Abstract
Background
Cryptococcus gattii has been the cause of an ongoing outbreak starting in 1999 on Vancouver Island, British Columbia and spreading to mainland Canada and the US Pacific Northwest. In the course of the outbreak, C. gattii has been identified outside of its previously documented climate, habitat, and host disease. Genotyping of C. gattii is essential to understand the ecological and geographical expansion of this emerging pathogen.
Methods
We developed and validated a mismatch amplification mutation assay (MAMA) real-time PCR panel for genotyping C. gattii molecular types VGI-VGIV and VGII subtypes a,b,c. Subtype assays were designed based on whole-genome sequence of 20 C. gattii strains. Publically available multilocus sequence typing (MLST) data from a study of 202 strains was used for the molecular type (VGI-VGIV) assay design. All assays were validated across DNA from 112 strains of diverse international origin and sample types, including animal, environmental and human.
Results
Validation revealed each assay on the panel is 100% sensitive, specific and concordant with MLST. The assay panel can detect down to 0.5 picograms of template DNA.
Conclusions
The (MAMA) real-time PCR panel for C. gattii accurately typed a collection of 112 diverse strains and demonstrated high sensitivity. This is a time and cost efficient method of genotyping C. gattii best suited for application in large-scale epidemiological studies.
Keywords: Cryptococcus gattii, Genotyping, Real-time PCR, Epidemiology
Background
Cryptococcosis, a potentially fatal fungal disease, has primarily been observed in immune-compromised individuals and mainly associated with Cryptococcus neoformans infection. It is now recognized that Cryptococcus gattii, once considered to be a variety of the Cryptococcus neoformans complex, is also capable of causing serious disease in immunocompetent individuals and animals [1,2]. C. gattii has been associated with a number of tree species in tropical and subtropical regions [3]. More recently, C. gattii caused an outbreak that began in 1999 on Vancouver Island, British Columbia and has spread to mainland Canada and the US Pacific Northwest [4]. This outbreak is unique in that it marked the identification of a Cryptococcus species in a new climatic region (from tropical to temperate), habitat (from tropical trees to temperate; e.g., Douglas Fir) and host disease (from primary neurologic to primary pulmonary) [3,5].
Recent epidemiological studies of C. gattii in North America provide insight into the organism’s geographical expansion as well as the distribution of molecular genotypes [6-9]. C. gattii has been classically classified into four molecular types by MLST/AFLP, VGI/AFLP4, VGII/AFLP6, VGIII/AFLP5, VGIV/AFLP7 [3,5], with additional molecular types recently identified [10]. Interestingly, molecular types have been associated with significant differences in disease type [3,5], antifungal susceptibilities [3,5,10], and severity and outcome [3,5].
Contemporary methods for genotyping C. gattii are PCR-restriction fragment length polymorphism (PCR-RFLP), amplified fragment length polymorphism (AFLP), multilocus microsatellite typing (MLMT), multilocus sequence typing (MLST), and most recent, matrix-assisted laser desorption ionization-time-of-flight mass spectrometry (MALDI-TOF MS) [11-14]. High resolution melting (HRM) is a method that has been used to identify the Cryptococcus neoformans-Cryptococcus gattii complex, though it has not been employed for genotyping within either species [15]. PCR-RFLP and AFLP require extensive lab work involving restriction enzyme digestion and gel electrophoresis [11]. Results are based on interpretation of gel electrophoresis profiles and as such, are not readily transferred or analyzed between laboratories. MLST, which requires DNA sequencing of seven housekeeping genes, is the preferred genotyping method for C. gattii and is easily transferrable between laboratories [16]. MLMT allows for finer genotype resolution than MLST and has high reproducibility between laboratories [14]. In some laboratories, real-time PCR is a preferable option to methods involving DNA sequencing (MLMT and MLST), which require either out-sourcing to a sequencing capable laboratory or investment in, and the maintenance of, an in-house instrument. Although MALDI-TOF MS shows promise as a new genotyping method, instrumentation is expensive and thus prohibitive for many public health laboratories. Conversely, real-time PCR instruments are becoming ubiquitous, easily maintained, and the use of unlabeled primers and no probe makes reagents inexpensive [17]. Therefore, real-time PCR is an accessible and increasing popular technology for widespread molecular epidemiological efforts.
Here, we present a panel of real-time PCR assays, based on mismatch amplification mutation assay (MAMA) methodology, for rapid and sensitive molecular genotyping of Cryptococcus gattii molecular types (VGI-VGIV) and the dominant North American VGII subtypes (VGIIa-c) [18,19]. MAMA, a form of allele-specific PCR (ASPCR), employs primers that are designed for SNP genotyping. We use known MLST sequences for the VGI-VGIV molecular type assay design and whole genome sequences of 20 strains to identify SNPs specific to each of the targeted VGII subtypes [9,20].
Methods
SYBR MAMA design
MAMA primers have an intentional penultimate mismatch nucleotide at the 3′ end; the ultimate base is always the SNP assay target and is a perfect match for the target SNP [18]. Mismatches decrease the efficiency of primer extension by Taq polymerase, such that if two mismatches are found together under the 3′ end of the primer, the efficiency of the PCR is significantly reduced. However, if a single mismatch at the penultimate base is present, extension occurs from the 3′ matched base, and efficiency of the PCR remains relatively high. Costly fluorogenic oligonucleotide probes are not needed to discriminate SNPs with this method. This discriminatory design results in a cost-efficient, powerful and simple method of SNP genotyping [17,21]. Separate PCR reactions are performed with a MAMA primer specific for only one of the two target SNPs and with one universal primer for amplification from the alternate direction. Comparison of cycle threshold (Ct) values will reveal which reaction is more efficient (has the smaller Ct value). The more efficient reaction corresponds to the SNP that is present in the sample.
MAMA design for MLST groups VGI, VGII, VGIII, and VGIV
The MLST SYBR MAMA design was informed by MLST data collected for 202 C. gatii strains from a worldwide collection [20]. The MLST library included sequences from 77, 75, 26, and 24 isolates of the VGI, VGII, VGIII, VGIV molecular types, respectively. The gene encoding mannitol-1-phosphate dehydrogenase (MPD1) was selected as the best candidate for assay design based on its sequence conservation within each of the four molecular types that allowed for design of assay primers with a minimum number of degenerate bases. All 15 of the known MPD1 allele sequences were aligned with SeqMan Pro v.9.0.4 (DNASTAR, Madison, WI). SNPs specific for each of the molecular types were identified in the sequence alignment. MAMA primers were manually designed in Primer Express 3.0 (Life Technologies, Carlsbad, CA) software with optimal mismatches chosen as suggested by Li et. al. [19] (Table 1).
Table 1.
MAMA real-time PCR assay sequences and targets for genotyping C. gattii
| Genotype | Assay Name | Gene (SNP position) | Base call match/mismatch | Universal Primer sequence 5′ -- > 3′ | Match MAMA Primer sequence 5′ -- > 3′ | Mismatch MAMA Primer sequence 5′ -- > 3′ |
|---|---|---|---|---|---|---|
| VGI |
VGI-MPD471 |
MPD1 (471) |
G/A |
AGACTGTCCCAATGTCAAGCTTTC |
GCCTTGTATGTGGTAACACCAGTG |
GWGCCTTGTATGTGGTAACACCAGTA |
| VGII |
VGII-MPD495 |
MPD1 (495) |
T/A |
AGACTGTCCCAATGTCAAGCTTTC |
ATTAACCTTAGTGTTGGAGACCTTGACT |
AACCTTAGTGTTGGAGACCTTGACA |
| VGIIa |
VGIIa-45211 |
hypothetical protein |
A/C |
CCCAGCAACCTTGATCTGGA |
AGCTGCTCTAAGAGACACATCATCA |
AGCTGCTCTAAGAGACACATCATCC |
| VGIIb |
VGIIb-502129 |
not annotated |
G/A |
AATCGCTCGTCCTCATATGACA |
GTAGGCGGTGGGATAAGGTG |
GGTAGGCGGTGGGATAAGGTA |
| VGIIc |
VGIIc-257655 |
non-coding region |
C/T |
CGTTAATTTGGTTGTTTGACAACCT |
AGCAACTCACGCAGAAACAGAC |
GAGCAACTCACGCAGAAACAGAT |
| VGIII |
VGIII-MPD198 |
MPD1 (198) |
T/A |
TGACATTGGGACAGTCTGCAAT |
ACTGCTGCTTCTCCCGTTGT |
CTGCTGCTTCTCCCGTTGA |
| VGIV | VGIV-MPD423 | MPD1 (423) | A/C | ACCCAGTCATTAACCTTAGTGTTGGA | CTCGTTCGTCAAYCACGTTAGA | TCGTTCGTCAAYCACGTTAGC |
MAMA design for VGIIa, VGIIb, and VGIIc subtypes
Whole genome sequence typing (WGST) analysis of 20 C. gattii strains from a previous study revealed canonical SNPs specific for each of the VGII a, b and c subtypes (n = 2720, 3547, and 3819, respectively) [9]. In order to minimize interference of adjacent mutations with primer design, the genotype-specific SNPs were sorted according to nearest neighboring mismatch within the sequence alignment; in short, the SNPs with the most-conserved flanking regions were the top candidates for assay design. Sequence from the R265 strain reference genome [GenBank: CH408164] [2] surrounding the genotype-specific SNPs was used for assay design. SYBR MAMA primers were designed using the same criteria as previously described for the MLST MAMA (Table 1).
Isolate selection
Initially, assays were validated with genomic DNA extracted from 57 C. gattii strains of North American origin and some historical isolates. The panel of isolates including: 13 VGIIa, 4 VGIIb, and 24 VGIIc, and 8 each of VGI and VGIII, was analyzed using each of the assays (Table 2). All DNAs were genotyped by MLST prior to screening. Further validation of the assays was accomplished by employing a more diverse isolate collection of 55 strains including isolates of international origin; this panel was comprised of 10 VGI, 10 VGIIa, 9 VGIIb, 8 VGIIc, 8 VGIII, and 10 VGIV molecular types (Table 3). The strains came from a variety of environmental, human and animal sources, including cats, a dog, an alpaca, a porpoise, a sheep and a cow.
Table 2.
C. gattii strains for initial assay validation
| Isolate ID | MLST | Year | Geographic origin | Source |
|---|---|---|---|---|
| B7488 |
VGI |
2009 |
Oregon |
Human |
| B7496 |
VGI |
2009 |
Hawaii |
Dolphin |
| B8551 |
VGI |
2010 |
Oregon |
Human |
| B8852 |
VGI |
2010 |
Oregon |
Human |
| B8886 |
VGI |
2010 |
Oregon |
Soil |
| B8887 |
VGI |
2010 |
Oregon |
Soil |
| B8990 |
VGI |
2010 |
California |
Human |
| B9009 |
VGI |
2011 |
Washington |
Human |
| B6864 |
VGIIa |
2004 |
Oregon |
Human |
| B7395 |
VGIIa |
2008 |
Washington |
Dog |
| B7422 |
VGIIa |
2009 |
Oregon |
Cat |
| B7436 |
VGIIa |
2009 |
California |
Alpaca |
| B7467 |
VGIIa |
2009 |
Oregon |
Porpoise |
| B8555 |
VGIIa |
2006 |
Washington |
Human |
| B8577 |
VGIIa |
2009 |
British Columbia |
Soil |
| B8793 |
VGIIa |
2010 |
Oregon |
Canine |
| B8849 |
VGIIa |
2010 |
Oregon |
Environmental |
| CA-1014 |
VGIIa |
unknown |
California |
Human |
| CBS-7750 |
VGIIa |
1990 |
California |
Environmental |
| ICB-107 |
VGIIa |
unknown |
Brazil |
Human |
| NIH-444 |
VGIIa |
1972 |
Washington |
Human |
| B7394 |
VGIIb |
2008 |
Washington |
Cat |
| B7735 |
VGIIb |
2009 |
Oregon |
Human |
| B8554 |
VGIIb |
2010 |
Oregon |
Dog |
| B8828 |
VGIIb |
2010 |
Washington |
Porpoise |
| B6863 |
VGIIc |
2005 |
Oregon |
Human |
| B7390 |
VGIIc |
2008 |
Idaho |
Human |
| B7432 |
VGIIc |
2009 |
Oregon |
Human |
| B7434 |
VGIIc |
2008 |
Oregon |
Human |
| B7466 |
VGIIc |
2008 |
Oregon |
Cat |
| B7491 |
VGIIc |
2009 |
Oregon |
Human |
| B7493 |
VGIIc |
2009 |
Oregon |
Sheep |
| B7641 |
VGIIc |
2008 |
Oregon |
Cat |
| B7737 |
VGIIc |
2009 |
Oregon |
Human |
| B7765 |
VGIIc |
2009 |
Oregon |
Dog |
| B8210 |
VGIIc |
2008 |
Oregon |
Human |
| B8214 |
VGIIc |
2009 |
Oregon |
Human |
| B8510 |
VGIIc |
2009 |
Oregon |
Human |
| B8549 |
VGIIc |
unknown |
Oregon |
Human |
| B8552 |
VGIIc |
unknown |
Oregon |
Human |
| B8571 |
VGIIc |
2009 |
Washington |
Human |
| B8788 |
VGIIc |
2010 |
Oregon |
Human |
| B8798 |
VGIIc |
2005 |
Oregon |
Human |
| B8821 |
VGIIc |
2010 |
Oregon |
Human |
| B8825 |
VGIIc |
2009 |
Oregon |
Human |
| B8833 |
VGIIc |
2010 |
Oregon |
Cat |
| B8838 |
VGIIc |
2010 |
Washington |
Human |
| B8843 |
VGIIc |
2010 |
Oregon |
Human |
| B8853 |
VGIIc |
2010 |
Oregon |
Cat |
| B7415 |
VGIII |
2009 |
California |
Alpaca |
| B7495 |
VGIII |
2009 |
California |
Human |
| B8212 |
VGIII |
2007 |
Oregon |
Human |
| B8260 |
VGIII |
2009 |
Washington |
Cat |
| B8262 |
VGIII |
1992 |
California |
Human |
| B8516/B8616 |
VGIII |
2009 |
Oregon |
Cat |
| B9143 |
VGIII |
2011 |
California |
Human |
| B9146 | VGIII | 2011 | California | Human |
Table 3.
C. gattii strains for additional assay validation
| Culture collection ID | Geographic origin | Sample type | MLST | Year of isolation |
|---|---|---|---|---|
| B4501 |
Australia |
Human |
VGI |
unknown |
| B4503 |
Australia |
Human |
VGI |
unknown |
| B4504 |
Australia |
Human |
VGI |
unknown |
| B4516 |
Australia |
Human |
VGI |
unknown |
| B5765 |
India |
Environmental |
VGI |
unknown |
| B9018 |
California |
Human |
VGI |
2011 |
| B9019 |
New Mexico |
Human |
VGI |
2011 |
| B9021 |
Rhode Island |
Human |
VGI |
2011 |
| B9142 |
Georgia |
Human |
VGI |
2011 |
| B9149 |
California |
Human |
VGI |
2011 |
| B8508 |
Oregon |
Human |
VGIIa |
2009 |
| B8512 |
Oregon |
Alpaca |
VGIIa |
2009 |
| B8558 |
Washington |
Human |
VGIIa |
2010 |
| B8561 |
Washington |
Human |
VGIIa |
2010 |
| B8563 |
Washington |
Human |
VGIIa |
2010 |
| B8567 |
Washington |
Dog |
VGIIa |
2010 |
| B8854 |
Washington |
Human |
VGIIa |
2010 |
| B8889 |
Oregon |
Environmental |
VGIIa |
2010 |
| B9077 |
Washington |
Environmental |
VGIIa |
2011 |
| B9296 |
British Columbia |
Environmental |
VGIIa |
2011 |
| B8211 |
Oregon |
Human |
VGIIb |
2009 |
| B8966 |
Oregon |
Horse |
VGIIb |
2010 |
| B9076 |
Washington |
Environmental |
VGIIb |
2011 |
| B9157 |
Washington |
Horse |
VGIIb |
2011 |
| B9170 |
Washington |
Porpoise |
VGIIb |
2011 |
| B9234 |
Washington |
Cat |
VGIIb |
2011 |
| B9290 |
British Columbia |
Cat |
VGIIb |
2011 |
| B9241 |
Oregon |
Human |
VGIIb |
2011 |
| B9428 |
Washington |
Cat |
VGIIb |
2012 |
| B9159 |
Washington |
Sheep |
VGIIc |
2011 |
| B9227 |
Oregon |
Cat |
VGIIc |
2011 |
| B9235 |
Oregon |
Human |
VGIIc |
2011 |
| B9244 |
Oregon |
Human |
VGIIc |
2011 |
| B9245 |
Oregon |
Human |
VGIIc |
2011 |
| B9295 |
British Columbia |
Environmental |
VGIIc |
2011 |
| B9302 |
Oregon |
Environmental |
VGIIc |
2011 |
| B9374 |
Oregon |
Human |
VGIIc |
2011 |
| B8965 |
New Mexico |
Human |
VGIII |
2010 |
| B9148 |
California |
Human |
VGIII |
2011 |
| B9151 |
Michigan |
Human |
VGIII |
2011 |
| B9163 |
New Mexico |
Human |
VGIII |
2011 |
| B9237 |
New Mexico |
Cat |
VGIII |
2011 |
| B9372 |
California |
Cow |
VGIII |
2011 |
| B9422 |
Oregon |
Cat |
VGIII |
2012 |
| B9430 |
Alaska |
Cat |
VGIII |
2012 |
| B7238 |
Botswana |
Human |
VGIV |
2005 |
| B7240 |
Botswana |
Human |
VGIV |
2005 |
| B7243 |
Botswana |
Human |
VGIV |
2005 |
| B7247 |
Botswana |
Human |
VGIV |
2005 |
| B7249 |
Botswana |
Human |
VGIV |
2005 |
| B7260 |
Botswana |
Human |
VGIV |
2006 |
| B7262 |
Botswana |
Human |
VGIV |
2006 |
| B7263 |
Botswana |
Human |
VGIV |
2006 |
| B7264 |
Botswana |
Human |
VGIV |
2006 |
| B7265 | Botswana | Human | VGIV | 2006 |
Isolate culturing and DNA extraction
Isolates were grown on Yeast Peptone Glucose (YPD) agar plus 0.5% NaCl at 37°C for 24 hours; and DNA was prepared using an UltraClean DNA Isolation Kit as described by the manufacturer, with some modifications (MO BIO Laboratories, Carlsbad, CA). Briefly, ~0.5 grams of microbial cells were suspended in lysis solution in a MicroBead tube and heated to 65°C for 15 minutes to increase lysis efficiency. The MicroBead tube was then secured horizontally using the MO BIO vortex adapter tube holder (MO BIO Laboratories, Carlsbad, CA) and vortexed at maximum speed for 10 minutes; post cell lysis, microtubes were immediately placed on ice for 5 minutes. After the lysis steps, DNA extraction was completed per manufacturer’s instructions. DNA was stored at −20°C.
Real-time PCR
Real-time PCR was performed on the ABI 7900HT real-time PCR System (Life Technologies, Carlsbad, CA). Reactions for both perfect match and mismatch primer sets were conducted in separate wells of a 384-well optical plate, and reactions for each primer set were run in triplicate. Reactions were 10 μL total volume composed of 1X Platinum SYBR Green qPCR SuperMix-UDG with ROX (Invitrogen, Grand Island, NY), 200 nM each of forward and reverse primers, and 1 μL DNA extract (diluted 1:10). Reactions were incubated for 3 min at 50°C for UDG digest followed by 3 min at 95°C for Taq polymerase activation. PCR consisted of 45 cycles of 15 s at 95°C for denaturation followed by 1 min at 60°C annealing and extension. Dissociation of PCR product was performed for 15 sec at 95°C, 15 sec at 60°C and 15 sec at 95°C as a quality assurance step to inspect reactions for primer-dimer. Dissociation curves were not used for isolate genotyping, rather to ensure amplification was specific for the targeted sequence and to preclude non-specific amplification associated with the ability of SYBR Green chemistry to bind any double-stranded DNA. Data were analyzed in Sequence Detection Systems 2.3 software (Life Technologies, Carlsbad, CA) for calculation of cycle threshold (Ct) values and interpretation of dissociation curves.
For MAMA results, the perfect match primer set will amplify earlier and yield the lowest Ct value, corresponding to the SNP genotype of the isolate; secondary delayed amplification plots with a higher Ct value, if present, are due to mismatch priming (Figure 1). An algorithm for genotype calling was implemented to expedite data analysis. The delta Ct value was calculated by subtracting the match primer mean Ct from the mismatch primer mean Ct. If the mismatch priming fails to yield a Ct value because it is beyond the instrument range, a Ct value = 40 is assigned in order to calculate a ΔCt.
Figure 1.
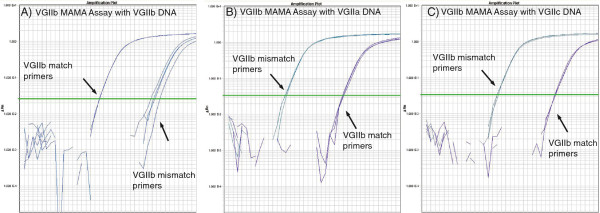
VGIIb MAMA plots with VGII DNA show the specificity of VGIIb MAMA for VGIIb DNA. (A) The VGIIb match primers amplify VGIIb DNA efficiently and yield a lower Ct value than the VGIIb mismatch primers, resulting in a VGIIb genotype call. (B) The VGIIb mismatch primers amplify VGIIa DNA more efficiently than the VGIIb match primers, resulting in a non-VGIIb genotype call. (C) VGIIb mismatch primers amplify VGIIc DNA more efficiently than the VGIIb match primers, again resulting in a non-VGIIb genotype call.
A negative ΔCt value indicates a mismatch allele, whereas a positive ΔCt indicates a match allele. A stringent threshold of |ΔCt| ≥ 3.3, approximately equivalent to one log10 difference in the dynamic range, was established to ensure accuracy of allele calls. If |ΔCt| < 3.3 is below the stringent threshold, this could result in an inaccurate genotype call. In this case, it is advisable to re-screen the sample across the failed assays.
Sensitivity and specificity of the assay panel were calculated as well as concordance with the known MLST type as determined by sequencing the MLST house keeping genes. Assay repeatability and reproducibility were tested by screening nine replicate reactions with the matching primer sets and DNA for each assay on three separate days. The lower limit of detection for each assay and its matching template pair was tested. Each matching template and assay pair was tested using six log10 serial dilutions of a single template DNA, starting with 0.5 ng/μl. Template DNA was quantified in triplicate by NanoDrop 3300 fluorospectrometer (NanoDrop Technologies, Wilmington, DE) using Quant-iT PicoGreen dsDNA Reagent (Life Technologies, Carlsbad, CA), according to manufacturer’s instructions. Real-time PCR reactions were performed in triplicate for each dilution.
Results
Initial validation revealed the assay panel was 100% sensitive; each assay appropriately identified the known isolate genotypes. The ΔCt values for our validation panel confirmed the stringent threshold ΔCt = 3.3 sufficient to discriminate the genotypes. In addition, the assay panel was 100% specific; no cross reactivity occurred between assays and non-matching genotypes. Further validation of the assay panel with additional strains revealed 100% sensitivity and specificity. A total of 112 strains were screened across the MLST assay panel and 100% sensitivity and specificity was observed (Table 4). A total of 68 previously genotyped strains were screened across the VGII subtyping assay panel with 100% sensitivity and specificity (Table 5). The assay coefficients of variation ranged from 0.22% to 4.33% indicating high assay repeatability and reproducibility within and between runs (Table 6). The assays were designed for genotyping of DNA from known C. gattii isolates, and are not validated for application to clinical specimens; they were able to detect DNA concentrations as low as 0.5 pg/μl (Table 7).
Table 4.
MLST SYBR MAMA Ct values and genotype assignments for VGI-VGIV
| |
VGI_MPD471 |
VGII_MPD495 |
VGIII_MPD198 |
VGIV_MPD423 |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Isolate ID | Strain type via MLST | VGI Ct Mean | non-VGI Ct Mean | Delta Ct | Type call via assay | VGII Ct Mean | non-VGII Ct Mean | Delta Ct | Type call via assay | VGIII Ct Mean | non-VGIII Ct Mean | Delta Ct | Type call via assay | VGIV Ct Mean | non-VGIV Ct Mean | Delta Ct | Type call via assay | Final Call |
| B7488 |
VGI |
17.0 |
29.0 |
11.9 |
VGI |
37.4 |
17.7 |
−19.7 |
non-VGII |
28.4 |
14.9 |
−13.5 |
non-VGIII |
32.4 |
16.3 |
−16.1 |
non-VGIV |
VGI |
| B7496 |
VGI |
18.2 |
28.0 |
9.8 |
VGI |
35.3 |
19.0 |
−16.3 |
non-VGII |
24.5 |
16.4 |
−8.1 |
non-VGIII |
31.7 |
17.9 |
−13.8 |
non-VGIV |
VGI |
| B8551 |
VGI |
17.3 |
29.6 |
12.3 |
VGI |
36.2 |
17.9 |
−18.3 |
non-VGII |
28.7 |
15.3 |
−13.4 |
non-VGIII |
39.0 |
16.7 |
−22.3 |
non-VGIV |
VGI |
| B8852 |
VGI |
21.1 |
30.9 |
9.8 |
VGI |
36.5 |
21.9 |
−14.6 |
non-VGII |
27.8 |
19.1 |
−8.8 |
non-VGIII |
32.0 |
20.6 |
−11.4 |
non-VGIV |
VGI |
| B8886 |
VGI |
18.9 |
29.2 |
10.3 |
VGI |
38.1 |
19.3 |
−18.8 |
non-VGII |
26.7 |
16.4 |
−10.3 |
non-VGIII |
32.3 |
17.9 |
−14.4 |
non-VGIV |
VGI |
| B8887 |
VGI |
15.9 |
28.3 |
12.4 |
VGI |
23.6 |
15.5 |
−8.1 |
non-VGII |
33.6 |
16.2 |
−17.4 |
non-VGIII |
34.1 |
15.5 |
−18.7 |
non-VGIV |
VGI |
| B8990 |
VGI |
18.8 |
30.9 |
12.1 |
VGI |
37.2 |
20.1 |
−17.1 |
non-VGII |
31.3 |
16.9 |
−14.3 |
non-VGIII |
40.0 |
19.3 |
−20.7 |
non-VGIV |
VGI |
| B9009 |
VGI |
21.6 |
31.0 |
9.4 |
VGI |
36.5 |
23.1 |
−13.4 |
non-VGII |
28.6 |
19.4 |
−9.2 |
non-VGIII |
40.0 |
21.1 |
−18.9 |
non-VGIV |
VGI |
| B4501 |
VGI |
16.1 |
26.7 |
10.6 |
VGI |
30.5 |
18.1 |
−12.4 |
non-VGII |
30.6 |
17.3 |
−13.3 |
non-VGIII |
29.4 |
16.4 |
−13.0 |
non-VGIV |
VGI |
| B4503 |
VGI |
15.9 |
27.2 |
11.2 |
VGI |
32.7 |
18.6 |
−14.1 |
non-VGII |
33.8 |
17.9 |
−15.9 |
non-VGIII |
28.7 |
16.1 |
−12.6 |
non-VGIV |
VGI |
| B4504 |
VGI |
15.6 |
27.2 |
11.5 |
VGI |
33.1 |
18.1 |
−15.1 |
non-VGII |
33.9 |
17.4 |
−16.4 |
non-VGIII |
28.7 |
15.8 |
−13.0 |
non-VGIV |
VGI |
| B4516 |
VGI |
15.3 |
26.8 |
11.5 |
VGI |
31.5 |
17.6 |
−13.9 |
non-VGII |
33.4 |
16.8 |
−16.6 |
non-VGIII |
29.7 |
15.3 |
−14.3 |
non-VGIV |
VGI |
| B5765 |
VGI |
17.2 |
28.0 |
10.8 |
VGI |
32.8 |
19.7 |
−13.0 |
non-VGII |
34.4 |
19.2 |
−15.2 |
non-VGIII |
29.0 |
16.3 |
−12.7 |
non-VGIV |
VGI |
| B9018 |
VGI |
17.7 |
30.0 |
12.3 |
VGI |
34.6 |
17.9 |
−16.7 |
non-VGII |
31.8 |
18.6 |
−13.2 |
non-VGIII |
35.0 |
18.3 |
−16.8 |
non-VGIV |
VGI |
| B9019 |
VGI |
16.9 |
26.1 |
9.2 |
VGI |
35.4 |
16.7 |
−18.7 |
non-VGII |
34.9 |
16.7 |
−18.2 |
non-VGIII |
30.5 |
16.8 |
−13.7 |
non-VGIV |
VGI |
| B9021 |
VGI |
21.4 |
32.9 |
11.5 |
VGI |
33.4 |
19.9 |
−13.5 |
non-VGII |
32.7 |
20.5 |
−12.2 |
non-VGIII |
35.5 |
20.4 |
−15.2 |
non-VGIV |
VGI |
| B9142 |
VGI |
16.0 |
26.3 |
10.3 |
VGI |
27.8 |
15.9 |
−11.9 |
non-VGII |
32.7 |
16.5 |
−16.2 |
non-VGIII |
31.7 |
16.6 |
−15.1 |
non-VGIV |
VGI |
| B9149 |
VGI |
17.7 |
26.8 |
9.1 |
VGI |
28.5 |
17.5 |
−11.0 |
non-VGII |
28.5 |
18.2 |
−10.3 |
non-VGIII |
31.0 |
18.3 |
−12.6 |
non-VGIV |
VGI |
| B6864 |
VGIIa |
27.8 |
17.5 |
−10.3 |
non-VGI |
19.3 |
33.1 |
13.8 |
VGII |
34.7 |
19.7 |
−15.0 |
non-VGIII |
40.0 |
16.1 |
−23.9 |
non-VGIV |
VGII |
| B7395 |
VGIIa |
28.9 |
18.8 |
−10.1 |
non-VGI |
21.3 |
32.6 |
11.3 |
VGII |
40.0 |
19.2 |
19.2 |
non-VGIII |
40.0 |
18.8 |
−21.2 |
non-VGIV |
VGII |
| B7422 |
VGIIa |
27.4 |
17.4 |
−10.0 |
non-VGI |
19.5 |
32.3 |
12.8 |
VGII |
35.4 |
19.1 |
−16.3 |
non-VGIII |
40.0 |
15.6 |
−24.4 |
non-VGIV |
VGII |
| B7436 |
VGIIa |
27.8 |
17.9 |
−9.9 |
non-VGI |
20.7 |
35.4 |
14.7 |
VGII |
36.5 |
16.9 |
−19.6 |
non-VGIII |
40.0 |
15.6 |
−24.4 |
non-VGIV |
VGII |
| B7467 |
VGIIa |
30.9 |
20.7 |
−10.1 |
non-VGI |
22.7 |
32.7 |
9.9 |
VGII |
37.7 |
23.4 |
−14.2 |
non-VGIII |
40.0 |
19.1 |
−20.9 |
non-VGIV |
VGII |
| B8555 |
VGIIa |
27.9 |
17.7 |
−10.2 |
non-VGI |
19.7 |
32.1 |
12.4 |
VGII |
34.6 |
20.8 |
−13.8 |
non-VGIII |
40.0 |
16.6 |
−23.4 |
non-VGIV |
VGII |
| B8577 |
VGIIa |
31.1 |
20.9 |
−10.2 |
non-VGI |
21.8 |
34.1 |
12.3 |
VGII |
33.1 |
23.4 |
−9.8 |
non-VGIII |
40.0 |
19.8 |
−20.2 |
non-VGIV |
VGII |
| B8793 |
VGIIa |
27.4 |
17.4 |
−10.0 |
non-VGI |
18.9 |
32.6 |
13.7 |
VGII |
39.0 |
24.9 |
−14.1 |
non-VGIII |
40.0 |
16.3 |
−23.7 |
non-VGIV |
VGII |
| B8849 |
VGIIa |
28.9 |
18.7 |
−10.1 |
non-VGI |
22.9 |
35.1 |
12.2 |
VGII |
36.0 |
22.7 |
−13.3 |
non-VGIII |
40.0 |
18.4 |
−21.6 |
non-VGIV |
VGII |
| CA-1014 |
VGIIa |
20.4 |
11.6 |
−8.8 |
non-VGI |
13.6 |
32.4 |
18.9 |
VGII |
31.1 |
12.8 |
−18.3 |
non-VGIII |
40.0 |
11.0 |
−29.0 |
non-VGIV |
VGII |
| CBS-7750 |
VGIIa |
27.2 |
17.3 |
−9.9 |
non-VGI |
18.8 |
33.1 |
14.3 |
VGII |
38.0 |
25.5 |
−12.5 |
non-VGIII |
40.0 |
15.8 |
−24.2 |
non-VGIV |
VGII |
| ICB-107 |
VGIIa |
28.1 |
18.2 |
−9.9 |
non-VGI |
20.0 |
34.7 |
14.8 |
VGII |
37.5 |
25.4 |
−12.1 |
non-VGIII |
40.0 |
15.6 |
−24.4 |
non-VGIV |
VGII |
| NIH-444 |
VGIIa |
24.9 |
14.9 |
−10.0 |
non-VGI |
17.0 |
33.2 |
16.2 |
VGII |
34.9 |
17.7 |
−17.2 |
non-VGIII |
40.0 |
13.3 |
−26.7 |
non-VGIV |
VGII |
| B8508 |
VGIIa |
23.7 |
14.8 |
−8.9 |
non-VGI |
17.4 |
30.4 |
13.0 |
VGII |
34.5 |
16.2 |
−18.2 |
non-VGIII |
29.1 |
14.9 |
−14.2 |
non-VGIV |
VGII |
| B8512 |
VGIIa |
23.5 |
14.6 |
−9.0 |
non-VGI |
16.7 |
30.6 |
13.9 |
VGII |
31.4 |
15.7 |
−15.6 |
non-VGIII |
29.7 |
14.8 |
−14.9 |
non-VGIV |
VGII |
| B8558 |
VGIIa |
22.5 |
13.7 |
−8.8 |
non-VGI |
15.9 |
29.9 |
14.0 |
VGII |
30.6 |
14.9 |
−15.7 |
non-VGIII |
30.1 |
14.3 |
−15.9 |
non-VGIV |
VGII |
| B8561 |
VGIIa |
26.5 |
17.7 |
−8.8 |
non-VGI |
20.3 |
34.2 |
14.0 |
VGII |
34.1 |
19.1 |
−15.0 |
non-VGIII |
33.2 |
22.2 |
−11.0 |
non-VGIV |
VGII |
| B8563 |
VGIIa |
24.4 |
16.0 |
−8.4 |
non-VGI |
18.4 |
32.8 |
14.4 |
VGII |
32.8 |
20.4 |
−12.4 |
non-VGIII |
32.2 |
17.3 |
−14.9 |
non-VGIV |
VGII |
| B8567 |
VGIIa |
25.6 |
17.0 |
−8.6 |
non-VGI |
19.4 |
34.1 |
14.7 |
VGII |
33.8 |
18.2 |
−15.6 |
non-VGIII |
35.1 |
16.8 |
−18.2 |
non-VGIV |
VGII |
| B8854 |
VGIIa |
24.7 |
15.8 |
−8.9 |
non-VGI |
18.1 |
32.7 |
14.6 |
VGII |
33.0 |
17.1 |
−15.9 |
non-VGIII |
33.2 |
15.8 |
−17.4 |
non-VGIV |
VGII |
| B8889 |
VGIIa |
28.0 |
17.6 |
−10.4 |
non-VGI |
20.3 |
33.1 |
12.7 |
VGII |
33.7 |
19.1 |
−14.6 |
non-VGIII |
32.4 |
17.5 |
−15.0 |
non-VGIV |
VGII |
| B9077 |
VGIIa |
33.6 |
17.8 |
−15.9 |
non-VGI |
15.4 |
28.6 |
13.2 |
VGII |
40.0 |
18.6 |
−21.5 |
non-VGIII |
40.0 |
18.6 |
−21.4 |
non-VGIV |
VGII |
| B9296 |
VGIIa |
27.3 |
19.8 |
−7.5 |
non-VGI |
18.6 |
34.0 |
15.4 |
VGII |
32.4 |
20.8 |
−11.6 |
non-VGIII |
34.9 |
19.2 |
−15.7 |
non-VGIV |
VGII |
| B7394 |
VGIIb |
31.9 |
22.5 |
−9.5 |
non-VGI |
23.5 |
33.5 |
10.0 |
VGII |
33.7 |
19.3 |
−14.4 |
non-VGIII |
40.0 |
20.2 |
−19.8 |
non-VGIV |
VGII |
| B7735 |
VGIIb |
26.9 |
17.8 |
−9.1 |
non-VGI |
18.3 |
33.3 |
15.0 |
VGII |
0.0 |
15.8 |
15.8 |
non-VGIII |
40.0 |
15.4 |
−24.6 |
non-VGIV |
VGII |
| B8554 |
VGIIb |
28.8 |
18.3 |
−10.5 |
non-VGI |
20.8 |
32.2 |
11.3 |
VGII |
35.5 |
22.0 |
−13.4 |
non-VGIII |
40.0 |
18.3 |
−21.7 |
non-VGIV |
VGII |
| B8828 |
VGIIb |
28.8 |
18.5 |
−10.3 |
non-VGI |
20.7 |
32.7 |
11.9 |
VGII |
35.9 |
19.2 |
−16.7 |
non-VGIII |
40.0 |
31.9 |
−8.1 |
non-VGIV |
VGII |
| B8211 |
VGIIb |
22.9 |
12.8 |
−10.1 |
non-VGI |
15.1 |
30.1 |
15.1 |
VGII |
33.0 |
13.9 |
−19.0 |
non-VGIII |
33.8 |
12.9 |
−21.0 |
non-VGIV |
VGII |
| B8966 |
VGIIb |
24.6 |
15.5 |
−9.0 |
non-VGI |
17.3 |
25.9 |
8.6 |
VGII |
29.3 |
15.6 |
−13.7 |
non-VGIII |
28.9 |
14.7 |
−14.2 |
non-VGIV |
VGII |
| B9076 |
VGIIb |
40.0 |
17.5 |
−22.5 |
non-VGI |
17.1 |
27.5 |
10.5 |
VGII |
40.0 |
18.4 |
−21.6 |
non-VGIII |
30.6 |
18.0 |
−12.6 |
non-VGIV |
VGII |
| B9157 |
VGIIb |
25.4 |
15.3 |
−10.2 |
non-VGI |
17.6 |
29.4 |
11.9 |
VGII |
31.2 |
16.1 |
−15.1 |
non-VGIII |
31.6 |
16.1 |
−15.5 |
non-VGIV |
VGII |
| B9170 |
VGIIb |
26.2 |
16.9 |
−9.3 |
non-VGI |
17.5 |
28.7 |
11.2 |
VGII |
29.5 |
17.6 |
−11.9 |
non-VGIII |
31.1 |
17.7 |
−13.4 |
non-VGIV |
VGII |
| B9234 |
VGIIb |
24.7 |
15.0 |
−9.6 |
non-VGI |
15.4 |
30.3 |
14.9 |
VGII |
30.2 |
15.7 |
−14.5 |
non-VGIII |
33.3 |
15.8 |
−17.5 |
non-VGIV |
VGII |
| B9290 |
VGIIb |
24.8 |
16.0 |
−8.8 |
non-VGI |
15.9 |
34.1 |
18.2 |
VGII |
30.6 |
20.8 |
−9.7 |
non-VGIII |
33.2 |
16.6 |
−16.6 |
non-VGIV |
VGII |
| B9241 |
VGIIb |
23.4 |
13.2 |
−10.3 |
non-VGI |
15.5 |
28.0 |
12.5 |
VGII |
30.0 |
13.9 |
−16.0 |
non-VGIII |
34.0 |
13.5 |
−20.5 |
non-VGIV |
VGII |
| B9428 |
VGIIb |
25.2 |
14.4 |
−10.7 |
non-VGI |
18.7 |
28.3 |
9.6 |
VGII |
30.2 |
15.5 |
−14.7 |
non-VGIII |
34.1 |
15.0 |
−19.1 |
non-VGIV |
VGII |
| B6863 |
VGIIc |
28.9 |
18.6 |
−10.2 |
non-VGI |
20.7 |
34.2 |
13.5 |
VGII |
33.2 |
22.7 |
−10.6 |
non-VGIII |
40.0 |
18.1 |
−21.9 |
non-VGIV |
VGII |
| B7390 |
VGIIc |
27.7 |
18.3 |
−9.5 |
non-VGI |
19.9 |
33.9 |
13.9 |
VGII |
39.5 |
24.7 |
−14.8 |
non-VGIII |
40.0 |
16.9 |
−23.1 |
non-VGIV |
VGII |
| B7432 |
VGIIc |
28.2 |
18.3 |
−9.9 |
non-VGI |
20.0 |
32.6 |
12.7 |
VGII |
34.8 |
18.0 |
−16.8 |
non-VGIII |
40.0 |
17.2 |
−22.8 |
non-VGIV |
VGII |
| B7434 |
VGIIc |
25.6 |
16.2 |
−9.4 |
non-VGI |
17.7 |
34.5 |
16.8 |
VGII |
34.4 |
17.9 |
−16.5 |
non-VGIII |
40.0 |
13.8 |
−26.2 |
non-VGIV |
VGII |
| B7466 |
VGIIc |
30.8 |
20.8 |
−10.0 |
non-VGI |
22.4 |
33.6 |
11.2 |
VGII |
37.4 |
23.7 |
−13.7 |
non-VGIII |
40.0 |
19.5 |
−20.5 |
non-VGIV |
VGII |
| B7491 |
VGIIc |
26.9 |
17.3 |
−9.6 |
non-VGI |
19.2 |
33.0 |
13.8 |
VGII |
0.0 |
16.8 |
16.8 |
non-VGIII |
40.0 |
16.7 |
−23.3 |
non-VGIV |
VGII |
| B7493 |
VGIIc |
27.1 |
17.4 |
−9.7 |
non-VGI |
18.6 |
33.6 |
15.1 |
VGII |
36.6 |
20.7 |
−15.8 |
non-VGIII |
40.0 |
16.1 |
−23.9 |
non-VGIV |
VGII |
| B7641 |
VGIIc |
26.0 |
17.3 |
−8.7 |
non-VGI |
18.7 |
32.3 |
13.7 |
VGII |
34.3 |
20.0 |
−14.3 |
non-VGIII |
40.0 |
15.6 |
−24.4 |
non-VGIV |
VGII |
| B7737 |
VGIIc |
28.0 |
18.5 |
−9.6 |
non-VGI |
20.1 |
34.3 |
14.2 |
VGII |
37.0 |
23.0 |
−14.0 |
non-VGIII |
40.0 |
18.0 |
−22.0 |
non-VGIV |
VGII |
| B7765 |
VGIIc |
22.5 |
13.0 |
−9.5 |
non-VGI |
14.5 |
34.1 |
19.6 |
VGII |
33.1 |
23.4 |
−9.7 |
non-VGIII |
40.0 |
12.9 |
−27.1 |
non-VGIV |
VGII |
| B8210 |
VGIIc |
27.8 |
18.1 |
−9.7 |
non-VGI |
19.6 |
33.3 |
13.7 |
VGII |
33.0 |
19.4 |
−13.5 |
non-VGIII |
40.0 |
16.8 |
−23.2 |
non-VGIV |
VGII |
| B8214 |
VGIIc |
27.1 |
17.7 |
−9.5 |
non-VGI |
19.8 |
34.9 |
15.1 |
VGII |
34.1 |
20.1 |
−14.0 |
non-VGIII |
40.0 |
16.1 |
−23.9 |
non-VGIV |
VGII |
| B8510 |
VGIIc |
26.8 |
17.6 |
−9.2 |
non-VGI |
18.8 |
33.2 |
14.5 |
VGII |
35.2 |
19.1 |
−16.1 |
non-VGIII |
40.0 |
15.6 |
−24.4 |
non-VGIV |
VGII |
| B8549 |
VGIIc |
26.8 |
16.2 |
−10.6 |
non-VGI |
18.7 |
33.5 |
14.8 |
VGII |
37.4 |
20.5 |
−16.9 |
non-VGIII |
40.0 |
29.6 |
−10.4 |
non-VGIV |
VGII |
| B8552 |
VGIIc |
27.1 |
17.0 |
−10.1 |
non-VGI |
18.6 |
33.2 |
14.6 |
VGII |
34.3 |
19.7 |
−14.6 |
non-VGIII |
40.0 |
16.6 |
−23.4 |
non-VGIV |
VGII |
| B8571 |
VGIIc |
28.8 |
19.4 |
−9.4 |
non-VGI |
21.5 |
33.4 |
11.9 |
VGII |
34.5 |
22.8 |
−11.8 |
non-VGIII |
40.0 |
19.5 |
−20.5 |
non-VGIV |
VGII |
| B8788 |
VGIIc |
26.0 |
16.0 |
−10.0 |
non-VGI |
18.5 |
29.5 |
11.0 |
VGII |
38.0 |
20.4 |
−17.6 |
non-VGIII |
40.0 |
16.6 |
−23.4 |
non-VGIV |
VGII |
| B8798 |
VGIIc |
36.0 |
24.7 |
−11.4 |
non-VGI |
26.5 |
33.3 |
6.8 |
VGII |
37.2 |
19.2 |
−18.0 |
non-VGIII |
40.0 |
22.5 |
−17.5 |
non-VGIV |
VGII |
| B8821 |
VGIIc |
30.5 |
20.5 |
−10.0 |
non-VGI |
22.3 |
33.0 |
10.7 |
VGII |
37.0 |
29.0 |
−8.0 |
non-VGIII |
40.0 |
18.7 |
−21.3 |
non-VGIV |
VGII |
| B8825 |
VGIIc |
27.4 |
17.8 |
−9.6 |
non-VGI |
19.6 |
33.7 |
14.1 |
VGII |
36.0 |
20.5 |
−15.5 |
non-VGIII |
40.0 |
17.5 |
−22.5 |
non-VGIV |
VGII |
| B8833 |
VGIIc |
29.2 |
20.7 |
−8.6 |
non-VGI |
19.5 |
33.4 |
13.9 |
VGII |
35.4 |
19.6 |
−15.8 |
non-VGIII |
40.0 |
15.5 |
−24.5 |
non-VGIV |
VGII |
| B8838 |
VGIIc |
29.2 |
19.1 |
−10.1 |
non-VGI |
21.5 |
32.8 |
11.3 |
VGII |
32.9 |
22.3 |
−10.6 |
non-VGIII |
40.0 |
18.5 |
−21.5 |
non-VGIV |
VGII |
| B8843 |
VGIIc |
29.5 |
19.4 |
−10.1 |
non-VGI |
21.5 |
33.7 |
12.2 |
VGII |
37.5 |
22.1 |
−15.4 |
non-VGIII |
40.0 |
19.1 |
−20.9 |
non-VGIV |
VGII |
| B8853 |
VGIIc |
33.3 |
23.1 |
−10.2 |
non-VGI |
24.8 |
33.7 |
8.9 |
VGII |
34.2 |
27.8 |
−6.4 |
non-VGIII |
40.0 |
21.5 |
−18.5 |
non-VGIV |
VGII |
| B9159 |
VGIIc |
29.6 |
17.5 |
−12.1 |
non-VGI |
19.1 |
29.9 |
10.7 |
VGII |
40.0 |
26.0 |
−14.0 |
non-VGIII |
40.0 |
18.0 |
−22.0 |
non-VGIV |
VGII |
| B9227 |
VGIIc |
24.4 |
15.3 |
−9.1 |
non-VGI |
15.5 |
28.1 |
12.6 |
VGII |
27.9 |
16.1 |
−11.9 |
non-VGIII |
31.0 |
16.3 |
−14.7 |
non-VGIV |
VGII |
| B9235 |
VGIIc |
24.6 |
15.1 |
−9.5 |
non-VGI |
15.3 |
28.9 |
13.7 |
VGII |
29.2 |
16.4 |
−12.7 |
non-VGIII |
31.2 |
15.9 |
−15.3 |
non-VGIV |
VGII |
| B9244 |
VGIIc |
27.3 |
18.4 |
−8.9 |
non-VGI |
18.5 |
31.8 |
13.3 |
VGII |
28.2 |
21.0 |
−7.2 |
non-VGIII |
30.6 |
18.8 |
−11.8 |
non-VGIV |
VGII |
| B9245 |
VGIIc |
26.8 |
17.9 |
−8.9 |
non-VGI |
18.0 |
33.5 |
15.5 |
VGII |
31.2 |
19.3 |
−11.9 |
non-VGIII |
34.2 |
18.5 |
−15.6 |
non-VGIV |
VGII |
| B9295 |
VGIIc |
28.6 |
19.5 |
−9.1 |
non-VGI |
19.9 |
40.0 |
20.1 |
VGII |
33.6 |
25.5 |
−8.1 |
non-VGIII |
34.4 |
20.3 |
−14.2 |
non-VGIV |
VGII |
| B9302 |
VGIIc |
24.6 |
14.1 |
−10.5 |
non-VGI |
16.9 |
26.7 |
9.8 |
VGII |
28.8 |
15.1 |
−13.7 |
non-VGIII |
31.5 |
14.1 |
−17.3 |
non-VGIV |
VGII |
| B9374 |
VGIIc |
24.8 |
14.2 |
−10.6 |
non-VGI |
18.2 |
27.3 |
9.1 |
VGII |
29.1 |
15.2 |
−13.9 |
non-VGIII |
32.8 |
14.4 |
−18.4 |
non-VGIV |
VGII |
| B7415 |
VGIII |
26.8 |
15.9 |
−10.9 |
non-VGI |
35.0 |
17.7 |
−17.3 |
non-VGII |
12.4 |
27.1 |
14.7 |
VGIII |
30.9 |
15.9 |
−15.0 |
non-VGIV |
VGIII |
| B7495 |
VGIII |
28.1 |
18.0 |
−10.1 |
non-VGI |
36.1 |
18.8 |
−17.3 |
non-VGII |
14.1 |
30.1 |
16.0 |
VGIII |
31.8 |
17.6 |
−14.2 |
non-VGIV |
VGIII |
| B8212 |
VGIII |
26.0 |
15.7 |
−10.3 |
non-VGI |
35.3 |
17.0 |
−18.3 |
non-VGII |
12.4 |
28.5 |
16.1 |
VGIII |
32.5 |
15.6 |
−16.9 |
non-VGIV |
VGIII |
| B8260 |
VGIII |
29.6 |
19.6 |
−10.0 |
non-VGI |
36.7 |
20.8 |
−15.9 |
non-VGII |
15.9 |
30.7 |
14.8 |
VGIII |
36.0 |
19.1 |
−16.9 |
non-VGIV |
VGIII |
| B8262 |
VGIII |
27.2 |
17.2 |
−10.0 |
non-VGI |
33.8 |
18.3 |
−15.5 |
non-VGII |
13.5 |
30.0 |
16.4 |
VGIII |
40.0 |
16.9 |
−23.1 |
non-VGIV |
VGIII |
| B8516/B8616 |
VGIII |
28.4 |
18.5 |
−9.9 |
non-VGI |
37.8 |
19.5 |
−18.3 |
non-VGII |
14.6 |
29.1 |
14.5 |
VGIII |
31.8 |
18.0 |
−13.8 |
non-VGIV |
VGIII |
| B9143 |
VGIII |
28.6 |
18.3 |
−10.3 |
non-VGI |
38.3 |
19.6 |
−18.7 |
non-VGII |
14.5 |
30.2 |
15.7 |
VGIII |
33.3 |
18.0 |
−15.3 |
non-VGIV |
VGIII |
| B9146 |
VGIII |
30.3 |
19.5 |
−10.8 |
non-VGI |
38.5 |
21.2 |
−17.3 |
non-VGII |
15.8 |
30.1 |
14.3 |
VGIII |
31.2 |
19.3 |
−11.9 |
non-VGIV |
VGIII |
| B8965 |
VGIII |
26.2 |
16.8 |
−9.4 |
non-VGI |
30.6 |
17.1 |
−13.5 |
non-VGII |
16.1 |
30.6 |
14.5 |
VGIII |
35.0 |
17.4 |
−17.6 |
non-VGIV |
VGIII |
| B9148 |
VGIII |
26.0 |
16.6 |
−9.4 |
non-VGI |
31.0 |
16.6 |
−14.4 |
non-VGII |
15.9 |
30.6 |
14.7 |
VGIII |
32.8 |
17.4 |
−15.4 |
non-VGIV |
VGIII |
| B9151 |
VGIII |
25.7 |
16.5 |
−9.3 |
non-VGI |
30.7 |
16.2 |
−14.4 |
non-VGII |
15.4 |
30.3 |
14.9 |
VGIII |
34.9 |
18.0 |
−17.0 |
non-VGIV |
VGIII |
| B9163 |
VGIII |
26.9 |
17.5 |
−9.4 |
non-VGI |
29.8 |
17.3 |
−12.5 |
non-VGII |
16.9 |
29.7 |
12.8 |
VGIII |
33.4 |
18.0 |
−15.4 |
non-VGIV |
VGIII |
| B9237 |
VGIII |
26.7 |
17.9 |
−8.9 |
non-VGI |
31.6 |
17.4 |
−14.2 |
non-VGII |
17.3 |
35.0 |
17.7 |
VGIII |
38.1 |
19.3 |
−18.9 |
non-VGIV |
VGIII |
| B9372 |
VGIII |
23.5 |
12.7 |
−10.9 |
non-VGI |
29.3 |
13.1 |
−16.1 |
non-VGII |
14.8 |
27.4 |
12.6 |
VGIII |
32.6 |
13.0 |
−19.6 |
non-VGIV |
VGIII |
| B9422 |
VGIII |
23.9 |
12.8 |
−11.1 |
non-VGI |
28.9 |
12.9 |
−15.9 |
non-VGII |
14.6 |
26.8 |
12.2 |
VGIII |
33.0 |
13.3 |
−19.7 |
non-VGIV |
VGIII |
| B9430 |
VGIII |
23.5 |
12.9 |
−10.6 |
non-VGI |
30.1 |
13.4 |
−16.8 |
non-VGII |
15.1 |
28.5 |
13.4 |
VGIII |
35.5 |
13.4 |
−22.0 |
non-VGIV |
VGIII |
| B7238 |
VGIV |
25.2 |
16.4 |
−8.8 |
non-VGI |
33.2 |
18.5 |
−14.7 |
non-VGII |
34.6 |
17.9 |
−16.7 |
non-VGIII |
16.3 |
27.4 |
11.1 |
VGIV |
VGIV |
| B7240 |
VGIV |
25.8 |
17.1 |
−8.8 |
non-VGI |
33.9 |
19.5 |
−14.5 |
non-VGII |
34.2 |
18.5 |
−15.7 |
non-VGIII |
17.0 |
28.8 |
11.8 |
VGIV |
VGIV |
| B7243 |
VGIV |
26.1 |
17.3 |
−8.8 |
non-VGI |
32.0 |
19.6 |
−12.4 |
non-VGII |
32.3 |
18.7 |
−13.6 |
non-VGIII |
16.8 |
27.1 |
10.2 |
VGIV |
VGIV |
| B7247 |
VGIV |
25.6 |
16.5 |
−9.1 |
non-VGI |
33.4 |
19.2 |
−14.2 |
non-VGII |
32.0 |
18.1 |
−13.9 |
non-VGIII |
16.3 |
28.4 |
12.1 |
VGIV |
VGIV |
| B7249 |
VGIV |
23.4 |
14.8 |
−8.6 |
non-VGI |
31.6 |
16.7 |
−14.9 |
non-VGII |
32.6 |
16.0 |
−16.6 |
non-VGIII |
14.5 |
31.1 |
16.5 |
VGIV |
VGIV |
| B7260 |
VGIV |
26.0 |
16.5 |
−9.4 |
non-VGI |
30.9 |
18.0 |
−13.0 |
non-VGII |
34.2 |
17.4 |
−16.8 |
non-VGIII |
15.7 |
27.0 |
11.2 |
VGIV |
VGIV |
| B7262 |
VGIV |
26.3 |
16.8 |
−9.5 |
non-VGI |
31.4 |
18.7 |
−12.7 |
non-VGII |
33.4 |
18.0 |
−15.4 |
non-VGIII |
15.8 |
27.5 |
11.6 |
VGIV |
VGIV |
| B7263 |
VGIV |
24.5 |
15.7 |
−8.9 |
non-VGI |
33.1 |
17.9 |
−15.3 |
non-VGII |
37.3 |
17.0 |
−20.3 |
non-VGIII |
15.8 |
28.0 |
12.2 |
VGIV |
VGIV |
| B7264 |
VGIV |
24.4 |
15.0 |
−9.4 |
non-VGI |
31.2 |
16.9 |
−14.3 |
non-VGII |
30.6 |
16.0 |
−14.6 |
non-VGIII |
14.8 |
26.8 |
12.0 |
VGIV |
VGIV |
| B7265 | VGIV | 27.5 | 17.3 | −10.2 | non-VGI | 34.1 | 19.6 | −14.5 | non-VGII | 32.1 | 18.8 | −13.3 | non-VGIII | 16.9 | 28.8 | 11.9 | VGIV | VGIV |
Table 5.
VGII subtyping SYBR MAMA Ct values and genotype assignments for VGIIa,b,c
| |
VGIIa_Assay_45211 |
VGIIb_Assay_502129 |
VGIIc_Assay_257655 |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Isolate ID | Strain type via MLST | VGIIa Ct Mean | non-VGIIa Ct Mean | Delta Ct | Type call via assay | VGIIb Ct Mean | non-VGIIb Ct Mean | Delta Ct | Type call via assay | VGIIc Ct Mean | non-VGIIc Ct Mean | Delta Ct | Type call via assay | Final Call |
| B6864 |
VGIIa |
17.2 |
30.5 |
13.3 |
VGIIa |
31.0 |
17.5 |
−13.5 |
non-VGIIb |
40.0 |
27.8 |
−12.2 |
non-VGIIc |
VGIIa |
| B7395 |
VGIIa |
19.8 |
33.5 |
13.7 |
VGIIa |
33.1 |
20.3 |
−12.9 |
non-VGIIb |
40.0 |
30.6 |
−9.4 |
non-VGIIc |
VGIIa |
| B7422 |
VGIIa |
18.3 |
33.6 |
15.4 |
VGIIa |
26.4 |
17.6 |
−8.8 |
non-VGIIb |
39.2 |
28.6 |
−10.6 |
non-VGIIc |
VGIIa |
| B7436 |
VGIIa |
18.6 |
31.7 |
13.1 |
VGIIa |
30.1 |
17.0 |
−13.2 |
non-VGIIb |
38.0 |
29.1 |
−8.9 |
non-VGIIc |
VGIIa |
| B7467 |
VGIIa |
20.5 |
37.3 |
16.8 |
VGIIa |
35.1 |
20.3 |
−14.7 |
non-VGIIb |
40.0 |
30.9 |
−9.1 |
non-VGIIc |
VGIIa |
| B8555 |
VGIIa |
17.1 |
31.2 |
14.1 |
VGIIa |
30.3 |
17.5 |
−12.8 |
non-VGIIb |
40.0 |
27.7 |
−12.3 |
non-VGIIc |
VGIIa |
| B8577 |
VGIIa |
20.8 |
36.8 |
16.0 |
VGIIa |
32.8 |
20.8 |
−12.1 |
non-VGIIb |
40.0 |
31.4 |
−8.6 |
non-VGIIc |
VGIIa |
| B8793 |
VGIIa |
15.1 |
29.8 |
14.7 |
VGIIa |
30.7 |
18.6 |
−12.1 |
non-VGIIb |
40.0 |
29.8 |
−10.2 |
non-VGIIc |
VGIIa |
| B8849 |
VGIIa |
19.8 |
34.4 |
14.6 |
VGIIa |
33.6 |
20.2 |
−13.4 |
non-VGIIb |
40.0 |
30.6 |
−9.4 |
non-VGIIc |
VGIIa |
| CA-1014 |
VGIIa |
13.1 |
27.3 |
14.2 |
VGIIa |
27.0 |
14.0 |
−13.0 |
non-VGIIb |
34.9 |
24.2 |
−10.7 |
non-VGIIc |
VGIIa |
| CBS-7750 |
VGIIa |
21.8 |
32.2 |
10.4 |
VGIIa |
33.4 |
21.5 |
−11.9 |
non-VGIIb |
40.0 |
34.1 |
−5.9 |
non-VGIIc |
VGIIa |
| ICB-107 |
VGIIa |
21.8 |
33.6 |
11.8 |
VGIIa |
33.2 |
21.2 |
−12.0 |
non-VGIIb |
40.0 |
33.8 |
−6.2 |
non-VGIIc |
VGIIa |
| NIH-444 |
VGIIa |
14.8 |
27.3 |
12.5 |
VGIIa |
28.5 |
15.3 |
−13.1 |
non-VGIIb |
36.1 |
25.7 |
−10.3 |
non-VGIIc |
VGIIa |
| B8508 |
VGIIa |
17.0 |
27.8 |
10.8 |
VGIIa |
26.5 |
17.3 |
−9.2 |
non-VGIIb |
31.7 |
22.7 |
−9.1 |
non-VGIIc |
VGIIa |
| B8512 |
VGIIa |
17.6 |
28.1 |
10.4 |
VGIIa |
26.3 |
18.0 |
−8.3 |
non-VGIIb |
33.2 |
24.2 |
−9.0 |
non-VGIIc |
VGIIa |
| B8558 |
VGIIa |
16.3 |
24.8 |
8.5 |
VGIIa |
27.3 |
15.3 |
−12.0 |
non-VGIIb |
29.4 |
20.0 |
−9.4 |
non-VGIIc |
VGIIa |
| B8561 |
VGIIa |
15.8 |
27.5 |
11.8 |
VGIIa |
25.0 |
16.9 |
−8.1 |
non-VGIIb |
33.4 |
23.2 |
−10.2 |
non-VGIIc |
VGIIa |
| B8563 |
VGIIa |
14.5 |
27.3 |
12.8 |
VGIIa |
23.9 |
15.6 |
−8.3 |
non-VGIIb |
31.7 |
21.7 |
−10.0 |
non-VGIIc |
VGIIa |
| B8567 |
VGIIa |
15.0 |
36.2 |
21.2 |
VGIIa |
24.5 |
16.0 |
−8.5 |
non-VGIIb |
31.8 |
22.2 |
−9.5 |
non-VGIIc |
VGIIa |
| B8854 |
VGIIa |
14.7 |
26.7 |
12.0 |
VGIIa |
24.1 |
15.1 |
−9.0 |
non-VGIIb |
31.4 |
22.2 |
−9.2 |
non-VGIIc |
VGIIa |
| B8889 |
VGIIa |
17.0 |
28.1 |
11.0 |
VGIIa |
25.9 |
17.3 |
−8.7 |
non-VGIIb |
33.2 |
23.8 |
−9.4 |
non-VGIIc |
VGIIa |
| B9077 |
VGIIa |
16.7 |
27.8 |
11.1 |
VGIIa |
25.6 |
16.7 |
−9.0 |
non-VGIIb |
32.9 |
24.4 |
−8.4 |
non-VGIIc |
VGIIa |
| B9296 |
VGIIa |
17.0 |
27.5 |
10.5 |
VGIIa |
25.5 |
17.3 |
−8.2 |
non-VGIIb |
32.9 |
24.8 |
−8.1 |
non-VGIIc |
VGIIa |
| B7394 |
VGIIb |
40.0 |
19.0 |
−21.0 |
non-VGIIa |
17.3 |
29.6 |
12.3 |
VGIIb |
40.0 |
29.0 |
−11.0 |
non-VGIIc |
VGIIb |
| B7735 |
VGIIb |
31.0 |
18.3 |
−12.8 |
non-VGIIa |
18.7 |
31.3 |
12.6 |
VGIIb |
38.1 |
28.9 |
−9.3 |
non-VGIIc |
VGIIb |
| B8554 |
VGIIb |
32.9 |
21.2 |
−11.7 |
non-VGIIa |
22.2 |
35.0 |
12.8 |
VGIIb |
40.0 |
30.4 |
−9.6 |
non-VGIIc |
VGIIb |
| B8828 |
VGIIb |
31.9 |
21.1 |
−10.8 |
non-VGIIa |
19.9 |
35.1 |
15.2 |
VGIIb |
40.0 |
30.5 |
−9.5 |
non-VGIIc |
VGIIb |
| B8211 |
VGIIb |
27.8 |
16.9 |
−10.9 |
non-VGIIa |
17.4 |
28.8 |
11.4 |
VGIIb |
32.3 |
22.3 |
−10.0 |
non-VGIIc |
VGIIb |
| B8966 |
VGIIb |
26.2 |
14.7 |
−11.5 |
non-VGIIa |
16.3 |
24.1 |
7.9 |
VGIIb |
31.8 |
23.2 |
−8.6 |
non-VGIIc |
VGIIb |
| B9076 |
VGIIb |
30.0 |
18.8 |
−11.2 |
non-VGIIa |
19.7 |
30.9 |
11.4 |
VGIIb |
39.1 |
27.0 |
−12.1 |
non-VGIIc |
VGIIb |
| B9157 |
VGIIb |
29.1 |
16.6 |
−12.4 |
non-VGIIa |
15.4 |
23.8 |
8.5 |
VGIIb |
30.3 |
21.3 |
−9.0 |
non-VGIIc |
VGIIb |
| B9170 |
VGIIb |
26.6 |
15.4 |
−11.2 |
non-VGIIa |
16.9 |
24.8 |
7.9 |
VGIIb |
31.0 |
22.7 |
−8.3 |
non-VGIIc |
VGIIb |
| B9234 |
VGIIb |
26.1 |
13.9 |
−12.2 |
non-VGIIa |
15.3 |
23.8 |
8.5 |
VGIIb |
30.2 |
21.2 |
−9.1 |
non-VGIIc |
VGIIb |
| B9290 |
VGIIb |
26.1 |
13.8 |
−12.3 |
non-VGIIa |
15.1 |
24.5 |
9.5 |
VGIIb |
30.6 |
21.2 |
−9.5 |
non-VGIIc |
VGIIb |
| B9241 |
VGIIb |
26.7 |
20.2 |
−6.5 |
non-VGIIa |
14.5 |
24.0 |
9.4 |
VGIIb |
30.5 |
21.4 |
−9.1 |
non-VGIIc |
VGIIb |
| B9428 |
VGIIb |
27.5 |
14.8 |
−12.6 |
non-VGIIa |
16.0 |
24.3 |
8.2 |
VGIIb |
32.0 |
22.4 |
−9.6 |
non-VGIIc |
VGIIb |
| B6863 |
VGIIc |
31.9 |
20.3 |
−11.5 |
non-VGIIa |
33.4 |
20.2 |
−13.2 |
non-VGIIb |
27.5 |
40.0 |
12.5 |
VGIIc |
VGIIc |
| B7390 |
VGIIc |
32.7 |
18.9 |
−13.8 |
non-VGIIa |
31.1 |
17.9 |
−13.2 |
non-VGIIb |
25.9 |
40.0 |
14.1 |
VGIIc |
VGIIc |
| B7432 |
VGIIc |
40.0 |
18.5 |
−21.5 |
non-VGIIa |
30.7 |
17.6 |
−13.1 |
non-VGIIb |
25.7 |
40.0 |
14.3 |
VGIIc |
VGIIc |
| B7434 |
VGIIc |
27.5 |
15.5 |
−12.0 |
non-VGIIa |
28.5 |
15.4 |
−13.1 |
non-VGIIb |
23.3 |
40.0 |
16.7 |
VGIIc |
VGIIc |
| B7466 |
VGIIc |
31.7 |
20.8 |
−10.9 |
non-VGIIa |
33.5 |
20.6 |
−12.8 |
non-VGIIb |
28.1 |
40.0 |
11.9 |
VGIIc |
VGIIc |
| B7491 |
VGIIc |
28.7 |
17.4 |
−11.2 |
non-VGIIa |
30.4 |
16.9 |
−13.5 |
non-VGIIb |
24.0 |
40.0 |
16.0 |
VGIIc |
VGIIc |
| B7493 |
VGIIc |
28.8 |
18.3 |
−10.6 |
non-VGIIa |
31.1 |
18.0 |
−13.1 |
non-VGIIb |
25.5 |
40.0 |
14.5 |
VGIIc |
VGIIc |
| B7641 |
VGIIc |
29.2 |
17.2 |
−12.0 |
non-VGIIa |
30.0 |
17.2 |
−12.8 |
non-VGIIb |
24.5 |
40.0 |
15.5 |
VGIIc |
VGIIc |
| B7737 |
VGIIc |
32.6 |
20.1 |
−12.5 |
non-VGIIa |
30.8 |
20.5 |
−10.4 |
non-VGIIb |
28.4 |
40.0 |
11.6 |
VGIIc |
VGIIc |
| B7765 |
VGIIc |
32.2 |
19.3 |
−12.8 |
non-VGIIa |
32.3 |
18.9 |
−13.3 |
non-VGIIb |
27.5 |
40.0 |
12.5 |
VGIIc |
VGIIc |
| B8210 |
VGIIc |
29.7 |
17.6 |
−12.0 |
non-VGIIa |
30.1 |
17.4 |
−12.7 |
non-VGIIb |
25.9 |
40.0 |
14.1 |
VGIIc |
VGIIc |
| B8214 |
VGIIc |
30.1 |
17.5 |
−12.5 |
non-VGIIa |
30.9 |
17.5 |
−13.4 |
non-VGIIb |
26.1 |
40.0 |
13.9 |
VGIIc |
VGIIc |
| B8510 |
VGIIc |
29.6 |
17.5 |
−12.0 |
non-VGIIa |
31.0 |
17.3 |
−13.7 |
non-VGIIb |
24.5 |
40.0 |
15.5 |
VGIIc |
VGIIc |
| B8549 |
VGIIc |
29.9 |
17.7 |
−12.1 |
non-VGIIa |
31.0 |
17.8 |
−13.2 |
non-VGIIb |
24.8 |
40.0 |
15.2 |
VGIIc |
VGIIc |
| B8552 |
VGIIc |
29.2 |
17.1 |
−12.0 |
non-VGIIa |
30.3 |
17.2 |
−13.1 |
non-VGIIb |
24.4 |
40.0 |
15.6 |
VGIIc |
VGIIc |
| B8571 |
VGIIc |
33.0 |
20.3 |
−12.7 |
non-VGIIa |
32.6 |
20.2 |
−12.5 |
non-VGIIb |
28.1 |
40.0 |
11.9 |
VGIIc |
VGIIc |
| B8788 |
VGIIc |
29.1 |
17.3 |
−11.7 |
non-VGIIa |
30.0 |
17.2 |
−12.8 |
non-VGIIb |
25.0 |
40.0 |
15.0 |
VGIIc |
VGIIc |
| B8798 |
VGIIc |
36.5 |
22.8 |
−13.7 |
non-VGIIa |
34.5 |
22.2 |
−12.3 |
non-VGIIb |
31.0 |
40.0 |
9.0 |
VGIIc |
VGIIc |
| B8821 |
VGIIc |
37.7 |
24.5 |
−13.2 |
non-VGIIa |
37.1 |
24.4 |
−12.7 |
non-VGIIb |
33.0 |
40.0 |
7.0 |
VGIIc |
VGIIc |
| B8825 |
VGIIc |
29.6 |
17.7 |
−11.9 |
non-VGIIa |
30.6 |
17.7 |
−12.9 |
non-VGIIb |
25.8 |
40.0 |
14.2 |
VGIIc |
VGIIc |
| B8833 |
VGIIc |
29.0 |
17.0 |
−12.0 |
non-VGIIa |
30.1 |
17.0 |
−13.1 |
non-VGIIb |
25.2 |
40.0 |
14.8 |
VGIIc |
VGIIc |
| B8838 |
VGIIc |
32.0 |
19.5 |
−12.5 |
non-VGIIa |
32.9 |
19.3 |
−13.7 |
non-VGIIb |
28.7 |
40.0 |
11.3 |
VGIIc |
VGIIc |
| B8843 |
VGIIc |
32.4 |
19.9 |
−12.5 |
non-VGIIa |
33.0 |
19.5 |
−13.5 |
non-VGIIb |
28.6 |
40.0 |
11.4 |
VGIIc |
VGIIc |
| B8853 |
VGIIc |
32.8 |
21.5 |
−11.3 |
non-VGIIa |
36.0 |
23.4 |
−12.6 |
non-VGIIb |
33.1 |
40.0 |
6.9 |
VGIIc |
VGIIc |
| B9159 |
VGIIc |
27.4 |
20.3 |
−7.1 |
non-VGIIa |
25.8 |
16.7 |
−9.1 |
non-VGIIb |
20.5 |
34.5 |
14.0 |
VGIIc |
VGIIc |
| B9227 |
VGIIc |
25.6 |
13.6 |
−12.0 |
non-VGIIa |
23.9 |
14.9 |
−9.0 |
non-VGIIb |
18.0 |
31.5 |
13.4 |
VGIIc |
VGIIc |
| B9235 |
VGIIc |
25.9 |
13.7 |
−12.1 |
non-VGIIa |
24.1 |
14.9 |
−9.2 |
non-VGIIb |
18.4 |
32.4 |
14.0 |
VGIIc |
VGIIc |
| B9244 |
VGIIc |
27.2 |
19.1 |
−8.1 |
non-VGIIa |
26.2 |
16.9 |
−9.2 |
non-VGIIb |
20.2 |
32.5 |
12.3 |
VGIIc |
VGIIc |
| B9245 |
VGIIc |
28.4 |
22.9 |
−5.5 |
non-VGIIa |
25.2 |
17.4 |
−7.8 |
non-VGIIb |
20.7 |
34.5 |
13.8 |
VGIIc |
VGIIc |
| B9295 |
VGIIc |
21.0 |
17.1 |
−3.8 |
non-VGIIa |
26.0 |
19.6 |
−6.4 |
non-VGIIb |
22.1 |
28.1 |
5.9 |
VGIIc |
VGIIc |
| B9302 |
VGIIc |
26.7 |
15.6 |
−11.1 |
non-VGIIa |
23.7 |
15.4 |
−8.3 |
non-VGIIb |
19.4 |
34.3 |
15.0 |
VGIIc |
VGIIc |
| B9374 | VGIIc | 27.4 | 21.6 | −5.8 | non-VGIIa | 24.0 | 15.3 | −8.7 | non-VGIIb | 19.4 | 33.4 | 14.0 | VGIIc | VGIIc |
Table 6.
Interassay and Intraassay for MLST and Subtyping MAMA
| Assay | interrun CV (%) | intrarun CV (%) |
|---|---|---|
| VGI |
4.33 |
1.56 |
| VGII |
2.35 |
0.22 |
| VGIII |
0.43 |
0.60 |
| VGIV |
1.37 |
1.08 |
| VGIIa |
0.22 |
0.50 |
| VGIIb |
1.27 |
0.92 |
| VGIIc | 1.61 | 0.32 |
Table 7.
Lower limit dynamic range for MLST and subtyping MAMA primer sets
| Primer set tested | Limit (pg) | Median Ct |
|---|---|---|
| VGI |
0.5 |
31.7 |
| non-VGI |
0.5 |
31.1 |
| VGII |
0.5 |
29.5 |
| non-VGII |
0.5 |
28.7 |
| VGIII |
0.5 |
28.5 |
| non-VGIII |
0.5 |
29.9 |
| VGIV |
0.5 |
33.7 |
| non-VGIV |
0.5 |
33.2 |
| VGIIa |
0.5 |
30.2 |
| non-VGIIa |
0.5 |
31.2 |
| VGIIb |
0.5 |
30.1 |
| non-VGIIb |
0.5 |
28.5 |
| VGIIc |
0.5 |
37.4 |
| non-VGIIc | 0.05 | 39.4 |
Discussion
C. gattii is an emerging pathogen in the US Pacific Northwest and British Columbia. Molecular and epidemiological investigations revealed the Vancouver Island, BC outbreak was attributed to a novel and seemingly hypervirulent VGIIa genotype [7,20,22]; moreover, the recent PNW outbreak was attributed to an additional novel genotype, VGIIc [23]. These apparent new genotypes (VGIIa and VGIIc), are responsible for greater than 90% of C. gattii infections in the BC/PNW region [7]. Given the increased virulence, varying antifungal susceptibilities and clinical outcomes caused by these genotypes, as compared to other C. gattii genotypes, it will be useful to conduct regular genotyping of C. gattii isolates for both clinical and epidemiological response purposes [5,7,9,16].
We have developed a MAMA real-time PCR panel for cost-efficient and rapid genotyping of C. gattii molecular types (I-IV) and VGII subtypes (a-c) as a means to better understand genotype distribution of C. gattii in North America. To validate the assays, we screened DNA from a diverse North American and international isolate collection of C. gattii isolates from human, environmental, and animal sources. All DNA had been previously typed by MLST. The assay panel performed with 100% sensitivity and specificity and was 100% concordant with MLST results. The VGII subtype specific assays may be more pertinent to the North American public health and medical communities; the molecular type (I-IV) specific assays will be useful for both North American and global genotyping. The assay is designed for screening in a cost-effective, step-wise manner. The molecular type-specific assays should be performed first on all isolates. In North America, the VGIV assay can be withheld for the first screen, as isolates of this molecular type have not yet been isolated from North America. For those North American isolates that are VGII by molecular type, the subtype-specific assays should be performed for typing VGIIa, VGIIb, or VGIIc. As we further our understanding of C. gattii populations around the world and their genotype-phenotype relationships, additional subtype specific assays can be similarly developed for local and global research purposes.
Conclusions
These PCR-based assays are an affordable, efficient, and sensitive means of genotyping C. gattii isolates. Both the assay methods and results can be easily transferred among laboratories. Assay results are based on real-time PCR cycle threshold values and are therefore objective and straightforward for local analysis. The assay panel presented here is a useful tool for conducting large-scale molecular epidemiological studies by public health and research laboratories.
Ethics statement
This study does not involve subjects or materials that would require approval by an ethics committee.
Abbreviations
MAMA: Mismatch amplification mutation assay; MLST: Multilocus sequence typing; PCR-RFLP: PCR-restriction fragment length polymorphism; AFLP: Amplified fragment length polymorphism; MLMT: Multilocus microsatellite typing; HRM: High resolution melting; MALDI-TOF MS: Matrix-assisted laser desorption ionization-time-of-flight mass spectrometry; ASPCR: Allele-specific PCR; SNP: Single nucleotide polymorphism; Ct: Cycle threshold; MPD1: Mannitol-1-phosphate dehydrogenase; WGST: Whole genome sequence typing.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
EK designed the assays, assisted with assay validation, data analysis and drafted the manuscript. EMD participated in the design and coordination of the study, data analysis and assisted with drafting the manuscript. KE performed assay validation and data analysis and assisted with drafting the manuscript. MB was involved in the study conception, design and coordination. JS and JG assisted with data analysis for study design. JT performed assay validation and assay data analysis. SL and ED assisted with study conception, design and coordination and manuscript review. PK assisted with study design, coordination and manuscript review. DE assisted with study conception, design, coordination, and drafting of the manuscript. All authors read and approved the final manuscript.
Contributor Information
Erin J Kelley, Email: ekelley@tgen.org.
Elizabeth M Driebe, Email: edriebe@tgen.org.
Kizee Etienne, Email: guf1@cdc.gov.
Mary E Brandt, Email: mbb4@cdc.gov.
James M Schupp, Email: jschupp@tgen.org.
John D Gillece, Email: jgillece@tgen.org.
Jesse S Trujillo, Email: jst46@email.arizona.edu.
Shawn R Lockhart, Email: gyi2@cdc.gov.
Eszter Deak, Email: edeak@mednet.ucla.edu.
Paul S Keim, Email: paul.keim@nau.edu.
David M Engelthaler, Email: dengelthaler@tgen.org.
Acknowledgements
The findings and conclusions of this article are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention.
The authors wish to thank the members of the Cryptococcus gattii Public Health Working Group for submission of many of the isolates used in this study.
This work was supported by funds from the National Institutes of Health: R21AI098059.
References
- Bovers M, Hagen F, Boekhout T. Diversity of the Cryptococcus neoformans-Cryptococcus gattii species complex. Rev Iberoam Micol. 2008;25(1):S4–S12. doi: 10.1016/S1130-1406(08)70019-6. [DOI] [PubMed] [Google Scholar]
- D’Souza CA, Kronstad JW, Taylor G, Warren R, Yuen M, Hu G, Jung WH, Sham A, Kidd SE, Tangen K, Lee N, Zeilmaker T, Sawkins J, McVicker G, Shah S, Gnerre S, Griggs A, Zeng Q, Bartlett K, Li W, Wang X, Heitman J, Stajich JE, Fraser JA, Meyer W, Carter D, Schein J, Krzywinski M, Kwon-Chung KJ, Varma A. et al. Genome variation in Cryptococcus gattii, an emerging pathogen of immunocompetent hosts. MBio. 2011;2:e00342–10. doi: 10.1128/mBio.00342-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockhart SR, Iqbal N, Bolden CB, DeBess EE, Marsden-Haug N, Worhle R, Thakur R, Harris JR. Epidemiologic cutoff values for triazole drugs in Cryptococcus gattii: correlation of molecular type and in vitro susceptibility. Diagn Microbiol Infect Dis. 2012;73(2):144–148. doi: 10.1016/j.diagmicrobio.2012.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephen CSL, Black W, Fyfe M, Raverty S. Multispecies outbreak of cryptococcosis on southern Vancouver Island, British Columbia. Can Vet J. 2002;43(10):792–794. [PMC free article] [PubMed] [Google Scholar]
- Iqbal N, DeBess EE, Wohrle R, Sun B, Nett RJ, Ahlquist AM, Chiller T, Lockhart SR. Correlation of genotype and in vitro susceptibilities of Cryptococcus gattii strains from the Pacific Northwest of the United States. J Clin Microbiol. 2010;48(2):539–544. doi: 10.1128/JCM.01505-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrnes EJ 3rd, Bildfell RJ, Frank SA, Mitchell TG, Marr KA, Heitman J. Molecular evidence that the range of the Vancouver Island outbreak of Cryptococcus gattii infection has expanded into the Pacific Northwest in the United States. J Infect Dis. 2009;199(7):1081–1086. doi: 10.1086/597306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrnes EJ 3rd, Li W, Lewit Y, Ma H, Voelz K, Ren P, Carter DA, Chaturvedi V, Bildfell RJ, May RC, Heitman J. Emergence and pathogenicity of highly virulent Cryptococcus gattii genotypes in the northwest United States. PLoS Pathog. 2010;6(4):e1000850. doi: 10.1371/journal.ppat.1000850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walraven CJ, Gerstein W, Hardison SE, Wormley F, Lockhart SR, Harris JR, Fothergill A, Wickes B, Gober-Wilcox J, Massie L, Ku TS, Firacative C, Meyer W, Lee SA. Fatal disseminated Cryptococcus gattii infection in New Mexico. PLoS One. 2011;6(12):e28625. doi: 10.1371/journal.pone.0028625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillece JD, Schupp JM, Balajee SA, Harris J, Pearson T, Yan Y, Keim P, DeBess E, Marsden-Haug N, Wohrle R, Engelthaler DM, Lockhart SR. Whole genome sequence analysis of Cryptococcus gattii from the Pacific Northwest Reveals unexpected diversity. PLoS One. 2011;6(12):e28550. doi: 10.1371/journal.pone.0028550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen F, Illnait-Zaragozi MT, Bartlett KH, Swinne D, Geertsen E, Klaassen CH, Boekhout T, Meis JF. In vitro antifungal susceptibilities and amplified fragment length polymorphism genotyping of a worldwide collection of 350 clinical, veterinary, and environmental Cryptococcus gattii isolates. Antimicrob Agents Chemother. 2010;54(12):5139–5145. doi: 10.1128/AAC.00746-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidrim JJ, Costa AK, Cordeiro RA, Brilhante RS, Moura FE, Castelo-Branco DS, Neto MP, Rocha MF. Molecular methods for the diagnosis and characterization of cryptococcus: a review. Can J Microbiol. 2010;56(6):445–458. doi: 10.1139/W10-030. [DOI] [PubMed] [Google Scholar]
- Firacative CTL, Meyer W. MALDI-TOF MS enables the rapid identification of the major molecular types within the Cryptococcus neoformans/C. Gattii species complex. PLoS One. 2012;7(5):e37566. doi: 10.1371/journal.pone.0037566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posteraro B, Vella A, Cogliati M, De Carolis E, Florio AR, Posteraro P, Sanguinetti M, Tortorano AM. Matrix-assisted laser desorption ionization-time of flight mass spectrometry-based method for discrimination between molecular types of Cryptococcus neoformans and Cryptococcus gattii. J Clin Microbiol. 2012;50(7):2472–2476. doi: 10.1128/JCM.00737-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanafy A, Kaocharoen S, Jover-Botella A, Katsu M, Iida S, Kogure T, Gonoi T, Mikami Y, Meyer W. Multilocus microsatellite typing for Cryptococcus neoformans var. grubii. Med Mycol. 2008;46(7):685–696. doi: 10.1080/13693780802027062. [DOI] [PubMed] [Google Scholar]
- Gago S, Zaragoza O, Cuesta I, Rodriguez-Tudela JL, Cuenca-Estrella M, Buitrago MJ. High-resolution melting analysis for identification of the Cryptococcus neoformans-Cryptococcus gattii complex. J Clin Microbiol. 2011;49(10):3663–3666. doi: 10.1128/JCM.01091-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer W, Aanensen DM, Boekhout T, Cogliati M, Diaz MR, Esposto MC, Fisher M, Gilgado F, Hagen F, Kaocharoen S, Litvintseva AP, Mitchell TG, Simwami SP, Trilles L, Viviani MA, Kwon-Chung J. Consensus multi-locus sequence typing scheme for Cryptococcus neoformans and Cryptococcus gattii. Med Mycol. 2009;47(6):561–570. doi: 10.1080/13693780902953886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birdsell DN, Pearson T, Price EP, Hornstra HM, Nera RD, Stone N, Gruendike J, Kaufman EL, Pettus AH, Hurbon AN, Buchhagen JL, Harms NJ, Chanturia G, Gyuranecz M, Wagner DM, Keim PS. Melt analysis of mismatch amplification mutation assays (Melt-MAMA): a functional study of a cost-effective SNP genotyping assay in bacterial models. PLoS One. 2012;7(3):e32866. doi: 10.1371/journal.pone.0032866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha RS, Zarbl H, Keohavong P, Thilly WG. Mismatch amplification mutation assay (MAMA): application to the c-H-ras gene. Genome Res. 1992;2(1):14–20. doi: 10.1101/gr.2.1.14. [DOI] [PubMed] [Google Scholar]
- Li B, Kadura I, Fu D-J, Watson DE. Genotyping with TaqMAMA. Genomics. 2004;83(2):311–320. doi: 10.1016/j.ygeno.2003.08.005. [DOI] [PubMed] [Google Scholar]
- Fraser JA, Giles SS, Wenink EC, Geunes-Boyer SG, Wright JR, Diezmann S, Allen A, Stajich JE, Dietrich FS, Perfect JR, Heitman J. Same-sex mating and the origin of the Vancouver Island Cryptococcus gattii outbreak. Nature. 2005;437(7063):1360–1364. doi: 10.1038/nature04220. [DOI] [PubMed] [Google Scholar]
- Liu CM, Driebe EM, Schupp J, Kelley E, Nguyen JT, McSharry JJ, Weng Q, Engelthaler DM, Keim PS. Rapid quantification of single-nucleotide mutations in mixed influenza A viral populations using allele-specific mixture analysis. J Virol Methods. 2010;163(1):109–115. doi: 10.1016/j.jviromet.2009.09.007. [DOI] [PubMed] [Google Scholar]
- Kidd SE, Hagen F, Tscharke RL, Huynh M, Bartlett KH, Fyfe M, Macdougall L, Boekhout T, Kwon-Chung KJ, Meyer W. A rare genotype of Cryptococcus gattii caused the cryptococcosis outbreak on Vancouver Island (British Columbia, Canada) Proc Natl Acad Sci U S A. 2004;101(49):17258–17263. doi: 10.1073/pnas.0402981101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva DC, Martins MA, Szeszs MW, Bonfietti LX, Matos D, Melhem MS. Susceptibility to antifungal agents and genotypes of Brazilian clinical and environmental Cryptococcus gattii strains. Diagn Microbiol Infect Dis. 2012;72(4):332–339. doi: 10.1016/j.diagmicrobio.2011.11.016. [DOI] [PubMed] [Google Scholar]


