Abstract
Regulatory changes rapidly accumulate between species, and interspecific hybrids often misexpress genes. Hybrid misexpression, expression levels outside the range of both parental species, can result from cis- and trans-acting regulatory changes that interact abnormally in hybrids. Thus, misexpressed genes may contribute to hybrid sterility. However, in the context of a whole organism, misexpression may not result directly from cis–trans interactions but rather indirectly from differences between hybrid and parental abundance of cell types. Here we eliminate the confounding effects of cell types by examining gene expression in a sterile interspecific yeast hybrid during meiosis. We investigated gene expression of the yeasts Saccharomyces cerevisiae, S. paradoxus, and their hybrid at multiple meiotic stages. Although the hybrid and parents exhibit similar changes in expression levels across meiosis, the hybrid meiotic program occurs earlier than either parent. The timing change produces a heterochronic pattern of misexpression during midmeiosis. Coincident with the timing of misexpression, we find a transition from predominantly trans-acting to cis-acting expression divergence and an increase in the number of opposing cis–trans changes. However, we find no direct relationship between opposing cis–trans changes and misexpression. Contrary to the notion that cis–trans interactions cause misexpression, a heterochronic shift in the normal meiotic gene expression program produces patterns of misexpression in an yeast hybrid. Our results imply that temporal dynamics of single cell types is important to understanding hybrid misexpression and its relationship to cis–trans interactions.
Keywords: speciation, hybrid, yeast, misexpression
Background
A major goal of evolutionary genetics is to understand the molecular basis of reproductive isolation (RI) between species. The relationship between regulatory evolution and RI is of particular interest as an abundance of gene expression divergence exists between species. Additionally, a variety of interspecific hybrids display extensive misexpression (Ranz et al. 2004; Landry et al. 2005; Haerty and Singh 2006; Moehring et al. 2006; Malone et al. 2007; Rottscheidt and Harr 2007; Renaut et al. 2009; Tirosh et al. 2009; Good et al. 2010; Lu et al. 2010; Llopart 2012), defined as hybrid expression levels outside the range of both parents’ expression levels (Ortíz-Barrientos et al. 2006). Novel interactions between cis- and trans-acting substitutions are thought to cause misexpression, and some direct evidence shows these interactions can contribute to RI at specific points of hybrid development (Bayes and Malik 2009; Maheshwari and Barbash 2012). However, we lack a general understanding of hybrid misexpression over the course of reproductive development.
Genome expression profiles of interspecific hybrids at single time points have revealed a number of suggestive associations between regulatory divergence and postzygotic isolation. For instance, disruption of gene expression in both sterile mice and flies has been observed (Good et al. 2010; Lu et al. 2010; Sackton et al. 2011; Sundararajan and Civetta 2011). Specifically, Drosophila hybrids disproportionately and ectopically misregulate genes that are mainly or solely expressed in males (Michalak and Noor 2003; Ranz et al. 2004). In the context of hybrid male sterility, many misregulated genes are related to spermatogenesis, although misexpression also occurs in other tissues (Graze et al. 2009). However, misexpression can also arise as a consequence of dysgenic phenotypes in the hybrid, such as gonadal atrophy, and so may be a consequence rather than a cause of hybrid sterilty (Ranz et al. 2004; Ortíz-Barrientos et al. 2006). Finally, misexpression may arise due to regulatory divergence independent of hybrid sterility (Ferguson et al. 2013).
Two questions are relevant to understanding misexpression in interspecific hybrids: when does misexpression occur during development and what causes misexpression? In Drosophila, misexpression is more pronounced in adult stages in comparison to larval stages (Moehring et al. 2006). Similarly, misexpression of a small number of genes in fish increases during development (Parker et al. 1985). However, another Drosophila study found more hybrid misexpression in larval and adult stages relative to the pupal stage (Artieri and Singh 2010), providing evidence against a cascading model of misexpression during development.
Studies of allele-specific expression in yeast and flies suggest that misexpression is a consequence of divergence in gene regulation, which may cause aberrant hybrid development. Both additive and nonadditive cis–trans interactions have been reported to cause novel expression in hybrids (Landry et al. 2005; Tirosh et al. 2009; Emerson et al. 2010; McManus et al. 2010; Llopart 2012). Furthermore, antagonistic cis–trans interactions have been associated with hybrid misexpression as an inherited mode of under- or overdominance (Landry et al. 2005; McManus et al. 2010; Schaefke et al. 2013), in which the hybrid expresses genes lower or higher than both parents, respectively. Of potential relevance, more cis- than trans-acting changes are found between species compared to within species. However, the cause of misexpression is hard to identify in mixed cell populations.
Yeast serves as an excellent model to not only understand hybrid misexpression but also relate misexpression to hybrid sterility. As yeasts are single cell organisms, hybrid misexpression is not confounded with the relative abundance of cell types or tissues. Furthermore, the progression of gene expression changes that occur over the course of meiosis has been well characterized for Saccharomyces cerevisiae (Chu et al. 1998; Primig and Esposito 2000). Additionally, multiple mechanisms are known to contribute to postzygotic isolation between S. cerevisiae and its sister species, S. paradoxus.
Two mechanisms are known to contribute to hybrid sterility in yeast. First, the mismatch repair (mmr) pathway recognizes a multitude of mismatches between interspecific homologous chromosomes in the hybrid and effectively prevents nonhomologous chromosomes from crossing over, which leads to aneuploidy and spore death. Deletions of mmr genes restore spore viability from less than 1% to 7–10%, supporting mmr pathway’s role in hybrid sterility (Hunter et al. 1996; Greig et al. 2003). Additionally tetraploid hybrids have almost fully rescued spore viability, which suggests that each chromosome in the tetraploid hybrid has a homologous chromosome with which to rearrange, thus allowing proper chromosome disjunction (Greig et al. 2002). Lastly, there is a linear correlation between intraspecific sequence divergence and spore inviability, suggesting that the mmr pathway may be involved in spore inviability within species of yeast (Liti et al. 2006). A second mechanism that contributes to RI is genetic incompatibilities. A special Dobzhansky–Muller incompatibility between nuclear and mitochondrial genomes contributes to postzygotic isolation in F2 hybrids that are derived from rare viable F1 hybrids (Lee et al. 2008; Chou et al. 2010). Additionally, Xu and He (2011) found that genetic incompatibilities affect sporulation efficiency in yeast F2 hybrids more than F2 hybrid growth.
In this study, we examine gene expression during sporulation of the budding yeasts S. cerevisiae, S. paradoxus, and their sterile hybrid. We find that hybrid gene misregulation is predominantly related to a heterochronic shift, whereby the hybrid meiotic gene expression program proceeds more rapidly than both parents. Coincident with this altered meiotic program, we find that expression divergence between the parental species is initially dominated by trans-acting changes and later dominated by cis-acting changes, between which we find more opposing cis–trans changes. We discuss these results in relation to known mechanisms of RI in yeast.
Results
Hybrid Sporulation Is Similar to S. paradoxus
To characterize sporulation of S. cerevisiae, S. paradoxus, and their hybrid, we monitored the cells’ progression through meiotic stages by 4′,6-diamidino-2-phenylindole (DAPI) staining of nuclei and fluorescent microscopy (see Materials and Methods). The number of nuclei in a cell indicates how many phases of meiosis the cell has completed. Tetranucleated cells have completed both meiosis I and II; binucleated cells have only completed meiosis I; and mononucleated cells are undifferentiated diploids.
We find that the hybrid progresses through meiosis I and II similarly to S. paradoxus but differently than S. cerevisiae (fig. 1 and supplementary fig. S1, Supplementary Material online). The parental mononucleate curves are significantly different than one another (analysis of variance [ANOVA], P = 0.01 and Materials and Methods for details), which indicates that the parents enter meiosis I at different times. However, the hybrid mononucleate curve is not different than either S. cerevisiae or S. paradoxus (ANOVA, P = 0.06 and P = 0.38, respectively), which indicates that the hybrid initially enters meiosis I intermediate of both parents. Compared with S. cerevisiae, S. paradoxus takes longer to complete both meiosis I and II, as measured by the formation of binucleates and tetranucleates, respectively. Binucleate formation is similar between the three strains, with only the hybrid being significantly different from S. cerevisiae (ANOVA, P = 0.04). Although S. paradoxus and the hybrid generate tetranucleates similarly (ANOVA, P = 0.54), S. cerevisiae forms tetranucleates differently than both the S. paradoxus and the hybrid (ANOVA, P = 0.003 and P = 0.009, respectively). Thus, the hybrid finishes meiosis I and II like S. paradoxus. On the basis of the observed progression through meiosis, we conclude that the sporulation program of S. paradoxus is largely dominant in the hybrid, with a slight indication of codominance at the earliest stages of meiosis (e.g., meiosis I).
Fig. 1.
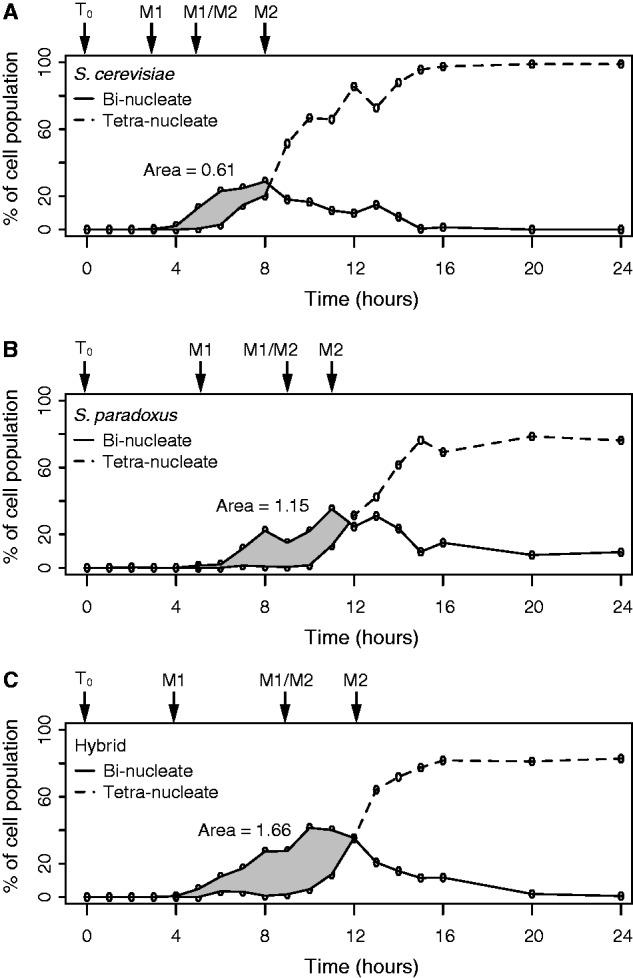
Profiles of meiotic divisions during sporulation. The frequency of binucleates (solid) and tetranucleates (dashed) is shown for S. cerevisiae (A), S. paradoxus (B), and their hybrid (C) over a 24-h time course. Each panel shows the average of three (parents) or four (hybrid) replicates. The associated error bars are removed for clarity but can be seen in supplementary fig. S1, Supplementary Material online. The area between the binucleate and tetranucleate curves is shown in gray and numerically labeled. The sampling of T0, M1, M1/M2, and M2 stages is shown by the arrows above each species’ graph.
The Hybrid Has a Longer Transition between Meiosis I and II Than Its Parents
To further understand differences between the hybrid and its parents, we measured the area between the bi- and tetranucleate curves from the beginning of sporulation to when the two curves intersect. The area between the bi- and tetranucleate curves can measure the speed of progression through meiosis, independent of when meiotic divisions begin or end (Galbraith et al. 1997); a larger area signifies a longer delay between meiosis I and II.
We measured the area between the bi- and tetranucleate curves for each sample and find that the hybrid has a larger area between its curves (fig. 1). The areas for S. cerevisiae and S. paradoxus are significantly different (t-test, P = 0.02), and the area for the hybrid is significantly different than those of both S. cerevisiae and S. paradoxus (t-test, P = 0.002 and P = 0.04, respectively). As the hybrid area is larger than both parents, the hybrid transitions from meiosis I and II over a longer period of time than both parents. To further characterize differences in how the two parents and their hybrid progress through meiosis, we performed an RNA-Seq study of gene expression differences during meiosis.
RNA-seq Profiling During Meiosis
To capture gene expression changes between the hybrid and its parents during meiosis, we defined four developmental stages based on the formation of binucleates and tetranucleates (supplementary fig. S1, Supplementary Material online). The four stages are defined by: T0 as the time at which we placed cells in sporulation media; M1 as an hour before binucleates appear; M1/M2 as an hour before tetranucleates appear; and M2 as an hour before the tetranucleates comprise the majority of the cell population. Thus, the T0, M1, M1/M2, and M2 stages correspond to: 0, 3, 5, and 8 h for S. cerevisiae; 0, 5, 9, and 11 h for S. paradoxus; and 0, 4, 9, and 12 for their hybrid (fig. 1).
We used RNA-Seq to measure gene expression in both parents as well as allele-specific and total gene expression in the hybrid. We obtained a total of 276 × 106 mapped reads with a median of 4.2 × 106 per sample (see Materials and Methods). We simultaneously mapped hybrid reads to both the S. cerevisiae and S. paradoxus genome and found the percentage of mapped reads from the hybrid to be similar to that of both parents. Of the hybrid reads, 49.4% mapped to the S. cerevisiae genome and 50.6% mapped to the S. paradoxus genome, suggesting minimal read mapping bias in the hybrid.
To characterize our developmental stages, we compared the S. cerevisiae expression profile to previously documented changes in gene expression (Chu et al. 1998). We found metabolic gene expression is high at T0 and rapidly decreases after M1; early sporulation gene expression begins to increase at M1; middle sporulation gene expression begins to increase at M1/M2; and late sporulation gene begins to increase at M2 (supplementary fig. S2, Supplementary Material online). Thus, our S. cerevisiae expression data cover the early and middle stages of meiosis and are consistent with previously reported patterns of gene expression during meiosis.
Heterochronic changes in the hybrid
To identify differentially expressed genes across stages and species, we compared two models (see Materials and Methods). Our null model has no explanatory variables and our alternative model has variables for species background (S. cerevisiae, S. paradoxus, and the hybrid), stage (T0, M1, M1/M2, M2), and an interaction between background and stage. Through this comparison, we found 1,083 out of 3,463 expressed genes are differentially expressed (P < 0.001). To refine this list to genes that exhibit an interaction between species’ background and developmental stage, we compared a model with and without this interaction term and found 352 genes (P < 0.05). The expression of these genes is of particular interest since they change over the course of meiosis in a species- or hybrid-specific pattern and cannot be explained by read mapping bias, which is expected to be constant across developmental stages.
To gain a general view of expression differences, we performed a principal coordinate analysis of the 352 genes differentially expressed across stages and species’ background (fig. 2). The first two coordinates explain 42% and 14% of the variation among samples and separate most of the samples from one another. The first coordinate orders the samples according to developmental stages. The second coordinate provides some additional resolution of the time points, particularly T0 compared with M1. At T0, hybrid expression lies in between those of S. cerevisiae and S. paradoxus on the second coordinate. However, at both the M1 and M1/M2 stages, hybrid expression is to the right of both S. cerevisiae and S. paradoxus expression on the first coordinate, indicative of a more advanced phase of the meiotic expression program (fig. 2). To a lesser degree, S. paradoxus expression is also more advanced than that of S. cerevisiae at the M1 and M1/M2 stages. The more advanced phase of S. paradoxus could be explained by our sampling at developmental stages rather than absolute time points, which were 2 (M1) and 4 (M1/M2) h later compared to S. cerevisiae. However, the more advanced phase of the hybrid cannot be explained by our sampling scheme, because both the M1 and M1/M2 stages were sampled at absolute times at or between those of the two parents. At the M2 stage, the expression of the hybrid is similar to both parents on the first coordinate and only slightly different on the second coordinate.
Fig. 2.
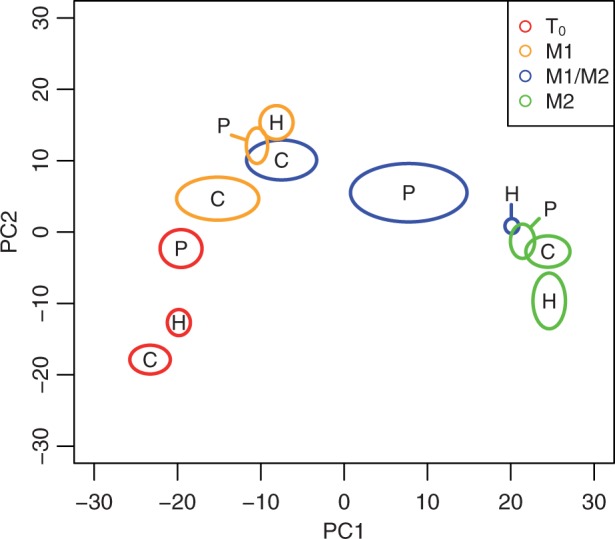
Principal coordinate analysis of 352 differentially expressed genes. Ovals show the 95% confidence interval of S. cerevisiae (C), S. paradoxus (P), and hybrid (H) principal coordinates at each meiotic stage and are centered on the mean values. Meiotic stages are T0 (red), M1 (orange), M1/M2 (blue), and M2 (green). The first and second principal coordinate explain 42% and 13% of the variation among samples, respectively.
For each stage, we identified genes whose expression in the hybrid occurs outside the expression range of the parents (table 1, t-test, P < 0.05). Most of the hybrid genes that are expressed outside the range of the parents occur at stages M1 and M1/M2. At M1, the hybrid expresses 21 genes lower than either parent, 11 of which are ribosomal proteins. Although the expression of these 21 genes declines during meiosis for both parents and the hybrid, hybrid expression begins to decline at M1 while the parents begin to decline at M2 (fig. 3). The genes that are expressed lower in the hybrid at M1/M2 are the same genes expressed lower at M1. At M1/M2, the hybrid expresses 26 genes higher than either parent, 10 of which are involved in middle and late sporulation. Although the expression of these 26 genes increases during meiosis for both parents and the hybrid, hybrid expression increases earlier than parental expression (fig. 3). Thus, genes that are misexpressed in the hybrid compared with both parents exhibit heterochronic changes, similar to changes seen in the composite analysis of all differentially expressed genes (fig. 2).
Table 1.
Number of Hybrid Genes Significantly Different from Both Parents.
| Hyrid expression relative to parenta | T0 | M1 | M1/M2 | M2 |
|---|---|---|---|---|
| Lower | 10 | 21 | 26 | 3 |
| Intermediate | 22 | 4 | 1 | 2 |
| Higher | 1 | 1 | 26 | 3 |
aHybrid expression is significantly different for both parents (t-test P < 0.05).
Fig. 3.
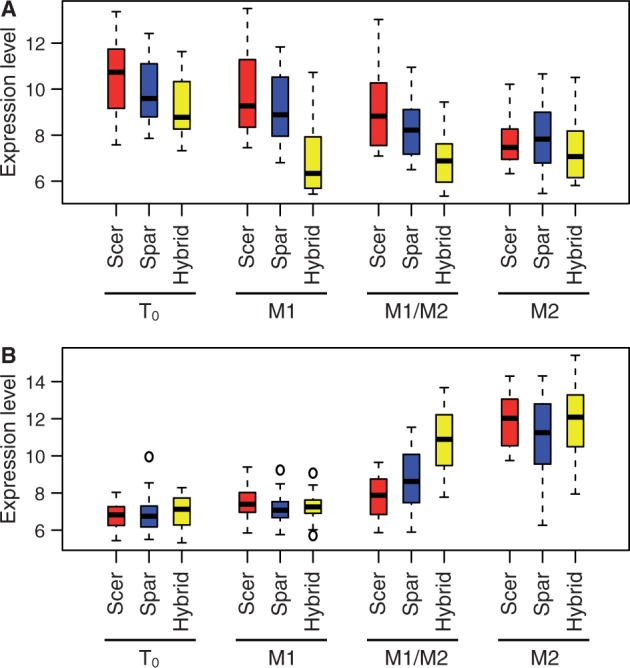
Temporal changes in misexpressed genes during sporulation. Box plots of 21 genes expressed lower in the hybrid than either parent at M1 and M1/M2 but similarly in the hybrid and both parents in M2 (A), and 26 genes expressed higher in the hybrid than in either parent at M1/M2 but similarly in the hybrid and both parents in M2 (B). Boxes indicate the span of the second and third quartiles and dashed lines indicate an estimate of the 95% confidence of the median. Boxes are shown for S. cerevisiae (Scer), S. paradoxus (Spar), and the hybrid at each stage (T0, M1, M1/M2, and M2). Expression levels are the normalized log 2 number of reads.
To further evaluate a difference in timing contributes to differences between the hybrid and the parents, we used a previously defined test for heterochrony (Somel et al. 2009) in the 352 differentially expressed genes (supplementary data file S1, Supplementary Material online). The test estimates the difference between timing and amplitude of expression differences of two expression patterns. The test finds no significant difference in expression levels between the hybrid and either parent. In contrast, the hybrid expresses 32 genes significantly faster than S. cerevisiae and 22 genes significantly faster than S. paradoxus (P < 0.05). If we look at patterns regardless of significance, the test finds differences in expression levels between the hybrid and S. cerevisiae, and between the hybrid and S. paradoxus for only 13 and 17 genes, respectively. In contrast, 339 genes are defined as accelerated in the hybrid versus both parents. Thus, an accelerated shift in timing rather than expression levels explains the expression differences between the hybrid and the parents.
Trans-Acting Factors Dominate Heterochronic Differences between the Hybrid and Its Parents
Gene expression differences can be produced by changes in cis-regulatory sequences, trans-acting factors, or a combination of the two. To determine whether the hybrid’s heterochronic changes are the result of cis- or trans-acting effects, we compared allele-specific expression of the hybrid to both parents’ expression. If gene expression patterns in the hybrid are dominated by trans-acting factors, S. cerevisiae and S. paradoxus alleles should exhibit similar patterns in the hybrid (McManus et al. 2010; Schaefke et al. 2013). Using the same set of 352 genes examined above, we find largely overlapping patterns of allele-specific expression in the hybrid, indicating that differences in the timing of gene expression between the hybrid and its parents are dominated by trans-acting factors (supplementary fig. S3, Supplementary Material online). However, the S. cerevisiae and S. paradoxus alleles are not identical in the hybrid, particularly at the later stages, indicating a contribution of cis-acting expression differences (supplementary fig. S3, Supplementary Material online).
Cis- and Trans-Acting Changes in Gene Expression
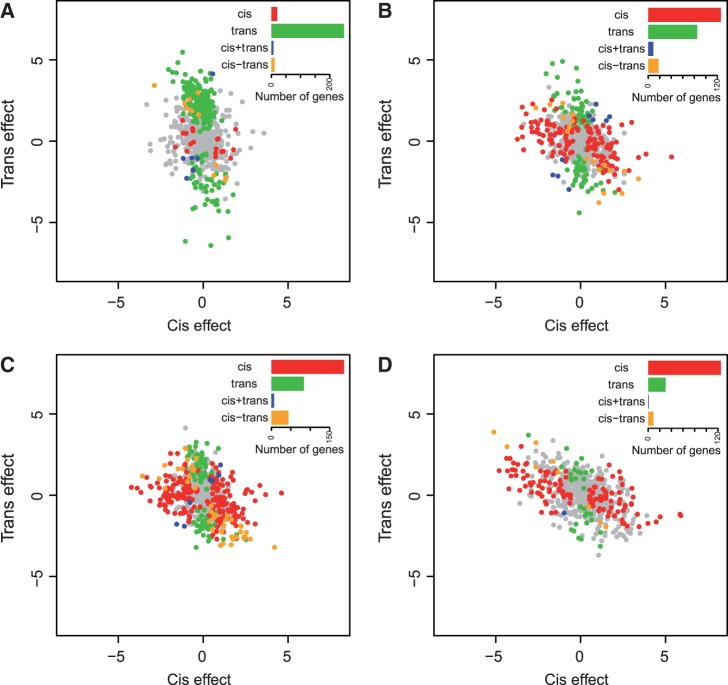
Altered gene regulation in the hybrid may not necessarily be in the form of misexpression; cis-and trans-acting changes can also lead to novel regulatory interactions. To identify genes exhibiting cis- and trans-acting expression differences, we compared allele-specific expression in the hybrid to both parents’ expression. Our null model has no explanatory variables and our alternative model has variables for stage (T0, M1, M1/M2, M2), species background (S. cerevisiae, S. paradoxus, and the hybrid), parental allele (S. cerevisiae or S. paradoxus), and interactions between at least two terms. Through this comparison, we find 1,102 differentially expressed genes (P < 0.001). Following previous work (Landry et al. 2005; McManus et al. 2010; Schaefke et al. 2013), we classified each gene’ expression divergence into four categories: cis-only, trans-only, cis + trans, and cis–trans (Materials and Methods). We found 677 genes that were classified into at least one of these categories at one stage (fig. 4). Additionally, we classified cis and trans divergence as in Wittkopp et al. (2004) and find similar results. However, the number of significant genes with cis and/or trans divergence is lower (supplementary data file S1, table S3, and figs. S3 and S4, Supplementary Material online). Thus, we will discuss our results using the method from Schaefke et al. (2013).
Fig. 4.
Genes classified into different categories of cis- or trans-acting expression differences. Each plot shows the log 2 normalized cis- (x-axis) or trans- (y-axis) effect for the T0 (A), M1 (B), M1/M2 (C), and M2 (D) stages. Each circle shows one of 677 genes that could be classified into four categories of expression divergence, which are: cis-only (red), trans-only (green), cis+trans (blue), and cis-trans (orange). Genes without significant expression differences are shown in gray. Inset within each panel shows the number of genes classified into each category.
Strikingly, we observe a transition from predominantly trans-acting expression divergence at T0 to predominantly cis-acting expression divergence between stages M1 to M2 (fig. 4, supplementary table S2 and data file S1, Supplementary Material online). Additionally we find there is an increase in the other categories: cis- and trans-effects in the same (cis + trans) or opposite (cis–trans) directions as meiosis progresses. Of particular interest are genes whose expression is due to opposing cis–trans interactions, as these genes could be candidates for genetic incompatibilities contributing to sterility in the hybrid. Many of these genes are essential for viability and involved in meiosis, rRNA processing and other translational processes, and mitochondrial functions (supplementary data file S1, Supplementary Material online).
Discussion
Interspecific hybrids express genes at levels outside of the range of either parental species. This pattern of misexpression can arise as a consequence of interactions between independent cis- or trans-acting changes along different lineages or due to abnormal development resulting in changes in the abundance of different cell types. In this study, we compared expression profiles of S. cerevisiae, S. paradoxus, and their sterile hybrid during sporulation and find that heterochronic changes in the meiotic expression program explain hybrid misexpression at individual time points. We also find an increased number of cis- and trans-acting changes with opposing cis–trans divergence at the same meiotic stages exhibiting a heterochronic shift in hybrid gene expression. However, interactions between cis- and trans-acting changes were not needed to explain misexpression in the hybrid, which we found to be primarily a result of the trans-acting hybrid environment. Our results demonstrate that in yeast temporal changes in gene expression represent an important component of regulatory differences between species and misexpression in interspecific hybrids.
Meiotic Divergence between S. cerevisiae and S. paradoxus
Our microscopy data show that S. cerevisiae and S. paradoxus progress through meiosis on different time scales. The temporal shift in meiosis is not the only heterochronic change that has been observed between these two species. Saccharomyces paradoxus both mates and germinates more slowly than S. cerevisiae (Murphy et al. 2006; Murphy and Zeyl 2012). In addition, mitotic gene expression profiles exhibit heterochronic divergence among S. cerevisiae strains and S. paradoxus (Simola et al. 2010). Given that S. cerevisiae is more thermophilic than S. paradoxus (Gonçalves et al. 2011), some heterochronic changes could be related to temperature preferences.
Additionally, the hybrid also displays novel temporal progress through meiosis in comparison to its parents. Although the hybrid takes longer to complete meiosis II, in a manner similar to S. paradoxus, it completes meiosis I in a manner that is intermediate of both parents. These meiotic differences result in a longer period between meiosis I and II in the hybrid (fig. 1). The cause of the longer meiotic transition in the hybrid may result from divergence between the two parents, whereby the hybrid follows the early completion of meiosis I in S. cerevisiae but the later completion of meiosis II in S. paradoxus. However, our data neither show a significant difference in the expression of known regulators of sporulation nor their targets.
The longer meiotic transition could also be a consequence of a well-known lack of recombination in the hybrid (Greig et al. 2002). Saccharomyces cerevisiae strains that cannot recombine homologous chromosomes progress more quickly through M1 than wild-type strains (Galbraith et al. 1997). These mutants produce binucleates quickly but tetranucleates at a wild-type pace, which creates a larger area between the bi- and tetranucleate curves. Likewise, both our phenotypic and expression data is consistent with the hybrid moving through meiosis I but not meiosis II more quickly than both parents. As hybrids have a lack of recombination and S. cerevisiae-recombinant mutants have accelerated meiosis I, our observation that the hybrid moves more quickly through meiosis I can be most easily explained by a lack of recombination. mmr mutant and allotetraploid hybrids have higher recombination rates, and an examination of those hybrids would be the next step in understanding how a lack of recombination in hybrids contributes to a longer meiotic transition than its parents.
Misexpression as a Consequence of Temporal Shifts between the Hybrid and Its Parents
Although misexpression has previously been observed in interspecific hybrids, the causes of misexpression are rarely known (Coolon and Wittkopp 2013; Ranz et al. 2013). The number of misexpressed genes we identified is not large due to our conservative analysis of differential expression. However, the overall pattern of differentially expressed genes from our principal coordinate analysis shows a heterochronic shift similar to misexpressed genes. The heterochronic shift does not occur at T0 where we find hybrid expression to lie between the parents’ expression, consistent with previous work (Tirosh et al. 2009). Thus, the simplest explanation for the shift in gene expression is that the normal meiotic expression program is activated earlier in the hybrid than either parent. A difference in developmental timing is of particular interest, as heterochrony has been believed to contribute to macroevolutionary changes between species (Gould 1977).
Previous studies have found hybrid misexpression changes during development, with adults showing more misexpression than earlier stages of development (Moehring et al. 2006; Artieri and Singh 2010). An increase in misexpression during development supports a cascading model of misexpression due to either evolved differences in gene regulation or changes in development. Regulatory divergence of genes expressed early during development can be propagated to extensive changes in expression later in development. Similarly, early changes in tissue abundance or cell types during development can be propagated to larger differences in adults. The observation that a pupal stage of Drosophila hybrids has fewer misexpressed genes than either larval or adult stages supplies evidence against either of the two cascading models (Artieri and Singh 2010) and suggests that pupal stages may be more immune to “developmental systems shift” (True and Haag 2001), perhaps due to the complexity of gene regulation during metamorphosis. Our results show that there is little to no misexpression outside the range of the normal temporal changes that occur during meiosis in an interspecific yeast hybrid. Although the observed temporal changes could be caused by misexpression of even a single master regulator of sporulation, the meiotic program does not appear to be altered other than its timing.
One surprising finding is a near absence of a shift in hybrid expression at the final M2 stage, where late sporulation genes involved in meiotic division and spore wall formation are turned on. The hybrid more closely resembles both parents at the M2 stage compared with the M1/M2 stage. Similarly, even though the completion of meiosis I and the formation of binucleates occurs at different time points, both parents and their hybrid reach their maximum percentage of tetrads produced at the same time point, ∼15 h. Thus, both the meiotic divisions and gene expression program complete at the same time, suggesting that the hybrid does not simply express its total meiotic program earlier than its parents.
Cis-Only and Trans-Only Changes between S. cerevisiae and S. paradoxus
We found a transition from trans- to cis-acting expression divergence over the course of meiosis. Previous studies have observed a fairly wide range in the proportion of expression differences attributable to cis-acting elements (Coolon and Wittkopp 2013). However, there is a tendency for cis-acting changes to be enriched between species compared with within species (Wittkopp et al. 2008; Emerson et al. 2010), and the proportion of cis-acting changes in yeast depends on the environment (Tirosh et al. 2009). For instance, Tirosh et al. (2009) mostly found cis changes between S. cerevisiae and S. paradoxus laboratory strains grown in synthetic complete media, while we mostly find trans changes between two respective wild isolates grown in YPD (1% yeast extract, 2% peptone, and 2% dextrose). Our observations of a transition from predominantly trans- to cis-acting expression differences between S. cerevisiae and S. paradoxus add time as another factor contributing to variability.
Previous work has also found associations between genes with both cis- and trans-acting divergence and whether a gene is misexpressed in an interspecific hybrid (Landry et al. 2005; McManus et al. 2010). Although we find an increase in opposing cis–trans changes at the M1 and M1/M2 stages, where we also see the most misexpressed genes (table 1), only six misexpressed genes have opposing cis–trans divergence. There may be some translational or meiotic consequences of this misexpression. Among the six genes, TEF1 is a translation elongation factor that help bind aminoacyl-tRNA to ribosomes, and CDC26 is a subunit of the anaphase-promoting complex (APC/C) involved in exit from mitosis. Although CDC26 function in meiosis is not known, the C. elegans homolog of CDC26 is required for the metaphase to anaphase transition through meiosis I (Dong et al. 2007).
Opposing Cis–Trans Changes between S. cerevisiae and S. paradoxus
During the M1 and M1/M2 stage, we observe an increase in the relative abundance of opposing cis–trans changes. The cis–trans changes are of particular interest, because they may only be present in the hybrid and could thus contribute to RI (Landry et al. 2005). A handful of genes are noteworthy. Four genes involved in later meiosis (CDC26, SPO12, HED1, and APC11) show an opposing cis–trans interaction between the parents. The hybrid expresses both alleles of these genes higher than the same allele in the parents. APC11 and CDC26 form the APC/C complex, which regulates the metaphase to anaphase transition of meiosis and the exit from both mitosis and meiosis to G1 phase. During mitosis, APC/C regulates SPO12, which regulates the exit of mitosis, and SPO12 may play a similar role during meiosis (Shah et al. 2001). HED1 is a suppressor of RED1, which is involved in the pachytene checkpoint. Hed1p suppresses Red1p when the recombination machinery is impaired (Tsubouchi and Roeder 2006). We can interpret this result as either the hybrid expressing HED1 to prevent and bypass recombination or to turn off early M1 genes.
Previous studies have shown that mitochondrial genes are involved in yeast hybrid sterility (Lee et al. 2008; Chou et al. 2010). Many of the genes for which we find opposing cis–trans changes are involved in mitochondrial maintenance and respiration. Although we did not specifically identify MRS1, which contributes to hybrid sterility between S. cerervisiae and S. paradoxus, we found another mitochondrial RNA splicing gene, MRS3. Overexpression of MRS3 overcomes splicing mutations in S. cerevisiae and may be involved in nuclear–mitochondrial incompatibilities much like MRS1.
Conclusions
In this study, we show that hybrid misexpression in yeast is a result of a heterochronic shift in the meiotic gene expression program. Although the cause of this shift remains unknown, it is consistent with yeast hybrids bypassing meiotic recombination. We find no direct relationship between genes exhibiting opposing cis–trans changes and misexpression. Yet both variables increase in number at the same stage of development. Although the extent to which hybrid misexpression in multicellular organisms is a consequence of changes in gene regulation, we provide an important insight into the causes of misexpression over a developmental pathway.
Materials and Methods
Strains
The strains used in this study are listed in supplementary table S1. We derived S. cerevisiae strains from YPS163 and S. paradoxus strains from N17. P. Sneigowski, University of Pennsylvania, provided initial strains. All genetic manipulations were created using a standard lithium acetate transformation and homologous recombination (Cubillos et al. 2009). We replaced the HO locus with dsdAMX4 in S. cerevisiae and NATMX4 in S. paradoxus and isolated haploid derivatives of the strains. For both parental species, we generated three independent diploid strains containing the double ho deletion. We generated interspecific hybrids by mating haploid strains from the two species and isolating their hybrids that contain the two dominant markers, dsdA and NAT. We generated two independent hybrids for each reciprocal cross.
Growth and sporulation conditions
We inoculated 100 ml of YPD (1% Bacto yeast extract, 2% Bacto peptone, and 2% dextrose) in 250-ml Erlenmeyer flasks and incubated the culture at 30 °C and 340 rpm for 15 h. To sporulate cells, we washed cells with water and resuspended the cells in 250 ml of complex sporulation media, or SPO (1% potassium acetate, 0.1% Bacto yeast extract, 0.05% dextrose) for a final concentration of 107 cells/ml. We incubated the cultures in 1-l baffled flasks at 30 °C at 340 rpm for 24 h. We used distilled water for all media.
Sampling
Once we resuspended cells in SPO, we sampled, washed, and snap-froze cells at every hour between 0 and 16 h and at 20 and 24 h. We fixed a subset of sampled cells in formaldehyde and ethanol, and stained fixed cells with DAPI (Galbraith et al. 1997). We counted nuclei using fluorescent microscopy. We defined developmental stages based on nuclei count. T0 is the time at which we placed cells in sporulation media: M1 is defined as an hour before we observe binucleates (when M1 is complete); M1/M2 (the transition between the end of M1 and the beginning of M2) is defined as an hour before tetranucleates appear; and the end of M2 is defined as an hour before the tetranucleates comprise the majority of the cell population.
Illumina Indexing Library, Alignments, and Mapping
We extracted total RNA from samples using Ambion RiboPure-Yeast Kit. We purified mRNA from total RNA using Ambion Micro PolyPurist Kit, reversed transcribed mRNA into cDNA, and sheered cDNA to 100 bp by sonication using a Bioruptor. We ligated indexed Illumina library adaptors to the sheared cDNA samples, size-selected ligated samples (250–350 bp), and then mixed at equal concentrations for a final concentration of 1 nM (Forsberg et al. 2012). The final library was sequenced by Genome Technology Access Center (GTAC) at Washington University using two Illumina HiSeq lanes at a run concentration of 6 pM.
We used bowtie (Langmead et al. 2009) to align each sample’s reads to the S. cerevisiae reference genome, S288c (Cherry et al. 2012), and S. paradoxus genome for strain N17 (Carter 2008), or the two genomes combined for reads from the hybrid. We required unique alignments with up to one mismatch and set all other options to default. We mapped aligned reads to 6,722 features shared between the annotated S288c and N17 genomes, of which 3,463 had one or more read. The group of 3,463 orthologs is composed of 2,894 open reading frames (ORFs), 189 autonomously replicating sequence (ARS), 105 tRNAs, 42 regulatory and chromosome maintenance RNAs (e.g. anti-sense RNA, snRNA), eight telomeres, two variants of 5s ribosomal subunit, and two centromeres. Only ORFs and ARSs have differential expression.
Differential Gene Expression Measurements
We used DESeq (Anders and Huber 2010) to measure differential expression. We estimated dispersions using the pooled-CR method for multivariate designed experiments and a separate model formula for each analysis, described below. As a control, we randomized each sample’s label and calculated the number of significant genes across 100 permutations. We calculated an empirical estimate of our false discovery rate (FDR) for each P-value cutoff used. This empirical FDR was found to be less than 1% for our following analyses.
To identify genes with differential expression between S. cerevisiae, S. paradoxus, and their hybrid over time, we used a generalized linear model (nbinomGLMTest) to compare a model without any explanatory variables to one with variables for developmental stage and species’ background: count ∼ stage*species, where the asterisk indicates the presence of both additive and interactive terms between the two variables. To identify genes that showed stage-specific differences between species’ background, we compared count ∼ stage + species to count ∼ stage*species. By comparing these two models, which only differ by a nonadditive interaction between stage and species, we ensure significant genes’ differential expression is due to biological significance rather than an artifact from any sequencing or read mapping bias between genomes because such biases are expected to be constant across stages.
To identify allele-specific expression differences in the hybrid and differences in allele expression in the hybrid versus parental background, we expanded our above two variable model to include a variable for allele-specific expression: count ∼ stage*species*allele. We compared our three-variable model to a null model without any explanatory variables and identified 1,102 genes. To identify genes that showed stage-specific differences between species’ background and allele, we compared count ∼ stage + species + allele to count ∼ stage*species*allele and identified 266 genes. Because the set of 266 and 1,102 genes showed very similar patterns of cis and trans expression categories (defined below and supplementary data file S1, Supplementary Material online), we presented the larger set in the results. The differentially expressed genes and P values for these analyses are provided in supplementary data file S1, Supplementary Material online.
Other Statistical Analyses
Differences in how S. cerevisiae, S. paradoxus, and their hybrid progressed through meiosis based on nuclei staining were tested through ANOVA. Each test was based on three S. cerevisiae or S. paradoxus replicates and four hybrid replicates with one factor for strain and one factor for time point. For simplicity, we conducted separate ANOVAs for each comparison, which involved two strains and 19 time points, and the reported P values are for the interaction between strain and time point. t-Tests were used for comparison of area between the bi- and tetranucleate time curves for each of the pairwise comparisons.
\We used variance-stabilized data from the above DESeq analyses for the remainder of our statistical analyses. To compare overall expression differences between the hybrid and parents, we applied principle coordinate analysis (PCoA) on the 352 genes whose expression level depended on an interaction between species and stage in our two-variable model. We obtained principle coordinates using the Euclidean distance between each pair of samples and the cmdscale function in the statistical package R, which represents the combined distance among all the samples in two-dimensional coordinates by ordination. We identified genes expressed by the hybrid outside of the range of the two parents’ expression using a two-tailed t-test with a P value cutoff of 0.05 (supplementary data file S1, Supplementary Material online). For our allele-specific analysis, we applied PCoA to the same 352 genes using variance-stabilized data from our three-variable model.
To test for heterochrony, we utilized the R functions in Somel et al. (2009). Briefly, we used the variance-stabilized data of the 352 genes whose expression level depended on an interaction between species and stage in our two-variable model. We found the average expression for S. cerevisiae, S. paradoxus, and the hybrid at each developmental stage, which for the test we defined numerically as 0, 1, 2, and 3 for T0, M1, M1/M2, and M2, respectively. Using absolute time did not change our results (data not shown). We used the heterochrony test to measure spline curves for one of the parents (reference species) and the hybrid and test whether transforming the hybrid spline curve by time or expression minimizes the differences between the hybrid and parental curves.
To measure cis- and trans-effects, we used the 1,102 genes whose expression level depended on one or more variables in our three-variable model. We tested for significant differences in expression (E) between the parents (P), S. cerevisiae (Sc), and S. paradoxus (Sp) for each gene (i), using a t-test with a P value cutoff of 0.05.
| (1) |
We tested for significant differences in expression (EH) between the S. cerevisiae and S. paradoxus alleles in the hybrid (H,Sc and H,Sp, respectively), using a t-test with a P value cutoff of 0.05.
| (2) |
To test whether the difference in expression between parental genes is of equal size as the allelic differences in the hybrid (EH-P), we used a t-test with a P value cutoff of 0.05.
| (3) |
Using these cutoffs, we define differential gene expression due to cis- and trans-effects followed the classification of Schaefke et al. (2013). Cis-only effects have expression differences between hybrid alleles (EH), but no expression differences between parent and hybrid alleles (EH-P). Trans-only effects have no expression differences between hybrid alleles (EH), but expression differences between the parent and hybrid alleles (EH-P). Cis+trans effects have both cis- and trans-effects in the same direction. Cis–trans effects have both cis- and trans-effects but in opposite directions. Results of these tests are provided in supplementary data file S1, Supplementary Material online.
Additional Data Files
The sequencing data are available from the NCBI Gene Expression Omnibus (GEO: GSE51809). Normalized expression levels of differentially expressed genes are also available as part of the supplementary data file S1, Supplementary Material online.
Supplementary Material
Supplementary data file S1, figures S1–S4, and tables S1–S3 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The authors thank Andrew Bergen and Elizabeth Engle for assistance in collecting RNA samples; Priya Sudarsanam and Bin Wang for advise with RNA-Seq; and Kara Powder and anonymous reviewers for their help in improving our work. This work was supported by a National Institutes of Health grant GM080669 to J.C.F. and a National Institutes of Health Cell and Molecular Biology training grant T32 GM007067 training grant to D.S.L.
References
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artieri CG, Singh RS. Molecular evidence for increased regulatory conservation during metamorphosis, and against deleterious cascading effects of hybrid breakdown in Drosophila. BMC Biol. 2010;8:26. doi: 10.1186/1741-7007-8-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayes JJ, Malik HS. Altered heterochromatin binding by a hybrid sterility protein in Drosophila sibling species. Science. 2009;326:1538–1541. doi: 10.1126/science.1181756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter D. 2008. Wellcome Trust Sanger Institute Saccharomyces Genome Resequencing Project: user manual. Cambridge (UK): The Sanger Institute. p. 1–32. Available from: ftp://ftp.sanger.ac.uk/pub/dmc/yeast/latest.
- Cherry JM, Hong EL, Amundsen C, Balakrishnan R, Binkley G, Chan ET, Christie KR, Costanzo MC, Dwight SS, Engel SR, et al. Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic Acids Res. 2012;40:D700–D705. doi: 10.1093/nar/gkr1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou J-Y, Hung Y-S, Lin K-H, Lee H-Y, Leu J-Y. Multiple molecular mechanisms cause reproductive isolation between three yeast species. PLoS Biol. 2010;8:e1000432. doi: 10.1371/journal.pbio.1000432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu S, DeRisi J, Eisen M, Mulholland J, Botstein D, Brown PO, Herskowitz I. The transcriptional program of sporulation in budding yeast. Science. 1998;282:699–705. doi: 10.1126/science.282.5389.699. [DOI] [PubMed] [Google Scholar]
- Coolon JD, Wittkopp PJ. Cis-and trans-regulation in Drosophila interspecific hybrids. In: Chen ZJ, Birchler JA, editors. Polyploid and hybrid genomics. Oxford: John Wiley, UK; 2013. pp. 37–58. [Google Scholar]
- Cubillos FA, Louis EJ, Liti G. Generation of a large set of genetically tractable haploid and diploid Saccharomyces strains. FEMS Yeast Res. 2009;9:1217–1225. doi: 10.1111/j.1567-1364.2009.00583.x. [DOI] [PubMed] [Google Scholar]
- Dong Y, Bogdanova A, Habermann B, Zachariae W, Ahringer J. Identification of the C. elegans anaphase promoting complex subunit Cdc26 by phenotypic profiling and functional rescue in yeast. BMC Dev Biol. 2007;7:19. doi: 10.1186/1471-213X-7-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson JJ, Hsieh LC, Sung HM, Wang TY, Huang CJ, Lu HHS, Lu MYJ, Wu SH, Li WH. Natural selection on cis and trans regulation in yeasts. Genome Res. 2010;20:826–836. doi: 10.1101/gr.101576.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson J, Gomes S, Civetta A. Rapid male-specific regulatory divergence and down regulation of spermatogenesis genes in Drosophila species hybrids. PLoS One. 2013;8:e61575. doi: 10.1371/journal.pone.0061575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsberg KJ, Reyes A, Wang B, Selleck EM, Sommer MOA, Dantas G. The shared antibiotic resistome of soil bacteria and human pathogens. Science. 2012;337:1107–1111. doi: 10.1126/science.1220761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbraith AM, Bullard SA, Jiao K, Nau JJ, Malone RE. Recombination and the progression of meiosis in Saccharomyces cerevisiae. Genetics. 1997;146:481–489. doi: 10.1093/genetics/146.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonçalves P, Valério E, Correia C, de Almeida JMGCF, Sampaio JP. Evidence for divergent evolution of growth temperature preference in sympatric Saccharomyces species. PLoS One. 2011;6:e20739. doi: 10.1371/journal.pone.0020739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good JM, Giger T, Dean MD, Nachman MW. Widespread over-expression of the X chromosome in sterile F(1)hybrid mice. PLoS Genet. 2010;6:e1001148. doi: 10.1371/journal.pgen.1001148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould SJ. Ontogeny and phylogeny. Cambridge (MA): Harvard University Press; 1977. [Google Scholar]
- Graze RM, McIntyre LM, Main BJ, Wayne ML, Nuzhdin SV. Regulatory divergence in Drosophila melanogaster and D. simulans, a genomewide analysis of allele-specific expression. Genetics. 2009;183:547–561. doi: 10.1534/genetics.109.105957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greig D, Louis EJ, Borts RH, Travisano M. Hybrid speciation in experimental populations of yeast. Science. 2002;298:1773–1775. doi: 10.1126/science.1076374. [DOI] [PubMed] [Google Scholar]
- Greig D, Travisano ME, Louis EJ, Borts RH. A role for the mismatch repair system during incipient speciation in Saccharomyces. J Evol Biol. 2003;16:429–437. doi: 10.1046/j.1420-9101.2003.00546.x. [DOI] [PubMed] [Google Scholar]
- Haerty W, Singh RS. Gene regulation divergence is a major contributor to the evolution of Dobzhansky–Muller incompatibilities between species of Drosophila. Mol Biol Evol. 2006;23:1707–1714. doi: 10.1093/molbev/msl033. [DOI] [PubMed] [Google Scholar]
- Hunter N, Chambers SR, Louis EJ, Borts RH. The mismatch repair system contributes to meiotic sterility in an interspecific yeast hybrid. EMBO J. 1996;15:1726–1733. [PMC free article] [PubMed] [Google Scholar]
- Landry CR, Wittkopp PJ, Taubes CH, Ranz JM, Clark AG, Hartl DL. Compensatory cis-trans evolution and the dysregulation of gene expression in interspecific hybrids of Drosophila. Genetics. 2005;171:1813–1822. doi: 10.1534/genetics.105.047449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H-Y, Chou J-Y, Cheong L, Chang N-H, Yang S-Y, Leu J-Y. Incompatibility of nuclear and mitochondrial genomes causes hybrid sterility between two yeast species. Cell. 2008;135:1065–1073. doi: 10.1016/j.cell.2008.10.047. [DOI] [PubMed] [Google Scholar]
- Liti G, Barton DBH, Louis EJ. Sequence diversity, reproductive isolation and species concepts in Saccharomyces. Genetics. 2006;174:839–850. doi: 10.1534/genetics.106.062166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llopart A. The rapid evolution of X-linked male-biased gene expression and the large-X effect in Drosophila yakuba, D. santomea, and their hybrids. Mol Biol Evol. 2012;29:3873–3886. doi: 10.1093/molbev/mss190. [DOI] [PubMed] [Google Scholar]
- Lu X, Shapiro JA, Ting CT, Li Y, Li C, Xu J, Huang H, Cheng YJ, Greenberg AJ, Li SH, et al. Genome-wide misexpression of X-linked versus autosomal genes associated with hybrid male sterility. Genome Res. 2010;20:1097–1102. doi: 10.1101/gr.076620.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maheshwari S, Barbash DA. Cis-by-trans regulatory divergence causes the asymmetric lethal effects of an ancestral hybrid incompatibility gene. PLoS Genet. 2012;8:e1002597. doi: 10.1371/journal.pgen.1002597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone JH, Chrzanowski TH, Michalak P. Sterility and gene expression in hybrid males of Xenopus laevis and X. muelleri. PLoS One. 2007;2:e781. doi: 10.1371/journal.pone.0000781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus CJ, Coolon JD, Duff MO, Eipper-Mains J, Graveley BR, Wittkopp PJ. Regulatory divergence in Drosophila revealed by mRNA-seq. Genome Res. 2010;20:816–825. doi: 10.1101/gr.102491.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalak P, Noor MAF. Genome-wide patterns of expression in Drosophila pure species and hybrid males. Mol Biol Evol. 2003;20:1070–1076. doi: 10.1093/molbev/msg119. [DOI] [PubMed] [Google Scholar]
- Moehring AJ, Teeter KC, Noor MAF. Genome-wide patterns of expression in Drosophila pure species and hybrid males. II. Examination of multiple-species hybridizations, platforms, and life cycle stages. Mol Biol Evol. 2006;24:137–145. doi: 10.1093/molbev/msl142. [DOI] [PubMed] [Google Scholar]
- Murphy HA, Kuehne HA, Francis CA, Sniegowski PD. Mate choice assays and mating propensity differences in natural yeast populations. Biol Lett. 2006;2:553–556. doi: 10.1098/rsbl.2006.0534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy HA, Zeyl CW. Prezygotic isolation between Saccharomyces cerevisiae and Saccharomyces paradoxus through differences in mating speed and germination timing. Evolution. 2012;66:1196–1209. doi: 10.1111/j.1558-5646.2011.01516.x. [DOI] [PubMed] [Google Scholar]
- Ortíz-Barrientos D, Counterman BA, Noor MAF. Gene expression divergence and the origin of hybrid dysfunctions. Genetica. 2006;129:71–81. doi: 10.1007/s10709-006-0034-1. [DOI] [PubMed] [Google Scholar]
- Parker HR, Philipp DP, Whitt GS. Gene regulatory divergence among species estimated by altered developmental patters in interspecific hybrids. Mol Biol Evol. 1985;2:217–250. doi: 10.1093/oxfordjournals.molbev.a040349. [DOI] [PubMed] [Google Scholar]
- Primig M, Esposito RE. The core meiotic transcriptome in budding yeasts. Nat Genet. 2000;6:415–423. doi: 10.1038/82539. [DOI] [PubMed] [Google Scholar]
- Ranz JM, Namgyal K, Gibson G, Hartl DL. Anomalies in the expression profile of interspecific hybrids of Drosophila melanogaster and Drosophila simulans. Genome Res. 2004;14:373–379. doi: 10.1101/gr.2019804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranz JM, Yeh S-D, Nyberg KG, Machado CA. Transcriptome profiling of Drosophila interspecific hybrids: insights into mechanisms of regulatory divergence and hybrid dysfunction. In: Chen ZJ, Birchler JA, editors. Polyploid and hybrid genomics. Oxford: John Wiley; 2013. pp. 15–36. [Google Scholar]
- Renaut S, Nolte AW, Bernatchez L. Gene expression divergence and hybrid misexpression between Lake Whitefish species pairs (Coregonus spp. Salmonidae) Mol Biol Evol. 2009;26:925–936. doi: 10.1093/molbev/msp017. [DOI] [PubMed] [Google Scholar]
- Rottscheidt R, Harr B. Extensive additivity of gene expression differentiates subspecies of the house mouse. Genetics. 2007;177:1553–1567. doi: 10.1534/genetics.107.076190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sackton TB, Montenegro H, Hartl DL, Lemos B. Interspecific Y chromosome introgressions disrupt testis-specific gene expression and male reproductive phenotypes in Drosophila. Proc Natl Acad Sci U S A. 2011;108:17046–17051. doi: 10.1073/pnas.1114690108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefke B, Emerson JJ, Wang TY, Lu MY, Hsieh LC, Li WH. Inheritance of gene expression level and selective constraints on trans- and cis-regulatory changes in yeast. Mol Biol Evol. 2013;30:2121–2133. doi: 10.1093/molbev/mst114. [DOI] [PubMed] [Google Scholar]
- Shah R, Jensen S, Frenz LM, Johnson AL, Johnston LH. The Spo12 protein of Saccharomyces cerevisiae: a regulator of mitotic exit whose cell cycle-dependent degradation is mediated by the anaphase-promoting complex. Genetics. 2001;159:965–980. doi: 10.1093/genetics/159.3.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simola DF, Francis C, Sniegowski PD, Kim J. Heterochronic evolution reveals modular timing changes in budding yeast transcriptomes. Genome Biol. 2010;11:R105. doi: 10.1186/gb-2010-11-10-r105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somel M, Franz H, Yan Z, Lorenc A, Guo S, Giger T, Kelso J, Nickel B, Dannemann M, Bahn S, et al. Transcriptional neoteny in the human brain. Proc Natl Acad Sci U S A. 2009;106:5743–5748. doi: 10.1073/pnas.0900544106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundararajan V, Civetta A. Male sex interspecies divergence and down regulation of expression of spermatogenesis genes in Drosophila sterile hybrids. J Mol Evol. 2011;72:80–89. doi: 10.1007/s00239-010-9404-5. [DOI] [PubMed] [Google Scholar]
- Tirosh I, Reikhav S, Levy AA, Barkai N. A yeast hybrid provides insight into the evolution of gene expression regulation. Science. 2009;324:659–662. doi: 10.1126/science.1169766. [DOI] [PubMed] [Google Scholar]
- True JR, Haag ES. Developmental system drift and flexibility in evolutionary trajectories. Evol Dev. 2001;3:109–119. doi: 10.1046/j.1525-142x.2001.003002109.x. [DOI] [PubMed] [Google Scholar]
- Tsubouchi H, Roeder GS. Budding yeast Hed1 down-regulates the mitotic recombination machinery when meiotic recombination is impaired. Genes Dev. 2006;20:1766–1775. doi: 10.1101/gad.1422506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittkopp PJ, Haerum BK, Clark AG. Evolutionary changes in cis and trans gene regulation. Nature. 2004;430:85–88. doi: 10.1038/nature02698. [DOI] [PubMed] [Google Scholar]
- Wittkopp PJ, Haerum BK, Clark AG. Independent effects of cis- and trans-regulatory variation on gene expression in Drosophila melanogaster. Genetics. 2008;178:1831–1835. doi: 10.1534/genetics.107.082032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, He X. Genetic incompatibility dampens hybrid fertility more than hybrid viability: yeast as a case study. PLoS One. 2011;6:e18341. doi: 10.1371/journal.pone.0018341. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.