Abstract
Detecting genes associated with milk fat composition could provide valuable insights into the complex genetic networks of genes underling variation in fatty acids synthesis and point towards opportunities for changing milk fat composition via selective breeding. In this study, we conducted a genome-wide association study (GWAS) for 22 milk fatty acids in 784 Chinese Holstein cows with the PLINK software. Genotypes were obtained with the Illumina BovineSNP50 Bead chip and a total of 40,604 informative, high-quality single nucleotide polymorphisms (SNPs) were used. Totally, 83 genome-wide significant SNPs and 314 suggestive significant SNPs associated with 18 milk fatty acid traits were detected. Chromosome regions that affect milk fatty acid traits were mainly observed on BTA1, 2, 5, 6, 7, 9, 13, 14, 18, 19, 20, 21, 23, 26 and 27. Of these, 146 SNPs were associated with more than one milk fatty acid trait; most of studied fatty acid traits were significant associated with multiple SNPs, especially C18:0 (105 SNPs), C18 index (93 SNPs), and C14 index (84 SNPs); Several SNPs are close to or within the DGAT1, SCD1 and FASN genes which are well-known to affect milk composition traits of dairy cattle. Combined with the previously reported QTL regions and the biological functions of the genes, 20 novel promising candidates for C10:0, C12:0, C14:0, C14:1, C14 index, C18:0, C18:1n9c, C18 index, SFA, UFA and SFA/UFA were found, which composed of HTR1B, CPM, PRKG1, MINPP1, LIPJ, LIPK, EHHADH, MOGAT1, ECHS1, STAT1, SORBS1, NFKB2, AGPAT3, CHUK, OSBPL8, PRLR, IGF1R, ACSL3, GHR and OXCT1. Our findings provide a groundwork for unraveling the key genes and causal mutations affecting milk fatty acid traits in dairy cattle.
Introduction
Fat is the major energy substance in milk and more than 50% milk total energy comes from milk fat, which accounts 3–5% of milk contents. Fat nutrition value depends on fatty acids. Monounsaturated fatty acids (MUFA) have a favourable effect on human health because of its cholesterol-declining properties. Polyunsaturated fatty acids (PUFA) of the n-6 and n-3 series are essential nutrients that exert an important influence on plasma lipids and serve cardiac and endothelial functions for prevention and treatment of coronary heart diseases [1]. Conjugated linoleic acid (CLA) has effects on bone formation and the immune system as well as fatty acids and lipid metabolism and gene expression in numerous tissues [2]. Saturated fatty acids (SFA) lead to increase the concentration of low density lipoprotein (LDL) cholesterol and cause cardio cerebral vascular disease [3]. Therefore, changing the proportions of dietary fat by decreasing SFA and increasing MUFA and PUFA is vital to Human health. It is suggested that the ideal balance would seem to approximate 1∶1.3∶1 for SFA∶MUFA∶PUFA [4].
From the genetics point of view, milk fatty acids are complex traits influenced by non-genetic factors, such as breed, herd, stage of lactation, etc [5], [6] and genetic factors [7]. Bovine milk fatty acids have been found to be heritable, with heritability estimates ranging from 0.22 to 0.71 [8], [9]. Short and medium chain C4 to C16 saturated and monounsaturated fatty acids, which are synthesized de novo in the mammary gland, have moderate to high heritability (0.4–0.6) [8], [9]. Long chain fatty acids (above C16) are derived from circulating plasma lipids, whereas have low to moderate heritability (about 0.2) [8], [9]. Identifying genes and loci responsible for the genetic variation is expected to contribute greatly to our understanding of milk fatty acids synthesis, and to develop a marker-assisted selection to improve fatty acids in dairy breeding program in future. In the past few years, candidate gene and quantitative trait locus (QTL) mapping approaches have been implemented to detect genes or QTLs for milk fatty acid traits. A few promising loci, e.g. DGAT1 p.Lys232Ala and SCD1 p.Ala293Val [10], [11], [12] and a large number of significant or suggestive genomic regions [13], [14] were identified. Although the above two methods have got a few prominent findings, identification of causal mutations is still a challenge due to the commonly existing limitations [15].
At present, genome-wide association study (GWAS) has become a powerful strategy to identify genetic variants associated with complex traits. Since the first GWAS was published in 2005 [16], a great number of relative studies were conducted in human and domestic animals. Of them, several GWASs have been applied to detect genes or loci for milk production traits [17], [18], [19], conformation traits [19], reproduction traits [20], [21], healthy traits [22], [23], etc, in dairy and beef cattle. However, only studies have been carried out for fatty acids in Dutch dairy cattle [24], [25]. We herein performed a GWAS for 22 milk fatty acid traits in a Chinese Holstein population to identify genes and chromosome segments with large effects on such traits.
Materials and Methods
The milk samples were collected along the regular quarantine inspection of the farms. The whole procedure for sample collection was carried out in strict accordance with the protocol approved by the Animal Welfare Committee of China Agricultural University (Permit Number: DK996).
Phenotypic data and traits
The Chinese Holstein population in this study comprised 784 cows, the daughters of 21 sire families. All cows in this study were from 18 farms of the Beijing Sanyuan Dairy Farm Center, where routine standard performance test, i.e. Dairy Herd Improvement system (DHI) have been carried out since 1999. A total of 50 ml milk sample was collected for each cow from the DHI laboratory of the Beijing Dairy Cattle Center during November to December, 2012. The procedure of milk sample collection was carried out corresponding to DHI sampling (dairy herd improvement). After DHI measure, the remaining milk samples were taken back to the laboratory within 4°C cooler and then stored at −20°C.
Phenotypic values of 16 kinds of main milk fatty acids were measured by gas chromatography at the Ministry of Agriculture Feed Industry Centre of China (http://www.mafic.ac.cn/intro/default.asp), which included SFA of C8:0, C10:0, C12:0, C14:0, C16:0, C18:0, C20:0, C22:0; MUFA of C14:1, C16:1, C18:1n9c; PUFA of CLA (cis-9, trans-11 C18:2), C18:3n3, C18:3n6, C18:2n6c and C20:5n3. Before measuring, milk samples should be done with pretreatment. First, total milk fat were extracted from approximately 2 ml of each milk sample. The specific procedure was as follows: 2 ml milk was mixed with 4 ml solution of N-hexane/isopropyl alcohol (3∶2) and 2 ml solution of Na2SO4, and centrifuged at 3,000×g for 20 min. The upper layer was collected into 20 ml hydrolysis tube and 200 µl of C19:0 methyl ester as the internal standard was mixed, and then the extracted fat was dried under nitrogen. Methyl esters of fat were performed in the next step. 2 ml of NaOCH3/Methanol was put into the above hydrolysis tube for 15 min water bath under 50°C, and was mixed with 2 ml of hydrochloric acid/methanol solution (1∶10) for 1.5 h water bath under 80°C. After the temperature fell to room temperature level, 3 ml of water and 6 ml of n-hexane were put into above hydrolysis tube, mixed, vortexed, and stratified. The upper layer was collected and dried under nitrogen, and finally dissolved in 1 ml of n-hexane. 1 ml methyl esters of fatty acids were prepared to be determined by gas chromatography using a gas chromatograph (6890N, Agilent) equipped with a flame-ionization detector and a high polar fused silica capillary column (SP™-2560, 100 m×0.25 mm ID, 0.20 µm film). About 1 µl sample was injected under the following gas chromatography conditions: Helium was used as the carrier gas at a flow of 45 ml/min. The split ratio was 100∶1. The oven temperature was programmed at 100°C and held for 10 min, then increased to 160°C at a rate of 6°C/min, held for 10 min, increased to 200°C with 5°C/min, held for 20 min, increased to 240°C at a rate of 4°C/min and held for 12 min. Both the injector temperature and the detector temperature were set on 260°C. Individual fatty acids were identified and quantified by comparing the methyl ester chromatograms of the milk fat samples with the chromatograms of pure fatty acids methyl ester standards (Supelco™ 37 Component FAME Mix), and were measured as the weight proportion of total fat weight (wt/wt%).
Based on the phenotypes of 16 milk fatty acids, 6 additional traits were obtained including SFA, UFA, SFA/UFA (the ratio of SFA to UFA), C14 index, C16 index and C18 index. The 3 indices were calculated as 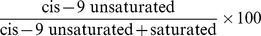 [26].
[26].
The descriptive statistics of these 22 fatty acid traits are presented in Table 1. Both SFA and UFA accounted for approximately 96% (wt/wt) of total fat.
Table 1. Descriptive statistics of the 22 fatty acid traits in Chinese Holstein.
| Traits | No. cows | Mean | Standard deviation | Variable coefficienta | Maximum | Minimum |
| C8:0 | 784 | 0.578 | 0.223 | 38.501 | 1.200 | 0.128 |
| C10:0 | 784 | 2.193 | 0.428 | 19.538 | 3.286 | 0.967 |
| C12:0 | 784 | 2.865 | 0.543 | 18.943 | 4.513 | 1.194 |
| C14:0 | 784 | 9.892 | 1.285 | 12.988 | 13.546 | 5.808 |
| C14:1 | 784 | 0.835 | 0.221 | 26.429 | 1.598 | 0.339 |
| C16:0 | 784 | 32.665 | 1.998 | 6.116 | 39.995 | 25.182 |
| C16:1 | 784 | 1.656 | 0.376 | 22.710 | 3.735 | 0.158 |
| C18:0 | 784 | 12.169 | 1.761 | 14.472 | 17.367 | 7.401 |
| C18:1n9c | 784 | 28.571 | 2.814 | 9.849 | 38.289 | 18.873 |
| C18:2n6c | 784 | 4.002 | 0.462 | 11.548 | 5.895 | 2.264 |
| C18:3n6 | 784 | 0.098 | 0.064 | 65.520 | 0.457 | 0.003 |
| C18:3n3 | 784 | 0.417 | 0.065 | 15.681 | 0.627 | 0.008 |
| CLA | 784 | 0.404 | 0.094 | 23.276 | 0.797 | 0.050 |
| C20:0 | 784 | 0.163 | 0.047 | 28.981 | 0.376 | 0.006 |
| C20:5n3 | 784 | 0.041 | 0.021 | 51.366 | 0.180 | 0.012 |
| C22:0 | 784 | 0.054 | 0.027 | 49.351 | 0.289 | 0.003 |
| C14 index | 784 | 7.763 | 1.677 | 21.596 | 14.624 | 3.460 |
| C16 index | 784 | 4.831 | 1.071 | 22.179 | 9.118 | 0.420 |
| C18 index | 784 | 70.144 | 3.335 | 4.754 | 80.298 | 55.891 |
| SFA | 784 | 62.134 | 3.066 | 4.934 | 72.604 | 47.670 |
| UFA | 784 | 36.481 | 3.044 | 8.345 | 46.678 | 26.156 |
| SFA/UFA | 784 | 1.722 | 0.225 | 13.092 | 2.776 | 1.113 |
Note:
Variable coefficient calculated as the ratio of standard deviation (SD) to the mean multiplied by 100.
Genotypes and quality control
The cows were genotyped using the Illumina BovineSNP50 BeadChip (Illumina Inc., San Diego, CA, US), of which, some individuals were genotyped with the 54K chip version1 containing 54,001 SNPs, and others were genotyped with the 54K chip version 2 including 54,609 SNPs. After genotype imputation by BEAGLE software (http://faculty.washington.edu/browning/beagle/beagle.html), the common SNP markers in both version chips were used in this study, as a result, the total number of SNPs in the panel was 52,340. The SNP positions were based on the bovinegenome assembly UMD_3.1.66 (http://www.ncbi.nlm.nih.gov/genome/guide/cow/).
The quality control procedure was as follows, 20 daughters were excluded due to low call rate (<90%), leading to 764 daughters remaining for the association analysis. On the other hand, 11,736 SNPs were removed for falling to meet the following requirements: 652 SNPs with <90% genotype call rate, 10,798 SNPs with a minor allele frequency (MAF) <0.05, 286 SNPs with extreme value of Hardy-Weinberg equilibrium statistics (P<10−6). Eventually, 40,604 SNPs passed these quality control filters, which was 77.6% of the SNPs in the panel. The average distance between adjacent markers was quite constant among different chromosomes. The shortest average distance was 56 kb on BTA25, and the longest average distance was 75 kb on BTA5 (except for 198 kb on BTAX).
Statistical analysis
The statistical tests followed a two-step analysis. For the first step, phenotypic values were corrected for fixed non-genetic effects by using SAS 9.1 general linear model (GLM) procedure. The statistical model was: 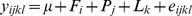 , where y
ijkl was the unadjusted phenotype; μ was the overall mean; Fi was the fixed effect of farm; Pj was the fixed effect of parity; Lk was the fixed effect of stage of lactation; e
ijkl was the random residual. In the second step, genome-wide association analyses were performed with quantitative trait procedure (additive model)of the PLINK software (v1.07) [27], and empirical p-values estimated based on the Wald-statistic. Individual pedigree of three generations was applied.
, where y
ijkl was the unadjusted phenotype; μ was the overall mean; Fi was the fixed effect of farm; Pj was the fixed effect of parity; Lk was the fixed effect of stage of lactation; e
ijkl was the random residual. In the second step, genome-wide association analyses were performed with quantitative trait procedure (additive model)of the PLINK software (v1.07) [27], and empirical p-values estimated based on the Wald-statistic. Individual pedigree of three generations was applied.
Manhattan plots of genome-wide association analyses were produced with R2.15.1 software (http://www.r-project.org/).
Significance level
Bonferroni correction was applied to adjust for multiple testing from the number of SNPs detected. A significant SNP at the genome-wise significance level was declared if a raw P value (unadjusted)<0.05/N, N is the number of SNP markers tested in analyses [28]. In the present study, Bonferroni genome-wise significance was 1.23E-06 (0.05/40604). As the Bonferroni correction threshold levels were strict and may lead to high false negatives, we calculated suggestive significant association threshold P-value as previously described [29], which was 2.46E-05 (1/40604).
Results
The global view of P-values for all SNPs of each trait was shown in Additional file 1. In total, 83 genome-wise significant SNPs (P<1.23E-06) and 314 suggestive significant SNPs (P<2.46E-05) were detected for 22 milk fatty acids on all chromosomes, ranged from 3 on BTA3, 4, 22 to 119 on BTA26 (Tables 2–6). For most of the studied fatty acids, significant associations were detected with more than one SNP, especially C14:1 (67 SNPs: 34 genome-wide, 33 suggestive), C18:0 (105 SNPs: 13 genome-wide, 92 suggestive), C14 index (84 SNPs: 49 genome-wide, 35 suggestive) and C18 index (93 SNPs: 14 genome-wide, 79 suggestive) (Table 7). The most significant SNP (BTB-00931481) was associated with both C14 index (P = 6.91E-17) and C14:1 (P = 7.08E-13) on BTA26. The top one common significant SNP (ARS-BFGL-NGS-4783) was associated with SFA, SFA/UFA and UFA. Besides, 146 SNPs were associated with multiple traits, especially ARS-BFGL-NGS-4939 on BTA14 for 9 traits. Further details on these associations are described as follows.
Table 2. Genome-wise and suggestive significant SNPs for short- and medium-chain saturated fatty acid traits (SCFA and MCFA).
| Trait | Ranka | SNP name | Chr. | Position(bp) | Nearest Gene/Candidate Gene | Distance(bp) | Raw P_value | P_value Bonferroni |
| C12:0 | 17 | BTA-120369-no-rs | 0 | 0 | NA | NA | 1.47E-05 | 0.599 |
| C14:0 | 19 | BTB-00046603 | 1 | 103185339 | SI | Within | 1.06E-05 | 0.432 |
| C14:0 | 26 | BTB-01201574 | 1 | 109284636 | SCHIP1 | Within | 1.84E-05 | 0.745 |
| C10:0 | 4 | ARS-BFGL-NGS-23583 | 1 | 132049236 | A4GNT | 14386 | 4.10E-06 | 0.166 |
| C12:0 | 9 | ARS-BFGL-NGS-23583 | 1 | 132049236 | A4GNT | 14386 | 6.09E-06 | 0.247 |
| C10:0 | 15 | BTB-00059412 | 1 | 132069357 | A4GNT | 1094 | 1.62E-05 | 0.658 |
| C10:0 | 10 | ARS-BFGL-NGS-37095 | 1 | 132497074 | SOX14 | 46530 | 1.23E-05 | 0.500 |
| C10:0 | 17 | BTA-51403-no-rs | 1 | 132518716 | SOX14 | 68172 | 1.91E-05 | 0.774 |
| C10:0 | 3 | ARS-BFGL-NGS-91327 | 2 | 12533969 | ZNF804A | 593727 | 3.70E-06 | 0.150 |
| C14:0 | 27 | ARS-BFGL-NGS-91327 | 2 | 12533969 | ZNF804A | 593727 | 2.19E-05 | 0.889 |
| C14:0 | 17 | ARS-BFGL-NGS-117409 | 2 | 37256390 | TANC1 | Within | 7.04E-06 | 0.286 |
| C14:0 | 10 | Hapmap42557-BTA-47352 | 2 | 38748216 | LOC101907729 | Within | 5.01E-06 | 0.204 |
| C14:0 | 8 | BTB-01053755 | 2 | 40313359 | NR4A2 | 295672 | 4.28E-06 | 0.174 |
| C12:0 | 21 | ARS-BFGL-NGS-30621 | 2 | 131539751 | HSPG2 | Within | 2.11E-05 | 0.855 |
| C12:0 | 16 | ARS-BFGL-NGS-20205 | 3 | 92427474 | SSBP3 | Within | 1.40E-05 | 0.569 |
| C14:0 | 20 | BTB-01477571 | 5 | 43481128 | CNOT2 | Within | 1.25E-05 | 0.509 |
| C14:0 | 1 | Hapmap49848-BTA-106779 | 5 | 45089737 | CPM | Within | 1.58E-07 | 6.42E-03 |
| C10:0 | 6 | Hapmap49071-BTA-17699 | 5 | 92618397 | PIK3C2G | 115016 | 7.53E-06 | 0.306 |
| C12:0 | 10 | BTB-01019973 | 7 | 79747454 | LOC101904982 | 440947 | 7.57E-06 | 0.308 |
| C12:0 | 7 | ARS-BFGL-NGS-67383 | 7 | 108307729 | EFNA5 | 738306 | 6.00E-06 | 0.244 |
| C10:0 | 1 | BTB-01556197 | 9 | 16892513 | HTR1B | 409905 | 5.89E-07 | 2.39E-02 |
| C12:0 | 3 | BTB-01556197 | 9 | 16892513 | HTR1B | 409905 | 2.66E-06 | 0.108 |
| C12:0 | 20 | ARS-BFGL-NGS-104719 | 9 | 18955509 | HMGN3 | 44851 | 2.10E-05 | 0.851 |
| C10:0 | 2 | ARS-BFGL-BAC-35400 | 9 | 21165167 | FAM46A | 594573 | 2.28E-06 | 0.093 |
| C14:0 | 18 | ARS-BFGL-BAC-35400 | 9 | 21165167 | FAM46A | 594573 | 7.23E-06 | 0.294 |
| C10:0 | 5 | ARS-BFGL-NGS-61979 | 9 | 23001645 | UBE3D | 42877 | 6.06E-06 | 0.246 |
| C12:0 | 6 | ARS-BFGL-NGS-61979 | 9 | 23001645 | UBE3D | 42877 | 5.66E-06 | 0.230 |
| C10:0 | 9 | Hapmap39984-BTA-21408 | 9 | 28538817 | LOC100848869 | 46056 | 1.10E-05 | 0.447 |
| C14:0 | 25 | Hapmap41109-BTA-93077 | 11 | 42713681 | BCL11A | 358295 | 1.83E-05 | 0.743 |
| C14:0 | 5 | ARS-BFGL-BAC-5848 | 12 | 68657690 | GPC6 | Within | 2.29E-06 | 0.093 |
| C10:0 | 20 | BTA-37592-no-rs | 15 | 72716471 | LRRC4C | 35911 | 2.17E-05 | 0.881 |
| C14:0 | 23 | BTB-00634528 | 16 | 31813761 | SMYD3 | Within | 1.35E-05 | 0.549 |
| C14:0 | 6 | UA-IFASA-8132 | 16 | 33607353 | C16H1orf100 | 7625 | 2.73E-06 | 0.111 |
| C10:0 | 21 | BTB-00648332 | 16 | 55421856 | PDPN | Within | 2.40E-05 | 0.973 |
| C14:0 | 16 | ARS-BFGL-NGS-102640 | 17 | 5963196 | PET112 | 17972 | 6.63E-06 | 0.269 |
| C14:0 | 2 | BTB-00669395 | 17 | 6266432 | FAM160A1 | Within | 2.09E-06 | 0.085 |
| C14:0 | 4 | Hapmap47945-BTA-41852 | 17 | 6295259 | FAM160A1 | Within | 2.26E-06 | 0.092 |
| C14:0 | 3 | BTB-00669586 | 17 | 6322271 | FAM160A1 | Within | 2.09E-06 | 0.085 |
| C14:0 | 13 | ARS-BFGL-NGS-20893 | 17 | 6669905 | SH3D19 | Within | 6.41E-06 | 0.260 |
| C14:0 | 14 | ARS-BFGL-NGS-115234 | 18 | 13136171 | JPH3 | 49852 | 6.48E-06 | 0.263 |
| C10:0 | 14 | Hapmap23685-BTA-132541 | 18 | 54271729 | STRN4/ SPHK2 | Within/1442497 | 1.59E-05 | 0.647 |
| C10:0 | 12 | UA-IFASA-7471 | 18 | 54311149 | SLC1A5/ SPHK2 | Within/1403077 | 1.41E-05 | 0.571 |
| C10:0 | 19 | ARS-BFGL-NGS-34500 | 19 | 15710458 | TMEM132E/ ACACA | 81903/1714400 | 2.11E-05 | 0.857 |
| C10:0 | 7 | ARS-BFGL-NGS-39328 | 19 | 51326750 | CCDC57/ FASN | Within/58172 | 8.54E-06 | 0.347 |
| C12:0 | 2 | ARS-BFGL-NGS-39328 | 19 | 51326750 | CCDC57/ FASN | Within/58172 | 1.16E-06 | 4.71E-02 |
| C14:0 | 11 | ARS-BFGL-NGS-39328 | 19 | 51326750 | CCDC57/ FASN | Within/58172 | 6.01E-06 | 0.244 |
| C14:0 | 21 | ARS-BFGL-NGS-87102 | 20 | 49859323 | CDH12 | 585652 | 1.26E-05 | 0.513 |
| C14:0 | 12 | ARS-BFGL-NGS-111676 | 20 | 51073910 | CDH12 | Within | 6.02E-06 | 0.244 |
| C12:0 | 13 | Hapmap53927-rs29025287 | 20 | 53303717 | CDH18 | 106159 | 1.03E-05 | 0.417 |
| C12:0 | 11 | BTB-00787949 | 20 | 53333822 | CDH18 | 76054 | 8.44E-06 | 0.343 |
| C14:0 | 9 | BTB-01583562 | 20 | 55425112 | LOC784462 | 71405 | 4.71E-06 | 0.191 |
| C10:0 | 16 | BTA-12468-no-rs | 21 | 9375095 | ARRDC4/ IGF1R | 436067/1107002 | 1.85E-05 | 0.749 |
| C12:0 | 12 | BTA-12468-no-rs | 21 | 9375095 | ARRDC4/ IGF1R | 436067/1107002 | 1.01E-05 | 0.411 |
| C10:0 | 8 | BTA-76414-no-rs | 21 | 9528223 | ARRDC4/ IGF1R | 589195/1260130 | 9.90E-06 | 0.402 |
| C12:0 | 1 | BTA-76414-no-rs | 21 | 9528223 | ARRDC4/ IGF1R | 589195/1260130 | 3.94E-07 | 1.60E-02 |
| C10:0 | 18 | ARS-BFGL-NGS-40159 | 21 | 21142616 | FANC1/RLBP1/ PLIN1 | Within/Within/360208 | 1.94E-05 | 0.786 |
| C12:0 | 14 | Hapmap26394-BTA-136497 | 22 | 27309084 | CNTN3 | Within | 1.05E-05 | 0.424 |
| C14:0 | 7 | Hapmap26394-BTA-136497 | 22 | 27309084 | CNTN3 | Within | 3.50E-06 | 0.142 |
| C10:0 | 11 | Hapmap57060-rs29023510 | 24 | 4805759 | FBXO15 | 246097 | 1.33E-05 | 0.539 |
| C12:0 | 19 | Hapmap57060-rs29023510 | 24 | 4805759 | FBXO15 | 246097 | 1.96E-05 | 0.795 |
| C12:0 | 4 | ARS-BFGL-NGS-78497 | 24 | 21330516 | SLC39A6 | Within | 2.89E-06 | 0.117 |
| C12:0 | 8 | BTB-00885512 | 24 | 30028775 | CHST9 | 8314 | 6.08E-06 | 0.247 |
| C10:0 | 13 | BTB-01077939 | 26 | 7685110 | PRKG1 | Within | 1.44E-05 | 0.586 |
| C12:0 | 22 | BTA-111275-no-rs | 26 | 9195089 | MINPP1/PRKG1 | Within/851454 | 2.39E-05 | 0.969 |
| C14:0 | 22 | ARS-BFGL-NGS-113226 | 27 | 97306 | LOC100335608 | Within | 1.34E-05 | 0.544 |
| C12:0 | 15 | BTB-01926888 | 27 | 16398882 | TRIML2/ ACSL1 | 303782/2110549 | 1.34E-05 | 0.545 |
| C12:0 | 5 | BTB-01603522 | 27 | 16421445 | TRIML2/ ACSL1 | 281219/2133112 | 3.55E-06 | 0.144 |
| C12:0 | 18 | ARS-BFGL-NGS-18922 | 29 | 21930571 | LUZP2 | 1125590 | 1.72E-05 | 0.696 |
| C14:0 | 24 | ARS-BFGL-NGS-19057 | 29 | 44196154 | CDC42EP2 | 1060 | 1.58E-05 | 0.640 |
| C14:0 | 15 | Hapmap60349-rs29021239 | X | 14062133 | ZNF280C | Within | 6.51E-06 | 0.264 |
Note:
Rank represents ranking of significant SNPs within each oftrait; The P_value with bold type represents the significance of genome-wise level; The gene name with bold type represents the nearest known gene to the significant SNPs; The gene name with bold type and underline represents the nearest novel candidate gene to the significant SNPs.
Table 6. Genome-wise and suggestive significant SNPs for sum of fatty acid traits.
| Trait | Rankb | SNP | Chr. | Position(bp) | Nearest Gene/Candidate Gene | Distance(bp) | Raw P_valuea | P_value Bonferroni |
| SFA | 13 | Hapmap42233-BTA-49670 | 1 | 82559884 | C1H3orf70/ EHHADH | 14106/38733 | 6.58E-06 | 0.267 |
| SFA/UFA | 14 | Hapmap42233-BTA-49670 | 1 | 82559884 | C1H3orf70/ EHHADH | 14106/38733 | 4.96E-06 | 0.202 |
| UFA | 33 | ARS-BFGL-NGS-13938 | 2 | 111650859 | MOGAT1 / ACSL3 | 19295/146311 | 2.25E-05 | 0.912 |
| SFA | 8 | Hapmap42304-BTA-73062 | 5 | 5049476 | KRR1/ OSBPL8 | 14567/770655 | 5.51E-06 | 0.224 |
| SFA/UFA | 12 | Hapmap42304-BTA-73062 | 5 | 5049476 | KRR1/ OSBPL8 | 14567/770655 | 4.17E-06 | 0.169 |
| UFA | 28 | Hapmap42304-BTA-73062 | 5 | 5049476 | KRR1/ OSBPL8 | 14567/770655 | 2.02E-05 | 0.819 |
| SFA/UFA | 32 | ARS-BFGL-NGS-38038 | 5 | 27992179 | NR4A1 | Within | 1.80E-05 | 0.731 |
| UFA | 26 | ARS-USMARC-624 | 5 | 28859701 | CSRNP2 | Within | 1.87E-05 | 0.760 |
| SFA | 10 | ARS-BFGL-NGS-8796 | 5 | 29095603 | LOC510716 | Within | 6.04E-06 | 0.245 |
| SFA/UFA | 8 | ARS-BFGL-NGS-8796 | 5 | 29095603 | LOC510716 | Within | 3.04E-06 | 0.123 |
| UFA | 3 | ARS-BFGL-NGS-8796 | 5 | 29095603 | LOC510716 | Within | 1.44E-06 | 0.058 |
| SFA/UFA | 22 | ARS-BFGL-NGS-35179 | 5 | 34325053 | SCAF11 | 62336 | 1.21E-05 | 0.493 |
| UFA | 20 | ARS-BFGL-NGS-35179 | 5 | 34325053 | SCAF11 | 62336 | 1.43E-05 | 0.579 |
| SFA | 17 | ARS-BFGL-NGS-69056 | 5 | 42285835 | CPNE8 | 135184 | 1.11E-05 | 0.449 |
| SFA/UFA | 29 | ARS-BFGL-NGS-69056 | 5 | 42285835 | CPNE8 | 135184 | 1.62E-05 | 0.657 |
| UFA | 8 | ARS-BFGL-NGS-69056 | 5 | 42285835 | CPNE8 | 135184 | 4.25E-06 | 0.173 |
| UFA | 27 | Hapmap52463-rs29025831 | 5 | 45473334 | LOC101905276 | 65002 | 1.93E-05 | 0.784 |
| SFA | 11 | Hapmap39862-BTA-74478 | 5 | 85672503 | BCAT1 | 74712 | 6.22E-06 | 0.253 |
| SFA/UFA | 6 | Hapmap39862-BTA-74478 | 5 | 85672503 | BCAT1 | 74712 | 2.26E-06 | 0.092 |
| UFA | 4 | Hapmap39862-BTA-74478 | 5 | 85672503 | BCAT1 | 74712 | 1.92E-06 | 0.078 |
| SFA/UFA | 16 | ARS-BFGL-NGS-99256 | 5 | 104714350 | VWF/ OLR1 | Within/4458707 | 5.47E-06 | 0.222 |
| UFA | 12 | ARS-BFGL-NGS-99256 | 5 | 104714350 | VWF/ OLR1 | Within/4458707 | 6.47E-06 | 0.263 |
| UFA | 16 | BTB-00246150 | 6 | 20993424 | PPA2 | Within | 9.44E-06 | 0.383 |
| SFA/UFA | 34 | Hapmap26001-BTC-038813 | 6 | 44926243 | PPARGC1A | Within | 1.97E-05 | 0.799 |
| UFA | 25 | Hapmap26001-BTC-038813 | 6 | 44926243 | PPARGC1A | Within | 1.85E-05 | 0.750 |
| UFA | 31 | BTB-00316291 | 7 | 64892251 | SPARC | Within | 2.18E-05 | 0.887 |
| SFA | 15 | BTB-00316348 | 7 | 64939808 | ATOX1 | 5940 | 8.62E-06 | 0.350 |
| SFA/UFA | 17 | BTB-00316348 | 7 | 64939808 | ATOX1 | 5940 | 6.58E-06 | 0.267 |
| UFA | 15 | BTB-00316348 | 7 | 64939808 | ATOX1 | 5940 | 9.25E-06 | 0.375 |
| SFA | 9 | BTB-00316650 | 7 | 65098028 | GLRA1 | Within | 6.02E-06 | 0.245 |
| SFA/UFA | 18 | BTB-00316650 | 7 | 65098028 | GLRA1 | Within | 7.85E-06 | 0.319 |
| UFA | 17 | BTB-00316650 | 7 | 65098028 | GLRA1 | Within | 9.98E-06 | 0.405 |
| UFA | 30 | BTB-02040446 | 8 | 30320626 | NFIB | 164574 | 2.16E-05 | 0.878 |
| UFA | 22 | Hapmap50126-BTA-83733 | 9 | 55449737 | LOC101907134 | 18763 | 1.73E-05 | 0.702 |
| SFA | 24 | BTB-01866513 | 10 | 70231445 | SLC35F4 | Within | 1.73E-05 | 0.701 |
| SFA | 29 | BTB-01203179 | 10 | 72694329 | DHRS7 | 53135 | 2.03E-05 | 0.825 |
| SFA/UFA | 28 | BTB-01203179 | 10 | 72694329 | DHRS7 | 53135 | 1.61E-05 | 0.653 |
| UFA | 10 | BTB-01203179 | 10 | 72694329 | DHRS7 | 53135 | 5.18E-06 | 0.210 |
| SFA | 2 | BTB-01332998 | 10 | 73466092 | SLC38A6 | 15901 | 8.08E-08 | 3.28E-03 |
| SFA/UFA | 2 | BTB-01332998 | 10 | 73466092 | SLC38A6 | 15901 | 3.54E-07 | 1.44E-02 |
| UFA | 2 | BTB-01332998 | 10 | 73466092 | SLC38A6 | 15901 | 4.87E-07 | 1.98E-02 |
| SFA | 1 | ARS-BFGL-NGS-4783 | 10 | 73487550 | SLC38A6 | 37359 | 6.07E-08 | 2.46E-03 |
| SFA/UFA | 1 | ARS-BFGL-NGS-4783 | 10 | 73487550 | SLC38A6 | 37359 | 2.64E-07 | 1.07E-02 |
| UFA | 1 | ARS-BFGL-NGS-4783 | 10 | 73487550 | SLC38A6 | 37359 | 4.01E-07 | 1.63E-02 |
| SFA | 7 | BTB-01501723 | 10 | 73655817 | TMEM30B | 6161 | 5.31E-06 | 0.216 |
| SFA/UFA | 37 | BTB-01501723 | 10 | 73655817 | TMEM30B | 6161 | 2.28E-05 | 0.924 |
| UFA | 34 | BTB-01501723 | 10 | 73655817 | TMEM30B | 6161 | 2.25E-05 | 0.912 |
| SFA | 19 | BTB-01079278 | 11 | 57078447 | REG3A | 435723 | 1.39E-05 | 0.565 |
| SFA/UFA | 36 | BTB-01079278 | 11 | 57078447 | REG3A | 435723 | 2.12E-05 | 0.859 |
| SFA | 23 | BTB-01079350 | 11 | 57107070 | REG3A | 464346 | 1.66E-05 | 0.675 |
| SFA | 3 | BTA-119672-no-rs | 11 | 102911946 | AK8 | Within | 1.34E-06 | 0.054 |
| SFA/UFA | 10 | BTA-119672-no-rs | 11 | 102911946 | AK8 | Within | 3.77E-06 | 0.153 |
| UFA | 7 | BTA-119672-no-rs | 11 | 102911946 | AK8 | Within | 4.03E-06 | 0.164 |
| UFA | 18 | BTB-01236909 | 12 | 50295112 | TBC1D4 | 382864 | 1.17E-05 | 0.476 |
| UFA | 38 | BTB-01980482 | 12 | 50451289 | TBC1D4 | 226687 | 2.38E-05 | 0.964 |
| SFA/UFA | 31 | ARS-BFGL-NGS-70206 | 13 | 48622655 | FERMT1 | Within | 1.65E-05 | 0.670 |
| UFA | 37 | ARS-BFGL-NGS-70206 | 13 | 48622655 | FERMT1 | Within | 2.31E-05 | 0.937 |
| SFA/UFA | 25 | ARS-BFGL-NGS-57820 | 14 | 1651311 | LOC100294916/ DGAT1 | Within/144114 | 1.41E-05 | 0.572 |
| SFA | 5 | ARS-BFGL-NGS-4939 | 14 | 1801116 | DGAT1 | Within | 2.80E-06 | 0.114 |
| SFA/UFA | 3 | ARS-BFGL-NGS-4939 | 14 | 1801116 | DGAT1 | Within | 1.15E-06 | 4.65E-02 |
| UFA | 9 | ARS-BFGL-NGS-4939 | 14 | 1801116 | DGAT1 | Within | 4.78E-06 | 0.194 |
| SFA/UFA | 21 | ARS-BFGL-NGS-107379 | 14 | 2054457 | LOC786966/ DGAT1 | 460/249619 | 1.17E-05 | 0.475 |
| SFA/UFA | 13 | ARS-BFGL-NGS-100480 | 14 | 4364952 | TRAPPC9 | Within | 4.68E-06 | 0.190 |
| UFA | 14 | ARS-BFGL-NGS-100480 | 14 | 4364952 | TRAPPC9 | Within | 7.52E-06 | 0.305 |
| SFA | 16 | ARS-BFGL-NGS-113293 | 14 | 77274386 | SLC2A5 | 3495 | 1.03E-05 | 0.419 |
| SFA/UFA | 5 | ARS-BFGL-NGS-113293 | 14 | 77274386 | SLC2A5 | 3495 | 1.41E-06 | 0.057 |
| UFA | 19 | ARS-BFGL-NGS-113293 | 14 | 77274386 | SLC2A5 | 3495 | 1.36E-05 | 0.551 |
| SFA | 22 | ARS-BFGL-NGS-15481 | 18 | 56611355 | AP2A1/ SPHK2 | Within/889940 | 1.55E-05 | 0.630 |
| SFA/UFA | 20 | ARS-BFGL-NGS-15481 | 18 | 56611355 | AP2A1/ SPHK2 | Within/889940 | 9.71E-06 | 0.394 |
| UFA | 24 | ARS-BFGL-NGS-15481 | 18 | 56611355 | AP2A1/ SPHK2 | Within/889940 | 1.79E-05 | 0.725 |
| SFA | 20 | ARS-BFGL-NGS-75390 | 19 | 4286783 | LOC790351 | 6533 | 1.44E-05 | 0.586 |
| SFA/UFA | 19 | ARS-BFGL-NGS-75390 | 19 | 4286783 | LOC790351 | 6533 | 9.64E-06 | 0.391 |
| SFA | 6 | ARS-BFGL-NGS-87368 | 19 | 7762820 | C19H17orf67 | 67914 | 3.15E-06 | 0.128 |
| SFA/UFA | 9 | ARS-BFGL-NGS-87368 | 19 | 7762820 | C19H17orf67 | 67914 | 3.72E-06 | 0.151 |
| UFA | 13 | ARS-BFGL-NGS-87368 | 19 | 7762820 | C19H17orf67 | 67914 | 6.64E-06 | 0.270 |
| SFA | 21 | ARS-BFGL-NGS-118339 | 20 | 3347138 | C20H5orf50 | 150804 | 1.54E-05 | 0.627 |
| SFA/UFA | 15 | ARS-BFGL-NGS-118339 | 20 | 3347138 | C20H5orf50 | 150804 | 5.31E-06 | 0.215 |
| UFA | 23 | ARS-BFGL-NGS-118339 | 20 | 3347138 | C20H5orf50 | 150804 | 1.75E-05 | 0.709 |
| SFA/UFA | 24 | BTA-50482-no-rs | 20 | 36336225 | EGFLAM | 157753 | 1.35E-05 | 0.548 |
| UFA | 36 | BTA-50482-no-rs | 20 | 36336225 | EGFLAM | 157753 | 2.31E-05 | 0.936 |
| SFA | 14 | ARS-BFGL-NGS-116806 | 20 | 36450009 | GDNF | 180881 | 7.68E-06 | 0.312 |
| SFA/UFA | 11 | ARS-BFGL-NGS-116806 | 20 | 36450009 | GDNF | 180881 | 3.92E-06 | 0.159 |
| UFA | 11 | ARS-BFGL-NGS-116806 | 20 | 36450009 | GDNF | 180881 | 5.23E-06 | 0.212 |
| SFA | 30 | ARS-USMARC-Parent-DQ990835-rs29012811 | 20 | 36570529 | GDNF | 60361 | 2.31E-05 | 0.939 |
| SFA/UFA | 27 | ARS-USMARC-Parent-DQ990835-rs29012811 | 20 | 36570529 | GDNF | 60361 | 1.56E-05 | 0.632 |
| UFA | 35 | ARS-USMARC-Parent-DQ990835-rs29012811 | 20 | 36570529 | GDNF | 60361 | 2.29E-05 | 0.931 |
| SFA | 27 | ARS-BFGL-NGS-17676 | 20 | 39017985 | PRLR | 55261 | 1.95E-05 | 0.790 |
| SFA/UFA | 23 | ARS-BFGL-NGS-17676 | 20 | 39017985 | PRLR | 55261 | 1.29E-05 | 0.523 |
| UFA | 39 | ARS-BFGL-NGS-17676 | 20 | 39017985 | PRLR | 55261 | 2.45E-05 | 0.994 |
| SFA/UFA | 38 | ARS-BFGL-NGS-55739 | 20 | 39787788 | C1QTNF3 | Within | 2.38E-05 | 0.965 |
| SFA | 25 | BTB-00783271 | 20 | 41201777 | SUB1 | 17036 | 1.73E-05 | 0.702 |
| SFA/UFA | 26 | BTB-00783271 | 20 | 41201777 | SUB1 | 17036 | 1.54E-05 | 0.626 |
| UFA | 29 | BTB-00783271 | 20 | 41201777 | SUB1 | 17036 | 2.09E-05 | 0.850 |
| SFA | 12 | BTB-01583562 | 20 | 55425112 | LOC101905359 | Within | 6.28E-06 | 0.255 |
| SFA | 4 | Hapmap44836-BTA-51861 | 21 | 20990602 | ABHD2/ PLIN1 | 8117/512222 | 1.76E-06 | 0.071 |
| SFA/UFA | 4 | Hapmap44836-BTA-51861 | 21 | 20990602 | ABHD2/ PLIN1 | 8117/512222 | 1.39E-06 | 0.056 |
| UFA | 6 | Hapmap44836-BTA-51861 | 21 | 20990602 | ABHD2/ PLIN1 | 8117/512222 | 2.09E-06 | 0.085 |
| UFA | 32 | Hapmap35708-SCAFFOLD316799_27843 | 21 | 25136856 | SH3GL3 | 52052 | 2.21E-05 | 0.896 |
| SFA/UFA | 39 | Hapmap33890-BES3_Contig418_1154 | 23 | 40236175 | ATXN1 | Within | 2.39E-05 | 0.968 |
| SFA | 28 | ARS-BFGL-BAC-28144 | 25 | 2606575 | LOC788915 | 21466 | 1.96E-05 | 0.795 |
| SFA/UFA | 7 | ARS-BFGL-BAC-28144 | 25 | 2606575 | LOC788915 | 21466 | 2.89E-06 | 0.117 |
| UFA | 5 | ARS-BFGL-BAC-28144 | 25 | 2606575 | LOC788915 | 21466 | 2.06E-06 | 0.083 |
| SFA | 26 | BTA-61650-no-rs | 26 | 41850719 | FGFR2 | Within | 1.91E-05 | 0.776 |
| SFA/UFA | 35 | BTA-61650-no-rs | 26 | 41850719 | FGFR2 | Within | 1.98E-05 | 0.802 |
| SFA/UFA | 33 | BTB-01926888 | 27 | 16398882 | TRIML2/ ACSL1 | 303782/2110549 | 1.96E-05 | 0.797 |
| UFA | 21 | BTB-01926888 | 27 | 16398882 | TRIML2/ ACSL1 | 303782/2110549 | 1.58E-05 | 0.641 |
| SFA | 18 | ARS-BFGL-NGS-106901 | 29 | 44372611 | SCYL1 | Within | 1.22E-05 | 0.495 |
| SFA/UFA | 30 | ARS-BFGL-NGS-106901 | 29 | 44372611 | SCYL1 | Within | 1.63E-05 | 0.663 |
Note: see note to Table 2.
Table 7. Numbers of significant SNPs with genome-wise and suggestive significance for 18 milk fatty acid traits.
| Trait | Genome-wise level | Suggestive level | Total |
| C10:0 | 1 | 20 | 21 |
| C12:0 | 2 | 20 | 22 |
| C14:0 | 1 | 26 | 27 |
| C14:1 | 34 | 33 | 67 |
| C16:0 | 0 | 18 | 18 |
| C16:1 | 0 | 6 | 6 |
| C18:0 | 13 | 92 | 105 |
| C18:1n9c | 0 | 20 | 20 |
| C18:2n6c | 3 | 3 | 6 |
| CLA | 0 | 7 | 7 |
| C14 index | 49 | 35 | 84 |
| C16 index | 0 | 10 | 10 |
| C18 index | 14 | 79 | 93 |
| SFA | 2 | 28 | 30 |
| UFA | 2 | 37 | 39 |
| SFA/UFA | 3 | 36 | 39 |
| C20:0 | 0 | 2 | 2 |
| C22:0 | 1 | 0 | 1 |
| Sum | 125 | 472 | 597 |
Note: Associations with C8:0, C18:3n3, C18:3n6 and C20:5n3 only reached chromosome-wise significance or non-significant, so they were not listed.
Table 3. Genome-wise and suggestive significant SNPs for long-chain saturated fatty acid traits (LCFA).
| Trait | Rankb | SNP | Chr. | Position(bp) | Nearest Gene/Candidate Gene | Distance(bp) | Raw P_valuea | P_value Bonferroni |
| C16:0 | 1 | ARS-BFGL-NGS-111692 | 0 | 0 | NA | NA | 1.84E-06 | 0.075 |
| C22:0 | 1 | ARS-BFGL-NGS-109692 | 1 | 15796320 | NCAM2 | 432009 | 6.70E-07 | 2.72E-02 |
| C18:0 | 78 | ARS-BFGL-NGS-111491 | 1 | 68457522 | PTPLB | 6964 | 1.46E-05 | 0.593 |
| C18:0 | 91 | Hapmap50666-BTA-34589 | 1 | 68533156 | PTPLB | Within | 1.88E-05 | 0.761 |
| C18:0 | 8 | ARS-BFGL-NGS-76111 | 1 | 103219606 | SI | 14878 | 7.51E-07 | 3.05E-02 |
| C18:0 | 25 | BTB-00048807 | 1 | 106245603 | OTOL1 | 454829 | 2.46E-06 | 0.100 |
| C18:0 | 26 | BTB-00048739 | 1 | 106295981 | OTOL1 | 404451 | 2.77E-06 | 0.112 |
| C18:0 | 16 | ARS-BFGL-NGS-111111 | 1 | 146302724 | HSF2BP/ AGPAT3 | 43214/402889 | 1.68E-06 | 0.068 |
| C18:0 | 41 | ARS-BFGL-NGS-109493 | 1 | 146354654 | HSF2BP/ AGPAT3 | Within/350959 | 5.28E-06 | 0.214 |
| C18:0 | 42 | BTA-56389-no-rs | 1 | 146384457 | HSF2BP/ AGPAT3 | Within/321156 | 5.28E-06 | 0.214 |
| C18:0 | 93 | Hapmap59917-rs29012418 | 2 | 24519348 | METAP1D | Within | 1.92E-05 | 0.778 |
| C18:0 | 88 | ARS-BFGL-BAC-2793 | 2 | 47449015 | KIF5C | Within | 1.79E-05 | 0.726 |
| C18:0 | 66 | Hapmap53388-rs29010903 | 2 | 63581955 | MGAT5 | 129790 | 1.18E-05 | 0.479 |
| C18:0 | 63 | BTB-01373917 | 2 | 79056259 | GYPC/ STAT1 | 199326/837973 | 1.12E-05 | 0.453 |
| C18:0 | 22 | ARS-BFGL-NGS-33744 | 2 | 79388083 | GYPC/ STAT1 | 83668/506149 | 2.26E-06 | 0.092 |
| C18:0 | 52 | ARS-BFGL-NGS-99030 | 2 | 98160191 | UNC80 | Within | 9.14E-06 | 0.371 |
| C18:0 | 11 | ARS-BFGL-NGS-45691 | 2 | 128484790 | RUNX3/ FABP3 | 144886/5700960 | 9.56E-07 | 3.88E-02 |
| C18:0 | 30 | ARS-BFGL-NGS-118924 | 2 | 128529102 | RUNX3/ FABP3 | 100574/5745272 | 3.22E-06 | 0.131 |
| C18:0 | 104 | ARS-BFGL-NGS-58955 | 2 | 133620177 | HTR6 | Within | 2.41E-05 | 0.978 |
| C18:0 | 10 | ARS-BFGL-NGS-45803 | 2 | 134246808 | IFFO2 | 46005 | 8.99E-07 | 3.65E-02 |
| C18:0 | 50 | ARS-BFGL-NGS-15882 | 3 | 991777 | MPZL1 | Within | 8.70E-06 | 0.353 |
| C18:0 | 55 | Hapmap34855-BES3_Contig373_1182 | 4 | 5549277 | IKZF1 | 31842 | 1.00E-05 | 0.407 |
| C18:0 | 79 | BTA-122414-no-rs | 4 | 34967013 | SEMA3D | 596525 | 1.50E-05 | 0.610 |
| C16:0 | 12 | Hapmap40292-BTA-71565 | 4 | 81400732 | C4H7orf10 | Within | 1.39E-05 | 0.564 |
| C18:0 | 7 | Hapmap30257-BTA-142970 | 5 | 25358659 | USP44 | 44459 | 6.91E-07 | 2.81E-02 |
| C18:0 | 60 | Hapmap41951-BTA-73168 | 5 | 28442563 | SLC4A8 | 25682 | 1.05E-05 | 0.425 |
| C18:0 | 73 | Hapmap49848-BTA-106779 | 5 | 45089737 | CPM | Within | 1.31E-05 | 0.533 |
| C18:0 | 12 | Hapmap50366-BTA-46960 | 5 | 68610818 | CHST11 | Within | 1.02E-06 | 4.14E-02 |
| C16:0 | 3 | Hapmap39862-BTA-74478 | 5 | 85672503 | BCAT1 | 74712 | 4.88E-06 | 0.198 |
| C16:0 | 7 | BTB-00316348 | 7 | 64939808 | ATOX1 | Within | 1.21E-05 | 0.490 |
| C18:0 | 82 | BTB-01553821 | 7 | 107940320 | EFNA5 | 1105715 | 1.63E-05 | 0.663 |
| C18:0 | 75 | BTB-00995040 | 8 | 22411464 | MIR31 | 123333 | 1.32E-05 | 0.536 |
| C18:0 | 18 | BTB-01709624 | 8 | 33060034 | LOC101904752 | 1103805 | 1.86E-06 | 0.075 |
| C18:0 | 43 | BTA-99986-no-rs | 8 | 33175156 | LOC101904752 | 988683 | 5.54E-06 | 0.225 |
| C18:0 | 61 | BTB-01973796 | 8 | 34146856 | LOC101904752 | 16983 | 1.07E-05 | 0.434 |
| C18:0 | 24 | BTB-01929442 | 8 | 34560427 | LOC101904752 | 52452 | 2.45E-06 | 0.099 |
| C18:0 | 70 | ARS-BFGL-NGS-17346 | 8 | 70653923 | PEBP4 | Within | 1.27E-05 | 0.514 |
| C18:0 | 67 | Hapmap57757-ss46526215 | 8 | 74949334 | BNIP3L | Within | 1.18E-05 | 0.481 |
| C18:0 | 46 | ARS-BFGL-NGS-105738 | 8 | 76089371 | APTX | 7156 | 6.77E-06 | 0.275 |
| C18:0 | 53 | BTB-00227581 | 8 | 76119002 | DNAJA1 | 1365 | 9.23E-06 | 0.375 |
| C18:0 | 29 | BTB-00359112 | 8 | 76403422 | NFX1 | Within | 3.14E-06 | 0.128 |
| C18:0 | 28 | ARS-BFGL-NGS-9052 | 8 | 78009328 | FRMD3 | 181107 | 3.10E-06 | 0.126 |
| C18:0 | 94 | BTB-01900316 | 8 | 78226921 | UBQLN1 | Within | 1.93E-05 | 0.783 |
| C18:0 | 15 | Hapmap23947-BTA-153013 | 8 | 78347212 | GKAP1 | Within | 1.66E-06 | 0.067 |
| C18:0 | 37 | Hapmap54400-rs29020952 | 8 | 79534851 | NTRK2 | Within | 4.82E-06 | 0.196 |
| C18:0 | 76 | Hapmap34874-BES3_Contig415_1312 | 8 | 79559282 | NTRK2 | Within | 1.34E-05 | 0.545 |
| C18:0 | 81 | ARS-BFGL-NGS-101844 | 9 | 13292880 | SLC17A5 | 12122 | 1.59E-05 | 0.644 |
| C18:0 | 96 | ARS-BFGL-NGS-101978 | 9 | 18123535 | HTR1B | 818548 | 2.04E-05 | 0.830 |
| C18:0 | 74 | ARS-BFGL-NGS-117379 | 9 | 97397264 | SOD2 | 1895 | 1.32E-05 | 0.535 |
| C18:0 | 45 | ARS-BFGL-NGS-66090 | 10 | 11104820 | SERINC5 | Within | 5.91E-06 | 0.240 |
| C18:0 | 39 | BTA-23031-no-rs | 10 | 27606965 | OR4K13 | 6376 | 4.93E-06 | 0.200 |
| C16:0 | 5 | ARS-BFGL-NGS-23492 | 10 | 34637715 | RASGRP1 | 356723 | 1.13E-05 | 0.460 |
| C16:0 | 10 | ARS-BFGL-NGS-503 | 10 | 34705386 | RASGRP1 | 424394 | 1.35E-05 | 0.547 |
| C16:0 | 11 | ARS-BFGL-NGS-4783 | 10 | 73487550 | SLC38A6 | 35850 | 1.37E-05 | 0.555 |
| C16:0 | 17 | ARS-BFGL-NGS-14667 | 10 | 81481148 | GALNTL1 | Within | 1.79E-05 | 0.728 |
| C16:0 | 8 | BTA-77299-no-rs | 10 | 81549606 | SLC39A9 | Within | 1.24E-05 | 0.503 |
| C16:0 | 16 | Hapmap44279-BTA-75297 | 10 | 84178157 | RGS6 | Within | 1.75E-05 | 0.712 |
| C18:0 | 102 | BTB-00471219 | 11 | 32709462 | NRXN1 | Within | 2.25E-05 | 0.915 |
| C18:0 | 103 | Hapmap25798-BTA-126388 | 11 | 32731961 | NRXN1 | Within | 2.40E-05 | 0.976 |
| C18:0 | 9 | Hapmap51531-BTA-98947 | 11 | 57522675 | REG3A | 880289 | 8.78E-07 | 3.57E-02 |
| C18:0 | 6 | BTB-01328920 | 11 | 57639084 | LRRTM4 | 792049 | 5.79E-07 | 2.35E-02 |
| C18:0 | 40 | BTA-33625-no-rs | 11 | 58306286 | LRRTM4 | 124847 | 5.07E-06 | 0.206 |
| C18:0 | 27 | Hapmap47549-BTA-25561 | 11 | 69506822 | LCLAT1 | 108007 | 2.85E-06 | 0.116 |
| C18:0 | 97 | BTB-00866714 | 12 | 65771899 | LOC101907906 | 250357 | 2.09E-05 | 0.850 |
| C18:0 | 62 | ARS-BFGL-BAC-5848 | 12 | 68657690 | GPC6 | Within | 1.10E-05 | 0.446 |
| C18:0 | 100 | Hapmap51198-BTA-27036 | 12 | 68703184 | GPC6 | Within | 2.15E-05 | 0.873 |
| C18:0 | 1 | ARS-BFGL-BAC-13788 | 12 | 69512332 | DCT | Within | 9.17E-09 | 3.72E-04 |
| C18:0 | 65 | Hapmap23511-BTA-119303 | 12 | 69768137 | SOX21 | 24828 | 1.14E-05 | 0.463 |
| C18:0 | 2 | ARS-BFGL-NGS-45730 | 12 | 70260457 | ABCC4 | 51353 | 1.05E-08 | 4.24E-04 |
| C18:0 | 101 | ARS-BFGL-NGS-40278 | 13 | 67122063 | BLCAP/ ACSS2 | Within/2280543 | 2.25E-05 | 0.914 |
| C16:0 | 4 | ARS-BFGL-NGS-4939 | 14 | 1801116 | DGAT1 | Within | 9.04E-06 | 0.367 |
| C18:0 | 57 | BTB-00557532 | 14 | 24643266 | XKR4 | 28554 | 1.03E-05 | 0.416 |
| C18:0 | 64 | ARS-BFGL-NGS-110022 | 14 | 40616098 | CRISPLD1 | 144633 | 1.12E-05 | 0.456 |
| C18:0 | 36 | ARS-BFGL-NGS-2025 | 14 | 65452336 | ZNF706 | 103905 | 4.72E-06 | 0.192 |
| C18:0 | 19 | Hapmap35102-BES3_Contig324_378 | 14 | 66523007 | RGS22 | Within | 1.87E-06 | 0.076 |
| C18:0 | 17 | ARS-BFGL-BAC-1991 | 14 | 82648300 | SNX16 | 491346 | 1.73E-06 | 0.070 |
| C18:0 | 34 | ARS-BFGL-NGS-12066 | 15 | 27291268 | BUD13 | 542743 | 3.98E-06 | 0.162 |
| C18:0 | 54 | BTB-01465034 | 15 | 49987359 | LOC784376 | 1153 | 9.65E-06 | 0.392 |
| C16:0 | 2 | ARS-BFGL-NGS-97658 | 15 | 68069900 | C15H11orf74 | 158748 | 2.72E-06 | 0.110 |
| C18:0 | 92 | BTA-37923-no-rs | 15 | 85011763 | GLB1L2 | Within | 1.88E-05 | 0.761 |
| C18:0 | 3 | ARS-BFGL-NGS-68533 | 16 | 60615012 | LOC101902340 | 35998 | 2.71E-07 | 1.10E-02 |
| C18:0 | 31 | ARS-BFGL-NGS-102798 | 16 | 61477887 | RALGPS2 | 82055 | 3.31E-06 | 0.134 |
| C18:0 | 95 | BTB-00653808 | 16 | 62179726 | AXDND1 | Within | 2.01E-05 | 0.817 |
| C18:0 | 87 | ARS-BFGL-NGS-117800 | 16 | 62871926 | LHX4 | Within | 1.75E-05 | 0.711 |
| C18:0 | 83 | ARS-BFGL-NGS-102835 | 17 | 3855180 | SFRP2 | 17042 | 1.67E-05 | 0.678 |
| C18:0 | 69 | ARS-BFGL-NGS-100229 | 17 | 4153159 | KIAA0922 | Within | 1.21E-05 | 0.493 |
| C18:0 | 14 | ARS-BFGL-NGS-77485 | 17 | 4926550 | TMEM154 | 39267 | 1.47E-06 | 0.059 |
| C18:0 | 38 | ARS-BFGL-NGS-118611 | 17 | 5357573 | FBXW7 | 23715 | 4.91E-06 | 0.199 |
| C18:0 | 89 | ARS-BFGL-NGS-38059 | 17 | 5829384 | PET112 | 151784 | 1.79E-05 | 0.728 |
| C18:0 | 84 | ARS-BFGL-NGS-114953 | 17 | 9753430 | NR3C2 | Within | 1.70E-05 | 0.688 |
| C18:0 | 98 | ARS-BFGL-NGS-111098 | 17 | 67193210 | PIWIL3 | 27876 | 2.10E-05 | 0.851 |
| C18:0 | 13 | ARS-BFGL-NGS-71116 | 17 | 68002540 | MYO18B | Within | 1.09E-06 | 4.41E-02 |
| C18:0 | 86 | ARS-BFGL-NGS-75816 | 19 | 36674728 | ANKRD40/ SREBF1 | Within/1424056 | 1.70E-05 | 0.692 |
| C18:0 | 71 | BTB-01790846 | 20 | 13263157 | SREK1 | 128568 | 1.27E-05 | 0.516 |
| C18:0 | 99 | Hapmap56230-rs29025779 | 20 | 25535571 | NDUFS4 | 12189 | 2.14E-05 | 0.869 |
| C18:0 | 105 | ARS-BFGL-NGS-118998 | 20 | 32030332 | GHR / OXCT1 | Within/653664 | 2.43E-05 | 0.988 |
| C18:0 | 48 | ARS-BFGL-NGS-87102 | 20 | 49859323 | CDH12 | 585652 | 7.71E-06 | 0.313 |
| C18:0 | 23 | BTB-01475042 | 20 | 51845309 | CDH12 | 220958 | 2.29E-06 | 0.093 |
| C18:0 | 20 | ARS-BFGL-NGS-51112 | 20 | 52816141 | CDH18 | 593735 | 1.89E-06 | 0.077 |
| C16:0 | 18 | BTB-00818669 | 21 | 40502782 | PRKD1 | 25243 | 1.95E-05 | 0.790 |
| C16:0 | 13 | BTB-01240884 | 21 | 41484660 | G2E3 | 128084 | 1.48E-05 | 0.600 |
| C16:0 | 9 | Hapmap24313-BTA-29957 | 21 | 51294895 | LRFN5 | 476940 | 1.30E-05 | 0.526 |
| C18:0 | 58 | ARS-BFGL-BAC-46707 | 23 | 5756655 | TINAG | 196853 | 1.03E-05 | 0.419 |
| C16:0 | 15 | BTA-56520-no-rs | 23 | 32581285 | LOC537017/ BTN1A1 | 59797/1214677 | 1.73E-05 | 0.700 |
| C18:0 | 33 | Hapmap58547-rs29023020 | 23 | 35911017 | LOC100847951/ PRL | Within/797267 | 3.81E-06 | 0.155 |
| C18:0 | 49 | BTA-58814-no-rs | 24 | 5828401 | CBLN2 | 207345 | 8.20E-06 | 0.333 |
| C18:0 | 80 | BTB-00878928 | 24 | 5883369 | CBLN2 | 262313 | 1.51E-05 | 0.612 |
| C18:0 | 59 | Hapmap34424-BES10_Contig566_926 | 24 | 6408329 | CBLN2 | 787273 | 1.04E-05 | 0.421 |
| C18:0 | 4 | ARS-BFGL-NGS-109955 | 24 | 11625175 | CDH7 | 411164 | 3.29E-07 | 1.33E-02 |
| C18:0 | 51 | BTA-24495-no-rs | 24 | 15527942 | PIK3C3 | 1180676 | 8.82E-06 | 0.358 |
| C18:0 | 56 | ARS-BFGL-NGS-13314 | 24 | 45775076 | SLC14A2 | Within | 1.01E-05 | 0.408 |
| C18:0 | 35 | BTA-59652-no-rs | 25 | 19315456 | LOC524391 | Within | 4.37E-06 | 0.178 |
| C16:0 | 6 | BTB-01619101 | 26 | 16035322 | CYP2C87 | Within | 1.16E-05 | 0.472 |
| C18:0 | 72 | ARS-BFGL-NGS-12828 | 26 | 37018466 | GFRA1 | Within | 1.29E-05 | 0.524 |
| C18:0 | 21 | ARS-BFGL-NGS-100468 | 26 | 38060272 | PDZD8 | 39152 | 2.10E-06 | 0.085 |
| C18:0 | 47 | ARS-BFGL-NGS-111901 | 26 | 41183634 | WDR11 | 28744 | 7.62E-06 | 0.309 |
| C18:0 | 68 | UA-IFASA-5698 | 26 | 42673967 | HTRA1 | Within | 1.21E-05 | 0.491 |
| C18:0 | 77 | Hapmap60810-rs29012623 | 27 | 12355411 | LOC100848735 | 130088 | 1.39E-05 | 0.565 |
| C20:0 | 2 | BTB-00965197 | 27 | 26813022 | NRG1 | Within | 1.93E-05 | 0.783 |
| C20:0 | 1 | ARS-BFGL-NGS-77002 | 27 | 30883081 | UNC5D | Within | 3.76E-06 | 0.153 |
| C18:0 | 85 | Hapmap35611-SCAFFOLD120249_17244 | 27 | 44005137 | ZNF385D | 5580 | 1.70E-05 | 0.691 |
| C16:0 | 14 | Hapmap27418-BTA-147969 | 29 | 23469370 | LOC540991 | 19676 | 1.66E-05 | 0.675 |
| C18:0 | 32 | ARS-BFGL-NGS-19057 | 29 | 44196154 | CDC42EP2 | 1060 | 3.65E-06 | 0.148 |
| C18:0 | 5 | ARS-BFGL-NGS-11681 | X | 3622615 | Gene desert | NA | 3.74E-07 | 1.52E-02 |
| C18:0 | 44 | Hapmap48540-BTA-97806 | X | 8700029 | ODZ1 | Within | 5.77E-06 | 0.234 |
| C18:0 | 90 | BTB-01492502 | X | 8724630 | ODZ1 | Within | 1.84E-05 | 0.746 |
Note: see note to Table 2.
Table 4. Genome-wise and suggestive significant SNPs for monounsaturated and polyunsaturated fatty acid traits (MUFA and PUFA).
| Trait | Rankb | SNP | Chr. | Position(bp) | Nearest Gene/Candidate Gene | Distance(bp) | Raw P_valuea | P_value Bonferroni |
| C14:1 | 34 | ARS-BFGL-NGS-15914 | 0 | 0 | NA | NA | 1.20E-06 | 4.87E-02 |
| CLA | 2 | ARS-BFGL-NGS-43953 | 1 | 113855984 | GPR149 | 288255 | 1.31E-05 | 0.531 |
| C14:1 | 39 | ARS-BFGL-BAC-42858 | 2 | 33401608 | LOC534542 | 231071 | 2.02E-06 | 0.082 |
| CLA | 4 | BTB-01649999 | 3 | 48062099 | RWDD3 | 336059 | 1.65E-05 | 0.672 |
| C18:1n9c | 9 | Hapmap42304-BTA-73062 | 5 | 5049476 | KRR1/ OSBPL8 | 14567/770655 | 1.25E-05 | 0.509 |
| C18:1n9c | 19 | ARS-BFGL-NGS-8796 | 5 | 29095603 | LOC510716 | Within | 2.25E-05 | 0.915 |
| C18:1n9c | 8 | ARS-BFGL-NGS-69056 | 5 | 42285835 | CPNE8 | 135184 | 1.18E-05 | 0.479 |
| C18:1n9c | 10 | Hapmap39862-BTA-74478 | 5 | 85672503 | BCAT1 | 74712 | 1.27E-05 | 0.515 |
| C18:1n9c | 6 | ARS-BFGL-NGS-99256 | 5 | 104714350 | VWF/ OLR1 | Within/4458707 | 8.88E-06 | 0.361 |
| C14:1 | 64 | ARS-BFGL-NGS-110361 | 7 | 15782979 | COL5A3 | Within | 2.03E-05 | 0.826 |
| C14:1 | 61 | ARS-BFGL-NGS-104050 | 7 | 62839580 | CSNK1A1 | 13832 | 1.48E-05 | 0.600 |
| C14:1 | 65 | Hapmap50476-BTA-79543 | 7 | 62889548 | CSNK1A1 | Within | 2.17E-05 | 0.882 |
| C18:1n9c | 18 | BTB-00316291 | 7 | 64892251 | SPARC | Within | 2.15E-05 | 0.874 |
| C18:1n9c | 12 | BTB-00316348 | 7 | 64939808 | ATOX1 | 5940 | 1.32E-05 | 0.534 |
| CLA | 6 | BTB-01541157 | 7 | 84582299 | ATP6AP1L | 15187 | 1.78E-05 | 0.722 |
| C18:2n6c | 5 | BTA-19330-no-rs | 7 | 93289032 | ARRDC3 | 35938 | 9.00E-06 | 0.366 |
| CLA | 5 | Hapmap27874-BTA-146513 | 9 | 53871363 | GPR63 | 53530 | 1.66E-05 | 0.676 |
| C18:1n9c | 15 | Hapmap50126-BTA-83733 | 9 | 55449737 | LOC101907134 | 18763 | 1.49E-05 | 0.606 |
| C16:1 | 6 | ARS-BFGL-NGS-115094 | 9 | 93183596 | TIAM2 | Within | 1.88E-05 | 0.762 |
| C16:1 | 4 | ARS-BFGL-NGS-57866 | 9 | 93246183 | TFB1M/TIAM2 | Within | 1.34E-05 | 0.545 |
| C18:1n9c | 7 | BTB-01332998 | 10 | 73466092 | SLC38A6 | 14392 | 1.02E-05 | 0.414 |
| C18:1n9c | 5 | ARS-BFGL-NGS-4783 | 10 | 73487550 | SLC38A6 | 35850 | 5.75E-06 | 0.233 |
| C16:1 | 5 | Hapmap38187-BTA-105082 | 12 | 69216840 | DCT | 286101 | 1.51E-05 | 0.614 |
| C16:1 | 2 | Hapmap53988-rs29024591 | 13 | 65855988 | EPB41L1/ ACSS2 | 23900/1014468 | 9.31E-06 | 0.378 |
| C16:1 | 3 | BPI-2 | 13 | 67833153 | LOC514211/ ACSS2 | Within/2991633 | 1.16E-05 | 0.470 |
| C18:2n6c | 4 | ARS-BFGL-NGS-57820 | 14 | 1651311 | LOC100294916/ DGAT1 | Within/144114 | 2.47E-06 | 0.100 |
| C14:1 | 60 | ARS-BFGL-NGS-4939 | 14 | 1801116 | DGAT1 | Within | 1.45E-05 | 0.590 |
| C18:1n9c | 2 | ARS-BFGL-NGS-4939 | 14 | 1801116 | DGAT1 | Within | 3.01E-06 | 0.122 |
| C18:2n6c | 1 | ARS-BFGL-NGS-4939 | 14 | 1801116 | DGAT1 | Within | 6.87E-08 | 2.79E-03 |
| C14:1 | 25 | Hapmap52798-ss46526455 | 14 | 1923292 | MAF1/ DGAT1 | Within/118454 | 2.32E-07 | 9.42E-03 |
| C18:2n6c | 3 | ARS-BFGL-NGS-107379 | 14 | 2054457 | LOC786966/ DGAT1 | 460/249619 | 1.13E-06 | 4.57E-02 |
| C18:1n9c | 1 | ARS-BFGL-NGS-100480 | 14 | 4364952 | TRAPPC9 | Within | 1.95E-06 | 0.079 |
| C14:1 | 43 | Hapmap32234-BTC-048199 | 14 | 7314869 | KHDRBS3 | 117233 | 3.48E-06 | 0.141 |
| C18:2n6c | 2 | ARS-BFGL-NGS-114448 | 16 | 32095335 | SMYD3 | Within | 4.72E-07 | 1.92E-02 |
| C16:1 | 1 | BTB-01090859 | 16 | 63248646 | XPR1 | 51157 | 4.29E-06 | 0.174 |
| C14:1 | 67 | ARS-BFGL-NGS-66923 | 17 | 18910914 | CCRN4L | Within | 2.36E-05 | 0.958 |
| C14:1 | 37 | ARS-BFGL-NGS-102933 | 17 | 19315294 | CCRN4L | 404368 | 1.46E-06 | 0.059 |
| C14:1 | 16 | BTA-91575-no-rs | 17 | 20099837 | SLC7A11 | 291555 | 3.42E-08 | 1.39E-03 |
| C14:1 | 47 | ARS-BFGL-NGS-25840 | 17 | 23694684 | LOC783956 | 163671 | 4.48E-06 | 0.182 |
| C14:1 | 32 | BTB-01585209 | 17 | 44910178 | ZNF605 | 53460 | 9.81E-07 | 3.98E-02 |
| C14:1 | 17 | ARS-BFGL-NGS-109854 | 17 | 44963124 | ZNF605 | 514 | 4.23E-08 | 1.72E-03 |
| C18:1n9c | 16 | ARS-BFGL-NGS-87368 | 19 | 7762820 | C19H17orf67 | 67914 | 1.76E-05 | 0.713 |
| C14:1 | 53 | BTA-117074-no-rs | 20 | 247319 | PANK3 | 42534 | 7.72E-06 | 0.313 |
| C14:1 | 62 | ARS-BFGL-NGS-114602 | 20 | 1052840 | SLIT3 | 979 | 1.52E-05 | 0.618 |
| C14:1 | 57 | ARS-BFGL-NGS-101925 | 20 | 1202954 | SLIT3 | 151093 | 8.96E-06 | 0.364 |
| C18:1n9c | 4 | ARS-BFGL-NGS-116806 | 20 | 36450009 | GDNF | 180881 | 4.66E-06 | 0.189 |
| C18:1n9c | 13 | ARS-BFGL-NGS-14031 | 20 | 36561330 | GDNF | 69560 | 1.46E-05 | 0.594 |
| C18:1n9c | 11 | ARS-USMARC-Parent-DQ990835-rs29012811 | 20 | 36570529 | GDNF | 60361 | 1.28E-05 | 0.519 |
| CLA | 1 | BTA-50525-no-rs | 20 | 36917645 | WDR70 | Within | 1.16E-05 | 0.469 |
| C18:1n9c | 20 | ARS-BFGL-NGS-111420 | 20 | 37553988 | SLC1A3 | 42121 | 2.30E-05 | 0.935 |
| C18:1n9c | 17 | Hapmap57276-ss46526009 | 20 | 37596667 | SLC1A3 | Within | 2.04E-05 | 0.829 |
| C18:2n6c | 6 | BTB-01340958 | 20 | 60400902 | DNAH5 | 840190 | 1.70E-05 | 0.689 |
| CLA | 7 | Hapmap26512-BTA-52638 | 21 | 55483469 | MIS18BP1 | 175 | 1.85E-05 | 0.752 |
| C14:1 | 54 | ARS-BFGL-BAC-46707 | 23 | 5756655 | TINAG | 196853 | 7.82E-06 | 0.318 |
| CLA | 3 | BTA-24495-no-rs | 24 | 15527942 | LOC783699 | 51814 | 1.57E-05 | 0.637 |
| C18:1n9c | 3 | ARS-BFGL-BAC-28144 | 25 | 2606575 | LOC788915 | 21466 | 4.34E-06 | 0.176 |
| C14:1 | 48 | Hapmap54064-rs29011996 | 26 | 5526925 | PCDH15 | Within | 4.58E-06 | 0.186 |
| C14:1 | 35 | ARS-BFGL-NGS-13746 | 26 | 9866940 | RNLS | 65137 | 1.42E-06 | 0.058 |
| C14:1 | 59 | BTA-61921-no-rs | 26 | 10255258 | LIPJ | 7406 | 1.42E-05 | 0.577 |
| C14:1 | 21 | ARS-BFGL-NGS-21794 | 26 | 10397362 | LOC100336557/ LIPK | 16309/35288 | 9.57E-08 | 3.88E-03 |
| C14:1 | 51 | BTA-08775-rs29022332 | 26 | 11201198 | SLC16A12 | 39720 | 6.64E-06 | 0.269 |
| C14:1 | 44 | ARS-BFGL-NGS-53115 | 26 | 11528933 | KIF20B | 61222 | 3.82E-06 | 0.155 |
| C14:1 | 24 | ARS-BFGL-NGS-63853 | 26 | 11942868 | MIR2895 | 67662 | 2.07E-07 | 8.42E-03 |
| C14:1 | 36 | ARS-BFGL-NGS-12381 | 26 | 12200948 | LOC100141242 | 24477 | 1.45E-06 | 0.059 |
| C14:1 | 20 | BTB-01908417 | 26 | 12268427 | LOC784522 | 15486 | 5.62E-08 | 2.28E-03 |
| C14:1 | 66 | ARS-BFGL-NGS-41148 | 26 | 12364119 | HTR7 | 10881 | 2.20E-05 | 0.892 |
| C14:1 | 28 | Hapmap52817-rs29011969 | 26 | 14155229 | HHEX | 29160 | 3.97E-07 | 1.61E-02 |
| C14:1 | 38 | ARS-BFGL-NGS-110475 | 26 | 15604631 | PLCE1 | Within | 1.54E-06 | 0.063 |
| C14:1 | 19 | ARS-BFGL-NGS-29299 | 26 | 16614068 | PDLIM1/ SORBS1 | Within/73061 | 5.42E-08 | 2.20E-03 |
| C14:1 | 23 | Hapmap41595-BTA-60800 | 26 | 16791783 | SORBS1 | Within | 2.07E-07 | 8.41E-03 |
| C14:1 | 40 | Hapmap58930-rs29010490 | 26 | 16822073 | SORBS1 | Within | 2.34E-06 | 0.095 |
| C14:1 | 42 | ARS-BFGL-NGS-41056 | 26 | 18906121 | CRTAC1 | Within | 2.70E-06 | 0.110 |
| C14:1 | 45 | ARS-BFGL-NGS-116902 | 26 | 18967997 | CRTAC1 | Within | 4.31E-06 | 0.175 |
| C14:1 | 55 | ARS-BFGL-NGS-25126 | 26 | 18994785 | CRTAC1 | Within | 7.84E-06 | 0.318 |
| C14:1 | 15 | ARS-BFGL-NGS-23064 | 26 | 20365711 | NKX2-3/ SCD1 | 34516/767033 | 2.93E-08 | 1.19E-03 |
| C14:1 | 14 | ARS-BFGL-NGS-77668 | 26 | 20393457 | NKX2-3/ SCD1 | 6770/744488 | 2.23E-08 | 9.05E-04 |
| C14:1 | 33 | BTB-00930925 | 26 | 20474308 | SLC25A28/ SCD1 | Within/658436 | 1.08E-06 | 4.39E-02 |
| C14:1 | 22 | ARS-BFGL-NGS-39397 | 26 | 20716721 | DNMBP/ SCD1 | Within/416023 | 1.37E-07 | 5.55E-03 |
| C14:1 | 27 | BTB-00930720 | 26 | 20903573 | LOC511498/ SCD1 | Within/244744 | 2.86E-07 | 1.16E-02 |
| C14:1 | 56 | Hapmap46411-BTA-15820 | 26 | 20984335 | CHUK / SCD1 | Within/148409 | 8.29E-06 | 0.336 |
| C14:1 | 31 | Hapmap31825-BTA-158647 | 26 | 21056547 | PKD2L1/ SCD1 | Within/76197 | 5.10E-07 | 2.07E-02 |
| C14:1 | 11 | Hapmap33073-BTA-162864 | 26 | 21180893 | SCD1 | 32576 | 1.31E-08 | 5.33E-04 |
| C14:1 | 1 | BTB-00931481 | 26 | 21226405 | WNT8B/ SCD1 | 14100/78088 | 7.08E-13 | 2.87E-08 |
| C14:1 | 12 | ARS-BFGL-NGS-110077 | 26 | 21322557 | HIF1AN/ SCD1 | 22399/174240 | 1.54E-08 | 6.23E-04 |
| C14:1 | 10 | ARS-BFGL-NGS-108305 | 26 | 21363670 | HIF1AN/ SCD1 | 63512/215353 | 8.07E-09 | 3.28E-04 |
| C14:1 | 9 | BTB-00931586 | 26 | 21409429 | PAX2/ SCD1 | 61334/261112 | 2.25E-09 | 9.12E-05 |
| C14:1 | 5 | ARS-BFGL-NGS-114149 | 26 | 21702714 | LZTS2/ SCD1 | 656/564769 | 5.50E-10 | 2.23E-05 |
| C14:1 | 6 | ARS-BFGL-NGS-116481 | 26 | 21977581 | LOC100847491/ SCD1 | 10062/829264 | 5.77E-10 | 2.34E-05 |
| C14:1 | 7 | Hapmap24832-BTA-138805 | 26 | 22016380 | BTRC/ SCD1 | Within/868063 | 7.18E-10 | 2.92E-05 |
| C14:1 | 8 | ARS-BFGL-NGS-6259 | 26 | 22059103 | BTRC/ SCD1 | Within/910786 | 7.23E-10 | 2.93E-05 |
| C14:1 | 4 | BTB-00932332 | 26 | 22118554 | BTRC/ SCD1 | Within/970237 | 3.56E-10 | 1.45E-05 |
| C14:1 | 3 | ARS-BFGL-NGS-107403 | 26 | 22889812 | NFKB2 | 1586 | 2.22E-12 | 9.02E-08 |
| C14:1 | 18 | Hapmap48222-BTA-122240 | 26 | 23641881 | C26H10orf26 | Within | 4.53E-08 | 1.84E-03 |
| C14:1 | 29 | Hapmap49372-BTA-91009 | 26 | 23689229 | C26H10orf26 | 3017 | 4.66E-07 | 1.89E-02 |
| C14:1 | 46 | BTA-60918-no-rs | 26 | 23853334 | CNNM2 | Within | 4.40E-06 | 0.179 |
| C14:1 | 13 | BTA-60935-no-rs | 26 | 23876476 | CNNM2 | Within | 1.56E-08 | 6.33E-04 |
| C14:1 | 52 | ARS-BFGL-NGS-2180 | 26 | 24477962 | SH3PXD2A | Within | 7.46E-06 | 0.303 |
| C14:1 | 49 | ARS-BFGL-NGS-1092 | 26 | 24531763 | SH3PXD2A | Within | 4.90E-06 | 0.199 |
| C14:1 | 2 | ARS-BFGL-NGS-118189 | 26 | 24786731 | SLK | Within | 1.69E-12 | 6.87E-08 |
| C14:1 | 58 | UA-IFASA-4715 | 26 | 25314352 | CCDC147 | 27285 | 9.16E-06 | 0.372 |
| C14:1 | 30 | Hapmap28763-BTA-162328 | 26 | 26757136 | SORCS3/ ECHS1 | 358845/891528 | 5.05E-07 | 2.05E-02 |
| C14:1 | 26 | BTA-87355-no-rs | 26 | 27251857 | SORCS1 | 558341 | 2.59E-07 | 1.05E-02 |
| C14:1 | 41 | BTA-10873-rs29016424 | 28 | 18317414 | RTKN2 | 41359 | 2.35E-06 | 0.095 |
| C14:1 | 63 | ARS-BFGL-NGS-118293 | 28 | 45409471 | CXCL12 | Within | 1.77E-05 | 0.718 |
| C18:1n9c | 14 | BTB-01088519 | 29 | 22161137 | CCDC179 | 85675 | 1.49E-05 | 0.605 |
| C14:1 | 50 | ARS-BFGL-NGS-27560 | X | 146596721 | STS | Within | 5.19E-06 | 0.211 |
Note: see note to Table 2.
Table 5. Genome-wise and suggestive significant SNPs for indices of fatty acid traits.
| Trait | Rankb | SNP | Chr. | Position(bp) | Nearest Gene/Candidate Gene | Distance(bp) | Raw P_valuea | P_value Bonferroni |
| C14index | 26 | ARS-BFGL-NGS-15914 | 0 | 0 | NA | NA | 2.59E-09 | 1.05E-04 |
| C14index | 38 | UA-IFASA-5862 | 0 | 0 | NA | NA | 8.04E-08 | 3.26E-03 |
| C18index | 65 | BTB-00270136 | 0 | 0 | NA | NA | 1.19E-05 | 0.482 |
| C18index | 91 | Hapmap60647-rs29027341 | 0 | 0 | NA | NA | 2.25E-05 | 0.912 |
| C18index | 23 | BTA-111771-no-rs | 1 | 37174447 | EPHA3 | 50365 | 2.86E-06 | 0.116 |
| C18index | 22 | ARS-BFGL-NGS-106725 | 1 | 41577955 | EPHA6 | Within | 2.74E-06 | 0.111 |
| C18index | 45 | ARS-BFGL-NGS-115763 | 1 | 66139653 | GTF2E1 | 67771 | 6.14E-06 | 0.249 |
| C18index | 35 | BTB-00032200 | 1 | 67764428 | DIRC2 | Within | 3.83E-06 | 0.156 |
| C18index | 33 | ARS-BFGL-NGS-35839 | 1 | 102589009 | BCHE | 178966 | 3.69E-06 | 0.150 |
| C18index | 24 | ARS-BFGL-NGS-111111 | 1 | 146302724 | HSF2BP/ AGPAT3 | 43214/402889 | 2.92E-06 | 0.118 |
| C18index | 12 | ARS-BFGL-NGS-109493 | 1 | 146354654 | HSF2BP/ AGPAT3 | Within/350959 | 1.13E-06 | 4.59E-02 |
| C18index | 13 | BTA-56389-no-rs | 1 | 146384457 | HSF2BP/ AGPAT3 | Within/321156 | 1.13E-06 | 4.59E-02 |
| C18index | 32 | ARS-BFGL-NGS-76347 | 1 | 146704618 | AGPAT3 | 995 | 3.66E-06 | 0.149 |
| C18index | 84 | Hapmap59917-rs29012418 | 2 | 24519348 | METAP1D | Within | 1.90E-05 | 0.771 |
| C18index | 21 | Hapmap53388-rs29010903 | 2 | 63581955 | MGAT5 | 129790 | 2.65E-06 | 0.107 |
| C18index | 3 | ARS-BFGL-NGS-33744 | 2 | 79388083 | GYPC/ STAT1 | 83687/506149 | 8.95E-08 | 3.63E-03 |
| C18index | 31 | Hapmap53419-rs29015159 | 2 | 88205436 | SATB2 | 43199 | 3.57E-06 | 0.145 |
| C18index | 5 | Hapmap33966-BES2_Contig368_774 | 2 | 88545567 | SATB2 | 90018 | 2.56E-07 | 1.04E-02 |
| C18index | 50 | ARS-BFGL-NGS-23872 | 2 | 95199905 | ADAM23 | Within | 7.34E-06 | 0.298 |
| C18index | 76 | ARS-BFGL-NGS-98354 | 2 | 95512347 | FASTKD2 | Within | 1.47E-05 | 0.598 |
| C18index | 55 | ARS-BFGL-NGS-99030 | 2 | 98160191 | UNC80 | Within | 8.85E-06 | 0.359 |
| C18index | 15 | ARS-BFGL-NGS-45691 | 2 | 128484790 | RUNX3/ FABP3 | 144886/5700960 | 1.41E-06 | 0.057 |
| C18index | 9 | ARS-BFGL-NGS-118924 | 2 | 128529102 | RUNX3/ FABP3 | 100574/5745272 | 6.04E-07 | 2.45E-02 |
| C18index | 28 | Hapmap30257-BTA-142970 | 5 | 25358659 | USP44 | 44459 | 3.24E-06 | 0.131 |
| C18index | 75 | ARS-BFGL-NGS-38038 | 5 | 27992179 | NR4A1 | Within | 1.45E-05 | 0.588 |
| C18index | 59 | Hapmap41951-BTA-73168 | 5 | 28442563 | SLC4A8 | 25682 | 9.43E-06 | 0.383 |
| C18index | 16 | ARS-BFGL-NGS-8796 | 5 | 29095603 | LOC510716 | Within | 1.82E-06 | 0.074 |
| C18index | 92 | ARS-BFGL-NGS-53488 | 5 | 41154328 | PRICKLE1 | 166915 | 2.35E-05 | 0.953 |
| C18index | 47 | ARS-BFGL-NGS-116897 | 5 | 95743746 | PLBD1/ OLR1 | Within/4500525 | 6.49E-06 | 0.264 |
| C18index | 73 | BTB-01685239 | 6 | 12282881 | UGT8 | 81408 | 1.44E-05 | 0.586 |
| C18index | 14 | BTB-00246150 | 6 | 20993424 | PPA2 | Within | 1.15E-06 | 4.68E-02 |
| C18index | 30 | Hapmap26001-BTC-038813 | 6 | 44926243 | PPARGC1A | Within | 3.54E-06 | 0.144 |
| C18index | 62 | Hapmap31284-BTC-039204 | 6 | 45096462 | PPARGC1A | 135929 | 1.09E-05 | 0.443 |
| C18index | 87 | Hapmap49746-BTA-76106 | 6 | 46140090 | LGI2/ PPARGC1A | Within/82649 | 1.97E-05 | 0.801 |
| C14index | 52 | ARS-BFGL-NGS-106015 | 6 | 61199572 | RBM47 | Within | 2.03E-06 | 0.082 |
| C14index | 54 | ARS-BFGL-NGS-80548 | 7 | 6434821 | C7H19orf44 | Within | 2.79E-06 | 0.113 |
| C14index | 42 | ARS-BFGL-NGS-110361 | 7 | 15782979 | COL5A3 | Within | 5.50E-07 | 2.23E-02 |
| C14index | 78 | ARS-BFGL-NGS-104050 | 7 | 62839580 | CSNK1A1 | 13832 | 1.70E-05 | 0.691 |
| C18index | 77 | BTB-00316650 | 7 | 65098028 | GLRA1 | Within | 1.67E-05 | 0.679 |
| C18index | 63 | BTB-01687547 | 8 | 20989026 | LOC101905651 | 328218 | 1.13E-05 | 0.458 |
| C18index | 48 | ARS-BFGL-NGS-9052 | 8 | 78009328 | FRMD3 | 181107 | 6.84E-06 | 0.278 |
| C18index | 90 | ARS-BFGL-NGS-106379 | 8 | 113159018 | TSSC1 | 90395 | 2.24E-05 | 0.909 |
| C18index | 52 | ARS-BFGL-NGS-15823 | 9 | 28887462 | PKIB | Within | 8.15E-06 | 0.331 |
| C18index | 10 | BTB-01332998 | 10 | 73466092 | SLC38A6 | 15901 | 6.76E-07 | 2.74E-02 |
| C18index | 29 | ARS-BFGL-NGS-4783 | 10 | 73487550 | SLC38A6 | 37359 | 3.30E-06 | 0.134 |
| C18index | 78 | ARS-BFGL-NGS-22113 | 10 | 73551579 | TMEM30B | 94651 | 1.68E-05 | 0.683 |
| C18index | 26 | BTB-00471219 | 11 | 32709462 | NRXN1 | Within | 3.14E-06 | 0.128 |
| C18index | 44 | Hapmap25798-BTA-126388 | 11 | 32731961 | NRXN1 | Within | 6.07E-06 | 0.247 |
| C18index | 6 | Hapmap40257-BTA-91916 | 11 | 32789048 | NRXN1 | Within | 3.25E-07 | 1.32E-02 |
| C18index | 93 | ARS-BFGL-NGS-85007 | 11 | 53430229 | CTNNA2 | 1292050 | 2.37E-05 | 0.964 |
| C18index | 72 | BTB-01079278 | 11 | 57078447 | REG3A | 436061 | 1.39E-05 | 0.563 |
| C18index | 60 | BTB-01079350 | 11 | 57107070 | REG3A | 464684 | 9.51E-06 | 0.386 |
| C18index | 1 | Hapmap51531-BTA-98947 | 11 | 57522675 | REG3A | 880289 | 2.08E-08 | 8.44E-04 |
| C18index | 2 | BTB-01328920 | 11 | 57639084 | LRRTM4 | 792049 | 4.34E-08 | 1.76E-03 |
| C18index | 54 | ARS-BFGL-NGS-114087 | 11 | 64057850 | SPRED2 | 293893 | 8.21E-06 | 0.333 |
| C18index | 25 | ARS-BFGL-NGS-43985 | 11 | 64184454 | SPRED2 | 420497 | 3.00E-06 | 0.122 |
| C18index | 40 | ARS-BFGL-NGS-91014 | 11 | 65493222 | C11H2orf66 | Within | 4.88E-06 | 0.198 |
| C16index | 8 | ARS-BFGL-NGS-110868 | 11 | 75329413 | KLHL29 | Within | 1.65E-05 | 0.670 |
| C18index | 89 | UA-IFASA-2295 | 11 | 97754180 | RALGPS1 | Within | 2.16E-05 | 0.878 |
| C18index | 27 | BTA-119672-no-rs | 11 | 102911946 | AK8 | Within | 3.21E-06 | 0.130 |
| C18index | 71 | BTB-00490466 | 12 | 47836570 | DIS3 | Within | 1.37E-05 | 0.557 |
| C14index | 81 | Hapmap42477-BTA-22799 | 12 | 48645720 | KLF12 | 181742 | 2.25E-05 | 0.914 |
| C18index | 83 | Hapmap57649-rs29022414 | 12 | 66068259 | GPC5 | 223887 | 1.85E-05 | 0.753 |
| C16index | 2 | Hapmap38187-BTA-105082 | 12 | 69216840 | DCT | 286101 | 7.38E-06 | 0.300 |
| C18index | 4 | ARS-BFGL-BAC-13788 | 12 | 69512332 | DCT | Within | 1.79E-07 | 7.27E-03 |
| C18index | 46 | ARS-BFGL-NGS-45730 | 12 | 70260457 | ABCC4 | 49496 | 6.28E-06 | 0.255 |
| C14index | 55 | ARS-BFGL-NGS-13252 | 12 | 81184160 | GGACT | 25567 | 3.04E-06 | 0.123 |
| C16index | 6 | Hapmap53988-rs29024591 | 13 | 65855988 | EPB41L1/ ACSS2 | 23900/1014468 | 1.19E-05 | 0.482 |
| C16index | 9 | Hapmap40712-BTA-33406 | 13 | 67101174 | BLCAP/ ACSS2 | 13538/2259654 | 1.74E-05 | 0.705 |
| C18index | 61 | ARS-BFGL-NGS-40278 | 13 | 67122063 | BLCAP/ ACSS2 | Within/2280543 | 9.81E-06 | 0.398 |
| C16index | 3 | BPI-2 | 13 | 67833153 | LOC514211/ ACSS2 | Within/2991633 | 7.51E-06 | 0.305 |
| C16index | 10 | ARS-BFGL-NGS-107113 | 13 | 67958189 | LOC514978/ ACSS2 | Within/3116669 | 1.83E-05 | 0.743 |
| C16index | 1 | Hapmap55254-rs29014939 | 13 | 69042143 | DHX35 | 573959 | 5.20E-06 | 0.211 |
| C18index | 41 | Hapmap30381-BTC-005750 | 14 | 1463676 | C14H8orf33/ DGAT1 | 23690/331749 | 5.62E-06 | 0.228 |
| C14index | 73 | ARS-BFGL-NGS-57820 | 14 | 1651311 | LOC100294916/ DGAT1 | Within/144114 | 1.03E-05 | 0.419 |
| C14index | 75 | ARS-BFGL-NGS-4939 | 14 | 1801116 | DGAT1 | Within | 1.15E-05 | 0.469 |
| C18index | 88 | ARS-BFGL-NGS-4939 | 14 | 1801116 | DGAT1 | Within | 2.00E-05 | 0.812 |
| C14index | 47 | Hapmap52798-ss46526455 | 14 | 1923292 | MAF1/ DGAT1 | Within/118454 | 1.08E-06 | 4.39E-02 |
| C14index | 80 | Hapmap30986-BTC-056068 | 14 | 10346734 | EFR3A | 64293 | 1.97E-05 | 0.799 |
| C18index | 51 | BTB-00557532 | 14 | 24643266 | XKR4 | 28554 | 7.35E-06 | 0.299 |
| C18index | 82 | ARS-BFGL-BAC-10245 | 14 | 31819743 | PDE7A | Within | 1.80E-05 | 0.731 |
| C18index | 34 | ARS-BFGL-NGS-2025 | 14 | 65452336 | ZNF706 | 103905 | 3.79E-06 | 0.154 |
| C18index | 8 | Hapmap35102-BES3_Contig324_378 | 14 | 66523007 | RGS22 | Within | 5.63E-07 | 2.29E-02 |
| C18index | 58 | UA-IFASA-4785 | 14 | 71096693 | C14H8orf37 | 300038 | 9.36E-06 | 0.380 |
| C18index | 79 | BTB-01296218 | 15 | 17344116 | ALKBH8 | 21050 | 1.77E-05 | 0.720 |
| C18index | 39 | ARS-BFGL-NGS-35704 | 15 | 18335423 | C15H11orf65 | Within | 4.83E-06 | 0.196 |
| C18index | 37 | ARS-BFGL-NGS-12066 | 15 | 27291268 | BUD13 | 542743 | 4.33E-06 | 0.176 |
| C18index | 43 | BTA-36518-no-rs | 15 | 32983903 | SORL1 | 228564 | 6.04E-06 | 0.245 |
| C18index | 81 | Hapmap56991-rs29010083 | 15 | 81595546 | LOC538839 | 52784 | 1.78E-05 | 0.724 |
| C18index | 68 | Hapmap46697-BTA-38171 | 16 | 2899256 | NUAK2 | Within | 1.24E-05 | 0.501 |
| C18index | 7 | ARS-BFGL-NGS-68533 | 16 | 60615012 | LOC101902340 | 35998 | 5.39E-07 | 2.19E-02 |
| C16index | 7 | BTB-01090859 | 16 | 63248646 | XPR1 | 51157 | 1.51E-05 | 0.615 |
| C18index | 64 | ARS-BFGL-NGS-36880 | 16 | 73736551 | SLC30A1 | 28425 | 1.14E-05 | 0.465 |
| C14index | 83 | ARS-BFGL-NGS-38696 | 17 | 18398611 | MGST2 | 29230 | 2.29E-05 | 0.929 |
| C14index | 64 | ARS-BFGL-NGS-66923 | 17 | 18910914 | CCRN4L | Within | 5.55E-06 | 0.225 |
| C14index | 50 | ARS-BFGL-NGS-102933 | 17 | 19315294 | CCRN4L | 404368 | 1.81E-06 | 0.074 |
| C14index | 39 | BTA-91575-no-rs | 17 | 20099837 | SLC7A11 | 291555 | 2.96E-07 | 1.20E-02 |
| C14index | 79 | Hapmap51443-BTA-40619 | 17 | 20996847 | PCDH18 | 380577 | 1.72E-05 | 0.698 |
| C14index | 53 | BTB-01585209 | 17 | 44910178 | ZNF605 | 53460 | 2.79E-06 | 0.113 |
| C14index | 35 | ARS-BFGL-NGS-109854 | 17 | 44963124 | ZNF605 | 514 | 6.46E-08 | 2.62E-03 |
| C18index | 19 | ARS-BFGL-NGS-71116 | 17 | 68002540 | MYO18B | Within | 2.41E-06 | 0.098 |
| C18index | 36 | ARS-BFGL-NGS-37725 | 17 | 68490453 | TPST2 | Within | 4.30E-06 | 0.175 |
| C18index | 74 | BTB-01790846 | 20 | 13263157 | SREK1 | 128568 | 1.44E-05 | 0.586 |
| C18index | 85 | ARS-BFGL-BAC-2469 | 20 | 33433160 | HEATR7B2/ OXCT1 | 22507/584434 | 1.93E-05 | 0.784 |
| C18index | 56 | ARS-BFGL-NGS-76756 | 20 | 33491273 | HEATR7B2/ OXCT1 | Within/642547 | 9.20E-06 | 0.373 |
| C18index | 49 | BTB-01423653 | 20 | 38578200 | SPEF2/ PRLR | Within/495046 | 6.96E-06 | 0.282 |
| C18index | 42 | BTB-01423676 | 20 | 38606353 | SPEF2/ PRLR | Within/466893 | 6.02E-06 | 0.244 |
| C18index | 20 | Hapmap30570-BTA-152778 | 20 | 38761711 | SPEF2/ PRLR | 154961/311535 | 2.55E-06 | 0.104 |
| C18index | 80 | ARS-BFGL-NGS-99716 | 21 | 63560239 | VRK1 | 216337 | 1.78E-05 | 0.723 |
| C18index | 53 | ARS-BFGL-NGS-39459 | 22 | 50474049 | CACNA2D2 | Within | 8.15E-06 | 0.331 |
| C18index | 69 | Hapmap54558-rs29009598 | 24 | 29187804 | CDH2 | Within | 1.27E-05 | 0.517 |
| C16index | 4 | ARS-BFGL-NGS-45679 | 24 | 42582505 | APCDD1 | Within | 7.82E-06 | 0.318 |
| C16index | 5 | Hapmap31260-BTC-015327 | 25 | 2224930 | ZG16B | Within | 8.51E-06 | 0.346 |
| C14index | 51 | Hapmap54064-rs29011996 | 26 | 5526925 | PCDH15 | Within | 2.02E-06 | 0.082 |
| C14index | 77 | BTB-01077939 | 26 | 7685110 | PRKG1 | Within | 1.35E-05 | 0.548 |
| C14index | 27 | ARS-BFGL-NGS-13746 | 26 | 9866940 | RNLS | 65137 | 3.71E-09 | 1.51E-04 |
| C14index | 72 | Hapmap58185-rs29022254 | 26 | 10002077 | RNLS | Within | 1.02E-05 | 0.415 |
| C14index | 36 | BTA-61921-no-rs | 26 | 10255258 | LIPJ | 7406 | 6.99E-08 | 2.84E-03 |
| C14index | 20 | ARS-BFGL-NGS-21794 | 26 | 10397362 | LOC100336557/ LIPK | 16309/35288 | 4.20E-10 | 1.70E-05 |
| C14index | 68 | Hapmap59335-rs29016866 | 26 | 10689379 | ACTA2 | 9731 | 8.36E-06 | 0.339 |
| C14index | 76 | BTA-111857-no-rs | 26 | 10815586 | FAS | 67560 | 1.17E-05 | 0.476 |
| C14index | 56 | BTB-00924013 | 26 | 10922061 | CH25H | 54132 | 3.05E-06 | 0.124 |
| C14index | 48 | BTA-08775-rs29022332 | 26 | 11201198 | SLC16A12 | 39720 | 1.09E-06 | 4.42E-02 |
| C14index | 57 | ARS-BFGL-NGS-53115 | 26 | 11528933 | KIF20B | 61222 | 3.10E-06 | 0.126 |
| C14index | 25 | ARS-BFGL-NGS-63853 | 26 | 11942868 | MIR2895 | 67662 | 2.50E-09 | 1.01E-04 |
| C14index | 61 | ARS-BFGL-NGS-12381 | 26 | 12200948 | LOC100141242 | 24477 | 4.76E-06 | 0.193 |
| C14index | 17 | BTB-01908417 | 26 | 12268427 | LOC784522 | 15486 | 1.71E-10 | 6.94E-06 |
| C14index | 67 | BTB-01841682 | 26 | 12295284 | LOC784522 | 42343 | 8.27E-06 | 0.336 |
| C14index | 84 | ARS-BFGL-NGS-41148 | 26 | 12364119 | HTR7 | 10881 | 2.32E-05 | 0.942 |
| C14index | 23 | Hapmap52817-rs29011969 | 26 | 14155229 | HHEX | 29160 | 1.34E-09 | 5.45E-05 |
| C14index | 69 | ARS-BFGL-NGS-85864 | 26 | 14532797 | CYP26A1 | 69001 | 8.38E-06 | 0.340 |
| C14index | 74 | ARS-BFGL-NGS-110475 | 26 | 15604631 | PLCE1 | Within | 1.04E-05 | 0.421 |
| C14index | 58 | BTB-00706838 | 26 | 15824141 | TBC1D12 | Within | 3.31E-06 | 0.134 |
| C14index | 70 | BTB-00927439 | 26 | 16315378 | CYP2C19 | 20815 | 9.08E-06 | 0.369 |
| C14index | 18 | ARS-BFGL-NGS-29299 | 26 | 16614068 | PDLIM1/ SORBS1 | Within/73061 | 2.06E-10 | 8.36E-06 |
| C14index | 49 | Hapmap41595-BTA-60800 | 26 | 16791783 | SORBS1 | Within | 1.09E-06 | 4.43E-02 |
| C14index | 28 | Hapmap58930-rs29010490 | 26 | 16822073 | SORBS1 | Within | 5.90E-09 | 2.40E-04 |
| C14index | 59 | ARS-BFGL-NGS-106959 | 26 | 17225652 | CC2D2B | Within | 3.80E-06 | 0.154 |
| C14index | 63 | ARS-BFGL-NGS-113660 | 26 | 17246984 | CC2D2B | Within | 5.20E-06 | 0.211 |
| C14index | 71 | ARS-BFGL-NGS-25217 | 26 | 17307507 | CCNJ | 13017 | 9.96E-06 | 0.404 |
| C14index | 66 | ARS-BFGL-NGS-114539 | 26 | 18808408 | SFRP5 | 21032 | 7.36E-06 | 0.299 |
| C14index | 82 | ARS-BFGL-NGS-97471 | 26 | 18882047 | CRTAC1 | 109 | 2.27E-05 | 0.921 |
| C14index | 34 | ARS-BFGL-NGS-41056 | 26 | 18906121 | CRTAC1 | Within | 4.78E-08 | 1.94E-03 |
| C14index | 41 | ARS-BFGL-NGS-116902 | 26 | 18967997 | CRTAC1 | Within | 4.05E-07 | 1.64E-02 |
| C14index | 43 | ARS-BFGL-NGS-25126 | 26 | 18994785 | CRTAC1 | Within | 5.96E-07 | 2.42E-02 |
| C14index | 62 | ARS-BFGL-NGS-71584 | 26 | 20290497 | GOT1/ SCD1 | Within/842247 | 5.11E-06 | 0.207 |
| C14index | 14 | ARS-BFGL-NGS-23064 | 26 | 20365711 | NKX2-3/ SCD1 | 34516/767033 | 2.95E-11 | 1.20E-06 |
| C14index | 15 | ARS-BFGL-NGS-77668 | 26 | 20393457 | NKX2-3/ SCD1 | 6770/744488 | 6.02E-11 | 2.44E-06 |
| C14index | 33 | ARS-BFGL-NGS-2464 | 26 | 20444634 | SLC25A28/ SCD1 | 21194/688110 | 3.96E-08 | 1.61E-03 |
| C14index | 19 | BTB-00930925 | 26 | 20474308 | SLC25A28/ SCD1 | Within/658436 | 2.61E-10 | 1.06E-05 |
| C14index | 24 | ARS-BFGL-NGS-39397 | 26 | 20716721 | DNMBP/ SCD1 | Within/416023 | 1.54E-09 | 6.25E-05 |
| C14index | 22 | BTB-00930720 | 26 | 20903573 | LOC511498/ SCD1 | Within/244744 | 6.89E-10 | 2.80E-05 |
| C14index | 31 | Hapmap46411-BTA-15820 | 26 | 20984335 | CHUK / SCD1 | Within/148409 | 1.10E-08 | 4.45E-04 |
| C14index | 16 | Hapmap31825-BTA-158647 | 26 | 21056547 | PKD2L1/ SCD1 | Within/76197 | 1.56E-10 | 6.35E-06 |
| C14index | 10 | Hapmap33073-BTA-162864 | 26 | 21180893 | SCD1 | 32576 | 3.04E-12 | 1.23E-07 |
| C14index | 1 | BTB-00931481 | 26 | 21226405 | WNT8B/ SCD1 | 14100/78088 | 6.91E-17 | 2.80E-12 |
| C14index | 13 | ARS-BFGL-NGS-110077 | 26 | 21322557 | HIF1AN/ SCD1 | 22399/174240 | 2.04E-11 | 8.28E-07 |
| C14index | 11 | ARS-BFGL-NGS-108305 | 26 | 21363670 | SCD1 /HIFIAN | 63512/215353 | 6.46E-12 | 2.62E-07 |
| C14index | 5 | BTB-00931586 | 26 | 21409429 | PAX2/ SCD1 | 61334/261112 | 3.39E-13 | 1.38E-08 |
| C14index | 4 | ARS-BFGL-NGS-114149 | 26 | 21702714 | LZTS2/ SCD1 | 656/564769 | 3.10E-13 | 1.26E-08 |
| C14index | 9 | ARS-BFGL-NGS-116481 | 26 | 21977581 | LOC100847491/ SCD1 | 10062/829264 | 1.20E-12 | 4.87E-08 |
| C14index | 7 | Hapmap24832-BTA-138805 | 26 | 22016380 | BTRC/ SCD1 | Within/868063 | 6.31E-13 | 2.56E-08 |
| C14index | 6 | ARS-BFGL-NGS-6259 | 26 | 22059103 | BTRC/ SCD1 | Within/910786 | 5.97E-13 | 2.42E-08 |
| C14index | 8 | BTB-00932332 | 26 | 22118554 | BTRC/ SCD1 | Within/970237 | 7.55E-13 | 3.06E-08 |
| C14index | 2 | ARS-BFGL-NGS-107403 | 26 | 22889812 | NFKB2 | 1586 | 2.62E-15 | 1.06E-10 |
| C14index | 21 | Hapmap48222-BTA-122240 | 26 | 23641881 | C26H10orf26 | Within | 5.68E-10 | 2.31E-05 |
| C14index | 30 | Hapmap49372-BTA-91009 | 26 | 23689229 | C26H10orf26 | 3017 | 6.47E-09 | 2.63E-04 |
| C14index | 32 | BTA-60918-no-rs | 26 | 23853334 | CNNM2 | Within | 3.34E-08 | 1.36E-03 |
| C14index | 12 | BTA-60935-no-rs | 26 | 23876476 | CNNM2 | Within | 1.98E-11 | 8.03E-07 |
| C14index | 40 | ARS-BFGL-NGS-2180 | 26 | 24477962 | SH3PXD2A | Within | 3.20E-07 | 1.30E-02 |
| C14index | 29 | ARS-BFGL-NGS-1092 | 26 | 24531763 | SH3PXD2A | Within | 6.21E-09 | 2.52E-04 |
| C14index | 65 | ARS-BFGL-NGS-18194 | 26 | 24575207 | SH3PXD2A | Within | 5.58E-06 | 0.227 |
| C14index | 3 | ARS-BFGL-NGS-118189 | 26 | 24786731 | SLK | Within | 6.55E-14 | 2.66E-09 |
| C14index | 46 | UA-IFASA-4715 | 26 | 25314352 | CCDC147 | 27285 | 9.22E-07 | 3.74E-02 |
| C14index | 44 | BTB-00935537 | 26 | 26585557 | SORCS3/ ECHS1 | 187266/719949 | 7.48E-07 | 3.04E-02 |
| C14index | 37 | Hapmap28763-BTA-162328 | 26 | 26757136 | SORCS3/ ECHS1 | 358845/891528 | 7.79E-08 | 3.16E-03 |
| C14index | 60 | BTA-87355-no-rs | 26 | 27251857 | SORCS1 | 558341 | 4.71E-06 | 0.191 |
| C14index | 45 | ARS-BFGL-NGS-1448 | 27 | 37357125 | HOOK3/ AGPAT6 | Within/1128138 | 7.66E-07 | 3.11E-02 |
| C18index | 11 | ARS-BFGL-NGS-110992 | 28 | 20421361 | REEP3 | 614577 | 9.76E-07 | 3.96E-02 |
| C18index | 18 | ARS-BFGL-NGS-4865 | 28 | 28011033 | CDH23 | Within | 2.38E-06 | 0.097 |
| C18index | 86 | ARS-BFGL-NGS-12970 | 29 | 40646639 | PPP1R32/ FADS1 | 2220/292226 | 1.94E-05 | 0.788 |
| C18index | 57 | ARS-BFGL-NGS-11681 | X | 3622615 | SLC25A43 | 24203 | 9.32E-06 | 0.378 |
| C18index | 67 | Hapmap48540-BTA-97806 | X | 8700029 | ODZ1 | Within | 1.23E-05 | 0.498 |
| C18index | 70 | Hapmap50046-BTA-58882 | X | 82022276 | MIR374B | 1635 | 1.33E-05 | 0.540 |
| C18index | 17 | Hapmap60551-rs29017241 | X | 82281306 | XIST | Within | 2.17E-06 | 0.088 |
| C18index | 38 | Hapmap60664-rs29017374 | X | 107043386 | CASK | 113889 | 4.61E-06 | 0.187 |
| C18index | 66 | Hapmap49563-BTA-30596 | X | 120716539 | IL1RAPL1 | Within | 1.21E-05 | 0.490 |
Note: see note to Table 2.
Short- and medium-chain saturated fatty acid traits (SCFA and MCFA)
For C10:0, C12:0 and C14:0, 21, 22 and 27 SNPs were detected, respectively. Of these 70 SNPs, 10 were associated with two or three traits. The most significant association of C10:0 (P = 5.89E-07), C12:0 (P = 3.94E-07), and C14:0 (P = 1.58E-07) were identified with BTB-01556197 on BTA9, BTA-76414-no-rs on BTA21 and Hapmap49848-BTA-106779 on BTA5, respectively. The SNP strongly associated with C10:0 (P = 8.54E-06), C12:0 (P = 1.16E-07) and C14:0 (P = 6.01E-06), ARS-BFGL-NGS-39328, is 58,172 bp close to the fatty acid synthase (FASN) gene on BTA19, which is well-known to affect fat composition of dairy cattle and beef.
Long-chain saturated fatty acid traits (LCFA)
A total of 126 significant SNPs for LCFA were detected mainly on BTA1, 2, 8, 10 and 17, including 105 for C18:0, 18 for C16:0, 2 for C20:0 and one for C22:0. The top one significant SNP (ARS-BFGL-BAC-13788) was associated with C18:0 (P = 9.17E-09) on BTA12. The strongest association of C22:0 (P = 6.70E-07) was identified with the SNP (ARS-BFGL-NGS-109692) on BTA1. The SNP (ARS-BFGL-NGS-4939) associated with C16:0 (P = 9.04E-06) on BTA14 is located within the diacylglycerol O-acyltransferase 1(DGAT1) gene, the major gene with large effect on milk fat in dairy cattle.
Monounsaturated and polyunsaturated fatty acid traits (MUFA and PUFA)
A total of 93 and 13 Significant SNPs for MUFA and PUFA were detected, respectively. Of them, 67, 6 and 20 SNPs were associated with C14:1, C16:1 and C18:1n9c, respectively. For C14:1, 29 out of 34 genome-wise significant SNPs on BTA26 were clustered within three regions: 6 fell in a 6.40 Mbp region (10.39∼16.79 Mbp), 16 fell in a 1.75 Mbp region (20.36∼22.11 Mbp) containing the stearoyl-CoA desaturase (SCD1) gene, and 7 SNPs fell in a 4.37 Mbp region (22.88∼27.25 Mbp). The SNP (BTB-00931481) on BTA26, 78,088 bp near to the SCD1 gene, showed the strongest effect (P = 7.08E-13). Though no SNPs for C16:1 and C18:1n9c reached genome-wise level, the second suggestive significant SNP (ARS-BFGL-NGS-4939) for C18:1n9c (P = 3.01E-06) is located within the DGAT1 gene on BTA14.
As for PUFA, 6 and 7 significant SNPs were detected for C18:2n6c and CLA, respectively. The most significant SNP (ARS-BFGL-NGS-4939) associated with C18:2n6c (P = 6.87E-08) is located within the DGAT1 gene on BTA14, while the most significant SNP (BTA-50525-no-rs) for CLA only reached suggestive level (P = 1.16E-05).
Indices of fatty acid traits (C14 index, C16 index, C18 index)
For indices of C14, C16 and C18, totally 84, 10 and 93 significant SNPs were detected, respectively. Forty-two SNPs associated with C14 index are located within a region of 16.89 Mbp on BTA26, which included four small segments: 6 in a 2.40 Mbp segment (9.86∼12.26 Mbp), 7 in a 4.84 Mbp segment (14.15∼18.99 Mbp), 18 in an 1.75 Mbp segment (20.36∼22.11 Mbp) containing the SCD1 gene, and 11 in a 3.87 Mbp segment (22.88∼26.75 Mbp). In addition, 56 common SNPs for C14 index and C14:1, 4 common SNPs for C18 index and C18:1n9c, 28 common SNPs for C18 index and C18:0, and 4 common SNPs for C16 index and C16:1were identified.
Sum of fatty acid traits (SFA, UFA, SFA/UFA)
A total of 108 significant associations mainly on BTA5, 10 and 20 with three sum of fatty acid traits were detected, which involved 52 distinct SNPs. Of them, 22 SNPs were simultaneously associated with three traits and 12 were common for two traits. The 0.96 Mbp region (72.69–73.65 Mbp) on BTA10 was associated with the three traits, in which the SNP (ARS-BFGL-NGS-4783) showed the strongest association for SFA (P = 6.07E-08), UFA (P = 4.01E-07) and SFA/UFA (P = 2.64E-07), respectively.
Discussions
To our knowledge, this is one of the first GWA study for milk fatty acids with high density SNP Chip. In this study, we detected a total of 83 genome-wise and 314 suggestive significant SNPs for 22 fatty acid traits. Among them, some SNPs are located within the QTL regions on BTA6, 14, 19 and 26 those have been reported by Stoop et al [13], Schenninket al [14] and Morris et al [30] for bovine milk fat composition. Sixteen SNPs on BTA14, 5 SNPs on BTA19, and 5 SNPs on BTA7 were consistent with the previous GWA study for fatty acid traits of dairy cattle [24]. However, associations of BTA19 with C16:1 and CLA were not found in this study. This is probably due to a different dairy population was tested. Several SNPs were found to be located within and/or close to genes that are known to have functions related to the milk composition. In addition, 20 novel prospective candidate genes affecting milk fatty acid traits were identified.
Chromosomes underlying novel promising candidate genes
On BTA1, 23 SNPs associated with 9 fatty acids (C10:0, C12:0, C14:0, CLA, C18:0, C18 index, C22:0, SFA and SFA/UFA) were detected. The SNP associated with SFA and SFA/UFA is 38,733 bp away from the 3-hydroxyacyl Coenzyme A dehydrogenase (EHHADH) gene. As a bi-functional enzyme, EHHADH is part of the classical peroxisomal fatty acid β-oxidation pathway, which is highly inducible via peroxisome proliferator-activated receptor α (PPARα) activation [31] and is essential for the production of medium-chain dicarboxylic acids [32]. Four SNPs for C18:0 and C18 index form an 0.40 Mbp region containing the 1-acylglycerol -3- phosphate O-acyltransferase 3 (AGPAT3) gene. AGPAT catalyzes the first step during de novo synthesis of triacylglycerol. AGPAT3 is a member of the acyltransferase family [33] and plays a key role in de novo phospholipid biosynthetic due to its function of converting lysophosphatidic acid into phosphatidic acid [34].
On BTA2, 21 SNPs showed associations with 7 traits (C10:0, C12:0, C14:0, C14:1, C18:0, C18 index and UFA). Two SNPs associated with C18:0 and C18 index are 0.50 Mbp away from the signal transducer and activator of transcription 1 (STAT1) gene, especially, one of them is the top 3 significant SNP for C18 index. STATs are transcription factors known to importance to cytokine signaling. STAT1 has a role in regulating the transcription of genes involved in milk protein synthesis and fat metabolism in Holstein [35]. The SNP associated with UFA is 0.14 Mbp and 19,295 bp away from the acyl-CoA synthetase long-chain family member 3 (ACSL3) gene and the monoacylglycerol O-acyltransferase 1 (MOGAT1) gene, respectively. ACSL3 is an isozyme of the long chain fatty acids coenzyme A ligase family that convert free long chain fatty acids into fatty acyl-CoA esters and has a substrate preference for PUFA [36]. Depletion of ACSL3 by RNAi causes a significant reduction in fatty acids uptake, thereby plays a key role in lipid biosynthesis and fatty acids degradation [37]. MOGAT1 catalyzes the synthesis of diacylglycerols, the precursor of triacylglycerol and phospholipids [38]. Two SNPs associated with C18:0 and C18 index are 5.70 Mbp and 5.74 Mbp away from the fatty acid binding protein 3 (FABP3) gene, respectively. FABP3 provides fatty acids for SCD, which is one of specific transporters for LCFA and one of the most abundant isoforms in bovine mammary tissue [39]. Eight contiguous SNPs associated with C18:0 and C18 index are located within a chromosome region of 63.58∼98.16 Mbp that overlaps a reported QTL region (67.56∼68.25 Mbp) for C14 index, C16 index, C18 index, SFA, MUFA, PUFA and SFA/UFA [40].
On BTA5, 17 SNPs showed association with 9 traits (C10:0, C14:0, C16:0, C18:0, C18:1n9c, C18 index, SFA, SFA/UFA and UFA). The top one significant SNP for C14:0 is within the carboxypeptidase M (CPM) gene. Up-regulation of CPM in macrophages (MAs) is associated with increased lipid uptake [41] and the highest expression of CPM was detected in human adipocyte cell [42]. The SNP associated with C18:1n9c, SFA, UFA and SFA/UFA is 0.77 Mbp away from the oxysterol binding protein-like 8 (OSBPL8) gene which encodes a member of the oxysterol-binding protein (OSBP) family, a group of intracellular lipid receptors. OSBPL8 has the capacity to modulate lipid homeostasis and SREBP activity probably through an indirect mechanism [43] and is a negative regulator of sequestering of triglyceride [44]. The chromosome region of 8.43 Mbp (95.74∼104.17 Mbp) associated with C18:1n9c, C18 index, UFA and SFA/UFA contains the oxidized low density lipoprotein (lectin-like) receptor 1 (OLR1) gene which can bind and degrade oxidized low-density lipoprotein [45], [46].
On BTA9, 13 SNPs showed association with 9 traits (C10:0, C12:0, C14:0, C16:1, C18:0, C18 index, C18:1n9c, CLA and UFA). The top one significant SNP for C10:0 and C12:0 and the SNP for C18:0 are 0.40, 0.81 Mbp away from the 5-hydroxytryptamine (serotonin) receptor 1B (HTR1B) gene, respectively. HTR1B is one of receptors for 5-hydroxytryptamine (serotonin). HTR1B gene knocked-out mice showed elevated aggression, higher food intake and impulsivity, indicating it possibly acts as a bridge between behavior and energy homeostasis [47]. Fatty acids, as energy signal, affect the activity of hypothalamic fat-sensitive neurons and impair nervous control of energy homeostasis [48]. HTR1B was also shown to affect milk production performance in Chinese Holstein [49].
On BTA20, 30 SNPs showed association with 11 traits (C12:0, C14:0, C14:1, C18:0, C18 index, C18:1n9c, C18:2n6c, CLA, SFA, SFA/UFA and UFA). The SNP associated with C18:0 is located within the growth hormone receptor (GHR) gene, the well-known major gene affecting milk fat trait [50]. Three SNPs associated with C18:0 and C18 index are within an 1.46 Mbp region containing the 3-oxoacid CoA transferase 1 (OXCT1) gene which has a major function to utilize ketone bodies by mammary [39]. The SNP associated with SFA, UFA and SFA/UFA is located within the prolactin receptor (PRLR) gene which activates the STAT5A (Signal transducer and activator of transcription 5A) expression [51] and is associated with milk composition traits [52].
On BTA21, 10 SNPs showed association with 8 traits (C10:0, C12:0, C16:0, C18 index, CLA, SFA, SFA/UFA and UFA). The 0.15 Mbp region (9.37∼9.52 Mbp) associated with C10:0 and C12:0 is 1.10 Mbp away from the insulin-like growth factor 1 receptor (IGF1R) gene. Furthermore, the top one significant SNP for C12:0 is located within such region. IGF1R was found to affect milk composition traits [53]. Two SNPs associated with C10:0, SFA, SFA/UFA and UFA are close to the lipin 1 (PLIN1) gene which plays a vital role in regulation on the expression of genes involved in milk fat synthesis [39].
On BTA26, 71 SNPs showed association with 8 traits (C10:0, C12:0, C14:1, C14 index, C16:0, C18:0, SFA and SFA/UFA). The nearest SNP is 32,576 bp close to the SCD1 gene which encodes key enzyme responsible for the conversion of SFA to MUFA in mammalian adipocytes [54] and were shown to be associated with milk fatty acids [10], [11], [12], [40], [55]. The SNP associated with C10:0 and C14 index is located within the protein kinase, cGMP-dependent, type I (PRKG1) gene which is a key regulator of adipokine secretion and browning of white fat depots [56] and brown fat cell differentiation [57]. The SNP associated with C12:0 is located within the multiple inositol-polyphosphate phosphatase 1 (MINPP1) gene. MINPP1 encodes multiple inositol polyphosphate phosphatase which converts 2, 3 bisphosphoglycerate (2,3-BPG) to 2-phosphoglycerate [58]. As known, 2,3-BPG is a key substrate for the triglyceride (TG) synthesis. Two SNPs associated with C14:1 and C14 index are 7,406 bp and 35,288 bp away from the lipase, family member J (LIPJ) gene and the lipase, family member K (LIPK) gene, respectively. The two genes belong to lipase family and take part in lipid catabolic process in human [59] and have an essential function in lipid metabolism of the most differentiated epidermal layers. The SNP for C14:1 and the one for C14:1 and C14 index are 0.71 Mbp and 0.89 Mbp away from the enoyl coenzyme A hydratase, short chain, 1(ECHS1) gene, respectively. ECHS1 takes part in fatty acid biosynthesis, elongation and metabolism and catalyzes the β-oxidation of fatty acid in human [60]. Two SNPs associated with C14:1 and C14 index are located within the sorbin and SH3 domain containing 1 (SORBS1) gene. SORBS1 is an important protein in the insulin-signaling pathway in the adipose depots of human [61] and has a positive regulation of lipid biosynthetic process [62]. The SNP associated with C14:1 and C14 index is located within the conserved helix-loop-helix ubiquitous kinase (CHUK) gene. CHUK takes part in mammary gland alveolus development, mammary gland epithelial cell proliferation [63] and lipogenesis through NF-κB activation pathway [64]. The SNP associated with C14:1 and C14 index is 1,586 bp away from the nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (NFKB2) gene which is essential for normal development of the mammary gland [65].
Chromosomes underlying known candidate genes
Apart from the seven chromosomes as mentioned above, other chromosome regions harboring several significant SNPs within or near to known genes involved in fatty acids synthesis were identified.
On BTA14, the SNP associated with 9 traits (C14:1, C16:0, C18:1n9c, C18:2n6c, C14 index, C18 index, SFA, UFA and SFA/UFA) is located within the DGAT1 gene, which has been confirmed to be the true QTL for milk fat composition in dairy cattle [66]. On BTA19, three SNPs associated with C10:0, C12:0 and C14:0 are 0.05, 1.71 and 1.42 Mbp away from FASN, ACACA (Acetyl-CoA carboxylase alpha) and SREBF1 (Sterol regulatory element binding transcription factor 1), respectively. FASN is a multifunctional enzyme with a central role in the de novo lipogenesis in mammals [45], [67]. ACACA catalyses biosynthesis of LCFA in mammalian cytosol [68]. SREBF1 is a transcription factor that regulates the expression of the SCD1 gene which is related to several genes of lipid metabolism [69]. On BTA6, the SNP associated with C18index is located within the peroxisome proliferator-activated receptor gamma, coactivator 1 alpha (PPARGC1A) gene which is involved in the regulation of fatty acids transcription and mammary gland metabolism [70]. Six SNPs on BTA13 associated with C16:1, C16 index, C18:0 and C18 index are located 1.01∼3.11 Mbp away from the acyl-CoA synthetase short-chain family member 2 (ACSS2) gene. ACSS2 provides activated acetate for de novo fatty acids synthesis [39]. Three SNPs within an 2.34 Mbp segment (54.27∼56.61 Mbp) on BTA18 associated with C10:0, SFA, SFA/UFA and UFA harbors the Sphingosine kinase 2 (SPHK2) gene. As a lipid mediator with both intra- and extracellular functions, SPHK2 has diacylglycerol kinase activity and involves the sphingolipid synthesis [39]. Two SNPs on BTA23 associated with C16:0 is located 1.21 Mbp and 0.79 Mbp away from the butyrophilin, subfamily 1, member A1 (BTN1A1) gene, which is essential for milk lipid droplet formation [39], and the PRL gene, which impacts milk fat composition through STAT5A [45], respectively.
The 0.03 Mbp region (16.39∼16.42 Mbp) on BTA27 associated with C12:0, SFA/UFA and UFA is 2.11 Mbp away from the acyl-CoA synthetase long-chain family member 1(ACSL1) gene. ACSL1 has a vital role in fatty acids activation for milk TAG [39]. The SNP for C14 index is 1.12 Mbp away from the 1-acylglycerol-3-phosphate O-acyltransferase 6 (AGPAT6) gene, a novel lipid biosynthetic gene required for triacylglycerol production in mammary epithelium, if AGPAT6 was knocked out, lactating mice failed to synthesize milk fat [71]. The SNP on BTA29 associated with C18 index is located 0.29 Mbp away from the fatty acid desaturase 1(FADS1) gene which catalyzes the synthesis of LCFA [72].
No significant SNPs were detected with C8:0, C18:3n3, C18:3n6 and C20:5n3, probably because these four traits have special population requirements.
Conclusions
The present genome-wide association study identified 83 genome-wide and 314 suggestive significant SNPs associated with 18 milk fatty acid traits. Some of these SNPs were located within or near to previously reported genes and QTL regions, while some of the SNPs were novel. Consequently, 20 novel promising candidate genes were identified for C10:0, C12:0, C14:0, C14:1, C14 index, C18:0, C18:1n9c, C18 index, SFA, UFA and SFA/UFA, such as HTR1B, CPM, PRKG1, MINPP1, LIPJ, LIPK, EHHADH, MOGAT1, ECHS1, STAT1, SORBS1, NFKB2, AGPAT3, CHUK, OSBPL8, PRLR, IGF1R, ACSL3, GHR and OXCT1. Our findings are helpful for follow-up studies to fine-mapping to unravel causal mutations for milk fatty acid traits in dairy cattle.
Supporting Information
Manhattan plots for each studied milk fatty acids trait. BTAX is represented by BTA30), the first line represents genome-wise significant level (raw P<1.23E-06), and the second line represents suggestive significant level (raw P<2.46E-05).
(PPTX)
Funding Statement
This work was supported by the High Technology Research and Development Program of China (2013AA102504), National Key Technologies R & D Program (2011BAD28B02, 2012BAD12B01), Beijing Dairy Industry Innovation Team, 948 Program (2011-G2A), Beijing Research and Technology program (D121100003312001), and Program for Changjiang Scholar and Innovation Research Team in University (IRT1191). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Rasmussen BM, Vessby B, Uusitupa M, Berglund L, Pedersen E, et al. (2006) Effects of dietary saturated, monounsaturated, and n-3 fatty acids on blood pressure in healthy subjects. Am J ClinNutr 83: 221–226. [DOI] [PubMed] [Google Scholar]
- 2. Belury MA (2002) Dietary conjugated linoleic acid in health: physiological effects and mechanisms of action. Annu Rev Nutr 22: 505–531. [DOI] [PubMed] [Google Scholar]
- 3. Mensink RP, Zock PL, Kester AD, Katan MB (2003) Effects of dietary fatty acids and carbohydrates on the ratio of serum total to HDL cholesterol and on serum lipids and apolipoproteins: a meta-analysis of 60 controlled trials. Am J ClinNutr 77: 1146–1155. [DOI] [PubMed] [Google Scholar]
- 4. Hayes KC (2002) Dietary fat and heart health: in search of the ideal fat. Asia Pacific J ClinNutr 11 (Suppl) S394–S400. [DOI] [PubMed] [Google Scholar]
- 5. Kay JK, Weber WJ, Moore CE, Bauman DE, Hansen LB, et al. (2005) Effects of week of lactation and genetic selection for milk yield on milk fatty acid composition in Holstein cows. J Dairy Sci 88: 3886–3893. [DOI] [PubMed] [Google Scholar]
- 6. Garnsworthy PC, Masson LL, Lock AL, Mottram TT (2006) Variation of milk citrate with stage of lactation and de novo fatty acid synthesis in dairy cows. J Dairy Sci 89: 1604–1612. [DOI] [PubMed] [Google Scholar]
- 7. Soyeurt H, Gillon A, Vanderick S, Mayeres P, Bertozzi C, et al. (2007) Estimation of heritability and genetic correlations for the major fatty acids in bovine milk. J Dairy Sci 90: 4435–4442. [DOI] [PubMed] [Google Scholar]
- 8. Bobe G, Minick Bormann JA, Lindberg GL, Freeman AE, Beitz DC (2008) Short communication: estimates of genetic variation of milk fatty acids in US Holstein cows. J Dairy Sci 91: 1209–1213. [DOI] [PubMed] [Google Scholar]
- 9. Stoop WM, van Arendonk JA, Heck JM, van Valenberg HJ, Bovenhuis H (2008) Genetic parameters for major milk fatty acids and milk production traits of Dutch Holstein-Friesians. J Dairy Sci 91: 385–394. [DOI] [PubMed] [Google Scholar]
- 10. Mele M, Conte G, Castiglioni B, Chessa S, Macciotta NP, et al. (2007) Stearoyl-coenzyme A desaturase gene polymorphism and milk fatty acid composition in Italian Holsteins. J Dairy Sci 90: 4458–4465. [DOI] [PubMed] [Google Scholar]
- 11. Schennink A, Heck JM, Bovenhuis H, Visker MH, van Valenberg HJ, et al. (2008) Milk fatty acid unsaturation: genetic parameters and effects of stearoyl-CoA desaturase (SCD1) and acyl CoA: diacylglycerolacyltransferase 1 (DGAT1). J Dairy Sci 91: 2135–2143. [DOI] [PubMed] [Google Scholar]
- 12. Conte G, Mele M, Chessa S, Castiglioni B, Serra A, et al. (2010) Diacylglycerolacyltransferase 1, stearoyl-CoA desaturase 1, and sterol regulatory element binding protein 1 gene polymorphisms and milk fatty acid composition in Italian Brown cattle. J Dairy Sci 93: 753–763. [DOI] [PubMed] [Google Scholar]
- 13. Stoop WM, Schennink A, Visker MH, Mullaart E, van Arendonk JA, et al. (2009) Genome-wide scan for bovine milk-fat composition. I. Quantitative trait loci for short- and medium-chain fatty acids. J Dairy Sci 92: 4664–4675. [DOI] [PubMed] [Google Scholar]
- 14. Schennink A, Stoop WM, Visker MH, van der Poel JJ, Bovenhuis H, et al. (2009) Short communication: Genome-wide scan for bovine milk-fat composition. II. Quantitative trait loci for long-chain fatty acids. J Dairy Sci 92: 4676–4682. [DOI] [PubMed] [Google Scholar]
- 15. Andersson L (2009) Genome-wide association analysis in domestic animals: a powerful approach for genetic dissection of trait loci. Genetica 136: 341–349. [DOI] [PubMed] [Google Scholar]
- 16. Klein RJ, Zeiss C, Chew EY, Tsai JY, Sackler RS, et al. (2005) Complement factor H polymorphism in age-related macular degeneration. Science 308: 385–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jiang L, Liu J, Sun D, Ma P, Ding X, et al. (2010) Genome wide association studies for milk production traits in Chinese Holstein population. PLoS One 5: e13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schopen GC, Visker MH, Koks PD, Mullaart E, van Arendonk JA, et al. (2011) Whole-genome association study for milk protein composition in dairy cattle. J Dairy Sci 94: 3148–3158. [DOI] [PubMed] [Google Scholar]
- 19. Cole JB, Wiggans GR, Ma L, Sonstegard TS, Lawlor TJ Jr, et al. (2011) Genome-wide association analysis of thirty one production, health, reproduction and body conformation traits in contemporary U.S. Holstein cows. BMC Genomics 12: 408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sahana G, Guldbrandtsen B, Bendixen C, Lund MS (2010) Genome-wide association mapping for female fertility traits in Danish and Swedish Holstein cattle. Anim Genet 41: 579–588. [DOI] [PubMed] [Google Scholar]
- 21. Schulman NF, Sahana G, Iso-Touru T, McKay SD, Schnabel RD, et al. (2011) Mapping of fertility traits in Finnish Ayrshire by genome-wide association analysis. Anim Genet 42: 263–269. [DOI] [PubMed] [Google Scholar]
- 22. Kirkpatrick BW, Shi X, Shook GE, Collins MT (2010) Whole-Genome association analysis of susceptibility to paratuberculosis in holstein cattle. Anim Genet [DOI] [PubMed] [Google Scholar]
- 23. Murdoch BM, Clawson ML, Laegreid WW, Stothard P, Settles M, et al. (2010) A 2cM genome-wide scan of European Holstein cattle affected by classical BSE. BMC Genet 11: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bouwman AC, Bovenhuis H, Visker MH, van Arendonk JA (2011) Genome-wide association of milk fatty acids in Dutch dairy cattle. BMC Genet 12: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bouwman AC, Visker MH, van Arendonk JA, Bovenhuis H (2012) Genomic regions associated with bovine milk fatty acids in both summer and winter milk samples. BMC Genet 13: 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kelsey JA, Corl BA, Collier RJ, Bauman DE (2003) The effect of breed, parity, and stage of lactation on conjugated linoleic acid (CLA) in milk fat from dairy cows. J Dairy Sci 86: 2588–2597. [DOI] [PubMed] [Google Scholar]
- 27. Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, et al. (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Han B, Kang HM, Eskin E (2009) Rapid and accurate multiple testing correction and power estimation for millions of correlated markers. PLoS Genet 5: e1000456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lander E, Kruglyak L (1995) Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet 11: 241–247. [DOI] [PubMed] [Google Scholar]
- 30. Morris CA, Cullen NG, Glass BC, Hyndman DL, Manley TR, et al. (2007) Fatty acid synthase effects on bovine adipose fat and milk fat. Mamm Genome 18: 64–74. [DOI] [PubMed] [Google Scholar]
- 31. Qi C, Zhu Y, Pan J, Usuda N, Maeda N, et al. (1999) Absence of spontaneous peroxisome proliferation in enoyl-CoA Hydratase/L-3-hydroxyacyl-CoA dehydrogenase-deficient mouse liver. Further support for the role of fatty acyl CoA oxidase in PPARalpha ligand metabolism. J BiolChem 274: 15775–15780. [DOI] [PubMed] [Google Scholar]
- 32. Houten SM, Denis S, Argmann CA, Jia Y, Ferdinandusse S, et al. (2012) Peroxisomal L-bifunctional enzyme (Ehhadh) is essential for the production of medium-chain dicarboxylic acids. J Lipid Res 53: 1296–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cao J, Li JL, Li D, Tobin JF, Gimeno RE (2006) Molecular identification of microsomal acyl-CoA:glycerol-3-phosphate acyltransferase, a key enzyme in de novo triacylglycerol synthesis. ProcNatlAcadSci U S A 103: 19695–19700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lu B, Jiang YJ, Zhou Y, Xu FY, Hatch GM, et al. (2005) Cloning and characterization of murine 1-acyl-sn-glycerol 3-phosphate acyltransferases and their regulation by PPARalpha in murine heart. Biochem J 385: 469–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cobanoglu O, Zaitoun I, Chang YM, Shook GE, Khatib H (2006) Effects of the signal transducer and activator of transcription 1 (STAT1) gene on milk production traits in Holstein dairy cattle. J Dairy Sci 89: 4433–4437. [DOI] [PubMed] [Google Scholar]
- 36. Van Horn CG, Caviglia JM, Li LO, Wang S, Granger DA, et al. (2005) Characterization of recombinant long-chain rat acyl-CoA synthetase isoforms 3 and 6: identification of a novel variant of isoform 6. Biochemistry 44: 1635–1642. [DOI] [PubMed] [Google Scholar]
- 37. Poppelreuther M, Rudolph B, Du C, Grossmann R, Becker M, et al. (2012) The N-terminal region of acyl-CoA synthetase 3 is essential for both the localization on lipid droplets and the function in fatty acid uptake. J Lipid Res 53: 888–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yen CL, Stone SJ, Cases S, Zhou P, Farese RV Jr (2002) Identification of a gene encoding MGAT1, a monoacylglycerolacyltransferase. ProcNatlAcadSci U S A 99: 8512–8517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bionaz M, Loor JJ (2008) Gene networks driving bovine milk fat synthesis during the lactation cycle. BMC Genomics 9: 366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rincon G, Islas-Trejo A, Castillo AR, Bauman DE, German BJ, et al. (2012) Polymorphisms in genes in the SREBP1 signalling pathway and SCD are associated with milk fatty acid composition in Holstein cattle. J Dairy Res 79: 66–75. [DOI] [PubMed] [Google Scholar]
- 41. Tsakiris I, Torocsik D, Gyongyosi A, Dozsa A, Szatmari I, et al. (2012) Carboxypeptidase-M is regulated by lipids and CSFs in macrophages and dendritic cells and expressed selectively in tissue granulomas and foam cells. Lab Invest 92: 345–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Denis CJ, Deiteren K, Hendriks D, Proost P, Lambeir AM (2013) Carboxypeptidase M in apoptosis, adipogenesis and cancer. ClinChimActa 415: 306–316. [DOI] [PubMed] [Google Scholar]
- 43. Zhou T, Li S, Zhong W, Vihervaara T, Beaslas O, et al. (2011) OSBP-related protein 8 (ORP8) regulates plasma and liver tissue lipid levels and interacts with the nucleoporin Nup62. PLoS One 6: e21078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yan D, Mayranpaa MI, Wong J, Perttila J, Lehto M, et al. (2008) OSBP-related protein 8 (ORP8) suppresses ABCA1 expression and cholesterol efflux from macrophages. J BiolChem 283: 332–340. [DOI] [PubMed] [Google Scholar]
- 45. Schennink A, Bovenhuis H, Leon-Kloosterziel KM, van Arendonk JA, Visker MH (2009) Effect of polymorphisms in the FASN, OLR1, PPARGC1A, PRL and STAT5A genes on bovine milk-fat composition. Anim Genet 40: 909–916. [DOI] [PubMed] [Google Scholar]
- 46. Imanishi T, Hano T, Sawamura T, Takarada S, Nishio I (2002) Oxidized low density lipoprotein potentiation of Fas-induced apoptosis through lectin-like oxidized-low density lipoprotein receptor-1 in human umbilical vascular endothelial cells. Circ J 66: 1060–1064. [DOI] [PubMed] [Google Scholar]
- 47. Bouwknecht JA, Hijzen TH, van der Gugten J, Maes RA, Hen R, et al. (2001) Absence of 5-HT(1B) receptors is associated with impaired impulse control in male 5-HT(1B) knockout mice. Biol Psychiatry 49: 557–568. [DOI] [PubMed] [Google Scholar]
- 48. Wolfgang MJ, Lane MD (2006) The role of hypothalamic malonyl-CoA in energy homeostasis. J BiolChem 281: 37265–37269. [DOI] [PubMed] [Google Scholar]
- 49. Zhang CL, Chen H, Wang YH, Zhang RF, Lan XY, et al. (2008) Serotonin receptor 1B (HTR1B) genotype associated with milk production traits in cattle. Res Vet Sci 85: 265–268. [DOI] [PubMed] [Google Scholar]
- 50. Blott S, Kim JJ, Moisio S, Schmidt-Kuntzel A, Cornet A, et al. (2003) Molecular dissection of a quantitative trait locus: a phenylalanine-to-tyrosine substitution in the transmembrane domain of the bovine growth hormone receptor is associated with a major effect on milk yield and composition. Genetics 163: 253–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Jiang S, Ren Z, Xie F, Yan J, Huang S, et al. (2012) Bovine prolactin elevates hTF expression directed by a tissue-specific goat beta-casein promoter through prolactin receptor-mediated STAT5a activation. BiotechnolLett 34: 1991–1999. [DOI] [PubMed] [Google Scholar]
- 52. Brym P, Kaminski S, Wojcik E (2005) Nucleotide sequence polymorphism within exon 4 of the bovine prolactin gene and its associations with milk performance traits. J Appl Genet 46: 179–185. [PubMed] [Google Scholar]
- 53. Bonakdar E, Rahmani HR, Edriss MA, Sayed Tabatabaei BE (2010) IGF-I gene polymorphism, but not its blood concentration, is associated with milk fat and protein in Holstein dairy cows. Genet Mol Res 9: 1726–1734. [DOI] [PubMed] [Google Scholar]
- 54. Ntambi JM, Miyazaki M (2004) Regulation of stearoyl-CoA desaturases and role in metabolism. Prog Lipid Res 43: 91–104. [DOI] [PubMed] [Google Scholar]
- 55. Moioli B, Contarini G, Avalli A, Catillo G, Orru L, et al. (2007) Short communication: Effect of stearoyl-coenzyme Adesaturase polymorphism on fatty acid composition of milk. J Dairy Sci 90: 3553–3558. [DOI] [PubMed] [Google Scholar]
- 56. Mitschke MM, Hoffmann LS, Gnad T, Scholz D, Kruithoff K, et al. (2013) Increased cGMP promotes healthy expansion and browning of white adipose tissue. FASEB J 27: 1621–1630. [DOI] [PubMed] [Google Scholar]
- 57. Haas B, Mayer P, Jennissen K, Scholz D, Berriel Diaz M, et al. (2009) Protein kinase G controls brown fat cell differentiation and mitochondrial biogenesis. Sci Signal 2: ra78. [DOI] [PubMed] [Google Scholar]
- 58. Cho J, King JS, Qian X, Harwood AJ, Shears SB (2008) Dephosphorylation of 2,3-bisphosphoglycerate by MIPP expands the regulatory capacity of the Rapoport-Luebering glycolytic shunt. ProcNatlAcadSci U S A 105: 5998–6003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Toulza E, Mattiuzzo NR, Galliano MF, Jonca N, Dossat C, et al. (2007) Large-scale identification of human genes implicated in epidermal barrier function. Genome Biol 8: R107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Takahashi M, Watari E, Shinya E, Shimizu T, Takahashi H (2007) Suppression of virus replication via down-modulation of mitochondrial short chain enoyl-CoA hydratase in human glioblastoma cells. Antiviral Res 75: 152–158. [DOI] [PubMed] [Google Scholar]
- 61. Yang WS, Lee WJ, Huang KC, Lee KC, Chao CL, et al. (2003) mRNA levels of the insulin-signaling molecule SORBS1 in the adipose depots of nondiabetic women. Obes Res 11: 586–590. [DOI] [PubMed] [Google Scholar]
- 62. Baumann CA, Ribon V, Kanzaki M, Thurmond DC, Mora S, et al. (2000) CAP defines a second signalling pathway required for insulin-stimulated glucose transport. Nature 407: 202–207. [DOI] [PubMed] [Google Scholar]
- 63. Cao Y, Bonizzi G, Seagroves TN, Greten FR, Johnson R, et al. (2001) IKKalpha provides an essential link between RANK signaling and cyclin D1 expression during mammary gland development. Cell 107: 763–775. [DOI] [PubMed] [Google Scholar]
- 64. Li QS, Pene V, Krishnamurthy S, Cha H, Liang TJ (2013) Hepattis C virus infection activates a novel innate pathway involving IKKα in lipogenesis and viral assembly. Nat Med 19: 722–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Solt LA, Madge LA, May MJ (2009) NEMO-binding domains of both IKKalpha and IKKbeta regulate IkappaB kinase complex assembly and classical NF-kappaB activation. J BiolChem 284: 27596–27608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Grisart B, Coppieters W, Farnir F, Karim L, Ford C, et al. (2002) Positional candidate cloning of a QTL in dairy cattle: identification of a missense mutation in the bovine DGAT1 gene with major effect on milk yield and composition. Genome Res 12: 222–231. [DOI] [PubMed] [Google Scholar]
- 67. Munoz G, Ovilo C, Noguera JL, Sanchez A, Rodriguez C, et al. (2003) Assignment of the fatty acid synthase (FASN) gene to pig chromosome 12 by physical and linkage mapping. Anim Genet 34: 234–235. [DOI] [PubMed] [Google Scholar]
- 68. Cronan JE Jr, Waldrop GL (2002) Multi-subunit acetyl-CoA carboxylases. Prog Lipid Res 41: 407–435. [DOI] [PubMed] [Google Scholar]
- 69. Harvatine KJ, Bauman DE (2006) SREBP1 and thyroid hormone responsive spot 14 (S14) are involved in the regulation of bovine mammary lipid synthesis during diet-induced milk fat depression and treatment with CLA. J Nutr 136: 2468–2474. [DOI] [PubMed] [Google Scholar]
- 70. Puigserver P, Spiegelman BM (2003) Peroxisome proliferator-activated receptor-gamma coactivator 1 alpha (PGC-1 alpha): transcriptional coactivator and metabolic regulator. Endocr Rev 24: 78–90. [DOI] [PubMed] [Google Scholar]
- 71. Beigneux AP, Vergnes L, Qiao X, Quatela S, Davis R, et al. (2006) Agpat6–a novel lipid biosynthetic gene required for triacylglycerol production in mammary epithelium. J Lipid Res 47: 734–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Rodriguez-Cruz M, Tovar AR, Palacios-Gonzalez B, Del Prado M, Torres N (2006) Synthesis of long-chain polyunsaturated fatty acids in lactating mammary gland: role of Delta5 and Delta6 desaturases, SREBP-1, PPARalpha, and PGC-1. J Lipid Res 47: 553–560. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Manhattan plots for each studied milk fatty acids trait. BTAX is represented by BTA30), the first line represents genome-wise significant level (raw P<1.23E-06), and the second line represents suggestive significant level (raw P<2.46E-05).
(PPTX)


