Abstract
Chemokine receptors and their ligands play a central role in cancer metastasis, inflammatory disorders, and viral infections. Viologen dendrimers (VGD) emerged recently as a promising class of synthetic polycationic ligands for chemokine receptor CXCR4. The objective of this study was to evaluate the potential of VGD as novel dual-function polycations capable of simultaneous CXCR4 antagonism and gene delivery. As part of our systematic studies, we have synthesized a library of VGD with differences in molecular architecture, number of positive charges, and type of capping group. The ability of VGD to condense DNA was evaluated, and physicochemical and biological properties of the resulting polyplexes were studied. We have evaluated the effect of VGD surface charge, size, capping group, and molecular architecture on physicochemical properties of polyplexes, transfection efficiency, CXCR4 antagonism, and cytotoxicity in human epithelial osteosarcoma (U2OS) and in human liver hepatocellular carcinoma (HepG2) cells. We found that properties and behavior of the polyplexes are most dependent on the number of positive charges and molecular weight of VGD and to a lesser extent on the type of a capping group. Using TNFα plasmid, we have demonstrated that VGD prevents CXCR4-mediated cancer cell invasion and facilitates TNFα-mediated cancer cell killing. Such dual-function carriers have potential to enhance the overall therapeutic outcomes of cancer gene therapy.
Introduction
Nonviral carriers for gene delivery are generally considered safer and better-controlled alternatives compared to viral vectors.1,2 Numerous carriers have shown success in compacting therapeutic DNA or other types of nucleic acids into nanosized particles and mediating transfection in target cells or tissues both in vitro and in vivo. These include cationic lipids and polymers such as DOTAP, poly(l-lysine) (PLL), poly(amido amine)s (PAMAM), poly(ethylene imine) (PEI), poly(propyleneimine) (PPI), or triazine dendrimers.3−9 However, low delivery efficiency of these nonviral carriers remains a major hurdle for progression into clinical applications.
Multiple attempts have been made to overcome the limitations of nonviral carriers. A recent concept suggests that therapeutic outcome of nonviral gene delivery can be enhanced by designing dual-function systems that not only deliver nucleic acids, but also function as pharmacologically active agents. The synergistic effect produced by combining pharmacological activity of the carrier and the therapeutic nucleic acids in one formulation can greatly enhance the overall therapeutic outcome. For example, Huang and colleagues developed a cationic lipid capable of delivering siRNA to knock down epidermal growth factor receptor (EGFR) while down-regulating the phosphorylated extracellular signal-regulated kinase (pERK) simultaneously to enhance anticancer activity.10 Rice and colleagues have designed a cationic polypeptide with intrinsic proteasome inhibitory function to enhance transfection activity of delivered plasmid DNA.11 We recently reported a novel type of bioreducible poly(amido amine)s that deliver plasmid DNA and inhibit chemokine receptor CXCR4.12,13
Chemokines, a family of 48 signaling proteins, control through chemotaxis the migration of cells that express the appropriate chemokine receptor. In tumors, a complex network of chemokines and chemokine receptors controls cell trafficking into and out of the tumor microenvironment.14 Importantly with respect to the present study, chemokines and chemokine receptors are directly involved in the molecular control of cancer metastasis and governing organ-specific homing of metastatic cancer cells. Although malignant cells from different types of cancer have different expression profiles of chemokine receptors, CXC receptor 4 (CXCR4) is the most widely expressed chemokine receptor in human cancers.
CXCR4 is a highly conserved transmembrane G protein-coupled receptor that exclusively binds its ligand SDF-1 (also named CXCL12). SDF-1, on the other hand, is known to bind also the CXCR7 receptor. While CXCR7 acts in part as a scavenger of SDF-1,15 recent reports suggest additional functions for the receptor in cancer progression.16 Binding of SDF-1 to CXCR4 activates several intracellular signaling transduction pathways that regulate proliferation, adhesion, and invasion of cancer cells and affect cell survival and migration.17−19 Over 25 different types of human cancer overexpress CXCR4, making it and the CXCR4/SDF-1 axis a promising target within the chemokine network for novel therapies.14 Evidence supporting the exploration of CXCR4 as a therapeutic target in cancer stems from experimental in vitro and in vivo studies as well as retrospective clinical studies.20−22 The studies have documented increased invasive and metastatic potential in CXCR4-expressing tumor cells. Furthermore, the CXCR4/SDF-1 axis in the tumor microenvironment appears to nurture local tumor growth.23−25 The following two mechanisms are distinct ways through which the CXCR4/SDF-1 axis promotes cancer progression: (i) CXCR4 facilitates the metastatic spread of the disease to sites where SDF-1 is highly expressed (e.g., lung, liver, bone marrow, and brain) and (ii) high expression of SDF-1 in the primary tumor enhances growth and inflammation of the tumor via local autocrine and paracrine mechanisms.26−28 In addition to its role in cancer, CXCR4 also functions as a co-receptor for HIV-1 entry into the host CD4+ T-cells, and its potential as a therapeutic target has been highlighted in many studies.29,30 CXCR4 also regulates the recruitment and invasion of immune cells like T-cells, macrophages, and neutrophils in multiple inflammatory conditions such as inflammatory bowel disease and lung inflammation.31−34
Inhibition of CXCR4 activity, whether by using small molecule antagonists, anti-CXCR4 antibodies, or anti-CXCR4 siRNA has already shown success in improving the treatment of multiple disease conditions.35−37 The first CXCR4 antagonist AMD3100 (Plerixafor) received FDA approval in 2008.38 Multiple other CXCR4 inhibitors are at various stages of preclinical and clinical development. Interestingly, dendrimers based on viologen (dialkylated 4,4′-bipyridinium salts) have been found to exhibit potent antagonistic activity against CXCR4.39 Previous studies systematically explored the structure–activity relationships of these viologen-based dendrimers (VGD) as HIV inhibitors. It was found that the HIV inhibition by VGD was controlled by the molecular architecture and number of positive charges in the dendrimer molecule. In addition, capping with thymine groups increased the binding affinity of VGD with the carboxylate residues in the binding site of the CXCR4 receptor by promoting formation of hydrogen bonds.39
In this study, we tested the hypothesis that, due to their polycationic nature, the VGD will be able to form complexes with plasmid DNA and mediate DNA transfection, while at the same time retaining their CXCR4 inhibiting activity. To test the hypothesis, we have examined the ability of a library of VGD with differences in molecular architecture, number of positive charges, and type of capping group to condense DNA and form VGD/DNA polyplexes. Transfection activity, cytotoxicity, and CXCR4 antagonism of the polycations and polyplexes were evaluated. The ability of the best performing VGD to simultaneously deliver therapeutic TNFα gene and inhibit invasion of cancer cells was then tested to establish their dual-function capability.
Results
Synthesis and Categorization of VGD
We have synthesized a series of 13 different VGD by conjugating different dendrons with various capping groups (methyl, ethyl, thymine) to two types of dendrimer cores to obtain different generations of comb-branched and spheroidal dendrimer architectures. These VGD were categorized into three groups based on their architecture and molecular weight (Scheme 1). The main characteristic features and properties are detailed in Table 1. Precursors VGD1–3 represent the main building blocks used in the synthesis of higher generation VGD and contain only 2 positive charges per molecule. Comb-branched VGD4–6 consist of generation 1 (G1) and 2 (G2) dendrimers with different capping groups and 10 or 26 positive charges. Spheroidal VGD consist of generation 0–3 dendrimers and carry 6, 18, 42, and 90 positive charges and are capped with different capping groups.
Scheme 1. Building Blocks and Design of Viologen Dendrimers (VGD).
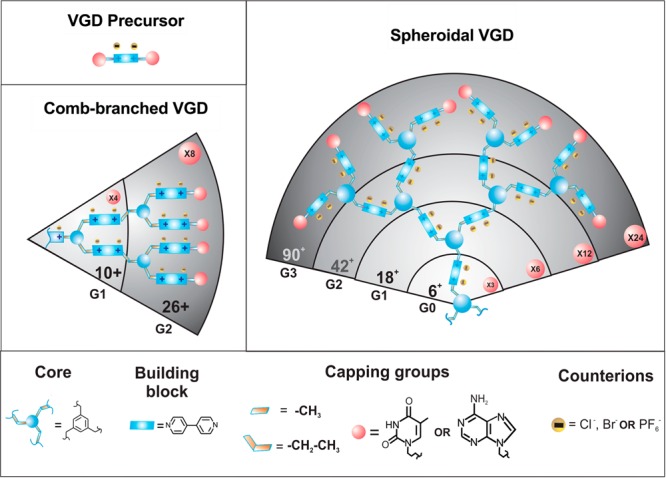
The numbers in the pink circles indicate total numbers of capping groups in a dendrimer generation.
Table 1. Properties of VGD.
| VGD | generation | capping group | number of charges | molecular weight | mass/charge | equivalent N/P ratioa |
|---|---|---|---|---|---|---|
| VGD Precursor | ||||||
| VGD1 | n/a | adenine | 2 | 640.3 | 320 | n/a |
| VGD2 | n/a | adenine and thymine | 2 | 645.3 | 323 | n/a |
| VGD3 | n/a | thymine | 2 | 780.5 | 390 | n/a |
| Comb-Branched VGD | ||||||
| VGD4 | G1 | ethyl | 10 | 2553 | 255 | 0.5 |
| VGD5 | G1 | thymine | 10 | 3105 | 311 | 1.0 |
| VGD6 | G2 | thymine | 26 | 7812 | 301 | 1.2 |
| Spheroidal VGD | ||||||
| VGD7 | G0 | Methyl | 6 | 1501 | 250 | 0.6 |
| VGD8 | G0 | thymine | 6 | 1300 | 217 | 0.7 |
| VGD9 | G1 | ethyl | 18 | 2687 | 149 | 1.0 |
| VGD10 | G1 | thymine | 18 | 4316 | 240 | 1.3 |
| VGD11 | G2 | thymine | 42 | 7947 | 189 | 1.0 |
| VGD12 | G3 | ethyl | 90 | 23 350 | 259 | 1.1 |
| VGD13 | G3 | thymine | 90 | 26 679 | 296 | 1.1 |
| Control | ||||||
| PEI | 25 000 (Mw) | 43.0 | 2.4 | |||
Minimum N/P ratio required to condense plasmid DNA.
DNA Condensation
DNA condensation capability of VGD was determined by EtBr exclusion assay (Figure 1). All comb-branched and spheroidal VGD were able to fully condense DNA in 10 mM HEPES buffer (pH 7.4). Equivalent N/P ratios required to fully condense DNA are summarized in Table 1. All these VGD displayed similar DNA condensation curves with a typical sigmoidal shape. DNA condensation was also confirmed by agarose gel electrophoresis. Absence of free DNA bands on the gel indicated complete DNA condensation and formation of stable polyplexes with all comb-branched and spheroidal VGD/DNA polyplexes prepared at N/P 4. VGD precursors (VGD1, VGD2, VGD3), however, failed to condense DNA into polyplexes even at N/P ratios as high as 20 (Figure S1 in Supporting Information).
Figure 1.
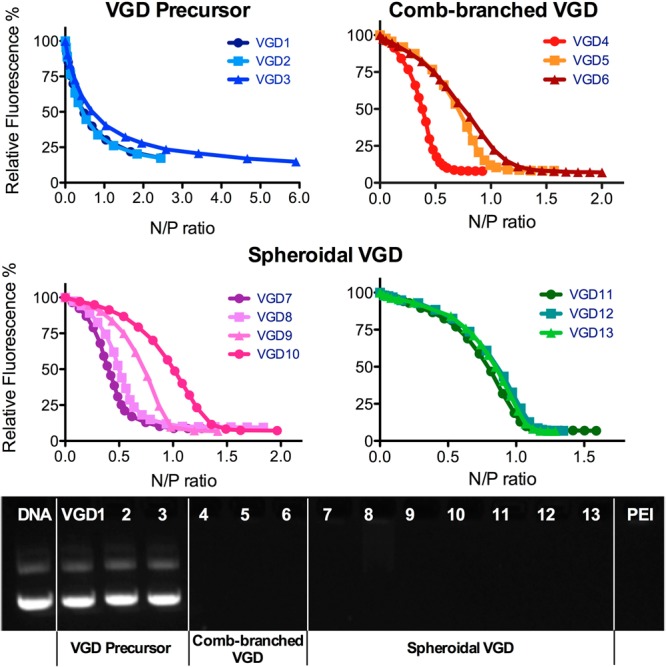
DNA condensation by VGD. DNA condensation ability of VGD was evaluated by EtBr exclusion assay in 10 mM HEPES buffer. Gel electrophoresis was conducted to confirm the condensed DNA and formation of VGD/DNA polyplexes (N/P 4).
Physicochemical Characterization of VGD/DNA Polyplexes
Hydrodynamic size and ζ-potential of all VGD/DNA polyplexes prepared at N/P 4 were measured by DLS (Figure 2). Consistent with their limited DNA condensing ability, the use of VGD precursors resulted in extensive aggregation of DNA and a negative surface charge of the aggregated particles (Figure S2). Polyplexes formed with the comb-branched VGD were positively charged with sizes ranging from 124 to 165 nm. In particular, generation 2 VGD6 formed significantly smaller polyplexes than generation 1 VGD4 and VGD5. Among the spheroidal VGD, the generation 0 VGD7 formed aggregated polyplexes with particle size >1 μm and a nearly neutral surface charge. Interestingly, VGD8 of the same generation formed polyplexes with smaller size of 320 nm and positive surface charge of 18 mV, suggesting better DNA condensation. Generation 1 dendrimers VGD9 and VGD10 formed DNA polyplexes which displayed small sizes around 50 nm and positive surface charge. Similarly, the sizes of the polyplexes formed using higher generations of dendrimers fell into a narrow range from 55 to 59 nm, with highly positively charged surfaces with ζ-potentials ranging from 20 to 25 mV.
Figure 2.
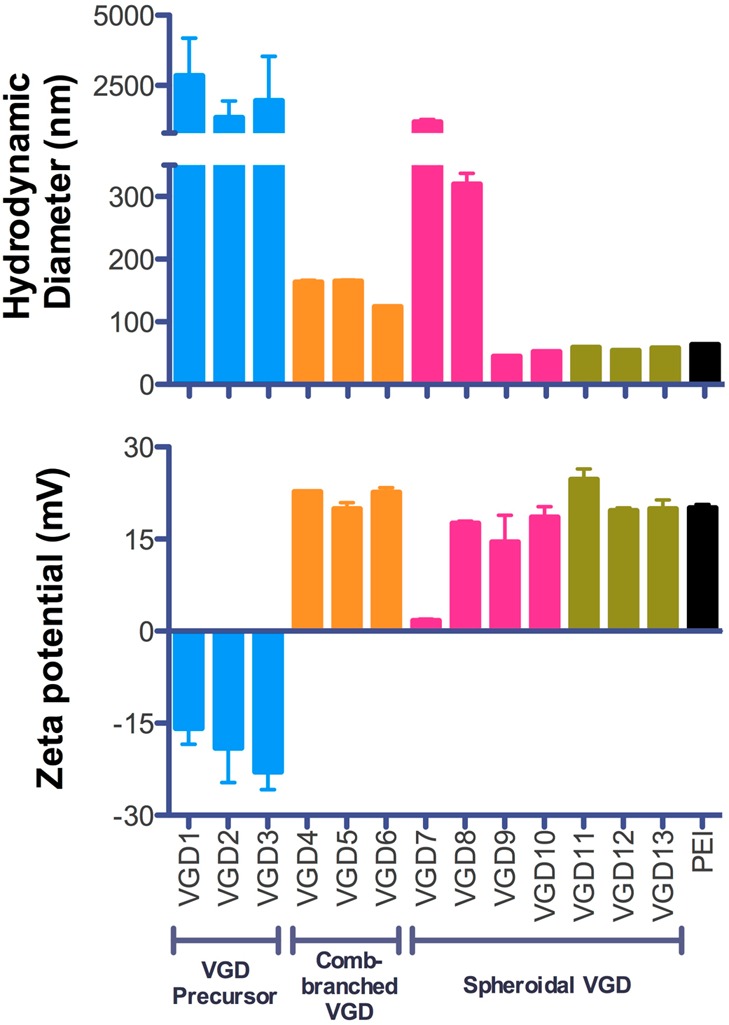
Size and ζ-potential of VGD/DNA polyplexes prepared at N/P 4 in 10 mM HEPES buffer (pH 7.4) and measured by DLS.
Cytotoxicity of VGD/DNA Polyplexes
Cytotoxicity of VGD/DNA polyplexes was first evaluated by MTS assay in U2OS cells (Figure 3). Cells were treated with polyplexes at different N/P ratios ranging from 4 to 40 for 4 h in the presence or absence of 10% FBS, followed by incubation in complete medium for another 24 h before measuring cell viability. In the presence of 10% FBS, polyplexes prepared with comb-branched VGD or VGD precursors showed no significant cytotoxicity. Polyplexes prepared with spheroidal VGD exhibited markedly higher cytotoxicity, except for generation 0 VGD7 and VGD8. VGD9 and VGD10 polyplexes showed toxicity at the highest N/P ratio (N/P 40) tested. Significant cytotoxicity was observed in VGD11–13 at N/P ratios above 15. In the absence of serum, the cytotoxicity of high-generation VGD polyplexes, both comb-branched VGD6 and spheroidal VGD9–13, increased significantly. For VGD11–13, only polyplexes prepared at N/P 4 showed acceptable cytotoxicity.
Figure 3.
Cytotoxicity of VGD/DNA polyplexes in U2OS and HepG2 cells. Cells seeded in 96-well plates were treated with polyplexes in medium (±10% FBS) followed by another 24 h incubation in complete culture medium before measuring cell viability using MTS assay.
The cytotoxicity of VGD/DNA polyplexes was then tested also in human liver hepatocellular carcinoma HepG2 cells. In general, all VGD/DNA polyplexes exhibited lower cytotoxicity in HepG2 cells than in U2OS cells. In the presence of serum, VGD9 and VGD10, which showed high cytotoxicity at N/P 40 in U2OS cells, did not show any significant cytotoxicity in HepG2 cells. Although the cytotoxicity profile of high-generation spheroidal VGD/DNA (VGD11–13) polyplexes was greatly improved in HepG2 cells, decreased cell viability at N/P 40 was still observed. In the absence of serum, cells treated with all the VGD/DNA polyplexes except for VGD11–13 maintained nearly 100% viability compared to untreated cells. VGD11–13 polyplexes displayed high cytotoxicity at N/P ratio above 15 in a dose-dependent manner. Cytotoxicity of the control polycation PEI exhibited similar cell line dependence as the VGD.
Transfection Efficiency of VGD/DNA Polyplexes
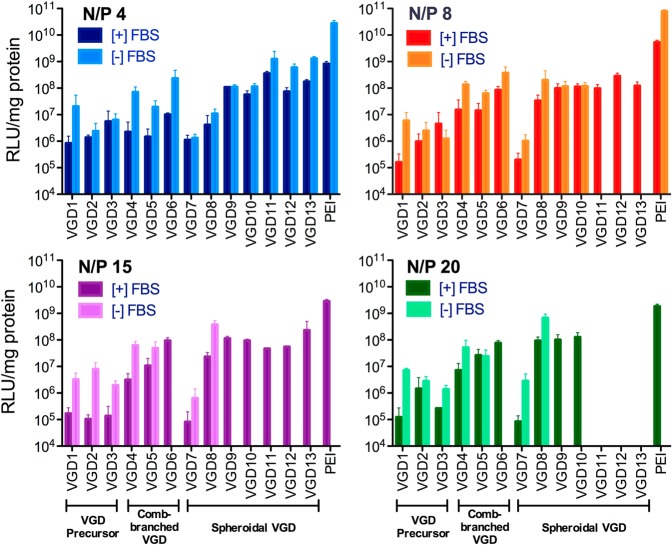
In vitro transfection activity of VGD/DNA polyplexes at different N/P ratios was evaluated in U2OS cells using luciferase plasmid DNA. Using the cytotoxicity data in Figure 3, only polyplex formulations that showed cell viability above 60% were tested for transfection. PEI/DNA polyplexes were used as the positive control (Figure 4). As expected, higher transfection efficiency was observed in serum-free conditions compared to conditions with 10% FBS. Using higher N/P ratios compromised the transfection efficiency in serum-free conditions due to increasing cytotoxicity. Polyplexes formed with the VGD precursors exhibited negligible transfection at all N/P ratios tested due to their poor ability to condense DNA. For comb-branched VGD polyplexes, VGD6 (G2) with higher molecular weight and more charges showed higher transfection efficiency in the presence of serum compared to VGD4 and VGD5 (G1). Likewise, the generation 0 VGD7 polyplexes exhibited marginal transfection efficiency that was similar to that of the VGD precursors. An exception was VGD8, which despite being a G0 dendrimer exhibited considerably higher transfection, especially at N/P > 8. Following a similar trend, VGD9 and VGD10 (G1) with higher molecular weight and more charges performed better than G0 VGD. Due to the high cytotoxicity of high-generation VGD (VGD11–13), only polyplexes prepared at N/P 4 were tested in serum-free conditions and only N/P 4, 8, and 15 were used in serum-containing conditions. Despite the cytotoxicity-imposed limit on the maximum dose, VGD11–13 polyplexes exhibited high transfection activity when compared with the other groups, especially at lower N/P ratios. Overall, comb-branched VGD6, spheroidal VGD8–13 exhibited high transfection activity that was in many cases comparable to the control PEI.
Figure 4.
Luciferase transfection of VGD/DNA polyplexes in U2OS cells. Transfection experiments were conducted in the presence or absence of 10% FBS. Transfection efficiency was expressed as RLU/mg protein ± SD (n = 3).
CXCR4 Antagonism of VGD/DNA Polyplexes
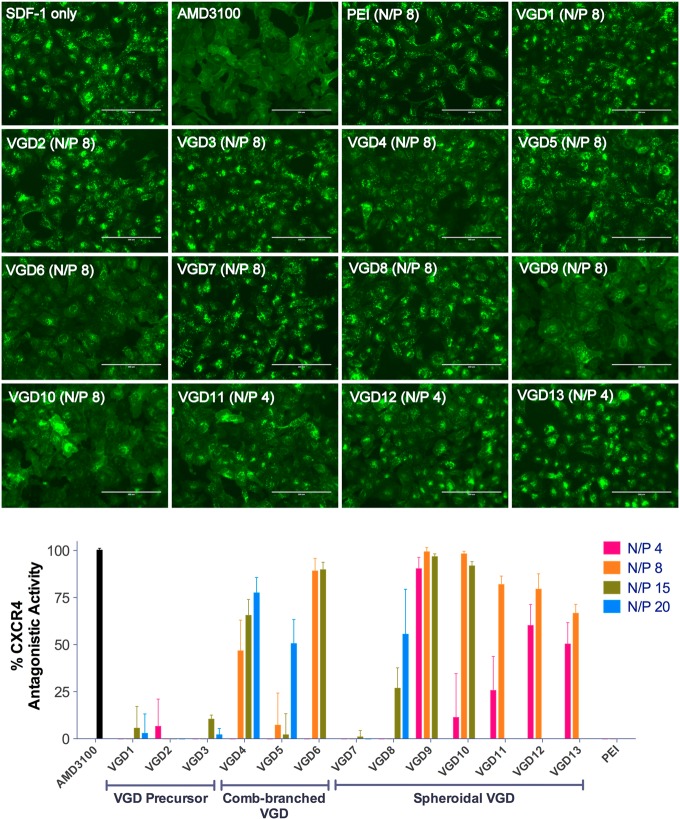
To examine the ability of VGD/DNA polyplexes to antagonize CXCR4, high-content analysis of receptor redistribution was performed. The analysis used U2OS cells stably expressing human CXCR4 receptor fused to the N-terminus of EGFP. During the analysis, cellular translocation of EGFP-CXCR4 upon stimulation with SDF-1 was monitored. As shown in Figure 5, we observed internalization of the CXCR4 receptor into endosomes in SDF-1-treated cells, as suggested by the punctate pattern of fluorescence distribution. Small-molecule CXCR4 antagonist AMD3100 (300 nM) inhibited the SDF-1-triggered receptor internalization, which was indicated by the diffuse pattern of the EGFP-CXCR4 fluorescence indicative of the presence of the receptor on the cell membrane surface. CXCR4 antagonistic activity was quantified by high-content analysis based on the number of fluorescent spots (i.e., endosomes) per cell using AMD3100 as a positive (100%) and SDF-1 only as a negative control (0%). Except for VGD precursors and generation 0 comb-branched VGD7, all VGD/DNA polyplexes inhibited SDF-1-triggered receptor internalization in a dose-dependent manner. Complete CXCR4 inhibition comparable to AMD3100 was observed in spheroidal VGD9 polyplexes at N/P 4, 8, and 15 and in VGD10 polyplexes at N/P 8 and 15. Among other dendrimers, comb-branched VGD6 polyplexes at N/P 8 and 15 showed >88% CXCR4 inhibition and VGD4 polyplexes exhibited 77% CXCR4 inhibition at N/P 20. High-generation spheroidal VGD11–13 polyplexes exhibited partial CXCR4 inhibition at lower N/P ratios. PEI/DNA polyplexes were used as negative control and showed no CXCR4 antagonism.
Figure 5.
CXCR4 antagonism of VGD/DNA polyplexes at different N/P ratios. The extent of CXCR4 antagonism was quantitated by high-content analysis of internalized receptors and compared with a positive (AMD3100, 100%) and a negative (SDF-1 only, 0%) control (scale bar = 200 μm).
Dual Functionality of VGD/DNA Polyplexes
The above screening results revealed that VGD6, VGD9, and VGD10 exhibit both high CXCR4 antagonism and high transfection activity. We have used these three dendrimers to evaluate the potential of VGD/DNA polyplexes as dual-function delivery systems with combined antimetastatic and antitumor activity. The potential for antimetastatic activity was evaluated by determining the ability of the polyplexes to inhibit CXCR4-mediated invasion of cancer cells. The antitumor (i.e., cell killing) activity was tested using TNFα as a therapeutic gene. Figure 6 (left) shows images of cells that migrated across the layer of extracellular matrix (Matrigel). In untreated cells without the SDF-1 chemokine gradient, only a small number of cells invaded through Matrigel, but the addition of SDF-1 triggered invasion of a large number of cells. Treatment with small molecule CXCR4 antagonist AMD3100 decreased the cell invasion to the background levels. All three VGD polyplexes displayed similar activity at inhibiting invasion of U2OS cancer cells when prepared at N/P ratio at which they exhibited full CXCR4 antagonism (see Figure 5), and under such conditions, the inhibitory activity was fully comparable to that of AMD3100. The VGD polyplexes were capable of delivering therapeutic TNFα plasmid (Figure 6 right). Depending on the type of VGD and N/P ratio, the treatment with the TNFα polyplexes resulted in 7% to 22% of TNFα-induced cell death 48 h after transfection.
Figure 6.
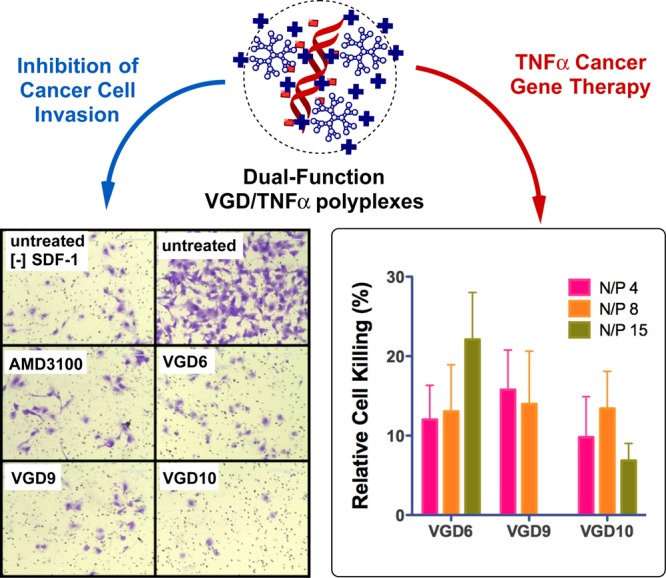
Simultaneous inhibition of cancer cell invasion (left) and TNFα gene delivery (right) by VGD polyplexes in U2OS cells. U2OS cells were treated with VGD/DNA polyplexes prepared with TNFα plasmid DNA. The VGD6 and VGD10 polyplexes were prepared at N/P 8 and VGD8 polyplexes were prepared at N/P 4. The cells were allowed to invade through a layer of Matrigel upon stimulation with SDF-1 for 16 h. AMD3100 (300 nM) was used as a positive control. TNFα gene delivery efficiency of VGD was determined from cell viability measured by MTS assay; normalized to cell viability observed with polyplexes formed with luciferase DNA.
Discussion
The main goal of this study was to synthesize viologen dendrimers capable of simultaneously functioning as gene delivery vectors and CXCR4 antagonists. In order to achieve the goal, we have used VGD that have been previously shown to prevent HIV infection by their inhibitory activity against CXCR4 receptor. The VGD dendrimers were expected to form polyplexes by electrostatic interactions between the polycationic scaffold of VGD and negatively charged DNA, while maintaining their CXCR4 antagonistic activity. Using different capping groups allowed us to adjust VGD binding to DNA. For example, using thymine capping group was expected to increase the binding affinity of VGD with DNA due to the complementary adenine-thymine base pair interactions. Multistep organic synthesis allowed us to obtain well-defined dendrimer structures needed to conduct the structure–activity relationship study. We have focused our attention on determining the influence of dendrimer molecular architecture, number of charges, and the type of capping group on DNA condensation ability, transfection activity, cytotoxicity, and CXCR4 antagonism.
Effective gene delivery carrier has to be able to condense DNA into nanosized polyplexes in order to facilitate cellular delivery of the DNA. The quantification of DNA condensation properties of VGD was performed using EtBr assay, which monitors the fraction of uncomplexed DNA that is accessible for intercalation with EtBr at a given N/P ratio.40 VGD precursors with the lowest number of positive charges were unable to condense DNA as indicated by the nonsigmoidal shape of the condensation curves and the presence of free DNA band in agarose gel electrophoresis. All the other VGD exhibited complete condensation of DNA, as suggested by results of both EtBr exclusion assay and agarose gel electrophoresis. These results reinforce previous findings with other cationic dendrimers and polymers that there is a minimum number of positive charges needed to achieve full condensation of large plasmid DNA molecules.
Physicochemical properties of polyplexes are critical for cell uptake and transfection activity. Thus, we analyzed the particle size and surface charge of the VGD/DNA polyplexes. We found that the total number of positive charges in VGD is overall the major factor that determines the physicochemical properties of the polyplexes. As suggested by the DNA condensation results, most generation 0 VGD failed to provide enough positive charges to condense DNA into nanosized particles. Interestingly, however, we found that a proper selection of a VGD capping group can compensate for the lack of positive charges and lead to DNA condensation. This phenomenon is well illustrated when comparing VGD7 with VGD8, which are both G0 VGD with six positive charges and similar molecular weight. We found that VGD8 was able to form polyplexes with small particle size and positive ζ-potential while VGD7 failed to do so. This could be explained by the thymine capping groups on VGD8, which provide additional hydrogen bond interactions with DNA due to the complementary A-T base pairing. In the case of VGD with more positive charges, the effect of thymine modification and hydrogen bond interactions on particle size and ζ-potential is not significant because of the dominant influence of cooperative electrostatic interactions.
Before testing transfection efficiency and CXCR4 inhibitory activity of VGD/DNA polyplexes, the cytotoxicity in U2OS and HepG2 cells was evaluated. HepG2 is a widely used and well-accepted in vitro model to predict liver toxicity. Although the mechanism of cytotoxicity caused by polyplexes is not fully understood, it is believed that interaction between the positively charged polyplexes and the negatively charged cell membrane is the initiating trigger.41−43 The interactions between polyplexes and cell membrane proteins and phospholipids disrupt the bilayer architecture and induce the formation of nanoscale holes.44 Once the polyplex concentration is above its toxic threshold, the membrane damage will eventually lead to cell death. Our results show that VGD/DNA polyplexes displayed a typical dose-dependent polyplex-caused cytotoxicity in cell culture. The cytotoxicity is highly correlated with the molecular weight and the cationic charge density of VGD, which are all well-recognized key parameters that determine the interaction with cell membranes. Low-molecular-weight VGD precursors (<2000) did not show any cytotoxicity under the conditions tested, while high-molecular-weight VGD (>7500) showed significant cytotoxicity, especially in U2OS cells. When comparing VGD with similar molecular weight, we found that the spheroidal VGD were considerably more toxic than the comb-branched type. This is most likely due to their higher charge density. Unlike the DNA condensation and particle size, no significant effect of the capping group on cytotoxicity was observed.
Based on the cytotoxicity results, we conducted a detailed evaluation of the dual functionality of VGD polyplexes by first studying gene delivery (Figure 4) and CXCR4 antagonism activity (Figure 5). The polyplexes were prepared at N/P ratios ranging from 4 to 20 and the transfection was conducted in conditions with or without serum. Results showed that transfection efficiency of VGD is strongly dependent on the molecular weight as well as the dendrimer architecture. VGD with higher molecular weight exhibit higher transfection activity than low-molecular-weight VGD. When using nontoxic doses (N/P 4), high-generation spheroidal VGD showed the highest transfection efficiency among all. The effect of N/P ratio on transfection activity was also observed. In general, VGD with low molecular weight exhibited significant dependence on the N/P ratio and their transfection activity increased with increasing N/P ratio. On the other hand, rising N/P ratio had a relatively minor effect on transfection activity of VGD with high molecular weight. Spheroidal VGD were likely to have better transfection ability than comb-branched VGD. As above, the type of capping group was important only in the case of VGD8 with small number of positive charges. The presence of a thymine capping group substantially increased transfection in comparison with VGD with a methyl capping group. VGD8 demonstrated 4- to 1000-fold higher transfection efficiency than VGD7 at various N/P ratios, suggesting that the contribution of extra hydrogen bond interactions with DNA leads to enhanced biological activity of the polyplexes. However, for high-molecular-weight VGD, the effect of thymine modification was insignificant, as the electrostatic interactions dominated the polyplex properties.
To evaluate the capability of VGD/DNA polyplexes to function as CXCR4 antagonists, we used a commercially available CXCR4-redistribution assay designed for screening CXCR4 antagonists (Figure 5). CXCR4 antagonists like AMD3100 competitively bind with the receptor and prevent SDF-1 binding, receptor endocytosis, and downstream signaling events induced by SDF-1. We used U2OS cells that stably express EGFP-tagged CXCR4. The extent of CXCR4 inhibition could be quantified by analyzing the fluorescent signal of the internalized EGFR-CXCR4 receptors into the endosomes. We found that CXCR4 antagonism of VGD also showed strong dependence on molecular weight. Unfortunately, the high cytotoxicity of the high-molecular-weight VGD restricted their use to low doses. Overall, after taking into account the cytotoxicity, spheroidal VGD with moderate molecular weight (VGD9 and VGD10) exhibited the highest potency among the tested formulations. These VGD showed activity fully comparable to the commercial small molecule CXCR4 antagonist AMD3100. Comb-branched VGD6 with higher molecular weight exhibited stronger CXCR4 antagonism than VGD4 and VGD5 with lower molecular weight. Interestingly, here we also found a noticeable difference between thymine-capped VGD8 and methyl-capped VGD7. VGD8 was able to achieve 50% CXCR4 antagonism, while VGD7 was nearly inactive at all the doses tested. We postulated in our previous study that this might be attributed to enhanced affinity with CXCR4 receptor due to the extra hydrogen bond interactions.
The CXCR4/SDF-1 axis is involved in migration and metastasis of multiple types of cancers, and CXCR4 antagonists like AMD3100 are known to inhibit invasion of CXCR4-expressing cancer cells. TNFα is a major inflammatory cytokine that has been used clinically for local treatment of several types of cancer, including locally advanced soft tissue sarcomas and metastatic melanomas.45 However, the mechanism of activity of TNFα is complex and it has become apparent that, under some conditions, TNFα can also have strong tumor-promoting effect that originates from its role in promoting cancer cell survival, migration and metastasis, and angiogenesis.46 Growing evidence indicates that of the factors involved in the tumor-promoting effect of TNFα is its ability to upregulate expression of CXCR4 in various cancers.47−49 We hypothesized that combining TNFα cancer gene therapy with simultaneous CXCR4 antagonism of VGD will improve the therapeutic outcome of TNFα monotherapy. We have evaluated the potential of VGD polyplexes to exert such combined effect by determining their ability to simultaneously inhibit cancer cell invasion and mediate TNFα transfection. We have selected the best-performing VGD based on their transfection activity and CXCR4 antagonism. All three tested VGD were capable of achieving cancer cell killing and blocking cancer cell invasion comparable to AMD3100. By combining the TNFα antitumor and CXCR4 antimetastatic functions in a single formulation, the VGD polyplexes represent promising delivery systems that can improve outcomes of TNFα-based anticancer treatments.
Conclusions
We have carried out a structure–activity relationship study of VGD. A series of VGD was synthesized and screened for the best candidates capable of fulfilling a dual function as CXCR4 antagonists and gene delivery vectors. VGD behavior strongly depends on the molecular weight, architecture, number of positive charges, and charge density. End group modification of low-molecular-weight VGD with thymine provides extra hydrogen bond interactions with DNA and CXCR4 receptor, which contributes to improved DNA condensation, transfection, and CXCR4 antagonism. Increasing the molecular weight and the number of positive charges improves transfection activity and CXCR4 antagonism; however, this is achieved at the expense of increased cytotoxicity. Hence, there is a fine balance between the desired biological activity and cytotoxicity. Comb-branched VGD6 (G2 with 26 positive charges) and spheroidal VGD9 and VGD10 (G1 with 18 positive charges) showed both high potency to antagonize CXCR4 and high transfection activity in vitro. Using TNFα plasmid, we have demonstrated that VGD simultaneously prevent CXCR4-mediated cancer cell invasion and facilitate TNFα-mediated cancer cell killing. Such dual-function carriers have potential to enhance the overall therapeutic outcomes of gene therapy for multiple diseases in which CXCR4 plays a prominent role, including cancer metastasis, HIV infection, and multiple inflammatory diseases.
Experimental Procedures
Materials
All chemicals used for VGD synthesis were of analytical grade and purchased from Sigma-Aldrich (St. Louis, MO) or Merck (Whitehouse Station, NJ). Ethyl acetate, diethyl ether, tetrahydrofuran (THF), methanol, and petroleum ether were distilled before use. Organic solutions were dried over anhydrous Na2SO4 or MgSO4 and concentrated with a rotary evaporator at reduced pressure. Reported yields are of purified products and were not optimized. Octahydrochloride of AMD3100 and 25 kDa branched poly(ethylene imine) (PEI) were purchased from Sigma-Aldrich. Plasmid DNA, gWiz high-expression luciferase (gWiz-Luc) containing luciferase reporter gene was from Aldevron (Fargo, ND). Plasmid DNA containing human TNFα gene was obtained from InvivoGen (San Diego, CA). Human stromal cell-derived factor 1 (SDF-1) was from Shenandoah Biotechnology, Inc. (Warwick, PA). Dulbecco’s Modified Eagle Medium (DMEM), Dulbecco’s Phosphate Buffered Saline (PBS), Fetal Bovine Serum (FBS), l-glutamine, and penicillin-streptomycin (Pen-Strep) solution were from Thermo Scientific (Waltham, MA). G418 sulfate and Minimum Essential Medium (MEM) were from Mediatech, Inc. (Manassas, VA). Cell culture inserts for 24-well plates (8.0 μm pores, Translucent PET Membrane) and BD Matrigel Basement Membrane Matrix were purchased from BD Biosciences (Franklin Lakes, NJ). All other reagents and chemicals were obtained from Fisher Scientific or VWR International unless otherwise noted.
VGD Synthesis
The synthesis of VGD followed previously published procedures and detailed description is provided in the Supporting Information.39,50−53 Briefly, the VGD precursors dialkylated 4,4′-bipyridinium units with nucleobases were synthesized by substitution reaction of alkylated N-heterocycle derivatives with the 4,4′-bipyridinium. VGD with polycationic scaffold carrying 6, 10, 18, 26, 42, and 90 positive charges per molecule (Scheme 1) were synthesized via a divergent strategy originated from the corresponding initiator core. To obtain the comb-branched dendrimers, VGD4 and VGD5 were synthesized by reacting a conjugated 4-fold nucleophilic core 1,1′-bis[3,5-bis(bromomethyl)phenyl]-4,4′-bipyridinium dihexafluorophosphate with monoalkylated 4,4′-bipyridinium unit with ethyl or thymine as capping group. The second-generation VGD6 was obtained from a 4-fold nucleophilic core by reaction with N-(3,5-di(hydroxymethyl)-benzyl)-4,4′-bipyridinium hexafluorophosphate followed by substitution of −OH by −Br with HBr/acetic acid at room temperature. The spheroidal VGD were synthesized from a trifunctional 1,3,5-tris[(-4,4′-bipyridinium)methyl]benzene trihexafluorophosphate core consisting of a mesityl derivative, linked to three 4,4′-bipyridine units.
Ethidium Bromide (EtBr) Exclusion Assay
The ability of VGD to condense DNA was determined by EtBr exclusion assay by measuring the changes in EtBr/DNA fluorescence. Luciferase plasmid DNA solution at a concentration of 20 μg/mL was prepared in 10 mM HEPES buffer (pH 7.4) and mixed with EtBr (1 μg/mL). Fluorescence intensity was measured and set to 100% (Ex 540 nm/Em 590 nm). Fluorescence readings were taken following a stepwise addition of VGD solutions. Condensation curve for each dendrimer was constructed and the equivalent nitrogen/phosphate (N/P) ratio for complete DNA condensation was established using the intersection point of the steepest tangent and the flattest tangent to the condensation curve.
Preparation and Physicochemical Characterization of VGD/DNA Polyplexes
DNA solution at a final concentration 20 μg/mL was prepared in 10 mM HEPES buffer (pH 7.4). VGD/DNA polyplexes were formed by adding predetermined volume of VGD to achieve the desired N/P ratio and mixed by vigorous vortexing for 10 s. Polyplexes were further allowed to stand at room temperature for 30 min prior to use. Hydrodynamic diameter and ζ-potential of VGD/DNA polyplexes were determined by dynamic light scattering (DLS) using a ZEN3600 Zetasizer Nano-ZS (Malvern Instruments Ltd., Worcestershire, UK). Results were expressed as mean ± standard error of the mean (SEM) of 3–10 experimental runs.
Agarose Gel Electrophoresis
DNA condensation by VGD was confirmed using agarose gel electrophoresis. VGD/DNA polyplexes (20 μL) prepared at N/P 4 were loaded onto a 0.8% agarose gel containing 0.5 μg/mL EtBr and run for 60 min at 120 V in 0.5× Tris/Borate/EDTA (TBE) buffer. The gel was then imaged under UV.
Cell Culture
Human hepatocellular carcinoma cell line HepG2 (ATCC 77400) was purchased from ATCC (Manassas, VA) and maintained in MEM supplemented with 10% FBS. Human epithelial osteosarcoma U2OS cells stably expressing human CXCR4 receptor fused to the N-terminus of enhanced green fluorescent protein (EGFP) (EGFP-CXCR4+ U2OS) were purchased form Fisher Scientific. The cells were cultured in DMEM supplemented with 2 mM l-glutamine, 10% FBS, 1% Pen-Strep, and 0.5 mg/mL G418.
Cytotoxicity of VGD/DNA Polyplexes
Cytotoxicity of VGD/DNA polyplexes was evaluated by MTS assay in both U2OS cells and HepG2 cells. The cells were seeded in 96-well microplates at a density of 6000 cells/well (U2OS) or 8000 cells/well (HepG2). After 24 h, culture medium in each well was replaced by 75 μL of antibiotic-free medium (±FBS) before adding 10 μL of VGD/DNA polyplexes prepared at different N/P ratios. After 4 h incubation, medium containing polyplexes was removed and the cells were incubated in complete culture medium for another 24 h. To measure cell viability, culture medium was aspirated and replaced with a mixture of 100 μL serum-free medium and 20 μL MTS reagent (CellTiter 96 AQueous Non-Radioactive Cell Proliferation Assay, Promega). After 1.5 h incubation, the absorbance was measured using SpectraMax M5e Multi-Mode Microplate Reader (Molecular Devices, CA) at λ = 490 nm. The relative cell viability (%) was calculated as [A]sample/[A]untreated × 100%.
Transfection Efficiency of VGD/DNA Polyplexes
Luciferase DNA transfections were conducted in 48-well microplates following previously published protocol. U2OS cells were seeded at a density of 20 000 cells/well 24 h prior to transfection. Culture medium in each well was removed and replaced with 150 μL of antibiotic-free medium (±10% FBS) before adding 20 μL of VGD/DNA polyplexes prepared at different N/P ratios (DNA dose 0.4 μg/well). After 4 h incubation, polyplexes were completely removed and the cells were cultured in complete culture medium for another 24 h prior to measuring luciferase expression. The medium was then discarded and the cells were lysed in 100 μL of 0.5× cell culture lysis reagent buffer (Promega, Madison, WI) for 30 min. To measure the luciferase content, 100 μL of 0.5 mM luciferin solution was automatically injected into each well containing 20 μL of cell lysate and the luminescence was integrated over 10 s using GloMax 96 Microplate Luminometer (Promega). Total cellular protein concentration in the cell lysate was determined by the Bicinchoninic acid protein assay using calibration curve constructed with standard bovine serum albumin solutions (Pierce, Rockford, IL). Transfection activity was expressed as RLU/mg cellular protein ± SD of triplicate samples.
Transfections with TNFα DNA were conducted in 96-well microplates. U2OS cells were seeded at a density of 3000 cells/well 24 h prior to transfection. Culture medium in each well was removed and replaced with 75 μL of antibiotic- and serum-free medium containing 25 μM chloroquine before adding 10 μL of polyplexes containing either luciferase or TNFα plasmid DNA prepared at different N/P ratios (DNA dose 0.2 μg/well). After 4 h incubation, polyplexes were completely removed and the cells were cultured in complete culture medium for another 48 h prior to measuring cell viability using MTS assay. The relative cell killing mediated by TNFα expression was determined by the following formula and expressed as mean ± SD of triplicate samples:
CXCR4 Antagonism of VGD/DNA Polyplexes
The CXCR4 antagonism of VGD/DNA polyplexes was evaluated by CXCR4 redistribution assay using high-content analysis. U2OS cells expressing functional EGFP-CXCR4 fusion protein were plated in black 96-well plates with optical bottom 18–24 h before the experiment at a seeding density of 8000 cells per well. The cells were washed twice with 100 μL assay buffer (DMEM supplemented with 2 mM l-glutamine, 1% FBS, 1% Pen-Strep, and 10 mM HEPES) and then incubated with different concentrations of VGD/DNA polyplexes or positive control AMD3100 (300 nM) in assay buffer containing 0.25% DMSO at 37 °C for 30 min. All the N/P ratios and final concentrations of VGD/DNA polyplexes were the same as in the luciferase transfection experiments. Human SDF-1 (SDF-1) was then added to each well to reach final concentration of 10 nM. Cells treated with SDF-1 alone were used as negative control. After 1 h incubation at 37 °C, the cells were fixed with 4% formaldehyde at room temperature for 20 min and washed 4 times with PBS. The cell nuclei were stained with 1 μM Hoechst in PBS containing 0.5% Triton X-100. Images were taken by EVOS fl microscope at 20×. For high-content analysis, internalization of the EGFP-tagged CXCR4 receptor was measured using Cellomics ArrayScan VT1 Reader and analyzed using SpotDetectorV3 BioApplication software. Percent CXCR4 internalization was calculated relative to the positive (AMD3100, 100%) and negative (SDF-1 only, 0%) controls. Image analysis algorithm was validated by dose–response curve of AMD3100.
Cancer Cell Invasion Assay
Modified Boyden chamber assay was used to determine the ability of VGD to inhibit invasion of cancer cells through a layer of extracellular matrix. Transwell inserts were coated with 40 μL Matrigel diluted 1:3 (v/v) with serum-free medium. The 24-well plates with coated inserts were then placed in 37 °C incubator for 2 h. CXCR4+ U2OS cells were trypsinized and resuspended with VGD polyplexes in serum-free medium for 30 min before adding to the inserts at a final concentration of 25 000 cells in 300 μL medium per insert. SDF-1 (20 nM in serum-free medium) was added as the chemoattractant to the wells in the companion plate. After 16 h, the noninvaded cells on the upper surface of the inserts were removed with a cotton swab. The invaded cells were then fixed by anhydrous methanol, stained by crystal violet dye, and visualized by EVOS xl inverted microscope.
Acknowledgments
We thank Dr. David Kelly in Molecular Biology Core Lab of Eppley Institute for Research in Cancer for the kind help with establishing a method for high-content analysis of CXCR4 redistribution assay. NIH support by a grant EB014570 is also acknowledged.
Supporting Information Available
Detailed description of VGD synthesis and results of agarose gel electrophoresis for VGD/DNA polyplexes prepared at N/P 20. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Davis M. E. (2002) Non-viral gene delivery systems. Curr. Opin. Biotechnol. 13, 128–31. [DOI] [PubMed] [Google Scholar]
- Leong K. W.; Mao H. Q.; Truong-Le V. L.; Roy K.; Walsh S. M.; August J. T. (1998) DNA-polycation nanospheres as non-viral gene delivery vehicles. J. Controlled Release 53, 183–93. [DOI] [PubMed] [Google Scholar]
- Merdan T.; Kopecek J.; Kissel T. (2002) Prospects for cationic polymers in gene and oligonucleotide therapy against cancer. Adv. Drug Delivery Rev. 54, 715–58. [DOI] [PubMed] [Google Scholar]
- Pack D. W.; Hoffman A. S.; Pun S.; Stayton P. S. (2005) Design and development of polymers for gene delivery. Nat. Rev. Drug Discovery 4, 581–93. [DOI] [PubMed] [Google Scholar]
- Navarro G.; Tros de Ilarduya C. (2009) Activated and non-activated PAMAM dendrimers for gene delivery in vitro and in vivo. Nanomedicine 5, 287–97. [DOI] [PubMed] [Google Scholar]
- Dufes C.; Keith W. N.; Bilsland A.; Proutski I.; Uchegbu I. F.; Schatzlein A. G. (2005) Synthetic anticancer gene medicine exploits intrinsic antitumor activity of cationic vector to cure established tumors. Cancer Res. 65, 8079–84. [DOI] [PubMed] [Google Scholar]
- Lim Y. B.; Kim T.; Lee J. W.; Kim S. M.; Kim H. J.; Kim K.; Park J. S. (2002) Self-assembled ternary complex of cationic dendrimer, cucurbituril, and DNA: noncovalent strategy in developing a gene delivery carrier. Bioconjugate Chem. 13, 1181–5. [DOI] [PubMed] [Google Scholar]
- Luo D.; Haverstick K.; Belcheva N.; Han E.; Saltzman W. M. (2002) Poly(ethylene glycol)-conjugated PAMAM dendrimer for biocompatible, high-efficiency DNA delivery. Macromolecules 35, 3456–3462. [Google Scholar]
- Merkel O. M.; Mintzer M. A.; Sitterberg J.; Bakowsky U.; Simanek E. E.; Kissel T. (2009) Triazine dendrimers as nonviral gene delivery systems: effects of molecular structure on biological activity. Bioconjugate Chem. 20, 1799–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.; Sen J.; Bathula S. R.; Yang Q.; Fittipaldi R.; Huang L. (2009) Novel cationic lipid that delivers siRNA and enhances therapeutic effect in lung cancer cells. Mol. Pharmaceutics 6, 696–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. E.; Rice K. G. (2008) A novel class of intrinsic proteasome inhibitory gene transfer peptides. Bioconjugate Chem. 19, 370–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.; Zhu Y.; Hazeldine S. T.; Li C.; Oupický D. (2012) Dual-function CXCR4 antagonist polyplexes to deliver gene therapy and inhibit cancer cell invasion. Angew. Chem., Int. Ed. Engl. 51, 8740–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.; Oupicky D. (2014) Effect of biodegradability on CXCR4 antagonism, transfection efficacy and antimetastatic activity of polymeric Plerixafor. Biomaterials 35, 5572–5579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balkwill F. R. (2012) The chemokine system and cancer. J. Pathol. 226, 148–157. [DOI] [PubMed] [Google Scholar]
- Rajagopal S.; Kim J.; Ahn S.; Craig S.; Lam C. M.; Gerard N. P.; Gerard C.; Lefkowitz R. J. (2010) β-arrestin-but not G protein-mediated signaling by the “decoy” receptor CXCR7. Proc. Natl. Acad. Sci. U.S.A. 107, 628–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X. Q.; Cheng G. C.; Hao M. G.; Zheng J. H.; Zhou X. M.; Zhang J. A.; Taichman R. S.; Pienta K. J.; Wang J. H. (2010) CXCL12/CXCR4/CXCR7 chemokine axis and cancer progression. Cancer Met. Rev. 29, 709–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L.; Karin M. (2001) Mammalian MAP kinase signalling cascades. Nature 410, 37–40. [DOI] [PubMed] [Google Scholar]
- Zlotnik A.; Burkhardt A. M.; Homey B. (2011) Homeostatic chemokine receptors and organ-specific metastasis. Nat. Rev. Immunol. 11, 597–606. [DOI] [PubMed] [Google Scholar]
- Burger M.; Glodek A.; Hartmann T.; Schmitt-Graff A.; Silberstein L. E.; Fujii N.; Kipps T. J.; Burger J. A. (2003) Functional expression of CXCR4 (CD184) on small-cell lung cancer cells mediates migration, integrin activation, and adhesion to stromal cells. Oncogene 22, 8093–8101. [DOI] [PubMed] [Google Scholar]
- Spoo A. C.; Lubbert M.; Wierda W. G.; Burger J. A. (2007) CXCR4 is a prognostic marker in acute myelogenous leukemia. Blood 109, 786–91. [DOI] [PubMed] [Google Scholar]
- Holm N. T.; Byrnes K.; Li B. D.; Turnage R. H.; Abreo F.; Mathis J. M.; Chu Q. D. (2007) Elevated levels of chemokine receptor CXCR4 in HER-2 negative breast cancer specimens predict recurrence. J. Surg. Res. 141, 53–9. [DOI] [PubMed] [Google Scholar]
- Chu Q. D.; Panu L.; Holm N. T.; Li B. D.; Johnson L. W.; Zhang S. (2010) High chemokine receptor CXCR4 level in triple negative breast cancer specimens predicts poor clinical outcome. J. Surg. Res. 159, 689–95. [DOI] [PubMed] [Google Scholar]
- Wald O.; Shapira O. M.; Izhar U. (2013) CXCR4/CXCL12 axis in non small cell lung cancer (NSCLC) pathologic roles and therapeutic potential. Theranostics 3, 26–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spano J.-P.; Andre F.; Morat L.; Sabatier L.; Besse B.; Combadiere C.; Deterre P.; Martin A.; Azorin J.; Valeyre D.; Khayat D.; Le Chevalier T.; Soria J.-C. (2004) Chemokine receptor CXCR4 and early-stage non-small cell lung cancer: pattern of expression and correlation with outcome. Ann. Oncol. 15, 613–617. [DOI] [PubMed] [Google Scholar]
- Oonakahara K.-i.; Matsuyama W.; Higashimoto I.; Kawabata M.; Arimura K.; Osame M. (2004) Stromal-Derived Factor-1α/CXCL12-CXCR 4 Axis Is Involved in the Dissemination of NSCLC Cells into Pleural Space. Am. J. Respir. Cell Mol. Biol. 30, 671–677. [DOI] [PubMed] [Google Scholar]
- Gangadhar T.; Nandi S.; Salgia R. (2010) The role of chemokine receptor CXCR4 in lung cancer. Cancer Biol. Ther. 9, 409–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wald O.; Izhar U.; Amir G.; Avniel S.; Bar-Shavit Y.; Wald H.; Weiss I. D.; Galun E.; Peled A. (2006) CD4+CXCR4highCD69+ T cells accumulate in lung adenocarcinoma. J. Immunol. 177, 6983–6990. [DOI] [PubMed] [Google Scholar]
- Wald O.; Izhar U.; Amir G.; Kirshberg S.; Shlomai Z.; Zamir G.; Peled A.; Shapira O. M. (2011) Interaction between neoplastic cells and cancer-associated fibroblasts through the CXCL12/CXCR4 axis: Role in non–small cell lung cancer tumor proliferation. J. Thorac. Cardiovasc. Surg. 141, 1503–1512. [DOI] [PubMed] [Google Scholar]
- Bleul C. C.; Wu L.; Hoxie J. A.; Springer T. A.; Mackay C. R. (1997) The HIV coreceptors CXCR4 and CCR5 are differentially expressed and regulated on human T lymphocytes. Proc. Natl. Acad. Sci. U. S. A. 94, 1925–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkhatib G. (2009) The biology of CCR5 and CXCR4. Curr. Opin. HIV AIDS 4, 96–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada M.; Kubo H.; Kobayashi S.; Ishizawa K.; He M.; Suzuki T.; Fujino N.; Kunishima H.; Hatta M.; Nishimaki K.; Aoyagi T.; Tokuda K.; Kitagawa M.; Yano H.; Tamamura H.; Fujii N.; Kaku M. (2011) The increase in surface CXCR4 expression on lung extravascular neutrophils and its effects on neutrophils during endotoxin-induced lung injury. Cell. Mol. Immunol. 8, 305–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukacs N. W.; Berlin A.; Schols D.; Skerlj R. T.; Bridger G. J. (2002) AMD3100, a CxCR4 antagonist, attenuates allergic lung inflammation and airway hyperreactivity. Am. J. Pathol. 160, 1353–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalo J. A.; Lloyd C. M.; Peled A.; Delaney T.; Coyle A. J.; Gutierrez-Ramos J. C. (2000) Critical involvement of the chemotactic axis CXCR4/stromal cell-derived factor-1 alpha in the inflammatory component of allergic airway disease. J. Immunol. 165, 499–508. [DOI] [PubMed] [Google Scholar]
- Danese S.; Gasbarrini A. (2005) Chemokines in inflammatory bowel disease. J. Clin. Pathol. 58, 1025–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes L. V.; Short S. P.; Neel N. F.; Salvo V. A.; Zhu Y.; Elliott S.; Wei Y.; Yu D.; Sun M.; Muir S. E.; Fonseca J. P.; Bratton M. R.; Segar C.; Tilghman S. L.; Sobolik-Delmaire T.; Horton L. W.; Zaja-Milatovic S.; Collins-Burow B. M.; Wadsworth S.; Beckman B. S.; Wood C. E.; Fuqua S. A.; Nephew K. P.; Dent P.; Worthylake R. A.; Curiel T. J.; Hung M. C.; Richmond A.; Burow M. E. (2011) Cytokine receptor CXCR4 mediates estrogen-independent tumorigenesis, metastasis, and resistance to endocrine therapy in human breast cancer. Cancer Res. 71, 603–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapteva N.; Yang A. G.; Sanders D. E.; Strube R. W.; Chen S. Y. (2005) CXCR4 knockdown by small interfering RNA abrogates breast tumor growth in vivo. Cancer Gene Ther. 12, 84–9. [DOI] [PubMed] [Google Scholar]
- Liang Z.; Yoon Y.; Votaw J.; Goodman M. M.; Williams L.; Shim H. (2005) Silencing of CXCR4 blocks breast cancer metastasis. Cancer Res. 65, 967–71. [PMC free article] [PubMed] [Google Scholar]
- Brave M.; Farrell A.; Lin S. C.; Ocheltree T.; Miksinski S. P.; Lee S. L.; Saber H.; Fourie J.; Tornoe C.; Booth B.; Yuan W. S.; He K.; Justice R.; Pazdur R. (2010) FDA review summary: mozobil in combination with granulocyte colony-stimulating factor to mobilize hematopoietic stem cells to the peripheral blood for collection and subsequent autologous transplantation. Oncology 78, 282–288. [DOI] [PubMed] [Google Scholar]
- Asaftei S.; Huskens D.; Schols D. (2012) HIV-1 X4 activities of polycationic ″viologen″ based dendrimers by interaction with the chemokine receptor CXCR4: study of structure-activity relationship. J. Med. Chem. 55, 10405–13. [DOI] [PubMed] [Google Scholar]
- Germershaus O.; Mao S.; Sitterberg J.; Bakowsky U.; Kissel T. (2008) Gene delivery using chitosan, trimethyl chitosan or polyethylenglycol-graft-trimethyl chitosan block copolymers: establishment of structure-activity relationships in vitro. J. Controlled Release 125, 145–54. [DOI] [PubMed] [Google Scholar]
- Fischer D.; Li Y.; Ahlemeyer B.; Krieglstein J.; Kissel T. (2003) In vitro cytotoxicity testing of polycations: influence of polymer structure on cell viability and hemolysis. Biomaterials 24, 1121–31. [DOI] [PubMed] [Google Scholar]
- Morgan D. M.; Clover J.; Pearson J. D. (1988) Effects of synthetic polycations on leucine incorporation, lactate dehydrogenase release, and morphology of human umbilical vein endothelial cells. J. Cell Sci. 91(Pt 2), 231–8. [DOI] [PubMed] [Google Scholar]
- Morgan D. M.; Larvin V. L.; Pearson J. D. (1989) Biochemical characterisation of polycation-induced cytotoxicity to human vascular endothelial cells. J. Cell Sci. 94(Pt 3), 553–9. [DOI] [PubMed] [Google Scholar]
- Hong S.; Leroueil P. R.; Janus E. K.; Peters J. L.; Kober M. M.; Islam M. T.; Orr B. G.; Baker J. R. Jr.; Banaszak Holl M. M. (2006) Interaction of polycationic polymers with supported lipid bilayers and cells: nanoscale hole formation and enhanced membrane permeability. Bioconjugate Chem. 17, 728–34. [DOI] [PubMed] [Google Scholar]
- van Horssen R.; Ten Hagen T. L.; Eggermont A. M. (2006) TNF-alpha in cancer treatment: molecular insights, antitumor effects, and clinical utility. Oncologist 11, 397–408. [DOI] [PubMed] [Google Scholar]
- Waters J. P.; Pober J. S.; Bradley J. R. (2013) Tumour necrosis factor and cancer. J. Pathol. 230, 241–8. [DOI] [PubMed] [Google Scholar]
- Rostasy K.; Gorgun G.; Kleyner Y.; Garcia A.; Kramer M.; Melanson S. M.; Mathys J. M.; Yiannoutsos C.; Skolnik P. R.; Navia B. A. (2005) Tumor necrosis factor alpha leads to increased cell surface expression of CXCR4 in SK-N-MC cells. J. Neurovirol. 11, 247–55. [DOI] [PubMed] [Google Scholar]
- Kulbe H.; Hagemann T.; Szlosarek P. W.; Balkwill F. R.; Wilson J. L. (2005) The inflammatory cytokine tumor necrosis factor-alpha regulates chemokine receptor expression on ovarian cancer cells. Cancer Res. 65, 10355–62. [DOI] [PubMed] [Google Scholar]
- Zhao C.; Lu X.; Bu X.; Zhang N.; Wang W. (2010) Involvement of tumor necrosis factor-alpha in the upregulation of CXCR4 expression in gastric cancer induced by Helicobacter pylori. BMC Cancer 10, 419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asaftei S.; De Clercq E. (2010) ″Viologen″ dendrimers as antiviral agents: the effect of charge number and distance. J. Med. Chem. 53, 3480–8. [DOI] [PubMed] [Google Scholar]
- Asaftei S. (2008) New hybrid compounds of nucleobases and organic redox molecules and their use, PCT/EP 2008/004527, WO2008/148560 A1, 1–56
- Asaftei S.; Lepadatu A. M.; Ciobanu M. (2011) Novel compounds with a viologen skeleton and N-heterocycles on the peripheries: electrochemical and spectroscopic properties. Helv. Chim. Acta 94, 1091–1101. [Google Scholar]
- Bongard D. (2008) PhD Thesis, University of Osnabrück. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.