Abstract
Little is known about how the immune system impacts human colorectal cancer invasiveness and stemness. Here we detected interleukin-22 (IL-22) in patient colorectal cancer tissues that was produced predominantly by CD4+ T cells. In a mouse model, migration of these cells into the colon cancer microenvironment required the chemokine receptor CCR6 and its ligand CCL20. IL-22 acted on cancer cells to promote activation of the transcription factor STAT3 and expression of the histone 3 lysine 79 (H3K79) methytransferase DOT1L. The DOT1L complex induced the core stem cell genes NANOG, SOX2 and Pou5F1, resulting in increased cancer stemness and tumorigenic potential. Furthermore, high DOT1L expression and H3K79me2 in colorectal cancer tissues was a predictor of poor patient survival. Thus, IL-22+ cells promote colon cancer stemness via regulation of stemness genes which negatively affects patient outcome. Efforts to target this network might be a strategy in treating colorectal cancer patients.
Keywords: Th22, IL-22, ILC22, DOT1L, STAT3, H3K79, Cancer stem cell, CCR6, epigenetics, colon cancer
Introduction
The interaction between tumor cells and immune elements may directly promote tumor development and progression (Ben-Neriah and Karin, 2011; Coussens et al., 2013), and/or result in immunoediting of the tumor which molds the cancer into either a dormant state (Dunn et al., 2002; Matsushita et al., 2012), or fosters tumor immune evasion (Pardoll, 2012; Zou, 2005). The role of tumor infiltrating CD8+ T cells and regulatory T (Treg) cells (Curiel et al., 2004; Galon et al., 2006) has been extensively studied in human cancers. IL-22+ immune cells are identified in humans and include both IL-22+CD4+ T (Th22) cells (Duhen et al., 2009; Trifari et al., 2009) and IL-22 expressing innate leukocytes (ILC22) (Cella et al., 2009;Spits and Cupedo, 2012), but the role of IL-22+ immune cells is poorly defined in the human cancer microenvironment.
IL-22-producing immune cells could have a role in molding cancer, particularly colon cancer. The cytokine IL-22 has been shown to protect intestinal epithelial cells from bacterial infection and inflammation damage in mice (Aujla et al., 2008; Basu et al., 2012; Hanash et al., 2012; Pickert et al., 2009; Sonnenberg et al., 2012; Sonnenberg et al., 2010; Zheng et al., 2008) and supports thymic repair (Dudakov et al., 2012). Recent mouse studies have revealed that IL-22+ cells stimulate tumor cell proliferation in a bacteria-induced colon cancer model (Kirchberger et al., 2013) and IL-22 binding protein (IL-22BP) reduces chemical carcinogen-induced colon cancer development (Huber et al., 2012). Interestingly, IL22 polymorphisms may be associated with an increased risk of colon carcinoma development (Thompson et al., 2010). This data suggests a potential link between IL-22+ cells and colorectal cancer development and progression in humans. However, the nature and clinical relevance of IL-22+ cells is poorly defined in patients with colorectal cancer. It is not known if and how IL-22+ cells impact human colon cancer.
It has been demonstrated that cancer-initiating cells or cancer stem cells play an important role in shaping the invasive cancer phenotype by contributing to tumor initiation, metastasis/relapse, and therapeutic resistance (Brabletz et al., 2005; Dean et al., 2005; Pardal et al., 2003; Reya et al., 2001; Vermeulen et al., 2012). The key issue in cancer stem cell biology is understanding the mechanisms that control cancer cell self-renewal and expansion. Recent evidence suggests some degree of external control from the microenvironment that defines the stem cell niche (Bendall et al., 2007; Cui et al., 2013; Scadden, 2006). Given that the protective role of IL-22 in epithelial cells (Aujla et al., 2008; Basu et al., 2012; Dudakov et al., 2012; Hanash et al., 2012; Pickert et al., 2009; Zheng et al., 2008) and its effects on bacteria (Huber et al., 2012) and chemical carcinogen (Kirchberger et al., 2013) induced cancer in mice, we hypothesized that colon cancer-infiltrating IL-22+ immune cells contribute to cancer stem cell renewal and expansion, reshape the tumor invasive phenotype, and affect colon cancer patient outcomes. In this work, we focused on the interaction between IL-22+ immune cells and cancer (stem) cells. We demonstrated that IL-22+CD4+ T cells promote colorectal cancer stemness via STAT3 transcription factor activation and induction of the methyltransferase DOT1L and that this is relevant for outcome in patients with colon cancer.
Results
IL-22 in the tumor environment promotes colon cancer stemness
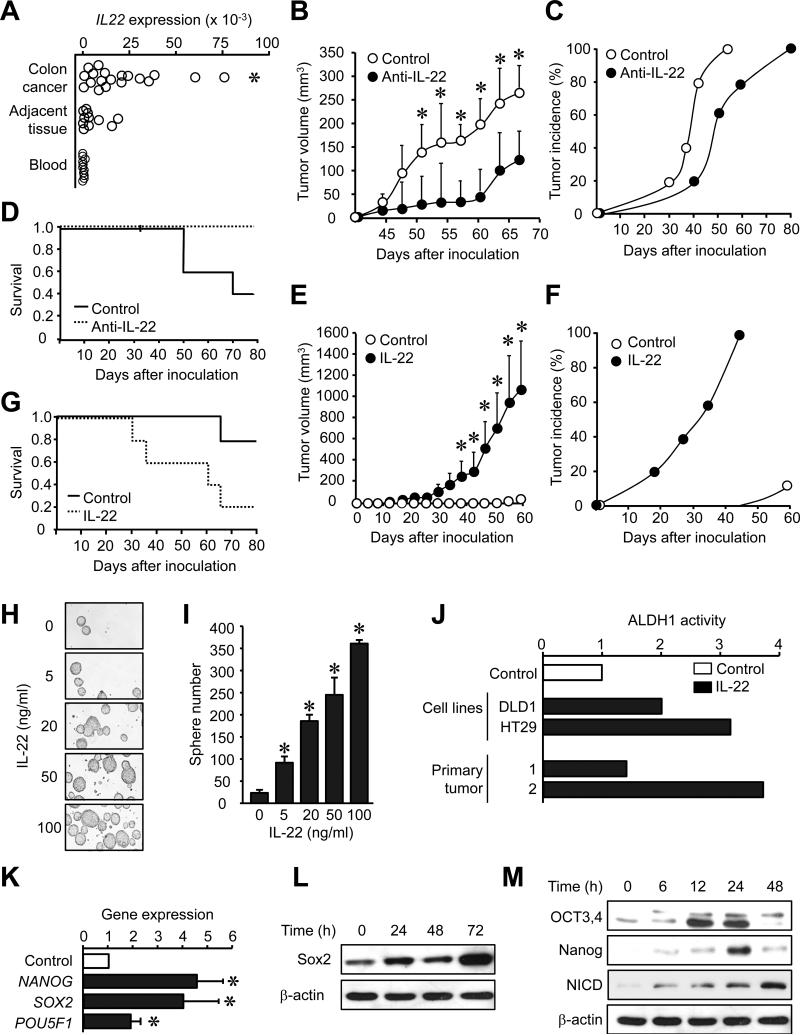
As IL-22 protects intestinal stem cells from immune-mediated tissue damage in mice (Hanash et al., 2012), we hypothesized that IL-22+ cells might support cancer stemness in patients with colon cancer. High amounts of IL-22 mRNA were detected in primary colon cancer tissues compared to peripheral blood and colon tissue adjacent to the cancer (Figure 1A). Next we examined the potential effects of endogenous IL-22 on primary tumor formation in a female NOD Shi-scid IL-2Rγnull (NSG) immune deficient mouse model (Cui et al., 2013; Curiel et al., 2004; Kryczek et al., 2012; Kryczek et al., 2011). To this end, single cell suspensions were made from fresh human colon cancer tissues. These cells contained all the primary cellular components in the colon cancer environment including CD3+ T cells within the CD45+ immune cell population, and lin-CD34−CD45−FSChighSSChigh primary colon cancer cells (Figure S1A). We equally divided this primary colon cancer tissue into two groups and injected the cells into NSG mice with a one-time treatment of either anti-human IL-22 monoclonal antibody (mAb) or isotype mAb. Anti-human IL-22 mAb dramatically reduced primary tumor volume (Figure 1B) and delayed tumor development (Figure 1C), and increased mouse survival (Figure 1D). Furthermore, we found that grafted colon cancer tissues (isolated from NSG mice) (Figure S1B) and original human colon cancer tissues (Figure 1A) and activated human peripheral mononuclear cells (PBMCs) expressed human IL-22, but not mouse IL-22 (Figure S1B). The data demonstrates that human, but not mouse, IL-22, in the human colon cancer environment promotes tumorigenesis in the NSG model in vivo.
Figure 1. IL-22 in the tumor microenvironment promotes colon cancer stemness.
(A) IL22 mRNA was detected by real-time PCR in colon cancer tissues, adjacent tissues and peripheral blood. *P < 0.05 compared to blood and adjacent tissues, 20 colon cancer patients.
(B-D) Single cells isolated from colon cancer tissue were mixed with anti-IL-22 antibody or control mAb, and then subcutaneously injected to NSG mice. Tumor growth (B, *P < 0.05, n = 5 per group), incidence (C, P = 0.037, n = 5 per group), and animal survival (D, P = 0.013, n = 5 per group) are shown.
(E-G) DLD-1 colon cancer cells (105) were pre-incubated with IL-22 (20 ng/ml) for 1 hour, and then subcutaneously injected to NSG mice. Tumor growth (E, *P < 0.05, n = 5 per group, incidence), incidence (F, P = 0.003, n = 5 per group), and animal survival (G, P = 0.013, n = 5 per group) are shown.
(H, I) DLD-1 colon cancer cells were cultured with IL-22. Sphere assay was performed with 2,000 cells. Representative image of spheres (H) and the mean numbers of spheres (I) are shown. *P < 0.05, n = 5.
(J) Colon cancer cell lines (DLD-1 and HT29) and two primary colon cancer cells (1, 2) were cultured with IL-22 for 24 hours. ALDH1 activity was determined by FACS based on aldefluor fluorescence. Cells treated with DEAB inhibitor were negative controls. Results are expressed as fold changes of aldefluor fluorescence. (P < 0.05 for all, n = 3 repeats per group).
(K-M) DLD-1 colon cancer cells were cultured with IL-22 (20 ng/ml). The mRNA of stem cell core gene was detected by real-time PCR (K) and proteins were detected by western blotting (L, M). (*P < 0.05, n = 5).
To confirm the tumorigenic potential of endogenous IL-22, we injected different concentrations of a colorectal adenocarcinoma cancer cell line, DLD-1 cells, into NSG mice to determine a nontumorigenic concentration. We found that 105 DLD-1 cells failed to form a tumor in the NSG mouse. However, exogenous IL-22 administration enabled tumor formation with 105 DLD-1 cells as shown by increased tumor volume (Figure 1E), accelerated tumor development (Figure 1F), and decreased mouse survival (Figure 1G). When 106 DLD-1 cells were inoculated into mice, IL-22 promoted tumor development (Figure S1C) and growth, as well (Figure S1D). Thus, IL-22 may enhance tumorigenesis by altering cancer stem cell properties.
In support of this notion, IL-22 promoted tumor sphere formation in a dose dependent manner (Figure 1H, I) and increased aldehyde dehydrogenase (ALDH1) activity in DLD-1, HT29 and two primary colon cancer cell lines (Figure 1J). ALDH1 is an operative marker of human cancer stem cells (Carpentino et al., 2009; Kryczek et al., 2012). Furthermore, IL-22 enhanced mRNA (Figure 1K) and protein (Figure 1L, M) expression of multiple core stem cell genes including NANOG, SOX2, and POU5F1 (OCT3/4), but had no effect on β-catenin and Wnt signaling (Figure S1E). Altogether, IL-22 stimulates expression of genes associated with core cancer stemness and promotes colon tumorigenicity.
IL-22+ cells are recruited into the tumor and promote cancer stemness via IL-22
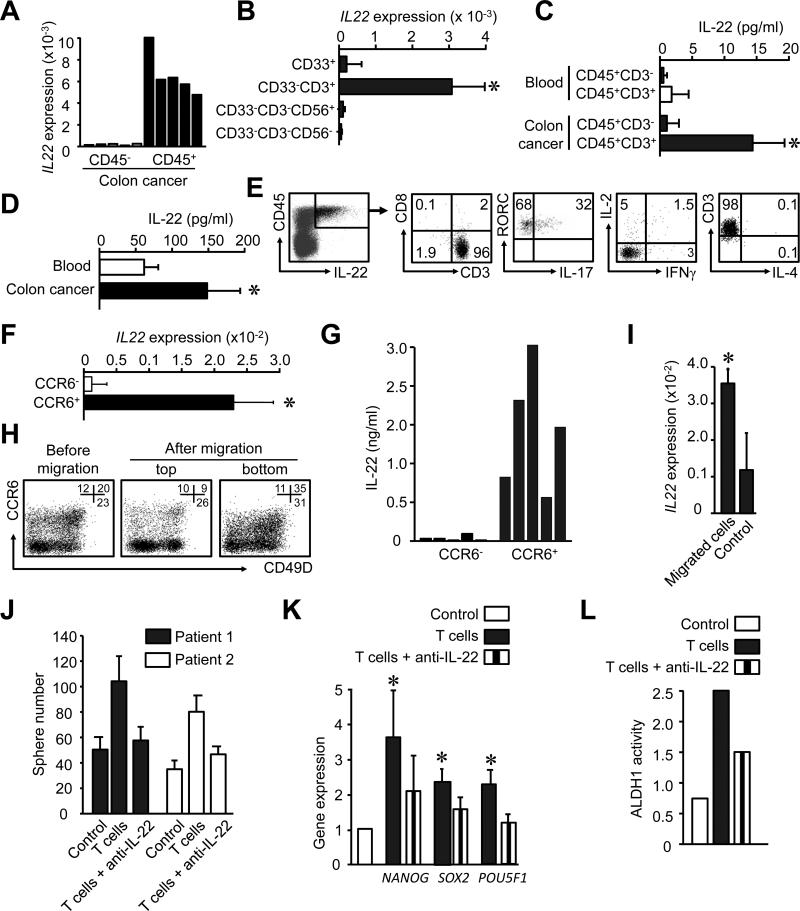
Given that IL-22 promotes colon cancer stemness, we examined the cellular source of IL-22 and the phenotype of IL-22+ cells in the human colon cancer environment. Real-time PCR revealed that IL-22 was expressed by CD45+ immune cells in the colon cancer environment (Figure 2A). Based on polychromatic flow cytometry analysis, CD3+ T cells were found to be the predominant cell type among CD45+ cells in the colon cancer microenvironment (Figure S2A, S2B). To further define the phenotype of the IL-22+ cells, we sorted colon cancer associated CD45+ cells into four populations: lineage negative cells (lin−), CD33−CD3−CD56+ cells (with NK or potentially ILC), CD33+ myeloid cells, and CD3+ T cells. We found that IL-22 mRNA expression was confined to CD3+ T cells (Figure 2B). Furthermore, sorted colon cancer-associated CD45+CD3+ T cells, but not colon-associated CD45+CD3− cells or blood CD45+CD3− cells, spontaneously released IL-22 (Figure 2C, D). We analyzed the cytokine profile of IL-22+ cells in the colon cancer. We found that IL-22+ cells were also CD3+CD8−CD4+ and expressed the transcription factor RORγ. Of the CD3+CD8−CD4+IL-22+ cells, 30% expressed IL-17, and whereas IL-4, IL-2 and IFNγ expression was detected in less than 5% of cells (Figure 2E). Thus, IL-22 is predominantly expressed by CD4+ T cells in the colorectal cancer microenvironment.
Figure 2. IL-22+ CD4+ T cells traffic into the tumor via CCR6-CCL20 and promote cancer stemenss via IL-22.
(A, B) Different cell populations were sorted from colon cancer tissues and IL-22 expression was measured by real time PCR. (5-10 donors, P < 0.05).
(C) Different CD45+ immune subsets (106/ml) were sorted from colon cancer and blood, and cultured for 12 hours. IL-22 was detected via ELISA. (n = 5, *P < 0.05).
(D) CD45+ subsets (106/ml) were sorted from colon cancer and blood, and were activated with anti-CD3 and anti-CD28 for 2 days. IL-22 was detected by ELISA (n = 5, *P < 0.05).
(E) Single colon cancer environmental cells were stained with anti-CD3, CD8, CD45, RORγc, IL-2, IL-4, IL-17, IL-22 and IFNγ antibodies. The phenotype and the expression of indicated cytokines were analyzed by FACS. Right panels were gated on IL-22+CD45+ cells. One of 3 independent experiments is shown.
(F, G) CD4+ T cells were sorted based on CCR6 expression and activated with anti-CD3 and anti-CD28 and antigen presenting cells. IL-22 mRNA was detected by real time PCR (F) and IL-22 protein by ELISA (G). (5 different donors, P < 0.05)
(H, I) Migration assay was conducted with CD4+ T cells for 4 hours in the presence of CCL20. The phenotype of migrated cells, non-migrated cells, and control (before migration) was analyzed by FACS (H). IL-22 expression was quantified with real-time PCR in the migrated and non-migrated cells. Results are expressed as the mean relative expression (I). (n = 3, P < 0.05).
(J) Sphere assay was performed with autologous colon tumor cells in the presence of activated colon cancer-associated T cells in a transwell system. Anti-IL-22 or isotype mAb was added in the assay. Results are shown as the mean numbers of spheres in triplicates. (2 of 5 patients are shown. P < 0.01).
(K, L) Primary colon cancer associated T cells were sorted and activated for 3 days. DLD-1 colon cancer cells were cultured with these T cell supernatants in the presence of anti-IL-22 or isotype mAbs. The mRNA of stem cell core genes was detected by real-time PCR after 6 hours (K) and ALDH activity was detected by FACS after 48 hours (L). (n = 5, *P < 0.05, compared to control and T cells with anti-IL-22).
We next examined how peripheral blood IL-22+CD4+ T cells traffic into the colon cancer microenvironment. We analyzed the expression of cell trafficking associated molecules including chemokine receptors and integrins on IL-22+CD4+ T cells in blood and colon cancer. We found that IL-22 was expressed by memory, but not naïve, CD4+ T cells in blood (Figure S2C, S2D). We sorted blood CD4+ T cells into C-C chemokine receptor type 6 (CCR6)− and CCR6+ populations, and subsequently examined IL-22 expression. We did not perform intracellular staining as this affects the detection of surface antigens. We found CCR6+, but not CCR6− cells expressed high amounts of IL-22 mRNA (Figure 2F) and protein (Figure 2G). CCR6+ T cells largely co-expressed CD49D integrin, the ligand for VCAM1 (Figure S2E). Among CCR6+ cells, IL-22 was predominantly expressed by primary (Figure S2F) and activated (Figure S2G) CD49D+ cells. Thus, IL-22+ cells are enriched in the CCR6+CD49D+ memory T cell pool.
We then asked whether IL-22+ T cells could migrate toward primary tumor tissues through chemokine (C-C motif) ligand 20 (CCL20), the ligand for CCR6. We observed that T cells efficiently migrated in response to CCL20 (Figure 2H), and that the migrating cells were enriched for IL-22+CD4+ T cells (Figure 2I), expressing CCR6 and CD49D (Figure 2H). We conducted an in vivo trafficking assay. CCR6+CD49+ T cells were treated with neutralizing anti-CCR6 mAb or isotype and were transfused into human colon cancer bearing NSG mice. After 48 hours, human T cells migrated into the grafted human colon cancer in NSG mice (Figure S2H, S2I). These T cells expressed IL-22, and CCR6 blockade reduced their migration (Figure S2H, S2I). Furthermore, high amounts of CCL20 (Figure S2J), and VCAM1 (Figure S2K) were detected in colon cancer tissues. Human T cells were found in the grafted human colon cancer in NSG mice (Figure S2L). The data suggests that CCR6 and CD49D signaling mediates homing of IL-22+CD4+ cells into the colon cancer microenvironment.
We further investigated the potential effects of primary colon cancer associated IL-22+CD4+ cells on colon cancer stemness. To this end, colon cancer cell sphere assay was performed with autologous colon cancer associated CD4+ T cells. We showed that these T cells enhanced primary colon cancer cell sphere formation while anti-IL-22 abrogated this effect (Figure 2J). In line with this, these T cells also increased core stem cell gene expression (Figure 2K) and ALDH1 activity (Figure 2L) in colon cancer cells. This increase in stemness was reduced with IL-22 blockade (Figure 2K, L). Thus, IL-22+CD4+ cells traffic into the tumor, and promote colon cancer stemness via secreting IL-22 in the colon cancer microenvironment.
IL-22 promotes colon cancer stemness via STAT3 activation
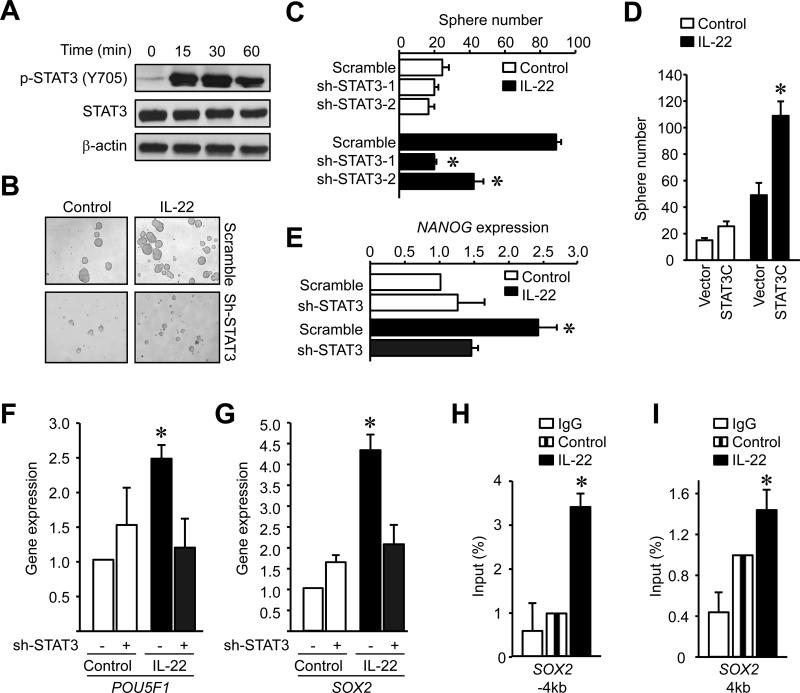
Next, we dissected the molecular mechanisms by which IL-22 promotes colon cancer stemness. Signal transducer and activator of transcription 3 (STAT3) plays a key role in the crosstalk between cancer and immune cells in the tumor microenvironment (Lee et al., 2009; Yu et al., 2007). In line with previous reports (Lejeune et al., 2002; Pickert et al., 2009), we observed that IL-22 activated STAT3 in colon cancer cells (Figure 3A). We next examined whether the effect of IL-22 on colon cancer stemness was STAT3-dependent. To this end, we manipulated STAT3 expression either with specific STAT3 knockdown (sh-STAT3) (Figure S3A) or forced expression of a STAT3 active domain (STAT3C) (Figure S3B) in colon cancer cells. Sh-STAT3 resulted in reduced colon cancer sphere numbers (Figure 3B, C). In the absence of IL-22, expression of STAT3C had minimal effects on colon cancer sphere formation, but addition of IL-22 increased colon cancer sphere formation in STAT3C-expressing cells compared with controls (Figure 3D). To determine if the effect of IL-22 was relatively STAT3-specific, we additionally evaluated the role of Notch on colon cancer sphere formation. IL-22 induced potent and equal sphere formation in colon cancer cells expressing vectors encoding: GFP (scramble), Notch active domain (Notch-IC), and Notch negative domain (Notch-DN) (Wang et al., 2011; Yamamoto et al., 2001) (Figure S3C). The data suggests that IL-22-mediated STAT3 activation is necessary and relatively specific in promoting colon cancer stemness. However, in the absence of IL-22, forced STAT3 activation alone is insufficient to strongly induce cancer stemness (Figure 3D). Consistent with this, IL-6 and IL-17 activate STAT3 but failed to promote colon cancer sphere formation (Figure S3D).
Figure 3. IL-22 promotes colon cancer stemness via STAT3 activation.
(A) Colon cancer cells were treated with IL-22 for different time points. The amount of phosphorylated STAT3 and STAT3 protein was detected by western blotting.
(B-D) Sphere assay was performed with shSTAT3 (B, C) or STAT3C (D) expressing colon cancer cells in the presence of IL-22. Results are shown as sphere images (B) and the mean numbers of spheres in triplicate (C, D). (n = 5. *P < 0.01).
(E-G) Colon cancer cells expressing shSTAT3 or scrambled vector were cultured with IL-22. Stem cell core gene mRNAs were detected by real-time PCR after 6 hours (n = 5, *P < 0.05).
(H, I) STAT3-ChIP assay was performed DLD-1 cells cultured with or without IL-22. (mean +/- SEM, n = 3, *P < 0.05).
We further explored the effects of IL-22-activated STAT3 in expression of stemness-related genes. STAT3 knockdown reduced core stem cell gene expression (Figure 3E-G). We speculated that STAT3 may directly bind to the promoters of core stem cell genes and subsequently induce their expression. We found several predictions for STAT3 binding on the SOX2 promoter region (http://www.sabiosciences.com/chipqpcrsearch.php). Chromatin immunoprecipitation (ChIP) demonstrated that IL-22 increased STAT3 binding in several sites on the SOX2 promoter area (Figure 3H, I) and suggests that STAT3 may directly activate stemness genes. STAT3 has been found to recruit p300 (Nakashima et al., 1999), enhancing target gene expression. We observed that IL-22 enhanced the binding of p300 to the SOX2 promoter (Figure S3H). As a positive control, we used FOS, which has been shown to be bound and activated by STAT3 (Figure S3E-G) (Yang et al., 2003). Thus, IL-22 promotes colon cancer stemness via STAT3 activation and its associated signaling genes.
DOT1L regulates IL-22 dependent colon cancer stemness via H3K79 methylation
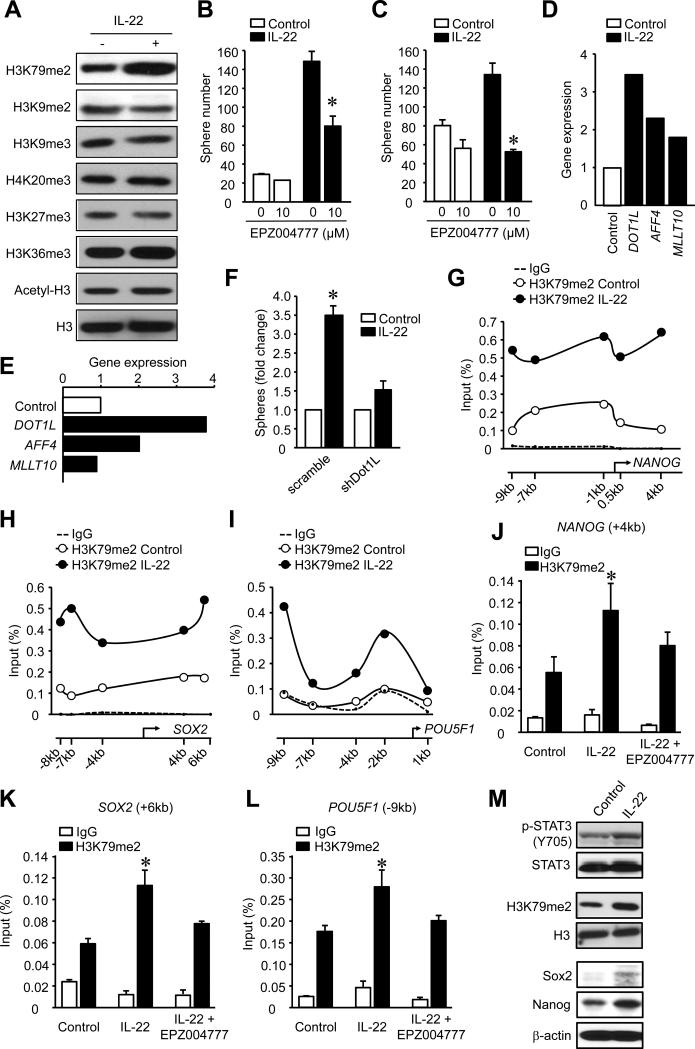
Epigenetic modifications of chromatin and their crosstalk with transcription factors play an important role in the regulation of gene expression. STAT3 binding to the promoters of core stem cell genes is highly context dependent (Hutchins et al., 2013) and not all STAT3 activating cytokines induced stemness (Figure S3D). Thus, STAT3 activation may not completely explain the increase in stemness. As transcription factors and epigenetic modifications often guide external signals to a specific genetic response, we wondered whether epigenetic control, including histone modifications, is involved in controlling IL-22-induced stemness gene expression. To this end, we examined the global changes in several histone marks in IL-22-treated colon cancer cells. Among several histone marks, we observed that IL-22 selectively increased the dimethylation of histone 3 lysine 79 (H3K79me2) (Figure 4A). Disruptor of telomeric silencing 1-like (DOT1L) is the sole H3K79 methyltransferase (Min et al., 2003; Ng et al., 2002). EPZ004777 (Daigle et al., 2011; Yu et al., 2012), a selective inhibitor of DOT1L, reduced H3K79me2 (Figure S4A) and suppressed DLD-1 (Figure 4B) and primary colon cancer (Figure 4C) sphere formation. Several proteins recruit DOT1L to mediate methylation of H3K79 including MCEF (AFF4), AF9 (MLLT3) and AF10 (MLLT10) (Mohan et al., 2010). IL-22 consistently promoted the expression of DOT1L, AFF4, but not MLLT10 in DLD-1 (Figure 4D) and primary colon cancer cells (Figure 4E). To further determine the role of DOT1L in colon cancer stemenss, we knocked down DOT1L expression with sh-DOT1L (Figure S4B) and forced ectopic DOT1L expression (Figure S4C). Similar to EPZ004777, DOT1L knockdown reduced colon cancer sphere formation (Figure 4F). Forced DOT1L expression led to an increase in NANOG (Figure S4D) and SOX2 (Figure S4E) mRNA in the absence of IL-22. However, IL-22 further increased stemness gene expression (Figure S4D, S4E) and sphere numbers (Figure S4F). IL-22 also stimulated c-Myc expression in colon cancer cells, but DOT1L overexpression had no effects on IL-22-stimulated c-Myc expression (Figure S4G). This data suggests that DOT1L signaling activation potentiates the colon cancer stemness program.
Figure 4. IL-22 promotes colon cancer stemness via DOT1L and H3K79me2.
(A) Colon cancer cells were treated with IL-22 for 48 hours. Histone modifications were analyzed by Western blotting.
(B, C) Colon cancer sphere assay was performed in the presence of IL-22 and the DOT1L inhibitor EPZ004777. Sphere numbers were recorded. DLD-1 cells (B), primary colon cancer cells (C). (n = 5, P < 0.05).
(D, E) Colon cancer cells were cultured for 12 hours with IL-22. Expression of the genes encoding members of the DOT1L complex was quantified by real-time PCR. DLD-1 cells (D), primary colon cancer cells (E). (n = 3, P < 0.05).
(F) Colon cancer sphere assay was performed with colon cancer cells expressing sh-DOT1L or vector in the presence of IL-22. Sphere numbers were recorded. (n = 5, P < 0.05).
(G-I) H3K79me2-ChIP assay was performed to examine H3K79me2 at the core stem cell genes promoters in DLD-1 colon cancer cells cultured with IL-22. One of 3 experiments is shown.
(J-L). H3K79me2 ChIP was performed to examine occupancy at core stem cell genes in DLD-1 colon cancer cells cultured with or without IL-22 and EPZ004777. One of three experiments is shown.
(M) IL-22 (0.5ug) was injected into the DLD-1 tumor. After 12-48 hours tumor tissues were extracted for Western blotting analysis for the STAT3, H3K79me2, SOX2 and NANOG proteins. One of 3 experiments with triplicate sections is shown.
We also explored whether IL-22 regulates H3K79me2 on core stem cell gene promoters. ChIP assays with H3K79me2 revealed an increase in H3K79me2 on the proximal promoter areas of NANOG (Figure 4G), SOX2 (Figure 4H) and POU5F1 (Figure 4I) in an IL-22 dependent manner. Moreover, EPZ004777 treatment resulted in reduced H3K79me2 on the stem cell gene promoter sites (Figure 4J-L). In support of these observations, IL-22 administration caused the activation of STAT3, H3K79 dimethylation, and core stemness gene activation in human colon cancer cells in the NSG model in vivo (Figure 4M). This data indicates that IL-22 regulates colon cancer stemness in a DOT1L- and H3K79me2-dependent manner.
STAT3 induces DOT1L expression and controls IL-22-induced cancer stemness
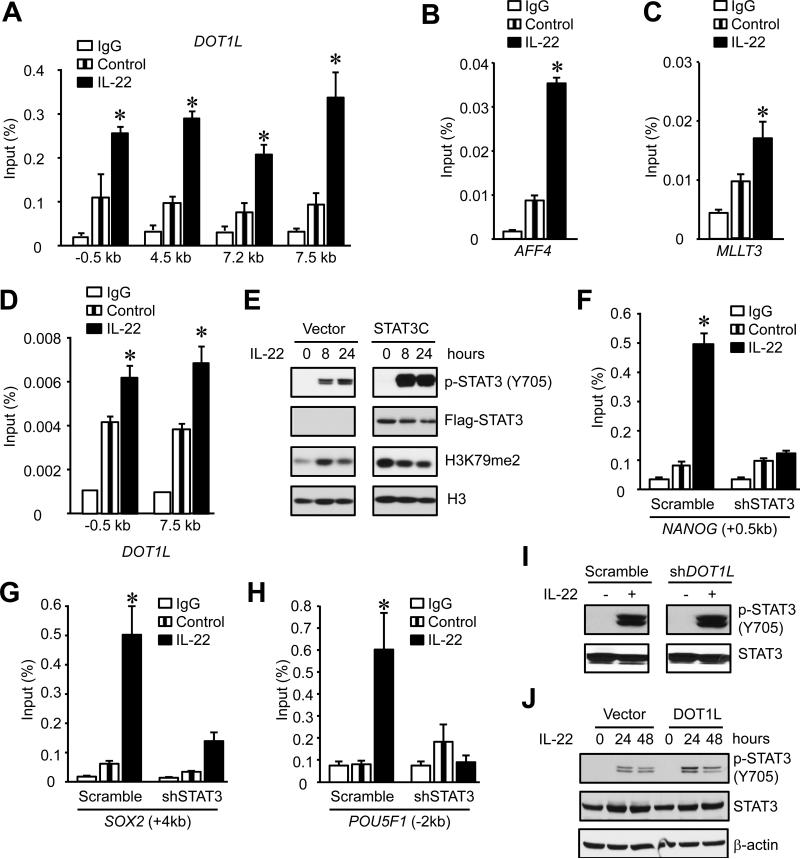
Both STAT3 (Figure 3) and DOT1L-H3K79 signaling (Figure 4) are involved in the control of IL-22-induced cancer stemness. We hypothesized that IL-22-activated STAT3 causes H3K79 methylation. The ENCODE ChIP-sequence (ChIP-Seq) database revealed STAT3 binding sites on the promoters of DOT1L, AFF4, MLLT3 and MLLT10 in a lymphoblastoid cell line (Access number: GSM935557) (Figure S5) (Birney et al., 2007). IL-22 treatment increased STAT3 binding to the DOT1L promoter area (Figure 5A) and to promoters of two other elements of the DOT1L complex, AFF4 (Figure 5B) and MLLT3 (Figure 5C), but not MLLT10 (data not shown). As a confirmatory experiment, we showed that IL-22 also augmented the binding of p300 to the promoter area of DOT1L (Figure 5D). Thus, IL-22 promotes the binding of the transcription factor STAT3 to the promoter area of the DOT1L complex and controls colon cancer stemness.
Figure 5. STAT3 stimulates DOT1L expression and promotes IL-22-induced cancer stemness.
(A-C) STAT3-ChIP assay was performed in DLD-1 colon cancer cells cultured with or without IL-22 to examine STAT3 occupancy at DOT1L (A), AFF4 (B), and MLLT3 (C) promoters. One of three experiments is shown.
(D). p300-ChIP assay was performed in DLD-1 colon cancer cells cultured with or without IL-22 to examine p300 occupancy at DOT1L promoter. (n = 3 with duplicates, *, P < 0.05).
(E). DLD-1 cells were transduced with STAT3C expressing or control lentiviral vectors, and cultured with IL-22. H3K79me2 was detected by Western blotting. One of 3 experiments is shown.
(F-H) H3K79me2-ChIP was performed in colon cancer cells cultured with IL-22 for 24 hours to examine H3K79me2 occupancy at the promoter areas of core stem cell genes. (n = 3 with duplicates, * P < 0.05).
(I, J) DLD-1 cells were transduced with shDOT1L (I) or DOT1L expressing (J) or control lentiviral vectors, and cultured with IL-22. STAT3 and STAT3 phosphorylation were detected by Western blotting. One of 3 experiments is shown.
In addition to the interaction of STAT3 on the DOT1L promoter, we explored whether STAT3 could directly impact the amount of H3K79me2 via DOT1L. Enhanced activation of STAT3 in STAT3C transduced cells caused a genome-wide increase in H3K79 dimethylation (Figure 5E), but no increase in sphere formation (Figure 3D) in the absence of IL-22. IL-22 treatment moderately augmented H3K79 dimethylation (Figure 5E) and dramatically increased sphere formation (Figure 3D). Accordingly, DOT1L gene expression was increased in colon cancer cells expressing STAT3C (Figure S3I). Thus, STAT3 activation could promote H3K79me2 via DOT1L.
To determine whether STAT3 regulates core stem cell gene expression through DOT1L-dependent H3K79 methylation, we performed ChIP with H3K79me2 in sh-STAT3 IL-22 treated colon cancer cells. STAT3 knockdown abrogated IL-22-induced H3K79 methylation on the stem cell core gene promoters of NANOG, SOX2 and POU5F1 (Figure 5F-H). The data further solidifies the notion that STAT3 is essential for H3K79 dimethylation and IL-22 potentiates potent cancer stemness via STAT3 mediated-H3K79 dimethylation. DOT1L knockdown (Figure 5I) or overexpression (Figure 5J) had no effects on IL-22-mediated STAT3 activation. This suggests that IL-22-induced STAT3 phosphorylation is DOT1L independent. Altogether, the results indicate that IL-22-activated STAT3 directly regulates DOT1L expression and subsequently induces H3K79 methylation at the stemness genes, facilitating and accelerating stemness gene activation.
High amounts of colon cancer H3K79-DOT1L predict poor patient survival
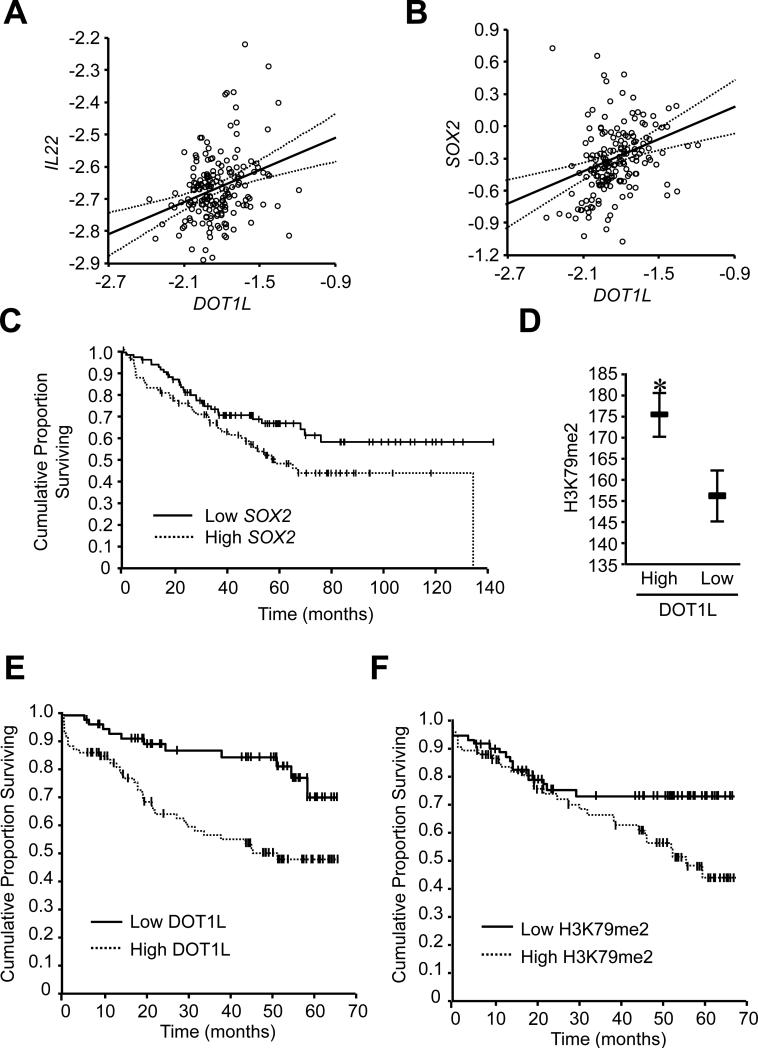
Finally, we examined the clinical relevance of the IL-22-DOT1L signaling pathway in colon cancer patients. To this end, we first analyzed the relationship between IL-22, DOT1L, and stem cell gene transcripts in patients with colorectal cancer from the National Center for Biotechnology Information Gene Expression Omnibus database (GSE17536) (Smith et al., 2010). The GSE17536 database includes 177 colorectal cancer patients with clinic and pathological information (Table S1). We found that IL-22 expression correlated with DOT1L (Figure 6A). Moreover, the expression of DOT1L correlated with SOX2 (Figure 6B). Furthermore, when we divided patients into “low” and “high” groups based on the median value of SOX2, we observed that high SOX2 expression was associated with poor patient survival (Figure 6C). Then, we quantified nuclear DOT1L (Figure S6A) and H3K79me2 (Figure S6B) via immunohistochemistry in paraffin-fixed colorectal cancer tissues from patients with available clinical and pathological information (Table S2). The expression of DOT1L correlated highly with that of H3K79me2 in the same tumor (Figure 6D). Furthermore, based on the median values of DOT1L intensity, we divided patients into “low” and “high” groups (Figure S6A). Overall survival was shorter in patients with high DOT1L staining compared to low DOT1L expression (Table S3, Figure 6E). Age and tumor stage (TNM) were important prognostic factors for colon cancer survival (Table S4). After adjusting for the clinical factors, overall survival remained shorter in patients with high DOT1L expression (Table S4). The data strongly suggest that increased tumor DOT1L abundance is a significant and independent predictor of poor survival in colorectal cancer. We further analyzed the relationship between tumor H3K79me2 expression and survival. Similar results were observed with H3K79 methylation (Figure S6B, Figure 6F). Overall survival was shorter in patients with high H3K79 dimethylation (Table S3, Figure 6F). Therefore, DOT1L expression and H3K79me2 could be a novel oncogenic predictor for poor survival in colorectal cancer. Altogether, IL-22+CD4+ T cells produce IL-22 and shape colon cancer stemness through a STAT3-DOT1L-mediated stem cell core signaling pathway.
Figure 6. High amounts of colon cancer H3K79-DOT1L predict poor patient survival.
(A, B) The correlation between DOT1L, IL22 and SOX2 transcripts in patients with colorectal cancer was examined analyzing 177 colorectal cancer patients (GSE17536). R=0.31, P =0.000032 (A); R=0.38, P =0.00000024 (B).
(C) The association between SOX2 transcripts and patient survival. The analyses were conducted in 177 colon cancer patients (GSE17536), P =0.046.
(D) The correlation between DOT1L and H3K79me2 in patients with colon cancer. n = 144, R=0.23, P = 0.005.
(E, F) The relationship between tumor DOT1L (E), H3K79me2 (F) expression and colon cancer overall survival was evaluated. n= 151, P = 0.001 (E), n = 144, P = 0.039 (F).
Discussion
The capacity of immune cells to modulate the cancer phenotype has been the subject of intensive investigation. The immune surveillance model proposes that immunoediting contributes to cancer dormancy in a mouse model (Dunn et al., 2002; Matsushita et al., 2012). It is also well known that immune cell subsets, when chronically activated, directly foster tumor development (Ben-Neriah and Karin, 2011; Coussens et al., 2013), and promote cancer progression (Ben-Neriah and Karin, 2011; Coussens et al., 2013; Cui et al., 2013). As a novel extension of this model, we demonstrate that IL-22+ immune cells select and sustain the invasive phenotype of human colon cancer: cancer stemness.
Th22 cells and IL-22 have been reported to be a protective immune element in infection, inflammation (Aujla et al., 2008; Basu et al., 2012; Hanash et al., 2012; Pickert et al., 2009; Zheng et al., 2008), and thymic repair (Dudakov et al., 2012). Colon cancer-associated IL-22+CD4+ T cells stimulate cancer stem cell core gene expression and promote cancer stemness. Cancer stemness and the invasive tumor phenotype are thought to be a cell-autonomous process specified by the genetic and epigenetic signature of cancer cells. Our data indicate that IL-22+CD4+ T cells, a crucial T cell immune component in the colon cancer microenvironment, functions as an environmental extrinsic signal, directly targets cancer cells, and defines their stemness. We focus on patients with advanced colon cancer due to practical and ethical reasons. Given the defined roles of IL-22 in cancer stemness, it is possible that ILC22 or/and other IL-22+ cells employ a similar mechanism and regulate cancer initiation and development in early phases of colon cancer. Nonetheless, it is reasonable to conclude that IL-22+CD4+ T cells participate in colon cancer stemness.
STAT3 activity is required for small-intestine crypt stem cell survival in mice (Matthews et al., 2011). We have demonstrated that both STAT3 activation and DOT1L-H3K79 signaling are essential for IL-22-induced cancer stemness. STAT3 and DOT1L facilitate core stem cell gene activation, and are indispensible in IL-22-induced cancer stemness. Although understanding the dynamic interaction between the transcription factor STAT3 and the histone mark H3K79me2 warrants further investigation, our work addresses a central question: do histone modifications cue and instruct transcription or support and correlate with gene activity (Henikoff and Shilatifard, 2011)? Our findings demonstrate an instructive role for methylated H3K79 in colon cancer stem cell gene activation stimulated by IL-22. Thus, our work reveals a relationship between a key transcription factor (STAT3) and an important epigenetic mark (H3K79) in determining cancer stemness.
DOT1L is the sole H3K79 methyltransferase (Min et al., 2003; Ng et al., 2002). We have shown that IL-22 regulates colon cancer stemness via DOT1L-H3K79. The amount of DOT1L expression in colon cancer independently predicts poor patient survival. Notably, it has been reported that DOT1L recruitment through Mixed Lineage Leukemia (MLL) fusion proteins is strongly associated with MLL transformation (Chang et al., 2010; Jo et al., 2011). It remains to be determined whether DOT1L is an oncogene in human epithelial cancer, and whether it is biochemically and functionally linked to the well-defined colon oncogenes. Nonetheless, our study is the first to demonstrate that DOT1L contributes to human epithelial carcinoma, including its involvement in colorectal tumorigenesis and its regulation by IL-22+CD4+ T cells. Therefore, our work has revealed novel epigenetic mechanisms by which IL-22+CD4+ T cells and IL-22 control human colon cancer stemness and tumorigenesis. Similar mechanisms may apply for other types of human cancers. As DOT1L inhibition is a proposed strategy for targeted therapy of leukemia with MLL translocation (Daigle et al., 2011), our work suggests that DOT1L may be a marker for colon cancer progression and targeting this pathway may be meaningful for colon cancer treatment.
Experimental procedures
Human subjects
Patients diagnosed with colon carcinomas were recruited in the study. All usage of human subjects in this study was approved by the local Institutional Review Board. 151 formalin-fixed, paraffin-embedded tumor tissue blocks were obtained during surgery. These patients underwent resection of the primary tumor at the 2nd Department of General Surgery in Medical University of Lublin between 2007 and 2008. The follow-up period was an average 2.8 years. Additional 177 patients with colon cancer were evaluated from the National Center for Biotechnology Information Gene Expression Omnibus database (GSE17536) (Smith et al., 2010). 36 fresh cancer tissues were collected from patients with colon cancer newly diagnosed at the University of Michigan and the University of Florida. Primary colon cancer cells, immune cell subsets and all the in vitro and in vivo functional assays were performed with single cells from fresh colon cancer tissues and peripheral blood.
Cell isolation and FACS analysis
Single cell suspensions were prepared from fresh colon cancer tissues as previously described (Curiel et al., 2004; Curiel et al., 2003; Kryczek et al., 2006; Zou et al., 2001). Immune cells and tumor cells were enriched using paramagnetic beads (StemCell Technology, Vancouver, Canada). Lin−CD45−EpCAM+ cells primary colon cancer cells and CD4+CD45+ T cells were sorted from stained single cell suspensions using a high speed cell sorter (FACSaria, Becton Dickinson Immunocytometry Systems, San Jose, CA). Cell purity was >98% as confirmed by flow cytometry (LSR II, BD). Cytokine profile was determined with intracellular staining and analyzed by LSRII (BD).
Cell culture and sphere formation
Three primary colon cancer cell lines (1, 2, 3) were established from fresh colon cancer tissues. DLD1 and HT29 cell lines (ATCC) were used in the experiments. Colon cancer cells were treated with recombinant IL-22 (R&D systems) and/or colon cancer infiltrating CD4+ T cells for different time points. The DOT1L inhibitor EPZ004777 (10 μM) and relevant antibodies were added in conventional or sphere culture (Kryczek et al., 2012). Tumor cell sphere formation and gene expression were examined (Kryczek et al., 2012).
Lentiviral transduction
Several lentiviral vectors were used to transduce colon cancer cells and establish stable cell lines. The lentiviral transduction efficiency was confirmed by GFP which was co-expressed by the lentiviral vector. The knockdown efficiency was assessed by Western blotting. The vectors included pGIPZ lentiviral vector encoding gene specific shRNAs for STAT3, DOT1L or scrambled shRNA (Puromycin resistant); lentiviral vectors encoding an active form of Notch (the transmembrane and intracellular domains, comprising residues 1,704–2,531) Notch-IC cDNA (Notch IC) (Wang et al., 2011; Yamamoto et al., 2001), or constitutively active STAT3 domain (STAT3C) (EF.STAT3C.Ubc.GFP) (Bromberg et al., 1999; Li and Shaw, 2006). Human DOT1L cDNA was cloned into the entry vector pENTR223.1 (Thermo Scientific Open Biosystem, OHS5894-202503093), and inserted into the destination/expressing vector pDEST26 by Gateway® Recombination Cloning Technique (Invitrogen). Human DOT1L expression vector (pDEST26-hDOT1L) was verified by restriction enzyme digestion and DNA sequencing.
Cytokine detection
The amount of cytokines protein was detected either by ELISA (R & D) or flow cytometry analyzer (FACS) as described previously (Curiel et al., 2004; Kryczek et al., 2011). All samples were acquired with LSR II (BD) and analyzed with DIVA software.
Real-Time reverse-transcriptase polymerase chain reaction (RT-PCR)
The mRNA was quantified by real-time RT-PCR. Specific primers are included in the supplementary information (Table S5). SYBR Green Master Mix was used to detect fluorescence. Relative expression was calculated according to the Ct value with normalization to GAPDH.
Western Blot
Western blotting was performed with specific antibodies against human STAT3 (9132, Cell Signaling), phosphorylated STAT3 (9138, Cell Signaling), Oct3/4 (sc-5279, Santa Cruz biotechnology), Nanog (ab21624, Abcam), Sox2 (MAB4343, Millipore), H3K79me2 (ab3594, Abcam), H3K9me2 (ab1220, Abcam), H3K9me3 (ab8898, Abcam), H4K20me3 (07-463, Millipore), H3K27me3 (07-449, Millipore), H3K36me3 (ab9050, Abcam), acetyl-Histone H3 (06-599, Millipore), Histone H3 (9715, Cell Signaling), β-Actin (A5441, Sigma), Dot1L (ab72454, Abcam), and Cleaved Notch1 (NICD, ab52301, Abcam). Signals were detected by ECL reagents (GE Healthcare, Buckinghamshire, UK).
In vitro and in vivo migration assays
In vitro migration assay was performed in a Transwell system with a polycarbonate membrane of 6.5-μm diameter with a 3-μm pore size as described (Curiel et al., 2004; Curiel et al., 2003). Purified T cell subsets were added to the upper chamber and CCL-20 (5 ng/ml, R&D) was added to the lower chamber. After 4h incubation at 37°C, the phenotype and number of T cells in the upper and lower chambers was determined by FACS. In vivo migration assay was performed in female NOD/Shi-scid/IL-2Rγnull (NSG) mice (6-8 weeks old, Jackson Lab, Bar Harbor, Maine) (Curiel et al., 2004; Curiel et al., 2003; Kryczek et al., 2012). Subcutaneous DLD-1 (106) tumor was established in NSG model. Human T cell subsets (5 ×106) were treated with anti-CCR6 and isotype antibody and were intravenously transferred into these NSG mice. After 48 hours, human T cells were analyzed in the tumors by FACS.
Chromatin immunoprecipitation (ChIP)
ChIP was performed according to the protocol with exceptions stated below (Upstate, Millipore; http://www.millipore.com/techpublications/tech1/mcproto407). Crosslinking was performed with 1% formaldehyde or 1% paraformaldehyde for 10 minutes. To enhance cell lysis, the lysate was run through a 27g needle three times and flash frozen in −80°C. Sonication was then performed with the Misonix 4000 water bath sonication unit at 15% amplitude for 10 minutes. Protein/DNA complex was precipitated with specific antibodies against H3K79me2 (abcam, ab3594), STAT3 (Santa Cruz, SC-482), p300 (Santa Cruz, SC-585), and IgG control (Millipore). DNA was then purified using a DNA Purification Kit (Qiagen). ChIP-enriched chromatin was used for Real-Time PCR with SYBR Green Master Mix, normalizing to input. Specific primers are listed in supplementary information (Table S6).
Immunohistochemistry (IHC)
Immunohistochemical staining on colon cancer tissue sections was performed on a DAKO Autostainer (DAKO, Carpinteria, CA) using DAKO LSAB+ and diaminobenzadine (DAB) as the chromogen. Serial sections of de-paraffinized tissue sections were labeled with rabbit polyclonal antibodies against human KMT4/Dot1L, H3K79me2 and CCL20 (AbCam), or mouse anti-human VCAM1 antibody (6G9, Abcam). H3K79me2 and DOT1L were localized in the nuclei, and were scored using the H-score method (Pirker et al., 2012). The H-score is a method of assessing the extent of nuclear immunoreactivity. The H score takes into account the percentage of positive cells (0-100%) in each intensity category (0-3+) and computes a final score, on a continuous scale between 0 and 300. The score is obtained by the formula: (3 × percentage of strongly staining nuclei) + (2 × percentage of moderately staining nuclei) + (1 × percentage of weakly staining nuclei), giving a range from 0 to 300 (Pirker et al., 2012). Any discrepancies were resolved by subsequent consultation with a diagnostic pathologist. The tissues were divided into high and low H3K79me2 and DOT1L expression based on the median value of H3K79me2 and DOT1L expression per tissue section.
In vivo tumor formation
Single cells were prepared from fresh colon cancer tissues. These colon cancer environmental cells contained all the primary cellular components in the colon cancer environment including CD3+ T cells within CD45+ immune cell population and lin−CD33-CD45−FSChighSSChigh primary colon cancer cells. These cells or colon cancer cells (102-5×106) were treated with anti-IL-22 or IL-22, and were subcutaneously injected into dorsal tissues of NSG mice (6-8 weeks old, Jackson Lab, Bar Harbor, Maine) (Curiel et al., 2004; Curiel et al., 2003; Kryczek et al., 2012). Tumor size was measured two times weekly using calipers fitted with a Vernier scale. Tumor volume was calculated based on three perpendicular measurements (Curiel et al., 2004; Curiel et al., 2003). Tumor incidence was monitored.
Statistical analysis
Wilcoxon rank-sum tests were used to compare two independent groups; for paired groups, Wilcoxon signed rank tests were used for comparison. Correlation coefficients (Spearman correlation, denoted by r, for ordinal data and Pearson correlation, denoted by r, for continuous data), together with a P-value (null hypothesis is that r is in fact zero), were computed to measure the degree of association between biomarkers. Log-rank test was used to compare time to tumor initiation between two groups. Overall patient survival was defined from date of diagnosis to disease related death. Data was censored at the last follow-up for patients who were disease-free or alive at the time of analysis. Survival functions were estimated by Kaplan-Meier methods. Cox's proportional hazards regression was performed to model survival (all classified as low and high based on the median value), after adjusting for age, grade and stage. The adequacy of the Cox regression model was assessed using graphical and numerical methods. All analyses were done using SAS 9.3 software. P < 0.05 was considered significant.
Supplementary Material
Acknowledgements
This work is supported (in part) by the National Institutes of Health grants (CA123088, CA099985, CA156685 and CA171306 for WZ; CA142808 and CA157663 for EH). We thank Wolf-Herve Fridman and Jay Hess for their intellectual support and valuable discussions, and Deborah Postiff, Michelle Vinco and Jackline Barikdar in the Tissue Procurement Core for their technical assistance. We are grateful for Drs. Weisheng Wu and Yongsheng Bai in the Bioinformatic Core for their help.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary Information:
Supplementary Information includes Figure legends, 6 supplementary Tables and 6 supplementary Figures.
The authors declare no competing financial interests.
References
- Aujla SJ, Chan YR, Zheng M, Fei M, Askew DJ, Pociask DA, Reinhart TA, McAllister F, Edeal J, Gaus K, et al. IL-22 mediates mucosal host defense against Gram-negative bacterial pneumonia. Nat Med. 2008;14:275–281. doi: 10.1038/nm1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu R, O'Quinn DB, Silberger DJ, Schoeb TR, Fouser L, Ouyang WJ, Hatton RD, Weaver CT. Th22 Cells Are an Important Source of IL-22 for Host Protection against Enteropathogenic Bacteria. Immunity. 2012;37:1061–1075. doi: 10.1016/j.immuni.2012.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Neriah Y, Karin M. Inflammation meets cancer, with NF-kappaB as the matchmaker. Nat Immunol. 2011;12:715–723. doi: 10.1038/ni.2060. [DOI] [PubMed] [Google Scholar]
- Bendall SC, Stewart MH, Menendez P, George D, Vijayaragavan K, Werbowetski-Ogilvie T, Ramos-Mejia V, Rouleau A, Yang J, Bosse M, et al. IGF and FGF cooperatively establish the regulatory stem cell niche of pluripotent human cells in vitro. Nature. 2007;448:1015–1021. doi: 10.1038/nature06027. [DOI] [PubMed] [Google Scholar]
- Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brabletz T, Jung A, Spaderna S, Hlubek F, Kirchner T. Opinion: migrating cancer stem cells - an integrated concept of malignant tumour progression. Nat Rev Cancer. 2005;5:744–749. doi: 10.1038/nrc1694. [DOI] [PubMed] [Google Scholar]
- Bromberg JF, Wrzeszczynska MH, Devgan G, Zhao Y, Pestell RG, Albanese C, Darnell JE., Jr. Stat3 as an oncogene. Cell. 1999;98:295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- Carpentino JE, Hynes MJ, Appelman HD, Zheng T, Steindler DA, Scott EW, Huang EH. Aldehyde dehydrogenase-expressing colon stem cells contribute to tumorigenesis in the transition from colitis to cancer. Cancer Res. 2009;69:8208–8215. doi: 10.1158/0008-5472.CAN-09-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cella M, Fuchs A, Vermi W, Facchetti F, Otero K, Lennerz JK, Doherty JM, Mills JC, Colonna M. A human natural killer cell subset provides an innate source of IL-22 for mucosal immunity. Nature. 2009;457:722–725. doi: 10.1038/nature07537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang MJ, Wu H, Achille NJ, Reisenauer MR, Chou CW, Zeleznik-Le NJ, Hemenway CS, Zhang W. Histone H3 lysine 79 methyltransferase Dot1 is required for immortalization by MLL oncogenes. Cancer Res. 2010;70:10234–10242. doi: 10.1158/0008-5472.CAN-10-3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coussens LM, Zitvogel L, Palucka AK. Neutralizing tumor-promoting chronic inflammation: a magic bullet? Science. 2013;339:286–291. doi: 10.1126/science.1232227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui TX, Kryczek I, Zhao L, Zhao E, Kuick R, Roh MH, Vatan L, Szeliga W, Mao Y, Thomas DG, et al. Myeloid-derived suppressor cells enhance stemness of cancer cells by inducing microRNA101 and suppressing the corepressor CtBP2. Immunity. 2013;39:611–621. doi: 10.1016/j.immuni.2013.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curiel TJ, Coukos G, Zou L, Alvarez X, Cheng P, Mottram P, Evdemon-Hogan M, Conejo-Garcia JR, Zhang L, Burow M, et al. Specific recruitment of regulatory T cells in ovarian carcinoma fosters immune privilege and predicts reduced survival. Nat Med. 2004;10:942–949. doi: 10.1038/nm1093. [DOI] [PubMed] [Google Scholar]
- Curiel TJ, Wei S, Dong H, Alvarez X, Cheng P, Mottram P, Krzysiek R, Knutson KL, Daniel B, Zimmermann MC, et al. Blockade of B7-H1 improves myeloid dendritic cell-mediated antitumor immunity. Nat Med. 2003;9:562–567. doi: 10.1038/nm863. [DOI] [PubMed] [Google Scholar]
- Daigle SR, Olhava EJ, Therkelsen CA, Majer CR, Sneeringer CJ, Song J, Johnston LD, Scott MP, Smith JJ, Xiao Y, et al. Selective killing of mixed lineage leukemia cells by a potent small-molecule DOT1L inhibitor. Cancer Cell. 2011;20:53–65. doi: 10.1016/j.ccr.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean M, Fojo T, Bates S. Tumour stem cells and drug resistance. Nat Rev Cancer. 2005;5:275–284. doi: 10.1038/nrc1590. [DOI] [PubMed] [Google Scholar]
- Dudakov JA, Hanash AM, Jenq RR, Young LF, Ghosh A, Singer NV, West ML, Smith OM, Holland AM, Tsai JJ, et al. Interleukin-22 drives endogenous thymic regeneration in mice. Science. 2012;336:91–95. doi: 10.1126/science.1218004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duhen T, Geiger R, Jarrossay D, Lanzavecchia A, Sallusto F. Production of interleukin 22 but not interleukin 17 by a subset of human skin-homing memory T cells. Nat Immunol. 2009;10:857–863. doi: 10.1038/ni.1767. [DOI] [PubMed] [Google Scholar]
- Dunn GP, Bruce AT, Ikeda H, Old LJ, Schreiber RD. Cancer immunoediting: from immunosurveillance to tumor escape. Nat Immunol. 2002;3:991–998. doi: 10.1038/ni1102-991. [DOI] [PubMed] [Google Scholar]
- Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A, Mlecnik B, Lagorce-Pages C, Tosolini M, Camus M, Berger A, Wind P, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313:1960–1964. doi: 10.1126/science.1129139. [DOI] [PubMed] [Google Scholar]
- Hanash AM, Dudakov JA, Hua G, O'Connor MH, Young LF, Singer NV, West ML, Jenq RR, Holland AM, Kappel LW, et al. Interleukin-22 protects intestinal stem cells from immune-mediated tissue damage and regulates sensitivity to graft versus host disease. Immunity. 2012;37:339–350. doi: 10.1016/j.immuni.2012.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Shilatifard A. Histone modification: cause or cog? Trends Genet. 2011;27:389–396. doi: 10.1016/j.tig.2011.06.006. [DOI] [PubMed] [Google Scholar]
- Huber S, Gagliani N, Zenewicz LA, Huber FJ, Bosurgi L, Hu B, Hedl M, Zhang W, O'Connor W, Jr., Murphy AJ, et al. IL-22BP is regulated by the inflammasome and modulates tumorigenesis in the intestine. Nature. 2012;491:259–263. doi: 10.1038/nature11535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins AP, Diez D, Takahashi Y, Ahmad S, Jauch R, Tremblay ML, Miranda-Saavedra D. Distinct transcriptional regulatory modules underlie STAT3's cell type-independent and cell type-specific functions. Nucleic Acids Res. 2013;41:2155–2170. doi: 10.1093/nar/gks1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo SY, Granowicz EM, Maillard I, Thomas D, Hess JL. Requirement for Dot1l in murine postnatal hematopoiesis and leukemogenesis by MLL translocation. Blood. 2011;117:4759–4768. doi: 10.1182/blood-2010-12-327668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchberger S, Royston DJ, Boulard O, Thornton E, Franchini F, Szabady RL, Harrison O, Powrie F. Innate lymphoid cells sustain colon cancer through production of interleukin-22 in a mouse model. The Journal of experimental medicine. 2013;210:917–931. doi: 10.1084/jem.20122308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryczek I, Liu S, Roh M, Vatan L, Szeliga W, Wei S, Banerjee M, Mao Y, Kotarski J, Wicha MS, et al. Expression of aldehyde dehydrogenase and CD133 defines ovarian cancer stem cells. Int J Cancer. 2012;130:29–39. doi: 10.1002/ijc.25967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryczek I, Zhao E, Liu Y, Wang Y, Vatan L, Szeliga W, Moyer J, Klimczak A, Lange A, Zou W. Human TH17 Cells Are Long-Lived Effector Memory Cells. Sci Transl Med. 2011;3:104ra100. doi: 10.1126/scitranslmed.3002949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryczek I, Zou L, Rodriguez P, Zhu G, Wei S, Mottram P, Brumlik M, Cheng P, Curiel T, Myers L, et al. B7-H4 expression identifies a novel suppressive macrophage population in human ovarian carcinoma. The Journal of experimental medicine. 2006;203:871–881. doi: 10.1084/jem.20050930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Herrmann A, Deng JH, Kujawski M, Niu G, Li Z, Forman S, Jove R, Pardoll DM, Yu H. Persistently activated Stat3 maintains constitutive NF-kappaB activity in tumors. Cancer Cell. 2009;15:283–293. doi: 10.1016/j.ccr.2009.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejeune D, Dumoutier L, Constantinescu S, Kruijer W, Schuringa JJ, Renauld JC. Interleukin-22 (IL-22) activates the JAK/STAT, ERK, JNK, and p38 MAP kinase pathways in a rat hepatoma cell line. Pathways that are shared with and distinct from IL-10. The Journal of biological chemistry. 2002;277:33676–33682. doi: 10.1074/jbc.M204204200. [DOI] [PubMed] [Google Scholar]
- Li L, Shaw PE. Elevated activity of STAT3C due to higher DNA binding affinity of phosphotyrosine dimer rather than covalent dimer formation. The Journal of biological chemistry. 2006;281:33172–33181. doi: 10.1074/jbc.M606940200. [DOI] [PubMed] [Google Scholar]
- Matsushita H, Vesely MD, Koboldt DC, Rickert CG, Uppaluri R, Magrini VJ, Arthur CD, White JM, Chen YS, Shea LK, et al. Cancer exome analysis reveals a T-cell-dependent mechanism of cancer immunoediting. Nature. 2012;482:400–404. doi: 10.1038/nature10755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews JR, Sansom OJ, Clarke AR. Absolute requirement for STAT3 function in small-intestine crypt stem cell survival. Cell death and differentiation. 2011;18:1934–1943. doi: 10.1038/cdd.2011.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min J, Feng Q, Li Z, Zhang Y, Xu RM. Structure of the catalytic domain of human DOT1L, a non-SET domain nucleosomal histone methyltransferase. Cell. 2003;112:711–723. doi: 10.1016/s0092-8674(03)00114-4. [DOI] [PubMed] [Google Scholar]
- Mohan M, Herz HM, Takahashi YH, Lin C, Lai KC, Zhang Y, Washburn MP, Florens L, Shilatifard A. Linking H3K79 trimethylation to Wnt signaling through a novel Dot1-containing complex (DotCom). Genes Dev. 2010;24:574–589. doi: 10.1101/gad.1898410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K, Yanagisawa M, Arakawa H, Kimura N, Hisatsune T, Kawabata M, Miyazono K, Taga T. Synergistic signaling in fetal brain by STAT3-Smad1 complex bridged by p300. Science. 1999;284:479–482. doi: 10.1126/science.284.5413.479. [DOI] [PubMed] [Google Scholar]
- Ng HH, Feng Q, Wang H, Erdjument-Bromage H, Tempst P, Zhang Y, Struhl K. Lysine methylation within the globular domain of histone H3 by Dot1 is important for telomeric silencing and Sir protein association. Genes Dev. 2002;16:1518–1527. doi: 10.1101/gad.1001502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardal R, Clarke MF, Morrison SJ. Applying the principles of stem-cell biology to cancer. Nat Rev Cancer. 2003;3:895–902. doi: 10.1038/nrc1232. [DOI] [PubMed] [Google Scholar]
- Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12:252–264. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickert G, Neufert C, Leppkes M, Zheng Y, Wittkopf N, Warntjen M, Lehr HA, Hirth S, Weigmann B, Wirtz S, et al. STAT3 links IL-22 signaling in intestinal epithelial cells to mucosal wound healing. The Journal of experimental medicine. 2009;206:1465–1472. doi: 10.1084/jem.20082683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirker R, Pereira JR, von Pawel J, Krzakowski M, Ramlau R, Park K, de Marinis F, Eberhardt WE, Paz-Ares L, Storkel S, et al. EGFR expression as a predictor of survival for first-line chemotherapy plus cetuximab in patients with advanced non-small-cell lung cancer: analysis of data from the phase 3 FLEX study. Lancet Onco. 2012;l13:33–42. doi: 10.1016/S1470-2045(11)70318-7. [DOI] [PubMed] [Google Scholar]
- Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- Scadden DT. The stem-cell niche as an entity of action. Nature. 2006;441:1075–1079. doi: 10.1038/nature04957. [DOI] [PubMed] [Google Scholar]
- Smith JJ, Deane NG, Wu F, Merchant NB, Zhang B, Jiang A, Lu P, Johnson JC, Schmidt C, Bailey CE, et al. Experimentally derived metastasis gene expression profile predicts recurrence and death in patients with colon cancer. Gastroenterology. 2010;138:958–968. doi: 10.1053/j.gastro.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenberg GF, Monticelli LA, Alenghat T, Fung TC, Hutnick NA, Kunisawa J, Shibata N, Grunberg S, Sinha R, Zahm AM, et al. Innate lymphoid cells promote anatomical containment of lymphoid-resident commensal bacteria. Science. 2012;336:1321–1325. doi: 10.1126/science.1222551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenberg GF, Nair MG, Kirn TJ, Zaph C, Fouser LA, Artis D. Pathological versus protective functions of IL-22 in airway inflammation are regulated by IL-17A. The Journal of experimental medicine. 2010;207:1293–1305. doi: 10.1084/jem.20092054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spits H, Cupedo T. Innate lymphoid cells: emerging insights in development, lineage relationships, and function. Annu Rev Immunol. 2012;30:647–675. doi: 10.1146/annurev-immunol-020711-075053. [DOI] [PubMed] [Google Scholar]
- Thompson CL, Plummer SJ, Tucker TC, Casey G, Li L. Interleukin-22 genetic polymorphisms and risk of colon cancer. Cancer Causes Control. 2010;21:1165–1170. doi: 10.1007/s10552-010-9542-5. [DOI] [PubMed] [Google Scholar]
- Trifari S, Kaplan CD, Tran EH, Crellin NK, Spits H. Identification of a human helper T cell population that has abundant production of interleukin 22 and is distinct from T(H)-17, T(H)1 and T(H)2 cells. Nat Immunol. 2009;10:864–871. doi: 10.1038/ni.1770. [DOI] [PubMed] [Google Scholar]
- Vermeulen L, de Sousa e Melo F, Richel DJ, Medema JP. The developing cancer stem-cell model: clinical challenges and opportunities. Lancet Oncol. 2012;13:e83–89. doi: 10.1016/S1470-2045(11)70257-1. [DOI] [PubMed] [Google Scholar]
- Wang Y, Liu Y, Malek SN, Zheng P. Targeting HIF1alpha eliminates cancer stem cells in hematological malignancies. Cell Stem Cell. 2011;8:399–411. doi: 10.1016/j.stem.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto N, Yamamoto S, Inagaki F, Kawaichi M, Fukamizu A, Kishi N, Matsuno K, Nakamura K, Weinmaster G, Okano H, Nakafuku M. Role of Deltex-1 as a transcriptional regulator downstream of the Notch receptor. The Journal of biological chemistry. 2001;276:45031–45040. doi: 10.1074/jbc.M105245200. [DOI] [PubMed] [Google Scholar]
- Yang E, Lerner L, Besser D, Darnell JE., Jr. Independent and cooperative activation of chromosomal c-fos promoter by STAT3. The Journal of biological chemistry. 2003;278:15794–15799. doi: 10.1074/jbc.M213073200. [DOI] [PubMed] [Google Scholar]
- Yu H, Kortylewski M, Pardoll D. Crosstalk between cancer and immune cells: role of STAT3 in the tumour microenvironment. Nat Rev Immunol. 2007;7:41–51. doi: 10.1038/nri1995. [DOI] [PubMed] [Google Scholar]
- Yu W, Chory EJ, Wernimont AK, Tempel W, Scopton A, Federation A, Marineau JJ, Qi J, Barsyte-Lovejoy D, Yi J, et al. Catalytic site remodelling of the DOT1L methyltransferase by selective inhibitors. Nat Commun. 2012;3:1288. doi: 10.1038/ncomms2304. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Valdez PA, Danilenko DM, Hu Y, Sa SM, Gong Q, Abbas AR, Modrusan Z, Ghilardi N, de Sauvage FJ, Ouyang W. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat Med. 2008;14:282–289. doi: 10.1038/nm1720. [DOI] [PubMed] [Google Scholar]
- Zou W. Immunosuppressive networks in the tumour environment and their therapeutic relevance. Nat Rev Cancer. 2005;5:263–274. doi: 10.1038/nrc1586. [DOI] [PubMed] [Google Scholar]
- Zou W, Machelon V, Coulomb-L'Hermin A, Borvak J, Nome F, Isaeva T, Wei S, Krzysiek R, Durand-Gasselin I, Gordon A, et al. Stromal-derived factor-1 in human tumors recruits and alters the function of plasmacytoid precursor dendritic cells. Nat Med. 2001;7:1339–1346. doi: 10.1038/nm1201-1339. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.