Summary
Objective
The TGF-β pathway plays a central role in joint development with polymorphism in TGF-β pathway genes implicated in osteoarthritis susceptibility. One association is to rs12901499, within intron 1 of SMAD3. Since rs12901499 is not in linkage disequilibrium with a non-synonymous polymorphism, it is likely the association is operating by influencing expression of SMAD3.
Design
Using tissues from the joints of primary osteoarthritis patients who had undergone joint replacement we measured the overall expression of SMAD3 by quantitative real-time PCR. We also measured allelic expression of SMAD3 using these tissues and vascular smooth muscle cells from patients with aneurysms and osteoarthritis syndrome, a rare condition featuring early-onset osteoarthritis. We tested the functional effect of SNPs in vitro using luciferase assays and assessed association with osteoarthritis using a large osteoarthritis case–control dataset.
Results
We observed that genotype at rs12901499 did not correlate with overall SMAD3 expression or allelic expression. However, genotype at a 3′UTR SNP, rs8031440, did correlate with SMAD3 expression in cartilage (P = 0.005) which was supported by allelic expression data showing that the G allele correlated with decreased SMAD3 expression in joint tissues and vascular smooth muscle cells. This G allele was underrepresented in osteoarthritis cases vs controls (P = 0.027, odds ratio = 0.921). rs8031440 is in perfect linkage disequilibrium with five other SMAD3 3′UTR SNPs and our luciferase analysis identified rs3743342 and rs12595334 as being functional.
Conclusion
SMAD3 is subject to cis-acting regulatory polymorphism in the tissues of relevance to both primary osteoarthritis and the aneurysms-osteoarthritis syndrome.
Keywords: Osteoarthritis, Aneurysm, Genetics, Susceptibility, SMAD3, Allelic expression
Introduction
The TGF-β signalling pathway plays a critical role in joint development and maintenance and there is accumulating evidence of involvement of this pathway in osteoarthritis (OA) aetiology1. Genetic studies have contributed to this evidence, with OA risk alleles reported in the genes GDF5, ASPN and SMAD32.
SMAD3 codes for the protein mothers against decapentaplegic homolog 3, an intracellular molecule that translocates the TGF-β signal to the nucleus instigating gene transcription. Smad3-knockout mice develop a degenerative joint disease resembling OA3 whilst nonsense, missense and frame-shift mutations of SMAD3 can result in the aneurysms and osteoarthritis syndrome (AOS), which involves thoracic aortic aneurysms and dissections with early-onset OA4, 5. In 2010 it was reported that the single nucleotide polymorphism (SNP) rs12901499, located in intron 1 of SMAD3, was associated with common primary hip and knee OA in a European cohort6, with P < 4.5 × 10−6.
As more susceptibility loci are identified for human diseases it has become apparent that the majority of risk alleles contribute to disease occurrence by influencing the expression of nearby genes, by either regulating transcription or the stability of their transcripts7. These are known as cis-acting expression quantitative trait loci or cis-eQTLs. Since rs12901499 is not in linkage disequilibrium (LD) with a non-synonymous polymorphism, we hypothesised that this OA association is marking a cis-eQTL that operates on SMAD3. In this manuscript we have used a number of experimental techniques to test this hypothesis. We were also keen to assess whether any such cis-eQTL was operating in tissues of relevance to AOS, thus potentially providing a pathological link between common OA and the syndrome.
Materials and methods
Patients
Joint tissues were obtained from patients undergoing total hip replacement (THR) or total knee replacement (TKR) for primary OA, as described previously8, 9. The radiological stage of the disease was a Kellgren and Lawrence grade of 2 or more. Cases of inflammatory arthritis, post-traumatic or post-septic arthritis or suggestive of skeletal or developmental dysplasias were excluded. The tissues collected were articular cartilage, infrapatellar fat pad, meniscus, and synovium. The Newcastle and North Tyneside research ethics committee granted ethical approval for the tissue collection (REC reference number 09/H0906/72) and informed consent was obtained from each donor. Additional patient details are listed in Supplementary Tables S1 and S2.
Nucleic acid extraction
On the day of surgery, tissue was snap-frozen at −80°C. Between 0.5 and 1.0 g of frozen tissue was ground to a powder using a Retsch mixermill 200 (Retsch Limited, Leeds, UK) under liquid nitrogen. DNA and RNA were extracted using an Omega EZNA DNA/RNA Isolation Kit (Omega Bio-Tek, Norcross, USA) and quantified using a NanoDrop ND-1000 Spectrophotometer (NanoDrop Technologies, Wilmington, USA).
Gene expression
Initially 2 μg of RNA was reverse transcribed using the SuperScript First-Strand cDNA synthesis kit (Invitrogen, Paisley, UK). SMAD3 expression was assessed by quantitative PCR and measured relative to the average expression of the housekeeping genes HPRT1, GAPDH and 18S. For each cDNA sample three pipetting replicates were performed for SMAD3 and the housekeeping genes. We combined the housekeeper cycle threshold (Ct) values and derived the mean Ct using this to compare against SMAD3 expression. Reactions were carried out using an ABI PRISM 7900HT Sequence Detection System (Applied Biosystems, Paisley, UK), and PrimeTime Mini qPCR Assays (Integrated DNA Technologies, Iowa, USA). Primer and probe sequences are listed in Supplementary Table S3(A). Relative expression levels compared to the housekeepers were calculated using the 2−ΔCt method.
Transcript SNP selection and SNP genotyping
Using public databases the transcript sequence of SMAD3 was searched for common SNPs (minor allele frequencies >10%). The 3′UTR SNP rs8031440 was chosen for analysis. This SNP has a minor allele frequency of 0.26 and pairwise r2 and D′ values of 0.01 and 0.21, respectively, relative to rs12901499. SNP genotyping was carried out by restriction fragment length polymorphism (RFLP) analysis. The primer sequences and the restriction enzymes are listed in Supplementary Table S3(B).
Allelic expression imbalance analysis (AEI)
cDNA from patients heterozygous at the transcript SNP rs8031440 were analysed for AEI using a readymade TaqMan genotyping assay (Applied Biosystems) for use with the ABI PRISM 7900HT Sequence Detection System (Applied Biosystems). The assay employs a probe labelled with FAM or VIC specific to each of the alleles of an SNP. Real-time PCR was carried out according to the manufacturer's instructions. The reactions were performed in five replicates and the allele ratios were calculated using the formula (2−FAM Ct/2−VIC Ct). The allelic ratios were calculated for genomic DNA and cDNA. The average allelic ratio of the genomic DNA (representing the assumed 1:1 ratio between alleles) was then used to normalise the cDNA allelic ratios and thus account for differences in fluorescent yield and incorporation specific to an assay. The capacity for the assay to discriminate between alleles was verified using a standard curve performed on cDNA and DNA of varying allelic ratios (data not shown).
Association with OA
We assessed SNP rs8031440 for evidence of association to OA using data from the arcOGEN study, a large GWAS conducted in the UK on population controls and cases with severe OA of the hip or knee, 80% of whom had undergone total joint replacement10. The array used was the Illumina 610 Quad array. rs8031440 is not on this array so SNP rs7166081, an SNP located downstream of SMAD3 and in perfect LD (r2 = 1, D′ = 1) with rs8031440, was used as a direct proxy. Since our expression studies were all performed on OA cases who had undergone total joint replacement we assessed association to rs8031440 using the 5804 arcOGEN OA cases who had also undergone this procedure.
Construction of luciferase reporter plasmids
To generate the constructs for use in the luciferase reporter gene assay, primers flanked by restriction enzyme sites were designed to amplify three DNA fragments encompassing the six 3′UTR SNPs; amplicon A containing rs8025774; amplicon B containing rs8031440, rs8031627, and rs2278670; and amplicon C containing rs12595334 and rs3743342. Primer sequences and restriction enzymes are listed in Supplementary Table S4(A). PCR products were purified using the QIAquick PCR purification kit (Qiagen, Hilden, Germany) and dephosphorylated with Antarctic phosphatase (GE Healthcare, Buckinghamshire, UK). The PCR products were cloned into the pMIR-report vector (Promega, Southampton, UK). Positive clones were sequenced to ensure the correct sequence of the construct.
Site-directed mutagenesis of luciferase reporter plasmids
To generate the necessary haplotypes primers were designed to force a base change at the relevant SNP (primer sequences are listed in Supplementary Table S4(B)). Mutagenesis was performed using the QuikChange II Site-Directed Mutagenesis Kit (Agilent Technologies, Berkshire, UK) following the manufacturer's instructions.
Cell cultures and luciferase assay
The SW1353 human chondrosarcoma cell line (ATCC, Middlesex, UK) and the MG63 human osteosarcoma cell line (ATCC) were used for transfection. Forty-eight hours before transfection cells were seeded at a density of approximately 17,500 cells per well in a 48-well plate. Cells were transfected with 500 ng DNA from the vector constructs and 15 ng Renilla luciferase control reporter vector pRL-TK (Promega), using ExGen 500 transfection reagent (Fermentas, Leicestershire, UK). After 24 h the cells were lysed and luciferase activity was determined using the Dual-Luciferase Reporter Assay System (Promega). Luminescence was measured using a MicroLumat Plus LB96V Microplate Luminometer (Berthold Technologies, Bad Wildbad, Germany). The luciferase activity was normalised to the Renilla activity. Six technical and three biological repeats were performed per construct.
Cell culture of aortic vascular smooth muscle cells (VSMCs)
VSMCs from aortic tissue were cultured in complete smooth muscle cell basal medium (SmBM; Lonza, Basel, Switzerland) supplemented with 5% FBS (Lonza), 0.1% human recombinant epidermal growth factor (rhEGF; Lonza), 0.1% human recombinant insulin, 0.2% human recombinant fibroblast growth factor B (hrFGF-B; Lonza), and 500 μl GA-100 (gentamicin sulfate/amphotericin-B; Lonza). Cells were maintained in 10 cm tissue culture dishes coated in 0.1% gelatin, in a humidified 5% CO2 incubator. Cells were passaged at 80% confluence. Nucleic acids were extracted from the cells as described above.
Molecular haplotyping
Using cDNA as a template we amplified a region of SMAD3 encompassing the Arg287Trp mutation and rs8025774, which is in perfect LD with rs8031440, in patients A and B. The primer sequences are listed in Supplementary Table S3(C). PCR was performed in a 50 μl reaction and the product was cloned into the pCR-4 TOPO vector using the TOPO TA cloning kit (Invitrogen). Colonies were picked, grown and the extracted plasmid DNA was directly sequenced to assess the molecular haplotype between Arg287Trp and rs8025774/rs8031440.
Bioinformatic analysis of microRNAs binding sites
PolymiRTS (http://compbio.uthsc.edu/miRSNP), TargetScan (http://www.targetscan.org/), MiRBase (http://www.mirbase.org/), PicTAR (http://pictar.mdc-berlin.de/) and miRWalk (http://www.umm.uni-heidelberg.de/apps/zmf/mirwalk/) were used to identify sequence variations in putative miRNA binding sites and to predict binding sites for miRNAs encompassing rs12595334 and rs3743342.
Statistical analysis
Differences in quantitative gene expression, and differences in luciferase activity between allelic and haplotypic forms, were assessed using a Mann–Whitney U Test and the GraphPad Prism software package (LaJolla, California, USA).
Results
SMAD3 expression in hip OA vs knee OA cartilage
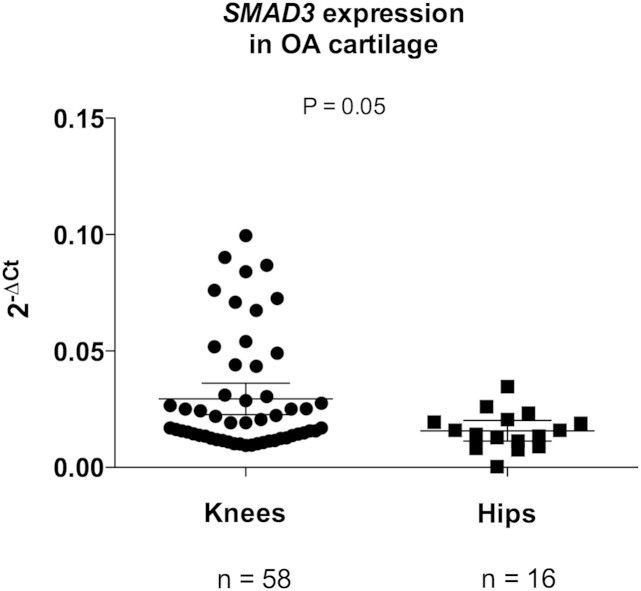
We initially demonstrated expression of SMAD3 in eight joint tissues collected from OA patients (Supplementary Figure S1). We then compared expression of SMAD3 between hip cartilage from 16 OA patients and knee cartilage from 58 OA patients [Fig. 1]. This revealed a significantly lower level of SMAD3 expression in the hip (P = 0.05).
Fig. 1.
Columnar scatter plots of the quantitative expression of SMAD3 in knee OA vs hip OA cartilage. n is the number of patients studied. The horizontal lines in each plot represent the mean and the 95% confidence interval. P-value was calculated using a Mann–Whitney U Test.
Quantitative expression of SMAD3 in cartilage stratified by genotype at rs12901499
To determine whether SMAD3 harboured cartilage eQTLs that correlated with the OA association signal we quantified levels of SMAD3 expression in our OA cartilage samples and then stratified the data by genotype at the OA associated SNP rs12902499 (Supplementary Figure S2(A)). There was no correlation between genotype and expression (P = 0.85). We repeated the analysis after separating the knee (Supplementary Figure S2(B)) and hip (Supplementary Figure S2(C)) cases. Again there was no correlation in the knee cartilage (P = 0.36) or the hip cartilage (P = 0.31).
AEI analysis of SMAD3 in cartilage
We further investigated the possible presence of SMAD3 eQTLs by AEI. Since rs12901499 is not a transcript SNP it could not be used in the AEI analysis. We therefore studied patients who were heterozygous for rs12901499 and for the 3′UTR SNP rs8031440, enabling us to indirectly test any allelic output that correlated with rs12901499, since if this were occurring we would expect to observe AEI in a high proportion of these compound heterozygotes. rs8031440 was chosen because it has a reasonably high minor allele frequency (0.26) ensuring that a large proportion of our patients will be heterozygous and therefore suitable for the AEI analysis. We also studied individuals who were homozygous for rs12901499 but heterozygous for rs8031440, thus enabling us to assess whether SMAD3 could also be subject to cis-eQTLs acting independently of the association signal marked by rs12901499.
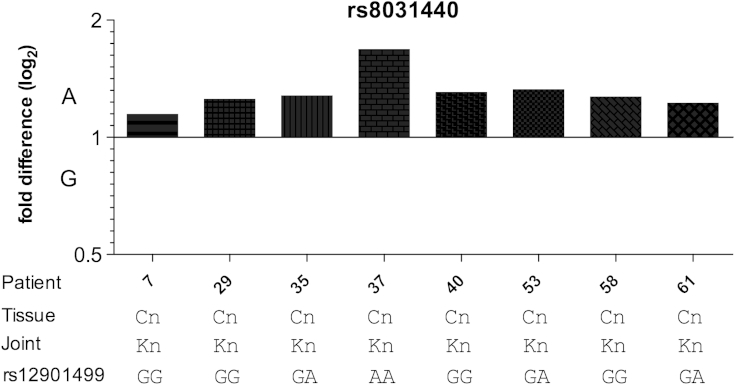
Cartilage from eight knee OA patients were analysed and each demonstrated AEI, with the G allele of rs8031440 showing reduced expression in all patients [Fig. 2 and Supplementary Table S5]. The average fold difference was 1.31, namely the A-allele of this SNP produced 31% more SMAD3 expression than the G allele. AEI was also observed in fat pad, synovium and meniscus samples from an additional six knee OA patients; the observed AEI is therefore not cartilage-specific (Supplementary Figure S3 and Supplementary Table S5). Seven of the 14 patients were heterozygous for rs12901499 whilst seven were not. The most likely explanation for our AEI result is that the 3′UTR SNP rs8031440 is functional and mediating AEI or that it is in very high LD with the functional SNP. It is unlikely that the OA associated SNP rs12901499 is responsible (either directly or indirectly) for the AEI observed.
Fig. 2.
Allelic expression analysis of the 3'UTR transcript SNP rs8031440 using cDNA from cartilage. The genotypes of the patients for the associated SNP rs12901499 are shown. The cDNA allelic ratio in each patient was compared to the 1:1 allelic ratio calculated from the DNA of that patient.
Quantitative expression of SMAD3 in cartilage stratified by genotype at rs8031440
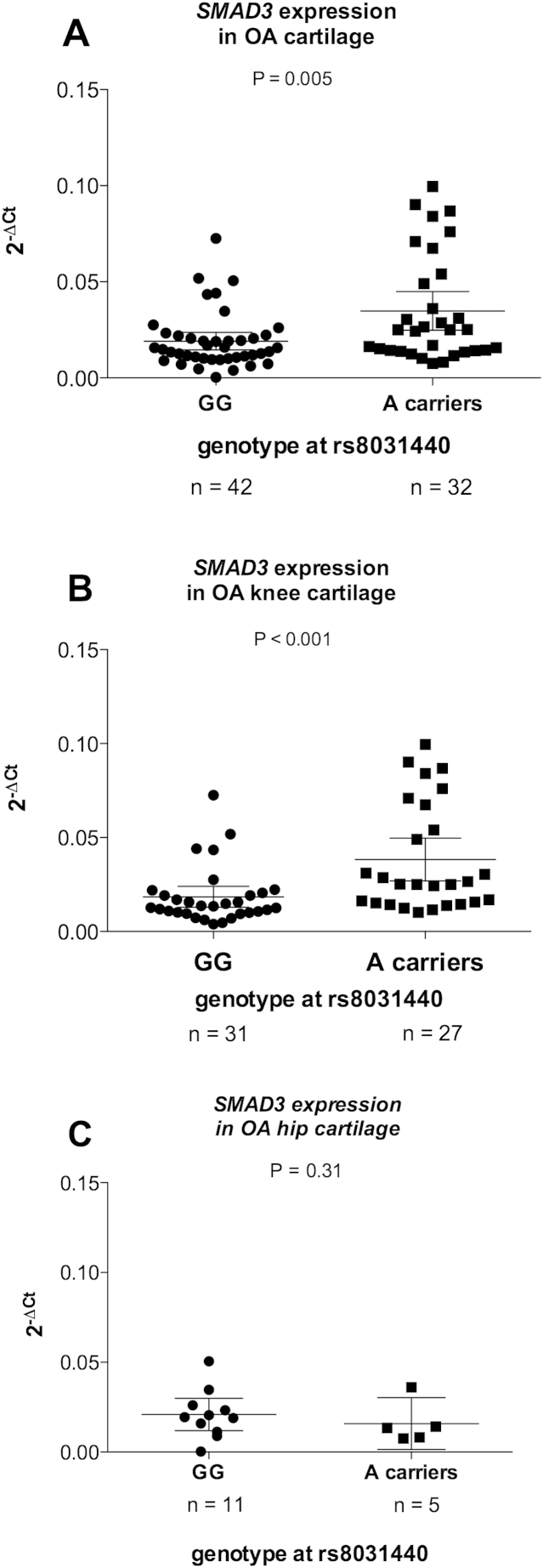
Having established that rs8031440 is highlighting a SMAD3 eQTL, we stratified our SMAD3 quantitative expression data by genotype at this SNP. We observed a significant correlation between SMAD3 expression in cartilage and rs8031440 genotype [Fig. 3(A)], with the G allele of the SNP correlating with reduced SMAD3 expression (P = 0.005). This G allele also correlated with reduced expression in our AEI analysis [Fig. 2]. Since this is the second time in which we have stratified this expression data by genotype at an SNP we applied a Bonferroni correction by multiplying the P by 2, giving a still significant P-value of 0.01. We stratified the OA patients into knee [Fig. 3(B)] and hip [Fig. 3(C)] cases and observed a significant correlation in the knee cases (P < 0.001). The number of hip cases was only 16, which may have limited our power to see an effect in this stratum.
Fig. 3.
Columnar scatter plots of the quantitative expression data of SMAD3. (A) shows OA cartilage, (B) shows OA knee cartilage, and (C) shows OA hip cartilage, with patients stratified by genotype at the 3′UTR transcript SNP rs8031440. Due to the low frequency of AA homozygotes at rs8031440 the analysis was performed between GG homozygotes and A-allele carriers (GA and AA combined). n is the number of patients studied. The horizontal lines in each plot represent the mean and the 95% confidence interval. P-values were calculated using a Mann–Whitney U Test.
rs3743342 and rs12595334 are functional
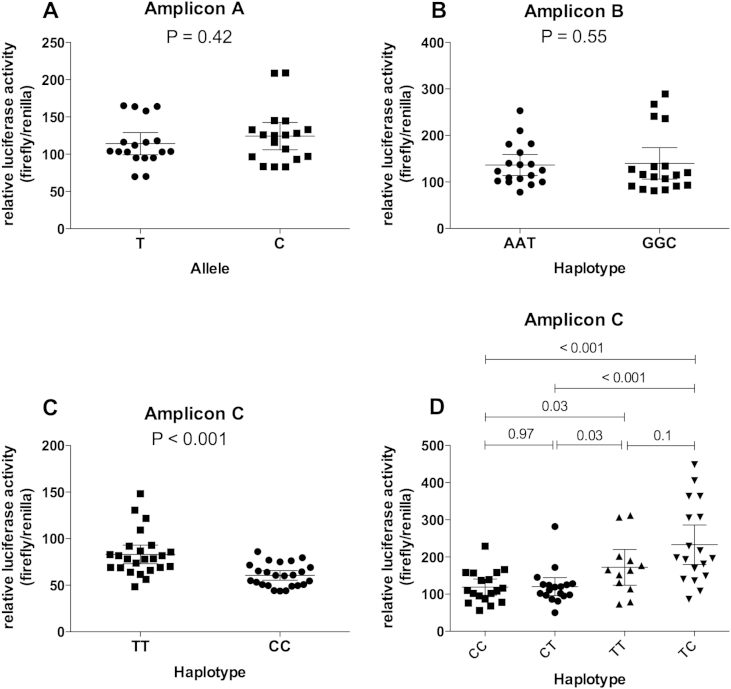
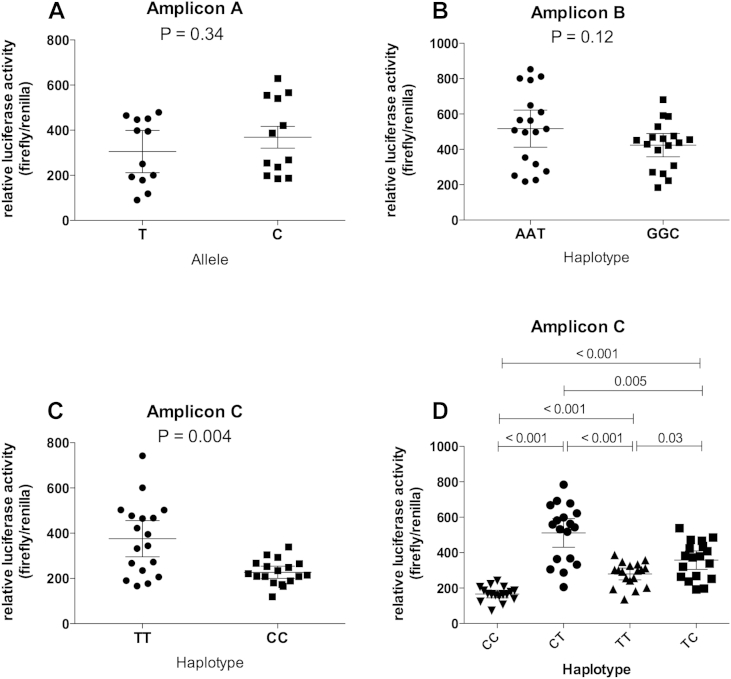
rs8031440 is in perfect LD (r2 = 1, D′ = 1) with five other SMAD3 3′UTR SNPs: rs8025774, rs8031627, rs2278670, rs12595334 and rs3743342. To assess whether rs8031440 or any of these five other SNPs are functional we cloned fragments of the 3′UTR into a luciferase plasmid and performed reporter gene expression assays using the SW1353 and the MG63 cell lines. We cloned three fragments: amplicon A, containing rs8025774; amplicon B, containing rs8031440, rs8031627 and rs2278670; and amplicon C, containing rs12595334 and rs3743342. For each amplicon we cloned both allelic forms. We did not observe any significant differences in luciferase gene expression between the alleles of rs8025774 or between the haplotypes of rs8031440-rs8031627-rs2278670 in either SW1353 [Fig. 4(A) and (B)] or MG63 [Fig. 5(A) and (B)] cells. We did however observe a significant difference between the haplotypes of rs12595334-rs3743342 in both cell types [P < 0.001 for SW1353, Fig. 4(C); P = 0.004 for MG63, Fig. 5(C)] indicating that one, or both, of these SNPs is influencing gene expression. Using site-directed mutagenesis we mutated each allele of these two SNPs to create the alternative rs12595334-rs3743342 haplotypes CT and TC and compared luciferase activity between these and the naturally occurring TT and CC haplotypes. Since the CC haplotype of rs12595334-rs3743342 is in LD with the low-expressing G allele of rs8031440, we focused our interpretation on changes to this haplotype. In SW1353 cells, mutating rs3743342 from a C to a T had no effect [P = 0.97, Fig. 4(D)] whereas mutating rs12595334 from a C to a T significantly increased luciferase activity [P < 0.001, Fig. 4(D)]. In MG63 cells, mutating rs3743342 from a C to a T did have an affect (unlike in SW1353), increasing activity [P < 0.001, Fig. 5(D)]. Mutating rs12595334 from a C to a T also significantly increased luciferase activity in the MG63 cells [P < 0.001, Fig. 5(D)]. Alternative effects were observed following changes to the TT haplotype. However, since the CC haplotype is in LD with the G allele of rs8031440, this luciferase data supports the patient tissue expression data. Both rs12595334 and rs3743342 are functional, with rs3743342 appearing to have a more cell-restricted effect.
Fig. 4.
Functional analysis of the six SMAD3 3'UTR SNPs in SW1353 cells. (A–C) show the results of the luciferase reporter assays for amplicons A, B and C, respectively. (D) shows the results of the luciferase reporter assay for the four possible haplotypes (CC, CT, TT, and TC) for rs12595334 and rs3743342. Data are the fold expression relative to Renilla activity and are shown as the mean and 95% confidence intervals from three biological repeats each with six technical replicates. P-values were calculated using a Mann–Whitney U Test.
Fig. 5.
Functional analysis of the six SMAD3 3'UTR SNPs in MG63 cells. (A–C) show the results of the luciferase reporter assays for amplicons A, B, and C, respectively. (D) shows the results of the luciferase reporter assay for the four possible haplotypes (CC, CT, TT, and TC) for rs12595334 and rs3743342. Data are the fold expression relative to Renilla activity and are shown as the mean and 95% confidence intervals from three biological repeats each with six technical replicates. P-values were calculated using a Mann–Whitney U Test.
3′UTRs can be the target site for gene expression regulation by microRNAs11. We therefore performed a database search of potential microRNA binding sites encompassing rs12595334 and rs3743342 but none were found for either allelic form of the SNPs.
Genetic association with OA
We assessed whether the eQTL marked by rs8031440, and potentially mediated by rs12595334 and rs3743342, was associated with OA. To do this we used the arcOGEN GWAS dataset, comprising 5804 OA cases who had undergone joint replacement and 11,009 population controls10. Neither rs8031440, rs12595334 or rs3743342 are on the Illumina 610 Quad array used by arcOGEN, nor are any of the other SMAD3 3′UTRs SNPs that are in perfect LD with them. The SNP rs7166081, which is located downstream of SMAD3, is however on the array and is also in perfect LD with rs8031440, rs12595334 and rs3743342. We therefore used the association data generated for rs7166081 as a direct proxy. This revealed that the SMAD3 eQTL showed marginal evidence of association with OA (P = 0.027; Table I). Stratification of the arcOGEN dataset by sex, by site of OA (hip or knee), and by sex combined with site did not enhance the association signal, with the most significant P-value being 0.001 (odds ratio of 0.915) for male knee cases. In all instances the C allele of rs7166081, which is equivalent to the G allele of rs8031440 correlating with decreased SMAD3 expression, was less common in OA cases.
Table I.
Association analysis of rs7166081 with OA using the arcOGEN data
| Stratum | Cases n | Controls n | C-allele frequency cases | C-allele frequency controls | P-value | Odds ratio* |
|---|---|---|---|---|---|---|
| All cases | 5804 | 11,009 | 0.220 | 0.235 | 0.027 | 0.921 (0.857–0.991) |
| Hip cases | 3039 | 11,009 | 0.214 | 0.233 | 0.002 | 0.896 (0.837–0.960) |
| Knee cases | 2164 | 11,009 | 0.221 | 0.233 | 0.077 | 0.932 (0.862–1.008) |
| Female cases | 3363 | 5515 | 0.217 | 0.235 | 0.030 | 0.904 (0.826–0.991) |
| Male cases | 2441 | 5494 | 0.211 | 0.232 | 0.020 | 0.883 (0.794–0.980) |
| Female hip cases | 1769 | 5515 | 0.222 | 0.235 | 0.183 | 0.931 (0.838–1.034) |
| Female knee cases | 1217 | 5515 | 0.215 | 0.232 | 0.014 | 0.903 (0.832–0.980) |
| Male hip cases | 1270 | 5494 | 0.220 | 0.232 | 0.232 | 0.931 (0.828–1.047) |
| Male knee cases | 947 | 5494 | 0.218 | 0.233 | 0.001 | 0.915 (0.867–0.965) |
(95% confidence intervals).
As noted in the introduction, one of the articles that stimulated our interest in SMAD3 reported an association of rs12901499 with hip and knee OA6, with P < 4.5 × 10−6. In the arcOGEN joint replacement cases this SNP had a P-value of 0.003.
AEI analysis of SMAD3 in AOS
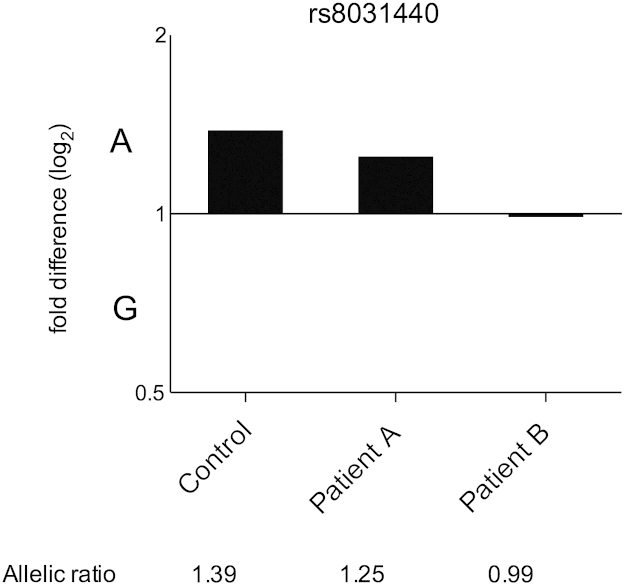
Finally we investigated the activity of the SMAD3 eQTL using vascular smooth muscle cells (VSMCs) derived from aortic tissue of AOS patients. This disease is caused by rare mutations in SMAD3 and we were keen at this stage to assess whether the eQTL was operating in a non-joint tissue of relevance to AOS. If so, it could potentially contribute to the interfamilial variability seen in this disease4. We studied two patients with AOS, caused by a missense Arg287Trp mutation in SMAD3, and a normal control individual. All three were heterozygous for rs8031440. The control (a male born in 1955) and patient A (a male born in 1979) both demonstrated AEI [Fig. 6], whereas patient B (a male born in 1947) did not. The AEI observed in the control and patient A was in the same direction as that observed in joint tissues, namely decreased expression of the G allele of rs8031440. The allelic ratios of 1.39 (control) and 1.25 (patient A) were also comparable to those seen in the primary OA patients (Supplementary Table S5). We conclude therefore that the SMAD3 eQTL is also active in a tissue that is of direct relevance to the aneurysm component of AOS.
Fig. 6.
Allelic expression analysis of the 3'UTR SNP rs8031440 using cDNA from vascular smooth muscle cells. The cDNA allelic ratio in each individual was compared to the 1:1 allelic ratio calculated from the DNA of that individual.
Using cDNA as a template we assessed the molecular haplotypes for the Arg287Trp mutation and rs8031440 in patients A and B. This revealed that in both patients the low-expressing G allele of rs8031440 was on the wild type allele. Patient B is the first cousin once removed of patient A and both have the heterozygous Arg287Trp mutation. The two patients have therefore inherited the G allele of rs8031440 from a recent common ancestor. The fact that this G allele shows reduced expression in patient B but not in patient A implies that its expression is likely to be modified by other cis-acting effects, which may be genetic or epigenetic.
A search of the PhenGenI (http://www.ncbi.nlm.nih.gov/gap/phegeni/) and RegulomeDB12 databases found no evidence that rs8031440 acts as an eQTL in other tissues, implying that the eQTL does not have a ubiquitous effect on SMAD3 expression.
Discussion
Using joint tissues from patients with primary OA we set out to assess whether the alleles of the OA associated SMAD3 SNP rs12901499 correlated with differences in expression of the gene, based on our hypothesis that this may be the mechanism through which the rs12901499 OA association is operating. We failed to find any evidence of such a correlation, implying that our hypothesis is incorrect. It is possible however that the association marked by rs12901499 is operating during development and that an investigation of the joint tissues of young individuals may reveal a correlation between rs12901499 and SMAD3 expression. The collection of a sufficiently large number of such tissues is however prohibitive, since young individuals do not present for joint replacement surgery.
During our investigations we did discover a SMAD3 eQTL that was operating in joint tissues, the first reported case of this as far as we are aware, and we were able to pinpoint the 3′UTR SNPs rs12595334 and rs3743342 as potential mediators of this effect. Using the arcOGEN data we were also able to show that this eQTL demonstrated nominal association to OA.
The allele that correlated with reduced expression of SMAD3, the G allele of rs8031440 (indirectly tested using the perfect proxy SNP rs7166081), was less common in the arcOGEN cases vs the arcOGEN controls. This genetic association data therefore implies that increased SMAD3 expression is an OA risk factor. However, since the P-value is modest it may be sensible to interpret it with caution at this time.
As noted in the introduction, AOS arises from rare heterozygous SMAD3 mutations that appear to be loss-of-function4, 5. Several involve frame-shift mutations that result in nonsense-mediated mRNA decay leading to little or no detectable transcript from the mutant allele, whilst another leads to the substitution of arginine for tryptophan at position 287 of the gene. This Arg287Trp mutation creates a protein that is unable to form SMAD3 protein homomers or heteromers with SMAD44. This will attenuate TGF-β signalling. We investigated the activity of the SMAD3 eQTL in VSMCs derived from the aortic tissue of two AOS patients, both of whom have the Arg287Trp mutation. The eQTL was operating in one of these patients, revealing that it is of direct relevance to this syndrome.
AOS has a high degree of interfamilial variability4, implying that it is likely to be influenced by the genetic background on which the wild type SMAD3 allele is operating; if the wild type allele happens to be on a low-expressing rather than a high-expressing copy of the gene then this may lead to an even greater reduction in functional SMAD3 and therefore a more severe phenotype. Comparing the inheritance of the eQTL between pedigrees in which AOS is segregating may therefore be informative in accounting for some of the phenotypic variability observed.
Since we have demonstrated that the SMAD3 eQTL is operating in aortic tissue, it may also be of value to compare, at the population level, the incidence of aneurysms between healthy individuals homozygous for high-expressing and low-expressing alleles of the eQTL; homozygotes for the low-expressing allele may be at greater risk of aortic abnormalities.
Overall, our molecular study of the expression of SMAD3 has highlighted allelic expression effects of relevance to both common and rare forms of OA. It also supports an investigation of the influence of these alleles in other conditions.
Author contributions
All authors were involved in drafting the article or revising it critically for important intellectual content, and all authors approved the final version to be published. Prof. Loughlin had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. This data can be accessed by emailing Prof. Loughlin.
Study conception and design. Raine, Reynard, Loughlin.
Acquisition of data. Raine.
Analysis and interpretation of data. Raine, Reynard, van de Laar, Bertoli-Avella, Loughlin.
Funding
Supported by Arthritis Research UK, by The European Union Seventh Framework Program (FP7/2007-2013) under grant agreement number 305815 (D-BOARD) and by the UK NIHR Biomedical Research Centre for Ageing and Age-related disease award to the Newcastle upon Tyne Hospitals NHS Foundation Trust.
Conflict of interest
There are no competing interests.
Acknowledgements
We thank the orthopaedic surgeons and research nurses of the Royal Victoria Hospital and the Freeman Hospital, Newcastle upon Tyne, for their assistance in the collection of the patient tissue samples used in this study. We also thank Catherine Syddall and Madhushika Ratnayake for technical advice. Our study used data that was generated by arcOGEN, funded by Arthritis Research UK (http://www.arthritisresearchuk.org/), grant number 18030.
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/8.1016/j.joca.2014.02.931.
Contributor Information
E.V.A. Raine, Email: e.v.a.raine@ncl.ac.uk.
J. Loughlin, Email: john.loughlin@ncl.ac.uk.
Appendix A. Supplementary data
The following are the supplementary data related to this article
References
- 1.van der Kraan P.M., Blaney Davidson E.N., van den Berg W.B. A role for age-related changes in TGFβ signaling in aberrant chondrocyte differentiation and osteoarthritis. Arthritis Res Ther. 2010;12:201. doi: 10.1186/ar2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Loughlin J. Genetic indicators and susceptibility to osteoarthritis. Br J Sports Med. 2011;45:278–282. doi: 10.1136/bjsm.2010.081059. [DOI] [PubMed] [Google Scholar]
- 3.Li T.F., Gao L., Sheu T.J., Sampson E.R., Flick L.M., Konttinen Y.T. Aberrant hypertrophy in Smad3-deficient murine chondrocytes is rescued by restoring transforming growth factor β-activated kinase 1/activating transcription factor 2 signaling: a potential clinical implication for osteoarthritis. Arthritis Rheum. 2010;62:2359–2369. doi: 10.1002/art.27537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van de Laar I.M., Oldenburg R.A., Pals G., Roos-Hesselink J.W., de Graaf B.M., Verhagen J.M. Mutations in SMAD3 cause a syndromic form of aortic aneurysms and dissections with early-onset osteoarthritis. Nat Genet. 2011;43:121–126. doi: 10.1038/ng.744. [DOI] [PubMed] [Google Scholar]
- 5.van de Laar I.M., van der Linde D., Oei E.H., Bos P.K., Bessems J.H., Bierma-Zeinstra S.M. Phenotypic spectrum of the SMAD3-related aneurysms-osteoarthritis syndrome. J Med Genet. 2012;49:47–57. doi: 10.1136/jmedgenet-2011-100382. [DOI] [PubMed] [Google Scholar]
- 6.Valdes A.M., Spector T.D., Tamm A., Kisand K., Doherty S.A., Dennison E.M. Genetic variation in the SMAD3 gene is associated with hip and knee osteoarthritis. Arthritis Rheum. 2010;62:2347–2352. doi: 10.1002/art.27530. [DOI] [PubMed] [Google Scholar]
- 7.Montgomery S.B., Dermitzakis E.T. From expression QTLs to personalized transcriptomics. Nat Rev Genet. 2011;12:277–282. doi: 10.1038/nrg2969. [DOI] [PubMed] [Google Scholar]
- 8.Wilkins J.M., Southam L., Price A.J., Mustafa Z., Carr A., Loughlin J. Extreme context specificity in differential allelic expression. Hum Mol Genet. 2007;16:537–546. doi: 10.1093/hmg/ddl488. [DOI] [PubMed] [Google Scholar]
- 9.Southam L., Rodriguez-Lopez J., Wilkins J.M., Pombo-Suarez M., Snelling S., Gomez-Reino J.J. An SNP in the 5'UTR of GDF5 is associated with osteoarthritis susceptibility in Europeans and with in vivo differences in allelic expression in articular cartilage. Hum Mol Genet. 2007;16:2226–2232. doi: 10.1093/hmg/ddm174. [DOI] [PubMed] [Google Scholar]
- 10.arcOGEN Consortium and arcOGEN Collaborators Identification of new susceptibility loci for osteoarthritis (arcOGEN): a genome-wide association study. Lancet. 2012;380:815–823. doi: 10.1016/S0140-6736(12)60681-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 12.Boyle A.P., Hong E.L., Hariharan M., Cheng Y., Schaub M.A., Kasowski M. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22:1790–1797. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.