitination by SCFSaf1. Since it was surprising that an F-box protein acted upon substrates targeted to the vacuole, we examined whether other SCF components were required. Mutations in either the cullin subunit (cdc53-1) or the SCF’s E2 (cdc34-2) eliminated Saf1 binding to polyubiquitinated Prb1 (Figure 6C). The reduced length of the polyubiquitin chains seen is due to the elevated temperature used for this experiment (Figure S3B). However, even at 30°C, Saf1 substrates appear ed less ubiquitinated than those of other SCF ligases (Figure S1), suggesting either that SCFSaf1 is less processive or that the UBA fusion interferes with the function of Saf1.
To determine whether the ubiquitination of Prb1 plays a role in the processing and translocation of preproPrb1, we examined the rate of processing of Prb1 in a saf1Δ strain. By pulsing Prb1, expressed under the control of the GAL1 promoter, one can see the initial accumulation of the preproPrb1 form, followed by the rapid accumulation of a size corresponding to proPrb1 (lacking P1) and finally, the appearance of mPrb1 (Figure 6D). This processing was unaffected in saf1Δ, suggesting that SAF1 is not required for processing or translocation (since the final step of processing is thought to occur in the vacuole). Purification of ubiquitinated Prb1 was intact in vam3Δ mutants, which are defective in translocation to the vacuole (Srivastava and Jones, 1998), suggesting that targeting of Prb1 occurs prior to reaching this organelle (Figures 6E). This was also the case for Prc1, shown using a vps10Δ mutant (Figure S3C), which fails to properly deliver Prc1 from the Golgi to the vacuole (Marcusson et al., 1994). Both a sec65-1 mutant, which blocks entry into the ER, and tunicamycin, which eliminates N-linked glycosylation, blocked Prb1 ubiquitination (Figures 6E and S3E). However, Prb1 could still be ubiquitinated in sec7-1 and sec23-1 mutants, both of which are defective in ER-to-Golgi traffic (Novick et al., 1980; Wolf et al., 1998) (Figure 6E).
To determine whether the selective targeting of unprocessed Prb1 requires all portions of the full length substrate, we made a series of deletion mutants of Prb1. Removal of Prb1’s signal sequence (ss) eliminates its ability to efficiently enter the ER, and this strongly reduced Saf1-mediated ubiquitination (Figures S3D and 6F). This is consistent with the absence of ubiquitination in the sec65-1 mutant (Figure 6E), suggesting that Prb1 must be ER-targeted before it is recognized. Moreover, eliminating either P1 or P2-3 blocked Saf1 targeting. At least in the case of the ΔP1 mutants, this is not due to a disruption in ER localization, as the ΔP1 and ΔP1ΔP2-3 peptides are altered in size after treatment with tunicamycin, suggesting that they have entered the ER (Figure S3D). These findings are consistent with Saf1 selectively targeting the unprocessed form.
The ubiquitination of ER proteins by Saf1 is reminiscent of the ERAD (ER-associated degradation) quality control pathway, a process whereby unfolded ER proteins are targeted for ubiquitination after their retrotranslocation from the ER into the cytoplasm (Tsai et al., 2002). ER proteins are thought to become ERAD substrates following redox stress, such as that induced by treatment with the reducing agent DTT, heat shock, or tunicamycin treatment. Saf1 ubiquitination of Prb1 was not increased by any of these treatments, but was in fact decreased (Figures S3B and S3E). It is possible that ubiquitination of Prb1 could decrease after this treatment because of competition with increased amounts of other substrates generated under these conditions. However, ubiquitination was decreased even after more modest treatments with these agents (Figure S3E). The Prc1 protein (a.k.a. CPY), identified by the Saf1 Ligase Trap, is a hallmark ERAD substrate when mutated at a single site, G255R, which is thought to cause its partial unfolding (Finger et al., 1993). We will refer to this point mutant as Prc1*. If Saf1 were part of the previously characterized ERAD pathway, its ubiquitination of Prc1 should increase significantly in a Prc1* strain. Despite the fact that there exists consistently higher levels of mutant Prc1* protein than wild type Prc1, we saw no increase in the percentage of ubiquitinated Prc1 associated with the Saf1 Ligase Trap (Figure 6G). (Note, the higher levels may reflect the fact that Prc1* is the only form of PRC1 expressed in these cells.) Interestingly, however, we do see a qualitative difference in that, in addition to the less ubiquitinated forms seen associated with Saf1, there is an additional lower mobility form. This suggests that a portion of the Prc1* that is targeted by Saf1 was previously targeted by another more processive (possibly ERAD) ligase. Together, these data suggest that Saf1 is not a component of the previously characterized ERAD quality control system, but rather, part of a ubiquitination pathway that specifically targets zymogens that cannot be processed.
Discussion
We have developed a method allowing us to trap a given E3 ubiquitin ligase in association with its substrates. This allows us to not only identify previously uncharacterized ubiquitin ligase substrates, but also provides a robust method by which we can determine whether a given ubiquitin ligase targets a particular protein in vivo. This is critical, since many substrates are not quantitatively degraded and, therefore, one cannot always see a strong selective stabilization of a substrate after mutation of the ligase in question. While we have carried out this proof of principle experiment on F-box proteins in yeast, this methodology should be readily applicable to other classes of ubiquitin ligases and other organisms.
We have used the Ligase Trapping technique on eight F-box proteins, and followed up candidate substrates for four of these in detail (Cdc4, Grr1, Ufo1, and Saf1). Many of our identified Grr1 and Cdc4 substrates are consistent with the known role of Grr1 in nutrient sensing and the role of both proteins in cell cycle regulation. While Ufo1 appears to target several proteins involved in translation, Saf1 emerges as an F-box protein with a focused role in targeting incompletely processed degradative enzymes bound for the vacuole.
The F-box substrates fell into three categories with respect to their stability. Some, such as Bud4 and Sfg1 were highly unstable proteins that were very strongly stabilized when the gene for the F-box protein targeting them (GRR1) was deleted. This corresponds to the simplest case in which a substrate is quantitatively targeted by a single ubiquitin ligase. Second, there were substrates, such as Tis11 and Pcl1, which were unstable and only partially stabilized in their respective F-box mutants, suggesting that they are redundantly targeted. Finally, there was a significant set of substrates that appeared stable even in wild type cells (e.g. Ykr045c and Rbg1). There are several possible explanations for this. These proteins may be targeted in only some subcellular locations or contexts (e.g. when part of a specific complex). Alternatively, they may be targeted in only a subset of cells that are either in a particular cell cycle phase or are under some stress. Importantly, the high molecular weight forms of Rbg1 and Ykr045c are lost upon cycloheximide treatment (Figure 4A and data not shown), suggesting that ubiquitination leads to degradation of the modified subset, although it is also possible that deubiquitination accounts for this. An advantage of Ligase Trapping is that, unlike many existing technologies, it allowed us to confirm that a given substrate was targeted by a particular ligase even when the targeting was redundant or did not lead to quantitative turnover.
Functional clusters of substrates identifies multilayer regulatory roles of Grr1, Cdc4 and Ufo1 in different aspects of cell biology
Live fluorescence microscopy showed that Grr1 transiently localizes at the bud neck during mitosis to disappear shortly after cytokinesis is completed (Blondel et al., 2005), where it controls the levels of Hof1 and Gic2. Bud4 also localizes at the bud neck during mitosis and its localization is required to mark the bud site and recruit downstream regulators of the cytokinetic ring (Kang et al., 2012). We show that Grr1 is responsible for the degradation of Bud4 in G1, suggesting that Grr1 promotes the down-regulation of Bud4 activity once this function is accomplished (Figure 3C). Grr1 also targets Sbe2, which is involved in cell wall integrity and has a putative role in establishing a correct polar budding pattern (Santos and Snyder, 2000) (Figure 3B). Interestingly, Sfg1 is a transcriptional repressor that targets many genes involved in mother-daughter cell separation and is thought to be a substrate of Cdk (White et al., 2009), which may target it for Grr1-mediated turnover.
Grr1 also regulates different metabolic pathways by promoting the degradation of substrates (including Pfk27, Tye7 and Mth1) in response to nutrient availability. We found that the PP1 subunit Gac1, which regulates glycogen storage, is regulated by Grr1. We find that Grr1 may also have a role in iron metabolism, as it targets Tis11 and Dre2. Tis11 is a highly conserved protein that interacts with AU-rich elements in the 3′ UTR of a specific group of mRNAs and promotes their turnover in conditions of iron starvation (Puig et al., 2005). Dre2 is an essential protein involved in the biogenesis of iron-sulfur proteins such as ribonucleotide reductase (Zhang et al., 2011). The regulation of the targeting of Dre2 by Grr1 was not clear, as bulk levels of Dre2 were largely stable. While Dre2 did not exhibit cell-cycle dependent regulation, Tis11 levels appear to fluctuate in the cell cycle. Thus, Grr1 might have a role in coordinating iron homeostasis with cell cycle progression or Tis11 may have an additional role in the targeting of messages encoding cell cycle related genes.
Despite the fact that the Cdc4 Ligase Traps resulted in somewhat reduced fitness, they identified several substrates, including the Swi/Snf transcription factor Swi1, the cyclin Pcl1, and the kinase Isr1. Surprisingly, two very strong Cdc4 hits, Osh3 and Ipt1, do not appear to be Cdc4 substrates (J. Lao, unpublished data). Myc-tagging these proteins may disrupt their ability to associate with Cdc4.
Three of the top four Ufo1 candidates identified were associated with translation: Rbg1, Yef3, and Rps2 (Table S1). Rps2 is a component of the small ribosomal subunit. Rbg1 and the elongation factor Yef3 are both components of polysomes (Fleischer et al., 2006; Hutchison et al., 1984; Wout et al., 2009). Rbg1 is conserved from E. coli to humans and, in bacteria, has been shown to be involved in the regulation of translation in response to nutrient deprivation (Wout et al., 2009). Moreover, a human homolog of Rbg1 (Drg2) undergoes SCF-mediated turnover (Chen et al., 2012), suggesting that this regulation is conserved.
Met30 is thought to regulate methionine biosynthesis pathways and the corresponding Ligase Trap only copurified its previously characterized target, Met4. Mdm30 regulates mitochondrial biology and we identified its known target, Fzo1 (a protein involved in mitochondrial fusion), and another mitochondrial protein, Yjl045w (Table S1). Intriguingly, Yjl045w is also a mitochondrial protein. While included in this study, it is unclear if Mfb1 and Skp2 are bona fide SCF components, since, unlike the other six F-box proteins examined here, neither of these identified the cullin Cdc53 in our purifications or in previously characterized proteomic or two hybrid analyses (Ho et al., 2002; Krogan et al., 2006; Seol et al., 2001). While Mfb1 identified no substrates, Skp2 did purify several proteins. Predominant among these were Dma1 and Dma2. These were previously known to be strong Skp2 interactors (Ho et al., 2002), but neither appeared to be direct substrates of Skp2 (data not shown). As these are themselves E3s, they are likely autoubiquitinated and thus purified in our second step.
Saf1 targets unprocessed Prb1
Of the substrates identified in our study, the most surprising by far were the identification of several vacuolar proteins. Because the background was so low with the Ligase Trapping system, we were confident enough to follow-up hits that seemed at first very unlikely SCF substrates. All four hits for the F-box protein Saf1 were vacuolar/lysosomal hydrolases. This is unanticipated for several reasons. First, these proteins are not thought to be present in the cytosol/nucleus, where the SCF is located. Second, they are stable proteins. Finally, the peptides corresponding to Prb1 matched the full-length preproPrb1, which is quite rare, suggesting that the unprocessed form was selectively ubiquitinated.
SCFSaf1 appears to target Prb1 that cannot be processed. This appears to require ER entry and is likely to occur after retrotranslocation of the protein back into the cytosol. However, Saf1 does not appear to function as part of an Hrd1-/Doa10-like ERAD pathway, as turnover of Prb1 is not promoted by its unfolding. As is the case for most quality control pathways, it does not appear that Saf1 targets a large percentage of preproPrb1, since steady state levels of the unprocessed protein are not altered upon deletion of SAF1. Prior to this study, only a single substrate of Saf1 had been identified. A purine salvage pathway protein, Aah1, was previously shown to be targeted by Saf1 during nitrogen starvation (Escusa et al., 2006). While Aah1 is not thought to be vacuolar, the fact that Aah1 is targeted under conditions that promote autophagy, a process during which nutrients are salvaged by targeting cellular structures to the vacuole/lysosome, is an intriguing connection between these Saf1 substrates. Prb1, Prc1 and Pho8 are all induced upon nutrient limitation (Hansen et al., 1977; Kaneko et al., 1985; Klar and Halvorson, 1975).
Interestingly, Saf1 contains neither LLR nor WD40 repeats, but instead has another beta-propeller domain composed of RCC1-like repeats (Escusa et al., 2007). Whether these RCC1-like repeats will, like WD40s and LRRs, recognize substrates only after phosphorylation is not yet known. An intriguing possibility is that Prb1 glycosylation is required. Prb1 is N-glycosylated (Mechler et al., 1982; Moehle et al., 1987) but while mutation of this site leads to much lower levels of Prb1, its relative level of ubiquitination is not strongly affected (Figure S3F). While there is typically not thought to be a direct correspondence between human and yeast F-box proteins, it is interesting to note that S. cerevisiae, S. pombe and humans all contain a single F-box protein with RCC1-like repeats (Jin et al., 2004). Whether this human F-box protein also targets lysosomal targets remains to be determined.
EXPERIMENTAL PROCEDURES
Yeast strains
Genotypes of yeast strains, including the specific experiments in which each was used, are detailed in Table S2. With the exception of Figures 6, S3 and S4 (W303 background), all strains are in the S288c background. Strains and plasmids were generated using standard techniques.
Plasmids and Western blotting
Plasmids and Western blotting are described in Supplemental Experimental Procedures.
One-step purification
One-step purification in Figure 1B, was performed using strains carrying pRS316 plasmids (pMS1-pMS4). Cells were grown to mid-log phase in C media lacking uracil (C-Ura) containing 2% raffinose. 2% galactose was added, and cultures were incubated an additional 3 hours. Cells were harvested from 100 ml culture at an optical density (OD600) of ~1.0, lysed in 700 μl HEPES lysis buffer (25 mM HEPES pH 7.5, 150 mM NaCl, 1 mM EDTA, 17 g/ml PMSF, 5 mM sodium fluoride, 80 mM β-glycerophosphate, 1 mM sodium orthovanadate and a Complete Proteasome Inhibitor Tablet (Roche Diagnostics) by bead beating in a cold block for 6 cycles of 1.5 minutes (alternated with 2 minutes on ice) and cleared by centrifugation at 4°C. Protein concentra tions were then quantified using the Bio-Rad Protein Assay, based on the method of Bradford and equal amounts of extract (2 mg) were incubated with 30 μl slurry of anti-Flag M2 Magnetic Beads (Sigma-Aldrich) while rotating at 4°C for 3 hours. Beads were collected on a magnetic rack and washed three times with 700 μl lysis buffer. Proteins were eluted by mild vortexing in 1x PBS buffer containing 500 ng/ml 3xFlag peptide (Sigma-Aldrich) for 30 minutes and analyzed by Western blotting against the Flag epitope on the Cdc4 proteins and the Myc tag or HA tag on Cdc6 and Far1.
Two-step purification
Two-step purifications were performed by harvesting cells at an optical density (OD600) of ~1.0. Cell cultures of 350 ml (small-scale) and 2–4 liters (large-scale) were used respectively for Western blot analysis and LC-MS/MS analysis. Cells were grown in YM-1 medium containing 2% raffinose and 2% galactose. Small-scale pellets were resuspended in 1.2 ml lysis buffer and lysed by bead beating as described in the one-step purification method. Large-scale pellets were resuspended in 3 ml lysis buffer, frozen in liquid nitrogen, grounded in a Retsch M301 ball mill and resuspended in additional 7 ml lysis buffer. Lysis buffer for small- and large-scale purifications were composed of (25 mM HEPES pH 7.5, 150 mM potassium acetate, 1 mM EDTA, 17 g/ml PMSF, 5 mM sodium fluoride, 80 mM β-glycerophosphate, 1 mM sodium orthovanadate, 0.02 mM MG132 (Sigma-Aldrich) and a Complete Proteasome Inhibitor Tablet (Roche Diagnostics). Cell lysates were cleared by centrifugation and incubated with 100 μl slurry (small-scale) and 200 μl slurry (large-scale) of anti-Flag M2 Magnetic Beads (Sigma-Aldrich) overnight while rotating at 4°C. Beads were collected on a magnetic rack and washed three times with PBS buffer, 0.1 % NP-40. Proteins were eluted by mild vortexing with five times beads volume of PBS buffer, 0.1 % NP-40 containing 500 ng/ml 3xFlag peptide (Sigma-Aldrich) for 45 minutes at room temperature. Eluted proteins (1st step) were denatured by adjusting elution buffer with 40 mM NaH2PO4, 5 mM Tris-Cl, 6M urea, pH 8 and incubated for 2 hours with 30 μl (small-scale) or 60 μl slurry (large-scale) of Ni-NTA agarose beads (Invitrogen) previously equilibrated with 100 mM NaH2PO4, 10 mM Tris-Cl, 8M urea, pH 8. Beads were washed three times with 100 mM NaH2PO4, 10 mM Tris-Cl, 8M urea, pH 8 and two times with PBS buffer. Proteins were eluted with three times beads volume of PBS buffer pH 8 containing 300 mM imidazole and 0.1 % RapiGest (Waters) by mild vortexing at room temperature for 30 minutes (2nd step). Relative to input, samples for SDS-PAGE were loaded at 170–800x and 1,300–2,800x for the Flag (1st) and His (2nd) elutions, respectively.
Sample preparation and mass spectrometry analysis
The sample preparation and mass spectrometry analysis are detailed in Supplemental Experimental Procedures.
Turnover analysis
Asynchronous cells were grown to mid-log phase in YM-1 containing 2% dextrose and treated with cycloheximide (50 μg/ml, Sigma-Aldrich). For cell-cycle experiments, cycloheximide was added to cells previously arrested with α factor (10 μg/ml, Elim Biopharm) or nocodazole (10 μg/ml, Sigma-Aldrich), both for 2 hours. Cell pellets were collected for western blotting at indicated time points.
GST pulldown assays
Strains carrying either pRS426 or pYES2-GRR1dF-FLAG-URA3 plasmids were grown to OD600 ~0.3 in 50ml. of synthetic media lacking uracil containing 2% raffinose. To induce, 2% galactose was added and cultures were grown for two doublings. Cells were lysed in a buffer containing 100 mM Tris-HCl, pH7.5, 300 mM NaCl, 2 mM EDTA, 0.2% NP-40 with a Roche Complete protease inhibitor tablet without EDTA (1 tablet/25 ml), 1 mM PMSF, and 4 Roche PhosSTOP phosphatase inhibitor tables (4 tablets/25 ml). Lysis was carried out by bead beating as described in the one-step purification method and cleared by centrifugation at 4°C. Lysates were incubated with a 25 μl slurry of anti-Flag M2 Magnetic Beads (Sigma-Aldrich) overnight while rotating at 4°C. Beads were washed three times with PBS buffer, 0.1 % NP-40. Proteins were eluted by mild vortexing with five times beads volume of PBS buffer, 0.1 % NP-40 containing 500 ng/ml 3xFlag peptide (Sigma-Aldrich) for 45 minutes at room temperature. Samples were loaded for SDS-PAGE at 0.06% of the total input and 20% of the Flag elution.
Supplementary Material
Mark_Highlights.
Fusing ubiquitin binding domains to F-box proteins allows capture of SCF substrates
A mass spectrometry-based Ligase Trap screen identified 18 candidate SCF targets
SCFSaf1 ubiquitinates the unprocessed zymogen form of vacuolar/lysosomal proteases
Acknowledgments
We would like to thank R. Aebersold, in whose laboratory A.M. performed the mass spectrometry analysis. We would also like to thank B. Topacio for technical assistance and T. Berens and J. Benanti for critical reading of the manuscript. We thank E. Wayner for providing anti-Ubiquitin antibody and C. Woolford (and the late B. Jones) for providing anti-Prb1 antibody. This work was supported by a fellowship from the American-Italian Cancer Foundation to M.S. and a grant (GM 70539) from the NIH to D.P.T.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper JW, Elledge SJ. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86:263–274. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- Baxter SM, Rosenblum JS, Knutson S, Nelson MR, Montimurro JS, Di Gennaro JA, Speir JA, Burbaum JJ, Fetrow JS. Synergistic computational and experimental proteomics approaches for more accurate detection of active serine hydrolases in yeast. Molecular & cellular proteomics: MCP. 2004;3:209–225. doi: 10.1074/mcp.M300082-MCP200. [DOI] [PubMed] [Google Scholar]
- Benanti JA, Cheung SK, Brady MC, Toczyski DP. A proteomic screen reveals SCFGrr1 targets that regulate the glycolytic-gluconeogenic switch. Nat Cell Biol. 2007;9:1184–1191. doi: 10.1038/ncb1639. [DOI] [PubMed] [Google Scholar]
- Blondel M, Bach S, Bamps S, Dobbelaere J, Wiget P, Longaretti C, Barral Y, Meijer L, Peter M. Degradation of Hof1 by SCF(Grr1) is important for actomyosin contraction during cytokinesis in yeast. EMBO J. 2005;24:1440–1452. doi: 10.1038/sj.emboj.7600627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busino L, Bassermann F, Maiolica A, Lee C, Nolan PM, Godinho SI, Draetta GF, Pagano M. SCFFbxl3 controls the oscillation of the circadian clock by directing the degradation of cryptochrome proteins. Science. 2007;316:900–904. doi: 10.1126/science.1141194. [DOI] [PubMed] [Google Scholar]
- Cardozo T, Pagano M. The SCF ubiquitin ligase: insights into a molecular machine. Nat Rev Mol Cell Biol. 2004;5:739–751. doi: 10.1038/nrm1471. [DOI] [PubMed] [Google Scholar]
- Chen J, Shen BY, Deng XX, Zhan Q, Peng CH. SKP1-CULLIN1-F-box (SCF)-mediated DRG2 degradation facilitated chemotherapeutic drugs induced apoptosis in hepatocellular carcinoma cells. Biochemical and biophysical research communications. 2012;420:651–655. doi: 10.1016/j.bbrc.2012.03.058. [DOI] [PubMed] [Google Scholar]
- Davis MA, Larimore EA, Fissel BM, Swanger J, Taatjes DJ, Clurman BE. The SCF-Fbw7 ubiquitin ligase degrades MED13 and MED13L and regulates CDK8 module association with Mediator. Genes & development. 2013;27:151–156. doi: 10.1101/gad.207720.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- Emanuele MJ, Elia AE, Xu Q, Thoma CR, Izhar L, Leng Y, Guo A, Chen YN, Rush J, Hsu PW, et al. Global identification of modular cullin-RING ligase substrates. Cell. 2011;147:459–474. doi: 10.1016/j.cell.2011.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escusa S, Camblong J, Galan JM, Pinson B, Daignan-Fornier B. Proteasome- and SCF-dependent degradation of yeast adenine deaminase upon transition from proliferation to quiescence requires a new F-box protein named Saf1p. Mol Microbiol. 2006;60:1014–1025. doi: 10.1111/j.1365-2958.2006.05153.x. [DOI] [PubMed] [Google Scholar]
- Escusa S, Laporte D, Massoni A, Boucherie H, Dautant A, Daignan-Fornier B. Skp1-Cullin-F-box-dependent degradation of Aah1p requires its interaction with the F-box protein Saf1p. The Journal of biological chemistry. 2007;282:20097–20103. doi: 10.1074/jbc.M702425200. [DOI] [PubMed] [Google Scholar]
- Finger A, Knop M, Wolf DH. Analysis of two mutated vacuolar proteins reveals a degradation pathway in the endoplasmic reticulum or a related compartment of yeast. Eur J Biochem. 1993;218:565–574. doi: 10.1111/j.1432-1033.1993.tb18410.x. [DOI] [PubMed] [Google Scholar]
- Finley D, Ulrich HD, Sommer T, Kaiser P. The ubiquitin-proteasome system of Saccharomyces cerevisiae. Genetics. 2012;192:319–360. doi: 10.1534/genetics.112.140467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischer TC, Weaver CM, McAfee KJ, Jennings JL, Link AJ. Systematic identification and functional screens of uncharacterized proteins associated with eukaryotic ribosomal complexes. Genes Dev. 2006;20:1294–1307. doi: 10.1101/gad.1422006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen RJ, Switzer RL, Hinze H, Holzer H. Effects of glucose and nitrogen source on the levels of proteinases, peptidases, and proteinase inhibitors in yeast. Biochimica et biophysica acta. 1977;496:103–114. doi: 10.1016/0304-4165(77)90119-2. [DOI] [PubMed] [Google Scholar]
- Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutilier K, et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- Hsiung YG, Chang HC, Pellequer JL, La Valle R, Lanker S, Wittenberg C. F-box protein Grr1 interacts with phosphorylated targets via the cationic surface of its leucine-rich repeat. Molecular and cellular biology. 2001;21:2506–2520. doi: 10.1128/MCB.21.7.2506-2520.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison JS, Feinberg B, Rothwell TC, Moldave K. Monoclonal antibody specific for yeast elongation factor 3. Biochemistry. 1984;23:3055–3063. doi: 10.1021/bi00308a032. [DOI] [PubMed] [Google Scholar]
- Jin J, Cardozo T, Lovering RC, Elledge SJ, Pagano M, Harper JW. Systematic analysis and nomenclature of mammalian F-box proteins. Genes Dev. 2004;18:2573–2580. doi: 10.1101/gad.1255304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonkers W, Rep M. Lessons from fungal F-box proteins. Eukaryot Cell. 2009;8:677–695. doi: 10.1128/EC.00386-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko Y, Tamai Y, Toh-e A, Oshima Y. Transcriptional and post-transcriptional control of PHO8 expression by PHO regulatory genes in Saccharomyces cerevisiae. Molecular and cellular biology. 1985;5:248–252. doi: 10.1128/mcb.5.1.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang PJ, Angerman E, Jung CH, Park HO. Bud4 mediates the cell-type-specific assembly of the axial landmark in budding yeast. J Cell Sci. 2012;125:3840–3849. doi: 10.1242/jcs.103697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplun L, Ivantsiv Y, Bakhrat A, Tzirkin R, Baranes K, Shabek N, Raveh D. The F-box protein, Ufo1, maintains genome stability by recruiting the yeast mating switch endonuclease, Ho, for rapid proteasome degradation. The Israel Medical Association journal: IMAJ. 2006;8:246–248. [PubMed] [Google Scholar]
- Kim W, Bennett EJ, Huttlin EL, Guo A, Li J, Possemato A, Sowa ME, Rad R, Rush J, Comb MJ, et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Molecular cell. 2011;44:325–340. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klar AJ, Halvorson HO. Proteinase activities of Saccharomyces cerevisiae during sporulation. J Bacteriol. 1975;124:863–869. doi: 10.1128/jb.124.2.863-869.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- Landry BD, Doyle JP, Toczyski DP, Benanti JA. F-box protein specificity for g1 cyclins is dictated by subcellular localization. PLoS genetics. 2012;8:e1002851. doi: 10.1371/journal.pgen.1002851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcusson EG, Horazdovsky BF, Cereghino JL, Gharakhanian E, Emr SD. The sorting receptor for yeast vacuolar carboxypeptidase Y is encoded by the VPS10 gene. Cell. 1994;77:579–586. doi: 10.1016/0092-8674(94)90219-4. [DOI] [PubMed] [Google Scholar]
- Mechler B, Muller M, Muller H, Meussdoerffer F, Wolf DH. In vivo biosynthesis of the vacuolar proteinases A and B in the yeast Saccharomyces cerevisiae. J Biol Chem. 1982;257:11203–11206. [PubMed] [Google Scholar]
- Moehle CM, Tizard R, Lemmon SK, Smart J, Jones EW. Protease B of the lysosomelike vacuole of the yeast Saccharomyces cerevisiae is homologous to the subtilisin family of serine proteases. Mol Cell Biol. 1987;7:4390–4399. doi: 10.1128/mcb.7.12.4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash P, Tang X, Orlicky S, Chen Q, Gertler FB, Mendenhall MD, Sicheri F, Pawson T, Tyers M. Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature. 2001;414:514–521. doi: 10.1038/35107009. [DOI] [PubMed] [Google Scholar]
- Novick P, Field C, Schekman R. Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell. 1980;21:205–215. doi: 10.1016/0092-8674(80)90128-2. [DOI] [PubMed] [Google Scholar]
- Orlicky S, Tang X, Willems A, Tyers M, Sicheri F. Structural basis for phosphodependent substrate selection and orientation by the SCFCdc4 ubiquitin ligase. Cell. 2003;112:243–256. doi: 10.1016/s0092-8674(03)00034-5. [DOI] [PubMed] [Google Scholar]
- Patton EE, Willems AR, Sa D, Kuras L, Thomas D, Craig KL, Tyers M. Cdc53 is a scaffold protein for multiple Cdc34/Skp1/F-box protein complexes that regulate cell division and methionine biosynthesis in yeast. Genes Dev. 1998;12:692–705. doi: 10.1101/gad.12.5.692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- Puig S, Askeland E, Thiele DJ. Coordinated remodeling of cellular metabolism during iron deficiency through targeted mRNA degradation. Cell. 2005;120:99–110. doi: 10.1016/j.cell.2004.11.032. [DOI] [PubMed] [Google Scholar]
- Raasi S, Varadan R, Fushman D, Pickart CM. Diverse polyubiquitin interaction properties of ubiquitin-associated domains. Nat Struct Mol Biol. 2005;12:708–714. doi: 10.1038/nsmb962. [DOI] [PubMed] [Google Scholar]
- Ravid T, Hochstrasser M. Diversity of degradation signals in the ubiquitin-proteasome system. Nat Rev Mol Cell Biol. 2008;9:679–690. doi: 10.1038/nrm2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed SI. Ratchets and clocks: the cell cycle, ubiquitylation and protein turnover. Nature reviews Molecular cell biology. 2003;4:855–864. doi: 10.1038/nrm1246. [DOI] [PubMed] [Google Scholar]
- Santos B, Snyder M. Sbe2p and sbe22p, two homologous Golgi proteins involved in yeast cell wall formation. Mol Biol Cell. 2000;11:435–452. doi: 10.1091/mbc.11.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seol JH, Shevchenko A, Deshaies RJ. Skp1 forms multiple protein complexes, including RAVE, a regulator of V-ATPase assembly. Nature cell biology. 2001;3:384–391. doi: 10.1038/35070067. [DOI] [PubMed] [Google Scholar]
- Sims JJ, Haririnia A, Dickinson BC, Fushman D, Cohen RE. Avid interactions underlie the Lys63-linked polyubiquitin binding specificities observed for UBA domains. Nature structural & molecular biology. 2009;16:883–889. doi: 10.1038/nsmb.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowyra D, Craig KL, Tyers M, Elledge SJ, Harper JW. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell. 1997;91:209–219. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- Srivastava A, Jones EW. Pth1/Vam3p is the syntaxin homolog at the vacuolar membrane of Saccharomyces cerevisiae required for the delivery of vacuolar hydrolases. Genetics. 1998;148:85–98. doi: 10.1093/genetics/148.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai B, Ye Y, Rapoport TA. Retro-translocation of proteins from the endoplasmic reticulum into the cytosol. Nature reviews Molecular cell biology. 2002;3:246–255. doi: 10.1038/nrm780. [DOI] [PubMed] [Google Scholar]
- White MA, Riles L, Cohen BA. A systematic screen for transcriptional regulators of the yeast cell cycle. Genetics. 2009;181:435–446. doi: 10.1534/genetics.108.098145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems AR, Schwab M, Tyers M. A hitchhiker’s guide to the cullin ubiquitin ligases: SCF and its kin. Biochim Biophys Acta. 2004;1695:133–170. doi: 10.1016/j.bbamcr.2004.09.027. [DOI] [PubMed] [Google Scholar]
- Wolf J, Nicks M, Deitz S, van Tuinen E, Franzusoff A. An N-end rule destabilization mutant reveals pre-Golgi requirements for Sec7p in yeast membrane traffic. Biochemical and biophysical research communications. 1998;243:191–198. doi: 10.1006/bbrc.1998.8084. [DOI] [PubMed] [Google Scholar]
- Wout PK, Sattlegger E, Sullivan SM, Maddock JR. Saccharomyces cerevisiae Rbg1 protein and its binding partner Gir2 interact on Polyribosomes with Gcn1. Eukaryot Cell. 2009;8:1061–1071. doi: 10.1128/EC.00356-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wunschmann J, Beck A, Meyer L, Letzel T, Grill E, Lendzian KJ. Phytochelatins are synthesized by two vacuolar serine carboxypeptidases in Saccharomyces cerevisiae. FEBS letters. 2007;581:1681–1687. doi: 10.1016/j.febslet.2007.03.039. [DOI] [PubMed] [Google Scholar]
- Yen HC, Elledge SJ. Identification of SCF ubiquitin ligase substrates by global protein stability profiling. Science. 2008;322:923–929. doi: 10.1126/science.1160462. [DOI] [PubMed] [Google Scholar]
- Yen JL, Flick K, Papagiannis CV, Mathur R, Tyrrell A, Ouni I, Kaake RM, Huang L, Kaiser P. Signal-induced disassembly of the SCF ubiquitin ligase complex by Cdc48/p97. Molecular cell. 2012;48:288–297. doi: 10.1016/j.molcel.2012.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu L, Wu X, An X, Stubbe J, Huang M. Investigation of in vivo diferric tyrosyl radical formation in Saccharomyces cerevisiae Rnr2 protein: requirement of Rnr4 and contribution of Grx3/4 AND Dre2 proteins. J Biol Chem. 2011;286:41499–41509. doi: 10.1074/jbc.M111.294074. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.