Abstract
Significantly more potent second generation 4-amino-7-chloroquinoline (4,7-ACQ) based inhibitors of the botulinum neurotoxin serotype A (BoNT/A) light chain were synthesized. Introducing an amino group at the C(3) position of the cholate component markedly increased potency (IC50 values for such derivatives ranged from 0.81 to 2.27 μM). Two additional subclasses were prepared: bis(steroidal)-4,7-ACQ derivatives and bis(4,7-ACQ)cholate derivatives; both classes provided inhibitors with nanomolar-range potencies (e.g., the Ki of compound 67 is 0.10 μM). During BoNT/A challenge using primary neurons, select derivatives protected SNAP-25 by up to 89%. Docking simulations were performed to rationalize the compounds’ in vitro potencies. In addition to specific residue contacts, coordination of the enzyme’s catalytic zinc and expulsion of the enzyme’s catalytic water were a consistent theme. With respect to antimalarial activity, the compounds provided better IC90 activities against chloroquine resistant (CQR) malaria than CQ, and seven compounds were more active than mefloquine against CQR strain W2.
Introduction
Antimalarial compounds consisting of two 4-amino-7-chloroquinoline (4,7-ACQ) components linked by flexible tethers were identified as some of the first small molecule botulinum neurotoxin serotype A light chain (BoNT/A LC) inhibitors.1 Shortly thereafter, we discovered that antimalarial agents composed of a 4,7-ACQ component coupled with either an adamantane or a cholic acid derived component provided effective dual inhibitors of the BoNT/A LC and Plasmodium falciparum strains D6 and W2 strains.42,43 This discovery led to a model in which 4,7-ACQ-based antimalarials are/have been modified with the intent of also generating novel BoNT/A LC inhibitors and vice versa.47 Accordingly, we continued to synthesize new derivatives to probe structure–activity relationships for such chemotypes and to generate more potent BoNT/A LC inhibitors that, because they incorporate a 4,7-ACQ moiety, would be expected to be prospective antimalarial agents.
Botulinum neurotoxins (BoNTs), exotoxins secreted by anaerobic, spore-forming bacterium Clostridium botulinum, are the most potent of biological toxins.2 BoNTs are responsible for the potentially fatal disease botulism, which is most commonly associated with food contamination, wound infection, and colonizing infection in infants. However, because of ease of production, dissemination,3 lethality,4 and amenability for use as biological weapons,5 BoNTs are classified as category A biothreat agents by the Centers for Disease Control and Prevention (CDC).
There are seven known BoNT serotypes (identified as A–G). Each cleaves a specific peptide bond in one or more of three proteins that form the soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE complex).6,7 The interaction of these three proteins facilitates the transport of acetylcholine into neuromuscular junctions. Botulinum neurotoxins are secreted as single polypeptide chains and, following post-translational processing, are composed of a heavy chain (HC) subunit and light chain (LC) subunit; the two subunits are covalently linked by a disulfide bridge.8,9 The HC, which comprises two domains of ∼50 kDa, is responsible for mitigating toxin internalization and release into the neuronal cytosol via an acidic endosome. The C-terminal domain (i.e., the ganglioside and protein receptor-binding domain) binds to the neuronal cell membrane and mediates the internalization of toxin containing endosomes; the N-terminal domain facilitates the release of the LC from the endosome into the cell cytosol. The LC is a zinc-dependent metalloprotease that cleaves SNARE proteins. This proteolysis eliminates neurotransmitter release into the synaptic cleft, thereby causing the flaccid paralysis that is medically diagnosed as botulism. BoNT serotypes A and E cleave synaptosomal-associated protein of 25 kDa (SNAP-25).10 Serotypes B, D, F,11 and G cleave vesicle-associated membrane protein,12 and serotype C1 cleaves both SNAP-25 and syntaxin.13
The BoNT/A LC is the most potent and longest acting of the BoNT LC serotypes in the neuronal cytosol.14 For example, the BoNT/A LC is 106-fold more potent than cobra toxin and 1011-fold more deadly than cyanide. The lethal dose of BoNT/A holotoxin is estimated to be between 1 and 5 ng kg–1 for humans.15 Currently, there are no small molecule therapeutics available to treat botulism. And while antibody-based vaccines are available,16 this avenue of treatment is limited by a short window of therapeutic opportunity, since such biomolecules cannot penetrate into the cell cytosol.4 Hence, developing small molecules that will effectively inhibit BoNT LC proteolytic activity postneuronal intoxication is of particular interest.
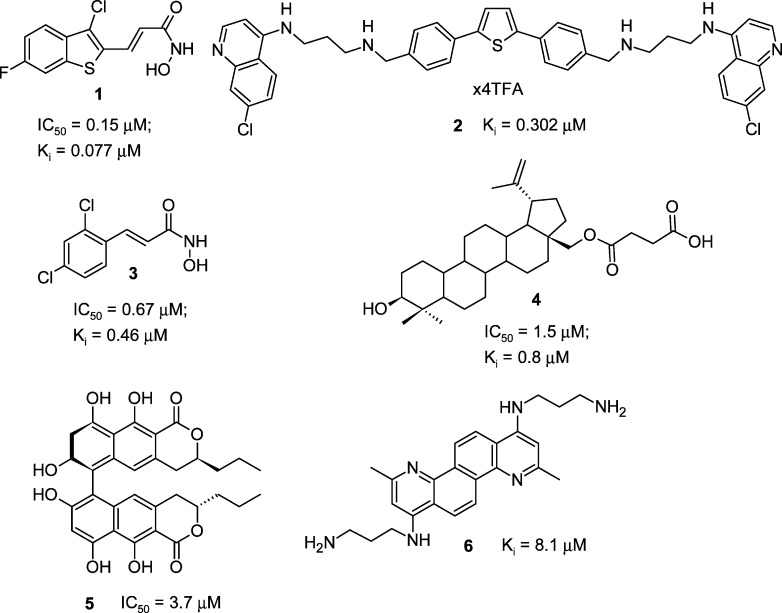
A variety of BoNT/A LC inhibitors have been reported17 and range from compounds bidentate coordinating the enzyme catalytic Zn2+18 to allosteric inhibitors.19 In general, the most potent BoNT/A LC inhibitors reported to date possess Ki values ranging from 0.1 to 10 μM and include hydroxamic acid based compounds 1 and 3,20 2,5-diphenylthiophene derivative 2,21 betulin derivative 4,22 naphthopyrone 5,23 and chrysene 6(24) (Chart 1).
Chart 1. Structures of Potent BoNT/A LC SMNPIs .
Malaria is a devastating global health threat, with nearly half of the world population at risk of being infected.25 Malaria is caused by five Plasmodium species: P. falciparum, P. ovale, P. vivax, P. malarie, and P. knowlesi, of which P. falciparum, which causes cerebral malaria, is the major cause of mortality. Malaria parasites contain an acidic food vacuole (FV) that digests hemoglobin (Hb), and it is generally accepted that the FV is the site of action for a number of aminoquinoline (ACQ) based drugs. The heme obtained from Hb degradation is toxic to the parasite and is therefore transformed into insoluble hemozoin pigment, while the globin is hydrolyzed into individual amino acids. Antimalarial drugs that are active within the FV appear to kill the parasite by either producing toxic free radicals26 or blocking hemozoin formation, as in the case of ACQ-based drugs.27 The development of widespread drug resistance to chloroquine (9, CQ, Chart 2), one of the most successful antimalarial drugs, has resulted in severe health issues for countries in malaria endemic regions. Although collected data indicate that mutations in the P. falciparum CQ-resistant transporter (PfCRT), multidrug resistance protein 1 (PfMDR1), and multidrug resistance-associated protein (PfMRP) are responsible for malaria parasite resistance to CQ and its analogues,28 additional analyses appear necessary to fully corroborate such data.29 Therefore, significant focus has been placed on the syntheses of peroxide antimalarials,30 as well as on the development of other chemotypes that prevent heme polymerization.31 Nevertheless, various ACQ-based derivatives are being investigated for their antimalarial activity, either since they appear highly active and nontoxic, such as pyrrolizidine-ACQ compounds32 and aminoquinoline AQ-13,33 or because of their contribution to the development of SAR, for example, compounds such as 4-N-, 4-S-, or 4-O-alkylaminoquinoline derivatives.34 Hybrid adducts of various biologically active compounds covalently bonded to the ACQ moiety,35 such as reversal agents,36 complexes with Au and Ru,37 antibiotic azithromycin,38 2-imidazolidine and ferrocenyl derivatives,39 and tetraoxanes,40 are of special interest. Very recently, a breakthrough in the antimalarial field has been achieved: quinolone antimalarials that target the parasite’s liver and blood stages, as well as forms that are crucial to disease transmission, have been reported.41
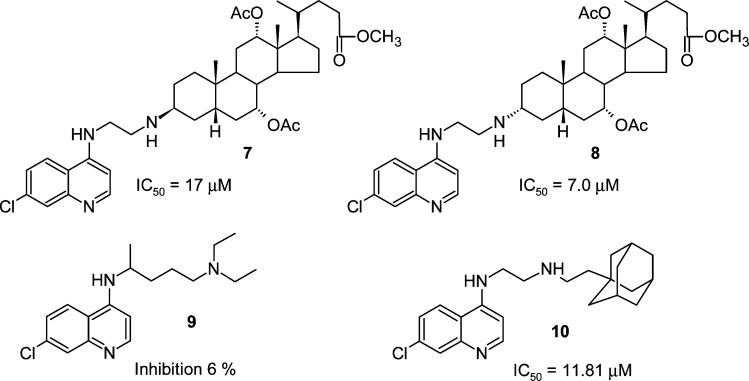
Chart 2. Structures and Activities of BoNT/A LC Inhibitors Possessing an ACQ Moiety.
As indicated above, we previously described the synthesis and inhibitory activity of BoNT/A LC inhibitors possessing a 4,7-ACQ moiety covalently tethered to either a cholic acid derived or an adamantane component. The compounds were database mined using a pharmacophore for BoNT/A LC inhibition42 and were originally synthesized as antimalarial agents. Examples include 7, 8, and 10 (Chart 2). In vitro testing indicated that the compounds inhibit the BoNT/A LC with IC50 values in the low micromolar range. Furthermore, it is important to note that several of the derivatives provide potent antimalarial activity against CQR strain W2 and multidrug resistant strain (MDR) TM91C235.43 For example, 7 was very active in vitro against P. falciparum CQR and CQ susceptible (CQS) strains, with a resistance index (RI (W2/D6)) of 0.65.43 This compound also cured mice infected with P. berghei in a Thompson test.44
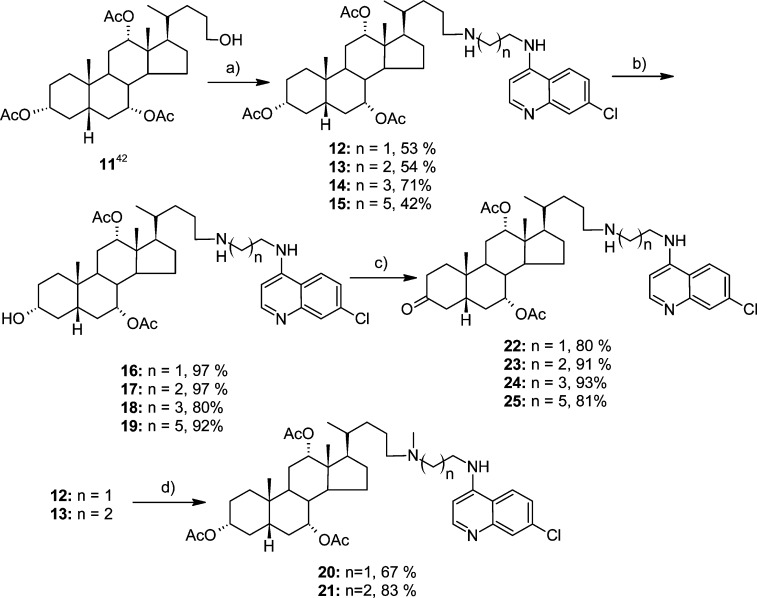
Herein, we present second generation 4,7-ACQ-cholate based inhibitors of the BoNT/A LC that are significantly more potent than initially discovered leads.42,43 In particular, we expanded our understanding of the SAR for this inhibitor chemotype via the generation of compounds with a variety of substitutions on the C(3) position of the cholic acid component. In addition, it was anticipated that such an approach would facilitate the synthesis of bis(4,7-ACQ)-cholic acid derivatives, which became desired target molecules (based on the promising activities of compounds 7, 8, and 12 (Scheme 1).42 Finally, because initially discovered derivatives of the 4,7-ACQ-cholic acid chemotype were originally developed as antimalarial agents, the compounds prepared during this study were also examined for potency against three P. falciparum strains. Overall, the unique capacity of this general chemotype to provide both inhibition of the BoNT/A LC and antimalarial activity provides a paradigm that facilitates the repositioning of derivatives based on target potency.
Scheme 1.
Results and Discussion
The ability of derivative 12 (Scheme 1) both to inhibit the BoNT/A LC42 and to act as an antimalarial agent43 prompted the development of a new generation of 4,7-ACQ-cholic acid based derivatives. The design of the compounds focused on investigating effects on potency resulting from (1) an additional basic group at position C(3) of the cholic acid component, (2) the incorporation of a second ACQ component, and (3) the length of the α,ω-diaminoalkylidene spacer between the steroid and 4,7-ACQ moiety(ies). Furthermore, functionalization of the C(3) basic group (acylation, mesylation, and alkylation), as well as changes in the basic character of the substituent (i.e., H-bond donor/acceptor strength and voluminosity), was examined in the context of inhibitory activity and metabolic stability. To aid in rationalizing differences in the in vitro potencies of the derivatives, detailed docking simulations were performed in the BoNT/A LC substrate cleft. In addition, assessment of the in vitro antimalarial activities of the new derivatives against the CQS and CQR strains of P. falciparum was also conducted, and the most active antimalarial of the series was examined in a P. berghei rodent model.
Synthesis
The syntheses of the target compounds are presented in Schemes 1–6. Derivatives 12–15 were synthesized using a previously described procedure.43 Hydrolysis of the acetate at C(3) afforded corresponding alcohols 16–19, which were further oxidized with IBX in DMSO in the presence of TFA to ketones 22–25 (Scheme 1). Derivatives 12 and 13 were selectively N-methylated using 37% formaldehyde and NaBH(OAc)3.
Scheme 6.
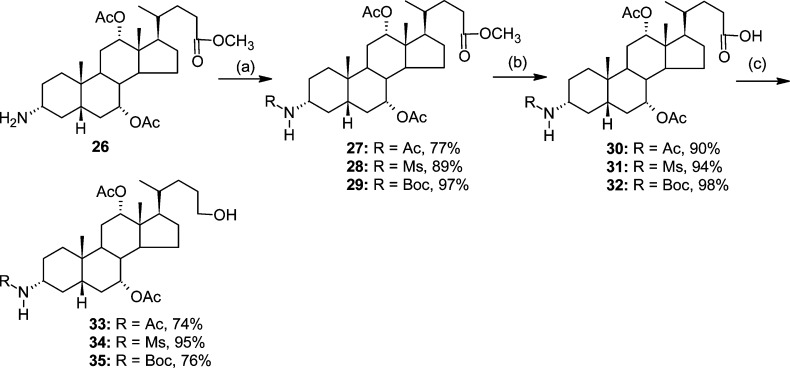
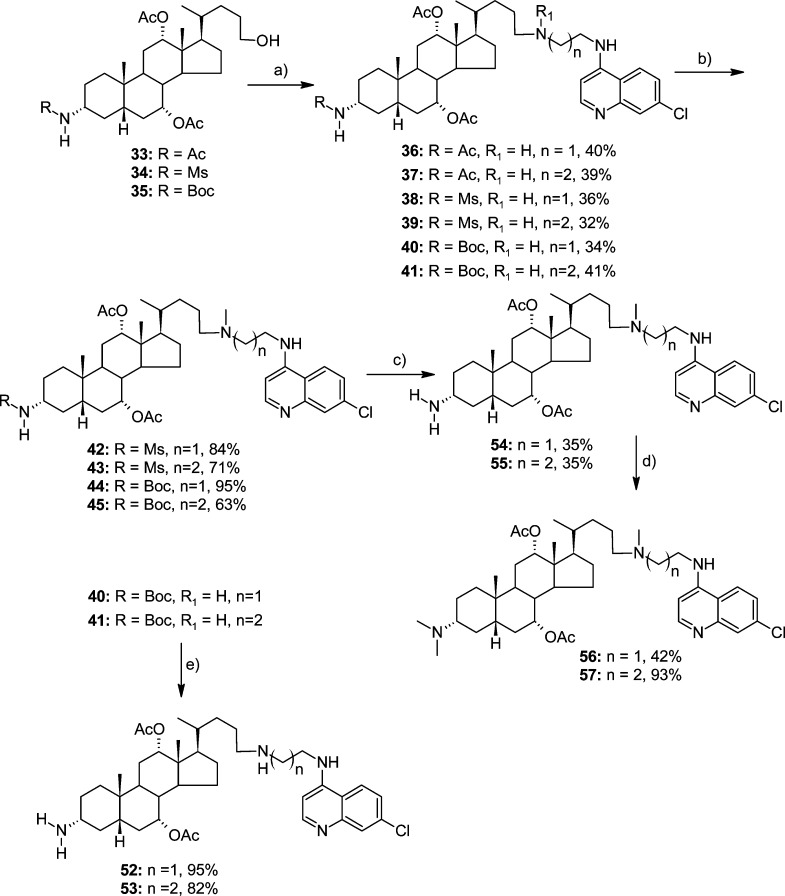
Key intermediates for the synthesis of N-alkylated congeners 36–57, i.e., compounds 33–35, were obtained from precursor 26(45) (Scheme 2). In the first step, the free amine was protected as an acetate, mesylate, or Boc derivative (27, 28, or 29, respectively). Subsequently, selective hydrolysis and reduction of the intermediate mixed anhydrides (reaction steps b and c, respectively) (Scheme 2) afforded compounds 33–35 in 54–80% overall yield. The three intermediate compounds were subsequently transformed (via the above indicated procedure) using N-(7-chloroquinolin-4-yl)ethane-1,2-diamine (ACQ2) or N-(7-chloroquinolin-4-yl)propan-1,3-diamine (ACQ3) into respective derivatives 36–41 (Scheme 3).
Scheme 2.
Scheme 3.
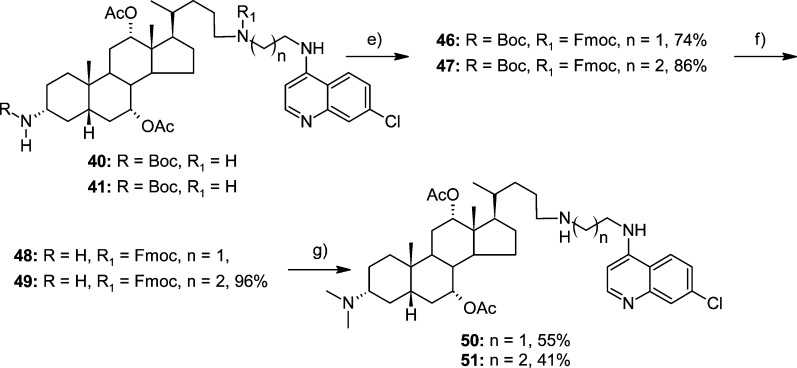
N-Mesyl and N-Boc derivatives 38–41 were methylated using 37% formaldehyde to yield 42–45, respectively. After removal of the N-Boc protecting group, permethylated products 56 and 57 were obtained in two steps from 44 and 45, respectively. On the other hand, 40 and 41 were transformed into 3α-(N,N-dimethyl) derivatives 50 and 51 by protective group manipulation using Fmoc and Boc protection groups (Scheme 4). Finally, derivatives 52 and 53, which contain primary amino groups, were obtained from 40 and 41 via the removal of the N-Boc group with 50% TFA in DCM (Scheme 3).
Scheme 4.
In the above reductive amination of the aldehyde derived from 35 with ACQ2 or ACQ3, bis-steroidal 4,7-ACQ derivatives 58 and 59 were also isolated as double alkylation byproducts (Scheme 5). Bis-steroidal 4-aminoquinoline derivatives 60 and 61, which possess free amino groups at position C(3), were obtained after the deprotection of 58 and 59, respectively.
Scheme 5.
Additional targets of the present work, bis-4,7-ACQ derivatives 62–68 (Scheme 6), were obtained as diastereomers at C(3) from 24-ACQ-ketones 22–25 (Scheme 1) via reaction with respective ACQ-amines. However, the products were obtained in rather low yield, and derivatives with shorter linkers, i.e., 62 and 63, having two and three methylene groups, respectively, were not separated into single isomers and were therefore tested in the biological assays as mixtures.
The structures of all synthesized compounds were confirmed by spectral and analytical techniques. The purity of all tested compounds was determined with Waters and Agilent HPLC instruments and was ≥95% for all. Analytical details are provided in the Experimental Section and Supporting Information.
Inhibition of the BoNT/A LC
The inhibitory efficacies of derivatives 12–68 at 20 μM, employing a well-established HPLC-based assay for BoNT/A LC inhibition,46 are shown in Table 1. The assay uses a synthetic substrate that contains the SNAP-25 scissile bond (SNAP-25 residues 187–203 of SNAP-25), and proteolytic activity is determined via comparison of the peak areas of the products versus the intact substrate.57 In the assay, CQ exhibited only marginal inhibition of the BoNT/A LC (6.51% at 20 μM), which clearly emphasizes the importance of the cholic acid component;47 the cholic acid component not only contributes to the overall amphiphilic character of the inhibitors but also results in additional interactions with amino acid residues in BoNT/A LC binding cleft (vide infra).
Table 1. In Vitro Inhibitory Activity of Tested Compounds against BoNT/A LC.
| BoNT/A LC |
|||
|---|---|---|---|
| compd | % inhibitiona | IC50 (μM) | Ki (μM)e |
| NSC 240898b | 73.31c | 2.62 | |
| 12 | 74.00 | 3.80 | 6.99 ± 0.46 |
| 13 | 81.43 | 2.28 | 5.67 ± 0.37 |
| 14 | 69.04 | ||
| 15 | 83.35 | ||
| 16 | 69.44 | 4.68 | |
| 17 | 69.88 | 5.18 | |
| 18 | 80.83 | ||
| 19 | 62.88 | ||
| 22 | 67.09 | 5.70 | |
| 23 | 72.86 | 2.32 | |
| 24 | 81.45 | ||
| 25 | 77.23 | ||
| 20 | 33.28 | 34.51 ± 4.72 | |
| 21 | 61.07 | 4.48 | 7.69 ± 0.39 |
| 36 | 73.95 | 2.46 | |
| 37 | 77.30 | 4.92 | |
| 38 | 74.38 | 5.09 | |
| 39 | 72.92 | 3.25 | |
| 40 | 67.37 | 4.90 | |
| 41 | 74.91 | 3.24 | |
| 42 | 33.75 | 24.97 | |
| 43 | 55.64 | 8.21 | |
| 44 | 29.97 | 6.78 | |
| 45 | 59.63 | 6.62 | |
| 50 | 93.51 | 1.02 | |
| 51 | 92.59 | 1.34 | |
| 52 | 89.34 | 0.81 | 3.22 ± 0.32 |
| 53 | 84.80 | 1.04 | 3.45 ± 0.35 |
| 54 | 89.48 | 1.14 | |
| 55 | 81.38 | 1.13 | |
| 56 | 85.54 | 2.27 | |
| 57 | 86.86 | 1.29 | |
| 60 | 82.22 | 3.05 | |
| 59 | 6.42 | ||
| 61 | 92.81 | 0.63 | |
| 62 | 96.47 | 0.341 ± 0.042 | |
| 63 | 97.24 | 0.171 ± 0.013 | |
| 64 | 96.68 | 0.300 ± 0.065 | |
| 65 | 89.57 | 0.389 ± 0.059 | |
| 66 | 95.46 | 0.285 ± 0.056 | |
| 67 | 95.43 | 0.103 ± 0.024 | |
| 68 | 93.70 | 0.300 ± 0.059 | |
| 9d | 6.51 | ||
Percent inhibition calculated at 20 μM. Percent inhibition measurements were performed in duplicate, and standard deviations were less than (25%) for all. IC50 calculations were determined by measuring enzyme activity at nine different SMNPI concentrations and in the absence of the small molecule. The small molecule concentrations used in the measurements were determined by estimating the IC50 value and moving in 1 log increments in either direction of the estimated value.
NSC240898, used as the control for comparison, displayed dose dependent inhibition of BoNT/A induced SNAP-25 cleavage in neurons with no toxicity at concentrations as high as 40 μM (ref (57)).
Average value from more than 20 measurements.
Tested as diphosphate salt.
For Ki determination, reaction velocity versus substrate concentration was plotted for multiple small molecule concentrations. These plots were analyzed using global kinetic analysis. Subsequently, the data were fit to a model of competitive inhibition and analyzed by nonlinear regression analysis.
The first set of inhibitors (12–15) was prepared to evaluate the effect on activity of increasing the length of the methylene bridge between the steroid core and the 4,7-ACQ component; the results indicated that increasing the length of the methylene bridge did not significantly affect inhibitory potency. Moreover, the results indicated that within the ACQ2 (compounds 12, 16, 22), ACQ3 (compounds 13, 17, and 23), and N-(7-chloroquinolin-4-yl)hexane-1,3-diamine (ACQ6, compounds 15, 19, and 25) series, transformations at C(3) from Ac to OH to C=O generally diminished the inhibitory activities of the respective compounds. N-(7-Chloroquinolin-4-yl)butane-1,3-diamine (ACQ4) derivatives (compounds 14, 18, and 24) were the exception. Among derivatives with nonbasic substituents at C(3) (Scheme 1), the most active were acetate 13 (IC50 = 2.28 μM) and keto derivative 23 (IC50 = 2.32 μM).
The introduction of a basic amino substituent at the C(3) position of the steroid component markedly increased inhibitory potency. Among this set of compounds, the most active was ACQ2 derivative 52, which contains an NH2 group at the C(3) position and a secondary amino group at the C(24) position (IC50 = 0.81 μM). The activity of this compound was followed by ACQ2 derivative 50 (IC50 = 1.02 μM), which possesses a tertiary amine at the C(3) position. The observed results suggest that for potent BoNT/A LC inhibitory activity the presence of an ionizable amino group at the C(3) position is critically important. The least active compounds are cholic acid derivatives with acidic NH groups (i.e., N-Ms and N-Boc) at C(3) (see 42 (IC50 = 24.97 μM) and 44 (IC50 = 6.78 μM)); both are ACQ2 derivatives. For derivatives in which amino groups at C(24) and/or C(3) were methylated, BoNT/A LC inhibitory activity was not significantly affected (for comparisons see 52 → 54, 53 → 55, 54 → 56, and 55 → 57).
It has been shown that the catalytic cleft of the BoNT/A LC is very plastic48 because of the mobility of flexible loop regions. Hence, we hypothesized that the cleft can conformationally adapt to accept large molecules,49,50 Consequently, we examined two additional subclasses: bis-steroidal 4,7-ACQ derivatives (58–61) and bis-4,7-ACQ cholic acid derivatives (62–68). Both groups of sterically demanding inhibitors substantially enhanced inhibition. A significant increase in inhibitory activity was observed after removal of the N-Boc protecting group, 59 (6%) vs 61 (93%, IC50 = 0.63 μM), which confirmed the importance of the presence of a basic amino group at C(3) for good activity. The bis-4,7-ACQ derivatives all inhibited the enzyme by ≥95% at 20 μM and retained comparable levels of potency at 10 μM (see Table 1S in Supporting Information). The Ki values of these derivatives were found to be lower than 0.4 μM, with compound 67 being the most potent derivative (Ki = 0.103 μM).
As an important step in further evaluating the inhibitors, three derivatives that were found to enter cells were examined for the ability to protect SNAP-25 during BoNT/A challenge in primary neurons (Table 2). Western blot analysis was used to determine the relative concentrations of intact and cleaved SNAP-25 in treated and control cells. The experiments were performed using 1.5 μL of a 10 mM concentration of ligand in the presence of 10 μL of a 50 nM solution of BoNT/A. The results indicate that 3-amino derivative 52 significantly protects SNAP-25 versus corresponding 3-(N-acetyl) derivative 36 (77 % vs 16 %, respectively), further confirming the importance of a basic amino substituent at cholic acid position C(3). Somewhat better results were obtained with bis-steroid derivative 60. In the presence of 5 μL of a 100 nM solution of antibiotic bafilomycin (Baf) SNAP-25 protection is slightly higher (77% with 52 only and 88% upon Baf addition vs 60 (89% alone and 96% upon 5 μL of Baf (100 nM) addition)). Using 25 μL of a 100 nM solution of Baf provided no further increase in protection. The results suggest that 4,7-ACQ-cholate-based inhibitors can successfully protect SNAP-25 in the presence of the BoNT/A holotoxin, with a slight additive effect when given in combination with Baf (a universal antagonist of all BoNTs in cell culture that acts by inhibiting the release of the LC into the neuronal cytosol3).
Table 2. Protection of SNAP-25 by Steroidal SMNPIsa.
| % of intact SNAP-25 |
|||
|---|---|---|---|
| compd | A | B | C |
| 36 | 16 | 28 | 36 |
| 60 | 89 | 96 | 93 |
| 52 | 77 | 88 | 81 |
| blank | 31b | 40 | 43 |
A: 488.5 μL of medium + 1.5 μL (10 mM) of compound + 10 μL of BONT-A (50 nM). B: 483.5 μL of medium + 1.5 μL (10 mM) of compound + 5 μL of Baf (100 nM) + 10 μL of BONT-A (50 nM). C: 463.5 μL of medium + 1.5 μL (10 mM) of compound + 25 μL of Baf (100 nM) + 10 μL of BONT-A (50 nM). Blank: 490 μL of medium + 10 μL if BONT-A (50 nM).
Average value from four measurements.
Docking Simulations
To rationalize the inhibitory potencies of the new derivatives, structure-based docking simulations were performed using Schrödinger Suite 2011 and the modules therein.51 The structure of the BoNT/A LC used in this study was obtained from X-ray crystal structure PDB code 3DS9.52
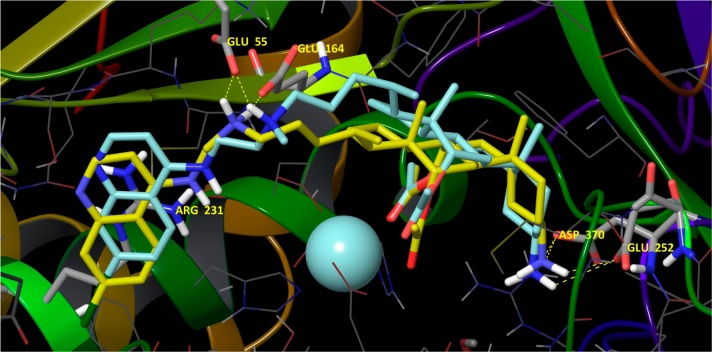
For compounds 12–57, which are composed of a single steroid and ACQ component, all docking studies indicated that a carbonyl oxygen from the acetoxy (AcO-) group at C(7) (steroid numbering) coordinates the enzyme’s Zn2+ (Figure 1) while concomitantly expelling the catalytic water. This effectively disables the enzyme’s proteolytic activity (see ligand interactions diagrams, Figures S1–S3). Importantly, this is a main difference versus previously described models in which no coordinate interaction with the Zn2+ was observed.42 Strong salt bridges and H-bonds between the N-C(24) ammonium cations of the ligands (i.e., from secondary or tertiary nitrogens) and Glu164 and/or Glu55 (Figure 1; see also Figure S1) were observed. Specifically, derivatives possessing secondary ammonium cations engaged in interactions with both Glu164 and Glu55, while derivatives with tertiary ammonium cations, because of steric hindrance, interacted with only with Glu55. These results appear to rationalize the improved inhibitory potencies of compounds possessing secondary ammonium cations versus derivatives with tertiary ammonium cations but which possess the same substituent at the C(3) position: 12 (74.00%) vs 20 (33.28%); 53 (84.80%, IC50 =1.04 μM) vs 55 (81.38%, IC50 =1.13 μM); 52 (89.34%, IC50 = 0.81 μM) vs 54 (89.48%, IC50 = 1.14 μM).
Figure 1.
Compounds 52 (yellow carbons) and 54 (turquoise carbons) docked in the catalytic cleft of the BoNT/A LC with key amino acid residues emphasized in stick. The carbonyl oxygen from the acetoxy group at the cholate C(7) (steroid numbering) coordinates the enzyme’s Zn2+ (not explicitly indicated). For full ligand interaction diagrams see Figure S1 in Supporting Information. Hydrogen bonding is shown for compound 52. The presence of a methyl group (cyan) on the C(24) nitrogen, 54, limits hydrogen bonding to only Glu55.
Another key interaction observed for inhibitors 12–57 was a strong cation−π interaction between the ligand’s 4,7-ACQ moiety and Arg231, with surrounding residues forming a pocket in which the aromatic component engages in hydrophobic collapse (Figures S1–S6).
Favorable interactions were also observed between substituents on the C(3) positions of the inhibitors’ cholic acid components and proximal residues. Importantly, these interactions appear to be heavily weighted with respect to rationalizing the inhibitory efficacies of ligands having neutral and basic C(3) substituents. Ligands with acetoxy and hydroxy groups can engage in an H-bond with Tyr366 (Figure S1 and Figure S2). Moreover, the methyl group from the acetoxy substituent may additionally stabilize the ligand–enzyme binding event through hydrophobic interactions with Phe369 and Tyr366. Hence, these interactions explain the more efficacious inhibition provided by the 3-acetoxy vs 3-hydroxy derivative (cf. 12 vs 17). Ligands possessing a C(3) amino group engage in favorable salt bridges and H-bonds with Glu252 and/or Asp370 (Figure 1; see also Figures S1 and S3). Ammonium cations derived from primary amino groups engage in interactions with both amino acid residues (52, 53, 54, and 55), while derivatives with tertiary nitrogens achieve interactions with only Asp370 (50, 51, 57, and 56). These strong electrostatic interactions could explain the increase in activity for the ligands with basic C(3) substituents when compared to noncharged substituents. Furthermore, ligands with primary ammonium cations can form two stabilizing interactions, while those with tertiary ammonium cations can engage in only one salt bridge. This is in accord with their activities and explains why compounds with the same substitution at C(24) follow the same relative activity patterns: 52 (IC50 = 0.81 μM) vs 50 (IC50 = 1.02 μM); 54 (IC50 = 1.14 μM) vs 56 (IC50 = 2.27 μM). The same relationship was also observed for compounds from the ACQ3 series, e.g., 53 (IC50 = 1.04 μM) vs 51 (IC50 = 1.34 μM).
The above observations can also explain the change in inhibitory activity in the sequence 52 → 50 → 56. In this series of ligands, the introduction of methyl groups on positions C(24) and C(3) leads to a decrease in interaction strength with acidic residues and consequently to a decrease in the activities of the ligands. The experimental results seem to further support the models based on observed decreases in the activities of ligands possessing BocNH or MeSO2NH groups (42, 44 vs 54, and 43, 45 vs 55).
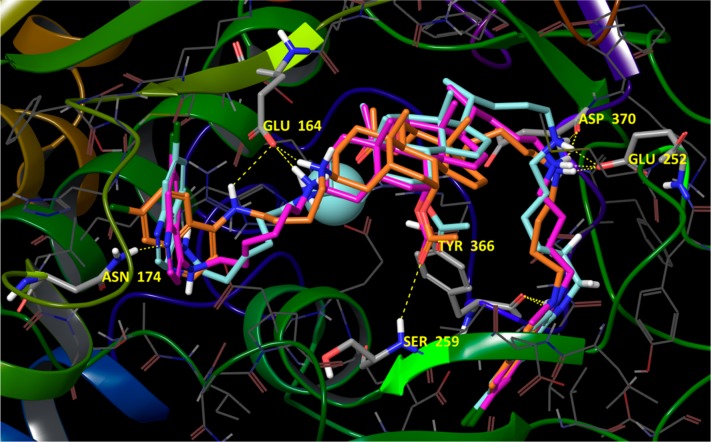
Next we examined derivatives possessesing 4,7-ACQ substituents linked to both positions C(3) and C(24) of the steroidal core. One series possesses an α-bonded ACQ substituent at C(3) (66 and 68, Figure S4), and the other possesses a β-bonded ACQ at C(3) (64, 65, and 67, Figure 2; see also Figure S6). The simulation results indicate that compounds of the α-C(3) series, i.e., 68 and 66, oriented similarly to the members of the mono-ACQ series but with some emerging differences. Most distinctive was a lack of any interactions by the amino group at C(3). For these ligands, three points of binding are most interesting: (1) the carbonyl oxygen of the α oriented acetoxy group at C(7) coordinates the enzyme’s Zn2+ (Figure S4); (2) a salt bridge interaction between the protonated amino group at C(24) and Glu164; (3) a subset involving favorable hydrophobic contacts between the 4,7-ACQ component tethered to the C(3) position of the cholic acid component and a subpocket formed by amino acids Tyr366, Tyr251, Val245, Phe243, Leu263, Phe273, and Leu367. Both 68 and 66 achieve further binding stabilization through π–π stacking with Tyr366, and for 68, an additional H-bond with the hydroxyl substituent of Tyr366 is predicted (Figure S4). In addition, the second 4,7-ACQ ring is stabilized via H-bonding with Thr176. Compound 68 is also stabilized through an H-bond with Ser259. Furthermore, model analysis indicates proximity between the side chain of Pro239 and the quinoline core of the C(24) 4,7-ACQ component. The role of proline interactions with aromatic moieties in the stabilization of small molecule binding has been reported53 (Figure S4).
Figure 2.
Compounds 64 (orange carbons), 65 (purple carbons), and 67 (turquoise carbons) docked in the catalytic cleft of BoNT/A LC with key amino acid residues emphasized in stick. Hydrogen bonding is shown for compound 64. The carbonyl oxygen from the acetoxy group at C(12) (steroid numbering) coordinates the enzyme’s Zn2+ (not explicitly indicated). For full ligand interaction diagrams see Figure S6 in Supporting Information.
β-C(3) derivatives are predicted to dock in the substrate binding cleft in the opposite direction versus α-C(3) derivatives. However, the β-C(3) derivatives retain the three major binding points indicated above: i.e., coordination of the enzyme’s catalytic Zn2+, a salt bridge with Glu164, and hydrophobic stabilization of the 4,7-ACQ aromatic core are all observed. In the β-C(3) series the coordination to Zn2+ is achieved via the carbonyl oxygen of AcO-C(12) and the salt bridge with Glu164 is formed via the protonated amino group at C(3) (Figure 2; see also Figure S6). Favorable hydrophobic interactions are observed between the 4,7-ACQ core linked to C(24) and the pocket composed of the same amino acid residues observed for α-C(3) derivatives. Additional interactions in this area are π–π stacking and H-bonding with Tyr366. New, stabilizing interactions include H-bonds or salt bridges between C(24) ammonium groups and Glu252 and Asp370. For 67 and 65, the second 4,7-ACQ component engages in H-bonding with Asn174; this interaction is possible because of a longer alkyl chain spacer compared to 64. The carbonyl oxygen from the acetate at C(7) of compounds 67, 64, and 65 is positioned within interaction distance with the backbone nitrogen atom of Ser259 (3.14 Å for 67, 2.90 Å for 64, and 3.87 Å for 65), and because of the relative flexibility of the binding site, it is feasible to assume that a hydrogen bond can be formed (Figure 2). The docking scores for these three derivatives followed the same trends as their Ki values (Table 4).
Table 4. Ki Values and Docking Scores and of Bis-AQ Derivatives.
| compd | Ki (μM) | docking scorea |
|---|---|---|
| 64 | 0.300 ± 0.065 | –14.481 |
| 65 | 0.389 ± 0.059 | –13.809 |
| 66 | 0.285 ± 0.056 | –12.932 |
| 67 | 0.103 ± 0.024 | –16.471 |
| 68 | 0.300 ± 0.059 | –13.224 |
A lower score indicates better binding of a given ligand in the substrate cleft.
Examination of an overlap of the docked structures of the α-C(3) series (66 and 68) and β-C(3) series (64, 65, and 67; Figure S5, 3α-derivatives in pink shades, 3β-derivatives in green shades) indicated that the latter is rotated 180° in the binding site around the midpoint of the C(8)–C(9) bond. However, the key functional groups of compounds in both series superimposed well. For example, superimpositions of the compounds’ aliphatic aminos, aromatic aminos, quinolone cores, and acetate groups that coordinate the enzyme’s Zn2+ ion are all uniform. Hence, it appears that differences caused by alterations in the stereochemistry at C(3) are compensated for by the length of the alkyl spacers connecting the 4,7-ACQ fragments and the central cholate components. Finally, the described overlap of the α- and β-series of bis-4,7-ACQ derivatives rationalizes the good activities of diastereomer mixtures 62 and 63.
Antimalarial Activity
All synthesized compounds were screened in vitro against three P. falciparum strains: D6 (CQ susceptible (CQS) strain), W2 (CQ resistant (CQR) strain), and TM91C235 (Thailand, a multidrug-resistant (MDR) strain), following well-established protocols.54 In brief, the malaria SYBR Green I based fluorescence (MSF) assay is a microtiter plate drug sensitivity assay that uses the presence of malarial DNA as a measure of parasitic proliferation in the presence of antimalarial drugs or experimental compounds. The intercalation of SYBR Green I dye, and its resulting fluorescence, is relative to parasite growth, and a compound that inhibits the growth of the parasite will result in lower fluorescence.
The compounds’ antimalarial activities are shown in Table 3. All of the compounds exhibited better IC90 activity against both CQR strains in comparison to CQ, and nine are more potent (IC90) against CQS strain D6 than CQ. When compared to MFQ, seven compounds are more active against CQR strain W2, and 19 are more active against MDR strain TM91C235 (IC90). In addition, three inhibitors are more active than artemisinin (ART) against the W2 and TM91C235 strains: 56, 54, 16, and 56, 54, 42 and 16, respectively (IC90).
Table 3. In Vitro Antimalarial Activities of Compounds Tested against P. falciparum Strains.
| W2a (nM) |
D6b (nM) |
TM91C235c (nM) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| compd | IC50 | IC90 | IC50 | IC90 | IC50 | IC90 | RId | HEPG2 IC50 (nM) | SIe | PBMC IC50 (nM) | HLM/MLM (min)f |
| 12 | 11.38 | 32.50 | 16.87 | 34.21 | 27.74 | 77.92 | 2.28/0.95 | ||||
| 13 | 9.42 | 18.98 | 9.72 | 13.80 | 14.54 | 31.85 | 2.31/1.38 | 980 | 104/101/67 | ||
| 14 | 23.92 | 46.52 | 54.49 | 2565 | 107/55/47 | ||||||
| 15 | 56.38 | 90.97 | 103.78 | 2643 | 47/29/25 | ||||||
| 16 | 6.58 | 10.70 | 2.91 | 5.13 | 6.71 | 13.26 | 2.58/2.08 | 2729 | 415/938/407 | 2814 | 7/30 |
| 17 | 12.76 | 18.79 | 9.30 | 16.46 | 11.92 | 30.27 | 1.84/1.14 | 1068 | 84/115/90 | 8/60 | |
| 18 | 33.78 | 52.08 | 56.31 | 1585 | 47/30/28 | ||||||
| 19 | 33.85 | 36.56 | 40.63 | 2044 | 60/56/50 | ||||||
| 22 | 6.47 | 14.11 | 3.38 | 4.56 | 17.64 | 36.75 | 8.06/3.09 | 1929 | 299/571/109 | 52240 | 6/13 |
| 23 | 6.89 | 12.10 | 6.12 | 10.02 | 9.19 | 18.15 | 1.81/1.21 | 1111 | 161/182/121 | 2607 | 14/6 |
| 24 | 174.92 | 211.63 | 213.74 | 1230 | 7/6/6 | ||||||
| 25 | 42.09 | 47.53 | 51.60 | 1281 | 30/27/25 | ||||||
| 20 | 18.55 | 38.63 | 7.47 | 19.06 | 22.97 | 32.90 | 1.73/2.03 | 1809 | 98/242/79 | ||
| 21 | 11.77 | 16.88 | 6.91 | 10.47 | 22.26 | 47.33 | 4.52/1.61 | 1580 | 134/229/71 | ||
| 36 | 15.21 | 62.21 | 16.59 | 31.80 | 15.21 | 34.56 | 1.09/1.96 | 1922 | 126/116/126 | ||
| 37 | 26.40 | 35.84 | 12.75 | 36.50 | 28.12 | 39.85 | 1.09/0.98 | 1448 | 55/114/51 | ||
| 38 | 44.58 | 71.12 | 19.44 | 53.02 | 26.40 | 45.62 | 0.86/1.34 | 5854 | 131/301/222 | ||
| 39 | 34.44 | 96.36 | 37.51 | 74.71 | 30.29 | 56.22 | 0.75/1.29 | 4463 | 130/119/147 | 11/47 | |
| 40 | 7.34 | 12.31 | 24.12 | 29.47 | 27.60 | 51.93 | 1.76/0.42 | 12797 | 1743/531/464 | ||
| 42 | 12.13 | 16.81 | 5.35 | 10.18 | 6.59 | 12.59 | 1.24/1.65 | 14480 | 1194/2707/2197 | 14/47 | |
| 43 | 30.47 | 54.74 | 31.67 | 55.88 | 20.33 | 28.56 | 0.51/0.98 | 2828 | 93/89/139 | 11/60 | |
| 44 | 8.05 | 11.69 | 8.93 | 10.94 | 16.34 | 22.63 | 2.07/1.07 | 2360 | 293/264/144 | ||
| 50 | 19.73 | 42.29 | 32.42 | 0.77/0.47g | 14096 | 714/333/435 | 60/60 | ||||
| 51 | 36.14 | 81.14 | 100.81 | 138.69 | 88.71 | 165.04 | 1.19/0.58 | 13823 | 382/137/156 | 7838 | |
| 52 | 161.44 | 234.83 | 143.83 | 176.12 | 146.77 | 220.15 | 1.25/1.33 | 5610 | 35/39/38 | 7881 | |
| 53 | 194.60 | 282.49 | 138.49 | 233.61 | 242.74 | 347.28 | 1.49/1.21 | 10419 | 54/75/43 | ||
| 54 | 2.88 | 3.74 | 3.60 | 4.46 | 6.18 | 9.20 | 2.06/0.84 | 14381 | 4993/3995/2327 | 2588 | 60/60 |
| 55 | 110.05 | 152.86 | 112.30 | 150.03 | 100.15 | 128.18 | 0.85/1.02 | 4972 | 45/44/50 | ||
| 56 | 1.35 | 1.35 | 2.35 | 2.63 | 2.90 | 3.32 | 1.26/0.51 | 9212 | 6824/3920/3176 | 2516 | 27/60 |
| 57 | 56.95 | 151.87 | 74.58 | 5278 | 93/35/71 | 60/60 | |||||
| 60 | 78.0 | 105.17 | 86.77 | 113.93 | 85.89 | 175.28 | 1.54/0.92 | 1648 | 21/19/19 | ||
| 59 | 368.93 | 368.93 | 368.93 | 35417 | 96/96/96 | ||||||
| 61 | 217.77 | 413.75 | 244.96 | 321.27 | 297.19 | 312.51 | 0.97/1.29 | 2130 | 10/9/7 | ||
| 52 | 98.19 | 119.64 | 124.15 | 3771 | 38/32/30 | 302 | |||||
| 63 | 108.31 | 143.32 | 194.74 | 4678 | 43/32/24 | ||||||
| 9 (CQ) | 456.20 | 697.97 | 12.27 | 16.11 | 138.82 | 373.25 | 23.17/43.32 | ||||
| MFQ | 4.93 | 15.25 | 15.70 | 39.09 | 36.50 | 134.07 | 3.43/0.39 | ||||
| ARTh | 6.70 | 11.50 | 9.00 | 12.80 | 13.40 | 17.40 | 1.36/0.90 | ||||
P. falciparum Indochina W2 clone.
P. falciparum African D6 clone.
P. falciparum multidrug resistant TM91C235 strain (Thailand).
Resistance index (RI) is defined as the ratio of the IC90 for the resistant versus sensitive strain, TM91C235/D6 and W2/D6, respectively.
Selectivity index (SI) is defined as ratio of the IC50 for HepG2/W2, HepG2/D6, and HepG2/TM91C235, respectively.
Stability of compounds during incubation with human (HLM) and mouse (MLM) liver microsomes.
For this compound index is defined as the ratio of the IC50 for the resistant versus sensitive strain, TM91C235/D6 and W2/D6, respectively;
Average of greater than eight replicates.
In general, ACQ2 derivatives are more active than ACQ3 derivatives, with an increase in the length of the alkyl chain resulting in a further decrease in antimalarial activity (e.g., 14, 15 vs 12, 13, and the compounds may further lose activity via the following set of transformations AcO–C(3) → HO–C(3) → O=C(3)). In contrast to BoNT/A LC inhibitory activity, further analysis of the compounds’ antimalarial activities did not establish a unique pattern of substituent influence on potency. Highly active (IC90 < 5 nM) and appreciably active (IC90 < 10 nM) compounds possess C(3)-amino, C(3)-OH, and C(3)-N-Boc substituents on the steroidal core; the same holds for the least active compounds (IC90 > 100 nM). However, alkylation of the secondary aliphatic amino group at C(24) (yielding a tertiary amine) favors antimalarial activity, as 56 and 54 are the most active derivatives. In general, the antimalarial behavior of the new derivatives suggests that the contribution of substituents to antimalarial activity is more complex than appears at first glance. In this regard, the compounds will be submitted for thorough analysis of their respective physicochemical parameters.
The resistance index (RI, Table 3) clearly indicates that most of the compounds are more active against the CQS D6 strain than against the CQ resistant strains. The above results clearly indicate that the mode of action of the steroid-derived aminoquinolines is similar to that of 9 (i.e., CQ), although the compounds are much more active than the parent compound, as shown in Table 3. This observation is supported by the results of a β-hematin inhibition experiment. Specifically, derivative 23 was selected for the evaluation of its ability to inhibit β-hematin formation using a β-hematin inhibitory activity assay (BHIA). Subsequently, it was found that 23 has an IC50 that is 3 times lower than that of 9 (CQ) (i.e., 23 (IC50 = 0.399) vs 9 (IC50 = 1.09)), thereby demonstrating a significantly better ability to interact with hematin than 9.24 Hence, these results indicate that our present compounds, like other ACQ-based antimalarials, most likely interfere with the heme detoxification mechanism of malaria.
In general, the examined derivatives showed low metabolic stability (Table 3). Out of the complete set of compounds, 57, 50, and 54 showed desirable in vitro stability, with t1/2 > 60 min in both human liver microsomes (HLMs) and mouse liver microsomes (MLMs). Somewhat less stable were derivatives 57, 43, and 17, which are stable against MLM but are moderately stable (56) or insufficiently stable (43 and 17) against HLM.
The toxicity of the compounds was estimated using human liver carcinoma cell line (HepG2) and peripheral blood mononuclear cells (PBMC). In general, the HEPG2 assay for cytotoxicity estimation revealed that almost all of the compounds are well tolerated by this cell line, possessing IC50 ≥ 1000 nM. In addition, high selectivity indices (SIs) were calculated for the most promising antimalarials 54 and 56 (SI > 2300). In addition, 50, 42, 40, 22, and 16 also displayed low toxicity (Table 3).
Of the tested compounds, the results indicate that 54 and 56 are the most prominent antimalarial agents. The only difference between the two compounds is substitution at the C(3) amino group: NH2 (54) and (CH3)2N (56). Derivative 56 showed somewhat better in vitro activity against all examined malarial strains (IC50, IC90); however, lower toxicity against HepG2 (54, IC50 = 14 380 nM, SI = 5000–2326; 56, IC50 = 9212 nM, SI = 6823–3177) and better metabolic stability against HLM led to the selection of 54 for in vivo testing.
Compound 54 was administrated po at 40 and 160 mg kg–1 day–1 on day 3, day 4, and day 5 after infection using the Thompson test in ICR female mice with 107 × 1 parasites infected with rodent P. berghei strain.24 Under both treatments mice died on day 13 postinfection. All mice died of malaria, and no signs of tissue damage caused by compound toxicity were observed upon necropsy. Unfortunately, this is one of many situations when compound activity in a living organism does not match excellent in vitro activity.
Conclusions
Second generation 4,7-ACQ-cholate-based inhibitors of the BoNT/A LC that are significantly more potent than initially discovered leads were described.42,43 The introduction of a basic amino group at the C(3) position of the cholic acid component markedly increased inhibitor potencies (IC50 = 0.81–2.27 μM). The observation that the substrate binding cleft of the BoNT/A LC is very flexible48 indicates that the enzyme can conformationally adapt to accept large molecules.49 This led us to evaluate two additional subclasses: bis(steroidal)-4,7ACQ derivatives and bis(4,7-ACQ)cholic acid derivatives. The bis(4,7-ACQ)cholic acid derivatives were found to possess Ki values of <0.4 μM, and compound 67 was the most potent derivative (inhibition (20 μM) of 95%, Ki = 0.103 μM). Compounds tested during BoNT/A challenge in primary neurons were found to protect SNAP-25 by up to 89%. In the presence of 5 μL of 100 nM solution of Baf, SNAP-25 protection was slightly higher; however, upon the addition of 25 μL of 100 nM solution of Baf, no further increase in protection was observed. These results suggest that our inhibitors can successfully protect SNAP-25 in the presence of the BoNT/A holotoxin, with only slight additive effects when given in combination with Baf. Docking simulations rationalized the BoNT/A LC inhibitory activities of the compounds in two main terms: (1) coordination of a carbonyl acetate oxygen to Zn2+ and the expulsion of the catalytic water and (2) formation of strong interactions between ligands and catalytically important amino acid residues, including salt bridges with Asp370 and Glu164 and π–π stacking/H-bonding with Tyr366.
With respect to antimalarial activity, all of the compounds exhibited better IC90 potencies against CQR strains versus CQ, and nine compounds are more potent (IC90) against CQS strain D6 than CQ. When compared to MFQ, seven compounds are more active against CQR strain W2, and 19 are more active against MDR strain TM91C235 (IC90). The two most active compounds exhibit very high in vitro activity against CQR strain W2 (1.35 nM and 3.74 nM, respectively); at the same time both compounds are very active against the BoNT/A LC (2.27 μM, 1.14 μM). However, it is disappointing that compound 54, which possesses excellent in vitro antimalarial activity, failed to protect/cure mice when administered at 40 and 160 mg kg–1 day–1 in a Thompson test.
Experimental Section
Chemistry
Melting points were determined on a Boetius PMHK or a Mel-Temp apparatus and were not corrected. Optical rotations were measured on a Rudolph Research Analytical automatic polarimeter, Autopol IV, in dichloromethane (DCM) or methanol (MeOH) as solvent. IR spectra were recorded on a Perkin-Elmer spectrophotometer FT-IR 1725X. 1H and 13C NMR spectra were recorded on a Varian Gemini-200 spectrometer (at 200 and 50 MHz, respectively) and on a Bruker Ultrashield Advance III spectrometer (at 500 and 125 MHz, respectively) employing indicated solvents (vide infra) using TMS as the internal standard. Chemical shifts are expressed in ppm (δ) values and coupling constants (J) in Hz. ESI mass spectra were recorded on an Agilent Technologies 6210 time-of-flight LC–MS instrument in positive ion mode with CH3CN/H2O 1/1 with 0.2% HCOOH as the carrying solvent solution. Samples were dissolved in CH3CN or MeOH (HPLC grade purity). The selected values were as follows: capillary voltage = 4 kV, gas temperature = 350 °C, drying gas = 12.l min–1, nebulizer pressure = 45 psig, fragmentator voltage = 70 V. The elemental analysis was performed on the Vario EL III C,H,N,S/O elemental analyzer (Elementar Analysensysteme GmbH, Hanau, Germany). Thin-layer chromatography (TLC) was performed on precoated Merck silica gel 60 F254 and RP-18 F254 plates. Column chromatography was performed on Lobar LichroPrep Si 60 (40–63 μm) and RP-18 (40–63 μm) columns coupled to a Waters RI 401 detector and on Biotage SP1system with UV detector and FLASH 12+, FLASH 25+, or FLASH 40+ columns charged with KP-SIL (40–63 μm, pore diameter 60 Å), KP-C18-HS (40–63 μm, pore diameter 90 Å), or KP-NH (40–63 μm, pore diameter 100 Å) as adsorbent. Compounds were analyzed for purity (HPLC) using a Waters 1525 HPLC dual pump system equipped with an Alltech Select degasser system, and dual λ 2487 UV–vis detector. For data processing, Empower software was used (methods A and B). Methods C and D involved use of Agylent Technologies 1260 liquid chromatograph equipped with quaternary pump (G1311B), injector (G1329B), 1260 ALS, TCC 1260 (G1316A), and detector 1260 DAD VL+ (G1315C). For data processing, LC OpenLab CDS ChemStation software was used. For details, see Supporting Information.
N1-(7-Chloroquinolin-4-yl)ethane-1,2-diamine (ACQ2), N1-(7-chloroquinolin-4-yl)propane-1,3-diamine (ACQ3), N1-(7-chloroquinolin-4-yl)butane-1,4-diamine (ACQ4), and N1-(7-chloroquinolin-4-yl)ethane-1,6-hexane (ACQ6) were prepared according to known procedures.55
N-(3α,7α,12α-Triacetoxy-5β-cholan-24-yl)-N′-(7′-chloroquinolin-4′-yl)ethane-1,2-diamine (12)
Alcohol 11 (1.08g, 2.07 mmol) was dissolved in DCM (100 mL). PCC (670 mg, 3.11 mmol) was added, and the mixture was stirred at room temperature for 3.5 h. Reaction mixture was filtered through a short column of SiO2 (eluent DCM/EA = 7/3). Crude aldehyde was dissolved in DCM (150 mL). ACQ2 (1.06 g, 4.78 mmol) was added, and mixture was stirred 15 min, followed by addition of NaBH(OAc)3 (1.01 g, 4.76 mmol), and stirring was continued at room temperature overnight. The mixture was transferred into a separatory funnel. Water was added, and following the intensive shaking, the water layer was adjusted to pH ≈ 12 with 0.5 M NaOH solution. Water layer was extracted 2 times more with DCM. Combined organic layers were rinsed with brine and dried over anhydrous Na2SO4. Solvent was removed under reduced pressure and crude mixture was prepared for column purification (dry flash, SiO2, eluent EA, EA/MeOH gradient 95/5 → 9/1, EA/MeOH/NH3 gradient 18/0.1/0.1 → 18/0.7/0.7 and flash chromatography (Biotage SP1 RP column, eluent MeOH/H2O gradient 8/2 → 9/1)]. Yield 793.4 mg (53%). Spectral and analytical data for compound 12 were identical to the ones already described.43 HPLC purity, method A: tR = 8.583, area 95.04%. Method B: tR = 8.723, area 96.87%.
N-(3α-Hydroxy-7α,12α-diacetoxy-5β-cholan-24-yl)-N′-(7′-chloroquinolin-4′-yl)ethane-1,2-diamine (16)
Mixture of 12 (501 mg, 0.69 mmol) and anhydrous K2CO3 (169 mg, 1.22 mmol) in dry MeOH (14 mL) was stirred at room temperature for 3 h and poured onto ice/water mixture. Solid was filtered off and dried in vacuum oven. Yield 460 mg (97%). Colorless foam softens at 118–121 °C. [α]D20 +0.040 (c 1.4 × 10–3 g/mL, DCM). IR (KBr): 3309w, 2930w, 2863w, 1715m, 1612w, 1580s, 1535w, 1449w, 1373m, 1332w, 1241s, 1138w, 1074m, 1019m, 967w, 937w, 897w, 878w, 850w, 805w, 767w cm–1. 1H NMR (500 MHz, CDCl3, δ): 8.51 (d, J = 5.1, H-C(2′)), 7.94 (d, J = 1.8, H-C(8′)), 7.74 (d, J = 9.1, H-C(5′)), 7.36 (dd, J1 = 9.1, J2 =2.1, H-C(6′)), 6.38 (d, J = 5.4, H-C(3′)), 5.95 (bs, H-N), 5.11–5.07 (m, H-C(12)), 4.92–4.88 (m, H-C(7)), 3.55–3.47 (m, H-C(3)), 3.36–3,30 (m, 2H-C(9′)), 3.02 (t, J = 5.6, 2H-C(10′)), 2.67–2.57 (m, 2H-C(24)), 2.09 (s, CH3COO), 2.07 (s, CH3COO), 0.90 (s, CH3-C(10)), 0.82 (d, J = 6.5, CH3-C(20)), 0.71 (s, CH3-C(13)). 13C NMR (125 MHz, CDCl3, δ): 170.61, 170.58, 151.97, 149.91, 149.03, 134.80, 128.58, 125.18, 121.34, 117.34, 99.12, 75.43, 71.56, 70.82, 49.75, 47.62, 47.48, 45.02, 43.39, 41.97, 41.01, 38.65, 37.74, 34.94, 34.84, 34.25, 33.28, 31.33, 30.49, 28.94, 27.29, 26.69, 25.55, 22.77, 22.52, 21.60, 21.40, 17.88, 12.20. (+)ESI-HRMS (m/z): [M + 2H]2+ 341.7034 (error, −2.20 ppm), [M + H]+ 682.3980 (error, −0.22 ppm). Combustion analysis (C39H56ClN3O5·H2O): calculated C 66.88, H 8.35, N 6.00; found C 66.51, H 7.82, N 6.10. HPLC purity, method A: tR = 8.651, area 96.19%. Method B: tR = 8.626, area 95.11%.
N-Methyl-N-(3α,7α,12α-triacetoxy-5β-cholan-24-yl)-N′-(7′-chloroquinolin-4′-yl)ethane-1,2-diamine (20)
Mixture of 12 (200 mg, 0.28 mmol), 37% formaldehyde (68.3 μL, 0.84 mmol), NaBH(OAc)3 (237 mg, 1.12 mmol) in DCM (9 mL) was stirred at room temperature until disappearance of starting compound (TLC, SiO2, EA/MeOH/NH3 = 9:1:1). Reaction mixture was transferred into separatory funnel. DCM (20 mL) and water (10 mL) were added. pH of water layer was adjusted to pH 12 using 0.5 M NaOH, and the mixture was worked up in the usual manner. Product was isolated after column chromatography (dry flash, SiO2, gradient hexane/EA 4:6 → EA). Yield 163 mg (80%). Colorless foam softens at 74–78 °C. [α]D20 +0.089 (c 1.3 × 10–3 g/mL, DCM). IR (KBr): 3276w, 2947m, 2872w, 1732s, 1678s, 1614m, 1456w, 1379m, 1333w, 1251s, 1201s, 1134m, 1025w, 966w, 889w, 832w, 801w cm–1. 1H NMR (500 MHz, CDCl3, δ): 8.53 (d, J = 5.0, H-C(2′)), 7.96 (d, J = 2.0, H-C(8′)), 7.68 (d, J = 9.0, H-C(5′)), 7.37 (dd, J1 = 2.0, J2 = 9.0, H-C(6′)), 6.37 (d, J = 5.5, H-C(3′)), 6.08 (bs, H-N, exchangeable with D2O), 5.10 (s, H-C(12)), 4.95–4.90 (m, H-C(7)), 4.61–4.54 (m, H-C(3)), 3.31–3.27 (m, 2H-C(9′)), 2.77–2.30, (m, 2H-C(10′)), 2.47–2.34 (m, 2H-C(24)), 2.28 (s, CH3-N), 2.08 (s, CH3COO), 2.07 (s, CH3COO), 2.05 (s, CH3COO), 0.92 (s, CH3-C(10)), 0.83 (d, J = 6.6, CH3-C(20)), 0.72 (s, CH3-C(13)). 13C NMR (125 MHz, CDCl3, δ): 170.52, 170.47, 170.35, 152.05, 149.85, 149.01, 134.79, 125.13, 121.24, 117.35, 99.25, 75.41, 74.04, 70.67, 57.85, 55.15, 47.93, 45.08, 43.52, 41.26, 40.91, 39.60, 37.73, 35.24, 34.64, 34.59, 34.28, 33.59, 31.89, 31.21, 29.66, 28.95, 27.42, 26.85, 25.60, 24.15, 22.80, 22.54, 21.58, 21.47, 21.34, 17.98, 12.23. (+)ESI-HRMS (m/z): [M + H]+ 738.42320 (error, −1.55 ppm). Combustion analysis for (C42H60ClN3O6·1.5H2O): calculated C 65.91, H 8.30, N 5.49; found C 65.86, H 8.33, N 5.40. HPLC purity, method B: tR = 8.703, area 96.21%. Method C: tR = 13.199, area 95.18%.
N-(3-Oxo-7α,12α-diacetoxy-5β-cholan-24-yl)-N′-(7′-chloroquinolin-4′-yl)ethane-1,2-diamine (22)
Solution of 16 (460 mg, 0.64 mmol) and IBX56 (896 mg, 3.2 mmol) was dissolved in DMSO. CF3CO2H (104.4 μL) was added, and the mixture was stirred at room temperature. After 6 h mixture was poured onto water and pH was adjusted to pH 11 using NaOH. The obtained solid was filtered off, washed with H2O, and dried in vacuo. Crude product was purified by column chromatography (dry flash, SiO2, eluent EA/MeOH gradient 95/5 → 9/1, EA/MeOH/NH3 gradient 18/1/1 → 9/1/1) and flash chromatography (Biotage SP1 RP column, eluent MeOH/H2O gradient 8/2 → 9/1). Yield 367 (80%). Colorless foam softens at 75–78 °C. [α]D20 +0.069 (c 1.3 × 10–3 g/mL, DCM). IR (ATR): 2933w, 2861w, 2610w, 1714m, 1609w, 1578m, 1532w, 1443w, 1374w, 1330w, 1239s, 1169w, 1137w, 1073w, 1023w, 964w, 877w, 847w, 813w, 767w cm–1. 1H NMR (500 MHz, CDCl3, δ): 8.52 (d, J = 5.3, H-C(2′)), 7.95 (d, J = 2.1, H-C(8′)), 7.71 (d, J = 8.9, H-C(5′)), 7.35 (dd, J1 = 8.9, J2 = 2.1, H-C(6′)), 6.38 (d, J = 5.5, H-C(3′)), 5.91 (bs, H-N), 5.14 (bs, H-C(12)), 5.03–4.96 (m, H-C(7)), 3.37–3.29 (m, 2H-C(9′)), 3.06–2.94 (m, Hα-C(4) and 2H-C(10′), 2.68–2.57 (m, 2H-C(24)), 2.09 (s, CH3COO), 2.07 (s, CH3COO), 1.02 (s, CH3-C(10)), 0.84 (d, J = 6.6, CH3-C(20)), 0.75 (s, CH3-C(13)). 13C NMR (125 MHz, CDCl3, δ): 212.05, 170.37, 170.09, 152.02, 149.84, 149.07, 134.74, 128.66, 125.13, 121.24, 117.33, 99.15, 75.24, 70.55, 49.69, 47.67, 47.45, 45.05, 44.52, 43.22, 42.22, 42.10, 41.91, 37.71, 36.57, 36.08, 34.94, 34.35, 33.27, 30.87, 29.77, 27.24, 26.74, 25.79, 22.77, 21.59, 21.42, 21.28, 17.90, 12.22. (+)ESI-HRMS (m/z): [M + 2H]2+ 340.6947 (error, −0.49 ppm), [M + H]+ 680.3812 (error, −1.86 ppm). Combustion analysis (C39H54ClN3O5): calculated C 68.85, H 8.00, N 6.18; found C 68.35, H 8.33, N 6.32. HPLC purity, method A: tR = 8.484, area 95.76%. Method B: tR = 7.775, area 95.05%. Method C: tR = 10.069, area 95.10%.
24-N-[(7-Chloroquinoline-4-yl)amino]ethylamino-3α-acetamido-7α,12α-diacetoxy-5β-cholane (36)
Mixture of alcohol 33 (200 mg, 0.38 mmol) and PCC (123 mg, 0.57 mmol) in dry DCM (40 mL) was stirred at room temperature for 3 h. Reaction mixture was filtered through a short chromatography column. Fractions that contained desired aldehyde were collected (dry flash, SiO2, eluent DCM/EA = 7/3). To the stirred mixture (10 min) of crude aldehyde obtained above, ACQ2 (193 mg, 0.874 mmol) in DCM (20 mL) at room temperature NaBH(OAc)3 (185 mg, 0.874 mmol) was added, and stirring was continued at room temperature for 6 h, followed by additional amount of NaBH(OAc)3 (2.75 mg, 13 mmol). After the reaction was completed Et3N was added, solvent was removed under reduced pressure, and product was isolated after column chromatography (dry flash, SiO2, EA, EA/MeOH gradient 95:5 → 9:1, EA/MeOH/NH3 = 18:0.1:0.1 → 18:0.7:0.7, Lobar Lichroprep B RP MeOH/H2O = 9:1). Yield 112.7 mg (40%). Colorless foam softness at 174–176 °C. [α]D20 +0.036 (c 1.0 × 10–3 g/mL, DCM). IR (KBr): 3273m, 2950m, 2869m, 1716m, 1634m, 1614m, 1582s, 1539m, 1449m, 1377m, 1333w, 1250m, 1142w, 1024w, 969w, 817w, 729w, cm–1. 1H NMR (200 MHz, CDCl3, δ): 8.31 (d, J = 5.7, H-C(2′)), 8.11 (d, J = 9.0, H-C(5′)), 7.77 (d, J = 1.7, H-C(8′)), 7.28 (dd, J1 = 1.6, J2 = 8.8, H-C(6′)), 6.29 (d, J = 5.7, H-C(3′)), 5.58 (d, J = 7.7, H-N), 5.06 (bs, H-C(12)), 4.89 (bs, H-C(7)), 3.60 (bs, H-C(3) and 2H-C(9′)), 3.23 (bs, 2H-C(10′)), 2.83 (bs, 2H-C(24)), 2.08 (s, CH3COO), 2.07 (s, CH3COO), 1.96 (s, CH3CONH), 0.90 (bs, CH3-C(10)), 0.80 (d, J = 6.2, CH3-C(20)), 0.69 (bs, CH3-C(13)). 13C NMR (50 MHz, CDCl3, δ): 170.38, 170.24, 169.33, 151.12, 149.39, 146.17, 136.03, 125.81, 123.30, 116.80, 98.63, 75.43, 70.77, 49.65, 49.42, 47.52, 46.69, 45.01, 43.32, 41.39, 40.84, 37.62, 36.07, 35.36, 34.76, 34.21, 32.90, 31.25, 28.86, 28.04, 27.26, 25.51, 24.56, 23.53, 22.67, 21.63, 21.36, 17.72, 12.15. 1H NMR (500 MHz, CDCl3, δ): 8.27 (d, J = 5.5, H-C(2′)), 8.24 (d, J = 8.9, H-C(5′)), 7.81(s, H-C(8′)), 7.35–7.30 (m, H-C(6′)), 6.27 (d, J = 6.0, H-C(3′)), 5.37 (d, J = 7.8, H-N), 5.07 (bs, H-C(12)), 4.90 (d, J = 2.3, H-C(7)), 3.90–3.40 (m, H-N, exchangeable with D2O, H-C(3) and 2H-C(9′)), 3.33–3.23 (m, 2H-C(10′)), 2.93–2.80 (m, 2H-C(24)), 2.10 (s, CH3COO), 2.08 (s, CH3COO), 1.97 (s, CH3CONH), 0.91 (bs, CH3-C(10)), 0.82 (d, J = 6.4, CH3-C(20)), 0.70 (bs, CH3-C(13)). 13C NMR (125 MHz, CDCl3, δ): 170.38, 170.20, 169.24, 151.49, 148.68, 145.44, 136.43, 126.05, 125.19, 123.61, 116.66, 98.55, 75.43, 70.79, 49.66, 49.45, 47.54, 46.55, 45.05, 43.37, 41.43, 40.82, 37.67, 36.14, 35.42, 34.81, 34.26, 32.92, 31.28, 29.64, 28.90, 28.09, 27.30, 25.56, 24.32, 23.59, 22.73, 21.68, 21.41, 17.76, 12.21. (+)ESI-HRMS (m/z): [M + H]+ 745.406 89 (error, −0.26 ppm). Combustion analysis (C41H59N4O5): calculated C 68.07, H 8.22, N 7.75; found C 68.17, H 8.19, N 7.51. HPLC purity, method A: tR = 2.088, area 98.43%. Method C: tR = 12.072, area 99.24%.
24-N-[(7-Chloroquinoline-4-yl)amino]ethylamino-3α-N′-tert-butylcarbamate-7α,12α-diacetoxy-5β-cholane (40) and N,N-Di(3α-N″-tert-butylcarbamate-7α,12α-diacetoxy-5β-cholan-24-yl)-N′-(7′-chloroquinoline-4′-yl)-1,3-ethanediamine (58)
Compounds 40 and 58 were obtained according to procedure described for 36, using alcohol 35 as starting material (271 mg, 0.468 mmol). Column chromatography (dry flash, SiO2, EA, EA/MeOH gradient 9:1 → 6:4, EA/MeOH/NH3 = 18:0.5:0.5 → 18:1.5:1.5, Lobar Lichroprep RP column MeOH/H2O = 9:1). Yield 123 mg (34%) of 40 and 168 mg (26%) of 58.
40. Colorless foam softens at 112–116 °C. [α]D20 +0.057 (c 1.0 × 10–3 g/mL, DCM). IR (KBr): 3343w, 2932m, 2866w, 1712s, 1610w, 1579s, 1527m, 1448m 1367s, 1330w, 1235s, 1167m, 1137m, 1062w, 1021m, 998m, 964w, 937w, 879w, 850w, 804w, 766w, 720w cm–1. 1H NMR (500 MHz, CDCl3, δ): 8.51 (d, J = 5.1, H-C(2′)), 7.94 (d, J = 2.2, H-C(8′)), 7.72 (d, J = 8.8, H-C(5′)), 7.35 (dd, J1 = 9.0, J2 = 2.0, H-C(6′)), 6.38 (d, J = 5.5, H-C(3′)), 5.95 (bs, N-H), 5.10–5.07 (m, H-C(12)), 4.91–4.89 (m, H-C(7)), 3.35–3.32 (m, 2H-C(9′)), 3.28 (bs, H-C(3)), 3.03 (t, J = 5.7, 2H-C(10′)), 2.70–2.55 (m, 2H-C(24)), 2.10 (s, CH3COO), 2.06 (s, CH3COO), 1.44 (s, Boc-N), 0.91 (s, CH3-C(10)), 0.82 (d, J = 6.2, CH3-C(20)), 0.71 (bs, CH3-C(13)). 13C NMR (125 MHz, CDCl3, δ): 170.37, 170.25, 151.99, 149.88, 149.04, 134.80, 128.63, 125.18, 121.29, 117.34, 99.15, 75.45, 70.86, 65.81, 49.71, 47.67, 47.43, 45.03, 43.36, 41.89, 41.53, 37.71, 36.40, 35.48, 34.96, 34.24, 33.30, 31.29, 28.85, 28.40, 27.31, 26.67, 25.50, 22.80, 22.69, 21.61, 21.37, 17.91, 15.24, 12.20. (+)ESI-HRMS (m/z): [M + H]+ 781.464 78 (error, −2.25 ppm). Combustion analysis for (C44H65ClN4O6·H2O): calculated C 66.10, H 8.45, N 7.01; found C 65.93, H 8.60, N 6.12. HPLC purity, method A: tR = 2.085, area 99.22%. Method B: tR = 8.856, area 95.25%.
58. Colorless foam softens at 148–153 °C. [α]D20 +0.106 (c 1.1 × 10–3 g/mL, DCM). IR (KBr): 3365w, 2934m, 2867w, 1712s, 1610w, 1580m, 1520mw, 1448w, 1366s, 1234s, 1166m, 1061w, 1021m, 998w, 965w, 938w, 880w, 851w, 804w, 721w cm–1. 1H NMR (500 MHz, CDCl3, δ): 8.54 (d, J = 5.1, H-C(2′)), 7.97 (d, J = 1.9, H-C(8′)), 7.67 (d, J = 8.6, H-C(5′)), 7.35 (dd, J1 = 8.8, J2 = 1.8, H-C(6′)), 6.37 (d, J = 5.4, H-C(3′)), 6.11 (bs, N-H), 5.08 (bs, 2 × H-C(12)), 4.92–4.87 (m, 2 × H-C(7)), 3.27 (bs, 2H-C(9′) and 2 × H-C(3)), 2.88–2.76 (m, 2H-C(10′)), 2.53–2.40 (m, 2 × 2H-C(24)), 2.06 (s, 2 × CH3COO), 2.05 (s, 2 × CH3COO), 1.45 (s, 2 × Boc-N), 0.91 (s, 2 × CH3-C(10)), 0.80 (d, J = 6.7, CH3-C(20)), 0.69 (bs, 2 × CH3-C(13)). 13C NMR (125 MHz, CDCl3, δ): 170.34, 170.25, 155.10, 151.98, 149.82, 148.93, 134.83, 128.65, 125.16, 121.18, 117.35, 99.27, 79.20, 75.39, 70.83, 53.89, 51.76, 50.79, 47.94, 45.06, 43.37, 41.53, 39.70, 37.69, 36.39, 35.48, 35.18, 34.24, 33.56, 31.28, 28.85, 28.40, 27.34, 25.51, 23.90, 22.78, 22.69, 21.61, 21.30, 17.97, 12.22. (+)ESI-HRMS (m/z): [M + 2H]2+ 670.929 92 (error, −0.94 ppm). Combustion analysis for (C77H118ClN5O12·H2O): calculated C 68.04, H 8.90, N 5.15; found C 67.84, H 9.05, N 5.02. HPLC purity, method C: tR = 11.618, area 99.15%. Method D: tR = 6.020, area 96.86%.
24-N-[(7-Chloroquinoline-4-yl)amino]propylamino-3α-N′-tert-butylcarbamate-7α,12α-diacetoxy-5β-cholane (41) and N,N-Di(3α-amino-tert-butylcarbamate-7α,12α-diacetoxy-5β-cholan-24-yl)-N′-(7′-chloroquinoline-4′-yl)-1,3-propanediamine (59)
Procedure with NaBH4
Mixture of alcohol 35 (3.96 g, 6.84 mmol) and PCC (2.12 g, 9.86 mmol) in dry DCM (349 mL) was stirred at room temperature for 3 h. Mixture was purified through a short chromatography column. Fractions containing aldehyde were collected (dry flash, SiO2, eluent DCM/EA = 7/3; TLC, SiO2, EA/Hex = 1/1, visualization by CAM). The obtained crude aldehyde was dissolved in dry MeOH (57 mL). ACQ3 (1.48 g 6.3 mmol) was added, and resulting mixture was stirred at room temperature for 12 h. NaBH4 (477 mg, 12.6 mmol) was added in one portion, and stirring was continued for 1 h. The solvent was removed under reduced pressure, and product was isolated upon column chromatography [dry flash, SiO2, eluent Hex/EA gradient 1/1 → 2/8, EA, EA/MeOH gradient 95/5 → 6/4, EA/MeOH/NH3 gradient 18/0.5/0.5 → 18/2.1/2.1 and flash chromatography (Biotage SP1 RP column, eluent MeOH/H2O gradient 7/3 → MeOH)].
41. Yield 2.24 g (41%). Colorless foam softness at 99–107 °C. [α]D20 +0.125 (c 1.4 × 10–3 g/mL, DCM). IR (KBr): 3313m, 2937m, 2868m, 1717s, 1613m, 1582s, 1536m, 1452m, 1372m, 1248s, 1211m, 1173m, 1137m, 1064w, 1024m, 985w, 848w, 852w, 803w, 768w, 608w cm–1. 1H NMR (200 MHz, CDCl3, δ): 8.50 (d, J = 5.6, H-C(2′)), 7.93 (d, J = 2.2, H-C(8′), 7.87 (s, H-N-Boc, exchangeable with D2O), 7.74 (d, J = 9.0, H-C(5′), 7.34 (dd, J1 = 9.0, J2 = 2.2 H-C(6′)), 6.31 (d, J = 5.6, H-C(3′)), 5.11 (bs, H-C(12)), 4.94/4.85 (m, H-C(7)), 4.39 (bs, H-N), 3.45–3.20 (m, 2H-C(9′) and H-C(3)), 3.00–2.85 (m, 2H-C(11′)), 2.73/2.56 (m, 2H-C(24)), 2.11 (s, CH3COO), 2.03 (s, CH3COO), 1.45 (s, (CH3)3C-N(Boc)), 0.91 (s, CH3-C(10)), 0.86 (d, J = 6.2, CH3-C(20)), 0.74 (s, CH3-C(13)). 13C NMR (50 MHz, CDCl3, δ): 170.49, 170.40, 155.20, 152.18, 150.57, 149.14, 134.53, 128.53, 124.65, 122.17, 117.60, 98.27, 79.22, 75.49, 70.79, 50.56, 49.65, 47.47, 45.01, 44.14, 43.50, 41.50, 37.65, 36.31, 35.45, 34.92, 34.20, 33.32, 31.23, 28.92, 28.39, 27.28, 27.19, 26.53, 25.53, 22.69, 21.54, 21.38, 17.90, 12.20. 1H NMR (500 MHz, CDCl3, δ): 8.49 (d, J = 5.3, H-C(2′)), 7.93 (d, J = 2.1, H-C(8′), 7.83 (s, H-N-Boc, exchangeable with D2O), 7.75 (d, J = 8.9, H-C(5′), 7.34 (dd, J1 = 8.8, J2 = 1.9 H-C(6′)), 6.31 (d, J = 5.5, H-C(3′)), 5.10 (bs, H-C(12)), 4.93–4.86 (m, H-C(7)), 4.39 (bs, H-N), 3.40 (t, J = 5.7, 2H-C(9′)), 3.28 (bs, H-C(3)), 2.97–2.89 (m, 2H-C(11′)), 2.73–2.56 (m, 2H-C(24)), 2.10 (s, CH3COO), 2.02 (s, CH3COO), 1.45 (s, (CH3)3C-N(Boc)), 0.91 (s, CH3-C(10)), 0.85 (d, J = 6.4, CH3-C(20)), 0.73 (s, CH3-C(13)). 13C NMR (125 MHz, CDCl3, δ): 170.42, 170.32, 151.94, 150.62, 148.93, 134.65, 128.35, 124.73, 122.20, 117.56, 98.26, 75.49, 70.82, 50.49, 49.46, 47.54, 45.06, 43.97, 43.50,41.55, 37.71, 36.36, 35.49, 34.97, 34.24, 33.35, 31.28, 28.94, 28.41, 27.32, 27.10, 26.44, 25.57, 22.78, 22.71, 21.56, 21.38, 17.93, 12.24. (+)ESI-HRMS (m/z): [M + H]+ 795.4822 (error, +0.06 ppm). Combustion analysis for (C45H67ClN4O6·0.5H2O): calculated C 67.18, H 8.52, N 6.96; found C 67.01, H 8.42, N 6.74. HPLC purity, method A: tR = 1.952, area 98.07%. Method C: tR = 15.915, area 98.61%.
Procedure with NaBH(OAc)3
Into stirred mixture of crude aldehyde (3.80 g, 6,6 mmol) and ACQ3 (3.11 g, 13 mmol) in DCM (126 mL) for 10 min at room temperature, NaBH(OAc)3 (2.8 mg, 13 mmol) was added. Stirring was continued for 12 h, followed by additional amount of NaBH(OAc)3 (2.8 mg) and stirring for 3 h. Then the solvent was removed under reduced pressure and products were isolated upon column chromatography [dry flash (SiO2, eluent EA, EA/MeOH gradient of 95/5 → 9/1, EA/MeOH/NH3 gradient of 18/0.5/0.5 → 9/1/1, and flash chromatography (Biotage SP1, RP column, eluent MeOH/H2O gradient 75/25 → 95/5, N-H column, eluent EA/Hex gradient 6/3 → EA). Yield 1.44 g of 41 (20%) and 800 mg of bis-steroidal amine 59 (13%).
59. Colorless foam softens at 149–153 °C. [α]D20 +0.131 (c 1.6 × 10–3 g/mL, DCM). IR (KBr): 3392m, 2950s, 2869m, 1735s, 1612m, 1581s, 1529m, 1450m, 1372s, 1244s, 1171s, 1065m, 1023m, 965m, 939w, 882w, 852w cm–1. 1H NMR (500 MHz, CDCl3, δ): 8.50 (d, J = 5.4, H-C(2′)), 7.93 (d, J = 1.9, H-C(8′)), 7.75–7.65 (m, H-C(5′) and H-N-Boc, exchangeable with D2O), 7.33 (dd, J1 = 8.8, J2 = 1.9, H-C(6′)), 6.32 (d, J = 5.4, H-C(3′)), 5.05 (bs, 2 × H-C(12)), 4.88 (d, i = 2.2, 2 × H-C(7)), 4.45 (bs, H-N), 3.45–3.20 (m, 2H-C(9′) and 2 × H-C(3)), 2.72–2.58 (m, 2H-C(11′)), 2.54–2.40 (m, 2 × 2H-C(24)), 2.04 (s, 4 × CH3COO), 1.44 (s, 2 × (CH3)3C-N(Boc)), 0.90 (s, 2 × CH3-C(10)), 0.75 (d, J = 6.4, 2 × CH3-C(20)), 0.67 (s, 2 × CH3-C(13)). 13C NMR (125 MHz, CDCl3, δ): 170.30, 170.24, 155.11, 152.06, 150.48, 149.08, 134.53, 128.47, 124.59, 121.98, 117.49, 98.41, 79.17, 75.36, 70.77, 54.98, 54.23, 50.71, 47.81, 44.98, 44.41, 43.31, 41.49, 37.63, 36.31, 35.44, 35.02, 35.18, 33.68, 31.23, 28.83, 28.36, 27.16, 25.47, 24.53, 23.59, 22.70, 22.64, 21.55, 21.26, 17.84, 12.15. (+)ESI-HRMS (m/z): [M + H]+ 1354.8695 (error, −0.02 ppm). Combustion analysis for C78H120ClN5O12: calculated C 69.13, H 8.92, N 5.17; found C 69.05, H 8.84, N 5.37. HPLC purity, method C: tR = 11,964, area 97.93%. Method D: tR = 5.892, area 96.83%.
24-N-Methyl-N-[(7-chloroquinoline-4-yl)amino]ethylamino-3α-methanesulfonamide-7α,12α-diacetoxy-5β-cholane (42)
Into stirred solution of 38 (121 mg, 0.16 mmol) and formaldehyde (37%, 54 μL, 0.72 mmol) in 5 mL of DCM, NaBH(OAc)3 (203.3 g, 0.96 mmol) was added. When starting 38 was completely consumed (TLC, SiO2, EA/MeOH/NH3 = 9:1:1), the reaction mixture was suspended in DCM/H2O mixture, pH was adjusted to 12 with 0.5 M NaOH, and transferred to separatory funnel and further worked-up in usual manner. Product was isolated after column chromatography (flash, Biotage SP1 NH column, eluent hexane/EA 4:6 → EA). Yield 103 mg (84%). Colorless foam softness at 110–114 °C. [α]D20 +0.048 (c 1.0 × 10–3 g/mL, DCM). IR (KBr): 3406m, 2949m, 2869m, 1731s, 1611w, 1582s, 1538w, 1452m, 1377m, 1320m, 1247s, 1150m, 1081w, 1023m, 969w, 880w, 849w, 812w, 763w, 733w cm–1. 1H NMR (500 MHz, CDCl3, δ): 8.54 (d, J = 5.5, H-C(2′)), 7.99 (d, J = 2.0, H-C(8′), 7.73 (d, J = 9.1, H-C(5′), 7.36 (dd, J1 = 8.9, J2 = 2.0 H-C(6′)), 6.37 (d, J = 5.5, H-C(3′)), 6.29 (bs, H-N, exchangeable with D2O), 5.09 (bs, H-C(12)), 4.95–4.90 (m, H-C(7)), 4.60–4.37 (m, H-N, exchangeable with D2O), 3.34 (bs, 2H-C(9′)), 3.20–3.10 (m, H-C(3)), 2.96 (s, CH3-N), 2.80 (t, J = 5.8, 2H-C(10′)), 2.52–2.40 (m, 2H-C(24)), 2.32 (s, CH3-SO2N), 2.07 (s, 2× CH3COO), 0.92 (s, CH3-C(10)), 0.82 (d, J = 6.6, CH3-C(20)), 0.71 (s, CH3-C(13)). 13C NMR (125 MHz, CDCl3, δ): 175.77, 170.41, 170.24, 151.41, 150.17, 148.33, 135.13, 127.96, 125.31, 121.49, 117.23, 99.02, 75.36, 70.66, 57.70, 54.89, 54.02, 47.84, 45.06, 43.38, 41.92, 41.61, 41.22, 39.53, 37.69, 37.36, 35.43, 35.16, 34.07, 33.44, 31.19, 29.40, 28.86, 27.37, 25.49, 23.64, 22.78, 22.61, 22.14, 21.57, 21.30, 17.94, 12.21. (+)ESI-HRMS (m/z): [M + H]+ 773.407 72 (error, +0.53 ppm). Combustion analysis for (C41H61ClN4O6S·2H2O): calculated C 60.83, H 8.09, N 6.92, S 3.96; found C 60.55, H 7.92, N 6.81, S 4.00. HPLC purity, method A: tR = 1.868, area 99.31%. Method B: tR = 1.859, area 98.31%.
24-N-Methyl-N-[(7-chloroquinoline-4-yl)amino]ethylamino-3α-N′-tert-butylcarbamate-7α,12α-diacetoxy-5β-cholane (44)
Into a stirred solution of 40 (1.00 g, 1.28 mmol) and formaldehyde (37%, 190 μL, 2.55 mmol) in dry MeOH (30 mL) was added a suspension of NaBH3CN (81 mg, 1.28 mmol) and anhydrous ZnCl2 (88 mg, 0.64 mmol) in MeOH (10 mL). After 2 h of stirring at room temperature the additional amount of NaBH3CN (41 mg, 0.64 mmol) and anhydrous ZnCl2 (44 mg, 0.32 mmol) were added as MeOH (10 mL) suspension and stirring was continued for 1 h. Solvent was evaporated under reduced pressure, and remaining residue was transferred into a separatory funnel as DCM suspension. Water was added and pH was adjusted to pH 12 with 0.5 M NaOH, and the organic layer was worked up in the usual manner. Product was obtained after flash column chromatography (Biotage SP NH column, eluent hexane/EA 4:6 → 2:6). Yield 968.4 mg (95%). Colorless foam softens at 96–101 °C. [α]D20 +0.163 (c 2.0 × 10–3 g/mL, DCM). IR (ATR): 3376w, 2949m, 2868w, 2802w, 1731s, 1611w, 1581s, 1528m, 1452m, 1374s, 1331w, 1246s, 1171m, 1063w, 1023m, 965w, 940w, 881w, 850w, 807w, cm–1. 1H NMR (500 MHz, CDCl3, δ): 8.46 (d, J = 5.4, H-C(2′)), 7.89 (s, J = 2.0, H-C(8′)), 7.71 (d, J = 8.9, H-C(5′)), 7.30 (dd, J1 = 8.8, J2 = 2.1, H-C(6′)), 6.33 (d, J = 5.4, H-C(3′)), 6.07 (s, H-N-Boc, exchangeable with D2O), 5.06 (s, H-C(12)), 4.92–4.88 (m, H-C(7)), 4.42 (bs, H-C(3)), 3.45 (s, CH3-N), 3.33 (bs, 2H-C(9′)), 3.05–2.95 (m, 2H-C(11′)), 2.70–2.43 (m, 2H-C(24) and H-N, exchangeable with D2O), 2.07 (s, CH3COO), 2.04 (s, CH3COO), 1.42 (s, (CH3)3C-N(Boc)), 0.88 (s, CH3-C(10)), 0.79 (d, J = 6.6, CH3-C(20)), 0.68 (s, CH3-C(13)). 13C NMR (125 MHz, CDCl3, δ): 170.34, 170.27, 151.80, 149.92, 148.87, 134.77, 128.36, 125.11, 121.43, 117.29, 99.98, 75.41, 70.82, 50.43, 49.66, 47.60, 47.33, 44.97, 43.30, 41.83, 41.48, 37.65, 36.35, 35.44, 34.89, 34.19, 33.23, 31.24, 28.79, 28.35, 27.25, 26.45, 25.45, 22.74, 22.63, 21.57, 21.31, 17.85, 12.15. (+)ESI-HRMS (m/z): [M + H]+ 795.481 85 (error, −0.43 ppm). Combustion analysis for C45H67ClN4O6: calculated C 67.94, H 8.49, N 7.04; found C 67.72, H 8.63, N 6.75. HPLC purity, method A: tR = 1.994, area 99.12%. Method C: tR = 9.936, area 98.20%.
24-N-[(7-Chloroquinoline-4-yl)amino]ethylamino-3α-N′,N′-dimethylamino-7α,12α-diacetoxy-5β-cholane (50)
Into a stirred mixture of crude 48 (817 mg, 0.90 mmol) and formaldehyde (37%, 0.2 mL, 2.8 mmol) in dry methanol (8 mL) at room temperature was added a suspension of anhydrous ZnCl2 (61 mg, 0.45 mmol) and NaBH3CN (56.8 mg, 0.90 mmol) in dry methanol (8 mL). The reaction mixture was stirred for 24 h, and the solvent was removed under reduced pressure. The residue was transferred into a separatory funnel as DCM solution, and water was added. pH was adjusted to pH 12 with 0.1 M NaOH. Layers were separated. Water layer was extracted with DCM (2 × 15 mL), and combined organic layers were washed with saturated NaHCO3 and brine and dried over anhydrous Na2SO4. Solution was filtered off. The solvent was removed under reduced pressure, and crude product (625 mg) was dissolved in solution of piperidine in DCM (20%, 27 mL). Reaction mixture was stirred for a further 24 h at room temperature, and the solvent was removed under reduced pressure, transferred into a separatory funnel as DCM solution, and worked up in the usual manner. The product was isolated after column chromatography purification [dry flash, SiO2, eluent MeOH, MeOH/NH3 gradient 95/5 → 9/1 and flash chromatography (Biotage SP1, eluent EA/Hex, gradient 6/4 → EA, gradient EA/MeOH 95/5 → 9/1)]. Yield 268 mg (55%). Colorless foam softens at 78–82 °C. [α]D20 +0.056 (c 1.8 × 10–3 g/mL, DCM). IR (KBr): 331w, 2935s, 2866m, 2772w, 1730s, 1610w, 1580s, 1534w, 1449m, 1375m, 1330w, 1242s, 1140w, 1069w, 1024w, 966w, 877w, 808w cm–1. 1H NMR (200 MHz, CDCl3, δ): 8.52 (d, J = 5.1, H–C(2′)), 7.94 (d, J = 2.2, H-C(8′)), 7.73 (d, J = 9.0, H-C(5′)), 7.35 (dd, J1 = 9.0, J2 = 2.2, H-C(6′)), 6.39 (d, J = 5.6, H-C(3′)), 5.94 (bs, H-N, exchangeable with D2O), 5.08 (bs, H-C(12)), 4.90–4.70 (m, H-C(7)), 3.40–3.26 (m, 2H-C(9′)), 3.08–2.96 (m, 2H-C(11′)), 2.70–2.54 (m, 2H-C(24)), 2.27 (s, (CH3)2N), 2.10 (s, CH3COO), 2.07 (s, CH3COO), 0,90 (s, CH3-C(10)), 0.82 (d, J = 6.2, CH3-C(20)), 0.71 (s, CH3-C(13)). 13C NMR (50 MHz, CDCl3, δ): 170.68, 170.58, 152.05, 149.86, 134.74, 128.63, 125.12, 121.28, 117.31, 99.12, 75.49, 70.94, 64.98, 49.71, 47.56, 47.43, 44.96, 43.30, 42.19, 41.90, 41.68, 37.64, 35.67, 34.89, 34.32, 33.23, 32.74, 31.50, 28.86, 27.24, 26.66, 25.40, 24.05, 22.67, 21.60, 21.40, 17.81, 12.13. (+)ESI-HRMS (m/z): [M + H]+ 709.446 54 (error, +1.59 ppm). Combustion analysis for C41H61ClN4O4·2H2O: calculated C 66.06, H 8.79, N 7.52; found C 66.22, H 8.98, N 7.38. HPLC purity, method A: tR = 1.925, area 98.15%. Method B: tR = 7.942, area 96.45%.
24-N-[(7-Chloroquinoline-4-yl)amino]ethylamino-3α-amino-7α,12α-diacetoxy-5β-cholane (52)
Compound 40 (100 mg, 0.128 mmol) was stirred at room temperature in DCM/TFA mixture (2 mL, 1:1, v/v) for 3 h. Solvent was removed under reduced pressure, and the residue was transferred as DCM solution to separatory funnel and worked up in the usual manner. Yield 83 mg (95.3%). Colorless foam softens at 94–97 °C. [α]D20 +0.046 (c 1.1 × 10–3 g/mL, DCM). IR (ATR): 3286w, 2932m, 2860m, 1724s, 1610w, 1578s, 1533w, 1448m, 1374m, 1330w, 1238s, 1135w, 1077w, 1021m, 964w, 936w, 878w, 847w, 806w, 766w cm–1. 1H NMR (500 MHz, CDCl3, δ): 8.51 (d, J = 5.4, H-C(2′)), 7.94 (d, J = 2.0, H-C(8′)), 7.72 (d, J = 8.7, H-C(5′)), 7.35 (dd, J1 = 8.9, J2 = 2.2, H-C(6′)), 6.37 (d, J = 5.4, H-C(3′)), 5.95 (bs, H-N,), 5.08 (bs, H-C(12)), 4.92–4.86 (m, H-C(7)), 3.37–3.30 (m, 2H-C(9′)), 3.03 (t, J = 5.7, 2H-C(11′)), 2.68–2.56 (m, 2H-C(24) and H-C(3)), 2.10 (s, CH3COO), 2.07 (s, CH3COO), 0.90 (s, CH3-C(10)), 0.82 (d, J = 6.4, CH3-C(20)), 0.71 (s, CH3-C(13)). 13C NMR (125 MHz, CDCl3, δ): 170.64, 170.60, 151.98, 149.88, 149.01, 134.80, 128.59, 125.17, 121.31, 117.33, 99.13, 75.49, 70.89, 51.64, 49.72, 47.62, 47.43, 45.03, 43.39, 41.89, 41.49, 39.41, 37.76, 35.50, 34.94, 34.36, 33.28, 31.42, 31.16, 28.98, 27.29, 26.64, 25.58, 22.78, 21.64, 21.46, 17.87, 12.19. (+)ESI-HRMS (m/z): [M + 2H]2+ 341.210 68 (error, −0.04 ppm). Combustion analysis for C39H57ClN4O4·H2O: calculated C 66.98, H 8.50, N 8.01; found C 66.84, H 8.32, N 7.92. HPLC purity, method A: tR = 2.004, area 98.23%. Method B: tR = 7.973, area 95.08%.
24-N-Methyl-N-[(7-chloroquinoline-4-yl)amino]ethylamino-3α-amino-7α,12α-diacetoxy-5β-cholane (54)
Compound 44 (800 mg, 1.0 mmol) was stirred at room temperature in DCM/TFA mixture (16 mL, 1:1, v/v) for 1.5 h. Then the solvent was removed under reduced pressure, and the residue was transferred as DCM solution into a separatory funnel and worked up in the usual manner. Product was isolated after flash column chromatography (Biotage SP, SiO2, eluent EA/MeOH, gradient 9:1 → 55/45). Yield 245 mg (35%). Colorless foam softens at 62–64 °C. [α]D20 +0.080 (c 2.0 × 10–3 g/mL, DCM). IR (ATR): 3409m, 2947s, 2865m, 1729m, 1668w, 1612w, 1582s, 1536w, 1451m, 1378m, 1331w, 1244s, 1157w, 1136w, 1077w, 1024m, 964w, 879w, 848w, 807w cm–1. 1H NMR (500 MHz, CDCl3, δ): 8.49 (d, J = 5.3, H-C(2′)), 7.92 (d, J = 2.0, H-C(8′)), 7.65 (d, J = 8.9, H-C(5′)), 7.31 (dd, J1 = 8.9, J2 = 2.1, H-C(6′)), 6.33 (d, J = 5.3, H-C(3′)), 6.02–6.96 (m, H-N, exchangeable with D2O), 5.08–5.03 (m, H-C(12)), 4.86 (d, J = 2.7, H-C(7)), 3.24 (q, J = 5.5, 2H-C(9′)), 2.70 (d, J = 5.9, 2H-C(11′)), 2.60–2.52 (m, H-C(3)), 2.43–2.30 (m, 2H-C(24)), 2.23 (s, CH3-N), 2.05 (s, 2 × CH3COO), 0.87 (s, CH3-C(10)), 0.79 (d, J = 6.6, CH3-C(20)), 0.68 (s, CH3-C(13)). 13C NMR (125 MHz, CDCl3, δ): 170.56, 152.05, 149.73, 149.04, 134.64, 128.64, 125.00, 121.19, 117.29, 99.18, 75.43, 70.84, 57.80, 55.11, 51.59, 47.78, 45.02, 43.38, 41.46, 41.25, 39.57, 39.49, 37.71, 35.49, 35.13, 34.29, 33.46, 31.36, 31.23, 28.94, 27.33, 25.54, 24.04, 22.73, 21.59, 21.37, 17.90, 12.15. (+)ESI-HRMS (m/z): [M + H]+ 695.429 22 (error, −0.78 ppm). Combustion analysis for C40H59ClN4O4·H2O: calculated C 67.34, H 8.62, N 7.85; found C 66.98, H 8.73, N 7.62. HPLC purity, method A: tR = 1.905, area 99.44%. Method C: tR = 11.413, area 96.83%.
24-N-Methyl-N-[(7-chloroquinoline-4-yl)amino]ethylamino-3α-N′,N′-dimethylamino-7α,12α-diacetoxy-5β-cholane (56)
Into a stirred solution of 54 (190 mg, 0.27 mmol) and formaldehyde (37%, 50 μL, 0.67 mmol) in dry MeOH (2 mL) was added a suspension of NaBH3CN (13 mg, 0.21 mmol) and anhydrous ZnCl2 (29 mg, 0.21 mmol) in dry MeOH (2 mL). After the reaction was completed (TLC, SiO2, EA/NH3/MeOH = 9:1:1) the solvent was evaporated under reduced pressure. The obtained residue was transferred into a separatory funnel as DCM suspension. Water was added. The pH was adjusted to pH 12 with 0.5 M NaOH, and the reaction was further worked up in the usual manner. Product was obtained after column chromatography (dry flash, SiO2, eluent EA, EA/MeOH gradient 9:1 → 8/2, EA/NH3/MeOH = 18:0.5:0.5 → 18:1.5:1.5). Yield 82 mg (42%). Colorless foam softens at 75–80 °C. [α]D20 +0.023 (c 2.0 × 10–3 g/mL, DCM). IR (ATR): 3424s, 2929s, 2867m, 1731s, 1612w, 1582s, 1531w, 1452m, 1377m, 1330w, 1244s, 1163w, 1070w, 1026m, 966w, 879w, 807w cm–1. 1H NMR (500 MHz, CDCl3, δ): 8.53 (d, J = 5.4, H-C(2′)), 7.95 (d, J = 2.0, H-C(8′)), 7.68 (d, J = 9.0, H-C(5′)), 7.35 (dd, J1 = 8.9, J2 = 2.1, H-C(6′)), 6.37 (d, J = 5.4, H-C(3′)), 6.00 (bs, H-N, exchangeable with D2O), 5.11–5.07 (m, H-C(12)), 4.92–4.87 (m, H-C(7)), 3.31–3.26 (m, 2H-C(9′)), 2.74 (t, J = 5.8, 2H-C(11′)), 2.46–2.35 (m, 2H-C(24)), 2.27 (s, (CH3)2N-C(3)), 2.26 (s, CH3-N), 2.08 (s, CH3COO), 2.07 (s, CH3COO), 0.91 (s, CH3-C(10)), 0.83 (d, J = 6.6, CH3-C(20)), 0.72 (s, CH3-C(13)). 13C NMR (125 MHz, CDCl3, δ): 170.64, 170.59, 152.15, 149.81, 149.14, 134.75, 128.78, 125.10, 121.22, 117.38, 99.28, 75.54, 70.99, 65.02, 57.86, 55.20, 47.80, 45.10, 43.43, 42.10, 41.81, 41.30, 39.62, 37.77, 35.77, 35.27, 34.44, 33.56, 32.61, 31.59, 28.96, 27.43, 25.50, 24.15, 23.93, 22.82, 22.76, 21.65, 21.40, 17.98, 12.23. (+)ESI-HRMS (m/z): [M + H]+ 723.460 48 (error, −0.80 ppm). Combustion analysis for C42H63ClN4O4: calculated C 69.73, H 8.78, N 7.74; found C 69.52, H 8.60, N 7.63. HPLC purity, method A: tR = 1.849, area 98.82%. Method D: tR = 5.179, area 98.67%.
N,N-Di(3α-amino-7α,12α-diacetoxy-5β-cholan-24-yl)-N′-(7′-chloroquinoline-4′-yl)-1,3-ethanediamine (60)
Compound 58 (140 mg, 0.104 mmol) was stirred at room temperature in DCM/TFA mixture (2 mL, 1:1, v/v) for 3 h. Solvent was removed under reduced pressure, and the residue was dissolved in DCM containing 5% MeOH and transferred into a separatory funnel. The organic layer was washed twice with saturated Na2CO3, once with brine, and dried over anhydrous Na2SO4. Solution was filtered off, and the solvent was removed under reduced pressure. Yield 75.7 mg (64%). Colorless foam softens at 128–132 °C. [α]D20 +0.032 (c 1.2 × 10–3 g/mL, DCM). IR (KBr): 2936m, 2864m, 1726s, 1611w, 1580m, 1526w, 1446w, 1375s, 1331w, 1238s, 1199.6m, 1132m, 1076w, 1021s, 964w, 937w, 878w, 832w, 801w, 765w, 720w cm–1. 1H NMR (500 MHz, CDCl3, δ): 8.53 (d, J = 5.3, H-C(2′)), 7.96 (d, J = 2.1, H-C(8′)), 7.64 (d, J = 9.2, H-C(5′) 7.35 (dd, J1 = 8.9, J2 = 2.1, H-C(6′)), 6.37 (d, J = 5.3, H-C(3′)), 6.03–5.99 (m, H-N, exchangeable with D2O), 5.07 (bs, 2 × H-C(12)), 4.91–4.86 (m, 2 × H-C(7)), 3.29–3.21 (m, 2H-C(9′) and 2 × H2N), 2.84–2.76 (m, 2H-C(11′)), 2.64–2.55 (m, 2 × H-C(3)), 2.51–2.37 (m, 2 × 2H-C(24)), 2.07 (s, 2 × CH3COO), 2.06 (s, 2 × CH3COO), 0.90 (s, 2 × CH3-C(10)), 0.80 (d, J = 6.6, 2 × CH3-C(20)), 0.69 (s, 2 × CH3-C(13)). 13C NMR (125 MHz, CDCl3, δ): 170.60, 152.16, 149.74, 149.13, 134.72, 128.81, 125.09, 121.06, 117.37, 99.33, 75.45, 70.90, 53.85, 51.74, 51.65, 47.93, 45.07, 43.39, 41.51, 39.73, 39.53, 35.17, 34.36, 33.58, 31.42, 31.28, 28.99, 27.33, 25.59, 23.97, 22.79, 21.65, 21.41, 17.95, 12.21. (+)ESI-HRMS (m/z): [M + 2H]2+ 570.878 12 (error, −1.85 ppm) [M + H]+ 1140.748 97 (error, −2.51 ppm). Combustion analysis for C67H102ClN5O8·H2O: calculated C 69.43, H 9.04, N 6.04; found C 69.39, H 9.27, N 5.91. HPLC purity, method A: tR = 1.838, area 97.10%. Method D: tR = 5.321, area 96.33%.
N,N′-Bis{2-[(7-chloroquinoline-4-yl)amino]ethyl}-7α,12α-diacetoxy-5β-cholane-3,24-diamine (62, Mixture of Diastereoisomers)
Mixture of 22 (385 mg, 0.57 mmol) and ACQ2 (251 mg, 1.13 mmol) in dry MeOH (9 mL) was stirred at room temperature for 12 h. Then NaBH3CN (45 mg, 0.72 mmol) was added in one portion, and after 1 h of stirring at room temperature glacial AcOH (3 drops) was added. Reaction mixture was stirred for further 48 h at room temperature. The reaction was quenched with Et3N (3 drops). Solvent was removed under reduced pressure, and product was isolated by column chromatography (dry flash, SiO2, eluent EA, EA/MeOH gradient 95/5 → 1/9, EA/MeOH/NH3 gradient 18/1/1 → 9/1/1, flash, Biotage SP1, RP column, eluent gradient MeOH/H2O 8/2 → 9/1). Yield 299 mg (59%). Colorless foam softens at 108–113 °C. [α]D20 +0.070 (c 1.4 × 10–3 g/mL, DCM). IR (ATR): 3379w, 2929m, 2863w, 1722m, 1611w, 1581s, 1535w, 1450w, 1375m, 1330w, 1248m, 1170w, 1137w, 1078w, 1024w, 965w, 879w, 849w, 807w cm–1. 1H NMR (200 MHz, CDCl3, δ): 8.50 (d, J = 5.6, 2 × H-C(2′)), 7.94 (d, J = 1.7, 2 × H-C(8′)), 7.78–7.68 (m, 2 × H-C(5′)), 7.37–7.26 (m, 2 × H-C(6′)), 6.42–6.32 (m, 2 × H-C(3′)), 6.11–5.89 (m, 2 × H-N exchangeable with D2O), 5.11(bs, H-C(12)), 4.90 (bs, H-C(7)), 3.33 (bs, 2 × 2H-C(9′)), 3.08–2.92 (m, 2 × 2H-C(10′) and H-C(3)), 2.70–2.55 (m, 2H-C(24)), 2.10 (s, CH3COO), 2.08 (s, CH3COO), 0.96 (s, CH3-C(10)), 0.83 (d, J = 6.2, CH3-C(20)), 0.72 (s, CH3-C(13)). 13C NMR (50 MHz, CDCl3, δ): 170.60, 170.51, 170.44, 151.96, 149.92, 148.97, 134.77, 128.56, 128.49, 125.18, 125.14, 121.39, 121.22, 121.13, 117.31, 99.16, 99.09, 75.51, 71.01, 70.90, 57.83, 52.04, 49.71, 47.60, 47.43, 44.96, 44.79, 43.30, 42.72, 42.32, 41.92, 41.44, 37.65, 36.96, 36.13, 35.34, 34.87, 34.71, 33.47, 33.25, 31.37, 31.10, 29.99, 28.93, 28.44, 28.26, 27.26, 26.62, 25.62, 24.87, 23.22, 22.74, 21.56, 21.29, 17.83, 12.13. 1H NMR (500 MHz, CDCl3, δ): 8.50 (d, J = 5.3, 2 × H-C(2′)), 7.97–7.91 (m, 2 × H-C(8′)), 7.76–7.64 (m, 2 × H-C(5′)), 7.37–7.30 (m, 2 × H-C(6′)), 6.41–6.34 (m, 2 × H-C(3′)), 6.07–5.84 (m, 2 × H-N exchangeable with D2O), 5.14–5.04 (m, H-C(12)), 4.93–4.86 (m, H-C(7)), 3.39–3.25 (m, 2 × 2H-C(9′)), 3.07–2.9 (m, 2 × 2H-C(10′)) 2.68–2.56 (m, H-C(3)), 2.26–2.13 (m, 2H-C(24)), 2.09 (s, CH3COO), 2.07 (s, CH3COO), 2.01 (s, CH3COO), 0.96 (s, CH3-C(10)), 0.92 (s, CH3-C(10)), 0.85–0.79 (m, CH3-C(20)), 0.72 (s, CH3-C(13)), 0.71 (s, CH3-C(13)). 13C NMR (125 MHz, CDCl3, δ): 170.44, 151.96, 149.91, 149.01, 134.79, 128.61, 128.55, 125.19, 125.16, 121.34, 121.09, 117.34, 99.18, 99.11, 75.54, 71.05, 57.87, 52.11, 49.71, 47.67, 47.47, 45.02, 44.87, 43.35, 42.75, 42.39, 41.96, 41.52, 37.73, 36.19, 34.93, 34.76, 33.54, 33.31, 31.16, 30.06, 28.99, 28.33, 27.31, 26.70, 25.67, 25.53, 24.95, 23.25, 22.79, 21.58, 21.31, 17.91, 12.19. (+)ESI-HRMS (m/z): [M + 2H]2+ 443.2334 (error, +0.03 ppm) [M + H]+ 885.4587 (error, +0.96 ppm). Combustion analysis (C50H66Cl2N6O4·1.5 H2O): calculated C 65.77, H 7.62, N 9.20; found C 65.30, H 7.24, N 9.07. HPLC purity, method B: tR = 1.788, area 98.83%. Method C: tR1 = 10.371, tR2 = 10 566, area 95.15%. Method D: tR1 = 4.947, tR2 = 5.036, area 96.07%.
Molecular Modeling
The structure of BoNT/A LC and binding site (BS) used in this study was obtained from previously published crystal structure of protein (PDB code 3DS9).52 All ligand structures were built in Maestro (Maestro, version 9.3; Schrödinger, LLC: New York, NY, 2012), and their structures were optimized in Semiempirical NNDO module of Schrödinger Suite 2012 (Schrödinger Suite 2012 Semiempirical NDDO Module; Schrödinger LLC: New York, NY, 2012), using the RM1 method. Ionization states for all ligands were examined using sequential pKa estimation in the Epik module (Epik, version 2.3; Schrödinger, LLC: New York, NY, 2012). Ligand log P were calculated using QikProp module (QikProp, version 3.5; Schrödinger, LLC: New York, NY, 2012). Grid docking of the ligands were performed using Glide ligand docking (Glide, version 5.8; Schrödinger, LLC: New York, NY, 2012), using standard precision and flexible ligand sampling, without any additional constraints. Generated docking structures were refined and used for the next step. All structures were docked in ionization state proposed by results of Epik pKa prediction for pH 7.0 ± 2.0, with protonated aliphatic nitrogen atoms in structure. For the next step, Induced Fit Docking was used (Schrödinger Suite 2012 Induced Fit Docking Protocol; Glide, version 5.8; Schrödinger, LLC: New York, NY, 2012; Prime, version 3.1; Schrödinger, LLC: New York, NY, 2012).
All modeling was performed within a pH range of 7.0 ± 1.0, which encapsulates the pH at which the in vitro experiments were conducted (i.e., pH 7.3). For the derivatives presented herein, computational predictions showed that the pKa values of the aliphatic nitrogen atoms range between 9.50 and 10.80 ± 1.0, while the pKa values of nitrogen atoms that are part of the ACQ moiety range between 7.40 and 8.00 ± 1.20. Accordingly, it is plausible to assume that all aliphatic nitrogen atoms in the ligand structures are protonated and that the aromatic ACQ nitrogens are only partially protonated (around 50%). Thus, simulations of ligand binding were conducted with protonated aliphatic nitrogen atoms and neutral aromatic nitrogens. Inhibitor docking was performed using the Glide module.51 The structures were further refined using Glide with “refine only” option.
Considering the relative inflexibility of the steroidal core and different conformational space of the α- and β-substituents at C(3), differences in binding modes could be expected as well. Taking into account the overall flexibility of the BoNT/A LC substrate cleft, it is clear that residues in the enzyme’s substrate cleft would undergo some additional conformational changes compared to smaller ligands. For these reasons, the substrate cleft was slightly relaxed via Induced Fit dockings, which was followed by Glide Grid docking. The highest scoring models were further optimized using the OPLS2005 force field with water as a solvent (the solvent effects were simulated using the analytical generalized Born/surface area mode), using Polak–Ribiere conjugate gradient with a maximum of 5000 iterations and gradient convergence threshold of 0.05 kJ/mol. During minimization, the backbone of the protein was constrained and the catalytic engine Zn2+ and His223, His227, and Glu262 were frozen in space. This treatment resulted in a slightly more open catalytic cleft. Most noticeable feature was the change in orientation of the Met253 residue. In previous binding simulations with smaller inhibitors, it was oriented into the binding cleft, thereby adding to the hydrophobic nature of the pocket. Now, Met253 reorients away from the binding cleft, thus providing enough space for the docking of a larger steroidal bis-ACQ system.
In Vitro Anti-BoNT/A LC Metalloprotease Activity57
The HPLC-based assay was used to evaluate small molecule mediated inhibition of the BoNT/A LC in vitro.42 In brief, this assay utilizes an N-terminal acetylated, C-terminal aminated synthetic peptide identical in sequence to residues 187–203 of SNAP-25. Compounds with intrinsic fluorescence quenching capability do not interfere with the activity measurements of this assay because substrate hydrolysis is determined by HPLC separation of the products from the substrate, followed by measurement of the peak areas. The assay mixture contained 40 mM HEPES–0.05% Tween (pH 7.3), BoNT/A recombinant LC, peptide substrate, 0.5 mg/mL bovine serum albumin, and various concentrations of small molecule. The assay was run at 37 °C, quenched by the addition of TFA, and analyzed using reverse-phase HPLC. Zinc (50 μM excess) was added to the assays to remove compounds that are zinc chelating agents.
Percent inhibition measurements were performed in duplicate, and standard deviations were less than 25% for all. IC50 calculations were determined by measuring enzyme activity at nine different SMNPI concentrations and in the absence of the small molecule. The small molecule concentrations used in the measurements were determined by estimating the IC50 value and moving in 1 log increments in either direction of the estimated value. The resulting kinetic data were subsequently fit to the Langmuir isotherm, Vi/Vo = 1/{1 + ([I]/IC50)h}, using nonlinear regression analysis. For Ki determination, reaction velocity versus substrate concentration was plotted for multiple small molecule concentrations. These plots were analyzed using global kinetic analysis. Subsequently, the data were fit to a model of competitive inhibition and analyzed by nonlinear regression analysis.
Primary Neuron Cell Culture
Ventral spinal cords were isolated from day 6 embryonic chicken embryos and cultured by the method of Kuhn58 as previously described.59 Spinal cords were removed from the embryos and the dorsal halves trimmed away. After trypsinization for 25 min, the cells were dissociated by trituration and preplated in Dulbecco’s modified Eagle medium for 1 h to enrich the neuronal population by adherence of non-neuronal cells to the culture dish. Cells remaining in suspension were centrifuged for 6 min at 1250 rpm, resuspended in Liebowitz’s L15 medium with 10% fetal bovine serum and N3 supplements, and plated onto six-well culture plates that were first coated with poly-l-lysine (1 mg/mL overnight) and then with laminin (7–10 μg/mL for 3–4 h). Neurons were incubated overnight at 37 °C in 5% CO2 before use.
Intoxication, Inhibitor Application, and Western Blot Analysis
Neurons were preincubated with inhibitor for 45 min. Afterward, the cells were incubated with 10 nM BoNT/A and inhibitor for 3.5 h. The cells were then rinsed with fresh culture medium, scraped, and collected into tubes. The tubes were centrifuged, and supernatant was removed and replaced with 30 μL of lysis buffer. After overnight storage at −80 °C, the tubes were thawed, vortexed briefly, and incubated on ice for 10 min. Lysates were centrifuged at high speed at 4 °C for 30 min to remove debris. Total protein was measured by the Bradford assay. 12% gels were used for SDS–PAGE, and proteins were blotted onto PVDF membranes. Western blots were probed with a 1:2500 dilution of SMI-81 anti-SNAP-25 antibody (Covance Research Products Inc., Berkeley, CA) and 1:20000 horseradish peroxidase (HRP) conjugated goat anti-mouse (Pierce, Rockford, IL). Western blots were developed using a chemiluminescent substrate and imaged with a BioRad Versadoc system. Protein bands were quantitated by densitometry with Quantity One software (BioRad, Hercules, CA).
In Vitro Antimalarial Activity
The in vitro antimalarial drug susceptibility screen is a modification of the procedures first published by Desjardins et al.,60 with modifications developed by Milhous et al.,54 and the details are given in ref (47). All synthesized aminoquinolines were screened in vitro against P. falciparum strains: CQ and MFQ susceptible strain D6 (clone of Sierra I/UNC isolate), CQ resistant but MFQ susceptible strain W2 (clone of Indochina I isolate), and CQ and MFQ resistant strain TM91C235 (clone of Southeast Asian isolate).
Toxicity: Assessment of SMNPI Toxicity in a HepG2 (Hepatocellular Carcinoma) Cell Line
The compounds were assayed for HepG2 toxicity according to protocol given in ref (47).
Toxicity: PBMC Cell Assay
Peripheral blood mononuclear cells (PBMC) were separated from whole heparinized blood of healthy volunteers. Blood was diluted with phosphate buffered saline (PBS) (1:1) and layered on Histopaque solution. After centrifugation, interface cells were collected and washed three times with PBS. After counting, cells were resuspended in nutrient medium. Nutrient medium was RPMI-1640 medium, supplemented with 10% fetal calf serum (FCS), 1% glutamine (200 mM), 1% β-mercaptoethanol (5 μM), 1% penicillin (10000 IU mL–1), and 1% streptomycin (10 mg mL–1). Cells were seeded (200 000 cells per well) in 96-well plates in nutrient medium with 0.5% phytohemagglutinin (PHA) (1 mg mL–1) and different concentrations of organic compounds in DMSO. Cells were incubated for 72 h at 37 °C in a humidified atmosphere with 6% CO2.
Absorbances were recorded at λ = 540 and 670 nm on a recorder (LKB 5060-006, LKB, Vienna, Austria). Absorbances corresponding to viability of cells were obtained as A540 – A670 differences. Percent of cell growth inhibition were calculated from formula (Asample – Aneg.control)/(Apos.control – Aneg.control) = X and (1 – X) × 100 = % inhibition. IC50 concentrations were defined as the concentration of a drug needed to inhibit cell survival by 50%, compared with vehicle-treated control.
Acknowledgments
The authors thank Dr. Nicoletta Basilico (Dipartimento di Sanità Pubblica–Microbiologia–Virologia, Università degli Studi di Milano, Italy) for β-hematin formation inhibition experiments. This research was supported by National Institute of Allergy and Infectious Diseases (U.S.) Grant 5-U01AI082051-02 and by the Ministry of Science and Technological Development of Serbia (Grant 172008). Furthermore, for J.C.B., the following statement is in compliance with Leidos Biomedical Research, Inc. contractual requirements: This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health (U.S.), under Contract HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services (U.S.) or the U.S. Army, nor does the mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government or the U.S. Army.
Glossary
Abbreviations Used
- ART
artemisinin
- BoNT/A LC
botulinum neurotoxin serotype A light chain
- 4,7-ACQ
4-amino-7-chloroquinoline
- CQ
chloroquine
- MFQ
mefloquine
- MLM
mouse liver microsome
- HLM
human liver microsome
- ACQn
N-(7-chloroquinolin-4-yl)alkane-1,n-diamine
- CQR
chloroquine resistant strain
- CQS
chloroquine susceptible strain
- MDR
multidrug resistant strain
Supporting Information Available
Synthetic procedures for a number of inhibitors; procedures for the determination of the purity of tested compounds; NMR spectra of all tested compounds; docking models of BoNT/A LC–ligand interaction diagrams for selected inhibitors. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Present Address
○ J.E.N.: Senior Scientist, The Geneva Foundation, 917 Pacific Avenue, Suite 600, Tacoma, WA 98402, U.S. Phone: 1-253-682-3816. E-mail: jnuss@genevausa.org.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Burnett J. C.; Schmidt J. J.; Stafford R. G.; Panchal R. G.; Nguyen T. L.; Hermone A. R.; Vennerstrom J. L.; McGrath C. F.; Lane D. J.; Sausville E. A.; Zaharevitz D. W.; Gussio R.; Bavari S. Novel Small Molecule Inhibitors of Botulinum Neurotoxin A Metalloprotease Activity. Biochem. Biophys. Res. Commun. 2003, 310, 84–93. [DOI] [PubMed] [Google Scholar]
- Burnett J. C.; Henchal E. A.; Schmaljohn A. L.; Bavari S. The Evolving Field of Biodefence: Therapeutic Developments and Diagnostics. Nat. Rev. Drug Discovery 2005, 4, 281–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paddle B. M. Therapy and Prophylaxis of Inhaled Biological Toxins. J. Appl. Toxicol. 2003, 23, 139–170. [DOI] [PubMed] [Google Scholar]
- Willis B.; Eubanks L. M.; Dickerson T. J.; Janda K. D. The Strange Case of the Botulinum Neurotoxin: Using Chemistry and Biology To Modulate the Most Deadly Poison. Angew. Chem., Int. Ed. 2008, 47, 8360–8379. [DOI] [PubMed] [Google Scholar]
- Wein L. M.; Liu Y. Analyzing a Bioterror Attack on the Food Supply: The Case of Botulinum Toxin in Milk. Proc. Natl. Acad. Sci. U.S.A. 2005, 102, 9984–9989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh B. R. Intimate Details of the Most Poisonous Poison. Nat. Struct. Biol. 2000, 7, 617–619. [DOI] [PubMed] [Google Scholar]
- Turton K.; Chaddock J. A.; Acharya K. R. Botulinum and Tetanus Neurotoxins: Structure, Function and Therapeutic Utility. Trends Biochem. Sci. 2002, 27, 552–558. [DOI] [PubMed] [Google Scholar]
- Lacy D. B.; Tepp W.; Cohen A. C.; DasGupta B. R.; Stevens R. C. Crystal Structure of Botulinum Neurotoxin Type A and Implications for Toxicity. Nat. Struct. Biol. 1998, 5, 898–902. [DOI] [PubMed] [Google Scholar]
- Swaminathan S.; Eswaramoorthy S. Structural Analysis of the Catalytic and Binding Sites of Clostridium Botulinum Neurotoxin B. Nat. Struct. Biol. 2000, 7, 693–699. [DOI] [PubMed] [Google Scholar]
- Binz T.; Blasi J.; Yamasaki S.; Baumeister A.; Link E.; Sudhof T. C.; Jahn R.; Niemann H. Proteolysis of SNAP-25 by Types E and A Botulinal Neurotoxins. J. Biol. Chem. 1994, 269, 1617–1620. [PubMed] [Google Scholar]
- Schmidt J. J.; Stafford R. G. Botulinum Neurotoxin Serotype F: Identification of Substrate Recognition Requirements and Development of Inhibitors with Low Nanomolar Affinity. Biochemistry 2005, 44, 4067–4073. [DOI] [PubMed] [Google Scholar]
- a Schiavo G.; Benfenati F.; Poulain B.; Rossetto O.; Polverino de Laureto P.; DasGupta B. R.; Montecucco C. Tetanus and Botulinum-B Neurotoxins Block Neurotransmitter Release by Proteolytic Cleavage of Synaptobrevin. Nature 1992, 359, 832–835. [DOI] [PubMed] [Google Scholar]; b Schiavo G.; Malizio C.; Trimble W. S.; Polverino de Laureto P.; Milan G.; Sugiyama H.; Johnson E. A.; Montecucco C.; Botulinum G. Neurotoxin Cleaves VAMP/Synaptobrevin at a Single Ala-Ala Peptide Bond. J. Biol. Chem. 1994, 269, 20213–20216. [PubMed] [Google Scholar]; c Schiavo G.; Rossetto O.; Catsicas S.; Polverino de Laureto P.; DasGupta B. R.; Benfenati F.; Montecucco C. Identification of the Nerve Terminal Targets of Botulinum Neurotoxin Serotypes A, D, and E. J. Biol. Chem. 1993, 268, 23784–23787. [PubMed] [Google Scholar]; d Schiavo G.; Shone C. C.; Rossetto O.; Alexander F. C.; Montecucco C. Botulinum Neurotoxin Serotype F is a Zinc Endopeptidase Specific for VAMP/Synaptobrevin. J. Biol. Chem. 1993, 268, 11516–11519. [PubMed] [Google Scholar]
- Blasi J.; Chapman E. R.; Yamasaki S.; Binz T.; Niemann H.; Jahn R. Botulinum Neurotoxin C1 Blocks Neurotransmitter Release by Means of Cleaving HPC-1/Syntaxin. EMBO J. 1993, 12, 4821–4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Foran P. G.; Davletov B.; Meunier F. A. Getting Muscles Moving Again after Botulinum Toxin: Novel Therapeutic Challenges. Trends Mol. Med. 2003, 9, 291–299. [DOI] [PubMed] [Google Scholar]; b Foran P. G.; Mohammed N.; Lisk G. O.; Nagwaney S.; Lawrence G. W.; Johnson E.; Smith L.; Aoki K. R.; Dolly J. O. Evaluation of the Therapeutic Usefulness of Botulinum Neurotoxin B, C1, E, and F Compared with the Long Lasting Type A. Basis for Distinct Durations of Inhibition of Exocytosis in Central Neurons. J. Biol. Chem. 2003, 278, 1363–1371. [DOI] [PubMed] [Google Scholar]
- Arnon S. S.; Schechter R.; Inglesby T. V.; Henderson D. A.; Bartlett J. G.; Ascher M. S.; Eitzen E.; Fine A. D.; Hauer J.; Layton M.; Lillibridge S.; Osterholm M. T.; O’Toole T.; Parker G.; Perl T. M.; Russell P. K.; Swerdlow D. L.; Tonat K. Botulinum Toxin as a Biological Weapon: Medical and Public Health Management. JAMA, J. Am. Med. Assoc. 2001, 285, 1059–1070. [DOI] [PubMed] [Google Scholar]
- Larsen J. C. U.S. Army Botulinum Neurotoxin (BoNT) Medical Therapeutics Research Program: Past Accomplishments and Future Directions. Drug Dev. Res. 2009, 70, 266–278. [Google Scholar]
- Eichhorn T.; Dolimbek B. Z.; Deeg K.; Efferth T.; Atassi M. Z. Inhibition in Vivo of the Activity of Botulinum Neurotoxin A by Small Molecules Selected by Virtual Screening. Toxicon 2012, 60, 1180–1190. [DOI] [PubMed] [Google Scholar]
- a Potavathri S.; Kantak A.; DeBoef B. Increasing Synthetic Efficiency via Direct C–H Functionalization: Formal Synthesis of an Inhibitor of Botulinum Neurotoxin. Chem. Commun. 2011, 47, 4679–4681. [DOI] [PubMed] [Google Scholar]; b Boldt G. E.; Kennedy J. P.; Janda K. D. Identification of a Potent Botulinum Neurotoxin A Protease Inhibitor Using in Situ Lead Identification. Chem. Org. Lett. 2006, 8, 1729–1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šilhár P.; Čapková K.; Salzameda N. T.; Barbieri J. T.; Hixon M. S.; Janda K. D. Botulinum Neurotoxin A Protease: Discovery of Natural Product Exosite Inhibitors. J. Am. Chem. Soc. 2010, 132, 2868–2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Čapek P.; Zhang Y.; Barlow D. J.; Houseknecht K. L.; Smith G. R.; Dickerson T. J. Enhancing the Pharmacokinetic Properties of Botulinum Neurotoxin Serotype A Protease Inhibitors through Rational Design. ACS Chem. Neurosci. 2011, 2, 288–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opsenica I.; Filipović V.; Nuss J. E.; Gomba L. M.; Opsenica D.; Burnett J. C.; Gussio R.; Šolaja B. A.; Bavari S. The Synthesis of 2,5-Bis(4-amidinophenyl)thiophene Derivatives Providing Submicromolar-Range Inhibition of the Botulinum Neurotoxin Serotype A Metalloprotease. Eur. J. Med. Chem. 2012, 53, 374–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šilhar P.; Alakurtti S.; Čapkova K.; Xiaochuan F.; Shoemaker C. B.; Yli-Kauhaluoma J.; Janda K. D. Synthesis and Evaluation of Library of Betulin Derivatives against the Botulinum Neurotoxin A Protease. Bioorg. Med. Chem. Lett. 2011, 21, 2229–2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardellina J. H.; Roxas-Duncan V. I.; Montgomery V.; Eccard V.; Campbell Y.; Hu X.; Khavrutskii I.; Tawa G. J.; Wallqvist A.; Gloer J. B.; Phatak N. L.; Höller U.; Soman A. G.; Joshi B. K.; Hein S. M.; Wicklow D. T.; Smith L. A. Fungal bis-Naphthopyrones as Inhibitors of Botulinum Neurotoxin Serotype A. ACS Med. Chem. Lett. 2012, 3, 387–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opsenica I.; Burnett J. C.; Gussio R.; Opsenica D.; Todorović N.; Lanteri C. A.; Sciotti R. J.; Gettayacamin M.; Basilico N.; Taramelli D.; Nuss J. E.; Wanner L.; Panchal R. G.; Šolaja B. A.; Bavari S. A Chemotype That Inhibits Three Unrelated Pathogenic Targets: The Botulinum Neurotoxin Serotype A Light Chain, P. falciparum Malaria, and the Ebola Filovirus. J. Med. Chem. 2011, 54, 1157–1169and references cited therein.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a TDR. http://www.who.int/tdr/diseases-topics/malaria/en/index.html (accessed September 30, 2013).; b Hay S. I.; Guerra C. A.; Tatem A. J.; Noor A. M.; Snow R. W. The Global Distribution and Population at Risk of Malaria: Past, Present, and Future. Lancet 2004, 4, 327–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Opsenica D. M.; Šolaja B. A. Antimalarial Peroxides. J. Serb. Chem. Soc. 2009, 74, 1155–1193and references sited therein. [Google Scholar]; b Opsenica I.; Terzić N.; Opsenica D.; Angelovski G.; Lehnig M.; Eilbracht P.; Tinant B.; Juranić Z.; Smith K. S.; Yang Z. S.; Diaz D. S.; Smith P. L.; Milhous W. K.; Đoković D.; Šolaja B. A. Tetraoxane Antimalarials and Their Reaction with Fe(II). J. Med. Chem. 2006, 49, 3790–3799. [DOI] [PubMed] [Google Scholar]
- O’Neill P. M.; Barton V. E.; Ward S. A.; Chadwick J.. 4-Aminoquinolines: Chloroquine, Amodiaquine and Next-Generation Analogues. In Treatment and Prevention of Malaria: Antimalarial Drug Chemistry, Action and Use; Staines H. M., Krishna S., Eds.; Springer: Basel, Switzerland, 2012; pp 19–44. [Google Scholar]
- Ecker A.; Lehane A. M.; Fidock D. A.. Molecular Markers of Plasmodium Resistance to Antimalarials. In Treatment and Prevention of Malaria: Antimalarial Drug Chemistry, Action and Use; Staines H. M., Krishna S., Eds.; Springer: Basel, Switzerland, 2012; pp 249–280. [Google Scholar]
- Roepe P. D. PfCRT-Mediated Drug Transport in Malarial Parasites. Biochemistry 2011, 50, 163–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Opsenica D. M.; Šolaja B. A.. Second-Generation Peroxides: The OZs and Artemisone. In Treatment and Prevention of Malaria: Antimalarial Drug Chemistry, Action and Use; Staines H. M., Krishna S., Eds.; Springer: Basel, Switzerland, 2012; pp 211–280. [Google Scholar]; b Slack R. D.; Jacobine A. M.; Posner G. H. Antimalarial Peroxides: Advances in Drug Discovery and Design. MedChemComm 2012, 3, 281–297. [Google Scholar]; c Kumar N.; Singh R.; Rawat D. S. Tetraoxanes: Synthetic and Medicinal Chemistry Perspective. Med. Res. Rev. 2012, 32, 581–610. [PubMed] [Google Scholar]
- a Kaur K.; Jain M.; Reddy R. P.; Jain R. Quinolines and Structurally Related Heterocycles as Antimalarials. Eur. J. Med. Chem. 2010, 45, 3245–3264. [DOI] [PubMed] [Google Scholar]; b Milner E.; McCalmont W.; Bhonsle J.; Caridha D.; Cobar J.; Gardner S.; Gerena L.; Goodine D.; Lanteri C.; Melendez V.; Roncal N.; Sousa J.; Wipf P.; Dow G. S. Anti-Malarial Activity of a Non-Piperidine Library of Next-Generation Quinoline Methanols. Malar. J. 2010, 9, 51. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Lanteri C. A.; Johnson J. D.; Waters N. C. Recent Advances in Malaria Drug Discovery. Recent Pat. Anti-Infect. Drug Discovery 2007, 2, 95–114. [DOI] [PubMed] [Google Scholar]
- Sparatore A.; Basilico N.; Casagrande M.; Parapini S.; Taramelli D.; Brun R.; Wittlin S.; Sparatore F. Antimalarial Activity of Novel Pyrrolizidinyl Derivatives of 4-Aminoquinoline. Bioorg. Med. Chem. Lett. 2008, 18, 3737–3740. [DOI] [PubMed] [Google Scholar]
- Fawaz Mzayek F.; Deng H.; Mather F. J.; Wasilevich E.; Huayin Liu H.; Hadi C. M.; Chansolme D. H.; Murphy H. A.; Melek B. H.; Tenaglia A. N.; Mushatt D. M.; Dreisbach A. W.; Lertora J. J. L.; Krogstad D. J. Randomized Dose-Ranging Controlled Trial of AQ-13, a Candidate Antimalarial, and Chloroquine in Healthy Volunteers. PLoS Clin. Trials 2007, 2, e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan J. K.; Alumasa J. N.; Yearick K.; Ekoue-Kovi K. A.; Casabianca L. B.; de Dios A. C.; Wolf C.; Roepe P. D. 4-N-, 4-S-, and 4-O-Chloroquine Analogues: Influence of Side Chain Length and Quinolyl Nitrogen pKa on Activity vs Chloroquine Resistant Malaria. J. Med. Chem. 2008, 51, 3466–3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouznetsov V. V.; Gómez-Barrio A. Recent Developments in the Design and Synthesis of Hybrid Molecules Based on Aminoquinoline Ring and Their Antiplasmodial Evaluation. Eur. J. Med. Chem. 2009, 44, 3091–3113. [DOI] [PubMed] [Google Scholar]
- a Burgess S. J.; Kelly J. X.; Shomloo S.; Wittlin S.; Brun R.; Liebmann K.; Peyton D. H. Synthesis, Structure–Activity Relationship, and Mode-of-Action Studies of Antimalarial Reversed Chloroquine Compounds. J. Med. Chem. 2010, 53, 6477–6489. [DOI] [PMC free article] [PubMed] [Google Scholar]; b October N.; Watermeyer N. D.; Yardley V.; Egan T. J.; Ncokazi K.; Chibale K. Reversed Chloroquines Based on the 3,4-Dihydropyrimidin-2(1H)-one Scaffold: Synthesis and Evaluation for Antimalarial, β-Haematin Inhibition, and Cytotoxic Activity. ChemMedChem 2008, 3, 1649–1653. [DOI] [PubMed] [Google Scholar]
- a Navarro M.; Castro W.; Martínez A.; Delgado R. A. S. The Mechanism of Antimalarial Action of [Au(CQ)(Pph3)]PF6: Structural Effects and Increased Drug Lipophilicity Enhance Heme Aggregation Inhibition at Lipid/Water Interfaces. J. Inorg. Biochem. 2011, 105, 276–282. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Beckford F.; Dourth D. Jr.; Shaloski M.; Didion J.; Thessing J.; Woods J.; Crowell V.; Gerasimchuk N.; Gonzalez-Sarrías A.; Seeram N. P. Half-Sandwich Ruthenium–Arene Complexes with Thiosemicarbazones: Synthesis and Biological Evaluation of [(η6-p-Cymene)Ru(piperonal thiosemicarbazones)Cl]Cl Complexes. J. Inorg. Biochem. 2011, 105, 1019–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perić M.; Fajdetić A.; Rupčić R.; Alihodžić S.; Žiher D.; Bukvić Krajačić M.; Smith K. S.; Ivezić-Schönfeld Z.; Padovan J.; Landek G.; Jelić D.; Hutinec A.; Mesić M.; Ager A.; Ellis W. Y.; Milhous W. K.; Ohrt C.; Spaventi R. Antimalarial Activity of 9a-N Substituted 15-Membered Azalides with Improved in Vitro and in Vivo Activity over Azithromycin. J. Med. Chem. 2012, 55, 1389–1401. [DOI] [PubMed] [Google Scholar]
- Musonda C. C.; Yardley V.; de Souza R. C. C.; Ncokazi K.; Egan T. J.; Chibale K. Antiplasmodial, B-Haematin Inhibition, Antitrypanosomal and Cytotoxic Activity in Vitro of Novel 4-Aminoquinoline 2-Imidazolines. Org. Biomol. Chem. 2008, 6, 4446–4451. [DOI] [PubMed] [Google Scholar]
- Opsenica I.; Opsenica D.; Lanteri C. A.; Anova L.; Milhous W. K.; Smith K. S.; Šolaja B. A. New Chimeric Antimalarials with 4-Aminoquinoline Moiety Linked to a Tetraoxane Skeleton. J. Med. Chem. 2008, 51, 6216–6219. [DOI] [PubMed] [Google Scholar]
- Nilsen A.; LaCrue A. N.; White K. L.; Forquer I. P.; Cross R. M.; Marfurt J.; Mather M. W.; Delves M. J.; Shackleford D. M.; Saenz F. E.; Morrisey J. M.; Steuten J.; Mutka T.; Li Y.; Wirjanata G.; Ryan E.; Duffy S.; Kelly J. X.; Sebayang B. F.; Zeeman A.-M.; Noviyanti R.; Sinden R. E.; Kocken C. H. M.; Price R. N.; Avery V. M.; Angulo-Barturen I.; Jiménez-Díaz M. B.; Ferrer S.; Herreros E.; Sanz L. M.; Gamo F.-J.; Bathurst I.; Burrows J. N.; Siegl P.; Guy R. K.; Winter R. W.; Vaidya A. B.; Charman S. A.; Kyle D. E.; Manetsch R.; Riscoe M. K. Quinolone-3-diarylethers: A New Class of Antimalarial Drug. Sci. Transl. Med. 2013, 5, 177ra37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnett J. C.; Opsenica D.; Sriraghavan K.; Panchal R. G.; G. Ruthel G.; Hermone A. R.; Nguyen T. L.; Kenny T. A.; Lane D. J.; McGrath C. F.; Schmidt J. J.; Vennerstrom J. L.; Gussio R.; Šolaja B. A.; Bavari S. A Refined Pharmacophore Identifies Potent 4-Amino-7-chloroquinoline-Based Inhibitors of the Botulinum Neurotoxin Serotype A Metalloprotease. J. Med. Chem. 2007, 50, 2127–2136. [DOI] [PubMed] [Google Scholar]
- Šolaja B. A.; Opsenica D.; Smith K. S.; Milhous W. K.; Terzić N.; Opsenica I.; Burnett J. C.; Nuss J.; Gussio R.; Bavari S. Novel 4-Aminoquinolines Active against Chloroquine-Resistant and Sensitive P. falciparum Strains That Also Inhibit Botulinum Serotype A. J. Med. Chem. 2008, 51, 4388–4391. [DOI] [PubMed] [Google Scholar]
- Recently, compound 7 was tested in vivo, P. berghei, Thompson test. At 320 mg kg–1 dose–1, 5/5 mice were cured. Detailed in vivo results will be published elsewhere.
- Albert D.; Feigel M. β-Loop, γ-Loop and Helical Peptide Conformations in Cyclopeptides Containing a Steroidal Pseudo-Amino Acid. Helv. Chim. Acta 1997, 80, 2168–2181. [Google Scholar]
- a Schmidt J. J.; Bostian K. A. Endoproteinase Activity of Type A Botulinum Neurotoxin: Substrate Requirements and Activation by Serum Albumin. J. Protein Chem. 1997, 16, 19–26. [DOI] [PubMed] [Google Scholar]; b Schmidt J. J.; Bostian K. A. Proteolysis of Synthetic Peptides by Type A Botulinum Neurotoxin. J. Protein Chem. 1995, 14, 703–708. [DOI] [PubMed] [Google Scholar]; c Schmidt J. J.; Stafford R. G. Fluorigenic Substrates for the Protease Activities of Botulinum Neurotoxins, Serotypes A, B, and F. Appl. Environ. Microbiol. 2003, 69, 297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Schmidt J. J.; Stafford R. G. A High-Affinity Competitive Inhibitor of Type A Botulinum Neurotoxin Protease Activity. FEBS Lett. 2002, 532, 423–426. [DOI] [PubMed] [Google Scholar]; e Schmidt J. J.; Stafford R. G.; Bostian K. A.; Type A. Botulinum Neurotoxin Proteolytic Activity: Development of Competitive Inhibitors and Implications for Substrate Specificity at the S10 Binding Subsite. FEBS Lett. 1998, 435, 61–64. [DOI] [PubMed] [Google Scholar]; f Schmidt J. J.; Stafford R. G.; Millard C. B. High-Throughput Assays for Botulinum Neurotoxin Proteolytic Activity: Serotypes A, B, D, and F. Anal. Biochem. 2001, 296, 130–137. [DOI] [PubMed] [Google Scholar]
- For contribution of non-steroidal increments to BoNT/A LC inhibitory activity, see the following:Opsenica I. M.; Tot M.; Gomba L.; Nuss J. E.; Sciotti R. J.; Bavari S.; Burnett J. C.; Solaja B. A. 4-Amino-7-chloroquinolines: Probing Ligand Efficiency Provides Botulinum Neurotoxin Serotype A Light Chain Inhibitors with Significant Antiprotozoal Activity. J. Med. Chem. 2013, 56, 5860–5871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson A. A.; Jiao G.-S.; Kim S.; Thai A.; Cregar-Hernandez L.; Margosiak S. A.; Johnson A. T.; Han G. W.; O’Malley S.; Stevens R. C. Structural Characterization of Three Novel Hydroxamate-Based Zinc Chelating Inhibitors of the Clostridium Botulinum Serotype A Neurotoxin Light Chain Metalloprotease Reveals a Compact Binding Site Resulting from 60/70 Loop Flexibility. Biochemistry 2011, 50, 4019–4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnett J. C.; Schmidt J. J.; McGrath C. F.; Nguyen T. L.; Hermone A. R.; Panchal R. G.; Vennerstrom J. L.; Kodukula K.; Zaharevitz D. W.; Gussio R.; Bavari S. Conformational Sampling of the Botulinum Neurotoxin Serotype A Light Chain: Implications for Inhibitor Binding. Bioorg. Med. Chem. 2005, 13, 333–341. [DOI] [PubMed] [Google Scholar]
- a Burnett J. C.; Wang C.; Nuss J. E.; Nguyen T. L.; Hermone A. R.; Schmidt J. J.; Gussio R.; Wipf P.; Bavari S. Pharmacophore-Guided Lead Optimization: The Rational Design of a Non-Zinc Coordinating, Sub-Micromolar Inhibitor of the Botulinum Neurotoxin Serotype A Metalloprotease. Bioorg. Med. Chem. Lett. 2009, 19, 5811–5813. [DOI] [PubMed] [Google Scholar]; b Nuss J. E.; Dong Y.; Wanner L. M.; Ruthel G.; Wipf P.; Gussio R.; Vennerstrom J. L.; Bavari S.; Burnett J. C. Pharmacophore Refinement Guides the Design of Nanomolar-Range Botulinum Neurotoxin Serotype A Light Chain Inhibitors. ACS Med. Chem. Lett. 2010, 1, 301–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- All ligand structures were drawn using in Maestro module (Suite 2012: Maestro, version 9.3, Schrödinger, LLC, New York, NY, 2012). Protonation and pKa of ligands were determined using Epik (Suite 2012: Epik, version 2.3, Schrödinger, LLC, New York, NY, 2012). Ligand docking was accomplished using Glide (Suite 2012: Glide, version 5.8, Schrödinger, LLC, New York, NY, 2012) and InducedFit (Suite 2012: Schrödinger Suite 2012 Induced Fit Docking Protocol; Glide, version 5.8, Schrödinger, LLC, New York, NY, 2012; Prime, version 3.1: Schrödinger, LLC: New York, NY, 2012). Additional energy minimizations were done using Impact (Suite 2012: Impact, version 5.8, Schrödinger, LLC, New York, NY, 2012) and MacroModel energy minimization (Suite 2012: MacroModel, version 9.9, Schrödinger, LLC: New York, NY, 2012).
- Zuniga J. E.; Schmidt J. J.; Fenn T.; Burnett J. C.; Arac D.; Gussio R.; Stafford R. G.; Badie S. S.; Bavari S.; Brunger A. T. A Potent Peptidomimetic Inhibitor of Botulinum Neurotoxin Serotype A Has a Very Different Conformation Than SNAP-25 Substrate. Structure 2008, 16, 1588–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biedermannova L.; Riley K. E.; Berka K.; Hobza P.; Vondrasek J. Another Role of Proline: Stabilization Interactions in Proteins and Protein Complexes Concerning Proline and Tryptophane. Phys. Chem. Chem. Phys. 2008, 10, 6350–6359. [DOI] [PubMed] [Google Scholar]
- Milhous W. K.; Weatherly N. F.; Bowdre J. H.; Desjardins R. E. In Vitro Activities of and Mechanisms of Resistance to Antifol Antimalarial Drugs. Antimicrob. Agents Chemother. 1985, 27, 525–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Musonda C. C.; Gut J.; Rosenthal P. J.; Yardley V.; Carvalho de Souza R. C.; Chibale K. Application of Multicomponent Reactions to Antimalarial Drug Discovery. Part 2: New Antiplasmodial and Antitrypanosomal 4-Aminoquinoline Gamma- and Delta-Lactams via a “Catch and Release” Protocol. Bioorg. Med. Chem. 2006, 14, 5605–5615. [DOI] [PubMed] [Google Scholar]; b Peck R. M.; Preston R. K.; Creech H. J. Nitrogen Mustard Analogs of Antimalarial Drugs. J. Am. Chem. Soc. 1958, 81, 3984–3989. [Google Scholar]; c Price C. C.; Leonard N. J.; Peel E. W.; Reitsem R. H. Some 4-Amino-7-chloroquinoline Derivatives. J. Am. Chem. Soc. 1946, 68, 1807–1808. [DOI] [PubMed] [Google Scholar]; d Singh C.; Malik H.; Puri S. K. Synthesis and Antimalarial Activity of a New Series of Trioxaquines. Bioorg. Med. Chem. 2004, 12, 1177–1182. [DOI] [PubMed] [Google Scholar]
- Frigerio M.; Santagostino M.; Sputore S.; Palmisano G. Oxidation of Alcohols with o-Iodoxybenzoic Acid (IBX) in DMSO: A New Insight into an Old Hypervalent Iodine Reagent. J. Org. Chem. 1995, 60, 7272–7276. [Google Scholar]
- Burnett J. C.; Ruthel G.; Stegmann C. M.; Panchal R. G.; Nguyen T. L.; Hermone A. R.; Stafford R. G.; Lane D. J.; Kenny T. A.; McGrath C. F.; Wipf P.; Stahl A. M.; Schmidt J. J.; Gussio R.; Brunger A. T.; Bavari S. Inhibition of Metalloprotease Botulinum Serotype A from a Pseudo-Peptide Binding Mode to a Small Molecule That Is Active in Primary Neurons. J. Biol. Chem. 2007, 282, 5004–5014. [DOI] [PubMed] [Google Scholar]
- Kuhn T. B. Growing and Working with Spinal Motor Neurons. Methods Cell Biol. 2003, 71, 67–87. [DOI] [PubMed] [Google Scholar]
- Stahl A. M.; Ruthel G.; Torres-Melendez E.; Kenny T. A.; Panchal R. G.; Bavari S. Primary Cultures of Embryonic Chicken Neurons for Sensitive Cell-Based Assay of Botulinum Neurotoxin: Implications for Therapeutic Discovery. J. Biomol. Screening 2007, 12, 370–377. [DOI] [PubMed] [Google Scholar]
- Desjardins R. E.; Canfield C. J.; Haynes D. E.; Chulay J. D. Quantitative Assessment of Antimalarial Activity in Vitro by a Semiautomated Microdilution Technique. Antimicrob. Agents Chemother. 1979, 16, 710. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.