Abstract
Detection of individual target cells among a large amount of blood cells is a major challenge in clinical diagnosis and laboratory protocols. Many researches show that two dimensional cells array technology can be incorporated into routine laboratory procedures for continuously and quantitatively measuring the dynamic behaviours of large number of living cells in parallel, while allowing other manipulations such as staining, rinsing, and even retrieval of targeted cells. In this study, we present a high-density cell self-assembly technology capable of quickly spreading over 300 000 cells to form a dense mono- to triple-layer cell arrangement in 5 min with minimal stacking of cells by the gentle incorporation of gravity and peripheral micro flow. With this self-assembled cell arrangement (SACA) chip technology, common fluorescent microscopy and immunofluorescence can be utilized for detecting and analyzing target cells after immuno-staining. Validated by experiments with real human peripheral blood samples, the SACA chip is suitable for detecting rare cells in blood samples with a ratio lower than 1/100 000. The identified cells can be isolated and further cultured in-situ on a chip for follow-on research and analysis. Furthermore, this technology does not require external mechanical devices, such as pump and valves, which simplifies operation and reduces system complexity and cost. The SACA chip offers a high-efficient, economical, yet simple scheme for identification and analysis of rare cells. Therefore, potentially SACA chip may provide a feasible and economical platform for rare cell detection in the clinic.
INTRODUCTION
Rare cell detection and identification is a fundamental and requisite technology in clinical diagnosis and laboratory protocols, such as the detection of inflammatory cells and pathogenic cells in immune diseases treatment,1, 2, 3 circulating tumor cells in cancer therapy,4, 5, 6, 7, 8 prenatal diagnosis,9, 10, 11, 12 and pluripotent stem cells researches in recent years.13, 14
Referring to Table TABLE I., cell identification research can be roughly categorized into 3 areas: physicochemical mechanism, applied technology, and associated apparatus. The same mechanism may be associated with different types of technologies and also carried out in active, passive, or stationary fluidic apparatus.
TABLE I.
Methods and technologies for cell identification/isolation.
| Common cell identification methods | ||||||
|---|---|---|---|---|---|---|
| Mechanism type | Differential attributes | Technology | Apparatus or applications | References | ||
| Physical | Osmotic tolerance | Hypotonic solution | RBC lysis buffer | 15 | ||
| Density | Zonal centrifugation | Mononuclear cell isolation buffer | 6, a 16 | |||
| Dimension | Inertial force | Size-dependent Hydrodynamic isolation | 31, a17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30 | |||
| Filtration | Microfluidic chip/membrane filters | 32, 33, 34, 35 | ||||
| Electric susceptibility | Dielectrophoresis/negative dielectrophoresis | DEP/n-DEP chip | 36, 37, 38, 39, 40 | |||
| Impedance | Impedance-test | Impedance-based cytometers | 41, 42, 43, a44a | |||
| Elasticity | Deformability-test | Deformability-based chip | 34, 45, 46, 47 | |||
| Optical-properties | Forward/side scattering | Flow cytometers | 65, 88, 89 | |||
| Raman scattering | Raman spectroscopy | 90, 91, 92, 93 | ||||
| Chemical/biological | DNA/RNA profiling | Polymerase chain reaction | Fluorescence labelling gel electrophoresis | Q-PCR/EP-PCR | 48, 49, 50, 51 | |
| Antibody specificity | Surface adhesion (trapping cells) | channels | Temperature/pH/light-sensitive gel | Trapping target cells at channel wall | 31, 58, 59, 60 | |
| structures | Trapping target cells at pillars | 14, a56, a57 | ||||
| objects | Trapping target cells at beads | 61a | ||||
| Magnetic beads | Magnetic beads | 17, 52, 53, 54, 55 | ||||
| Immuno-fluorescence | Flow cytometry | Flow cytometers | 64, 65, 66 | |||
| Microscopy | Cell arrays | 67, 68, 69, 70, 71, 72, 73, 74, 75 | ||||
| Fluidic cell chips | 6, 76, 77a | |||||
| Bio-capability | To form spheroids, endocytose, balance electrolytes | cell chips | 57, 94 | |||
| Integrated system | Using a variety of techniques | Automatic/semi-automatic system | 78, 79, a80, 81 | |||
Research paper using clinical cancer patient blood.
Mechanisms can be classified as physical and chemical/biological. The most common physical attributes for differentiating cells are their osmotic tolerances,15 densities,16 and dimensions.17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35 Common technologies using the dimension attribute to separate cells include fluidic force17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31 and filtration,32, 33, 34, 35 typically coupled with fluidic systems for cell sorting. Especially, the dimension-based micro inertial fluidic force separation technology has attracted a lot of attention in recent years.26, 27, 28, 29, 30, 31 Other physical mechanisms used to separate cells include electric susceptibility,36, 37, 38, 39, 40 impedance,41, 42, 43, 44 and elasticity.45, 46, 47 However, the major issue encountered by physical mechanisms is their lack of specificity.
On the other hand, chemical/biological methods can offer better selectivity for cell type identification. Common methods include DNA/RNA profiling48, 49, 50, 51 and antibody specific recognition. Antibody recognition is the most commonly used and has been coupled with various technologies for cell identification such as surface adhering, magnetic bead capturing,52, 53, 54, 55 or fluorescence indicating. In surface adhesion, antibodies are employed to immobilize target cells onto specific areas, such as micro rods array,56, 57 channel walls,58, 59, 60 or glass beads.61 In addition, the captured cells can be released back to the fluidic systems by utilizing temperature-sensitive, pH-sensitive, or light-sensitive gel.59, 61 In immunofluorescence reporting, the combination of antibody mechanism and fluorescence technology is very popular in clinical and laboratory research.62, 63 This method typically utilizes flow cytometry or microscopy for cell identification.64, 65, 66 Comparing with flow cytometry, microscopy is another relatively low cost and readily available technology. Hence, there has been much research in developing cell arrays67, 68, 69, 70, 71, 72, 73, 74, 75 and fluidic cell chips76, 77 in conjunction with microscopes. The common methods and technologies for cell identification are listed in Table TABLE I.. Multiple methods may be used in an integrated semi-automated or automated system in clinical applications. For example, CELLSEARCH® (Johnson and Johnson Plaza New Brunswick, NJ),78 HD-CTC,79, 80, 81 etc. However, high setup cost is usually associated with such highly integrated systems.
Polymerase chain reaction (PCR) and flow cytometry with immunofluorescence have been developed for identifying rare cells and are widely used among large medical research centers or research agencies.7, 8, 62, 82, 83 However, these two expensive technologies are difficult to be utilized in developing countries and remote or rural areas due to their high setup cost and lengthy operational procedures. To simplify and improve the process for identifying rare cells while managing costs, two dimensional (2D) cell arrays were developed.4, 84 The key elements of 2D cell array technology include arrangement of cells into a mono-layer, labeling target cells with an immuno-fluorescence marker and detecting labeled cells with a fluorescent microscope. 2D cell arrays, unlike PCR, do not require cell lysis. Therefore, the cells' geometric information, with the exception of the binding immuno-fluorescence marker, is retained in the analysis. The traditional smear technique is essentially an old-fashioned 2D cell array. It involves spreading a large quantity of cells on a flat surface to ease the observation by microscope. However, the smear technique is not reliable when the ratio of target cells to peripheral blood cells is low. Because cells are not spread evenly, the technique leaves gaps between cells while many cells aggregate or stack, inhibiting the observation of target cells.85 As a result, technologies for more controllable formation of 2D cell array were developed.
In general, modern 2D cell arrays can be classified as active or passive (i.e., stationary) (shown in Table TABLE II.). In active 2D cell arrays, cells are carried in fluid and trapped in micro-structures to form cell arrays.67, 68, 69, 70 This method has the advantage of positioning cells in precise locations within the array, but the structures and microfluidic channels occupy much of the limited microscopic field. Consequently, the number of cells within the microscopic field may not be large enough and result in low detection efficiency. Moreover, in order to maintain a stable fluidic field, external pumps and valves are needed to control flow discharge and velocity. The inherent complexity of active 2D cell arrays limits their ability to meet the “ease of use” objective of clinical medicine.
TABLE II.
Technology comparison of 2D cell arrays or chip.
| Cell array/array-like arrangement | |||||
|---|---|---|---|---|---|
| Type | Cell density on the retaining area | Area for single array or chip | Maximum cell number in single array or chip | References | |
| Active cell array | ∼200 cells/mm2 | ∼56 mm2 | (7 mm × 8 mm) | ∼1.0 × 104 cells/chip | 67 |
| ∼330 cells/mm2,a | ∼ 7.5 mm2 | (2.5 mm × 3 mm) | ∼800 cells/chip | 69 | |
| Passive or stationary cell array | ∼1800 cells/mm2 | ∼12 mm2 | (3 mm × 4 mm) | ∼3.3 × 104 cells/dish | 73 |
| ∼2500 cells/mm2 | ∼3.14 mm2 | (2 mm circle) | ∼7.8 × 103 cells/chip | 72 | |
| ∼3450 cells/mm2 | ∼9 mm2 | (3 mm × 3 mm) | ∼3.1 × 104 cells/array | 75 | |
| Self-assembled cell chip | ∼14 300 cells/mm2 | ∼19.63 mm2 | (5 mm circle) | ∼3.0 × 105 cells/chip human peripheral blood mononuclear cells | This study (SACA) |
∼330 cells/mm2 in the picture, and ∼3300 cells/mm2 in the article.
Typical passive and stationary 2D cell arrays take the form of microwell arrays and offer simplified fabrication and operational procedures as well as improved application practicality.1 The space between cells is smaller and the density of cells in the viewing field is higher, increasing the detection efficiency for rare cells.71, 72, 73, 74, 75 However, such microwell arrays still have difficulties in ensuring the uniform distribution of cells.84 Although such arrays have the advantage of simple operation and low cost, the random distribution of cells results in low detection efficiency. In addition, most 2D cell arrays do not allow for extraction of the identified target cells for further analysis in the living state. To culture target rare cells after purification, traditional clinical methods extract target cells using high pressure flow cytometry, a costly technology that still results in low cell viability.66
Therefore, a practical 2D cell array technology must overcome two conflicting goals. First, the cells cannot be too close, resulting in stacking together which will cause the immunofluorescence signal to be blocked in the microscopic field. Second, the cells should not be excessively dispersed resulting in reduction of the number of cells in the observation field and decreasing the detection efficiency. The objective of this research is to develop a self-assembled cell arrangement (SACA) chip, with glass slides, to uniformly distribute cells in a microscopic field without complex trapping structures or external power sources, such as a pump. SACA chip should be capable of arranging cells densely in a 2D array-like arrangement while minimizing cell stacking.71 In addition, samples should not undergo fierce flow field changes and should retain viability for in situ culture directly in the SACA chip for follow-on research and analysis. Technologies and commercial products for rapid scanning of large surfaces have been well established; however, such technology or product may not be available in areas with limited resources. SACA of this research requires only a standard fluorescence microscope for investigation without expensive or complicated fabrication and operation. High cell density on SACA allows manual screening of rare cells with microscopes in a reasonable amount of time.
The SACA system should be fast, simple, reliable, convenient, and economical to facilitate its use in developing countries and areas with limited medical resources.
MATERIALS AND METHODS
SACA chip design and fabrication
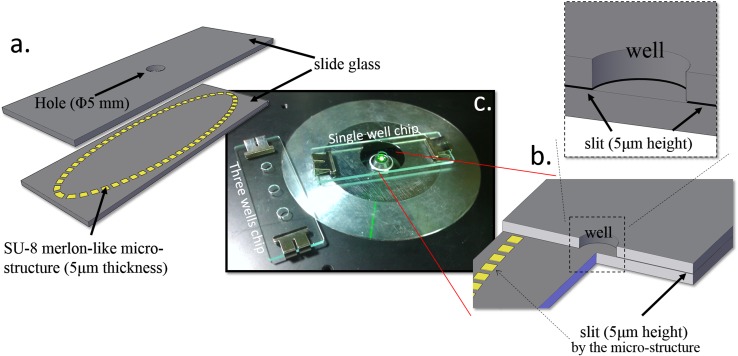
The SACA chip consists of two standard microscope glass slides (76 mm × 26 mm × 1 mm). The upper slide contains a hole with 5 mm diameter. It is coated with fluoro-octyltrichlorosilane (FOTS) as an anti-adhesion layer.86 On the bottom slide, there is a 5 μm-thick SU-8 3005 photoresist ring shaped with a merlon-like microstructure (Figure 1a) (SU-8 3005, Microchem Corporation, 3000 rpm spin speed, 30 s). Top and bottom glass slides are clapped together in Phosphate-Buffered Saline (PBS, Gibco, Carlsbad, CA) solution. After the assembly of slides, the hole in the top slide serves as a sample loading well for forming the 2D cell arrangement (Figure 1b). A 5 μm slit is formed between two glass slides separated by the SU-8 spacers, and micro flow is allowed to radiate outward at the bottom of the well through microstructure gaps. Cells in a drop of solution will be directly loaded into the center well and the self-assembly process will occur automatically on cells through the action from gravity in vertical direction and capillary flow radially in the lateral direction, as shown in Figure 2. In general, the well diameter/liquid height ratio should be designed roughly similar to the cell spreading/descending ratio to allow enough time for cells to spread over the well. Since the slit need to be designed smaller than cell as 5 μm, the outward flow speed, governed by evaporation of liquid at the slit outmost boundary, was measured about 10 μm/s. To accommodate as many cells as possible, we would hope the diameter can be in the range of cm, however, the liquid height need also to be increased to cm, which is a little too high for fabrication. As a result, the diameter of the well is finally set as 5 mm for the best optimization among flow speed ratio, slit size limitation, and fabrication capability in the current study. Wells with larger size are still under development for accommodating cells in the order of 106–107.
Figure 1.
The SACA. (a) Component schematic diagram. (b) The positional relationship of the well and the micro slit channel (chip sectional view). (c) The actual photos of chips on the inverted fluorescent microscope.
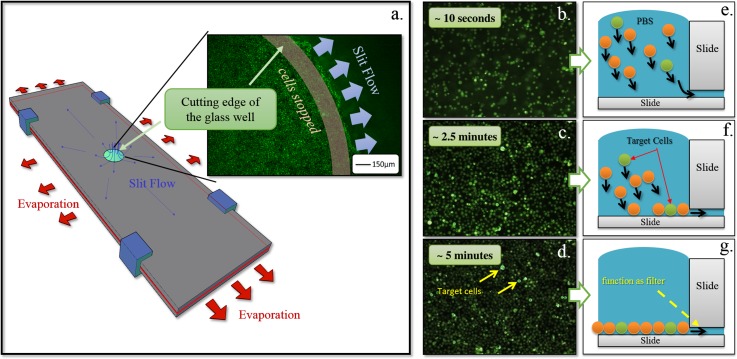
Figure 2.
(a) Evaporation drives radial slit flow and cell dispersion. (b)–(d) The bottom view of the settling and organization of cells over 5 min. (e)–(g) The corresponding illustrated cross-sectional side view of (b)–(d) shows the settling and organization of cells over 5 min. If there are gaps between cells at the bottom, cells above will try to fill the gaps and eventually settle at the bottom of the well.
Cell culture and preparation
Several kinds of cells were used in this study.
HL-60 cells
HL-60 (human promyelocytic leukemia) cell line (from ATCC, American Type Culture Collection, Bethesda, MD, USA) was used to study the flow field in the chip. These cells were maintained by passaging twice weekly with Roswell Park Memorial Institute (RPMI) 1640 medium supplemented with 20% fetal bovine serum (FBS) under 5% CO2 at 37 °C.
HeLa cells
HeLa (human cervical carcinoma) cell line (from ATCC) was used to emulate rare target cells in the test of sensitivity and reliability of the chip in detecting rare cells. The cells were maintained by passaging twice weekly with Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% FBS under 5% CO2 at 37 °C.
BT-474 cells
BT-474 (Human breast carcinoma) cell line (from ATCC) was used to emulate rare target cells in the test of sensitivity and reliability of the chip when detecting rare cells. The cells were maintained by passaging three times weekly with RPMI-1640 supplemented with 10% FBS under 5% CO2 at 37 °C.
Purification of human peripheral blood mononuclear cells (PBMCs)
Human PBMCs were used to simulate the clinical specimen by mixing with different concentrations of target cells in this research. Human peripheral blood samples from volunteers contained a large number of red blood cells (RBCs). To avoid an excessive number of cells, interference from hemoglobin, and other issues, RBCs must be removed before the detection. Hence, RBC lysis buffer (ACK Lysing Buffer, Invitrogen, Grand Island, NY, USA) is used to remove RBCs from the samples. After this treatment, white blood cells (WBCs) become the main component in the PBMC sample which also contains target rare cells. (In this research, the human blood samples were obtained with the full consent of donors. Safety precautions were taken and no donors were harmed.)
SACA chip operation overview
Before loading into the SACA chip, adherent cells, HeLa and BT-474, were detached from 100 mm diameter culture dishes after adding 1 ml trypsin EDTA (0.25%, Gibco, Carlsbad, CA). 10× volume of DMEM + FBS was then added to deactivate remaining trypsin. Cells were then centrifuged to a pellet and re-suspended in PBS with pH 7.4 for further use.
PBS solution containing cells was loaded into the chip well, which was also filled with PBS solution. In Figure 1c, the sample in the chip will be observed by inverted fluorescence microscope (IX71 inverted fluorescence microscope, Olympus Corporation, Tokyo, Japan).
EXPERIMENT
Cell spreading and self-assembly in SACA chip (E1)
In order to observe and analyze the phenomena and organizing processes of the self-assembled 2D cell chip, HL-60 cells were first used in the tracking cell dynamics experiments. The HL-60 cells were stained with Alexa Fluor® 488 conjugate of Wheat Germ Agglutinin (WGA) (Invitrogen, Grand Island, NY, USA) and observed by inverted fluorescence microscope (IX71 inverted fluorescence microscope, Olympus Corporation, Tokyo, Japan). PBS solution containing cells was loaded into the well, which was also filled with PBS solution (see Figure 1c).
Numerical simulation and cell dynamic tracking (E2)
Numerical simulations are intended to mimic experiments, to extend the implications of the experiments, and to identify the physical significance for further studies by solving an equation set based on conservation laws. Governing equations adopted in the calculations of this research include mass conservation equations, momentum conservation equations, and volume fraction equations. The complete set of equations was solved using the finite-volume numerical method (CFD-ACE+ code from the CFD Research Corporation, ESI group, France) based on the configuration and boundary/initial conditions in the experiments. At the same time, the arrangement of the cells over time was tracked in the bottom-view video, recorded by CCD camera mounted on the inverted microscope as shown in supplementary Figure S1.87
Validation of rare cells identification by SACA chip (E3)
Validation of the functionality of the cell arrangement was conducted by comparing experimental cell detection data with the calculated data based on known ratio of target cells and background cells.
HeLa cells were divided into two groups and stained with two types of WGA antibody fluorescence. HeLa cells with Alexa Fluor 488 (green) fluorescence and HeLa cells with Alexa Fluor® 647 (red) fluorescence, mixed at 1:10, 1:100, 1:1000, and 1:10 000 ratios, were loaded into the chip for microscopic inspection. The same experiments were repeated five times to assure the statistical significance of final experimental data.
Rare cell detection from human blood by SACA chip (E4)
In order to approach a clinical detection process, a simulated clinical experiment was conducted. BT474 breast cancer cells were used to act as rare target cells and mixed with human whole blood samples. Samples were prepared in different ratios per experimental requirements. Usually, RBCs are removed by RBCs lysis buffer to avoid detection difficulties due to the vast amount of red blood cells in clinical laboratory.52 However, apoptotic debris of RBCs may be left in the solution containing PBMCs and rare target cells. This experiment verified whether the apoptotic debris and blood platelets would interfere the formation of the cell arrangement. As target cells, BT-474 cells, a type of carcinoma derived from epithelial tissues, can be identified with EpCAM antibody conjugated with FITC. Other cells, as background cells, were marked with CD45 antibody conjugated with Alexa Fluor 647 (red). After these two staining steps, samples were loaded into the chip to be inspected using a microscope.
Cells culture on SACA chip (E5)
After experiment E2, the cells in the chip were cultured in RPMI-1640 supplemented with 10% FBS under 5% CO2 at 37 °C for 3 days. Then the chip was un-clapped and slightly washed with PBS solution to remove the primary PBMCs that did not adhere to the well bottom. Because many rare but important cells in peripheral blood are adhesion cells, such as epithelial cells, progenitor cells, pluripotent stem cells, or some immune cells, they can be easily separated from the PBMCs, which are suspension cells.
RESULTS AND DISCUSSION
Cell spreading and self-assembly in SACA chip (E1)
Solution flows through the slit at the bottom of the well and reaches the outer edges of glass slides by capillary and evaporation effects. Driven by capillary force, solution in the well will continue to flow outward to replace the vaporized water. This forms an outwardly radiating flow field which distributes cells laterally (see Figure 2a). In Figure 2, the photographs ((b)–(d)) are taken by a camera mounted under the bottom of the well. The illustrations ((e)–(g)) on the right display the cross-sectional view of the well. Carried by the current towards the slit at bottom of the well, cells gradually sink to the bottom while flowing laterally towards the well wall in this flow field. After approximately 5 min, all cells are spread at the bottom of the well. In the process, cell arrangement and fluidic field are controlled by the following mechanisms:
-
1.
Gravity: forces cells to sink to the bottom of well and finally all cells settle on the same plane. It allows simultaneous inspection of large volume of cells.
-
2.
Well wall: without the containment of well wall, cells would be carried by flow field and spread freely and unrestrictedly on the glass slide. Hence, cell distribution would fail to form high density and high volume cell arrangements (see Figures 2e, 2f).
-
3.
Slit at the well bottom: the height of the slit is slightly smaller than the average cell diameter. The slit functions as a filter to prevent cells escaping from the well. Because cells are not rigid, they may be squeezed into the slit to relieve pressure when there are excessive cells jamming at the bottom of the well (see Figure 2a photograph). When a cell enters the slit, it is unlikely to flow too far because it is dragged by friction and cell-glass adhesion. Since the glass is transparent, we can easily observe the cells in the slit. This phenomenon is demonstrated in Figure 2a, which is an enlarged picture of the edge of the well from bottom view.
-
4.
Lateral driving force: besides functioning as a filter, the slit at the well bottom provides a lateral driving force in the fluidic field. This driving force is due to evaporation of fluid at the outer edges of glass slides (see red arrows in Figure 2a).
Because the total surface tension force of the glass slide at the outer edges is larger than that of the well, it causes fluid in the well to continuously flow toward the edge of the well to replace water lost through evaporation. Effectively, this forms a stable lateral driving force field without a mechanical pump. For the purpose of stabilizing flow, the evaporation actuation can also be replaced by arranging a sponge at the outer edge of the well.
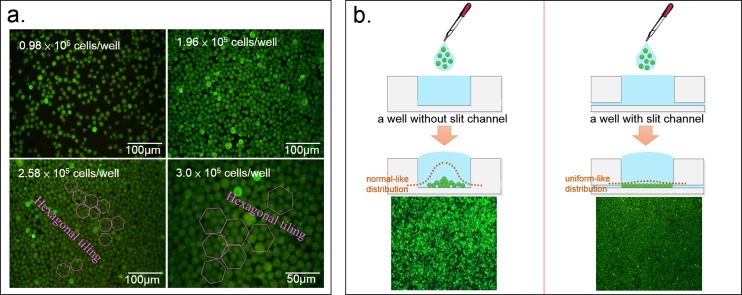
In Figure 3a, in the well, the organization of the cell arrangement changes as the total number of cells in the well changes. When the total number of cells exceeds 2.58 × 105, the cell arrangement begins to form hexagonal tiling which is the highest density organization in a 2D arrangement. However, because of the individual differences of the cells and cell sizes, there are still defects in hexagonal tiling. When there are more than 3 × 105 HL-60 cells in the well, it reaches saturation. The central part of the cell chip will begin to form double or even triple layers, rather than monolayer. Still, using fluorescence detection, two to three layers of stacked cells can be detected. In addition to density, the cells arrangement is also affected by the consistency of cells size. Figure 3b shows that the slit channel significantly impacts the sinking and organization of the cell arrangement. The slit flow drives cells to spread evenly and to form a tight, typically monolayer and array-like arrangement. In the absence of the slit channel, the cells are much more likely to accumulate in the center than at the border because of the hemispherical shape of the droplet containing cells, thereby forming a bell-curve-like stack.
Figure 3.
(a) Different numbers of cells in the well leads to different types of cell organization. When the cell number exceeds 258 000 in the well, the cell arrangement begins to form hexagonal tiling, the highest density in 2D arrangement. (b) The bottom slit channel has a significant impact to the organization of the cell arrangement. It allows the cells to be distributed more evenly.
Numerical simulation and cell dynamic tracking (E2)
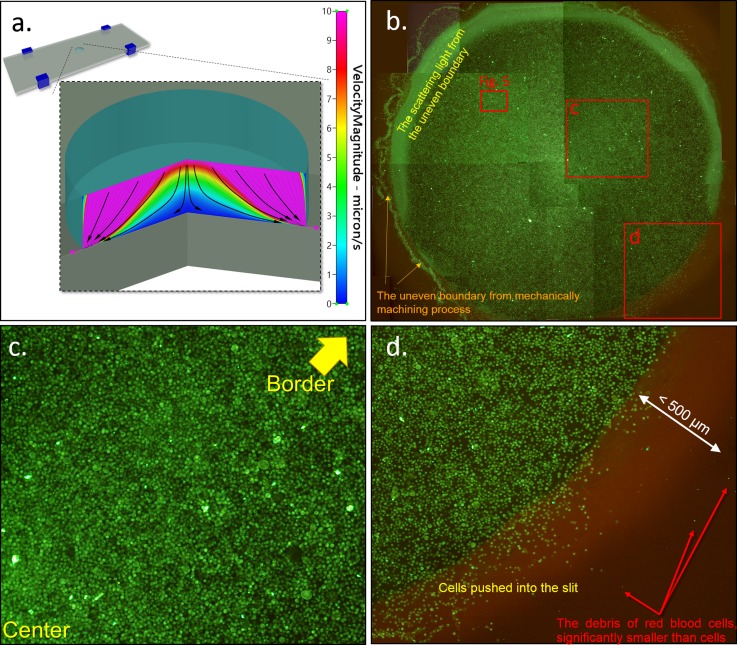
CFD-RC is used to simulate the flow field in the self-assembled chip. In the simulation result (Figure 4a), the slow velocity area is shown in blue, while the high velocity area is shown in pink. Streamlines are also shown in Figure 4a. Lateral velocity is higher when it is closer to the top and to the side of the flow field. Lateral velocity is low when it is near the central bottom of the well. Lateral velocity would increase with slight elevation as illustrated by the streamline distribution. But even in the bottom edge region of the well, the fastest velocity in the fluidic field is only about 10 μm/s and decrease with time. Figure 4b shows the full map of the well, which contains 2.5 × 105 cells, by combining 12 photos. The distribution of cells is similar to the prediction from the numerical simulation result of flow speed distribution. Since the outward velocity in the center region is lower than that at the border in the simulation, the cell would be easier to stop in the center than at the border by well-surface friction. As a result, the cell density in the center will be somehow higher than that at the border. This phenomenon was verified by experiments from the cell distribution images across the whole diameter of the well. However, as long as the cells are not stack to one another, individual cells can still be clearly imaged. The scattering light from the border of the hole is due to the uneven boundary formed during machining process of the well on glass substrate, and the light scattering does not affect much on the observation of the cell arrangement underneath the border region (Figure 4b). Since the outflow is very slow (about 10 μm/s) in the micro slit, it is unlikely that the cells can flood into the micro slit from the center, and which is verified from the experiment result: we can see more scattered cells distribution at the border than that in the center by the phenomenon that average fluorescence intensity is higher in the center than that at the border of the assembled cells (refer to Figures 4b, 4c, 3b). Even a very small number of cells really be pushed into the slit, due to viscous friction between the micro slit and cells as well as the gradually smaller flow velocity in radiation type flow, the cells will not slide for far distance and stop at a final position at the border region. The average stopping distance from the picture can be identified as about 200 μm, as the region shown between the yellow and red dashed lines in Figure 4d. This part of cells occupies only less than 1% of the total cells, and standard fluorescence observation under microscope can still recognize the target cells because the slit is merely the height of a single cell and the bottom glass slide is transparent without any problem.
Figure 4.
(a) Numerical simulation results by CFD-RC illustrating the flow field in the self-assembly cell chip when the velocity of slip flow is 10 μm/s. The results show that the lateral velocity is higher when it is closer to the top or edge of the well. (b) Photograph of the cell arrangement in the well (diameter of 5 mm). The distribution of cells is similar to the results of numerical simulation. About 250 000 cells, 12 photos combined. (c) The region near center. (d) The border region.
The amount of fluid that flows through the slit is affected by the peripheral evaporation surrounding the chip. Due to the increase of cross-sectional area of the slit's outer edge and the adherence of the bottom glass slide, the flow velocity decreases as the cells flow towards the outer edge of the slit. Few cells entered the slit and those cells would stop within a few hundred microns. Furthermore, those cells can be easily observed.
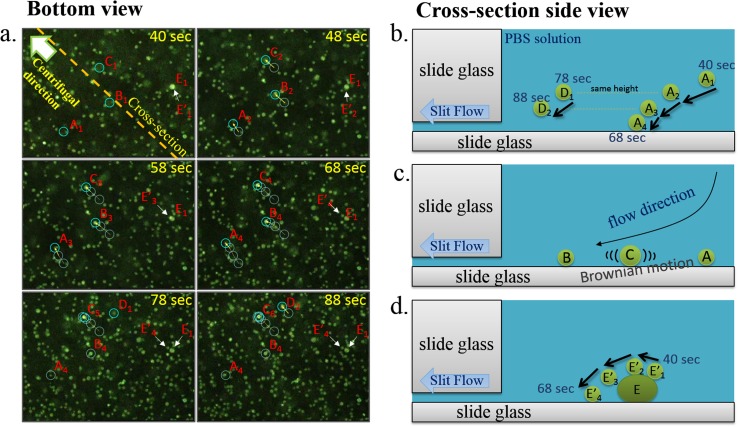
Figure 5a (the enlarged area of red-lined box in Figure 4b) is filmed with an inverted fluorescence microscope aimed at the bottom of the well. The timer starts when the cell-PBS solution is loaded into the well. The lower right corner of the picture is close to the center of the well, while the upper left corner of the picture is close to the outer edge. Cells showing a sharp image are the cells that have settled or almost settled at the bottom of the well. Cells appearing as foggy light spots are not quite near the bottom of the well. Further away from the bottom, those cells fall outside the working distance of the object lens and are completely immersed in the green light background produced by all the other cells' fluorescence.
Figure 5.
(a) Snapshots from bottom-view videos from 40 s to 88 s after the PBS solution containing cells were loaded onto the chip. (b) As a cell sinks from the top to bottom of the well, its lateral velocity deceases. Cell D at 78 s is about the same height as cell A at 48 s, and their lateral movement is about the same over 10 s. (c) As shown in cell C, some cells may still move slightly after they settle at the bottom of the well due to Brownian movement. This phenomenon provides an additional mechanism for tighter cell organization. (d) The movement of the E and E′ bright spot between 40 and 68 s shows that cells stacked on top of other cells will continue to move in fluidic fields and find gaps or voids among lower cells. This is consistent with numerical analysis. This phenomenon shows why the cells do not stack and ultimately form tight organization on the chip.
When loading into the well containing PBS solutions, cells would randomly disperse in a bell curve shape if there were no slit at bottom of the well. The descending process of cells was shown in supplementary Figure S2.87 With the slit, cells are also dispersed by the lateral flow field. The three phenomena in the flow field are illustrated in Figure 5 (also refer to the supplementary figures87).
Radial dispersion of cells
At 40 s, cells A, B, and C enter the microscopic field, although their images are slightly blurred. Between 40 and 68 s, tacking of these three cells show that they all moved towards upper left corner as they are carried by the flow field and their images gradually become clear as they get closer to the bottom of the well. The movement nearly comes to a halt when they touch down at the bottom at 68 s mark (also shown in Figure 5b).
Self-assembly of cell monolayer
Some cells can still slide even after they touch the bottom of the well. This sliding provides additional opportunity for cells to create a tighter formation in the self-assembly process. For example, cell C sank and entered the microscopic field with similar lateral movement as cells A and B, finally showing a sharp image when it touches the well bottom. However, the follow-up tracking shows that at the 78-s and 88-s marks, cell C's position continued to move in random directions. In fact, as the cell arrangement formation is near completion after 5 min, many cells are still moving in random directions similar to cell C unless blocked by obstacles. In addition to not adhering to the glass surface at the bottom, there are two possible factors for this phenomenon. One factor is Brownian motion and another is that larger cells are receiving larger lateral fluidic field force. These two factors continue to push cells into voids in the cell arrangement (also shown in Figure 5c).
Height/lateral velocity correlation
In tracking the drift of cells A, B, and C, we observed that the higher the cell position (they are less clear and less bright in the photos) above the well bottom, the longer the distance they travel as shown in sequence of photos. Cells closer to the well bottom (they are more clear and bright) move a shorter distance. This phenomenon is consistent with the numerical simulation (Figure 4a) results.
The height/lateral velocity correlation is further demonstrated by the observation of cell D's position changes between 78 and 88 s. Cell D at 78 s is about the same clarity as cells A, B, and C at 48 s. It implies that cell D at 78 s is at about the same height in well as cells A, B, and C at 48 s. The observed travel distance of cell D in 10 s between 78 and 88 s is about the same as cells A, B, and C in 10 s between 48 and 58 s. This is consistent with the numerical analysis results. In adjacent areas, cells at the same height receive similar flow forces.
Another observation is that cell E has stopped at the well bottom at 40 s. Although cell E is quite bright, from 40 s to 58 s, it can be seen that the brightest spot (E′) around cell E is moved from the lower right corner to the upper left corner of the cell over time. At 68 s, the reason for its exceptional brightness is finally revealed: there was another cell E′ on top of Cell E. Cell E′ was driven by the fluidic field and flew over Cell E to its upper left corner at 68 s. Seen as a sharp bright spot in a microscopy field, cell E′ was already close to the well bottom and was directly on top of cell E. This phenomenon proves that even if there is only one cell height difference, upper cells will bear greater thrust, pass over the lower cells and fall to the well bottom (also shown in Figure 5d).
These three phenomena facilitate the continuous cell movement from the center of the well to the outer region of the well, until the upper cells find gaps among bottom cells and settle at the well bottom, eventually forming a tight cell arrangement.
Validation of rare cell identification by SACA chip (E3)
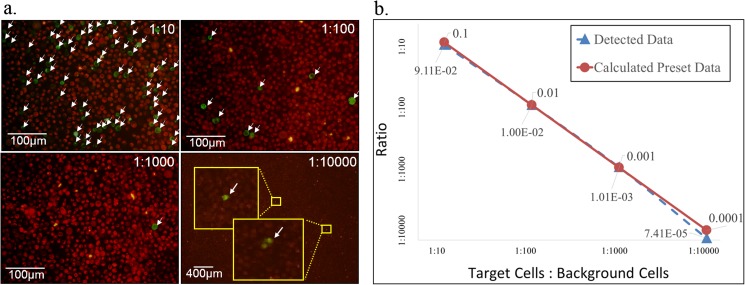
As shown in Figure 6a, green fluorescent target cells can be easily identified in all four samples of different ratios. With straightforward calculations, the experimental detection data and calculated preset data match quite well. Experimental detection data only deviate slightly from the calculated preset data at a ratio of 1:10 000 target cells to background cells. This discordance is likely the result of errors in preparing the solutions and in taking photographs of the samples due to the extremely low number of target cells. This demonstrates the reliability of the cell arrangement in detecting target cells (Figure 6b). The identification method is based on full cell image scanning, containing many information, such as fluorescent colors, cell size, and cell shape. This is a fully developed and widely used technique. All the cells dropped into SACA chip are completely preserved in the well, and even a small amount of leaking cells on the well boundary can still be imaged. Therefore, a very low detection error rate is anticipated when inspecting the entire sample at single cell level in this high-density cell arrangement. The only concern is that the arrangement is in fact not a monolayer and that stacked cells would shield fluorescent signals. However, the results prove that there is little data loss due to aggregation of cells that block each other's fluorescent signals. The data demonstrate that the self-assembled chip not only can assemble cells in a dense formation but also creates very few stacking layers.
Figure 6.
(a) The target cells (green) are HeLa cells with Alexa Fluor 488 fluorescence, and the background cells (red) are HeLa cells with Alexa Fluor 647 fluorescence. The target cells can be detected easily in all four different ratios of mixtures. (b) The comparison of the detected data and calculated pre-set data with different ratios of targeted cells to background cells.
Rare cell detection from human blood by SACA chip (E4)
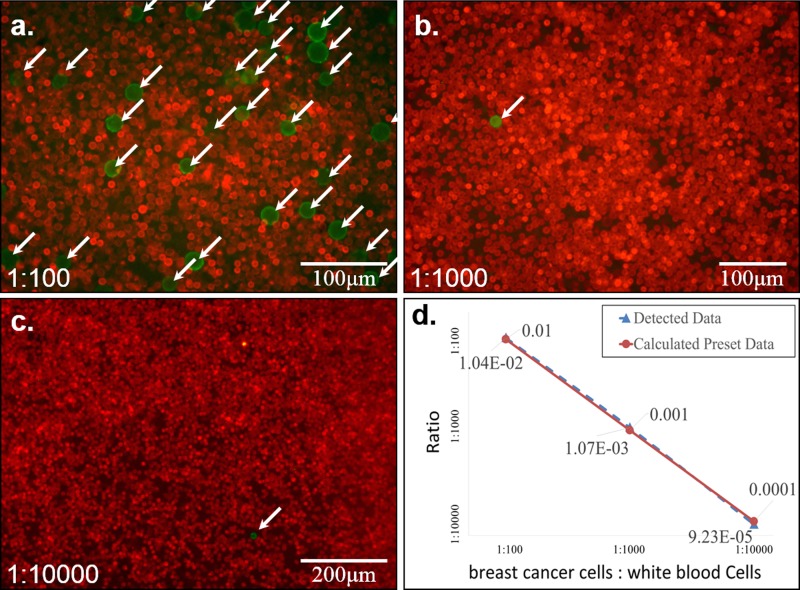
Figure 7 shows the results of experiment E4. Two ratios of target cells to background cells, 1:100 and 1:10 000, were used in this experiment. For both ratios, the target cells can easily be identified among the WBCs. Microscopic photos show that apoptotic debris, resulting from lysis of RBCs, did not affect the cell arrangement operation.
Figure 7.
(a) The target cells (green) are BT-474 cells (breast carcinoma) labeled with EpCAM antibody conjugated with FITC fluorescence, and the background cells (red) are white blood cells labeled with CD45 antibody conjugated with Alexa Fluor 647 fluorescence. The target cells can be detected easily in all three ratios of mixtures. (b) The comparison of the detected data and calculated preset data with different ratios of breast cancer cells (BT-474) to white blood cells.
The experiment shows that the WBCs are constrained in the well while the apoptotic debris and blood platelet would flow out of the well through the 5 μm slit. This process would reduce their interference in the formation of the cell arrangement.
Figures 7a, 7b, 7c show that target cells can be easily detected in spite of some level of stacking in mono- to triple layers of WBCs. This phenomenon is confirmed with confocal microscope (Nikon IX71 Inverted microscope with the Cooke Corporation electron multiplication digital 12 bit CCD camera system). As shown in Figure 7d, the detection data are very consistent with the calculated preset data. It confirms that moderate stacking of WBC cells does not interfere with the detection of target cells.
In practical applications, if the number of target cells is very low, such as less than 10 cells in one viewing field, counting target cells by human eyes will be faster than software analysis of a CCD-derived image. The results of this experiment demonstrate that the SACA chip indeed can be used to detect cell line cancer cells in real human blood and it is compatible with common techniques and equipment for removing RBCs. The system provides a mechanism for cell array-like arrangement that does not interfere with identification of target cells in a relatively short period of time.
In this experiment, a well of 5 μm diameter can fit into the viewing field of a 4× objective (4× objective + 10× Eyepiece). In the case of low numbers of target cells, the entire well can be quickly analyzed by human eyes in 30 s after the samples have been loaded into the SACA chip. If it is operated with a CCD camera, higher magnification at 10 × objective is required in order to resolve the rarer fluorescence signal. The total analysis time will be increased to about 2 min because the well image is segmented into twelve 1392 × 1024 fields due to the constraints of camera's resolution (Pixelfly, PCO AG, Donaupark, Germany). (The time estimate does not include the time spent on pre-treatment and preparation of samples.)
Although the SACA chip utilizes a highly manual operation in this study, it demonstrates the feasibility of such platform for rare target cell detection in the future. The SACA chip, as a tool for quickly spreading sample cells in a high density monolayer-like arrangement and with appropriate design augmentation, may assist other automated analytical system, particularly the full imaging cell identification systems, such as HD-CTC system presented by Kuhn et al.79, 80, 81 The low cost and passive design of SACA chips may have the potential to be expanded to clinical diagnosis with the aid of MEMS technology.
Cell culture on SACA chip (E5)
As shown in Figure 8, after 3 days culture, bt-474 cells are adhered to the bottom of the well, while blood cells are merely touching, not adhering to, the bottom of the well. Therefore, separation and purification of target cells are achieved by slightly washing culture medium with PBS solutions to remove most of the primary lymphocytes. The identified bt-474 cells can also be picked-up by pipette and easily transferred to a more suitable environment for further investigation or incubation. The experimental results demonstrate that this chip not only detects and measures the quality of adhesion rare target cells but also purifies and cultivates them. The system provides practical use for the follow-on research and analysis of rare cells.
Figure 8.
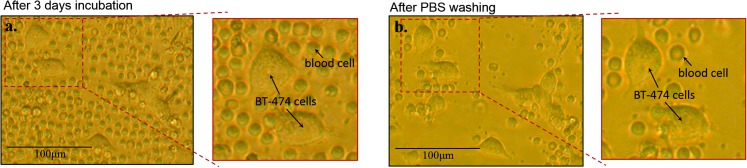
(a) After self-assembly of human blood/cancer cells on the SACA chip, the cells on the chip were cultured for 3 days. (b) Most primary lymphocyte (blood cells) on the chip can be easily removed by slight washing with PBS solution. Some primary lymphocytes which are lying on the bottom of the well can be removed by slightly stronger washing.
CONCLUSION
In summary, we have developed a simple method for forming a 2D self-assembled monolayer (SAM) cell arrangement by employing gravity, micro capillary flow, and cell filter design, without using external mechanical devices such as a pump. The formation of the 2D cell arrangement can be accomplished within 5 min with no loss of sample cells. Confirmed by successful experiments in detecting cell line cancer cells in real human blood with a ratio fewer than 1/100 000 in a 5 μm well containing more than 3 × 105 cells, this technology is potentially suitable for clinical applications to both adhesive and suspension cells. The identified cells can also be cultured for further analysis or applications. The self-assembled cell chip requires only a standard fluorescence microscope for investigation without expensive or complicated fabrication and operation. This low cost but highly accurate 2D cell self-assembly technology is particularly suitable for developing countries and areas with scarce medical resources.
With appropriate design augmentation, the low-cost and simple passive design of the SACA chip may have the potential to be expanded to clinical research in rare cell detection.
ACKNOWLEDGMENTS
The authors thank the National Nanoscience and Nanotechnology Program, National Science Council, Taiwan for their support through Grant Nos. NSC-101-2811-E-20 007-012 and NSC-101-2120-M-007-001.
References
- Autebert J., Coudert B., Bidard F.-C., Pierga J.-Y., Descroix S., Malaquin L., and Viovy J.-L., Methods 57, 297 (2012). 10.1016/j.ymeth.2012.07.002 [DOI] [PubMed] [Google Scholar]
- Gijs M. A. M., Lacharme F., and Lehmann U., Chem. Rev. 110, 1518 (2010). 10.1021/cr9001929 [DOI] [PubMed] [Google Scholar]
- Mach A. J., Adeyiga O. B., and Di Carlo D., Lab Chip 13, 1011 (2013). 10.1039/c2lc41104k [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Carlo D. and Lee L. P., Anal. Chem. 78, 7918 (2006). 10.1021/ac069490p [DOI] [PubMed] [Google Scholar]
- Maheswaran S. and Haber D. A., Curr. Opin. Genet. Dev. 20, 96 (2010). 10.1016/j.gde.2009.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ts'o P. O. P., Pannek J., Wang Z.-P. P., Lesko S. A., Steven Bova G., Partin A. W., and Bova G. S., Urology 49, 881 (1997). 10.1016/S0090-4295(97)00157-X [DOI] [PubMed] [Google Scholar]
- Alunni-Fabbroni M. and Sandri M. T., Methods 50, 289 (2010). 10.1016/j.ymeth.2010.01.027 [DOI] [PubMed] [Google Scholar]
- Doyen J., Alix-Panabières C., Hofman P., Parks S. K., Chamorey E., Naman H., and Hannoun-Lévi J.-M., Crit. Rev. Oncol. Hematol. 81, 241 (2012). 10.1016/j.critrevonc.2011.05.004 [DOI] [PubMed] [Google Scholar]
- Steele C. D., Wapner R. J., Smith J. B., Haynes M. K., and Jackson L. G., Clin. Obstet. Gynecol. 39, 801 (1996). 10.1097/00003081-199612000-00009 [DOI] [PubMed] [Google Scholar]
- Hromadnikova I., Houbova B., Hridelova D., Voslarova S., Kofer J., Komrska V., and Habart D., Prenat. Diagn. 23, 235 (2003). 10.1002/pd.556 [DOI] [PubMed] [Google Scholar]
- Mohamed H., Turner J. N., and Caggana M., J. Chromatogr. A 1162, 187 (2007). 10.1016/j.chroma.2007.06.025 [DOI] [PubMed] [Google Scholar]
- D'Souza E., Kulkarni S., Colah R. B., and Mohanty D., Hemoglobin 31, 39 (2007). 10.1080/03630260601057047 [DOI] [PubMed] [Google Scholar]
- Hyun K.-A. and Jung H.-I., Electrophoresis 34, 1028 (2013). 10.1002/elps.201200417 [DOI] [PubMed] [Google Scholar]
- Nagrath S., Sequist L. V., Maheswaran S., Bell D. W., Irimia D., Ulkus L., Smith M. R., Kwak E. L., Digumarthy S., Muzikansky A., Ryan P., Balis U. J., Tompkins R. G., Haber D. A., and Toner M., Nature 450, 1235 (2007). 10.1038/nature06385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron I. L. and Fullerton G. D., Biochem. Cell Biol. 68, 894 (1990). 10.1139/o90-132 [DOI] [PubMed] [Google Scholar]
- Hibino N., Nalbandian A., Devine L., Martinez R. S., McGillicuddy E., Yi T., Karandish S., Ortolano G. A., Shin'oka T., Snyder E., and Breuer C. K., Tissue Eng. Part C 17, 993 (2011). 10.1089/ten.tec.2011.0110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J. H., Krause S., Tobin H., Mammoto A., Kanapathipillai M., and Ingber D. E., Lab Chip 12, 2175 (2012). 10.1039/c2lc40072c [DOI] [PubMed] [Google Scholar]
- Sun J., Li M., Liu C., Zhang Y., Liu D., Liu W., Hu G., and Jiang X., Lab Chip 12, 3952 (2012). 10.1039/c2lc40679a [DOI] [PubMed] [Google Scholar]
- Tanaka T., Ishikawa T., Numayama-Tsuruta K., Imai Y., Ueno H., Matsuki N., and Yamaguchi T., Lab Chip 12, 4336 (2012). 10.1039/c2lc40354d [DOI] [PubMed] [Google Scholar]
- Zhou J. and Papautsky I., Lab Chip 13, 1121 (2013). 10.1039/c2lc41248a [DOI] [PubMed] [Google Scholar]
- Parichehreh V. and Sethu P., Lab Chip 12, 1296 (2012). 10.1039/c2lc21034g [DOI] [PubMed] [Google Scholar]
- Bhagat A. A. S., Bow H., Hou H. W., Tan S. J., Han J., and Lim C. T., Med. Biol. Eng. Comput. 48, 999 (2010). 10.1007/s11517-010-0611-4 [DOI] [PubMed] [Google Scholar]
- Gossett D. R., Weaver W. M., Mach A. J., Hur S. C., Tse H. T. K., Lee W., Amini H., and Di Carlo D., Anal. Bioanal. Chem. 397, 3249 (2010). 10.1007/s00216-010-3721-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhagat A. A. S., Kuntaegowdanahalli S. S., Kaval N., Seliskar C. J., and Papautsky I., Biomed. Microdevices 12, 187 (2010). 10.1007/s10544-009-9374-9 [DOI] [PubMed] [Google Scholar]
- Park J.-S. and Jung H.-I., Anal. Chem. 81, 8280 (2009). 10.1021/ac9005765 [DOI] [PubMed] [Google Scholar]
- Di Carlo D., Irimia D., Tompkins R. G., and Toner M., Proc. Natl. Acad. Sci. U.S.A. 104, 18892 (2007). 10.1073/pnas.0704958104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mach A. J. and Di Carlo D., Biotechnol. Bioeng. 107, 302 (2010). 10.1002/bit.22833 [DOI] [PubMed] [Google Scholar]
- Hur S. C., Mach A. J., and Di Carlo D., Biomicrofluidics 5, 22206 (2011). 10.1063/1.3576780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhagat A. A. S., Hou H. W., Li L. D., Lim C. T., and Han J., Lab Chip 11, 1870 (2011). 10.1039/c0lc00633e [DOI] [PubMed] [Google Scholar]
- Hur S. C., Tse H. T. K., and Di Carlo D., Lab Chip 10, 274 (2010). 10.1039/b919495a [DOI] [PubMed] [Google Scholar]
- Stott S. L., Hsu C.-H., Tsukrov D. I., Yu M., Miyamoto D. T., Waltman B. A., Rothenberg S. M., Shah A. M., Smas M. E., Korir G. K., Floyd F. P., Gilman A. J., Lord J. B., Winokur D., Springer S., Irimia D., Nagrath S., Sequist L. V., Lee R. J., Isselbacher K. J., Maheswaran S., Haber D. A., and Toner M., Proc. Natl. Acad. Sci. U.S.A. 107, 18392 (2010). 10.1073/pnas.1012539107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H., Chueh B., Wu H., Hall E. W., Li C., Schirhagl R., Lin J.-M., and Zare R. N., Lab Chip 11, 238 (2011). 10.1039/c0lc00121j [DOI] [PubMed] [Google Scholar]
- Kim M. S., Sim T. S., Kim Y. J., Kim S. S., Jeong H., Park J.-M., Moon H.-S., Il Kim S., Gurel O., Lee S. S., Lee J.-G., and Park J. C., Lab Chip 12, 2874 (2012). 10.1039/c2lc40065k [DOI] [PubMed] [Google Scholar]
- McFaul S. M., Lin B. K., and Ma H., Lab Chip 12, 2369 (2012). 10.1039/c2lc21045b [DOI] [PubMed] [Google Scholar]
- Lim L. S., Hu M., Huang M. C., Cheong W. C., Gan A. T. L., Looi X. L., Leong S. M., Koay E. S.-C., and Li M.-H., Lab Chip 12, 4388 (2012). 10.1039/c2lc20750h [DOI] [PubMed] [Google Scholar]
- Gascoyne P., Mahidol C., Ruchirawat M., Satayavivad J., Watcharasit P., and Becker F. F., Lab Chip 2, 70 (2002). 10.1039/b110990c [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han K.-H. and Frazier A. B., Lab Chip 8, 1079 (2008). 10.1039/b802321b [DOI] [PubMed] [Google Scholar]
- Han K.-H., Han S.-I., and Frazier A. B., Lab Chip 9, 2958 (2009). 10.1039/b909753h [DOI] [PubMed] [Google Scholar]
- Shafiee H., Sano M. B., Henslee E. A., Caldwell J. L., and Davalos R. V., Lab Chip 10, 438 (2010). 10.1039/b920590j [DOI] [PubMed] [Google Scholar]
- Moon H.-S., Kwon K., Kim S.-I., Han H., Sohn J., Lee S., and Jung H.-I., Lab Chip 11, 1118 (2011). 10.1039/c0lc00345j [DOI] [PubMed] [Google Scholar]
- Cheung K., Gawad S., and Renaud P., Cytometry A 65, 124 (2005). 10.1002/cyto.a.20141 [DOI] [PubMed] [Google Scholar]
- Sun T. and Morgan H., Microfluid. Nanofluid. 8, 423 (2010). 10.1007/s10404-010-0580-9 [DOI] [Google Scholar]
- Holmes D., Pettigrew D., Reccius C. H., Gwyer J. D., van Berkel C., Holloway J., Davies D. E., and Morgan H., Lab Chip 9, 2881 (2009). 10.1039/b910053a [DOI] [PubMed] [Google Scholar]
- Choi H., Kim K. B., Jeon C. S., Hwang I., Lee S., Kim H. K .H. C., and Chung T. D., Lab Chip 13, 970 (2013). 10.1039/c2lc41376k [DOI] [PubMed] [Google Scholar]
- Kuo J. S., Zhao Y., Schiro P. G., Ng L., Lim D. S. W., Shelby J. P., and Chiu D. T., Lab Chip 10, 837 (2010). 10.1039/b922301k [DOI] [PubMed] [Google Scholar]
- Hur S. C., Henderson-MacLennan N. K., McCabe E. R. B., and Di Carlo D., Lab Chip 11, 912 (2011). 10.1039/c0lc00595a [DOI] [PubMed] [Google Scholar]
- Preira P., Grandné V., Forel J.-M., Gabriele S., Camara M., and Theodoly O., Lab Chip 13, 161 (2013). 10.1039/c2lc40847c [DOI] [PubMed] [Google Scholar]
- Davies R. T., Kim J., Jang S. C., Choi E.-J., Gho Y. S., and Park J., Lab Chip 12, 5202 (2012). 10.1039/c2lc41006k [DOI] [PubMed] [Google Scholar]
- Guo M. T., Rotem A., Heyman J. A., and Weitz D. A., Lab Chip 12, 2146 (2012). 10.1039/c2lc21147e [DOI] [PubMed] [Google Scholar]
- Deininger M., Buchdunger E., and Druker B. J., Blood 105, 2640 (2005). 10.1182/blood-2004-08-3097 [DOI] [PubMed] [Google Scholar]
- Bustin S., J. Mol. Endocrinol. 29, 23 (2002). 10.1677/jme.0.0290023 [DOI] [PubMed] [Google Scholar]
- Hoshino K., Huang Y.-Y., Lane N., Huebschman M., Uhr J. W., Frenkel E. P., and Zhang X., Lab Chip 11, 3449 (2011). 10.1039/c1lc20270g [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A., Byvank T., Chang W.-J., Bharde A., Vieira G., Miller B. L., Chalmers J. J., Bashir R., and Sooryakumar R., Lab Chip 13, 1172 (2013). 10.1039/c2lc41201b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lien K.-Y., Chuang Y.-H., Hung L.-Y., Hsu K.-F., Lai W.-W., Ho C.-L., Chou C.-Y., and Lee G.-B., Lab Chip 10, 2875 (2010). 10.1039/c005178k [DOI] [PubMed] [Google Scholar]
- Issadore D., Shao H., Chung J., Newton A., Pittet M., Weissleder R., and Lee H., Lab Chip 11, 147 (2011). 10.1039/c0lc00149j [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleghorn J. P., Pratt E. D., Denning D., Liu H., Bander N. H., Tagawa S. T., Nanus D. M., Giannakakou P. A., and Kirby B. J., Lab Chip 10, 27 (2010). 10.1039/b917959c [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bichsel C. A., Gobaa S., Kobel S., Secondini C., Thalmann G. N., Cecchini M. G., and Lutolf M. P., Lab Chip 12, 2313 (2012). 10.1039/c2lc40130d [DOI] [PubMed] [Google Scholar]
- Cheung L. S. L., Zheng X., Stopa A., Baygents J. C., Guzman R., Schroeder J. A., Heimark R. L., and Zohar Y., Lab Chip 9, 1721 (2009). 10.1039/b822172c [DOI] [PubMed] [Google Scholar]
- Gurkan U. A., Anand T., Tas H., Elkan D., Akay A., Keles H. O., and Demirci U., Lab Chip 11, 3979 (2011). 10.1039/c1lc20487d [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., Cheung L. S.-L., Schroeder J. A., Jiang L., and Zohar Y., Lab Chip 11, 3269 (2011). 10.1039/c1lc20331b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y., Liu Y., Allen P. B., Asghar W., Mahmood M. A. I., Tan J., Duhon H., Kim Y., Ellington A. D., and Iqbal S. M., Lab Chip 12, 4693 (2012). 10.1039/c2lc21251j [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantel K., Brakenhoff R. H., and Brandt B., Nat. Rev. Cancer 8, 329 (2008). 10.1038/nrc2375 [DOI] [PubMed] [Google Scholar]
- Arya S. K., Lim B., and Rahman A. R. A., Lab Chip 13, 1995 (2013). 10.1039/c3lc00009e [DOI] [PubMed] [Google Scholar]
- Mao X., Nawaz A. A., Lin S.-C. S., Lapsley M. I., Zhao Y., McCoy J. P., El-Deiry W. S., and Huang T. J., Biomicrofluidics 6, 24113 (2012). 10.1063/1.3701566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dive C., Gregory C. D., Phipps D. J., Evans D. L., Milner A. E., and Wyllie A. H., Biochim. Biophys. Acta: Mol. Cell Res. 1133, 275 (1992). 10.1016/0167-4889(92)90048-G [DOI] [PubMed] [Google Scholar]
- Suh T. K., Schenk J. L., and Seidel G. E., Theriogenology 64, 1035 (2005). 10.1016/j.theriogenology.2005.02.002 [DOI] [PubMed] [Google Scholar]
- Tan W.-H. and Takeuchi S., Proc. Natl. Acad. Sci. U.S.A. 104, 1146 (2007). 10.1073/pnas.0606625104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan W.-H. and Takeuchi S., Lab Chip 8, 259 (2008). 10.1039/b714573j [DOI] [PubMed] [Google Scholar]
- Di Carlo D., Aghdam N., and Lee L. P., Anal. Chem. 78, 4925 (2006). 10.1021/ac060541s [DOI] [PubMed] [Google Scholar]
- Di Carlo D., Wu L. Y., and Lee L. P., Lab Chip 6, 1445 (2006). 10.1039/b605937f [DOI] [PubMed] [Google Scholar]
- Chen T.-J., Chang J.-C., Chang Y.-C., and Tseng F.-G., in IEEE 26th International Conference on Micro Electro Mechanical Systems ( IEEE, 2013), pp. 923–926.
- Deutsch M., Deutsch A., Shirihai O., Hurevich I., Afrimzon E., Shafran Y., and Zurgil N., Lab Chip 6, 995 (2006). 10.1039/b603961h [DOI] [PubMed] [Google Scholar]
- Rettig J. R. and Folch A., Anal. Chem. 77, 5628 (2005). 10.1021/ac0505977 [DOI] [PubMed] [Google Scholar]
- Yamamura S., Kishi H., Tokimitsu Y., Kondo S., Honda R., Rao S. R., Omori M., Tamiya E., and Muraguchi A., Anal. Chem. 77, 8050 (2005). 10.1021/ac0515632 [DOI] [PubMed] [Google Scholar]
- Wang Y., Shah P., Phillips C., Sims C. E., and Allbritton N. L., Anal. Bioanal. Chem. 402, 1065 (2012). 10.1007/s00216-011-5535-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S.-B., Wu M.-H., Lin Y.-H., Hsieh C.-H., Yang C.-L., Lin H.-C., Tseng C.-P., and Lee G.-B., Lab Chip 13, 1371 (2013). 10.1039/c3lc41256c [DOI] [PubMed] [Google Scholar]
- Alyassin M. A., Moon S., Keles H. O., Manzur F., Lin R. L., Hæggstrom E., Kuritzkes D. R., and Demirci U., Lab Chip 9, 3364 (2009). 10.1039/b911882a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristofanilli M., Hayes D. F., Budd G. T., Ellis M. J., Stopeck A., Reuben J. M., Doyle G. V., Matera J., Allard W. J., Miller M. C., Fritsche H. A., Hortobagyi G. N., and Terstappen L. W. M. M., J. Clin. Oncol. 23, 1420 (2005). 10.1200/JCO.2005.08.140 [DOI] [PubMed] [Google Scholar]
- Ben Hsieh H., Marrinucci D., Bethel K., Curry D. N., Humphrey M., Krivacic R. T., Kroener J., Kroener L., Ladanyi A., Lazarus N., Kuhn P., Bruce R. H., and Nieva J., Biosens. Bioelectron. 21, 1893 (2006). 10.1016/j.bios.2005.12.024 [DOI] [PubMed] [Google Scholar]
- Kuhn P. and Bethel K., Phys. Biol. 9, 010301 (2012). 10.1088/1478-3975/9/1/010301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieva J., Wendel M., Luttgen M. S., Marrinucci D., Bazhenova L., Kolatkar A., Santala R., Whittenberger B., Burke J., Torrey M., Bethel K., and Kuhn P., Phys. Biol. 9, 016004 (2012). 10.1088/1478-3975/9/1/016004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantel K., Alix-Panabières C., and Riethdorf S., Nat. Rev. Clin. Oncol. 6, 339 (2009). 10.1038/nrclinonc.2009.44 [DOI] [PubMed] [Google Scholar]
- Economos C., Morrissey C., and Vessella R. L., Curr. Opin. Urol. 22, 190 (2012). 10.1097/MOU.0b013e3283519b58 [DOI] [PubMed] [Google Scholar]
- Nilsson J., Evander M., Hammarström B., and Laurell T., Anal. Chim. Acta 649, 141 (2009). 10.1016/j.aca.2009.07.017 [DOI] [PubMed] [Google Scholar]
- Anthony Fauci J. L., Braunwald E., Kasper D., Hauser S., Longo D., and Jameson J., in Harrison's Manual of Medicine, 17th ed. (McGraw Hill Professional, 2009), p. 1264. [Google Scholar]
- Hsieh H.-Y., Wang P.-C., Wu C.-L., Huang C.-W., Chieng C.-C., and Tseng F.-G., Anal. Chem. 81, 7908 (2009). 10.1021/ac900552v [DOI] [PubMed] [Google Scholar]
- See supplementary material at http://dx.doi.org/10.1063/1.4874716 for schematics of cells descending process.
- von Dassow P., van den Engh G., Iglesias-Rodriguez D., and Gittins J. R., J. Plankton Res. 34, 1011 (2012). 10.1093/plankt/fbs061 [DOI] [Google Scholar]
- de Grooth B. G., Terstappen L. W., Puppels G. J., and Greve J., Cytometry 8, 539 (1987). 10.1002/cyto.990080602 [DOI] [PubMed] [Google Scholar]
- Premasiri W. R., Moir D. T., Klempner M. S., Krieger N., Jones G., and Ziegler L. D., J. Phys. Chem. B 109, 312 (2005). 10.1021/jp040442n [DOI] [PubMed] [Google Scholar]
- Nolan J. P., Duggan E., Liu E., Condello D., Dave I., and Stoner S. A., Methods 57, 272 (2012). 10.1016/j.ymeth.2012.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster K., Urlaub E., and Gapes J., J. Microbiol. Methods 42, 29 (2000). 10.1016/S0167-7012(00)00169-X [DOI] [PubMed] [Google Scholar]
- Kneipp K., Haka A. S., Kneipp H., Badizadegan K., Yoshizawa N., Boone C., Shafer-Peltier K. E., Motz J. T., Dasari R. R., and Feld M. S., Appl. Spectrosc. 56, 150 (2002). 10.1366/0003702021954557 [DOI] [Google Scholar]
- Pamme N. and Wilhelm C., Lab Chip 6, 974 (2006). 10.1039/b604542a [DOI] [PubMed] [Google Scholar]