Abstract
Cystic fibrosis (CF) airways are characterized by bacterial infections, excess mucus production and robust neutrophil recruitment. The main CF airway pathogen is Pseudomonas aeruginosa. Neutrophils are not capable of clearing the infection. Neutrophil primary granule components, myeloperoxidase (MPO) and human neutrophil elastase (HNE), are inflammatory markers in CF airways, and their elevated levels are associated with poor lung function. Identifying the mechanism of MPO and HNE release from neutrophils is of high clinical relevance for CF. Here we show that human neutrophils release large amounts of neutrophil extracellular traps (NETs) in the presence of Pseudomonas aeruginosa. Bacteria are entangled in NETs and co-localize with extracellular DNA. MPO, HNE and citrullinated histone H4 are all associated with DNA in Pseudomonas-triggered NETs. Both, laboratory standard strains and CF isolates of Pseudomonas aeruginosa induce DNA, MPO and HNE release from human neutrophils. The increase in peroxidase activity of neutrophil supernatants after Pseudomonas exposure indicates that enzymatically active MPO is released. Pseudomonas aeruginosa induces a robust respiratory burst in neutrophils that is required for extracellular DNA release. Inhibition of the cytoskeleton prevents Pseudomonas-initiated superoxide production and DNA release. NADPH oxidase inhibition suppresses Pseudomonas-induced release of active MPO and HNE. Blocking MEK/ERK signaling results in only minimal inhibition of DNA release induced by Pseudomonas. Our data describe in vitro details of DNA, MPO and HNE release from neutrophils activated by Pseudomonas aeruginosa. We propose that Pseudomonas-induced NET formation is an important mechanism contributing to inflammatory conditions characteristic of CF airways.
Keywords: Neutrophil, cystic fibrosis, NET, myeloperoxidase, elastase
Introduction
Cystic fibrosis (CF) is an inherited disorder resulting from mutations in the cystic fibrosis transmembrane conductance regulator (CFTR) anion channel (1). Several organs are affected in CF, but lung complications remain the main cause of disease morbidity and mortality (2). Lack of apical plasma membrane expression of a fully functional CFTR in respiratory epithelial cells leads to altered fluid absorption, ion secretion and mucus retention within the airways. As a result, CF airways become obstructed and infected with bacteria leading to robust neutrophil infiltration and chronic inflammation (3). A wide variety of bacterial species have been detected in CF airways, but Pseudomonas aeruginosa represents the predominant pathogen infecting most CF patients (4–6).
The main pharmacological approaches used in the clinical management of CF airway disease focus on treating bacterial infections with antibiotics, promoting airway clearance by lysing DNA within mucus plugs, and rehydrating the airway surface (7–10). Survival and quality of life have improved significantly for people with CF over the last two decades; however, for most patients the disease still cannot be adequately managed, let alone cured, and more effective treatments are urgently needed (8). Recently, promising new drug candidates have emerged that address the core problem in CF, the dysfunctional CFTR membrane protein (11–13). Although these CFTR “protein-assist” therapies are very promising, currently there is only one FDA-approved drug that potentiates CFTR function, in the case of defective channel gating, and no drugs have been approved to date to correct the defective cellular trafficking of the common CFTR F508del mutation (13).
Airway inflammation in CF is the product of a complex set of innate immune interactions. The neutrophil is a pivotal cellular player influencing the outcome of these interactions. Neutrophil density in CF sputum has been shown to correlate with CF disease severity (measured as forced expiratory volume in 1 second, FEV1) (14, 15). Sputum and blood concentrations of human neutrophil elastase (HNE) and myeloperoxidase (MPO) in CF patients are associated with declines in lung function (14–18). Interleukin-8 (IL-8), a major neutrophil chemoattractant that both airway epithelial cells and neutrophils produce, has also been associated with CF lung function decline (15, 16). Interlekuin-1beta (IL-1β), a proinflammatory cytokine mainly produced by macrophages but also secreted by activated neutrophils, has also been linked to CF lung damage (14). These data obtained from clinical samples of CF patients clearly show that neutrophil recruitment and ‘activation’ are major contributors to lung damage.
It is therefore important to understand why recruited neutrophils release their granule contents in CF. Formation of neutrophil extracellular traps (NETs) offers a possible mechanism (19). In NETs, extracellular DNA is associated with neutrophil granule components (HNE, MPO) and histones (19, 20). Neutrophils release NETs in response to bacterial and inflammatory stimuli (19, 21). Negative correlations were found between CF sputum extracellular DNA concentrations and lung function measures (16, 22). Neutrophils undergoing NET formation were detected in CF sputum samples (23–25). Histone citrullination, a histone modification characteristic for NET formation was also detected in CF sputum samples (23).
P. aeruginosa is likely to activate neutrophils in CF airways. This organism is associated with diminished pulmonary function in CF, and similar to many other pathogens, triggers robust NET release in vivo (15, 26) and in vitro (27, 28). Pyocyanin, a major toxin of P. aeruginosa, that can be relatively abundant in the CF airway, enhances NET formation and has been linked to lung function decline in this disorder (26, 28–30). Pseudomonas and neutrophils are found in close proximity and neutrophils have been shown to phagocytose bacteria in airway samples of CF patients (31). These data suggest that Pseudomonas-neutrophil interactions are highly relevant to CF airway pathophysiology. The major objective of this study was to provide insight into the mechanism of Pseudomonas-induced NET formation, a process highly relevant to neutrophil dysfunction in the CF airway. Our hypothesis was that Pseudomonas aeruginosa-stimulated NET formation contributes to release of MPO, HNE and extracellular DNA. Our approach to test this hypothesis was to quantify NET formation, MPO and HNE release and activities after exposure of human neutrophils in vitro to laboratory standard strains and CF isolates of Pseudomonas aeruginosa.
Materials and Methods
Human subjects
The Institutional Review Board of the University of Georgia approved the human subject study to collect peripheral blood from healthy volunteers (UGA# 2012-10769-06). Enrolled healthy volunteers were older than 18 years of age, non-pregnant, heavier than 110 pounds without any infectious disease complication and signed written informed consent form (32).
Human subjects also included CF patients enrolled in an observational study of CF lung disease severity, “Genetics of CF Lung Disease” (Seattle Children’s Hospital institutional review board approved protocol 10855 and Partners Healthcare Systems/Massachusetts General Hospital institutional review board approved protocol 2011P000544). The protocols and informed consent procedures were approved by the Institutional Review Board of the Seattle Children’s Hospital, and Partners Healthcare Systems / Massachusetts General Hospital, and the studies were performed in accordance with the ethical guidelines of the Declaration of Helsinki. All 10 CF patients in this study were homozygous for the F508del allele of the CFTR gene, and were categorized as having “severe” lung disease: they were in the lowest quintile for age of airway obstruction, as assessed by their median forced expiratory volume during the initial second of exhalation (for more details: (28)).
X-linked (gp91phox-deficient) and autosomal (p47phox-deficient) chronic granulomatous disease (CGD) patients and healthy control subjects were recruited under the National Institute of Allergy and Infectious Diseases Institutional Review Board-approved protocol NIH#05-I-0213, “Evaluation of Patients with Immune Function Abnormalities”. Human subjects provided written informed consent for participation. 10 ml blood was drawn from CGD patients and was processed in parallel with healthy volunteer’s blood. Two X-CGD patients (Patient #2 and 3) and one p47phox-deficient CGD patient participated in the study; all subjects were characterized in previous studies with regards to their genetic defects and residual NADPH oxidase activities.(33) Patient #2 (1.7 nmol/106 cells/hr, gp-146, 0.75%) and patient #3 (2.38 nmol/106 cells/hr (1.05%).(33) The p47phox−/− patient (p47-16) had NADPH oxidase activity of 2.34 nmol/106 cells/hr (1.04%) and superoxide production in healthy neutrophils was 226 nmol/106 cells/hr (100 %) (28, 33).
Human neutrophil isolation
Neutrophils were isolated as described previously (32). Briefly, whole blood was drawn at the Health Center of University of Georgia (Athens, GA, USA) or at the Department of Transfusion Medicine of the National Institutes of Health (Bethesda, MD, USA) and coagulation was prevented with heparin. Red blood cells were removed by Dextran sedimentation (GE Healthcare, UK). Leukocytes were layered on top of a 5-step Percoll gradient (65, 70, 75, 80 and 85%, Sigma, St. Louis, MO) and the 70–75–80% Percoll layers containing neutrophils were harvested. Cells were stored in a mix of autologous serum and RPMI-1640 medium (Life Technologies, Grand Island, NY) until use at room temperature. Trypan blue dye extrusion was used to determine cell viability (>99%). Purity of the neutrophil preparations was confirmed by cytospin. Serum was prepared from coagulated blood by centrifugation and sterile filtration. Calcium- and magnesium-containing HBSS (Mediatech, Manassas, VA) containing 1% autologous serum, 5 mM glucose and 10 mM HEPES was used as assay buffer.
Pseudomonas aeruginosa strains
The following Pseudomonas aeruginosa strains were used in this study: PA14 (kind gift from Dr. Frederick Ausubel, Massachussetts General Hospital, Boston, MA), PAO1 (purchased, ATCC), PA2192 and GFP-expressing PAO1 (kindly provided by Dr. Joanna Goldberg, Emory University, GA) and PA10145 (purchased, ATCC 10145). Cystic fibrosis isolates of Pseudomonas aeruginosa were obtained as described previously (28). Bacteria were cultured in LB broth overnight. Late exponential phase cultures were washed twice and suspended in HBSS. Bacterial density was set to 109/ml using OD=0.6 at 600 nm.
Quantification of neutrophil extracellular traps
NETs were quantified essentially as described with some modifications (32). Neutrophils were attached to poly D-lysine coated 96-well black transparent bottom plates (Thermo Scientific, Rochester, NY) in assay medium containing 10 μM Sytox Orange (Life Technologies, Grand Island, NY, USA). For inhibitor tests, cells were pretreated with specific inhibitors for 15 minutes prior to bacterial infection. Neutrophils were infected with 1 to 50 MOI Pseudomonas aeruginosa PA14 for dose-dependent extracellular DNA release assay or 10 MOI for inhibitor experiments. Fluorescence (excitation: 530 nm, emission: 590 nm) was measured in a fluorescence plate reader (Varioskan Ascent, Thermo Scientific) for up to 5 hours at 37 °C. Fluorescence of samples containing 500 μg/mL saponin (Sigma, St. Louis, MO) with neutrophils was used as the maximal signal for DNA release (100%). Rise in fluorescence was referred to as “extracellular DNA release” and expressed as relative fluorescence units (RFU) or normalized on saponin-treated as maximal signal (referred to as “% of max”). The following inhibitor concentrations were used: DPI (diphenylene iodonium, NADPH oxidase inhibitor, 10 μM, Sigma), U0126 (MEK1/2 inhibitor, 25 μM, Sigma), MEK162 (MEK1/2 inhibitor, 50 μM, Sigma), PD98059 (MEK1 inhibitor, 20 μM, Sigma) and cytochalasin-D (cytoskeleton inhibitor, 10 μM, Sigma).
Immunostaining and fluorescent microscopy
Neutrophils were seeded on 12 mm glass cover slips (VWR International, Radnor, PA) in 24-well plates (Thermo Scientific, Rochester, NY). PA14 (opsonized with 10% autologous serum, 37°C, 30min) was added to neutrophils and incubated for 3 hours at 37°C. After fixation with 4% paraformaldehyde (Affymetrix, Celeveland, OH) samples were blocked with 5% Normal Donkey serum (Jackson ImmunoResearch Laboratories INC, West Grove, PA) and 0.1% saponin (Sigma, St.Louis, MO) in PBS for 30 min at RT. The following antibodies were used: monoclonal mouse anti-human myeloperoxidase/FITC antibody (1:500, Dako, Clone MPO-7), polyclonal rabbit anti-histone H4 (citrulline 3) (1:1000, Millipore, Billerica, MA) and rabbit anti-human neutrophil elastase (1:1000) overnight at 4°C. For HNE and citH4 staining, Alexa Fluor 594-labelled donkey anti-rabbit secondary antibody was used for 1 hr (1:2000, Molecular Probes, Grand Island, NY). Samples were stained with DAPI (2min, RT, 1:20000, Molecular Probes, Grand Island, NY) and washed in PBS twice. Specimens mounted with ProLong Antifade Kit (Molecular Probes, Grand Island, NY) were analyzed with Zeiss AxioCam HRM fluorescence microscope (Axioplan2 imaging software).
Measurement of superoxide production
Superoxide release was assessed by Diogenes chemiluminescence superoxide-detection kit (National Diagnostics, Atlanta, GA). Adherent neutrophils were stimulated by Pseudomonas aeruginosa PA14 (50 MOI), PMA (100 nM) or were left unstimulated. Chemiluminescence was measured by Varioskan Flash luminescence microplate reader (Thermo Scientific, Logan, UT) for 90 min. Data are shown as kinetics of representative curves (relative luminescence units, RLU) or integral superoxide production values (int. RLU) obtained by multiplying accumulated luminescence for the entire duration of the measurement with the ratio of the measurement cycle length and integration time.
Supernatant collection for ELISA and enzyme activity measurements
Supernatants of neutrophils were used to measure secretion and activities of MPO and HNE. 100,000 neutrophils/well were seeded into 96-well poly-D-lysine coated transparent plates (Thermo Scientific, Rochester, NY), stimulated with 100 nM PMA or Pseudomonas aeruginosa PA14 (50 MOI), respectively. After 3hrs of incubation at 37°C in HBSS including 1% serum, cell supernatants were collected, centrifuged for 10 min at 10,000g to remove debris or bacteria and stored at −80 °C for further analysis or used immediately. Each supernatant was diluted 1:30 for HNE ELISA, 1:100 for MPO ELISA or left undiluted for MPO activity test.
MPO and HNE ELISA
Concentrations of human MPO in neutrophil supernatants were measured by commercial ELISA kit (R&D Systems, Minneapolis, MN). Serial dilutions prepared from MPO standard provided in the kit (stock: 125ng/ml) were used to quantify MPO concentrations of unknown samples. HNE release was assessed by ELISA. Supernatant samples diluted with coating buffer (25mM carbonate, 25mM bicarbonate, pH 9.6) were incubated overnight at 4°C in 96-well high binding microloan ELISA plates (Greiner bio-one, Germany). After blocking with 1% BSA for 1hr, anti-HNE rabbit polyclonal antibody (1:500 in PBS, Calbiochem, 481001, EMD Millipore, MA) was added for 2hrs at RT. After repeated washes, samples were incubated with horse radish peroxidase-linked donkey anti-rabbit antibody (1:2000 in PBS, NA934V, GE Healthcare, UK) for 1hr. Blue coloration developed in the presence of TMB (Thermo Scientific, Rockford, IL) peroxidase substrate solution. Reaction was stopped by adding 1N hydrochloric acid (Sigma, St. Louis, MO) and absorption was read at 450nm wavelength with Eon microplate photometer (BioTek, Winooski, VT). Purified HNE standard (Cell Sciences, Canton, MA; stock: 1mg/ml) was used to determine HNE concentrations in unknown samples.
Measurement of peroxidase activity
Peroxidase activity was measured by hydrogen peroxide-dependent oxidation of Amplex Red. 50 μl undiluted neutrophil supernatants were added to 96 well non-transparent black microplates (Costar, Corning, NY) and mixed with assay solution containing 100 μM Amplex Red (Molecular Probes INC, Eugene, OR) and 100 μM hydrogen peroxide (Sigma, St. Louis, MO). Production of the fluorescent product was measured with fluorescence plate reader (Varioskan Ascent, Thermo Scientific) after 30min at 560 nm excitation and 590nm emission wavelengths. Standards with known MPO concentrations (R&D Systems, stock: 125 ng/ml MPO) were used to determine peroxidase activities of unknown samples. Results are expressed as “equivalent ng/ml MPO activity”. This way of data presentation was chosen (instead of showing unit enzymatic activities per time and volume) to enable comparison between MPO concentration and activity in neutrophil supernatants.
Measurement of “NET-free” and “NET-linked” bacterial numbers
PA14 was added to human neutrophils (100.000/well) attached to poly D-lysine coated transparent 96-well plates (Thermo Scientific, Rochester, NY) in HBSS containing 1% autologous serum, 5mM glucose (Sigma, St. Louis, MO) and 10 mM HEPES (Cellgro, Tewksbury, MA). Bacteria were added at two different doses, PA14:PMN MOI=10:1 or 50:1. After 3 hrs supernatants were carefully collected and referred to as “NET-free” fraction. Absence of DNA in this fraction was confirmed by DNA gel electrophoresis. Equal volume of assay medium containing 30 μg/ml DNAse1 (Roche Applied Science, Indianapolis, IN) was added back to neutrophils and incubated for 15 min – this volume was referred to as “NET-linked” fraction. After centrifugation, concentration of living PA14 bacteria in each sample was determined by a microplate-based bacterial growth assay as described (34).
Statistical analysis
Results were analyzed by Student’s t-test or one-way ANOVA with Tukey post-test for multiple comparisons. Each experiment was independently performed at least three times with neutrophils isolated from different donors. Correlation was determined by computing the Pearson correlation coefficient (r). The significance of correlation was (two-tailed probability) determined using the correlation coefficient and the sample size (20) using an online statistical software (Statistics Calculators, version 3.0 Beta). Statistically significant differences were considered as *, p<0.05; **, p<0.01; ***, p<0.001).
Results
Human neutrophils release extracellular DNA in response to Pseudomonas aeruginosa
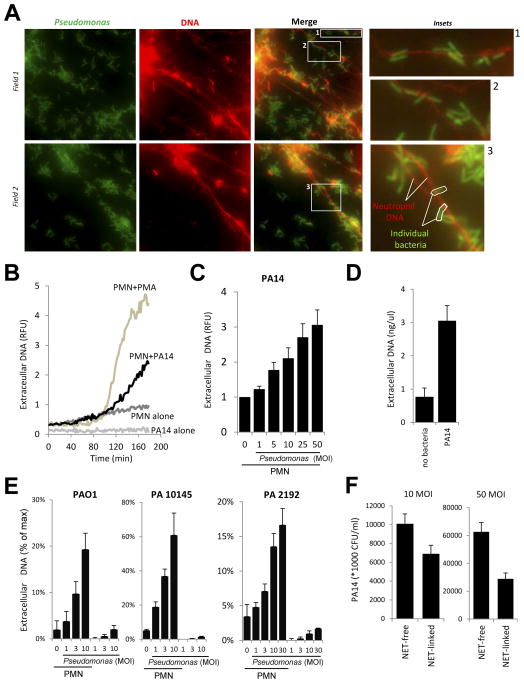
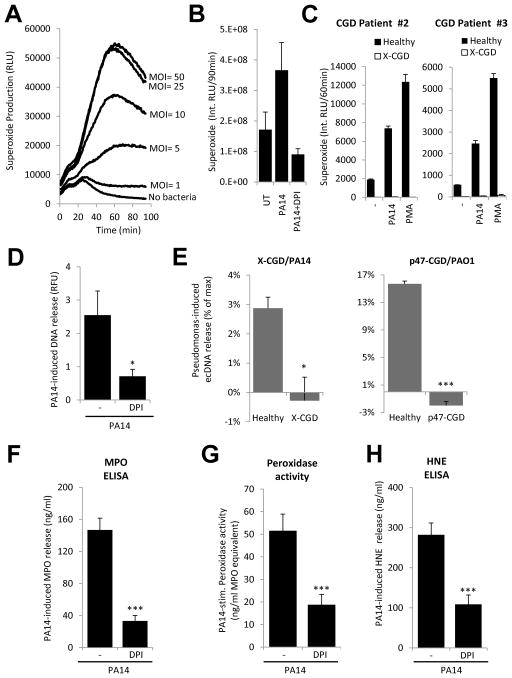
We show that human neutrophils release extracellular DNA in response to GFP-expressing Pseudomonas aeruginosa PAO1 (Fig. 1A). Bacteria are entrapped in NETs (Fig. 1A and insets). Neutrophils also release DNA in response to another Pseudomonas aeruginosa strain, PA14 (Fig. 1B–C). PA14-induced DNA release starts at the same time when induced by the positive control, phorbol myristate acetate (PMA) (Fig. 1B). Bacteria (PA14) alone do not release any DNA (Fig. 1B). DNA release from adherent human neutrophils is induced in a dose-dependent manner by Pseudomonas aeruginosa strain PA14 (Fig. 1B). Quantitation reveals an average release of 3.01 ng/μl DNA from 100.000 neutrophils upon PA14-exposure (Fig. 1D). PAO1 and two other Pseudomonas aeruginosa strains (PA10145, PA2192) also stimulate DNA release in a dose-dependent manner (Fig. 1E). We next tried to estimate the proportion of live bacteria attached to NETs after a 4hr incubation time. The supernatant of Pseudomonas-neutrophil suspensions was very carefully removed after incubation, did not contain NETs and was therefore termed “NET-free” (Fig. 1F). Absence of DNA in the NET-free fraction was confirmed by running the samples on DNA agarose gels (data not shown). An equal volume of new assay medium containing DNAse1 was added back and was referred to as “NET-linked”. As shown in Fig. 1F, we found that 41.3+/−3.4% of live bacteria (mean+/−S.D., n=3) were associated with NETs when a MOI of 10:1 was used and 29.7+/−13.6% (mean+/−S.D., n=3) with a MOI of 50:1. These measurements reveal that in vitro NETs are capable of trapping large amounts of Pseudomonas aeruginosa.
FIGURE 1. Pseudomonas aeruginosa induces extracellular DNA release in human neutrophils.
(A) Adherent neutrophils were incubated with GFP-expressing Pseudomonas aeruginosa PAO1 (3hrs). Sytox Orange extracellular DNA-binding dye was added and unfixed samples were imaged with a fluorescence microscope (one representative experiment, n=4). Original magnification ×1000 (insets, ×4000) 10 MOI. (B) Kinetics of Pseudomonas aeruginosa PA14-stimulated extracellular DNA release by neutrophils (Sytox Orange fluorescence) was compared with PA14 alone (50 MOI), neutrophils alone or the positive control PMA (100 nM). One representative result (n=3). (C) PA14 triggers dose-dependent extracellular DNA release in neutrophils (Sytox Orange fluorescence, 3hrs, normalized to unstimulated control as 100%). Mean±SEM, n=5. D) DNA release from neutrophils upon PA14 exposure (3 hrs) was quantitated using Sytox Orange and calf thymus DNA serial dilutions. n=3. E) Dose-dependent DNA release in human neutrophils exposed to Pseudomonas aeruginosa strains PAO1, PA 10145 and PA2192 (0–30 MOI, 3hrs, Sytox Orange fluorescence, mean+/−S.E.M., n=3). F) Numbers of NET-associated (NET-linked) and NET-independent (NET-free) living PA14 bacteria were assessed after 3 hrs of incubation with neutrophils at two different doses (10 and 50 MOI) according to details found in Methods section. Mean+/−S.E.M., n=3.
Pseudomonas-induced NETs contain citrullinated histone H4
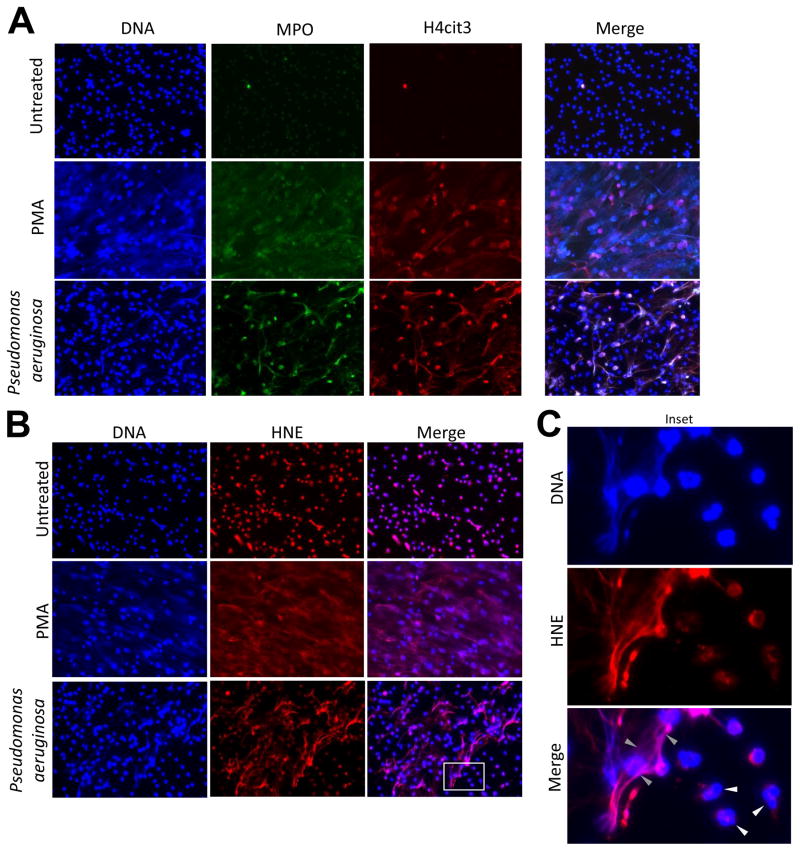
The signaling steps leading from stimulation to NET formation in human neutrophils have not been well defined, but citrullination of histones (H3 and H4) has been implicated and confirmed by several groups (35–37). Citrullinated histone H3 was detected in CF sputum samples by immunofluorescence (23). Here we show that histone H4 is citrullinated on amino acid residue 3 and co-localizes with extracellular DNA after Pseudomonas exposure (Fig. 2A).
Figure 2. NETs stimulated by Pseudomonas aeruginosa contain citrullinated histone H4, myeloperoxidase and human neutrophil elastase.
A) MPO (green) and H4cit3 (red) co-localize with extracellular DNA in PA14-induced NETs. Adherent human neutrophils were stimulated by PA14 (50:1 MOI), 100 nM PMA or were left unstimulated (3hrs). Neutrophils were fixed and stained with anti-MPO-FITC antibody or anti-H4cit3 primary and donkey anti-rabbit 594 secondary antibodies. DNA was counterstained by DAPI. One representative experiment, n=5. Original magnification ×400. B) Localization of HNE in PA14- or PMA-stimulated human neutrophils was detected by immunofluorescence (rabbit anti-HNE primary antibody followed by donkey anti-rabbit 594-linked secondary antibody). Both intracellular and extracellular DNA was stained with DAPI. One representative measurement, n=4. Original magnification ×400. C) Inset of the HNE immunostaining (B) reveals fine details of HNE localization in NETs after PA14 exposure (gray arrowheads). As expected, HNE did not overlap with intact nuclear DNA within resting neutrophils (white arrowheads). Representative images, n=5. Original magnification ×2000.
Myeloperoxidase is associated with Pseudomonas-triggered extracellular DNA traps
MPO is stored in azurophilic granules in neutrophils, becomes associated with extracellular DNA in NETs and is also implicated in the progress of NET formation (19, 20). Our indirect immunofluorescence analysis detects co-localization of MPO with extracellular DNA and citrullinated histone H4 in Pseudomonas-stimulated NETs (Fig. 2A). These data prove that MPO released from neutrophils in presence of Pseudomonas aeruginosa is associated with NETs.
Neutrophil elastase is bound to NETs after Pseudomonas challenge
HNE is a major inflammatory marker of CF airways (14–16, 38). HNE is associated with extracellular DNA in NETs stimulated in vitro by PMA and in CF sputum samples (19, 24). While HNE is stored in primary granules in resting neutrophils, it becomes associated with extracellular DNA after stimulation with Pseudomonas aeruginosa PA14 (Fig. 2B, C). Thus, the main CF airway pathogen Pseudomonas aeruginosa activates human neutrophils to release NETs that contain extracellular DNA, HNE, MPO and cit3H4.
MPO and HNE are released after neutrophil exposure to Pseudomonas aeruginosa
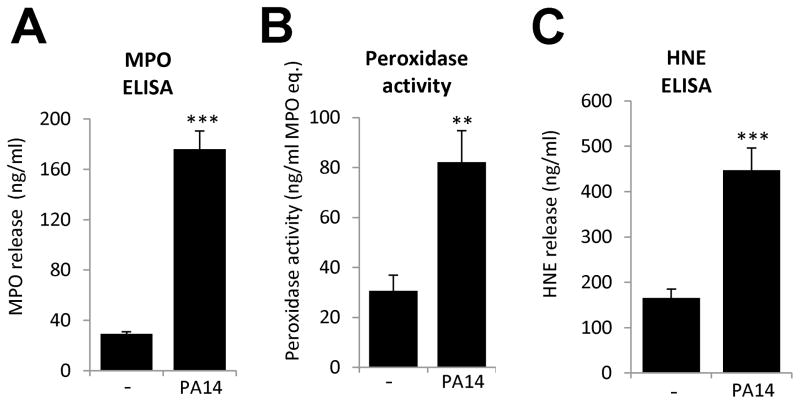
To confirm our immunofluorescence data, we measured MPO and HNE release in supernatants of Pseudomonas-stimulated human neutrophils by ELISA. Throughout the entire manuscript we measured total MPO or HNE concentrations and MPO activity in our samples. We did not distinguish between NET-associated and NET-free proteins. Quantitation of MPO release by ELISA detected 176.0 +/− 14.5 ng/ml MPO in neutrophil supernatants after Pseudomonas (PA-14) stimulation and only 29.2+/−1.7 ng/ml with basal release (mean+/−S.E.M., n=8) (Fig. 3A). We also observed an increased peroxidase activity in supernatants of Pseudomonas-activated neutrophils (basal: 28.9+/−6.0, bacterial stimulus: 77.7+/−11.9 (ng/ml MPO activity, mean+/−S.E.M., n=8) (Fig. 3B). A similarly large increase was observed in HNE release upon exposure to PA14 (Fig. 3C). Background release of HNE (165.1+/−19.8 ng/ml) was enhanced 2.7-fold by bacterial challenge (447.2+/−40.4 ng/ml) (mean+/−S.E.M., n=8). We did not detect any significant peroxidase activity with bacteria alone (data not shown).
Figure 3. MPO and HNE are detected in neutrophil supernatants after Pseudomonas aeruginosa challenge.
Adherent human neutrophils were exposed to Pseudomonas aeruginosa PA14 (50 MOI) for 2.5 to 3 hrs. Cell supernatants were assayed for MPO release, HNE release and peroxidase activity according to the Materials and Methods. (A) PA14-induced MPO release in neutrophils measured by ELISA. Mean+/−S.E.M., n=8. (B) Pseudomonas increases extracellular peroxidase activity in neutrophils. Peroxidase activity was determined by hydrogen peroxide-dependent Amplex Red oxidation in neutrophil supernatants. Mean+/−S.E.M., n=8. (C) HNE concentrations were measured by ELISA in Pseudomonas-exposed neutrophil supernatants. Mean+/−S.E.M., n=8. Mean+/−S.E.M., n=8. Significance was evaluated by student’s two tailed T-test (**P<0.01, ***P<0.001)
Human neutrophils release DNA, MPO and HNE in presence of CF isolates of Pseudomonas aeruginosa
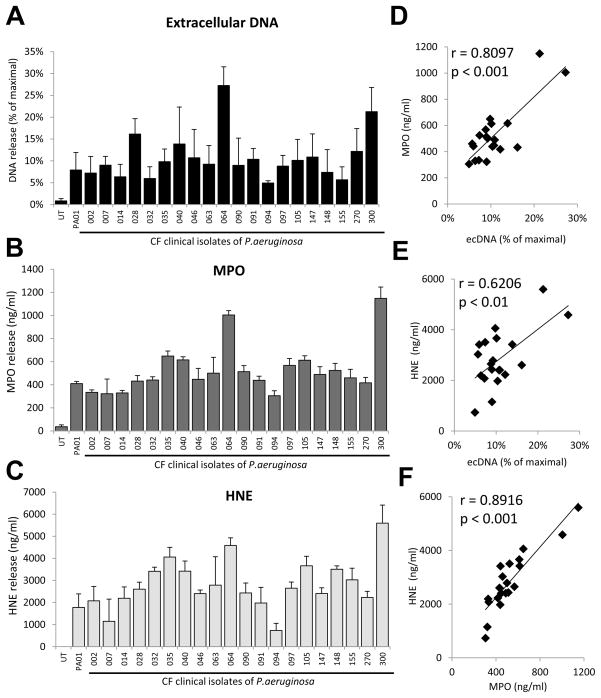
To confirm that our previous findings are not limited to Pseudomonas laboratory strains, we exposed human neutrophils to CF clinical isolates of P. aeruginosa and measured DNA, MPO and HNE releases. All clinical isolates induced release of extracellular DNA (Fig. 4A), MPO (Fig. 4B) and HNE (Fig. 4C) at comparable levels than PAO1. This confirms that clinical isolates behave similarly to the laboratory strains and that PAO1 and PA14 are good models to study Pseudomonas-initiated neutrophil activation. DNA, MPO and HNE release data show similar patterns among isolates. To determine the extent of correlation among the parameters we measured, we plotted ecDNA, MPO, and HNE release of clinical isolates against each other (Fig. 4 D–F). We calculated the Pearson correlation coefficients (r) to measure the strength of a linear association and the significance between any of these two variables. MPO vs. HNE: r= 0.89157226975503, p= 0.00000013; MPO vs. DNA: r= 0.80978489595945, p= 0.00001517; HNE vs. DNA: r= 0.62058387267863, p= 0.00350493. Correlations in each combination were determined to be significant.
Figure 4. Cystic fibrosis isolates of Pseudomonas aeruginosa stimulate release of DNA, MPO and HNE from human neutrophils.
Attached neutrophils were incubated with twenty P. aeruginosa CF isolates (10 MOI, 3hrs). Supernatants were assayed for release of extracellular DNA (A), MPO (B) and HNE (C). DNA release was measured by Sytox Orange fluorescence. MPO and HNE releases were quantitated by ELISA. Mean+/−S.E.M., n=4. D) Plotting MPO data against extracellular DNA data obtained in A–C. E) Plotting HNE data against extracellular DNA data obtained in A–C. F) Plotting MPO data against HNE data obtained in A–C. In D–F each dot represents a CF isolate of P. aeruginosa. Pearson correlation coefficient (r).
The neutrophil respiratory burst is activated by Pseudomonas aeruginosa
NADPH oxidase activity is required for NET formation triggered by bacteria and PMA (39, 40). The requirement of neutrophil respiratory burst activity for Pseudomonas-induced NETs has not been studied. First, we wanted to see whether adherent human neutrophils respond to Pseudomonas aeruginosa with superoxide production. Neutrophils stimulated by PA14 released large amounts of superoxide that was dependent on the dose of bacterial challenge (Fig. 5A). As expected, the flavoenzyme inhibitor diphenylene iodonium (DPI) diminished the Pseudomonas-initiated respiratory burst (Fig. 5B). We also exposed neutrophils of X-linked chronic granulomatous diseases (CGD) patients to PA14 and measured the chemiluminescence signal. CGD neutrophils were entirely unresponsive to the bacterium, whereas oxidase-competent control neutrophils responded with large respiratory burst activity (Fig. 5C). Thus, human neutrophils respond to P. aeruginosa with oxidative activity.
Figure 5. The NADPH oxidase is required for Pseudomonas-stimulated NET formation, MPO and HNE release.
Respiratory burst activity of neutrophils was measured by Diogenes luminescence superoxide assay. (A) Pseudomonas aeruginosa PA14 induces superoxide production in adherent human neutrophils in a dose-dependent manner (1 to 50 MOI). Kinetics are shown, chemiluminescence was measured for 90 min. One representative measurement, n=4. (B) PA14-triggered NADPH oxidase activity is blocked by DPI (10μM). Inhibitor was added 15 min prior to stimulation. PA14:neutrophil MOI=10. Integrated chemiluminescence for 90 min is shown. Mean+/−S.E.M., n=4. (C) X-CGD neutrophils do not respond with superoxide production to Pseudomonas aeruginosa PA14 or PMA (100 nM). Experimental data obtained on both patients are shown. Mean+/−S.D. D) Extracellular DNA release (increase in Sytox Orange fluorescence, 3 hrs) from human neutrophils induced by Pseudomonas aeruginosa PA14 (10 MOI) is inhibited by DPI (10 μM). Mean+/−S.E.M., n=5 (E) CGD neutrophils (X-CGD, left; p47-deficient, right) do not release DNA in response to Pseudomonas aeruginosa (PA14 or PAO1). One experiment/CGD donor, assays were performed in triplicates. Mean+/−S.D. Pseudomonas aeruginosa PA14-induced MPO release (F), peroxidase activity (G) and HNE secretion (H) were inhibited by 10 μM DPI. Mean +/−S.E.M., n=8. Significance was evaluated by student’s two tailed T-test (*P<0.05, ***P<0.001)
Reactive oxygen species are required for neutrophil activation by Pseudomonas aeruginosa
We then examined whether treating neutrophils with the NADPH oxidase inhibitor DPI before Pseudomonas challenge would affect extracellular DNA release and observed a significant reduction by prior DPI-treatment (Fig. 5D). Neutrophils from both a gp91phox- and a p47phox-deficient patient remained unresponsive to PAO1 or PA14, further confirming the requirement for a functional NADPH oxidase in Pseudomonas-induced NET formation (Fig. 5E). DPI pretreatment also greatly reduced Pseudomonas-induced total MPO release (Fig. 5F) and extracellular peroxidase activity (Fig. 5G). PA14-triggered MPO release (146.8+/−14.8 ng/ml) was reduced by up to 77.5% by DPI (33.2+/−7.0 ng/ml) (mean+/−.S.E.M., n=8) (Fig. 5F). PA14-stimulated extracellular peroxidase activity (243.6+/−34.7) was inhibited by up to 63.6% by NADPH oxidase inhibition (88.6+/−21.6) (RFU, mean+/−S.E.M., n=8) (Fig. 5G). PA14-triggered secretion of total HNE (282.1+/−29.4 ng/ml) was also reduced by 61.5% following DPI pretreatment (108.7+/−23.2 ng/ml) (mean+/−S.E.M., n=8) (Fig. 5H). Our results demonstrate that NADPH oxidase mediates Pseudomonas-induced NET formation and this process is responsible for the majority of MPO and HNE release from neutrophils upon Pseudomonas challenge.
Pseudomonas-stimulated respiratory burst and DNA release requires an intact cytoskeleton
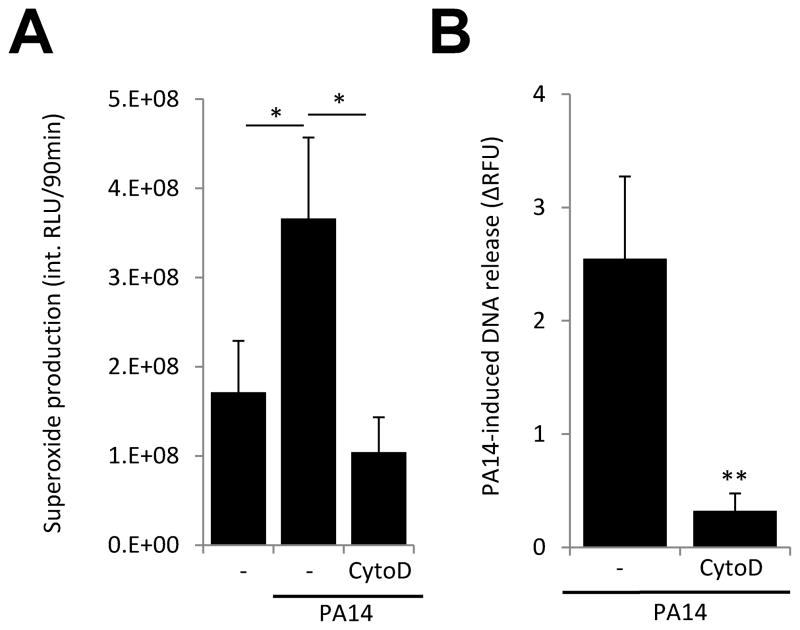
Human neutrophils phagocytose bacteria; however, the extent to which this process is required to Pseudomonas-activated NET formation is unknown. We stimulated human neutrophils with P. aeruginosa PA14 in presence of the cytoskeleton inhibitor cytochalasin-D and measured superoxide production and NET formation. Cytochalasin-D treatment blocked both the Pseudomonas-triggered respiratory burst (Fig. 6A) and DNA release (Fig. 6B).
Figure 6. Cytochalasin-D inhibits Pseudomonas-stimulated respiratory burst and DNA release.
A) Human neutrophils were stimulated with PA14 (10 MOI, 3 hrs) in the presence or absence of 10 μM cytochalasin-D. Superoxide production was measured with Diogenes superoxide-specific chemiluminescence kit. Mean+/−S.E.M., n=3. ANOVA, Tukey’s post-hoc analysis. B) Extracellular DNA release (Sytox Orange fluorescence) was measured in neutrophils exposed to PA14 (10 MOI, 3 hrs) in presence of 10 μM cytochalasin-D. PA14-stimulated values are shown after subtraction of background activity. Values are expressed as % of maximal DNA release. Mean+/−S.E.M., n=3. Student’s t-test, *P<0.05, **P<0.01.
Role of MEK/ERK signaling in Pseudomonas-triggered NET formation
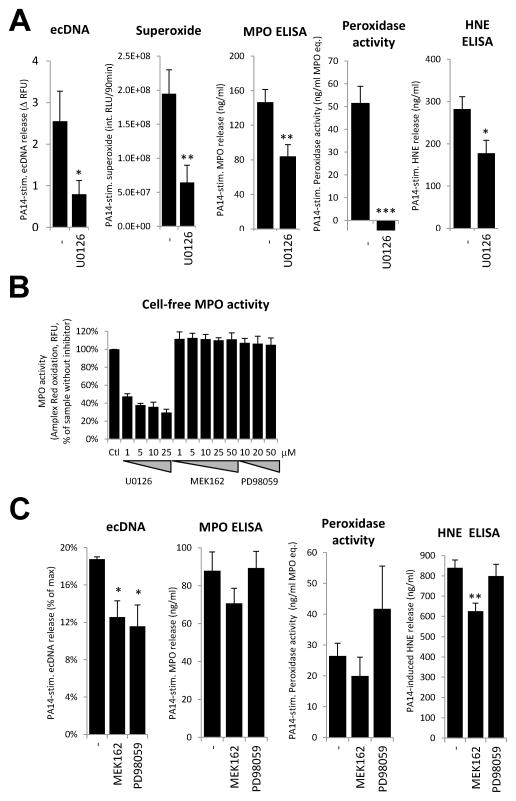
Activation of the MEK/ERK signaling pathway has been implicated in PMA-stimulated NET formation (41, 42). We tested the effect of U0126, a compound with known inhibitory effect on MEK1/2, on Pseudomonas-triggered neutrophil activation. U0126 reduced PA14-induced extracellular DNA release and caused a 67.0% drop in PA14-stimulated superoxide production (Fig. 7A). U0126 had an inhibitory action of 42.6% on Pseudomonas-triggered MPO release, but entirely blocked peroxidase activity in the supernatant (Fig. 7A). The fact that U0126 blocked MPO activity but had only moderate-medium inhibitory action on the other parameters raised concerns that U0126 can interfere with MPO activity (Fig. 7A). We therefore tested U0126 and two other, well-characterized MEK inhibitors (MEK162 and PD98059) in a cell-free system measuring MPO activity. Surprisingly we found that U0126 strongly inhibited in vitro MPO function whereas MEK162 and PD98059 were without effect (Fig. 7B).
Figure 7. Minimal contribution of MEK/ERK signaling to Pseudomonas-induced DNA release.
(A) Human neutrophils were exposed to PA14 (50 MOI) for 3hrs in the presence or absence of U0126 (25 μM) and the following parameters were measured according to the detailed descriptions found in the Methods section: extracellular DNA release (Sytox Orange, n=5), superoxide production (Diogenes, n=4), MPO release (ELISA, n=8), peroxidase activity (Amplex Red, n=8) and HNE release (ELISA, n=8). Mean+/−S.E.M. Student’s t-test (B) Enzymatic activity of 500 μM MPO (Amplex Red oxidation) was measured in a cell-free system in the presence of indicated concentrations of U0126, MEK-162 and PD98059. Results are normalized on the inhibitor-free sample (mean+/−S.D., n=2). (C) Neutrophils were exposed to PA14 (50 MOI) for 3hrs in the presence or absence of MEK-162 (50 μM) or PD98059 (20 μM) and the following parameters were measured: extracellular DNA release (Sytox Orange, n=4), MPO release (ELISA, n=4), peroxidase activity (Amplex Red, n=4) and HNE release (ELISA, n=4). Mean+/−S.E.M. ANOVA, Tukey’s post-hoc analysis. *P<0.05, **P<0.01, ***P<0.001.
We next studied the effects of MEK162 and PD98059 on neutrophils. Both inhibitors had minor but significant inhibition of Pseudomonas-triggered extracellular DNA release: MEK162 inhibited 33% of DNA release (n=4), and PD98059 inhibited 38% of DNA release (n=4) (Fig. 7C, first panel). MEK162 and PD98059 did not influence release or activity of MPO (Fig. 7C). MEK162 (but not PD98059) mildly but significantly reduced HNE release (Fig. 7C). These data confirm that whereas MEK/ERK signaling has minor contribution to ecDNA secretion, it does not generally influence release of MPO and HNE.
Discussion
Neutrophil airway recruitment and activation in CF is poorly understood and has not been targeted therapeutically. In this study we explored details of neutrophil responses to the main CF airway pathogen, P. aeruginosa. This mechanism is of high importance to understand CF lung inflammation because both the presence of P. aeruginosa and neutrophil activation have been strongly linked to diminished lung function in CF (14–18, 26, 38, 43–45).
We found that the main effector response of neutrophils in response to PA14 in vitro is NET formation (Fig. 1–3). The backbone of NETs is extracellular DNA (19). DNA has been detected in CF airways, and its presence correlates with accelerated decline of lung function in CF patients.(16, 22) Moreover, DNAse treatment is routinely used to ease CF respiratory symptoms (16, 22, 46, 47). Our data show that neutrophils stimulated by different P. aeruginosa strains (PA14, PAO1, PA10145 and PA2192) release large amounts of DNA (Fig. 1). This is consistent with data obtained by several other groups (25, 27, 28, 48–50). Our data allow comparison of different bacterial strains used in Pseudomonas research and found that PA14 and PA10145 are the strongest NET inducers, whereas PAO1 and PA 2192 release NETs to a lesser extent (Fig. 1). Similarly, CF isolates show a wide variation in NET induction: most strains stimulate DNA release at levels comparable to that of PAO1, while a few (#28, #64, #300) provoke marked responses from human neutrophils (Fig. 4A). Although our data used human neutrophils isolated from peripheral blood of normal volunteers, these data confirm that P. aeruginosa is a strong inducer of DNA release from neutrophils and suggest that P. aeruginosa is likely to stimulate NET formation in CF airways.
Neutrophil azurophilic granule components MPO and HNE are found in large amounts in CF airways and contribute to tissue destruction through oxidative stress and proteolytic damage (14, 16, 18). Recently, HNE was found in the airways of CF infants, where its presence was associated with persistent bronchiectasis (38). To define the mechanism of DNA release as NET formation, co-localization of neutrophil granule proteins (HNE, MPO) and histones with DNA is required (19, 40). Citrullination of histones (H3, H4) by peptidyl arginine deiminase 4 (PAD4) is required for in vitro and in vivo NET release (35–37, 51). Citrullinated histone H3 has previously been detected in CF sputum samples (23). Pseudomonas-stimulated NETs contain citrullinated histone H4 (citH4), MPO, and HNE that co-localize with extracellular DNA, as detected by immunofluorescence (Fig. 2). Our data show that when neutrophils are in presence of Pseudomonas in vitro, DNA release occurs mainly through NET formation. Physical association of DNA, MPO, and HNE was found previously in CF sputum samples (23, 24). Absolute quantitation of total MPO release detected by ELISA versus enzymatic activity data suggests that about 45–50% of released MPO is enzymatically active after Pseudomonas challenge (Fig. 3, 5 F–H, 7 A,C). MPO has been shown to remain active in its NET-bound state (52).
Our survey of CF isolates of P. aeruginosa shows for the first time that these clinical strains stimulate release of extracellular DNA, MPO, and HNE from human neutrophils (Fig. 4). Previously, only NET-mediated killing of CF isolates of P. aeruginosa was measured (27). Linear correlation between extracellular DNA, MPO, and HNE indicate that their release is not independent but coordinated, through mechanisms such as NET formation (Fig. 4 D–F). All clinical isolates tested trigger DNA/MPO/HNE release, but to different extent. Future studies are required to determine whether the extent of NET induction by the clinical isolates correlates with clinical measures of the CF patients from whom they were isolated.
Oxidative stress has been implicated in the pathogenesis of CF (53). In the inflammatory CF lung, neutrophils are the main source of released reactive oxygen species (ROS). P. aeruginosa stimulates a robust neutrophil respiratory burst that is required for DNA release and contributes to MPO and HNE release (Fig. 5). Our data confirm previous findings that DPI inhibits extracellular DNA release from neutrophils stimulated by a clinical isolate of P. aeruginosa of unknown origin (49). Beyond producing tissue-damaging ROS, neutrophil NADPH oxidase is required for maximal release of DNA, MPO and HNE after Pseudomonas challenge. This suggests that NADPH oxidase-dependent NET formation stimulated by Pseudomonas occurs in CF airways. The need for NADPH oxidase to mediate NET formation could depend on the nature of the stimuli (54). The observation that DPI blocks DNA/NET release in Pseudomonas-treated neutrophils suggests that DPI treatment can distinguish between NET-dependent and NET-independent release mechanisms (Fig. 5). Incubation with DPI inhibited a major part of both MPO and HNE release (Fig. 5F–I), suggesting that NETs are the main mechanism of neutrophil inflammatory marker release.
Our previous data indicate that NET formation triggered by calcium pyrophosphate dihydrate crystals requires an intact cytoskeleton (32). Similarly, bacteria represent microscopic particles that neutrophils routinely phagocytose. The observation that cytochalasin-D inhibits Pseudomonas-stimulated superoxide production and NET formation further suggests that NET release requires an intact cytoskeleton (Fig. 6). Cytochalasin-B treatment has a similar effect on Staphylococcus aureus-triggered NET formation (55). It is not clear whether or not phagocytosis of microbes is required or not for microbe-stimulated NET formation. One school of thoughts argues that neutrophils kill microbes either by phagocytosis (and consequent intracellular killing) or by extracellular NET formation, but not using both mechanisms (56).
By studying the effect of the MEK/ERK pathway in Pseudomonas-triggered NET formation, to our surprise we found that U0126 was very efficient at blocking neutrophil effector functions, whereas MEK162 and PD98059 were not (Fig. 7). The fact that U0126 is an efficient blocker of MPO activity could explain this difference. This raises concerns about using U0126 as a MEK inhibitor in neutrophil studies since it clearly blocks MPO activation as well. U0126 has been previously used to implicate the MEK-ERK signaling pathway in PMA- or Helicobacter pylori-stimulated NET formation (41). On the other hand, this supports that MPO activity is required for NET formation stimulated by PMA but also by P. aeruginosa (55, 57). The requirement of MPO for Pseudomonas-induced NETs is, however, controversial since other studies found that MPO-deficient human neutrophils or murine neutrophils don’t have impaired NET release.(48, 49) The mechanism of inhibitory action of U0126 on MPO function is not known. Its chemical structure (four primary amines and two disulfide bonds) suggests that U0126 could be an excellent scavenger of hypochlorous ions (55, 58). MEK162- and PD98059-mediated blockade of the MEK-ERK pathway decreased Pseudomonas-stimulated DNA release but had no significant effect on MPO or HNE activities (except for MEK162 on HNE ELISA) (Fig. 7). This suggests that the MEK/ERK pathway contributes to Pseudomonas-induced NET release to a similar extent as in PMA induction (41). However, this minor effect on NET inhibition does not appear to contribute significantly to MPO or HNE release, as NET formation is not their only release mechanism.
P. aeruginosa acquires resistance to NET-mediated killing in CF airways (27). Clinical data clearly indicate that neutrophils are unable to remove Pseudomonas effectively from the CF lung, as both NET-mediated and classical intracellular killing mechanisms fail. Neutrophils instead release their dangerous granule contents into the airway lumen. Although the CF airway environment is very complex and multiple mechanisms can be responsible for neutrophil dysfunction, our detailed in vitro characterization of Pseudomonas-induced NET formation (DNA associated with active MPO and HNE) presents a likely mechanism for excess inflammatory mediator release from neutrophils in CF. Our study helps to understand neutrophil dysfunction, a clinically relevant mechanism of CF airway disease.
Acknowledgments
We would like to thank the staff of the UGA Health Center: Angela Standridge and Vickie Cromer for phlebomoty; Adam Davis, Houston Taylor and Dr. Ronald L. Forehand for their continuous support. We thank Sandra Anaya-O’Brien for CGD patient recruitment and Dr. Douglas Kuhns for archived patient data (NIH, NIAID). We are grateful to Drs. Brian Condie and Kristina Buac (Department of Genetics, UGA) for providing access to and help with the AxioZeiss fluorescence microscope. We also thank Dr. Joanna B. Goldberg (Emory University, Atlanta, GA) for providing the GFP-expressing PAO1 and the PA2192 strains.
This work was supported by startup funds provided to B. Rada from the Office of the Vice President for Research, University of Georgia) and by the Intramural Research Program of the NIH, National Institute of Allergy and Infectious Diseases.
References
- 1.Kim Chiaw P, Eckford PD, Bear CE. Insights into the mechanisms underlying CFTR channel activity, the molecular basis for cystic fibrosis and strategies for therapy. Essays in biochemistry. 2011;50:233–248. doi: 10.1042/bse0500233. [DOI] [PubMed] [Google Scholar]
- 2.Ciofu O, Hansen CR, Hoiby N. Respiratory bacterial infections in cystic fibrosis. Current opinion in pulmonary medicine. 2013;19:251–258. doi: 10.1097/MCP.0b013e32835f1afc. [DOI] [PubMed] [Google Scholar]
- 3.Cohen TS, Prince A. Cystic fibrosis: a mucosal immunodeficiency syndrome. Nature medicine. 2012;18:509–519. doi: 10.1038/nm.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Foweraker J. Recent advances in the microbiology of respiratory tract infection in cystic fibrosis. British medical bulletin. 2009;89:93–110. doi: 10.1093/bmb/ldn050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.West SE, Zeng L, Lee BL, Kosorok MR, Laxova A, Rock MJ, Splaingard MJ, Farrell PM. Respiratory infections with Pseudomonas aeruginosa in children with cystic fibrosis: early detection by serology and assessment of risk factors. JAMA : the journal of the American Medical Association. 2002;287:2958–2967. doi: 10.1001/jama.287.22.2958. [DOI] [PubMed] [Google Scholar]
- 6.Gilligan PH. Microbiology of airway disease in patients with cystic fibrosis. Clinical microbiology reviews. 1991;4:35–51. doi: 10.1128/cmr.4.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ratjen F. Treatment of early Pseudomonas aeruginosa infection in patients with cystic fibrosis. Current opinion in pulmonary medicine. 2006;12:428–432. doi: 10.1097/01.mcp.0000245712.51514.a1. [DOI] [PubMed] [Google Scholar]
- 8.Gaspar MC, Couet W, Olivier JC, Pais AA, Sousa JJ. Pseudomonas aeruginosa infection in cystic fibrosis lung disease and new perspectives of treatment: a review. European journal of clinical microbiology & infectious diseases : official publication of the European Society of Clinical Microbiology. 2013;32:1231–1252. doi: 10.1007/s10096-013-1876-y. [DOI] [PubMed] [Google Scholar]
- 9.Pisi G, Chetta A. Airway clearance therapy in cystic fibrosis patients. Acta bio-medica : Atenei Parmensis. 2009;80:102–106. [PubMed] [Google Scholar]
- 10.Cohen-Cymberknoh M, Shoseyov D, Kerem E. Managing cystic fibrosis: strategies that increase life expectancy and improve quality of life. American journal of respiratory and critical care medicine. 2011;183:1463–1471. doi: 10.1164/rccm.201009-1478CI. [DOI] [PubMed] [Google Scholar]
- 11.Accurso FJ, Rowe SM, Clancy JP, Boyle MP, Dunitz JM, Durie PR, Sagel SD, Hornick DB, Konstan MW, Donaldson SH, Moss RB, Pilewski JM, Rubenstein RC, Uluer AZ, Aitken ML, Freedman SD, Rose LM, Mayer-Hamblett N, Dong Q, Zha J, Stone AJ, Olson ER, Ordonez CL, Campbell PW, Ashlock MA, Ramsey BW. Effect of VX-770 in persons with cystic fibrosis and the G551D-CFTR mutation. The New England journal of medicine. 2010;363:1991–2003. doi: 10.1056/NEJMoa0909825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu H, Burton B, Huang CJ, Worley J, Cao D, Johnson JP, Jr, Urrutia A, Joubran J, Seepersaud S, Sussky K, Hoffman BJ, Van Goor F. Ivacaftor potentiation of multiple CFTR channels with gating mutations. Journal of cystic fibrosis : official journal of the European Cystic Fibrosis Society. 2012;11:237–245. doi: 10.1016/j.jcf.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 13.Ramsey BW, Davies J, McElvaney NG, Tullis E, Bell SC, Drevinek P, Griese M, McKone EF, Wainwright CE, Konstan MW, Moss R, Ratjen F, Sermet-Gaudelus I, Rowe SM, Dong Q, Rodriguez S, Yen K, Ordonez C, Elborn JS, Group VXS. A CFTR potentiator in patients with cystic fibrosis and the G551D mutation. The New England journal of medicine. 2011;365:1663–1672. doi: 10.1056/NEJMoa1105185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sagel SD, Wagner BD, Anthony MM, Emmett P, Zemanick ET. Sputum biomarkers of inflammation and lung function decline in children with cystic fibrosis. American journal of respiratory and critical care medicine. 2012;186:857–865. doi: 10.1164/rccm.201203-0507OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mayer-Hamblett N, Aitken ML, Accurso FJ, Kronmal RA, Konstan MW, Burns JL, Sagel SD, Ramsey BW. Association between pulmonary function and sputum biomarkers in cystic fibrosis. American journal of respiratory and critical care medicine. 2007;175:822–828. doi: 10.1164/rccm.200609-1354OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim JS, Okamoto K, Rubin BK. Pulmonary function is negatively correlated with sputum inflammatory markers and cough clearability in subjects with cystic fibrosis but not those with chronic bronchitis. Chest. 2006;129:1148–1154. doi: 10.1378/chest.129.5.1148. [DOI] [PubMed] [Google Scholar]
- 17.Regelmann WE, Siefferman CM, Herron JM, Elliott GR, Clawson CC, Gray BH. Sputum peroxidase activity correlates with the severity of lung disease in cystic fibrosis. Pediatric pulmonology. 1995;19:1–9. doi: 10.1002/ppul.1950190102. [DOI] [PubMed] [Google Scholar]
- 18.Garner HP, Phillips JR, Herron JG, Severson SJ, Milla CE, Regelmann WE. Peroxidase activity within circulating neutrophils correlates with pulmonary phenotype in cystic fibrosis. The Journal of laboratory and clinical medicine. 2004;144:127–133. doi: 10.1016/j.lab.2004.04.010. [DOI] [PubMed] [Google Scholar]
- 19.Brinkmann V, Reichard U, Goosmann C, Fauler B, Uhlemann Y, Weiss DS, Weinrauch Y, Zychlinsky A. Neutrophil extracellular traps kill bacteria. Science. 2004;303:1532–1535. doi: 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- 20.Papayannopoulos V, Metzler KD, Hakkim A, Zychlinsky A. Neutrophil elastase and myeloperoxidase regulate the formation of neutrophil extracellular traps. J Cell Biol. 2010;191:677–691. doi: 10.1083/jcb.201006052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brinkmann V, Laube B, Abu Abed U, Goosmann C, Zychlinsky A. Neutrophil extracellular traps: how to generate and visualize them. J Vis Exp. 2010 doi: 10.3791/1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carswell F, Robinson DW, Ward CC, Waterfield MR. Deoxyribonucleic acid output in the sputum from cystic fibrosis patients. European journal of respiratory diseases. 1984;65:53–57. [PubMed] [Google Scholar]
- 23.Manzenreiter R, Kienberger F, Marcos V, Schilcher K, Krautgartner WD, Obermayer A, Huml M, Stoiber W, Hector A, Griese M, Hannig M, Studnicka M, Vitkov L, Hartl D. Ultrastructural characterization of cystic fibrosis sputum using atomic force and scanning electron microscopy. Journal of cystic fibrosis : official journal of the European Cystic Fibrosis Society. 2012;11:84–92. doi: 10.1016/j.jcf.2011.09.008. [DOI] [PubMed] [Google Scholar]
- 24.Papayannopoulos V, Staab D, Zychlinsky A. Neutrophil elastase enhances sputum solubilization in cystic fibrosis patients receiving DNase therapy. PloS one. 2011;6:e28526. doi: 10.1371/journal.pone.0028526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dubois AV, Gauthier A, Brea D, Varaigne F, Diot P, Gauthier F, Attucci S. Influence of DNA on the activities and inhibition of neutrophil serine proteases in cystic fibrosis sputum. Am J Respir Cell Mol Biol. 2012;47:80–86. doi: 10.1165/rcmb.2011-0380OC. [DOI] [PubMed] [Google Scholar]
- 26.Hunter RC, Klepac-Ceraj V, Lorenzi MM, Grotzinger H, Martin TR, Newman DK. Phenazine content in the cystic fibrosis respiratory tract negatively correlates with lung function and microbial complexity. American journal of respiratory cell and molecular biology. 2012;47:738–745. doi: 10.1165/rcmb.2012-0088OC. [DOI] [PubMed] [Google Scholar]
- 27.Young RL, Malcolm KC, Kret JE, Caceres SM, Poch KR, Nichols DP, Taylor-Cousar JL, Saavedra MT, Randell SH, Vasil ML, Burns JL, Moskowitz SM, Nick JA. Neutrophil extracellular trap (NET)-mediated killing of Pseudomonas aeruginosa: evidence of acquired resistance within the CF airway, independent of CFTR. PLoS One. 2011;6:e23637. doi: 10.1371/journal.pone.0023637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rada B, Jendrysik MA, Pang L, Hayes CP, Yoo DG, Park JJ, Moskowitz SM, Malech HL, Leto TL. Pyocyanin-enhanced neutrophil extracellular trap formation requires the NADPH oxidase. PLoS One. 2013;8:e54205. doi: 10.1371/journal.pone.0054205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilson R, Sykes DA, Watson D, Rutman A, Taylor GW, Cole PJ. Measurement of Pseudomonas aeruginosa phenazine pigments in sputum and assessment of their contribution to sputum sol toxicity for respiratory epithelium. Infection and immunity. 1988;56:2515–2517. doi: 10.1128/iai.56.9.2515-2517.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rada B, Leto TL. Pyocyanin effects on respiratory epithelium: relevance in Pseudomonas aeruginosa airway infections. Trends Microbiol. 2013;21:73–81. doi: 10.1016/j.tim.2012.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Williams BJ, Dehnbostel J, Blackwell TS. Pseudomonas aeruginosa: host defence in lung diseases. Respirology. 2010;15:1037–1056. doi: 10.1111/j.1440-1843.2010.01819.x. [DOI] [PubMed] [Google Scholar]
- 32.Pang L, Hayes CP, Buac K, Yoo DG, Rada B. Pseudogout-associated inflammatory calcium pyrophosphate dihydrate microcrystals induce formation of neutrophil extracellular traps. J Immunol. 2013;190:6488–6500. doi: 10.4049/jimmunol.1203215. [DOI] [PubMed] [Google Scholar]
- 33.Kuhns DB, Alvord WG, Heller T, Feld JJ, Pike KM, Marciano BE, Uzel G, DeRavin SS, Priel DA, Soule BP, Zarember KA, Malech HL, Holland SM, Gallin JI. Residual NADPH oxidase and survival in chronic granulomatous disease. The New England journal of medicine. 2010;363:2600–2610. doi: 10.1056/NEJMoa1007097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rada BK, Geiszt M, Kaldi K, Timar C, Ligeti E. Dual role of phagocytic NADPH oxidase in bacterial killing. Blood. 2004;104:2947–2953. doi: 10.1182/blood-2004-03-1005. [DOI] [PubMed] [Google Scholar]
- 35.Rohrbach AS, Slade DJ, Thompson PR, Mowen KA. Activation of PAD4 in NET formation. Front Immunol. 2012;3:360. doi: 10.3389/fimmu.2012.00360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leshner M, Wang S, Lewis C, Zheng H, Chen XA, Santy L, Wang Y. PAD4 mediated histone hypercitrullination induces heterochromatin decondensation and chromatin unfolding to form neutrophil extracellular trap-like structures. Front Immunol. 2012;3:307. doi: 10.3389/fimmu.2012.00307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Y, Li M, Stadler S, Correll S, Li P, Wang D, Hayama R, Leonelli L, Han H, Grigoryev SA, Allis CD, Coonrod SA. Histone hypercitrullination mediates chromatin decondensation and neutrophil extracellular trap formation. J Cell Biol. 2009;184:205–213. doi: 10.1083/jcb.200806072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sly PD, Gangell CL, Chen L, Ware RS, Ranganathan S, Mott LS, Murray CP, Stick SM, Investigators AC. Risk factors for bronchiectasis in children with cystic fibrosis. The New England journal of medicine. 2013;368:1963–1970. doi: 10.1056/NEJMoa1301725. [DOI] [PubMed] [Google Scholar]
- 39.Almyroudis NG, Grimm MJ, Davidson BA, Rohm M, Urban CF, Segal BH. NETosis and NADPH oxidase: at the intersection of host defense, inflammation, and injury. Frontiers in immunology. 2013;4:45. doi: 10.3389/fimmu.2013.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fuchs TA, Abed U, Goosmann C, Hurwitz R, Schulze I, Wahn V, Weinrauch Y, Brinkmann V, Zychlinsky A. Novel cell death program leads to neutrophil extracellular traps. J Cell Biol. 2007;176:231–241. doi: 10.1083/jcb.200606027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hakkim A, Fuchs TA, Martinez NE, Hess S, Prinz H, Zychlinsky A, Waldmann H. Activation of the Raf-MEK-ERK pathway is required for neutrophil extracellular trap formation. Nat Chem Biol. 2011;7:75–77. doi: 10.1038/nchembio.496. [DOI] [PubMed] [Google Scholar]
- 42.Keshari RS, Verma A, Barthwal MK, Dikshit M. Reactive oxygen species-induced activation of ERK and p38 MAPK mediates PMA-induced NETs release from human neutrophils. Journal of cellular biochemistry. 2013;114:532–540. doi: 10.1002/jcb.24391. [DOI] [PubMed] [Google Scholar]
- 43.Carlsson M, Eriksson L, Erwander I, Wieslander J, Segelmark M. Pseudomonas-induced lung damage in cystic fibrosis correlates to bactericidal-permeability increasing protein (BPI)-autoantibodies. Clinical and experimental rheumatology. 2003;21:S95–100. [PubMed] [Google Scholar]
- 44.Parad RB, Gerard CJ, Zurakowski D, Nichols DP, Pier GB. Pulmonary outcome in cystic fibrosis is influenced primarily by mucoid Pseudomonas aeruginosa infection and immune status and only modestly by genotype. Infection and immunity. 1999;67:4744–4750. doi: 10.1128/iai.67.9.4744-4750.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Emerson J, Rosenfeld M, McNamara S, Ramsey B, Gibson RL. Pseudomonas aeruginosa and other predictors of mortality and morbidity in young children with cystic fibrosis. Pediatric pulmonology. 2002;34:91–100. doi: 10.1002/ppul.10127. [DOI] [PubMed] [Google Scholar]
- 46.Hodson ME. Aerosolized dornase alfa (rhDNase) for therapy of cystic fibrosis. American journal of respiratory and critical care medicine. 1995;151:S70–74. doi: 10.1164/ajrccm/151.3_Pt_2.S70. [DOI] [PubMed] [Google Scholar]
- 47.Pilsczek FH, Salina D, Poon KK, Fahey C, Yipp BG, Sibley CD, Robbins SM, Green FH, Surette MG, Sugai M, Bowden MG, Hussain M, Zhang K, Kubes P. A novel mechanism of rapid nuclear neutrophil extracellular trap formation in response to Staphylococcus aureus. Journal of immunology. 2010;185:7413–7425. doi: 10.4049/jimmunol.1000675. [DOI] [PubMed] [Google Scholar]
- 48.Akong-Moore K, Chow OA, von Kockritz-Blickwede M, Nizet V. Influences of chloride and hypochlorite on neutrophil extracellular trap formation. PLoS One. 2012;7:e42984. doi: 10.1371/journal.pone.0042984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Parker H, Dragunow M, Hampton MB, Kettle AJ, Winterbourn CC. Requirements for NADPH oxidase and myeloperoxidase in neutrophil extracellular trap formation differ depending on the stimulus. J Leukoc Biol. 2012;92:841–849. doi: 10.1189/jlb.1211601. [DOI] [PubMed] [Google Scholar]
- 50.Brinkmann V, Goosmann C, Kuhn LI, Zychlinsky A. Automatic quantification of in vitro NET formation. Front Immunol. 2012;3:413. doi: 10.3389/fimmu.2012.00413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li P, Li M, Lindberg MR, Kennett MJ, Xiong N, Wang Y. PAD4 is essential for antibacterial innate immunity mediated by neutrophil extracellular traps. J Exp Med. 2010;207:1853–1862. doi: 10.1084/jem.20100239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Parker H, Albrett AM, Kettle AJ, Winterbourn CC. Myeloperoxidase associated with neutrophil extracellular traps is active and mediates bacterial killing in the presence of hydrogen peroxide. J Leukoc Biol. 2012;91:369–376. doi: 10.1189/jlb.0711387. [DOI] [PubMed] [Google Scholar]
- 53.Galli F, Battistoni A, Gambari R, Pompella A, Bragonzi A, Pilolli F, Iuliano L, Piroddi M, Dechecchi MC, Cabrini G F. Working Group on Inflammation in Cystic. . Oxidative stress and antioxidant therapy in cystic fibrosis. Biochimica et biophysica acta. 2012;1822:690–713. doi: 10.1016/j.bbadis.2011.12.012. [DOI] [PubMed] [Google Scholar]
- 54.Parker H, Winterbourn CC. Reactive oxidants and myeloperoxidase and their involvement in neutrophil extracellular traps. Front Immunol. 2012;3:424. doi: 10.3389/fimmu.2012.00424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Palmer LJ, Cooper PR, Ling MR, Wright HJ, Huissoon A, Chapple IL. Hypochlorous acid regulates neutrophil extracellular trap release in humans. Clin Exp Immunol. 2012;167:261–268. doi: 10.1111/j.1365-2249.2011.04518.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lu T, Kobayashi SD, Quinn MT, Deleo FR. A NET Outcome. Front Immunol. 2012;3:365. doi: 10.3389/fimmu.2012.00365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Metzler KD, Fuchs TA, Nauseef WM, Reumaux D, Roesler J, Schulze I, Wahn V, Papayannopoulos V, Zychlinsky A. Myeloperoxidase is required for neutrophil extracellular trap formation: implications for innate immunity. Blood. 2011;117:953–959. doi: 10.1182/blood-2010-06-290171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pereira WE, Hoyano Y, Summons RE, Bacon VA, Duffield AM. Chlorination studies. II. The reaction of aqueous hypochlorous acid with alpha-amino acids and dipeptides. Biochim Biophys Acta. 1973;313:170–180. doi: 10.1016/0304-4165(73)90198-0. [DOI] [PubMed] [Google Scholar]