Abstract
In mammals, the KIBRA locus has been associated with memory performance and cognition by genome-wide single nucleotide polymorphism screening. Genetic studies in Drosophila and human cells have identified KIBRA as a novel regulator of the Hippo signaling pathway, which plays a critical role in human tumorigenesis. Recent studies also indicated that KIBRA is involved in other physiological processes including cell polarity, membrane/vesicular trafficking, mitosis and cell migration. At the biochemical level, KIBRA protein is highly phosphorylated by various kinases in epithelial cells. Here, we discuss the updates concerning the function and regulation of KIBRA in the brain and beyond.
Keywords: KIBRA, WWC family, memory performance, phosphorylation, Hippo pathway, cell polarity, cell migration
1. KIBRA gene and WWC family
The gene KIBRA was first described in 2003 and the name was given for its predominant mRNA expression in kidney and brain (1). The human KIBRA gene localizes on the positive strand of chromosome 5q34. The coding region encompasses about 180,000 base pairs and has 23 exons. The full-length transcripts express proteins with 1119 (isoform 1) or 1118 (isoform 2) or 1113 (isoform 3) amino acids due to alternative in-frame splice sites in the 3′ coding region. Human KIBRA contains two WW domains (a short protein module of approximately 40 amino acids that has two highly conserved tryptophans) at the N-terminus (amino acids 7-39 and 54-86). WW domains are responsible for recognizing proteins with proline rich motifs such as PPxY (x represents any amino acid). The C2 domain (amino acids 655-783) contains two four-stranded β-sheets that are responsible for a Ca2+-sensitive interaction with phospholipids (2). Additionally, KIBRA also contains several coiled-coil structures, a glutamic acid-rich domain, a class III PDZ (PSD95/Dlg/ZO-1) binding motif and an atypical protein kinase C (aPKC) binding region (Figures 1 and 2).
Fig. 1.
Sequence alignment (Clustal 2.1) and domain features of human WWC family proteins. The NCBI accession numbers for each protein are: NP_001155133 (KIBRA/WWC1), NP_079225 (WWC2) and AGV22437 (WWC3). Color legends: yellow for WW domains; green for potential coiled-coil domains; red for C2 domain; pink for glutamic-rich region; blue for PDZ-binding motif.
Fig. 2.
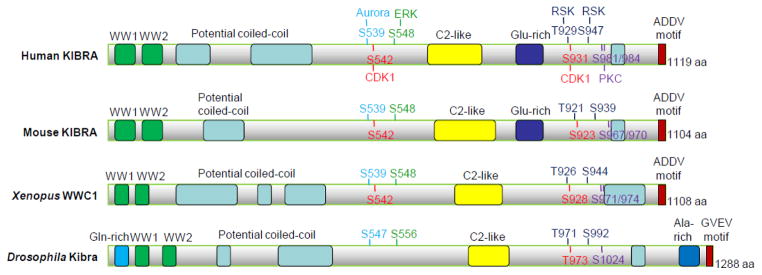
KIBRA/WWC1 orthologs and phosphorylation sites. Various domains are marked with different colors. The known phosphorylation sites and their corresponding kinases (with matched colors) are also indicated.
KIBRA (also known as WWC1) belongs to the WWC (WW and C2 domain containing) family, which comprises two additional highly similar paralogs, WWC2 and WWC3, in addition to KIBRA/WWC1 (Figure 1) (3). WWC2 and WWC3 share high structural similarity with KIBRA/WWC1 except that the glutamic acid-rich domain is specific for KIBRA/WWC1. Besides brain and kidney, WWC2 and WWC3 are preferentially expressed in thyroid, immune cells, reproductive tissues, liver and lung. The functions of WWC2 and WWC3 are not well studied yet.
The WWC family is evolutionarily conserved. KIBRA has been identified in many species ranging from insects to all vertebrates, but does not exist in yeast and worm (Figure 2). However, not all species express all three WWC family proteins. For example, lower organisms including Drosophila only have KIBRA. While fishes encode only two WWC genes, most other vertebrates including frog, rat and human have all three WWC members (3). Notably, due to a chromosomal translocation event in the evolution of the mouse lineage, Mus musculus expresses only KIBRA/WWC1 and WWC2 but lacks WWC3 (4). Whether there is functional interplay among the WWC proteins is almost completely unknown. However, a recent study showed that WWC2 expression is upregulated in the developing brain of the KIBRA knockout mice, indicating a possible compensatory function of these WWC family members (5).
So far, five transcription starting sites (TSS) have been identified in the region of the KIBRA gene (6). The TSS1b and TSS1c are located 153 and 415 bp upstream of the earlier annotated TSS1a, while the TSS2 and TSS3 are located in the first intron of KIBRA. The TSS1b and TSS1c are constitutively used in kidney and brain cells, resulting in transcripts for full-length KIBRA. The TSS2 and TSS3 are exclusively used in kidney cells, initiating transcripts for KIBRA isoforms without WW domains. Accordingly, their promoters, which are also located in the first intronic region, are specifically activated in kidney cells. The transcription factor 7-like 2 (TCF7L2) is believed to regulate KIBRA promoters, and binding sites for TCF7L2 have been identified near the promoters (6).
2. Expression patterns of KIBRA
KIBRA mRNA is highly enriched in human kidney, brain and testes (1). Gene expression studies and immunohistological staining have shown that KIBRA is expressed in memory-related regions of the brain, such as hippocampus and cortex, as well as in the cerebellum and the hypothalamus (7, 8). In the kidney, KIBRA is expressed in glomerular podocytes, tubules and the collecting ducts (9). In human normal breast tissue, KIBRA mRNA can be found at all stages of gland development and KIBRA protein has been detected in the luminal epithelium surrounding the ducts (10). In normal gastric tissue, KIBRA is expressed at the apical and cell-cell junction regions, but in gastric cancer tissue, increased expression of KIBRA can be detected not only in apical and junctional regions but also in the cytoplasm (11).
At the subcellular level, KIBRA was mainly cytoplasmic in green monkey kidney (CV1) cells (1). In hippocampal neurons, KIBRA shows a somatodendritic distribution with a perinuclear enrichment, and KIBRA is also a component of the postsynaptic density in rat brain (8). In breast cancer cells, although KIBRA is mainly present in the cytoplasmic fraction, it can also be detected in the nuclear fraction (12). KIBRA can also be detected in the heart and colon. In cultured cells, KIBRA protein is readily detected in epithelial cells of mammary, pancreatic and prostate origin (L. Z. and J.D., unpublished observations).
3. Binding partners of KIBRA
As described above, KIBRA protein has multiple binding motifs for interacting with other proteins to exert its functions. About twenty interacting partners of KIBRA have been identified in recent years. KIBRA was first identified by yeast two hybrid screens as a binding partner of human dendrin (1), which contains two PPxY motifs that bind to the WW domains of KIBRA. Another interacting partner, synaptopodin, also contains PPxY and sequence homology with dendrin in the PPxY surrounding area, thus also binding to the WW domains (9). Dendrin and synaptopodin are both actin-cytoskeleton proteins, so their binding to KIBRA is considered to be involved in cell polarity and motility, by KIBRA linking them to the polarity proteins (9). An example of the polarity proteins is the PALS1-associated tight junction protein (PATJ), which binds to the ADDV motif at the extreme C-terminus of KIBRA. Through its WW domains, KIBRA was also reported to bind to another PPXY motif-containing protein, discoidin domain receptor family member 1 (DDR1), an epithelial-specific collagen-activated receptor tyrosine kinase, in mammary epithelium (10, 13). The association between KIBRA and DDR1 was attenuated by collagen-induced DDR1 phosphorylation (10).
KIBRA specifically binds to the catalytic domain of PKC isoform zeta, but not to other PKC family members (15). The amino acids 953-996 of KIBRA are responsible for the PKCζ binding. PKMζ contains a catalytic domain similar to that of PKCζ but without the regulatory domain (16), so it is likely that it binds to KIBRA as well (9, 17). The constitutively active kinase PKMζ is exclusively expressed in the brain and is necessary for maintenance of long-term potentiating (18). Therefore, KIBRA may regulate memory through interacting with PKCζ/PKMζ. Additionally, the sequences between the WW domains and the C2 domain (amino acids 129-525) of KIBRA was shown to bind to Sec3, a component of Exocyst, thus mediating the aPKC-Exocyst interaction during migration of normal rat kidney (NRK) cells (19).
In a study with mass spectroscopic analysis of the dynein light chain 1 (DLC1)-associated proteins from breast cancer cells, KIBRA was found to interact with DLC1, a cytoskeleton signaling component, although the interacting site on KIBRA was not determined (12). In the same study, KIBRA was also demonstrated to associate with histone H3 via its glutamic acid-rich region, indicating a bridging function of KIBRA between DLC1 and chromatin. Since DLC1 is also known as estrogen receptor (ER)-binding protein and enhances ER transactivation, the DLC1-KIBRA-histone-3 binding presumably facilitates the ER-chromatin interaction. In another study, KIBRA was revealed to interact with sorting nexin-4 (SNX4), which regulates transportation from peripheral early endosomes to the juxtanuclear endocytic recycling compartment (20). SNX-4 coordinates endosome sorting through association with the minus end-directed microtubule motor dynein, which is mediated by KIBRA binding to both SNX4 and DLC1 (20).
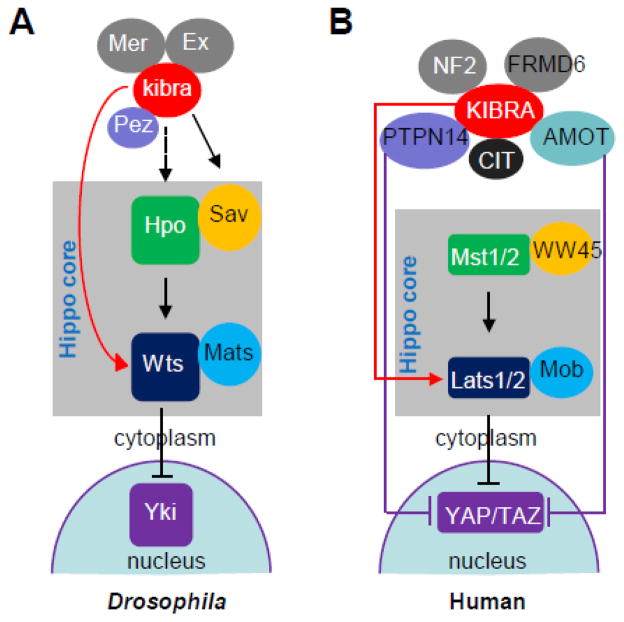
Recent studies have identified KIBRA/kibra (KIBRA in human and mouse and kibra in Drosophila) as an upstream regulator of the Hippo signaling network which acts by binding to multiple signaling molecules in this pathway (21–24). In Drosophila, kibra complexes with tumor suppressors Merlin (Mer) and Expanded (Ex), two upstream regulators of the Hippo pathway (21–23). Ex contains three PPxY motifs and the RxPPxY motif which was shown to bind to the first WW domain of kibra (22). Mer also associates with kibra (21–23), and both N- and C-terminal part of kibra are sufficient to associate with Mer (22). Salvador (Sav) is one of the core components of the Hippo pathway functioning downstream of the kibra-Mer-Ex complex and contains WW domains for homodimerization (25). By yeast two hybrid assays and co-immunoprecipitation, it has been shown that the WW domains of kibra and Sav can interact with each other to form kibra/Sav heterodimer (23). In cultured Drosophila cells, kibra also co-immunoprecipitates with warts, another core member of the Hippo pathway (22) and the immunoglobulin domain-containing cell adhesion molecule Echinoid (26). The association between KIBRA and neurofibromatosis type 2 (NF2, ortholog of Drosophila Merlin) seems to be conserved in human cells (22,24,27). However, unlike the case with Drosophila, KIBRA does not associate with FERM domain containing 6 (FRMD6, ortholog of Expanded) in mammalian cells (22). Association between large tumor suppressor (Lats1/2, orthologs of Drosophila warts) kinases and KIBRA was also reported to be mediated through the WW domains of KIBRA and the PPxY motifs of Lats (24). Moreover, KIBRA was also shown to interact with protein tyrosine phosphatase, non-receptor type 14 (PTPN14) and angiomotin family proteins (AMOT) (28), all of which are known regulators of YAP phosphorylation and localization. Therefore, KIBRA associates with multiple upstream components of the Hippo pathway from Drosophila to human cells, suggesting that KIBRA functions as a signal integrator in the Hippo pathway and indicating crosstalk between KIBRA and other upstream complexes in the regulation of Hippo-YAP activity (28) (Figure 3).
Fig. 3.
KIBRA in Hippo pathway. (A) In Drosophila, kibra forms complexes with Mer and Ex and functions upstream of the Hpo-Sav complex; kibra also interacts with Wts and Pez (PTPN14). (B) In human cells, KIBRA associates with multiple proteins and regulates the Hippo-YAP signaling activity independent of Mst1/2. Arrows and blunted ends indicate positive (activation) and negative regulation (inhibition), respectively. Dashed arrow indicates unknown interaction.
Posttranslational modifications (phosphorylations), and thus regulation of KIBRA function, are mediated through an association with a group of kinases including aPKC (15) and its neuronal isoform PKMζ [13], citron kinase (CIT) (28), Aurora kinases (29), cyclin-dependent kinase 1 (CDK1) (30), extracellular signal-regulated kinases (ERK) and p90 ribosomal S6 kinase (RSK) (31) as well as their cellular counterplayers protein phosphatase 1 (PP1) (29), cell division cycle phosphatase 14 (CDC14) (30) and PTPN14 (28, 32) (see also section 7 below). In addition to these binding partners, several other molecules have also been reported to interact with KIBRA, such as the protein interacting with C-kinase 1 (PICK1), which regulates the major excitatory neurotransmitter receptor α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor (AMPAR) in brains (5), KIAA0513, a marker in brain tissue for schizophrenia (33) and the axonal fasciculation regulator fasciculation and elongation protein zeta1 (FEZ1) (34) (Table 1).
Table 1.
Binding partners of KIBRA/kibra
| Name | Binding domains (or other influences on binding) | In vitro/In vivo | Ref. |
|---|---|---|---|
| Dendrin | WW domains of KIBRA | In vitro (Y2H) | (1) |
| PKCζ | The catalytic domain of PKCζ; C-terminus of KIBRA | In vitro (Y2H)/In vivo (co-localization) | (15) |
| Histone H3; DLC1 | Glutamic acid-rich region of KIBRA | In vitro (GST)/In vivo (IP) | (12) |
| SNX4 | Not determined | In vitro (Y2H)/In vivo (co-localization) | (20) |
| DDR1 | WW domains of KIBRA; the PPxY motif in DDR1 | In vivo (IP) | (10) |
| PATJ Synaptopodin |
The ADDV (PDZ) motif of KIBRA binds to the eighth PDZ domain of PATJ; WW domains of KIBRA bind to synaptopodin | In vitro (Y2H; GST)/In vivo (co-localization) | (9) |
| PKMζ | C-terminal region of KIBRA | In vivo (IP) | (9, 17) |
| Exocyst Sec 3 | Amino acids 129-525 of KIBRA | In vitro (Y2H)/In vivo (IP) | (19) |
| Lats1/2 | The N-terminal 86 amino acids of KIBRA and the PPxY motif in Lats2 | In vivo (IP) | (24) |
| Aurora-A | Ser 539 phosphorylation of KIBRA and amino acids 354-403 of Aurora-A. | In vivo (IP) | (29) |
| PP1 | The catalytic activity of PP1 is required for binding to KIBRA. | In vivo (IP) | (29) |
| CDC14A/B | CDK1 phosphorylation of KIBRAis involved | In vivo (IP) | (30) |
| RSK1/2 | Phosphorylation of KIBRA is required for interacting with RSK1, but not RSK2 | In vivo (IP) | (31) |
| FEZ1 | Coiled-coil domain of KIBRA | In vitro (GST; Y2H) | (34) |
| PAR3/PAR6β | Not determined | In vivo (IP; co-localization) | (14) |
| Citron (CIT) | PPxY motif of C-terminus of CIT kinase with WW domains of KIBRA | In vivo (IP) | (28) |
| AMOT | Not determined | In vivo (IP) | (28) |
| PICK1 | Not determined | In vitro (Y2H)/In vivo (IP; co-localization) | (5) |
| Merlin/(NF2); Expanded | The first WW domain of kibra and the ExRxPPxY motif | In vivo (IP) | (21–24, 27) |
| PTPN14 (Pez) | Not determined | In vitro (Y2H)/In vivo (IP) | (28,32) |
| Salvador (Sav) | WW domains of kibra and Sav | In vitro (Y2H)/In vivo (IP) (Drosophila) | (23) |
| Echinoid (Ed) | Intracellular domain of Ed | In vivo (IP) (Drosophila) | (26) |
Abbreviations: Y2H: yeast two hybrid; IP: immunoprecipitation; GST: Glutathione-S-Transferase pulldown
4. KIBRA function in the brain and kidney
Since KIBRA’s discovery, most studies of KIBRA have been focused on its role in learning and memory and other neurological disorders (For a review see ref.(35)). KIBRA is mainly expressed in memory-related regions of the human and rat brains including cortex and hippocampus (7, 8). In the hippocampus, KIBRA is mainly distributed in the somatodendritic region and enriched at the postsynaptic density (8). A single-nucleotide polymorphism (SNP) rs17070145 within the ninth intron of KIBRA was implicated in human cognition from three independent, cognitively normal cohorts (7). The T-allele carriers (TT or CT) perform better than T-allele non-carriers (CC) in memory performance. Functional magnetic resonance imaging (fMRI) detected the allele-dependent differences in hippocampus activations during memory retrievals; the T-allele non-carriers needed more activation to reach the same level of epitope memory as T-allele carriers (7). Another study also replicated the KIBRA effect on episodic memory, but found the T allele carriers had increased hippocampal activation, suggesting that the enhanced memory performance of T allele carriers is due to hippocampal activity, but not the negative compensation (36). The role of the rs17070145 SNP on the cognitive performance of normal individuals has also been demonstrated by many other studies (37–42). However, non-replications have also been reported in some cohorts (43–45). These different conclusions may come from differences in genetic background (39), ages (46), genders and life style (44), disease status (38, 47) and tobacco uses (48). A recent meta-analysis including more than thirteen thousand subjects reported a significant association between rs17070145 and both episodic and working memory (49).
The molecular consequences of the identified intronic SNP on KIBRA expression and function are not clear. However, a recent report demonstrated an almost complete linkage disequilibrium between rs17070145 and two exonic SNPs (rs3822660G/T and rs3822659T/G) that lead to a replacement of two adjacent amino acids (M734I and S735A) within the KIBRA C2 domain (2). The encoded KIBRA variants display different binding preferences for phosphatidylinositols on endosomal vesicles, which may affect their specific physiological role in higher brain functions and in neurological diseases such as Alzheimer dementia or schizophrenia. Interestingly, a so far uncharacterized exonic SNP (rs139606423; R969W) within the KIBRA gene leads to a mutated binding region (and likely an altered binding affinity) for PKMζ, an important regulator of long term memory storage (50). Further studies will be necessary to analyze a putative linkage between rs139606423 and cognitive performance.
Evidence for association between KIBRA and memory performance also came from animal studies. The RhoA/ROCK/Rac pathway has been implicated in cognitive function (51, 52). This pathway is an upstream modulator of PKCζ and thus may alter the activity of KIBRA (53). Peripheral delivery of the ROCK inhibitor hydroxyfasudil was revealed to improve spatial learning and working memory in rats (54). More direct evidence for the involvement of KIBRA in episodic memory is that both hippocampal knock-down of KIBRA in rats and KIBRA knock-out in mice reduced learning and memory performance in spatial memory tasks, accompanied with decreased PKMζ level (17). PKMζ is a brain-specific protein kinase and is involved in memory maintenance (50). KIBRA stabilizes PKMζ and prevents it from proteasomal degradation by direct interaction between a short sequence motif near the C-terminus of KIBRA and the kinase, suggesting that KIBRA may affect memory maintenance by regulating synaptic PKMζ level (17). More recently, training rats in the T-maze task significantly increased the performance accuracy of rats and also increased the expression levels of KIBRA and PKMζ in the prefrontal cortex, supporting the notion that KIBRA and PKMζ is closely related to reference memory in rats (55). Additionally, KIBRA may regulate higher brain function by regulating AMPAR trafficking and synaptic plasticity (5). Adult KIBRA knockout mice showed reduced long-term potentiating and long-term depression as well as deficits in contextual fear learning and memory (5).
Associations between the KIBRA locus and several neuropsychiatric diseases were also evidenced. The first study on KIBRA and Alzheimer disease (AD) in 2009 revealed that the T allele of KIBRA rs17070145 was significantly associated with increased risk for very-late onset AD (OR = 2.89) (56). However, a later study showed the opposite conclusion; the T allele non-carriers showed increased risk of late onset AD in 2 cohorts (57). In the latter study, microarray assay on laser-captured microdissected neurons exhibited up-regulation of KIBRA in AD-affected brain regions, including the hippocampal, posterior cingulate and temporal cortex regions. The rs17070145 T allele non-carriers have reduced glucose metabolism in AD-affected regions in comparison to T allele carriers, as detected by positron emission tomography. In contrast, in an Asian cohort, the rs17070145 SNP was not associated with the late-onset AD but associated with the young AD patients (58).
In some patients, the KIBRA genotype affects memory differently compared to healthy subjects. For example, in patients with traumatic brain injury and subjective memory complaints, the rs17070145 T allele non-carriers perform better than T allele carriers (59, 60). This suggests that the C allele may have positive functions in pathological conditions. No association between KIBRA genetic polymorphism and mild cognitive impairment or recurrent depressive disorders was found (41, 61).
KIBRA is highly expressed in kidney, which points to a crucial role of this protein in renal functions (1). Indeed, a disturbed expression of KIBRA in kidney podocytes affects their migration activities and processes involved in cell polarity (9). Furthermore, patients suffering from focal segmental glomerulosclerosis (FSGS) display an upregulated KIBRA expression in glomeruli, indicating that KIBRA is crucial for normal kidney physiology (62).
5. KIBRA in cell polarity and trafficking
By linking the tight junction protein PATJ and cytoskeleton protein synaptopodin, KIBRA positively modulates the directional migration of podocytes (9). In migrating podocytes, KIBRA accumulates and co-localizes with PATJ and synaptopodin in the leading edge and modulates cell migration (9). This study also provided new information regarding the function of KIBRA: it serves as a linker molecule between polarity proteins (e.g. PATJ and aPKC) and components of the cytoskeleton (e.g. synaptopodin and dendrin) to regulate cell migration. Similarly, interaction between aPKC and Exocyst is required for proper cell migration in NRK cells and KIBRA mediates this interaction by linking aPKC and Sec3, a component of Exocyst (19). KIBRA knockdown in NRK cells inhibited aPKC localization at the leading edge and thus inhibited cell migration. In Drosophila, the apical Hippo pathway complex (including kibra) localizes to cell-cell-contacts and signals through hippo and warts to regulate the polarization of actin and promote migration (63). Kibra is also required for oocyte polarity in Drosophila (23).
KIBRA can negatively regulate cell polarity by suppressing apical exocytosis in epithelial cells. The partitioning defective 3 (PAR3)-aPKC-PAR6 complex plays fundamental roles in cell polarity (64). KIBRA is localized in the same position (apical domain and cell-cell junctions) with aPKC in polarized epithelial cells, and is directly associated with aPKC (64, 65). Knockdown of KIBRA expanded the apical domain of Madin-Darby canine kidney (MDCK) cysts, suggesting that KIBRA suppresses apical domain expansion during cyst formation (14). This process was elucidated to be independent of Hippo pathway (14). KIBRA also suppresses the formation of apical-containing vacuoles through enhanced de novo apical exocytosis (14). Interestingly, the abnormal phenotypes in KIBRA knockdown cells were rescued by aPKC inhibition, indicating that KIBRA regulates apical domain development by inhibiting the kinase activity of aPKC (14). The overexpression of KIBRA in epithelial cells failed, but the overexpression of the aPKC binding site of KIBRA delayed the re-establishment of cell-to-cell contacts in a calcium switch assay (14). Since KIBRA expression is highly enriched in the brain, it is surprising that the role of KIBRA in the polarity of neurons (which are highly polarized cells) has not been determined.
Recent studies also showed that KIBRA plays a role in vesicular trafficking. For example, KIBRA is involved in trafficking of AMPAR, which is the major excitatory neurotransmitter receptor in the brain, and which is assembled by four subunits (GluA 1-4) (66). KIBRA associates with AMPAR and its partner PICK1. KIBRA knockdown does not affect internalization of the AMPAR, but accelerates the rate of GluA recycling to the membrane, indicating that KIBRA helps retain AMPAR in cell plasma after internalization (5). Makuch et al. speculated that the loss of KIBRA can be compensated by the homologous protein WWC2, explaining the unaffected basal transmission and surface expression of AMPAR receptors in the KIBRA knockout mice (5). KIBRA is also necessary for transferrin receptor (TfnR) trafficking in HeLa cells (20). Although knockdown of KIBRA does not affect the internalization of TfnR, it retains TfnR in the endosomal sorting compartment and inhibits the trafficking of TfnR to the endosomal recycling compartment, thus increasing the lysosomal-mediated degradation of TfnR (20).
6. KIBRA in growth control and human cancer
Although numerous studies have defined the roles of KIBRA in the brain, its physiological function in non-neuronal cells is relatively less understood. In Drosophila, kibra was shown to function as a tumor suppressor that regulates the Hippo signaling pathway, which controls tissue growth and organ size (21–23). Kibra associates with Mer and Ex and directly binds to Hippo-Sav complex to regulate the Hippo signaling pathway. Loss of kibra results in imaginal disc overgrowth, oogenesis defects and increased target gene expression of Hippo signaling (23). Human KIBRA functions together with NF2 to stimulate Lats1/2 phosphorylation, thus inducing activation of the Hippo pathway to suppress the transcriptional activity of YAP (yes-associated protein), which is the downstream effector of Hippo signaling (23,24), indicating that the tumor suppressive function of KIBRA may be conserved in the mammalian system. Indeed, loss of KIBRA expression in immortalized breast epithelial cells results in epithelial-to-mesenchymal transition (EMT) features which are concomitant with decreased phosphorylation levels of Lats and YAP, but not mammalian sterile-20 like (Mst), and reduced expression of KIBRA in breast cancer specimens of Claudin-low subtypes correlates with poor prognosis (67).
The KIBRA promoter contains a well-defined CpG island (68). Hypermethylation in this region was detected in 70% of B-cell acute lymphocytic leukemias, but almost no methylation was found in common epithelial cancers including breast, colorectal, kidney, lung, and prostate (68). The reason for this highly cell type-specific inactivation of KIBRA is not known at this time. However, there is no obvious CpG island in the human WWC2 and WWC3 locus. Interestingly, epigenetic inactivation (downregulation) of KIBRA was shown to be correlated with malignant state of B-cell acute lymphocytic leukemia (68). Furthermore, KIBRA promoter methylation status was also revealed to be associated with poor prognosis of chronic lymphocytic leukemia patients, including high CD38 expression and immunoglobulin heavy chain variable genes (IGHV) unmutated status (69). However, a role of KIBRA in human cancer (including leukemia) development has not been firmly established. The KIBRA null-allele mice are available and these mice exhibit overall normal development and growth (5,17). It is very possible that WWC2 compensates for KIBRA function during development, thus the double knockouts of both KIBRA/WWC2 will be needed to elucidate the physiological function of KIBRA (and WWC2) during normal and potential cancer development.
Although the above studies implicate a tumor suppressive function of KIBRA, a very recent study reported that overexpression of KIBRA in low aPKC-expressing gastric cancer correlates with enhanced lymphatic invasion and poor prognosis (11), indicating the positive role of the KIBRA-aPKC axis in promoting the progression of gastric cancer. In addition, downregulation of KIBRA significantly reduced cell proliferation and motility in breast cancer cells (31). In line with these studies, several previous studies have also demonstrated the positive role of KIBRA in regulating cell migration and proliferation (9, 10, 12, 14, 19). Thus, the dual function (suppressive or promoting) of KIBRA in cell proliferation/migration may be cell or tissue-type specific and more systematic investigations are required for clarity.
7. KIBRA regulation/phosphorylation
KIBRA is a phosphoprotein and multiple kinases have been identified to phosphorylate KIBRA. aPKCζ interacts with and phosphorylates KIBRA at Ser 975 and Ser 978 in vitro. This phosphorylation does not influence the cellular localization of KIBRA, but it may regulate KIBRA dimerization (15). However, it is not known whether these phosphorylations occur in cells. Recent studies demonstrated that several members/regulators of the Hippo pathway were involved in mitotic-related processes (70–74) and regulated during mitosis (75–78). Interestingly, as an upstream regulator of Hippo signaling, KIBRA is also regulated during mitosis. KIBRA Ser 539 is the primary phosphorylation site for Aurora-A and Aurora-B kinases both in vitro and in vivo, and this phosphorylation plays a role in mitotic progression (29). Aurora phosphorylation of KIBRA is dephosphorylated by PP1 during mitotic exit. Since Aurora kinases and PP1 play important roles in mitotic-related events such as spindle assembly and centrosome formation (79–82), KIBRA may also be a component of the mitotic apparatus (29). Further studies showed that KIBRA is required for full activation of Aurora kinases during mitosis. KIBRA also promotes the phosphorylation of Lats2 on Ser 83 through activating Aurora-A. Knockdown of KIBRA causes mitotic abnormalities, including mitotic spindle defects and chromosome misalignment (83). It is possible that KIBRA-Aurora-Lats2 all regulate the activities of each other to control proper mitotic events during cell cycle progression (83). In addition to the Ser 539 site, KIBRA is also phosphorylated at two highly conserved serine residues (Ser 542 and Ser 931) by CDK1 during spindle-damaging agents-induced mitotic arrest (30). Elimination of CDK1-mediated phosphorylation of KIBRA promoted cell exit from Taxol-arrested G2/M phase, suggesting a role of KIBRA and its mitotic phosphorylation in spindle checkpoint activation (30). In yeast, the phosphatase Cdc14 triggers mitotic exit by antagonizing Cdk-mediated phosphorylation of their substrates (84). Interestingly, the human phosphatases CDC14A/B associate with and dephosphorylate the CDK1-mediated phosphorylation of KIBRA (30), however a role of KIBRA in mitotic exit has not been established. Mitotic phospho-regulation of KIBRA does not affect the Hippo-YAP signaling activity (29, 30) and is likely independent of the Hippo pathway.
Our very recent study showed that KIBRA is also phosphorylated by the ERK-RSK cascade. ERK1/2 phosphorylates KIBRA at Ser 548 both in vitro and in vivo, and this phosphorylation is required for KIBRA-mediated cell proliferation in breast cancer cells (31). Interestingly, previous reports indicated that KIBRA is required for collagen-induced ERK signaling activation (10) and KIBRA knockdown abolished the ERK activity in migrating NRK cells (19). It is not clear at this time (and will be interesting to explore) whether ERK phosphorylation is involved in regulating KIBRA-mediated ERK activation. RSK1/2 are critical downstream mediators of ERK1/2 kinases, and were shown to associate with and specifically phosphorylate KIBRA at Thr 929 and Ser 947 (31). RSK phosphorylation positively modulates KIBRA activity in both proliferation and migration in breast cancer cells (31). The mitotic phosphorylation sites (Ser 539 and Ser 931; except for Ser 542, which only exists in vertebrates) and ERK-RSK sites (Ser 548, Thr 929 and Ser 947) are evolutionarily conserved from Drosophila to human (29–31) (Figure 2) and all these sites also exist in both human WWC2 and WWC3 (Figure 4). The aPKC sites are less conserved (Figures 2 and 4). However, there is currently no published study concerning the phospho-regulation of WWC2 and WWC3.
Fig. 4.

Conservation of phosphorylation sites among human WWC family proteins. Sequences are downloaded from NCBI as in Figure 1. Blue marks the Aurora phosphorylation site; red marks the CDK1 sites; green highlights the ERK site; the RSK sites are in dark blue and the PKC sites are indicated with purple.
In addition to its regulation by phosphorylation, KIBRA expression is greatly induced in response to progestin in progesterone-responsive human breast cancer cell lines (10), suggesting that KIBRA is also a hormonal-related protein, although the biological significance of this induction/regulation is not known.
8. Conclusions and future directions
Many reports from both human and mouse genetics have firmly established KIBRA’s function in the brain. However, it remains elusive whether or how the other two paralogs WWC2 and WWC3 play a role in brain physiology. Furthermore, how KIBRA is regulated in the neurons is still unclear. For example, is there any KIBRA phosphorylation site playing a role in the neuron/brain? Moreover, more data are needed to confirm the association of KIBRA and AD or other brain-related disorders. Such studies may ultimately demonstrate KIBRA as a highly attractive target for the treatment of neurological diseases, including AD and dementia (35). Beyond the brain, KIBRA is also attractive for its function in cancer cell migration and proliferation. However, most conclusions are based on cell culture models and few studies involve animal and human patients.
Highlights.
KIBRA is a memory-associated protein
KIBRA also plays roles in migration, polarity and growth control in epithelial cells
KIBRA is phosphorylated by various kinases
KIBRA functions as an adaptor protein to exercise its functions by interacting with other proteins.
Acknowledgments
We thank Dr. Joyce Solheim for critical reading and comments on the manuscript. Research in the laboratory of J.D. is supported in part by National Institutes of Health grants P20 GM103489 and R01 GM109066. L. Z. is a graduate student supported by fellowships from University of Nebraska Medical Center Graduate Studies and China Scholarship Council.
Abbreviations
- AD
Alzheimer disease
- AMPAR
α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor
- CDC14A/B
cell division cycle 14A/B
- CDK1
cyclin-dependent kinase 1
- DDR1
discoidin domain receptor family 1
- DLC1
dynein light chain 1
- EMT
epithelial-to-mesenchymal transition
- ERK
extracellular signal-regulated kinases
- ER
estrogen receptor
- FEZ1
fasciculation and elongation protein zeta1
- FRMD6
FERM (Band 4.1, Ezrin, Radixin and Moesin) domain containing 6
- MDCK
Madin-Darby canine kidney
- Mst1/2
mammalian sterile-20 like 1/2
- NF2
neurofibromatosis type 2
- NRK
normal rat kidney
- Lats1/2
large tumor suppressor kinase 1/2
- PAR
partitioning defective
- PATJ
PALS1(protein associated with lin-seven 1)-associated tight junction protein
- PDZ
postsynaptic density 95 (PSD95)/disc large (Dlg)/zonula occludens-1 (ZO-1)
- PICK1
protein interacting with C-kinase 1
- aPKC
atypical protein kinase C
- PKM
protein kinase M
- PP1
protein phosphatase 1
- PTPN14
protein tyrosine phosphatase, non-receptor type 14
- RSK1/2
p90 ribosomal S6 kinase 1/2
- SNP
single nucleotide polymorphism
- SNX4
sorting nexin 4
- TCF7L2
transcription factor 7-like 2
- TfnR
transferrin receptor
- WWC
WW and C2 domain containing
- YAP
yes-associated protein
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kremerskothen J, Plaas C, Buther K, Finger I, Veltel S, Matanis T, Liedtke T, Barnekow A. Characterization of KIBRA, a novel WW domain-containing protein. Biochem Biophys Res Commun. 2003;300(4):862–7. doi: 10.1016/s0006-291x(02)02945-5. [DOI] [PubMed] [Google Scholar]
- 2.Duning K, Wennmann DO, Bokemeyer A, Reissner C, Wersching H, Thomas C, Buschert J, Guske K, Franzke V, Floel A, et al. Common exonic missense variants in the C2 domain of the human KIBRA protein modify lipid binding and cognitive performance. Transl Psychiatry. 2013;3:e272. doi: 10.1038/tp.2013.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yoshihama Y, Chida K, Ohno S. The KIBRA-aPKC connection: A potential regulator of membrane trafficking and cell polarity. Commun Integr Biol. 2012;5(2):146–51. doi: 10.4161/cib.18849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nguyen DK, Yang F, Kaul R, Alkan C, Antonellis A, Friery KF, Zhu B, de Jong PJ, Disteche CM. Clcn4-2 genomic structure differs between the X locus in Mus spretus and the autosomal locus in Mus musculus: AT motif enrichment on the X. Genome research. 2011;21(3):402–9. doi: 10.1101/gr.108563.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Makuch L, Volk L, Anggono V, Johnson RC, Yu Y, Duning K, Kremerskothen J, Xia J, Takamiya K, Huganir RL. Regulation of AMPA receptor function by the human memory-associated gene KIBRA. Neuron. 2011;71(6):1022–9. doi: 10.1016/j.neuron.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guske K, Schmitz B, Schelleckes M, Duning K, Kremerskothen J, Pavenstadt HJ, Brand SM, Brand E. Tissue-specific differences in the regulation of KIBRA gene expression involve transcription factor TCF7L2 and a complex alternative promoter system. J Mol Med (Berl) 2013 doi: 10.1007/s00109-013-1089-y. [DOI] [PubMed] [Google Scholar]
- 7.Papassotiropoulos A, Stephan DA, Huentelman MJ, Hoerndli FJ, Craig DW, Pearson JV, Huynh KD, Brunner F, Corneveaux J, Osborne D, et al. Common Kibra alleles are associated with human memory performance. Science. 2006;314(5798):475–8. doi: 10.1126/science.1129837. [DOI] [PubMed] [Google Scholar]
- 8.Johannsen S, Duning K, Pavenstadt H, Kremerskothen J, Boeckers TM. Temporal-spatial expression and novel biochemical properties of the memory-related protein KIBRA. Neuroscience. 2008;155(4):1165–73. doi: 10.1016/j.neuroscience.2008.06.054. [DOI] [PubMed] [Google Scholar]
- 9.Duning K, Schurek EM, Schluter M, Bayer M, Reinhardt HC, Schwab A, Schaefer L, Benzing T, Schermer B, Saleem MA, et al. KIBRA modulates directional migration of podocytes. J Am Soc Nephrol. 2008;19(10):1891–903. doi: 10.1681/ASN.2007080916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hilton HN, Stanford PM, Harris J, Oakes SR, Kaplan W, Daly RJ, Ormandy CJ. KIBRA interacts with discoidin domain receptor 1 to modulate collagen-induced signalling. Biochim Biophys Acta. 2008;1783(3):383–93. doi: 10.1016/j.bbamcr.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 11.Yoshihama Y, Izumisawa Y, Akimoto K, Satoh Y, Mizushima T, Satoh K, Chida K, Takagawa R, Akiyama H, Ichikawa Y, et al. High expression of KIBRA in low atypical protein kinase C-expressing gastric cancer correlates with lymphatic invasion and poor prognosis. Cancer Sci. 2013;104(2):259–65. doi: 10.1111/cas.12066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rayala SK, den Hollander P, Manavathi B, Talukder AH, Song C, Peng S, Barnekow A, Kremerskothen J, Kumar R. Essential role of KIBRA in co-activator function of dynein light chain 1 in mammalian cells. The Journal of biological chemistry. 2006;281(28):19092–9. doi: 10.1074/jbc.M600021200. [DOI] [PubMed] [Google Scholar]
- 13.Vogel WF, Aszodi A, Alves F, Pawson T. Discoidin domain receptor 1 tyrosine kinase has an essential role in mammary gland development. Mol Cell Biol. 2001;21(8):2906–17. doi: 10.1128/MCB.21.8.2906-2917.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoshihama Y, Sasaki K, Horikoshi Y, Suzuki A, Ohtsuka T, Hakuno F, Takahashi S, Ohno S, Chida K. KIBRA suppresses apical exocytosis through inhibition of aPKC kinase activity in epithelial cells. Current biology: CB. 2011;21(8):705–11. doi: 10.1016/j.cub.2011.03.029. [DOI] [PubMed] [Google Scholar]
- 15.Buther K, Plaas C, Barnekow A, Kremerskothen J. KIBRA is a novel substrate for protein kinase Czeta. Biochem Biophys Res Commun. 2004;317(3):703–7. doi: 10.1016/j.bbrc.2004.03.107. [DOI] [PubMed] [Google Scholar]
- 16.Hernandez AI, Blace N, Crary JF, Serrano PA, Leitges M, Libien JM, Weinstein G, Tcherapanov A, Sacktor TC. Protein kinase M zeta synthesis from a brain mRNA encoding an independent protein kinase C zeta catalytic domain. Implications for the molecular mechanism of memory. The Journal of biological chemistry. 2003;278(41):40305–16. doi: 10.1074/jbc.M307065200. [DOI] [PubMed] [Google Scholar]
- 17.Vogt-Eisele A, Kruger C, Duning K, Weber D, Spoelgen R, Pitzer C, Plaas C, Eisenhardt G, Meyer A, Vogt G, et al. KIBRA (KIdney/BRAin protein) regulates learning and memory and stabilizes Protein kinase Mzeta. J Neurochem. 2013 doi: 10.1111/jnc.12480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yao Y, Kelly MT, Sajikumar S, Serrano P, Tian D, Bergold PJ, Frey JU, Sacktor TC. PKM zeta maintains late long-term potentiation by N-ethylmaleimide-sensitive factor/GluR2-dependent trafficking of postsynaptic AMPA receptors. J Neurosci. 2008;28(31):7820–7. doi: 10.1523/JNEUROSCI.0223-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosse C, Formstecher E, Boeckeler K, Zhao Y, Kremerskothen J, White MD, Camonis JH, Parker PJ. An aPKC-exocyst complex controls paxillin phosphorylation and migration through localised JNK1 activation. PLoS Biol. 2009;7(11):e1000235. doi: 10.1371/journal.pbio.1000235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Traer CJ, Rutherford AC, Palmer KJ, Wassmer T, Oakley J, Attar N, Carlton JG, Kremerskothen J, Stephens DJ, Cullen PJ. SNX4 coordinates endosomal sorting of TfnR with dynein-mediated transport into the endocytic recycling compartment. Nature cell biology. 2007;9(12):1370–80. doi: 10.1038/ncb1656. [DOI] [PubMed] [Google Scholar]
- 21.Baumgartner R, Poernbacher I, Buser N, Hafen E, Stocker H. The WW domain protein Kibra acts upstream of Hippo in Drosophila. Developmental cell. 2010;18(2):309–16. doi: 10.1016/j.devcel.2009.12.013. [DOI] [PubMed] [Google Scholar]
- 22.Genevet A, Wehr MC, Brain R, Thompson BJ, Tapon N. Kibra is a regulator of the Salvador/Warts/Hippo signaling network. Developmental cell. 2010;18(2):300–8. doi: 10.1016/j.devcel.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu J, Zheng Y, Dong J, Klusza S, Deng WM, Pan D. Kibra functions as a tumor suppressor protein that regulates Hippo signaling in conjunction with Merlin and Expanded. Developmental cell. 2010;18(2):288–99. doi: 10.1016/j.devcel.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiao L, Chen Y, Ji M, Dong J. KIBRA regulates Hippo signaling activity via interactions with large tumor suppressor kinases. The Journal of biological chemistry. 2011;286(10):7788–96. doi: 10.1074/jbc.M110.173468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohnishi S, Guntert P, Koshiba S, Tomizawa T, Akasaka R, Tochio N, Sato M, Inoue M, Harada T, Watanabe S, et al. Solution structure of an atypical WW domain in a novel beta-clam-like dimeric form. FEBS letters. 2007;581(3):462–8. doi: 10.1016/j.febslet.2007.01.008. [DOI] [PubMed] [Google Scholar]
- 26.Yue T, Tian A, Jiang J. The cell adhesion molecule echinoid functions as a tumor suppressor and upstream regulator of the Hippo signaling pathway. Developmental cell. 2012;22(2):255–67. doi: 10.1016/j.devcel.2011.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang N, Bai H, David KK, Dong J, Zheng Y, Cai J, Giovannini M, Liu P, Anders RA, Pan D. The Merlin/NF2 tumor suppressor functions through the YAP oncoprotein to regulate tissue homeostasis in mammals. Developmental cell. 2010;19(1):27–38. doi: 10.1016/j.devcel.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang W, Li X, Huang J, Feng L, Dolinta KG, Chen J. Defining the protein-protein interaction network of the human hippo pathway. Molecular & cellular proteomics: MCP. 2014;13(1):119–31. doi: 10.1074/mcp.M113.030049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiao L, Chen Y, Ji M, Volle DJ, Lewis RE, Tsai MY, Dong J. KIBRA protein phosphorylation is regulated by mitotic kinase aurora and protein phosphatase 1. The Journal of biological chemistry. 2011;286(42):36304–15. doi: 10.1074/jbc.M111.246850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ji M, Yang S, Chen Y, Xiao L, Zhang L, Dong J. Phospho-regulation of KIBRA by CDK1 and CDC14 phosphatase controls cell-cycle progression. Biochem J. 2012;447(1):93–102. doi: 10.1042/BJ20120751. [DOI] [PubMed] [Google Scholar]
- 31.Yang S, Ji M, Zhang L, Chen Y, Wennmann DO, Kremerskothen J, Dong J. Phosphorylation of KIBRA by the extracellular signal-regulated kinase (ERK)-ribosomal S6 kinase (RSK) cascade modulates cell proliferation and migration. Cell Signal. 2013;26(2):343–51. doi: 10.1016/j.cellsig.2013.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Poernbacher I, Baumgartner R, Marada SK, Edwards K, Stocker H. Drosophila Pez acts in Hippo signaling to restrict intestinal stem cell proliferation. Current biology: CB. 2012;22(5):389–96. doi: 10.1016/j.cub.2012.01.019. [DOI] [PubMed] [Google Scholar]
- 33.Lauriat TL, Dracheva S, Kremerskothen J, Duning K, Haroutunian V, Buxbaum JD, Hyde TM, Kleinman JE, McInnes LA. Characterization of KIAA0513, a novel signaling molecule that interacts with modulators of neuroplasticity, apoptosis, and the cytoskeleton. Brain Res. 2006;1121(1):1–11. doi: 10.1016/j.brainres.2006.08.099. [DOI] [PubMed] [Google Scholar]
- 34.Assmann EM, Alborghetti MR, Camargo ME, Kobarg J. FEZ1 dimerization and interaction with transcription regulatory proteins involves its coiled-coil region. The Journal of biological chemistry. 2006;281(15):9869–81. doi: 10.1074/jbc.M513280200. [DOI] [PubMed] [Google Scholar]
- 35.Schneider A, Huentelman MJ, Kremerskothen J, Duning K, Spoelgen R, Nikolich K. KIBRA: A New Gateway to Learning and Memory? Front Aging Neurosci. 2010;2(4) doi: 10.3389/neuro.24.004.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kauppi K, Nilsson LG, Adolfsson R, Eriksson E, Nyberg L. KIBRA polymorphism is related to enhanced memory and elevated hippocampal processing. J Neurosci. 2011;31(40):14218–22. doi: 10.1523/JNEUROSCI.3292-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yasuda Y, Hashimoto R, Ohi K, Fukumoto M, Takamura H, Iike N, Yoshida T, Hayashi N, Takahashi H, Yamamori H, et al. Association study of KIBRA gene with memory performance in a Japanese population. World J Biol Psychiatry. 2010;11(7):852–7. doi: 10.3109/15622971003797258. [DOI] [PubMed] [Google Scholar]
- 38.Vassos E, Bramon E, Picchioni M, Walshe M, Filbey FM, Kravariti E, McDonald C, Murray RM, Collier DA, Toulopoulou T. Evidence of association of KIBRA genotype with episodic memory in families of psychotic patients and controls. J Psychiatr Res. 2010;44(12):795–8. doi: 10.1016/j.jpsychires.2010.01.012. [DOI] [PubMed] [Google Scholar]
- 39.Preuschhof C, Heekeren HR, Li SC, Sander T, Lindenberger U, Backman L. KIBRA and CLSTN2 polymorphisms exert interactive effects on human episodic memory. Neuropsychologia. 2010;48(2):402–8. doi: 10.1016/j.neuropsychologia.2009.09.031. [DOI] [PubMed] [Google Scholar]
- 40.Bates TC, Price JF, Harris SE, Marioni RE, Fowkes FG, Stewart MC, Murray GD, Whalley LJ, Starr JM, Deary IJ. Association of KIBRA and memory. Neurosci Lett. 2009;458(3):140–3. doi: 10.1016/j.neulet.2009.04.050. [DOI] [PubMed] [Google Scholar]
- 41.Almeida OP, Schwab SG, Lautenschlager NT, Morar B, Greenop KR, Flicker L, Wildenauer D. KIBRA genetic polymorphism influences episodic memory in later life, but does not increase the risk of mild cognitive impairment. J Cell Mol Med. 2008;12(5A):1672–6. doi: 10.1111/j.1582-4934.2008.00229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schaper K, Kolsch H, Popp J, Wagner M, Jessen F. KIBRA gene variants are associated with episodic memory in healthy elderly. Neurobiology of aging. 2008;29(7):1123–5. doi: 10.1016/j.neurobiolaging.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 43.Sedille-Mostafaie N, Sebesta C, Huber KR, Zehetmayer S, Jungwirth S, Tragl KH, Fischer P, Krugluger W. The role of memory-related gene polymorphisms, KIBRA and CLSTN2, on replicate memory assessment in the elderly. J Neural Transm. 2012;119(1):77–80. doi: 10.1007/s00702-011-0667-9. [DOI] [PubMed] [Google Scholar]
- 44.Wersching H, Guske K, Hasenkamp S, Hagedorn C, Schiwek S, Jansen S, Witte V, Wellmann J, Lohmann H, Duning K, et al. Impact of common KIBRA allele on human cognitive functions. Neuropsychopharmacology. 2011;36(6):1296–304. doi: 10.1038/npp.2011.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Need AC, Attix DK, McEvoy JM, Cirulli ET, Linney KN, Wagoner AP, Gumbs CE, Giegling I, Moller HJ, Francks C, et al. Failure to replicate effect of Kibra on human memory in two large cohorts of European origin. Am J Med Genet B Neuropsychiatr Genet. 2008;147B(5):667–8. doi: 10.1002/ajmg.b.30658. [DOI] [PubMed] [Google Scholar]
- 46.Schuck NW, Doeller CF, Schjeide BM, Schroder J, Frensch PA, Bertram L, Li SC. Aging and KIBRA/WWC1 genotype affect spatial memory processes in a virtual navigation task. Hippocampus. 2013 doi: 10.1002/hipo.22148. [DOI] [PubMed] [Google Scholar]
- 47.Hayashi N, Kazui H, Kamino K, Tokunaga H, Takaya M, Yokokoji M, Kimura R, Kito Y, Wada T, Nomura K, et al. KIBRA genetic polymorphism influences episodic memory in Alzheimer’s disease, but does not show association with disease in a Japanese cohort. Dement Geriatr Cogn Disord. 2010;30(4):302–8. doi: 10.1159/000320482. [DOI] [PubMed] [Google Scholar]
- 48.Zhang H, Kranzler HR, Poling J, Gruen JR, Gelernter J. Cognitive flexibility is associated with KIBRA variant and modulated by recent tobacco use. Neuropsychopharmacology. 2009;34(12):2508–16. doi: 10.1038/npp.2009.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Milnik A, Heck A, Vogler C, Heinze HJ, de Quervain DJ, Papassotiropoulos A. Association of KIBRA with episodic and working memory: a meta-analysis. Am J Med Genet B Neuropsychiatr Genet. 2012;159B(8):958–69. doi: 10.1002/ajmg.b.32101. [DOI] [PubMed] [Google Scholar]
- 50.Sacktor TC. How does PKMzeta maintain long-term memory? Nat Rev Neurosci. 2011;12(1):9–15. doi: 10.1038/nrn2949. [DOI] [PubMed] [Google Scholar]
- 51.Woo S, Gomez TM. Rac1 and RhoA promote neurite outgrowth through formation and stabilization of growth cone point contacts. J Neurosci. 2006;26(5):1418–28. doi: 10.1523/JNEUROSCI.4209-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Loudon RP, Silver LD, Yee HF, Jr, Gallo G. RhoA-kinase and myosin II are required for the maintenance of growth cone polarity and guidance by nerve growth factor. J Neurobiol. 2006;66(8):847–67. doi: 10.1002/neu.20258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Van Kolen K, Slegers H. Atypical PKCzeta is involved in RhoA-dependent mitogenic signaling by the P2Y(12) receptor in C6 cells. FEBS J. 2006;273(8):1843–54. doi: 10.1111/j.1742-4658.2006.05205.x. [DOI] [PubMed] [Google Scholar]
- 54.Huentelman MJ, Stephan DA, Talboom J, Corneveaux JJ, Reiman DM, Gerber JD, Barnes CA, Alexander GE, Reiman EM, Bimonte-Nelson HA. Peripheral delivery of a ROCK inhibitor improves learning and working memory. Behav Neurosci. 2009;123(1):218–23. doi: 10.1037/a0014260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang DC, Liu PC, Hung HS, Chen TJ. Both PKMzeta and KIBRA are closely related to reference memory but not working memory in a T-maze task in rats. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 2013 doi: 10.1007/s00359-013-0862-2. [DOI] [PubMed] [Google Scholar]
- 56.Rodriguez-Rodriguez E, Infante J, Llorca J, Mateo I, Sanchez-Quintana C, Garcia-Gorostiaga I, Sanchez-Juan P, Berciano J, Combarros O. Age-dependent association of KIBRA genetic variation and Alzheimer’s disease risk. Neurobiology of aging. 2009;30(2):322–4. doi: 10.1016/j.neurobiolaging.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 57.Corneveaux JJ, Liang WS, Reiman EM, Webster JA, Myers AJ, Zismann VL, Joshipura KD, Pearson JV, Hu-Lince D, Craig DW, et al. Evidence for an association between KIBRA and late-onset Alzheimer’s disease. Neurobiology of aging. 2010;31(6):901–9. doi: 10.1016/j.neurobiolaging.2008.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang HF, Tan L, Yu JT, Ma XY, Liu QY, Wang W. Age-dependent association of KIBRA gene polymorphism with Alzheimer’s disease in Han Chinese. Mol Biol Rep. 2013;40(12):7077–82. doi: 10.1007/s11033-013-2830-x. [DOI] [PubMed] [Google Scholar]
- 59.Wagner AK, Hatz LE, Scanlon JM, Niyonkuru C, Miller MA, Ricker JH, Conley YP, Ferrell RE. Association of KIBRA rs17070145 polymorphism and episodic memory in individuals with severe TBI. Brain Inj. 2012;26(13–14):1658–69. doi: 10.3109/02699052.2012.700089. [DOI] [PubMed] [Google Scholar]
- 60.Nacmias B, Bessi V, Bagnoli S, Tedde A, Cellini E, Piccini C, Sorbi S, Bracco L. KIBRA gene variants are associated with episodic memory performance in subjective memory complaints. Neurosci Lett. 2008;436(2):145–7. doi: 10.1016/j.neulet.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 61.Galecki P, Szemraj J, Florkowski A, Talarowska M, Bienkiewicz M, Galecka E, Lewinski A. Single nucleotide polymorphism of the KIBRA gene in recurrent depressive disorders. Neuro Endocrinol Lett. 2010;31(1):97–102. [PubMed] [Google Scholar]
- 62.Bennett MR, Czech KA, Arend LJ, Witte DP, Devarajan P, Potter SS. Laser capture microdissection-microarray analysis of focal segmental glomerulosclerosis glomeruli. Nephron Experimental nephrology. 2007;107(1):e30–40. doi: 10.1159/000106775. [DOI] [PubMed] [Google Scholar]
- 63.Lucas EP, Khanal I, Gaspar P, Fletcher GC, Polesello C, Tapon N, Thompson BJ. The Hippo pathway polarizes the actin cytoskeleton during collective migration of Drosophila border cells. The Journal of cell biology. 2013;201(6):875–85. doi: 10.1083/jcb.201210073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Suzuki A, Ohno S. The PAR-aPKC system: lessons in polarity. Journal of cell science. 2006;119(Pt 6):979–87. doi: 10.1242/jcs.02898. [DOI] [PubMed] [Google Scholar]
- 65.Yoshihama Y, Hirai T, Ohtsuka T, Chida K. KIBRA Co-localizes with protein kinase Mzeta (PKMzeta) in the mouse hippocampus. Biosci Biotechnol Biochem. 2009;73(1):147–51. doi: 10.1271/bbb.80564. [DOI] [PubMed] [Google Scholar]
- 66.Shepherd JD, Huganir RL. The cell biology of synaptic plasticity: AMPA receptor trafficking. Annu Rev Cell Dev Biol. 2007;23:613–43. doi: 10.1146/annurev.cellbio.23.090506.123516. [DOI] [PubMed] [Google Scholar]
- 67.Moleirinho S, Chang N, Sims AH, Tilston-Lunel AM, Angus L, Steele A, Boswell V, Barnett SC, Ormandy C, Faratian D, et al. KIBRA exhibits MST-independent functional regulation of the Hippo signaling pathway in mammals. Oncogene. 2013;32(14):1821–30. doi: 10.1038/onc.2012.196. [DOI] [PubMed] [Google Scholar]
- 68.Hill VK, Dunwell TL, Catchpoole D, Krex D, Brini AT, Griffiths M, Craddock C, Maher ER, Latif F. Frequent epigenetic inactivation of KIBRA, an upstream member of the Salvador/Warts/Hippo (SWH) tumor suppressor network, is associated with specific genetic event in B-cell acute lymphocytic leukemia. Epigenetics. 2011;6(3):326–32. doi: 10.4161/epi.6.3.14404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shinawi T, Hill V, Dagklis A, Baliakas P, Stamatopoulos K, Agathanggelou A, Stankovic T, Maher ER, Ghia P, Latif F. KIBRA gene methylation is associated with unfavorable biological prognostic parameters in chronic lymphocytic leukemia. Epigenetics. 2012;7(3):211–5. doi: 10.4161/epi.7.3.19222. [DOI] [PubMed] [Google Scholar]
- 70.Shimizu T, Ho LL, Lai ZC. The mob as tumor suppressor gene is essential for early development and regulates tissue growth in Drosophila. Genetics. 2008;178(2):957–65. doi: 10.1534/genetics.107.081570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Florindo C, Perdigao J, Fesquet D, Schiebel E, Pines J, Tavares AA. Human Mob1 proteins are required for cytokinesis by controlling microtubule stability. Journal of cell science. 2012;125(Pt 13):3085–90. doi: 10.1242/jcs.097147. [DOI] [PubMed] [Google Scholar]
- 72.Mardin BR, Lange C, Baxter JE, Hardy T, Scholz SR, Fry AM, Schiebel E. Components of the Hippo pathway cooperate with Nek2 kinase to regulate centrosome disjunction. Nature cell biology. 2010;12(12):1166–76. doi: 10.1038/ncb2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hergovich A, Kohler RS, Schmitz D, Vichalkovski A, Cornils H, Hemmings BA. The MST1 and hMOB1 tumor suppressors control human centrosome duplication by regulating NDR kinase phosphorylation. Current biology: CB. 2009;19(20):1692–702. doi: 10.1016/j.cub.2009.09.020. [DOI] [PubMed] [Google Scholar]
- 74.Yabuta N, Okada N, Ito A, Hosomi T, Nishihara S, Sasayama Y, Fujimori A, Okuzaki D, Zhao H, Ikawa M, et al. Lats2 is an essential mitotic regulator required for the coordination of cell division. The Journal of biological chemistry. 2007;282(26):19259–71. doi: 10.1074/jbc.M608562200. [DOI] [PubMed] [Google Scholar]
- 75.Toji S, Yabuta N, Hosomi T, Nishihara S, Kobayashi T, Suzuki S, Tamai K, Nojima H. The centrosomal protein Lats2 is a phosphorylation target of Aurora-A kinase. Genes to cells: devoted to molecular & cellular mechanisms. 2004;9(5):383–97. doi: 10.1111/j.1356-9597.2004.00732.x. [DOI] [PubMed] [Google Scholar]
- 76.Morisaki T, Hirota T, Iida S, Marumoto T, Hara T, Nishiyama Y, Kawasuzi M, Hiraoka T, Mimori T, Araki N, et al. WARTS tumor suppressor is phosphorylated by Cdc2/cyclin B at spindle poles during mitosis. FEBS letters. 2002;529(2–3):319–24. doi: 10.1016/s0014-5793(02)03360-4. [DOI] [PubMed] [Google Scholar]
- 77.Praskova M, Xia F, Avruch J. MOBKL1A/MOBKL1B phosphorylation by MST1 and MST2 inhibits cell proliferation. Current biology: CB. 2008;18(5):311–21. doi: 10.1016/j.cub.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yang S, Zhang L, Liu M, Chong R, Ding SJ, Chen Y, Dong J. CDK1 phosphorylation of YAP promotes mitotic defects and cell motility and is essential for neoplastic transformation. Cancer research. 2013;73(22):6722–33. doi: 10.1158/0008-5472.CAN-13-2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bischoff JR, Plowman GD. The Aurora/Ipl1p kinase family: regulators of chromosome segregation and cytokinesis. Trends Cell Biol. 1999;9(11):454–9. doi: 10.1016/s0962-8924(99)01658-x. [DOI] [PubMed] [Google Scholar]
- 80.Meraldi P, Nigg EA. Centrosome cohesion is regulated by a balance of kinase and phosphatase activities. Journal of cell science. 2001;114(Pt 20):3749–57. doi: 10.1242/jcs.114.20.3749. [DOI] [PubMed] [Google Scholar]
- 81.Fernandez A, Brautigan DL, Lamb NJ. Protein phosphatase type 1 in mammalian cell mitosis: chromosomal localization and involvement in mitotic exit. The Journal of cell biology. 1992;116(6):1421–30. doi: 10.1083/jcb.116.6.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cheng A, Dean NM, Honkanen RE. Serine/threonine protein phosphatase type 1gamma1 is required for the completion of cytokinesis in human A549 lung carcinoma cells. The Journal of biological chemistry. 2000;275(3):1846–54. doi: 10.1074/jbc.275.3.1846. [DOI] [PubMed] [Google Scholar]
- 83.Zhang L, Iyer J, Chowdhury A, Ji M, Xiao L, Yang S, Chen Y, Tsai MY, Dong J. KIBRA regulates aurora kinase activity and is required for precise chromosome alignment during mitosis. The Journal of biological chemistry. 2012;287(41):34069–77. doi: 10.1074/jbc.M112.385518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Visintin R, Craig K, Hwang ES, Prinz S, Tyers M, Amon A. The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk-dependent phosphorylation. Molecular cell. 1998;2(6):709–18. doi: 10.1016/s1097-2765(00)80286-5. [DOI] [PubMed] [Google Scholar]