Abstract
We used whole transcriptome microarrays to assess changes in gene expression and monitored mortality rates and epicuticular hydrocarbons (CHCs) in response to desiccation stress in four natural populations of Drosophila mojavensis from Baja California and mainland Mexico. Desiccation had the greatest effect on gene expression, followed by biogeographical variation at regional and population levels. Genes involved in environmental sensing and cuticular structure were up-regulated in dry conditions, while genes involved in transcription itself were down-regulated. Flies from Baja California had higher expression of reproductive and mitochondrial genes, suggesting that these populations have greater fecundity and higher metabolic rates. Host plant differences had a surprisingly minor effect on the transcriptome. In most cases, desiccation-caused mortality was greater in flies reared on fermenting cactus tissues than laboratory media. Water content of adult females and males was significantly different, and was lower in Baja California males. Different groups of CHCs simultaneously increased and decreased in amounts due to desiccation exposure of 9 and 18 hr and were population-specific and dependent on larval rearing substrates. Overall, we observed that changes in gene expression involved a coordinated response of behavioral, cuticular and metabolic genes. Together with differential expression of cuticular hydrocarbons, this study revealed some of the mechanisms that have allowed D. mojavensis to exploit its harsh desert conditions. Certainly, for D. mojavensis that uses different host plants, population-level understanding of responses to stressors associated with future climate change in desert regions must be evaluated across geographical and local ecological scales.
Keywords: cuticular hydrocarbons, desert, desiccation, gene expression, genome, host plant, microarray, transcriptome
Introduction
Over 30% of the earth's landmass consists of arid or semi-arid areas (Okin et al. 2006). Desertification is expected to increase as a result of global climate change and anthropogenic influence (Karl & Trenberth 2003). There is therefore a critical need to understand how organisms will respond to these environmental changes, and the study of desert species constitutes an important first step towards that goal. Many studies have examined the physiology and ecology of desert organisms, but our understanding of genetic and genome-level adaptation to deserts is minimal. The only desert organism whose genome has been sequenced so far is Drosophila mojavensis (12 Genomes 2007), a well-studied cactophilic species that presents a unique opportunity to obtain an integrated understanding of genomic responses to the harsh abiotic conditions that occur in arid lands. It is also a member of the well studied cactus-yeast-Drosophila model system, providing a rich background for ecological genomic studies (Barker & Starmer 1982; Barker et al. 1990). Drosophila mojavensis has diverged into ecologically diverse desert habitats (Etges et al. 1999; Heed 1978), has undergone adaptation to different host plants (Etges 1989b, 1990, 1993), and some geographically isolated populations are now considered incipient species (Etges 1998; Etges & Ahrens 2001; Etges et al. 2007). Thus, D. mojavensis is an excellent model for understanding the genomics of adaptation to arid environments and reproductive isolation.
In the New World, about half of the ca. 100 species in the large D. repleta group use fermenting cactus tissues to carry out their life cycles (Heed 1982; Oliveira et al. 2012; Wasserman 1992). Within the mulleri species complex, D. mojavensis, D. arizonae and D. navojoa form a monophyletic group that is endemic to Mexico and the southwestern United States (Ruiz et al. 1990). In the Sonoran and Mojave Deserts and adjacent arid lands, D. mojavensis was isolated from the ancestor of its closest relative, D. arizonae, on the mainland in association with tectonic drift of present-day peninsular Baja California (Garrick et al. 2009; Gastil et al. 1975; Riddle et al. 2000). Derived mainland Mexico and Arizona populations of D. mojavensis use organ pipe, Stenocereus thurberi, and sina cactus, S. alamosensis, and diverged ca 250 kya (Reed et al. 2007; Smith et al. 2012) from Baja California populations that primarily use pitaya agria cactus, S. gummosus, with occasional use of cochal cactus, Myrtillocactus cochal (Fellows & Heed 1972; Heed & Mangan 1986).
Mainland Mexico and southern Arizona populations have undergone genetic differentiation in allozyme and chromosome inversion frequencies (Etges et al. 1999; Zouros 1974), host plant-specific life histories (Etges 1989b, 1990, 1993; Etges & Heed 1987; Etges et al. 1999), and physiological adaptation to alternate cactus host species (Etges & Klassen 1989; Starmer et al. 1977). Further, Baja California, mainland Mexico and Arizona D. mojavensis exhibit low, but significant sexual isolation (Markow 1991; Zouros & d'Entremont 1974) and postmating, prezygotic isolation (Knowles & Markow 2001) among populations with no observed postmating hybrid inviability/sterility. These rearing substrate shifts have also influenced sex-specific epicuticular hydrocarbon (CHC) differences that mediate premating isolation (Etges & Ahrens 2001; Stennett & Etges 1997), in addition to courtship song differences (Etges et al. 2007; Etges et al. 2006).
Epicuticular hydrocarbons also serve as a barrier to cuticular water loss (Gibbs 2002; Gibbs and Rajpurohit 2010), because differences in the amount and composition of CHCs affect the permeability of the cuticle (Johnson & Gibbs 2004). In D. mojavensis, CHCs are composed of long chain (C29 – C50) alkanes, alkenes, branched alkenes, alkadienes, alkatrienes, and alkatetraenes (Etges & Jackson 2001; Yew et al. 2011) that can change with age and environmental temperature (Gibbs et al. 1998; Toolson et al. 1990). Wild-caught adults and flies reared from cactus rots have significantly lower amounts of CHCs than flies reared on cactus in the lab (Etges 2002), suggesting that natural environmental conditions may significantly affect water balance. While the underlying genetic pathways for some hydrocarbon components are partially understood in some drosophilids (reviewed in Gleason et al. 2009), genetic and environmental influences on hydrocarbon production and deposition are poorly known for most species.
In comparison to other drosophilids, D. mojavensis is tolerant of desert conditions, including high temperatures (Krebs 1999; Stratman & Markow 1998) and low humidity (Gibbs & Matzkin 2001; Gibbs et al. 2003b; Matzkin and Markow 2009; Kellermann et al, 2012). It is adapted to feed on necrotic cactus tissues rich in secondary compounds, e.g. phytosterols, fatty acids, and triterpene glyosides toxic to other insects (Fogleman & Danielson 2001; Fogleman et al. 1986; Fogleman & Heed 1989). Furthermore, adult flies can metabolize volatile compounds like ethanol vapor and other cactus fermentation by-products (Etges 1989a; Etges & Klassen 1989; Starmer et al. 1977). Thus, D. mojavensis thrives under a wide variety of environmental stressors and represents an ideal species in which to identify genomic mechanisms of stress resistance.
Here, we take advantage of the sequenced genome of D. mojavensis to investigate transcriptional responses to desiccation stress, the first of several studies from our group using whole genome microarrays to uncover functional genomic responses to ecological variation in desert conditions, host plant use, and geographical variation. We show that exposure to low humidity conditions causes transcriptional changes in over a thousand of genes, in addition to significant geographic differences in adult mortality, water loss, and shifts in cuticular hydrocarbon profiles. Overall, greater than 90% of all predicted genes in D. mojavensis were differentially expressed in response to one or more environmental or biogeographical factors, or interactions between them.
Material and Methods
Origin of stocks and husbandry
Populations of D. mojavensis were collected from cactus rots and over baits in the field, returned to the lab, and mass reared on banana food (Brazner & Etges 1993) in 35 ml shell vials at 20-22°C. Two populations from Baja California, San Quintin (SQ) and Punta Prieta (PP), were collected in 2008. A mainland Sonora population from Punta Onah (PO) was collected in 2007, and 20 isofemale lines collected in 2007 from Organ Pipe National Monument (OPNM), Arizona, were donated by S. Castrezana (Table 1).
Table 1.
Origins of the five populations of Drosophila mojavensis in this study and estimates of the numbers of flies used to establish laboratory populations. All flies were collected over banana baits in nature unless otherwise noted.
| Population | Stock number | Latitude Longitude | Number of founders |
|---|---|---|---|
| Punta Onah, Sonora | PO07 | 29° 5“23.15”N 112°10“15.59”W |
472† |
| Organ Pipe National Monument, AZ | OPNM08 | 31°58“4.88”N 112°46“5.75”W |
20 isofemale lines* |
| San Quintin, Baja California | SQ08 | 30°30“41.88”N 115°53“34.51”W |
372# |
| Punta Prieta, Baja California | PP08 | 28°52“43.48”N 114° 7“28.90”W |
465 |
includes ca 80 adults aspirated from agria rots
10 to 20 adults from each isofemale line collected in May 2007 were combined and mass reared for at least 6 generations to form this stock. Lines were provided by S. Castrezana
includes 355 adults that emerged from 3 agria rots
After several generations in the laboratory to increase population sizes, thousands of adult flies from each population were introduced into 12,720 cm3 plexiglass population chambers and allowed to mate for 7-10 days. For each of the four populations, eggs were collected in food cups attached to the cages and distributed into six 250-ml milk bottles containing banana food. Bottle cultures were established at moderate larval densities to minimize nutritionally caused maternal/environmental effects from the vial cultures and maintained in an incubator programmed on a 14:10 LD photoperiod that cycled from 27°C to 17°C. All bottle-reared adults were separated by sex on the day of eclosion and aged to maturity on lab food in vials in the incubator.
Approximately 100 adults of each sex from each population were then introduced into separate oviposition chambers, and eggs were collected daily for 10 h in 5.5 cm diameter Petri dishes containing agar-cactus media. Eggs from replicate chambers were washed in deionized water, 70% ethanol, again in sterile deionized water, counted into groups of 200, transferred to a 1 cm2 piece of sterilized filter paper, and placed on fermenting cactus in an incubator programmed as described above. Fermenting cactus cultures were set up in plugged half pint bottles with 75 g of aquarium gravel at the bottom covered with a 5.5-cm diameter piece of filter paper. Bottles were then autoclaved, 60 g of either agria or organ pipe tissues were placed in the bottles, and the bottles were autoclaved again for 8 min at low pressure. After cooling to room temperature, each culture was inoculated with 0.5 mL of a pectolytic bacterium, Erwinia cacticida (Alcorn et al. 1991) and 1.0 mL of a mixture of seven yeast species common in natural agria and organ pipe rots (Starmer 1982): Dipodascus starmeri, Candida sonorensis, C. valida, Starmera amethionina, Pichia cactophila, P. mexicana, and Sporopachydermia cereana. All unhatched eggs were counted to allow calculation of egg to adult viability in order to monitor culture conditions, and all eclosed adults from each replicate culture were counted daily under CO2 anesthesia, separated by sex, and kept on banana food in vials in the incubator until flies were ready for the experiments. Females were aged until they were sexually mature, i.e. 8 days, before the desiccation experiments began.
Desiccation Treatments
Groups of 24 eight day-old females were placed in 35-ml pre-sterilized glass vials containing 10 g of Drierite desiccant in an incubator set at 25°C with constant illumination. Flies were restricted to the bottom of a vial (1.5 cm) with a foam stopper, desiccant was added above the sponge, and the vial was sealed with Parafilm®. Preliminary experiments with cactus-reared flies exposed to low humidity conditions for 0 (control), 12, and 24 hours (after Gibbs & Matzkin 2001) revealed that many flies were already dead by 24 hours (see below), so we used exposure periods of 0, 9 and 18 hours. Flies were removed after each time period, frozen in liquid nitrogen, and stored at -80° C prior to RNA extraction. A total of 24 treatments (4 populations × 2 host cacti × 3 desiccation times) were included with four-fold replication (in most cases; 3 or 5 samples were collected for a few treatments), resulting in 95 groups of 24 females each for RNA extraction and microarray analysis. Additional replicates were included, so that we could directly estimate mortality rates due to desiccation exposure, changes in mass and water content in both females and males, and assess cuticular hydrocarbon profiles.
Desiccation time and mortality rates
We directly assayed desiccation-caused mortality rates with male and female adults reared on lab food and males reared on fermenting agria or organ pipe cactus, in order to investigate the causes for increased mortality rates in cactus–reared flies. For the latter experiments, female flies were used for transcriptome experiments (see below). Ten flies per vial were exposed to the desiccant as previously described or kept in empty (control) vials until all flies were dead at 25°C with constant illumination. The number of dead flies was counted every four hours, four times a day with a 12 hour interval during the evening. The average time to death in hours per vial was calculated by linear interpolation of numbers of dead flies observed at each time point. To investigate possible bias in mortality due to fly age, we included fly age when exposure to low humidity began as a factor by starting experiments with flies aged 0, 3, 6, 9, and 12 days. Thus, a total of 80 treatment combinations were included for flies reared on lab food (4 populations × 2 sexes × 5 ages × 2 desiccation times) and 80 treatments for males reared on cacti (4 populations × 5 ages × 2 hosts × 2 desiccation times). Females reared on fermenting cactus were used for RNA extractions (see below). In both cases, three replicates were performed for each treatment. All data were analyzed by ANOVA in SAS (SAS-Institute 2004).
Body Mass and Water Content
We assessed body mass and water content differences as possible factors contributing to desiccation resistance. Adult fly water content was estimated as described by Gibbs et al. (2003a). Three groups of ten flies aged 8-12 days from the 16 treatments (4 populations × 2 cacti × 2 sexes) were frozen briefly at -80°C, thawed, and weighed on a Mettler microbalance. Flies were re-weighed after drying overnight at 50°C. Water content was calculated as the difference between wet and dry weight. Fly age ranged from 8-12 days.
Cuticular hydrocarbons
We sampled CHCs from five cactus-reared adult females from each of 24 treatments (4 populations × 2 hosts × 3 desiccation times) in order to reveal the dynamics of desiccation-caused CHC expression. Total CHCs were extracted by immersing single 8 day old flies in hexane for 20 min in a 300 μl glass vial insert (Microliter Analytical Supplies, Suwanee, GA), evaporating off all hexane in a 40°C heating block, and freezing each sample at -20°C until analysis. Individual CHC extracts were redissolved in 5 μl of heptane containing a known amount of docosane (C22) as an internal standard. One μl of each sample was analyzed by capillary gas-liquid chromatography using an automated Shimadzu GC-17A (Shimadzu Scientific Instruments, Columbia, MD) fitted with a 15 m (ID = 0.22 mm) Rtx-5 fused-silica column (Restek Corporation, Bellefont, PA). Injector and detector temperatures were set at 290° C and 345° C, respectively, with the injector port in split mode (ratio = 3:1), and the column was heated from 200°C to 345°C at 15°C/min, holding at 345° C for 4 min. Amounts of the CHCs were expressed as ng/fly and analyzed by ANOVA, Principal Components Analysis, and Canonical Discriminant Function Analysis in SAS (SAS Institute, Cary NC, USA, 2004).
RNA isolation, cDNA synthesis, microarray hybridization and visualization
RNA was isolated from groups of 24 female flies using RNeasy mini-kits (Qiagen, Valencia, California USA). After extraction, RNA was stored at -80°C until microarray hybridizations were performed. Invitrogen Superscript Double-Stranded cDNA Synthesis kits were used to synthesize double-stranded cDNA. Each cDNA sample was quantified using a NanoDrop spectrophotometer (NanoDrop Technologies) to verify that all samples met concentration and purity requirements (concentration ≥ 100 ng/ul; A260/A280 ≥ 1.8; A260/A230 ≥ 1.8). cDNA samples were labeled with Cy3 using a NimbleGen (Roche Diagnostics) One Color DNA Labeling kit. Hybridizations with custom NimbleGen 12-plex microarrays were performed with a NimbleGen Hybridization System 4. Probe selection for the microarrays was based on predicted transcripts from the genome assembly for D. mojavensis (http://flybase.net/genomes/Drosophila_mojavensis/current/fasta/dmoj-all-transcript-r1.3.fasta.gz), downloaded April 14, 2009. Nine probes per transcript were generated for each of 14,528 predicted transcripts. Other spots in the design included negative (random probe) controls, controls for contamination from adjacent subarrays, and blank (buffer) controls. Array scanning was performed with a GenePix 4000B scanner (Molecular Devices) and associated software at 532 nm, with photomultiplier gain settings that resulted in less than one in 105 normalized counts at saturation.
Processing of hybridization data
We normalized all gene expression data in a quantile manner as suggested by Bolstad et al. (2003) using NimbleScan v2.5. Gene ‘*.calls’ files were generated using the Robust Multichip Average (RMA) algorithm (Irizarry et al. 2003), and analysis and visualization of fluorescence data were performed using ArrayStar v3 software. Linear fluorescence intensities were used for ANOVA and false discovery rate (FDR) analyses in SAS (SAS Institute, 2004). All microarray data were MIAME compliant, and can be retrieved from the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo) under the series access number GSE43220.
Gene Annotation
We performed reciprocal BLAST searches between the predicted transcriptomes of D. melanogaster and D. mojavensis to investigate the functions of genes exhibiting statistically significant changes in expression. A total of 9,114 putative D. melanogaster orthologs were found after submission of 14,528 predicted D. mojavensis transcripts, i.e. only ∼ 63% of D. mojavensis genes possessed substantial transcript similarity with D. melanogaster. We assumed that these genes had conserved functions across the Drosophila genomes, so that annotations assigned to D. melanogaster genes were applicable to D. mojavensis. These annotations were used in further analyses, including Gene Ontology (GO) term enrichment, gene expression clustering, and pathway analysis. Supplementary Table S1 provides a list of FBtr IDs for D. mojavensis transcripts and FBgn IDs for their putative orthologs in D. melanogaster.
Statistical analyses
Our microarray experimental design included 3-5 replicates for each combination of four populations (PO, OPNM, PP and SQ), two host diets (AG and OP) and three desiccation treatments (0, 9 and 18 hours) for a total of 95 whole transcriptome hybridizations. The normalized fluorescence for each microarray probe set was subjected to a replicated 4-way mixed model ANOVA in SAS using the model:
where μ is the grand mean, Ri is the effect of geographic region (Baja California vs. the mainland), Pj is the effect of population nested within regions, Hk is the effect of host cactus, Dl is the effect of desiccation, IR×H is the interaction between region and cactus, IR×D is the interaction between region and dry air treatment, IP×H is the interaction between population and cactus nested within region, etc., and Eijk is the error term. To correct for multiple comparisons, we calculated false-discovery rates for the overall ANOVA data and for all pair-wise comparisons between treatments (Benjamini & Hochberg 1995).
Bioinformatic analysis
We used DAVID v6.7 (Huang et al. 2009) for gene enrichment analyses. Because the annotation for our transcripts was derived from a subset of the transcriptome of D. melanogaster, the background gene list consisted of these 9114 putative orthologs instead of the entire D. melanogaster transcriptome. We used the medium stringency defaults of DAVID for analyses to minimize exclusion of potentially interesting Gene Ontology (GO) terms. Gene Ontology GO_FAT and KEGG pathway terms were used to identify functionally related clusters of GO categories. Clusters with enrichment scores less than 1.3, corresponding to P > 0.05, were excluded from further analyses. We then submitted GO terms from these clusters to GO-Module, a web-based tool that identifies false positives included because of the hierarchical nature of the gene ontology (Yang et al. 2011).
Because we were also interested in temporal patterns of gene expression as desiccation progressed, we performed expression profiling using the Short Time-series Expression Miner, STEM (Ernst & Bar-Joseph 2006). Based on STEM profiles, genes showing up or down-regulation were grouped separately and submitted to DAVID and GO-Module for clustering and pathway analysis. We used Cytoscape (www.cytoscape.org) to visualize heat maps and perform clustering analyses.
Results
Mean ± 1 SE percent egg to adult viability and development time across the four populations reared on both cacti (n = 24 cultures) was 79.2 ± 1.7 % and 15.9 ± 0.3 da, respectively. Thus, our cactus rearing conditions produced flies of consistent viability, development times, and resulting adult body sizes as in previous studies (cf. Etges 1998).
Desiccation tolerance, mortality, and water loss
Adult mortality varied significantly due to low humidity exposure, age, and population for flies reared on lab food (Table S2, Figures S1, S2). No mortality differences due to sex were observed, so the data were pooled; however, a significant Sex × Desiccation interaction (P = 0.011, Table S2) was caused by a greater increase in desiccation-caused mortality in males than females (results not shown). Mean age at death (hr) ± 1 SE of control adults, 66.52 ± 0.97 hr, was significantly greater than that of adults exposed to zero humidity, 41.29 ± 0.93 hr, but was not uniform across all populations (Supplementary Figure S1). Adults from both mainland populations survived longer than Baja California adults under control conditions, i.e. they had greater starvation resistance in ambient humidity, consistent with previous studies (Starmer et al. 1977; Etges and Klassen 1989). However, there was no evidence for a mainland vs. Baja California difference in adult survivorship under low humidity conditions. Age at death declined almost monotonically with age in both control and desiccation treatments (Supplementary Figure S2) except for control 0, 3 and 6 day and desiccated 9 and 12 day-old adults: these pair-wise differences were not significantly different (P > 0.05). Thus, the starting age of flies exposed to low humidity had rather minor effects on subsequent survivorship under either control or experimental conditions.
We could only compare effects of preadult rearing substrates, cactus vs. lab food, on male desiccation tolerance, because cactus-reared females were used for microarray analyses. There were significant differences in desiccation tolerance due to population, food, desiccation, and age as well as significant Population × Food, Population × Desiccation, Population × Age, and Food × Age interactions (Figure 1, Table 2). Mortality differences among populations were consistent with the first experiment performed on lab food (Supplementary Figure S1), but the Population × Food interaction demonstrated that desiccation effects on mortality of lab food-reared males differed significantly from that of flies reared on fermenting cactus (Figure 1, Table 2).
Figure 1.
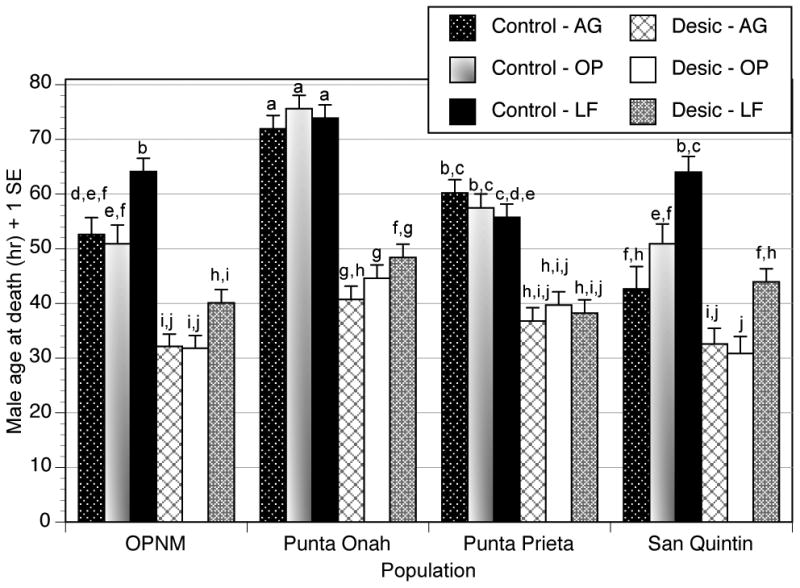
Age at death of adult male D. mojavensis reared on cactus or lab food from four populations that were of different ages when placed in control (ambient humidity) or zero humidity conditions to start the experiment. Ages were not graphed to emphasize rearing substrate and population differences. Females were not included in this analysis because they were used for microarray analysis. Letters above the error bars indicate posthoc groupings from Duncan's Multiple Range test (P < 0.05) – least square means were unavailable for some treatment combinations due to unequal sample sizes.
Table 2.
ANOVA results for survival time (hr) between control and zero humidity for male D. mojavensis reared on three substrates (food); agria cactus, organ pipe cactus, and laboratory media.
| Source of variation | df | Type III SS | F Value | Pr > F |
|---|---|---|---|---|
| Population | 3 | 9837.657 | 36.94 | < 0.0001 |
| Food | 2 | 2218.595 | 12.50 | < 0.0001 |
| Population × food | 6 | 2509.596 | 4.71 | 0.0002 |
| Desiccation | 1 | 32163.290 | 362.33 | < 0.0001 |
| Population × desiccation | 3 | 1815.137 | 6.82 | 0.0002 |
| Food × desiccation | 2 | 20.138 | 0.11 | 0.893 |
| Population × food × desiccation | 6 | 837.621 | 1.57 | 0.156 |
| Age | 4 | 13758.417 | 38.75 | < 0.0001 |
| Population × age | 12 | 2465.595 | 2.31 | 0.009 |
| Food × age | 8 | 3625.428 | 5.11 | < 0.0001 |
| Population × food × age | 20 | 2036.042 | 1.15 | 0.304 |
| Desiccation time × age | 4 | 183.908 | 0.52 | 0.723 |
| Population × desiccation × age | 12 | 1071.280 | 1.01 | 0.445 |
| Food × desiccation × age | 8 | 2026.800 | 2.85 | 0.005 |
| Population × food × desiccation × age | 20 | 1321.758 | 0.74 | 0.777 |
| Error | 217 | 19262.75 |
Overall, rearing substrate effects were diminished under desiccation as compared to controls. Males reared on lab food tended to survive significantly longer than those reared on cactus substrates except for Punta Prieta, Baja California males (Figure 1), and a significant Population × Desiccation interaction was similar to that seen in the lab food-only study (Table S2, Figure S1). Age effects were also similar to those in the first experiment with lab food-reared flies (results not shown). Therefore, cactus rearing substrates tended to increase adult mortality under desiccation compared with lab food. This may explain the higher survivorship of adults in previous lab food studies (Gibbs & Matzkin 2001; Matzkin et al. 2007; Matzkin and Markow 2009; Kellermann et al. 2012).
Wet and dry mass and adult body water content significantly differed among populations (Table 3, Figure 2). Wet mass differences between males and females were population specific as shown by a Sex × Population interaction (F = 6.87, P = 0.001). Water content also showed a Sex × Population interaction (Table 3), where female water content tended to be much more uniform across populations than male water content. Punta Onah, Sonora males had significantly higher water content than the other populations (Fig. 2) consistent with the greater survivorship of Punta Onah males in low humidity (Figure 1). There were no significant rearing substrate effects, agria vs. organ pipe cactus, on body water content (Table 3), consistent with the general lack of cactus-induced differences in mortality in control or desiccation treatments (Figure 1).
Table 3.
ANOVA results for variation in body mass for groups of 8-12 day old adult D. mojavensis before (wet mass), after drying overnight at 50° C (dry mass), and adult body water content (delta mass). Males and females from the four populations reared on both host cacti were included.
| Wet mass | Dry mass | Delta mass | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| Type III | Type III | Type III | ||||||||
| Source of variation | df | SS | F | P | SS | F | P | SS | F | P |
| Population | 3 | 66.136 | 10.46 | < 0.0001 | 2.404 | 4.6 | 0.009 | 44.238 | 7.13 | 0.0009 |
| Sex | 1 | 26.223 | 12.44 | 0.001 | 18.601 | 106.9 | < 0.0001 | 0.653 | 0.32 | 0.578 |
| Population × sex | 3 | 43.449 | 6.87 | 0.001 | 0.711 | 1.36 | 0.273 | 33.826 | 5.45 | 0.004 |
| Cactus | 1 | 6.192 | 2.94 | 0.097 | 0.346 | 1.99 | 0.169 | 3.611 | 1.75 | 0.196 |
| Population × cactus | 3 | 11.167 | 1.77 | 0.175 | 0.336 | 0.64 | 0.594 | 8.758 | 1.41 | 0.259 |
| Sex × cactus | 1 | 0.150 | 0.07 | 0.791 | 0.346 | 1.99 | 0.169 | 0.953 | 0.46 | 0.503 |
| Population × sex × cactus | 3 | 1.817 | 0.29 | 0.834 | 0.313 | 0.6 | 0.620 | 2.829 | 0.46 | 0.715 |
Figure 2.
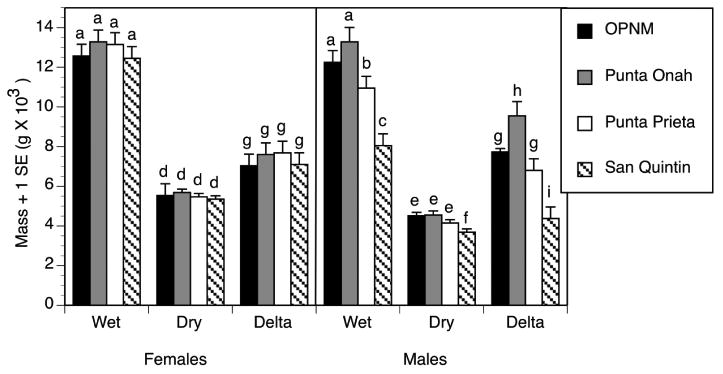
Differences in wet and dry mass of male and female D. mojavensis from the four populations in this study that were reared on agria and organ pipe cactus. Adult body water content, delta mass, is the difference between wet and dry mass after drying adults overnight at 50° C. Different letters above the error bars indicate significant least square mean differences between groups (P < 0.05).
Cuticular hydrocarbon responses to desiccation
Female CHCs varied significantly between populations and cactus rearing substrates, as anticipated from previous studies (Etges and Ahrens 2001), as well as in response to desiccation exposure and its interaction with population and cactus substrates (Supplementary Table S3). Thus, short-term CHC responses to low humidity were population-specific and depended on larval rearing substrates. Different groups of CHCs increased or decreased in amounts with increasing exposure to desiccation, even though total CHCs per fly did not change (Table 4). Among those that responded to low humidity, all but 31-methyldotricont-8-ene and 8,24-tritricontadiene (both C33) are typically minor CHC components in D. mojavensis (Etges & Jackson 2001), and these two CHCs increased in amounts with increasing exposure to low humidity. There was little evidence that specific kinds of CHCs increased or decreased in amount, i.e. methylalkanes, alkenes, methylalkenes, or alkadienes. The 12 CHCs that increased quantitatively included C31, C32, C34, C35, C36, C38, C39, and C40 components (Table 4).
Table 4.
Comparisons of least square means of the 31 epicuticular hydrocarbon components in female D. mojavensis included in this study in the three experimental desiccation treatments: 0 hr (control), 9 hr, and 18 hr in zero humidity. Mean hydrocarbon amounts are expressed in ng/fly. P values are from ANOVA.
| Hydrocarbon | ECL1 | 0 hr vs 9 hr vs 18 hr | P |
|---|---|---|---|
| 2-methyloctacosane | C28.65 | 40.278, 42.241, 41.322 | ns |
| 2-methyltricontane | C30.65 | 104.294, 106.515, 97.931 | ns |
| 7- and 9-hentricontene | C30.78 | 7.171 > 5.984 < 7.565 | 0.03 |
| Unknown | C32 | 2.922, 3.529 < 4.578 | 0.009 |
| Unknown alkene | C33br1 | 0.533, 0.470, 0.581 | ns |
| 11-and 13-methyldotricontane | C33br2 | 7.247, 6.391 > 5.761 | 0.03 |
| Unknown alkene | C33br3 | 5.553 > 4.739 > 3.928 | < 0.0001 |
| 31-methyldotricont-8-ene | C32.47 | 36.560, 32.386 > 26.352 | < 0.0001 |
| 31-methyldotricont-6-ene | C32.56 | 3.124, 3.271 > 2.771 | 0.007 |
| 8,24-tritricontadiene | C32.63 | 32.574, 35.527 > 30.313 | 0.03 |
| 7,25-tritricontadiene | C32.70 | 35.223, 37.0793, 35.5903 | ns |
| 10-, 12-, and 14-tritricontene | C32.79 | 9.767, 8.411 < 12.454 | 0.008 |
| Unknown | C32.86 | 1.318, 1.092, 1.011 | ns |
| 8,26-tetratricontadiene | C34diene1 | 7.481, 6.099 < 7.528 | 0.007 |
| 6,24- and 6,26-tetracontadiene | C34diene2 | 15.267, 16.282 < 17.176 | 0.03 |
| 10-, 12-, and 14 tetretricontene | C34ene | 5.422, 6.802 < 8.420 | < 0.0001 |
| 33-methlytetratricont-10-ene | C35alk1 | 10.417, 12.638, 12.649 | ns |
| 33-methlytetratricont-8-ene | C35alk2 | 12.464 < 14.773 > 12.746 | 0.03 |
| Unknown alkene | C35alk3 | 25.122, 28.061 > 23.930 | 0.009 |
| 9,25-pentatricontadiene | C34.59 | 88.664, 88.664, 89.458 | ns |
| 8,26-pentatricontadiene | C34.66 | 306.191, 280.118, 295.936 | ns |
| 7,27-pentatricontadiene | C34.73 | 32.792, 33.538 < 42.955 | 0.011 |
| Unknown diene | C36a | 8.137, 6.923, < 9.542 | 0.023 |
| Unknown alkene | C36b | 10.649, 12.348 < 15.087 | 0.004 |
| 35-methylhexatricont-10-ene | C37br | 2.610, 2.755, 2.710 | ns |
| 9,27-heptatricontadiene | C36.5 | 26.253, 29.090, 25.998 | ns |
| 8,28-heptatricontadiene | C36.6 | 67.738, 66.200, 76.153 | ns |
| 14-, 16-, and 12-hexatricontene | C36.7 | 41.698, 37.238, 42.248 | ns |
| Unknown alkene | C38 | 5.084, 5.096 < 6.927 | 0.008 |
| Unknown alkene | C39 | 4.647 < 6.340, 6.790 | 0.04 |
| Unknown alkene | C40 | 3.447, 2.903 < 3.854 | 0.009 |
| Total hydrocarbons per fly | 960.646, 962.261, 970.177 | ns |
These covarying CHCs that increased in quantity with exposure to low humidity showed negative correlations along the largest axis of variation from Canonical Discriminant Function (CDF) analysis (Table 5). Inspection of the largest +/- scores for CV 1 revealed complete correspondence with the +/- changes in CHC amounts (Table 4): negative CV scores corresponded to those CHCs that increased in amounts and positive scores corresponded to those that decreased in amounts. Decreases in average CHC centroid scores from 0, 9, to 18 hr of zero humidity exposure were significantly different from each other (Figure 3; all Euclidean distances, P < 0.05). CV 2 scores showed similar patterns of CHC covariation (Table 5), although mean CV 2 scores increased from controls at 9 hr and then decreased at 18 hr. Overall, significant increases in CHC amounts in response to 9 and 18 h of exposure to zero humidity involved mostly a variety of less abundant female D. mojavensis CHCs with fewer CHCs decreasing in amounts.
Table 5.
Total structure of the first 5 canonical variates (CV) for the 31 female cuticular hydrocarbons in this study. CV 1 and 2 loadings in bold correspond to the hydrocarbons that covaried (+/-) and showed significant shifts in amounts due to desiccation exposure. See Table 4.
| Cuticular Hydrocarbon | ECL1 | CV 1 | CV 2 | CV 3 | CV 4 | CV 5 |
|---|---|---|---|---|---|---|
| 2-methyloctacosane | C28.65 | -0.057 | 0.022 | -0.011 | 0.001 | 0.013 |
| 2-methyltricontane | C30.65 | 0.152 | 0.155 | -0.008 | 0.043 | 0.049 |
| 7- and 9-hentricontene | C30.78 | -0.103 | -0.290 | 0.020 | -0.108 | -0.033 |
| Unknown | C32 | -0.291 | 0.034 | 0.074 | -0.101 | -0.002 |
| Unknown alkene | C33br1 | 0.065 | 0.059 | -0.005 | 0.003 | -0.018 |
| 11-and 13-methyldotricontane | C33br2 | 0.202 | -0.109 | 0.059 | -0.082 | -0.080 |
| Unknown alkene | C33br3 | 0.318 | -0.105 | -0.001 | 0.016 | 0.014 |
| 31-methyldotricont-8-ene | C32.47 | 0.429 | -0.078 | 0.020 | 0.029 | 0.031 |
| 31-methyldotricont-6-ene | C32.56 | 0.244 | 0.139 | -0.018 | 0.064 | 0.055 |
| 8,24-tritricontadiene | C32.63 | 0.099 | 0.185 | 0.066 | 0.016 | -0.066 |
| 7,25-tritricontadiene | C32.70 | -0.056 | -0.033 | 0.020 | -0.037 | -0.026 |
| 10-, 12-, and 14-tritricontene | C32.79 | -0.240 | -0.169 | -0.068 | 0.182 | 0.102 |
| Unknown | C32.86 | 0.047 | 0.103 | -0.064 | 0.052 | 0.080 |
| 8,26-tetratricontadiene | C34diene1 | -0.136 | -0.168 | -0.012 | -0.029 | 0.031 |
| 6,24- and 6,26-tetracontadiene | C34diene2 | -0.181 | -0.070 | 0.091 | -0.053 | 0.032 |
| 10-, 12-, and 14 tetretricontene | C34ene | -0.343 | 0.110 | -0.045 | -0.003 | 0.134 |
| 33-methlytetratricont-10-ene | C35alk1 | -0.050 | 0.202 | -0.003 | 0.031 | 0.043 |
| 33-methlytetratricont-8-ene | C35alk2 | 0.099 | 0.210 | -0.025 | 0.131 | -0.009 |
| Unknown alkene | C35alk3 | 0.176 | 0.181 | 0.023 | 0.080 | 0.086 |
| 9,25-pentatricontadiene | C34.59 | -0.018 | 0.023 | 0.033 | 0.010 | -0.011 |
| 8,26-pentatricontadiene | C34.66 | 0.052 | -0.097 | 0.036 | 0.010 | 0.018 |
| 7,27-pentatricontadiene | C34.73 | -0.222 | 0.112 | -0.008 | 0.156 | -0.015 |
| Unknown alkene | C36a | -0.181 | -0.059 | 0.069 | 0.196 | 0.042 |
| Unknown alkene | C36b | -0.256 | -0.063 | -0.014 | -0.028 | -0.069 |
| 35-methylhexatricont-10-ene | C37br | 0.056 | 0.132 | -0.011 | 0.081 | -0.005 |
| 9,27-heptatricontadiene | C36.5 | 0.006 | -0.015 | -0.027 | 0.017 | -0.070 |
| 8,28-heptatricontadiene | C36.6 | -0.043 | -0.049 | 0.052 | -0.037 | -0.012 |
| 14-, 16-, and 12-hexatricontene | C36.7 | -0.006 | 0.010 | 0.012 | 0.027 | 0.069 |
| Unknown alkene | C38 | -0.342 | -0.089 | -0.129 | 0.007 | 0.047 |
| Unknown alkene | C39 | -0.210 | 0.147 | 0.378 | 0.119 | 0.043 |
| Unknown alkene | C40 | -0.228 | -0.247 | 0.127 | 0.018 | 0.076 |
Equivalent chain length for each hydrocarbon component.
Figure 3.
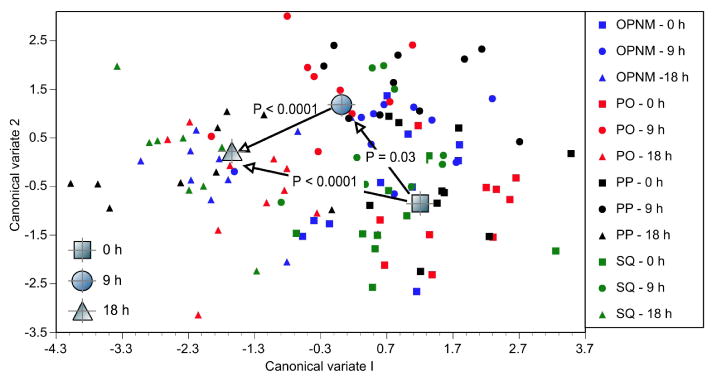
Canonical Discriminant Function biplot showing shifts in cuticular hydrocarbon profiles of female D. mojavensis from the four populations in this study exposed to zero humidity. 0, 9, and 18 hr mean centroids for CV 1 and 2 are shown, and P values refer to significantly different Euclidean distances between means. Cactus differences were not included. OPNM = Organ Pipe National Monument, Arizona; PO = Punta Onah, Sonora; PP = Punta Prieta, Baja California; SQ = San Quintin, Baja California.
Transcriptome results: Summary of differentially expressed transcripts
Our mixed-model nested ANOVA design included four main effects and seven interaction effects with populations nested within regions. At a false-discovery rate (FDR) of P < 0.01, the ANOVA revealed a total of 768,808 statistically significant pair-wise differences between treatments (Table 6). The magnitude of many effects was small (below 5% in some cases), so for some analyses we set an arbitrary cut-off of 1.5-fold expression change for inclusion in further analyses, reducing the number of differences analyzed by nearly two-thirds (Table 6). Our objective was to identify broad patterns of changes in gene expression, while also allowing us to identify specific gene categories that were especially affected by experimental variables.
Table 6.
Summary of gene expression differences.
| Treatments | No. of Pairwise Differences; FDR<0.01 | No. of Unique Genes | No. of Genes with Dmel Orthologs | No. of Pairwise Differences; 1.5× fold change cut-off | No. of Unique Genes | No. of Genes with Dmel Orthologs |
|---|---|---|---|---|---|---|
| Main effects: | ||||||
| Region (R) | 3098 | 3098 | 1963 | 184 | 184 | 64 |
| Population (P) | 11926 | 5263 | 3417 | 1182 | 514 | 217 |
| Host cactus (H) | 18 | 18 | 15 | 0 | 0 | 0 |
| Desiccation (D) | 16794 | 8917 | 6089 | 1152 | 828 | 507 |
| Interaction Effects: | ||||||
| 5. R × H | 6896 | 2556 | 1594 | 790 | 249 | 96 |
| 6. R × D | 56983 | 11467 | 7401 | 8625 | 2825 | 1520 |
| 7. P × H | 25097 | 5041 | 3198 | 4609 | 835 | 346 |
| 8. P × D | 152055 | 12351 | 7941 | 50539 | 5731 | 3062 |
| 9. H × D | 43037 | 8507 | 5865 | 5266 | 1413 | 813 |
| 10. R × H × D | 131779 | 11148 | 7231 | 35836 | 4454 | 2364 |
| 11. P × H × D | 321125 | 12607 | 8011 | 161234 | 8336 | 4572 |
| Column totals: | 768808 | 80973 | 52725 | 269417 | 25369 | 13561 |
| Number of unique genes: | 13342 | 8481 | 8394 | 4587 |
For the four main effects, desiccation affected the largest number of transcripts (8917; Table 6), while the expression of only 18 genes was affected by host plant differences. In the latter case, the largest statistically significant difference was < 25%, so that no diet-related genes passed the 1.5-fold change cut-off. Region and population (nested within region) affected a total of 3098 and 5262 transcripts, respectively. There were five two-factor interactions and two three-factor interactions included in our statistical model. When interaction terms were included, over 90% of predicted genes were differentially expressed under some combination of experimental variables (Table 6).
We used Cytoscape v2.8.2 (Smoot et al. 2011) to generate and visualize gene expression networks with the ExpressionCorrelation plugin (www.baderlab.org/Software/ExpressionCorrelation), which creates networks of gene expression similarities based on the Pearson correlation coefficient. For this analysis, we included all 8,394 genes with at least a 50% fold change in expression for one or more main or interaction effects. Gene expression clustered into three main subgroups based on desiccation time (Figure 4). Fly samples desiccated for 9 or 18 hr clustered with with each other; within these treatments samples from Baja California and mainland Mexico and Arizona tended to cluster with each other. Thus, desiccation had the greatest effect on global gene expression, followed by geographic region, with little effect of host plant.
Figure 4.
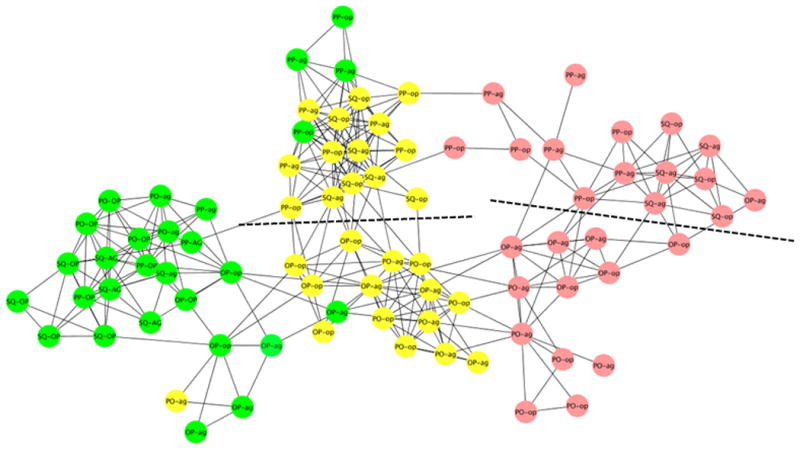
Gene expression network generated using Cytoscape. All genes whose expression differed by at least 50% in at least one pairwise comparison of experimental treatments (Table 8) were included in the analysis. Dashed lines separate samples from the mainland and Baja California after 9 and 18 hr of desiccation.
Desiccation effects
Analyses using DAVID are restricted to lists of 3000 or fewer genes, but >6000 genes affected by desiccation stress had orthologs in D. melanogaster. We therefore used STEM (Ernst & Bar-Joseph 2006) to investigate desiccation-related patterns of expression in these genes. STEM classified our data into 16 temporal patterns, six of which were highly overrepresented relative to chance (Chi-squared test, P < 10-14 for each pattern). These included two patterns each in which expression increased or decreased consistently over time. In the other two patterns, gene expression increased or decreased early in desiccation, then did not change between 9 and 18 hr.
We performed separate DAVID and GO-Module analyses for the up-regulated and down-regulated gene lists. For up-regulated genes, eight clusters of GO terms were uncovered with enrichment scores greater than 3, corresponding to P < 0.001. Summaries of these categories are listed in Table 7A, and more detailed information is provided in Supplementary Table S4. Two of these groups, including that with the second greatest enrichment, contained genes related to sensing of the environment. The third- and fourth-most enriched clusters included genes encoding cuticular components, and two others involved genes involved in ion transport and neuropeptides. Ten down-regulated GO clusters included four related to transcription, chromatin organization and development (Table 9B, Supplementary Table S3).
Table 7.
Summary of functional information for Gene Ontology categories upregulated or downregulated during desiccation stress. Only categories with enrichment scores >3.0 (corresponding to P<0.001) are included. More information on genes included in these categories and GO clusters with lower enrichment scores are included in Supplementary Tables S2 and S3.
| A. Clusters of GO categories that were upregulated during desiccation stress. | |||
|---|---|---|---|
| Cluster No. | General GO categories | Enrichment | Total No. Genes |
| 1 | membrane proteins | 14.60 | 273 |
| 2 | perception of smell | 10.65 | 119 |
| 3 | structural constituent of chitin-based cuticle | 8.37 | 42 |
| 4 | chitin metabolism | 7.52 | 93 |
| 5 | cation transport | 4.99 | 90 |
| 6 | plasma membrane | 4.65 | 77 |
| 7 | detection of chemical stimulus | 3.73 | 23 |
| 8 | neuropeptide hormone, signaling | 3.44 | 15 |
| B. Clusters of GO categories that were downregulated during desiccation stress. | |||
|---|---|---|---|
| Cluster No. | General GO categories | Enrichment | Total No. Genes |
| 1 | zinc ion binding | 12.04 | 216 |
| 2 | regulation of transcription | 5.57 | 240 |
| 3 | chromatin organization and modification | 4.86 | 62 |
| 4 | positive regulation of transcription | 4.42 | 62 |
| 5 | cell migration | 3.92 | 64 |
| 6 | neuron differentiation and morphogenesis | 3.82 | 121 |
| 7 | nucleoplasm, transcription | 3.58 | 75 |
| 8 | metamorphosis, development | 3.56 | 82 |
| 9 | tracheal development | 3.29 | 43 |
| 10 | DNA repair | 3.02 | 56 |
To further characterize desiccation-related changes in gene expression, we separately analyzed sets of genes that were up- or down-regulated, during early (0 to 9 hr) and late (9 to 18 hr) desiccation stress. In the first nine hours, four up-regulated GO clusters had enrichment scores >3, corresponding to P < 0.001, and included genes related to mitochondrial respiration, ribosome components, fatty acid biosynthesis and photoreception (Supplementary Table S6). Lower enrichment clusters included several composed of genes related to mitochondria, chitin metabolism and intermediary metabolism. Between 9 and 18 hours, broadly similar categories of genes continued to show increased expression, except for mitochondrial respiration.
Clusters of genes that were down-regulated during the first 9 hours of desiccation stress included a wide array of cellular processes, many related to chromosome structure and gene transcription (Supplementary Table S7). Note that the third most highly enriched cluster (EASE score > 12), which initially included several GO terms related to cation binding, was reduced to a single term (GO:0008270; zinc ion binding) after elimination of false positives with GO-Module. Several clusters were related to oogenesis, suggesting reduced reproductive effort. Broadly similar responses were seen in the subsequent nine hours of desiccation.
In analyzing these data, we noticed that 19 genes were included on lists of up- and down-regulated genes. Expression of 10 genes decreased from 0 to 9 hours of desiccation, then increased from 9 to 18 hours. Six had putative orthologs in D. melanogaster, and interestingly, 4 of these 6 had been annotated as structural components of the chorion. Of the 9 genes (3 with D. melanogaster orthologs) whose expression increased, then decreased, two are annotated as structural components of the vitelline membrane. The other annotated gene was phantom (phm), which encodes a cytochrome P450 catalyzing an early step in ecdysone biosynthesis (Warren et al. 2004).
Geographic variation
3098 genes (1963 with putative orthologs in D. melanogaster) differed in expression between populations from Baja California and mainland Mexico and Arizona, but only 187 differed by at least 50%. Of these, only 64 had putative orthologs in D. melanogaster. One cluster of functionally related genes with enrichment scores greater than 1.3, i.e. a type 1 error of 0.1, was recovered with DAVID. This cluster (11 GO categories, enrichment score 1.54) contained several GO terms related to female reproduction (e.g. formation of gametes, ovarian follicular cells, and the eggshell). Because of overlap among these categories, this cluster represented only ten genes, nine of which were more highly expressed in flies from Baja California (Figure 5; 2-tailed sign test, P < 0.022).
Figure 5.
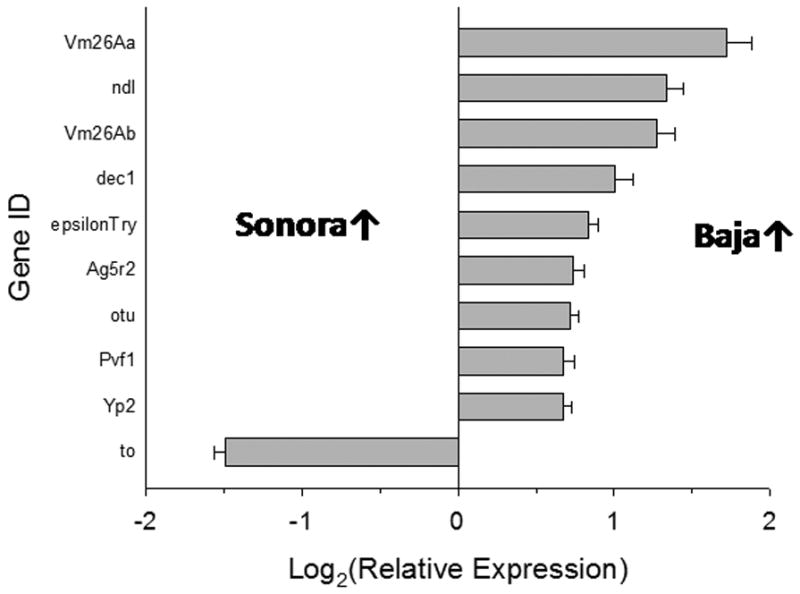
Relative expression of genes associated with reproduction in flies from the mainland and Baja California. These genes were identified by DAVID from a list of genes with at least a 50% difference in expression between these geographic regions.
To investigate broader transcriptome differences between Baja California and mainland flies, we relaxed the 1.5-fold change cutoff restriction and re-analyzed the larger data set containing all 1963 genes with orthologs in D. melanogaster and FDR P < 0.01. Four of the six most enriched GO term clusters (enrichment scores 1.97-4.18) included categories associated with mitochondria and oxidative phosphorylation (Supplementary Table S8). Inspection of pair-wise differences for individual genes indicated that mitochondrial protein genes, especially those related to electron transport and oxidative phosphorylation, were more highly expressed in flies from Baja California. The most highly overrepresented GO term cluster included 129 genes, 86 of which were expressed at higher levels in flies from Baja California (2-tailed sign test, P < 0.002). Figure 6 shows the relative expression of 43 differentially expressed genes in the category GO:0006119; oxidative phosphorylation. Forty-one of these were expressed at higher levels in flies from Baja California (2-tailed sign test, P < 0.0001). Thus, Baja California females exhibited higher expression of transcripts associated with mitochondrial energy production under low humidity conditions.
Figure 6.
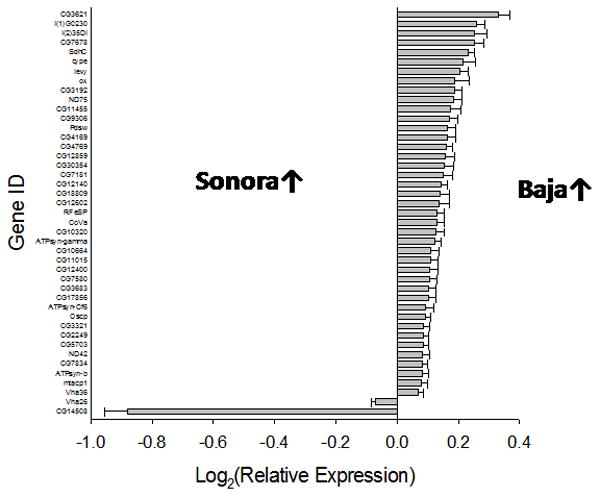
Relative expression of genes associated with oxidative phosphorylation (GO:0006119) in flies from the mainland and Baja California.
Additional overrepresented GO clusters were related to aging (23 genes) and immune function (42 genes). Because of their potential relevance to life history and survival differences between mainland and Baja California populations, we investigated these for potential geographic patterns. No consistent geographic differences were detected for aging-related genes (2-tailed sign test, P > 0.6). Immune function genes were not differentially expressed between regions overall (P > 0.8), but there was a tendency for higher expression of innate immune response genes (GO:0045087) in mainland flies, and humoral immune response genes (GO:0006959) in flies from Baja California. Because of overlap between these categories, however, overall expression did not differ significantly between regions (2-tailed sign tests, P > 0.3).
5262 genes differed in expression among populations (nested within geographic region). This group included all of the 3098 genes differentially expressed between mainland and Baja California regions, and >85% of significant pair-wise differences were between one of the mainland populations and one of the Baja Californian populations. Because >3000 of these genes had orthologs in D. melanogaster, we used DAVID to identify overrepresented GO terms in the subset of genes whose expression differed by at least 50%. Our DAVID analysis revealed four clusters with enrichment scores greater than 2 (P < 0.01). The most overrepresented cluster included GO terms related to female reproduction.
Host plant and population-by-host plant interactions
As noted above, only 18 genes (15 with orthologs in D. melanogaster) differed in expression at FDR < 0.01. Interestingly, 16 of these were expressed at higher levels in flies reared on organ pipe cactus than on agria. Clustering analysis using DAVID found one poorly enriched cluster (enrichment score = 0.54) consisting of genes annotated as binding cations. When we further relaxed our criteria for inclusion to allow FDR < 0.05, 106 genes (62 with orthologs in D. melanogaster) were differentially expressed. Analysis using DAVID uncovered a single overrepresented cluster (enrichment score = 2.08), including five genes annotated as having anion transport activity.
Host plant and population have been shown to interact in affecting traits such as CHC and reproductive isolation between populations (Etges & Ahrens 2001; Stennett & Etges 1997), so we were particularly interested in identifying genes that might contribute to these effects. 835 genes with significant Population × Cactus interactions differed in expression by >50% for at least one pair-wise comparison. Thus, there were significant differences among populations (nested within region) in gene expression when reared on agria or organ pipe cactus. This set included all 247 genes with significant Region × Cactus interactions. Inspection of functional clusters revealed that these were similar to the categories revealed by analysis of regional or population effects alone. Only one Region × Cactus cluster was identified at P < 0.05 (enrichment score > 1.3), including GO terms associated with female reproduction (Supplementary Table S9). For Population × Cactus interactions, eight clusters were identified (Supplementary Table S10), including GO terms related to female reproduction, immune function and lipid metabolism.
Over 4000 genes exhibited significant Region × Desiccation × Host Plant effects, but only 2354 had putative orthologs in D. melanogaster. DAVID analyses of these genes produced 17 clusters of enriched GO terms (Supplementary Table S11). In general, the categories represented overlapped with those returned in main effect and two-way interaction analyses. This was likely because all of the genes included in this analysis also appeared as differentially regulated in region and desiccation analyses or in pair-wise interactions between each other or with host plant.
In another attempt to identify GO categories associated with host plant-by-population interactions, we considered all genes exhibiting statistically significant host-by-region interactions. We then removed all genes exhibiting host or region effects, to leave a list of genes specific to Host Plant × Region interactions, and analyzed the remaining list using DAVID. We performed a similar analysis for Population × Host Plant interactions. The goal was to identify GO terms specifically associated with interactions between host plants and geographic location. In both cases, no overrepresented GO term clusters were identified.
Discussion
Desert insects, like D. mojavensis, are often exposed to extremes in heat, cold and desiccation, and are especially vulnerable to desiccation because of their large surface area to volume ratio. Among drosophilids, D. mojavensis is the most desiccation resistant species studied to date (Gibbs & Matzkin 2001; Matzkin & Markow 2009; Kellermann et al. 2012). Desiccation exposure in D. mojavensis resulted in manifold effects on adult mortality, directional increases and decreases in CHC quantities, and region, population, host plant, and desiccation dependent variation in gene expression. The relatively large number of observed differentially expressed genes caused by treatment interactions vs. main effects (Table 6), made possible by a completely replicated experimental design, suggests that population-level understanding of genomic responses to relevant environmental stressors like desiccation must be evaluated across geographical and local ecological scales.
Effects of desiccation
An important aspect of survival during desiccation stress is water conservation, and two highly enriched clusters of up-regulated genes included chitin metabolism and cuticle constituents (Table 7). Similar results have been obtained in desiccation-stressed mosquitoes (Wang et al. 2011). The cuticular constituent cluster included over 30 genes with orthologs in D. melanogaster coding for known cuticular proteins. This suggests that D. mojavensis can improve the water retaining properties of the cuticle during desiccation stress by increasing production of cuticular proteins (Gibbs & Rajpurohit 2010). It is important to note that this occurs even as overall protein synthesis is being reduced, as several highly enriched clusters of down-regulated genes involved transcription (Table 7). Our data indicate that D. mojavensis generally decreases protein synthesis and overall biosynthesis when desiccated, but increases the relative expression of genes involved in processes that protect against water stress.
Cuticular hydrocarbons (CHCs) provide a hydrophobic layer that is the main barrier to water loss (Gibbs 2002). We found that total CHC content was not affected by desiccation stress, but CHC composition did change. Gleason et al. (2009) identified 30 potential elongases and desaturases thought to be involved in CHC biosynthesis in D. melanogaster, eighteen of which have orthologs in D. mojavensis. Ten of these were differentially expressed as a result of desiccation, with six consistently up-regulated from 0 to 18 hr [Cyt-b5-r, desat2, CG2781 (fatty acid elongase), CG9743 (stearoyl-CoA 9-desaturase), CG5278, and CG33110 (both elongases), and two consistently down-regulated, CG17928 (Δ5 desaturase) and CG5326 (elongase)]. A particularly interesting candidate gene was desat2 (FBtr0161209), whose expression increased 65% in the first 9 hr, then declined by 28% at 18 hr. As there were significant shifts in groups of CHCs with increasing desiccation exposure (Supplementary Table S3, Table 4), particularly significant increases in 12 CHCs comprising monoenes and dienes from C31 to C40, changes in desaturase and elongase expression may be responsible. We note that these changes in CHC amounts and profiles occurred rapidly, suggesting that CHC pools are dynamic and respond to environmental conditions. Increases in expression of genes associated with fatty acid biosynthesis and desaturation during the first 9 hours of desiccation (Supplementary Table S6) support this interpretation.
Our data also suggest an important behavioral component to desiccation responses. Highly enriched upregulated gene clusters identified by STEM analysis include sensory perception (chemical and visual), neuropeptides, and ion transporters that may act in signal processing by the nervous system. When D. mojavensis is first exposed to low humidity (in the absence of chemical or visual cues), it becomes immobile for ∼12 hr, then becomes active (Gibbs et al. 2003a). This behavior may be very different if flies are provided with a potential water source during desiccation stress. Increases in ion transporter expression (Table S4) could also reflect increased water retention by the Malpighian tubules and cells throughout the body. Conversely, reduced expression of tracheal morphogenesis genes (Table S5) may help to reduce respiratory water loss by reducing turnover and remodeling of the tracheal system. Our data are consistent with findings in desiccation-selected D. melanogaster, in which genes associated with respiratory system development were highly overrepresented in single nucleotide polymorphisms under selection (Telonis-Scott et al. 2011).
Given the behavioral changes described above, it is surprising that the GO cluster with the greatest overrepresentation during short-term desiccation was enriched in terms related to mitochondrial respiration (Table S6), suggesting an increase in metabolism. Previous work suggested that D. mojavensis primarily uses carbohydrates when desiccated, but also metabolizes proteins (Marron et al. 2003). Lipids are not metabolized to a significant extent, although we did observe increased expression of lipid biosynthetic enzymes in the first 9 hr, followed by increased expression of triacylglycerol lipases between 9 and 18 hr. A potential explanation for these observations is that flies utilize glycolysis to metabolize carbohydrates during early desiccation stress and release bound water of hydration, while synthesizing fatty acids from the resulting acetyl-CoA, some of which may be used in CHC turnover (Figures 1, 2). As desiccation stress progresses, flies may switch fuels to metabolize lipids. Lipid metabolism is obligately aerobic, so upregulation of mitochondrial genes during the early stages of desiccation may serve to allow later fatty acid oxidation.
Numerous genes involved in amino acid transport and metabolism were expressed at higher levels during desiccation stress (P < 0.01; Supplementary Table S4). In addition, genes involved in pyridoxal phosphate (vitamin B6) increased in expression from 9 to 18 hr of desiccation. Pyridoxal phosphate is an essential cofactor for transaminases that interconvert amino acids and related compounds, so this finding supports an overall increase in amino acid metabolism. Inspection of the up-regulated genes directly associated with amino acid metabolism suggests that catabolism of amino acids increases during desiccation. Amino acid catabolism generates small carbohydrates such as oxaloacetate and fumarate, which can be metabolized by the Krebs cycle to generate ATP. We also note that several amino acids are used as compatible osmolytes to protect cellular function during water stress in many organisms (Yancey, 2005). Changes in amino acid metabolism and transport are consistent with the idea that D. mojavensis uses some amino acids to protect its tissues from dehydration.
Expression patterns of genes related to reproduction showed several interesting features. During both early and late desiccation stress, GO categories related to reproduction were significantly overrepresented among down-regulated genes. An additional 19 genes exhibited complicated expression patterns. Nine were expressed at higher levels after 9 hr, followed by significantly lower expression after 18 hr. One of these was phm, two encoded vitelline membrane proteins, and 6 were unannotated. Ten genes decreased in expression, then increased, including 4 chorion structural proteins. These results suggest that desiccation causes dynamic changes in ovarian function. Increasing egg production seems counterintuitive during a period of resource stress, but late increases in egg structural components suggest that water-stressed female D. mojavensis may try to lay eggs while they still can.
We compared our desiccation-responsive genes to a database of 263 potential desiccation-related genes (in D. melanogaster) available from the Center for Environmental and Stress and Adaptation Research (CESAR; cesar.org.au). 160 of these had putative orthologs in D. mojavensis, but only eight of these (including desat2) were differentially expressed in response to desiccation in our study. Our low success rate may reflect the fact that the majority of the genes in this database were identified in desiccation-selected populations, rather than flies that been directly exposed to desiccation (Sørensen et al. 2007). Thus, laboratory selection may have acted primarily on the constitutive expression of genes that do not respond directly to desiccation. Other genes identified as differentially expressed in D. melanogaster either do not have a putative ortholog in D. mojavensis (Smp-30; Sinclair et al. 2007) or did not respond to desiccation in our study (desiccate; Kawano et al. 2010; FBtr0163396 in D. mojavensis).
Matzkin and Markow (2009) concluded that desiccation stress in D. mojavensis was associated with a decrease in metabolic rates, based largely on changes in expression of several metabolic enzymes. For comparison, we specifically investigated four central metabolism genes these authors identified as being differentially expressed during desiccation stress: Adh-2, GAPDH, Tal and PEPCK (Matzkin & Markow 2009). Our results confirmed a decrease in Adh-2 expression of ∼30%, but desiccation had no effect on expression of GAPDH or Tal in our experiments. Phosphoenolpyruvate carboxykinase (PEPCK) decreased in expression by ∼30% from 0 to 9 hr, then doubled from 9 to 18 hr. There are several reasons why our results may differ from previous work. We exposed our populations to a maximum of 18 hours of desiccation stress, while Matzkin and Markow (2009) used a 40 hr desiccation treatment. Their flies suffered significant mortality, while ours did not. Flies in our experiments were, however, significantly stressed, as preliminary experiments revealed that many flies were dead by 24 hours, causing us to alter our experimental design to exposure times of 9 and 18 hr (see Methods). These differences in desiccation resistance could reflect geographical differences in source populations (Matzkin et al. 2007) or rearing conditions. We observed significant increases in desiccation-related adult mortality when flies were reared on natural host plants vs. lab food (Figure 1, Table 2), whereas Matzkin and Markow (2009) reared their flies on banana-Opuntia medium only. Preadult rearing substrates, cactus vs. lab food, also influence adult desiccation responses, in addition to other adult phenotypes such as mating behavior, CHCs, and body size (Etges 1990, 1992; Etges & Ahrens 2001).
Population and host plant effects
Populations of D. mojavensis from Baja California and the mainland differed in expression of numerous genes related to reproduction, particularly chorion structural proteins. These and additional related genes also exhibited significant region-by-host and population-by-host interactions. These results are consistent with previous studies demonstrating inter- and intrapopulation variation in fecundity (Etges & Heed 1992; Etges & Klassen 1989) and host-mediated reproductive isolation among populations (Etges and Ahrens 2001). We also observed differential expression of reproduction genes during desiccation stress (see above).
Expression of only 18 genes differed between host plant treatments, and none of the differences exceeded 25%. Our results contrast sharply with those obtained for third instar larvae, in which >500 genes were found to be differentially expressed on agria and organ pipe (Matzkin et al., 2006; Matzkin, 2012). This may not be surprising, because we assayed transcription in adult females that had been reared on fermenting cactus through eclosion and then matured on lab food. Although 15 of the differentially expressed genes we detected had putative orthologs in D. melanogaster, no experimental evidence was available relating to the functions of any of them. Electronic annotations indicated two potential odorant binding proteins, which could be involved in host plant recognition.
Reproductive isolation between mainland and peninsular populations is affected by the host cactus on which flies are reared (Etges & Ahrens 2001). We therefore examined genes with significant region-by-host and population-by-host interactions to try to identify candidates that might be involved. Genes involved with reproduction were overrepresented, as were heme binding proteins. A total of 19 differentially expressed genes included 7 cytochrome P450 genes, some of which may be involved in detoxifying host secondary compounds. One of the P450 genes was phantom, whose product catalyzes an early step in ecdysone synthesis (Warren et al. 2004). Ecdysteroids act to regulate oogenesis in adult females, so phm may have a role in host-mediated differences in fecundity. In general, however, multiple approaches did not suggest any transcriptional component to host plant effects on flies from different populations. Post-transcriptional processes (e.g. post-translational modifications of gene products) may be more important.
A critical limitation of this study (and any functional ‘omic research) is the poor annotation of most genomes. Whether RNA sequencing or microarrays, the latter still accurate and useful tools (Malone & Oliver 2011), are used to assess variation in transcription, lack of functional, lineage-specific gene functional information will constrain interpretation of results. Approximately 75 of the 14,528 predicted genes in D. mojavensis have formal annotations entered into FlyBase, and these are primarily accessory gland proteins and odorant binding proteins. We were therefore forced to rely on the annotation of putative orthologs from D. melanogaster. Over one third (5414) of the predicted genes in D. mojavensis did not have a clear ortholog and so were excluded from our analysis. Of those that did, frequently the functional annotation for D. melanogaster was based on sequence similarity to a gene of known function, rather than direct experimental evidence. For historical reasons, Drosophila geneticists have concentrated on developmentally-regulated genes that yield a morphological phenotype (or death) when disrupted. The annotated fraction of the transcriptome is therefore highly biased towards genes expressed during pre-adult stages. Our findings that many such genes are affected by desiccation in adult flies clearly demonstrate that these genes are active in adults, but their adult functions are largely unknown. Genome annotation remains a very weak link in our understanding of how gene expression is integrated to affect organismal physiology.
Supplementary Material
Supplemental Figure S1. Age at death of lab food-reared adult D. mojavensis, sexes pooled, from four populations in control (ambient humidity) or zero humidity conditions. Different letters above the error bars indicate significant least square mean differences between groups (P < 0.05).
Supplemental Figure S2. Age at death of lab food-reared adult D. mojavensis from four populations that were of different ages when placed in control (ambient humidity) or zero humidity conditions to start the experiment. Different letters above the error bars indicate significant least square mean differences between groups (P < 0.05).
Acknowledgments
We thank A. J. Marlon and G. Almeida for excellent technical assistance and C. Ross for statistical help. Funding was provided by National Science Foundation grant EF-0723930 to AGG and WJE. The UNLV Genomics Core Facility was supported by grants from the National Center for Research Resources (5P20RR016464-11) and the National Institute of General Medical Sciences (8P20GM103440-11).
Footnotes
Data Accessibility.: Microarray data: Gene Expression Omnibus accession GSE43220.
Body_mass_data: doi:10.5061/dryad.rp187
CHC data: doi:10.5061/dryad.rp187
Mortality Data: doi:10.5061/dryad.rp187
Viability data: doi:10.5061/dryad.rp187
Contributor Information
Subhash Rajpurohit, Email: rsubhash@sas.upenn.edu.
Cássia C. Oliveira, Email: cassia.oliveira@lyon.edu.
William J. Etges, Email: wetges@uark.edu.
Allen G. Gibbs, Email: allen.gibbs@unlv.edu.
Literature Cited
- Alcorn SM, Orum TV, Steigerwalt AG, et al. Taxonomy and pathogenicity of Erwinia cacticida sp. nov. Int J Syst Bacteriol. 1991;41:197–212. doi: 10.1099/00207713-41-2-197. [DOI] [PubMed] [Google Scholar]
- Barker JSF, Starmer WT. Ecological Genetics and Evolution: The Cactus-Yeast-Drosophila Model System. Academic Press; Sydney: 1982. [Google Scholar]
- Barker JSF, Starmer WT, MacIntyre RJ. Ecological and Evolutionary Genetics of Drosophila. Plenum Press; New York: 1990. [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society B. 1995;57:289–300. [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on bias and variance. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Brazner JC, Etges WJ. Pre-mating isolation is determined by larval rearing substrates in cactophilic Drosophila mojavensis. II. Effects of larval substrates on time to copulation, mate choice, and mating propensity. Evolutionary Ecology. 1993;7:605–624. [Google Scholar]
- Drosophila 12 Genomes Consortium. Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- Ernst J, Bar-Joseph Z. STEM: a tool for the analysis of short time series gene expression data. BMC Bioinformatics. 2006;7:191. doi: 10.1186/1471-2105-7-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etges WJ. Divergence in cactophilic Drosophila: The evolutionary significance of adult ethanol metabolism. Evolution. 1989a;43:1316–1319. doi: 10.1111/j.1558-5646.1989.tb02579.x. [DOI] [PubMed] [Google Scholar]
- Etges WJ. Evolution of developmental homeostasis in Drosophila mojavensis. Evolutionary Ecology. 1989b;3:189–201. [Google Scholar]
- Etges WJ. Direction of life history evolution in Drosophila mojavensis. In: Barker JSF, Starmer WT, MacIntyre RJ, editors. Ecological and Evolutionary Genetics of Drosophila. Plenum; New York: 1990. pp. 37–56. [Google Scholar]
- Etges WJ. Premating isolation is determined by larval rearing substrates in cactophilic Drosophila mojavensis. Evolution. 1992;46:1945–1950. doi: 10.1111/j.1558-5646.1992.tb01180.x. [DOI] [PubMed] [Google Scholar]
- Etges WJ. Genetics of host-cactus response and life-history evolution among ancestral and derived populations of cactophilic Drosophila mojavensis. Evolution. 1993;47:750–767. doi: 10.1111/j.1558-5646.1993.tb01231.x. [DOI] [PubMed] [Google Scholar]
- Etges WJ. Premating isolation is determined by larval rearing substrates in cactophilic Drosophila mojavensis. IV. Correlated responses in behavioral isolation to artificial selection on a life history trait. The American Naturalist. 1998;152:129–144. doi: 10.1086/286154. [DOI] [PubMed] [Google Scholar]
- Etges WJ. Divergence in mate choice systems: does evolution play by rules? Genetica. 2002;116:151–166. [PubMed] [Google Scholar]
- Etges WJ, Ahrens MA. Premating isolation is determined by larval rearing substrates in cactophilic Drosophila mojavensis. V. Deep geographic variation in epicuticular hydrocarbons among isolated populations. The American Naturalist. 2001;158:585–598. doi: 10.1086/323587. [DOI] [PubMed] [Google Scholar]
- Etges WJ, de Oliveira CC, Gragg E, et al. Genetics of incipient speciation in Drosophila mojavensis. I. Male courtship song, mating success, and genotype × environment interactions. Evolution. 2007;61:1106–1119. doi: 10.1111/j.1558-5646.2007.00104.x. [DOI] [PubMed] [Google Scholar]
- Etges WJ, Heed WB. Sensitivity to larval density in populations of Drosophila mojavensis: Influences of host plant variation on components of fitness. Oecologia. 1987;71:375–381. doi: 10.1007/BF00378710. [DOI] [PubMed] [Google Scholar]
- Etges WJ, Heed WB. Remating effects on the genetic structure of female life histories in populations of Drosophila mojavensis. Heredity. 1992;68:515–528. doi: 10.1038/hdy.1992.74. [DOI] [PubMed] [Google Scholar]
- Etges WJ, Jackson LL. Premating isolation is determined by larval rearing substrates in cactophilic Drosophila mojavensis. VI. Epicuticular hydrocarbon variation in Drosophila mojavensis cluster species. Journal of Chemical Ecology. 2001;27:2125–2149. doi: 10.1023/a:1012203222876. [DOI] [PubMed] [Google Scholar]
- Etges WJ, Johnson WR, Duncan GA, Huckins G, Heed WB. Ecological genetics of cactophilic Drosophila. In: Robichaux R, editor. Ecology of Sonoran Desert Plants and Plant Communities. University of Arizona Press; Tucson: 1999. pp. 164–214. [Google Scholar]
- Etges WJ, Klassen CS. Influences of atmospheric ethanol on adult Drosophila mojavensis: Altered metabolic rates and increases in fitness among populations. Physiological Zoology. 1989;62:170–193. [Google Scholar]
- Etges WJ, Over KF, de Oliveira CC, Ritchie MG. Inheritance of courtship song variation among geographically isolated populations of Drosophila mojavensis. Animal Behaviour. 2006;71:1205–1214. [Google Scholar]
- Fellows DP, Heed WB. Factors affecting host plant selection in desert-adapted cactiphilic Drosophila. Ecology. 1972;53:850–858. [Google Scholar]
- Fogleman JC, Danielson PB. Chemical interactions in the cactus-microorganism-Drosophila model system of the Sonoran Desert. American Zoologist. 2001;41:877–889. [Google Scholar]
- Fogleman JC, Duperret SM, Kircher HW. The role of phytosterols in host plant utilization by cactophilic Drosophila. Lipids. 1986;21:92–96. doi: 10.1007/BF02534309. [DOI] [PubMed] [Google Scholar]
- Fogleman JC, Heed WB. Columnar cacti and desert Drosophila: the chemistry of host plant specificity. In: Schmidt JO, editor. Special Biotic Relationships of the Southwest. Univ. New Mexico Press; Albuquerque: 1989. pp. 1–24. [Google Scholar]
- Garrick RC, Nason JD, Meadows CA, Dyer RJ. Not just vicariance: phylogeography of a Sonoran Desert euphorb indicates a major role of range expansion along the Baja peninsula. Molecular Ecology. 2009;18:1916–1931. doi: 10.1111/j.1365-294X.2009.04148.x. [DOI] [PubMed] [Google Scholar]
- Gastil RG, Phillips RP, Allison EC. Reconnaissance geology of the state of Baja California. Geological Society of America Memoir. 1975;140:1–170. [Google Scholar]
- Gibbs AG. Water balance in desert Drosophila: lessons from non-charismatic microfauna. Comparative Biochemistry and Physiology. 2002;133:781–789. doi: 10.1016/s1095-6433(02)00208-8. [DOI] [PubMed] [Google Scholar]
- Gibbs AG, Fukuzato F, Matzkin LM. Evolution of water conservation mechanisms in desert Drosophila. Journal of Experimental Biology. 2003a;206:1183–1192. doi: 10.1242/jeb.00233. [DOI] [PubMed] [Google Scholar]
- Gibbs AG, Louie AK, Ayala JA. Effects of temperature on cuticular lipids and water balance in a desert Drosophila: Is thermal acclimation beneficial? Journal of Experimental Biology. 1998;201:71–80. doi: 10.1242/jeb.201.1.71. [DOI] [PubMed] [Google Scholar]
- Gibbs AG, Matzkin LM. Evolution of water balance in the genus Drosophila. Journal of Experimental Biology. 2001;204:2331–2338. doi: 10.1242/jeb.204.13.2331. [DOI] [PubMed] [Google Scholar]
- Gibbs AG, Perkins MC, Markow TA. No place to hide: microclimates of Sonoran Desert Drosophila. Journal of Thermal Biology. 2003b;28:353–362. [Google Scholar]
- Gibbs AG, Rajpurohit S. Water-proofing properties of cuticular lipids. In: Blomquist GJ, Bagneres AG, editors. Insect Lipids; Biology, Biochemistry and Chemical Biology. Cambridge University Press; 2010. pp. 100–120. [Google Scholar]
- Gleason JM, James RA, Wicker-Thomas C, Ritchie MG. Identification of quantitative trait loci function through analysis of multiple cuticular hydrocarbons differing between Drosophila simulans and Drosophila sechellia females. Heredity. 2009;103:416–424. doi: 10.1038/hdy.2009.79. [DOI] [PubMed] [Google Scholar]
- Heed WB. Ecology and genetics of Sonoran Desert Drosophila. In: Brussard PF, editor. Ecological Genetics: The Interface. Springer-Verlag; New York: 1978. pp. 109–126. [Google Scholar]
- Heed WB. The origin of Drosophila in the Sonoran Desert. In: Barker JSF, Starmer WT, editors. Ecological Genetics and Evolution: The Cactus-Yeast-Drosophila Model System. Academic Press; Sydney: 1982. pp. 65–80. [Google Scholar]
- Heed WB, Mangan RL. Community ecology of the Sonoran Desert Drosophila. In: Ashburner M, Carson HL, Thompson JN, editors. The Genetics and Biology of Drosophila. Academic Press; New York: 1986. pp. 311–345. [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nature Protocols. 2009;4(1):44–45. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Johnson RA, Gibbs AG. Effects of mating stage on water balance, metabolism and cuticular hydrocarbons of the desert harvester ant, Pogonomyrmex barbatus. Journal of Insect Physiology. 2004;50:943–953. doi: 10.1016/j.jinsphys.2004.07.006. [DOI] [PubMed] [Google Scholar]
- Karl TR, Trenberth KE. Modern global climate change. Science. 2003;302:1719–1723. doi: 10.1126/science.1090228. [DOI] [PubMed] [Google Scholar]
- Kawano T, Shimoda M, Matsumoto H, et al. Identification of a gene, Desiccate, contributing to desiccation resistance in Drosophila melanogaster. Journal of Biological Chemistry. 2010;285:38889–38897. doi: 10.1074/jbc.M110.168864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellermann V, Loeschcke V, Hoffmann AA, Kristensen TN, Flojgaard C, David JR, Svenning JC, Overgaard J. Phylogenetic constraints in key functional traits behind species' climate niches: patterns of desiccation and cold resistance across 95 Drosophila species. Evolution. 2012;66:3377–3389. doi: 10.1111/j.1558-5646.2012.01685.x. [DOI] [PubMed] [Google Scholar]
- Knowles LL, Markow TA. Sexually antagonistic coevolution of a postmating-prezygotic reproductive character in desert Drosophila. Proceedings of the National Academy of Sciences (USA) 2001;98:8692–8696. doi: 10.1073/pnas.151123998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs RA. A comparison of Hsp70 expression and thermotolerance in adults and larvae of three Drosophila species. Cell Stress & Chaperones. 1999;4:243–249. doi: 10.1379/1466-1268(1999)004<0243:acohea>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone J, Oliver B. Microarrays, deep sequencing and the true measure of the transcriptome. BMC Biology. 2011;9:34. doi: 10.1186/1741-7007-9-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markow TA. Sexual isolation among populations of Drosophila mojavensis. Evolution. 1991;45:1525–1529. doi: 10.1111/j.1558-5646.1991.tb02656.x. [DOI] [PubMed] [Google Scholar]
- Marron MT, Markow TA, Kain KJ, Gibbs AG. Effects of starvation and desiccation on energy metabolism in desert and mesic Drosophila. Journal of Insect Physiology. 2003;49:261–270. doi: 10.1016/s0022-1910(02)00287-1. [DOI] [PubMed] [Google Scholar]
- Matzkin LM. Population transcriptomics of cactus host shifts in Drosophila mojavensis. Molecular Ecology. 2012;21:2428–2439. doi: 10.1111/j.1365-294X.2012.05549.x. [DOI] [PubMed] [Google Scholar]
- Matzkin LM, Markow TA. Transcriptional regulation of metabolism associated with the increased desiccation resistance of the cactophilic Drosophila mojavensis. Genetics. 2009;182:1279–1288. doi: 10.1534/genetics.109.104927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzkin LM, Watts TD, Bitler BG, Machado CA, Markow TA. Functional genomics of cactus host shifts in Drosophila mojavensis. Molecular Ecology. 2006;15:4635–4643. doi: 10.1111/j.1365-294X.2006.03102.x. [DOI] [PubMed] [Google Scholar]
- Matzkin LM, Watts TD, Markow TA. Desiccation resistance in four Drosophila species: Sex and population effects. Fly. 2007;1:268–273. doi: 10.4161/fly.5293. [DOI] [PubMed] [Google Scholar]
- Okin GS, Gillette DA, Herrick JE. Multi-scale controls on and consequences of Aeolian processes in landscape change in arid and semi-arid environments. Journal of Arid Environments. 2006;65:253–275. [Google Scholar]
- Oliveira DCSG, Almeida FC, O'Grady PM, et al. Monophyly, divergence times, and evolution of host plant use inferred from a revised phylogeny of the Drosophila repleta species group. Molecular Phylogenetics and Evolution. 2012;64:533–544. doi: 10.1016/j.ympev.2012.05.012. [DOI] [PubMed] [Google Scholar]
- Reed LK, Nyboer M, Markow TA. Evolutionary relationships of Drosophila mojavensis geographic host races and their sister species Drosophila arizonae. Molecular Ecology. 2007;16:1007–1022. doi: 10.1111/j.1365-294X.2006.02941.x. [DOI] [PubMed] [Google Scholar]
- Reich M, Liefeld T, Gould J, et al. GenePattern 2.0. Nat Genet. 2006;38:500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- Riddle BR, Hafner DJ, Alexander LF, Jaeger JR. Cryptic vicariance in the historical assembly of a Baja California Peninsular Desert biota. Proceedings of the National Academy of Sciences (USA) 2000;97:14438–14443. doi: 10.1073/pnas.250413397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz A, Heed WB, Wasserman M. Evolution of the mojavensis cluster of cactophilic Drosophila with descriptions of two new species. Journal of Heredity. 1990;81:30–42. doi: 10.1093/oxfordjournals.jhered.a110922. [DOI] [PubMed] [Google Scholar]
- SAS-Institute. SAS/STAT 9.1.2. SAS Institute, Inc.; Cary, NC: 2004. [Google Scholar]
- Sinclair BJ, Gibbs AG, Roberts SP. Gene transcription during exposure to, and recovery from, cold and desiccation stress in Drosophila melanogaster. Insect Molecular Biology. 2007;16:435–443. doi: 10.1111/j.1365-2583.2007.00739.x. [DOI] [PubMed] [Google Scholar]
- Smith G, Lohse K, Etges WJ, Ritchie MG. Model-based comparisons of phylogeographic scenarios resolve the intraspecific divergence of cactophilic Drosophila mojavensis. Molecular Ecology. 2012;21:3293–3307. doi: 10.1111/j.1365-294X.2012.05604.x. [DOI] [PubMed] [Google Scholar]
- Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen J, MM N, V L. Gene expression profile analysis of Drosophila melanogaster selected for resistance to environmental stressors. Journal of Evolutionary Biology. 2007;20:1624–1636. doi: 10.1111/j.1420-9101.2007.01326.x. [DOI] [PubMed] [Google Scholar]
- Starmer WT. Associations and interactions among yeasts. In: Barker JSF, Starmer WT, editors. Ecological Genetics and Evolution The Cactus-Yeast-Drosophila Model System. Academic Press; Sydney: 1982. pp. 159–174. [Google Scholar]
- Starmer WT, Heed WB, Rockwood-Sluss ES. Extension of longevity in Drosophila mojavensis by environmental ethanol: Differences between subraces. Proceedings of the National Academy of Sciences (USA) 1977;74:387–391. doi: 10.1073/pnas.74.1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stennett MD, Etges WJ. Pre-mating isolation is determined by larval rearing substrates in cactophilic Drosophila mojavensis. III. Epicuticular hydrocarbon variation is determined by use of different host plants in Drosophila mojavensis and Drosophila arizonae. Journal of Chemical Ecology. 1997;23:2803–2824. [Google Scholar]
- Stratman R, Markow TA. Resistance to thermal stress in desert Drosophila. Functional Ecology. 1998;12:965–970. [Google Scholar]
- Telonis-Scott M, Gane M, DeGaris S, Sgro CM, Hoffmann AA. High resolution mapping of candidate alleles for desiccation resistance in Drosophila melanogaster under selection. Molecular Biology and Evolution. 2012;29:1335–1351. doi: 10.1093/molbev/msr294. [DOI] [PubMed] [Google Scholar]
- Toolson EC, Markow TA, Jackson LL, Howard RW. Epicuticular hydrocarbon composition of wild and laboratory-reared Drosophila mojavensis Patterson and Crow (Diptera: Drosophilidae) Annals of the Entomological Society of America. 1990;83:1165–1176. [Google Scholar]
- Wang MH, Marinotti O, Vardo-Zalik A, Boparai R, Yan G. Genome-wide transcriptional analysis of genes associated with acute desiccation stress in Anopheles gambiae. PLoS ONE. 2011;6:e26011. doi: 10.1371/journal.pone.0026011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren JT, Petryk A, Marqués G, et al. Phantom encodes the 25-hydroxylase of Drosophila melanogaster and Bombyx mori: a P450 enzyme critical in ecdysone biosynthesis. Insect Biochemistry and Molecular Biology. 2004;34:991–1010. doi: 10.1016/j.ibmb.2004.06.009. [DOI] [PubMed] [Google Scholar]
- Wasserman M. Cytological evolution of the Drosophila repleta species group. In: Krimbas CB, Powell JR, editors. Drosophila Inversion Polymorphism. CRC Press; Boca Raton: 1992. pp. 455–552. [Google Scholar]
- Yancey PH. Organic osmolytes as compatible, metabolic and counteracting cytoprotectants in high osmolarity and other stresses. Journal of Experimental Biology. 2005;208:2819–2830. doi: 10.1242/jeb.01730. [DOI] [PubMed] [Google Scholar]
- Yang X, Li J, Lee Y, Lussier YA. GO-Module: functional synthesis and improved interpretation of Gene Ontology patterns. Bioinformatics. 2011;27:1444–1446. doi: 10.1093/bioinformatics/btr142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yew JY, Dreisewerd K, de Oliveira CC, Etges WJ. Male-specific transfer and fine scale spatial differences of newly identified cuticular hydrocarbons and triacylglycerides in a Drosophila species pair. PLoS ONE. 2011;6:e16898. doi: 10.1371/journal.pone.0016898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zouros E. Genic differentiation associated with the early stages of speciation in the mulleri subgroup of Drosophila. Evolution. 1974;27:601–621. doi: 10.1111/j.1558-5646.1973.tb00709.x. [DOI] [PubMed] [Google Scholar]
- Zouros E, d'Entremont CJ. Sexual isolation among populations of Drosophila mojavensis race B. Drosophila Information Service. 1974;51:112. doi: 10.1111/j.1558-5646.1980.tb04830.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure S1. Age at death of lab food-reared adult D. mojavensis, sexes pooled, from four populations in control (ambient humidity) or zero humidity conditions. Different letters above the error bars indicate significant least square mean differences between groups (P < 0.05).
Supplemental Figure S2. Age at death of lab food-reared adult D. mojavensis from four populations that were of different ages when placed in control (ambient humidity) or zero humidity conditions to start the experiment. Different letters above the error bars indicate significant least square mean differences between groups (P < 0.05).


