Abstract
Background
Lower levels of genomic DNA methylation in blood DNA has been associated with risk of different cancers and several cancer risk factors. To understand the use of genomic methylation measures as biomarkers of cancer risk, data are needed on within-individual changes over time.
Methods
Using information from 77 subjects with blood collected at 2 visits on average 8 years apart, we examined whether levels of DNA methylation change with time and if so, whether selected cancer risk factors predict these changes. We measured DNA methylation levels in peripheral blood mononuclear cells (PBMC) using three assays that have been used in epidemiologic studies: (i) luminometric methylation assay (LUMA)(ii) LINE-1 by pyrosequencing, and (iii) Sat2 by MethyLight.
Results
Close to a third of all individuals had large changes over time (≥10%) in LUMA with 19.5% increasing and 13.0% decreasing. For Sat2, two-thirds of individuals had large changes with 40% increasing and 26% decreasing over time. In contrast, only 3.9% of individuals had large changes in LINE-1 over time. The degree of change in PBMC DNA methylation was statistically significantly inversely associated with methylation levels at baseline; greater decreases were observed in individuals with higher baseline values for each assay.
Conclusions
These data, if replicated, suggest that changes in DNA methylation over time are highly associated with baseline values of the assay and vary by assay type.
Impact
These findings suggest that assays that change more over time may warrant consideration for studies that measure later life exposures.
Introduction
Genomic methylation measured in peripheral blood has been associated with a number of different cancer types, including colon (1, 2), bladder (3–5), stomach (6), breast (7), and head and neck (8). Genomic DNA methylation levels in blood have also been associated with selected risk factors including age, gender, dietary folate status, and cigarette smoking (reviewed in ref. 9). Although these lines of evidence are intriguing, there remain several limitations in overall inference, particularly as many epidemiologic studies rely primarily on a DNA methylation measured at a single time and generally use samples collected after the disease has been diagnosed.
Total 5-methylcytosine (5mC) content has been observed to be associated with age (10, 11), but this association has not been entirely consistent (5) and has mostly been observed through cross-sectional studies rather than within-individual changes over time. There are few longitudinal studies on the effect of aging on changes in blood DNA methylation (12, 13). Measuring DNA methylation by the luminometric methylation assay (LUMA) in blood collected on average 11 years apart in individuals ranging in age from 69 to 96 years, Bjornsson and colleagues (12) observed large variability in levels of DNA methylation over time with 29% of individuals having greater than or equal to 10% methylation change (with a range of −30%–26% for the whole study). Studies that have attempted to more completely examine the age effect however, suggest that age may account for a small proportion of variation over time (13, 14).
Understanding within-individual differences over time is critical for understanding whether environmental factors, in addition to or apart from age, can influence DNA methylation changes. For example, a study showing greater differences in 5mC in blood DNA in older twins compared with younger twins (11) has been used to give strong support to the hypothesis that the environmental changes (either exogenous or endogenous) across the life course may relate to changes in DNA methylation. However, this study did not directly test within-individual changes over time, but rather compared cross-sectionally older versus younger twins. Environmental factors such as benzene (15) and arsenic (16) have been associated with genomic DNA methylation markers in blood but again these studies have mainly been cross-sectional in nature. Only a few studies have specifically examined whether selected exposures (primarily folate intervention studies) are associated with within-individual changes in genomic DNA methylation over time (reviewed in ref. 9). These studies primarily examine within-individual changes over very short periods of time.
Using information from 77 individuals with 2 blood specimens in the New York site of Breast Cancer Family Registry (BCFR), we examined whether genomic DNA methylation measured by 3 commonly used assays in epidemiologic studies (LUMA, LINE-1, and Sat2) in peripheral blood mononuclear cells (PBMC) changed over time and whether these changes were associated with selected risk factors for cancer.
Materials and Methods
We examined 77 individuals (21 males and 56 females) with at least 2 blood specimens at 2 different time points (average of 8.6 years between blood draws) participating in the New York site of the BCFR (17). Each individual completed an epidemiologic questionnaire. We extracted genomic DNA from PBMCs by a salting out procedure (18). This study was approved by Columbia University’s Institutional Review Board.
The luminometric methylation assay (LUMA) is based on digestion of genomic DNA with methylation-sensitive and methylation-insensitive restriction enzymes, followed by methylation quantification by pyrosequencing as described (12). We measured LINE-1 methylation status by pyrosequencing using primers and PCR condition as described (15). Methylation quantification was done using the PyroMark Q24 1.010 software (18). We measured Sat2 using the sequences of probes and forward and reverse primers of Sat2-M1 described in Weisenberger and colleagues (19). PCR was carried out using the PCR program as described in Wu and colleagues (18). Assays were run on an ABI Prism 7900 Sequence Detection System (Perkin-Elmer). For the MethyLight and pyrosequencing assays, aliquots of DNA (500 ng) were bisulfite treated with the EZ DNA methylation kit (Zymo Research). The interassay coefficients of variation (CV) were 2.8, 0.9, and 5.8 for LUMA, LINE-1, and Sat2, respectively.
Statistical methods
We calculated Spearman correlation coefficients for each marker with age at baseline and time since baseline (years of follow up). We used multivariable linear regression models to examine the associations between the absolute change in each methylation marker over time with the following variables: age at baseline blood draw, gender, time between blood draws, cancer status at the follow up (time 2 blood draw), BRCA1/2 mutation status, body mass index (BMI) at baseline, smoking status at baseline and baseline methylation levels for each respective marker. We carried out sensitivity analyses for these models to see whether our findings were robust to outliers and to examine whether the findings were materially altered when we removed the 15 subjects who reported a prior cancer at one of the time points.
Results
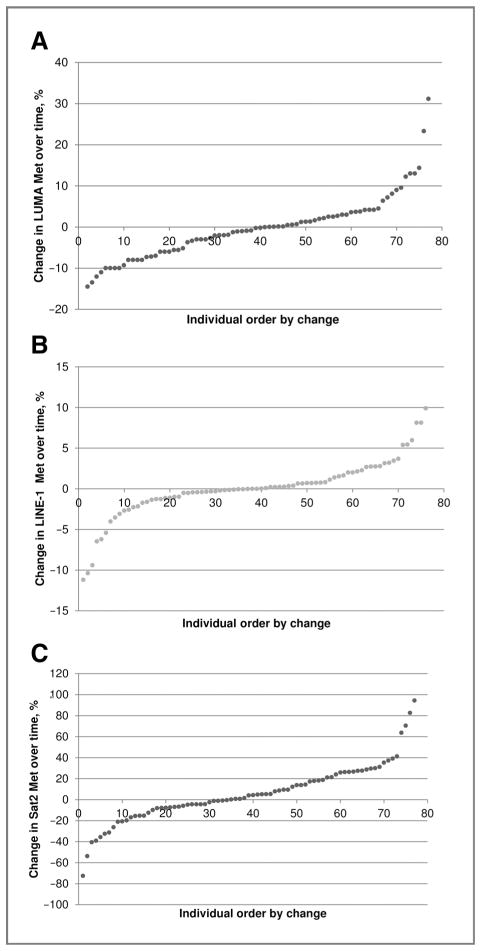
The range between the 2 blood draws was 1.3 to 13.2 years (mean =8.6 years, SD =2.8). Nine individuals had a history of cancer at the baseline and 6 subjects were diagnosed with any cancer during the follow up. A total of 13 and 7 individuals had mutations in BRCA1 and BRCA2, respectively. There were no cross-sectional associations between levels of PBMC DNA methylation measured by the 3 assays and age at the time of the blood draw (range from 18–84 years). Figure 1 presents the associations of absolute changes in DNA methylation over time for each methylation assays. The mean and SD of within-individual difference for LUMA, LINE-1, and Sat2 was −0.3 (7.8), 0.1 (3.5), and 5.8 (27.6), respectively. There were 19.5% and 13.0% of individuals whose LUMA values decreased or increased by ≥10%. Only 3.9% of individuals had LINE-1 methylation values that decreased or increased by ≥10%. For Sat2, 40% of individuals had values that increased and 26% of individuals had values that decreased more than 10%. Overall, Sat2 methylation increased over time with the Spearman correlation coefficient of r = 0.24 (P = 0.04).
Figure 1.
Overall range of absolute changes in percentage of DNA methylation over time by methylation assay. A, change in LUMA methylation over time (mean = −0.3, SD = 7.8, greatest gain = 31.2; greatest loss = −14.5). B, change in LINE-1 methylation over time (mean = 0.1, SD = 3.5, greatest gain =9.9; greatest loss = −11.2). C, change in Sat2 methylation over time (mean = 5.8, SD = 27.6, greatest gain = 94.4, greatest loss = −72.6).
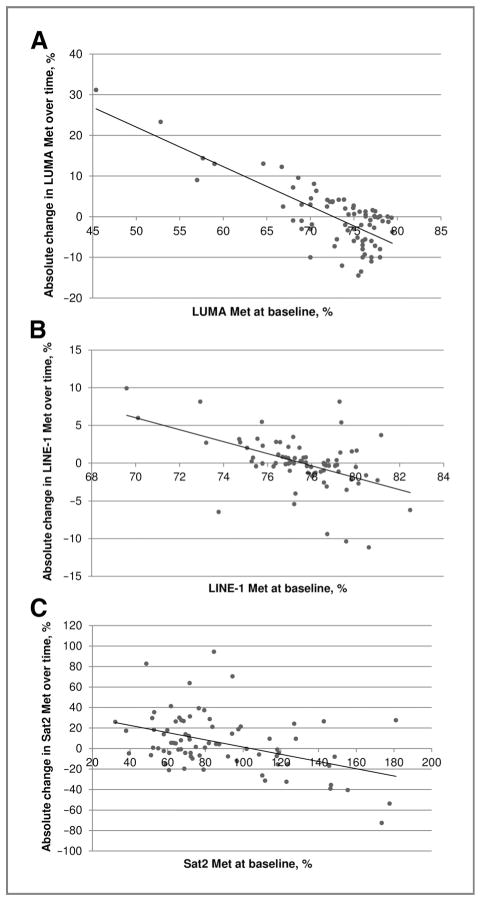
Figure 2 shows the associations between absolute change in each DNA methylation assay with baseline values for that assay. Changes in DNA methylation over time were significantly inversely associated with levels of DNA methylation at baseline. Spearman correlation coefficients were −0.53 (P < 0.0001), −0.46 (P < 0.0001), and −0.35 (P = 0.0002) for LUMA, LINE-1, and Sat2, respectively. Smoking status at baseline was not associated with levels of DNA methylation or change in DNA methylation over time (data not shown).
Figure 2.
Associations between absolute change in DNA methylation by baseline value of DNA methylation marker. A, the correlation of absolute change in LUMA methylation with baseline LUMA methylation (r = −0.53, P < 0.0001). B, the correlation of absolute change in LINE1 methylation with baseline LINE1 methylation (r = −0.46, P < 0.0001). C, the correlation of absolute change in Sat2 methylation with baseline Sat2 methylation (r = −0.35, P = 0.002).
The levels of Sat2 methylation at both time points were statistically lower in subjects with a BRCA1 mutation compared with those without. The mean levels of Sat2 methylation were 64.3 at baseline and 73.7% at follow-up among subjects with a BRCA1 mutation. The corresponding mean levels were 92.3% and 97.4% for subjects without a BRCA1 mutation. The mean levels of Sat2 methylation were 77.0 and 99.2% for those with and without mutations in either gene, respectively (P = 0.001).
The mean levels of LUMA methylation at baseline were 67.9% and 73.7% for subjects with and without a history of cancer at baseline, respectively (P = 0.005). The mean levels of LINE-1 methylation at baseline were higher in subjects with a history of cancer at baseline compared with those without a history of cancer (79.5% vs. 77.3%, P = 0.007). Compared with individuals without any cancer, individuals with either a history of cancer or new cancer during the follow-up period had greater decreases in LINE1 and Sat2 methylation over time. The mean changes in LINE-1 were −3.1% and 0.8% for those with and without cancer, respectively. The corresponding levels of Sat2 were −8.7% with and 9.3% without for those with and without cancer, respectively (P = 0.02).
The associations of change in DNA methylation over time were inversely correlated with baseline levels, with standardized β values of −0.78, −0.41, and −0.43 for LUMA, LINE-1, and Sat2, respectively (Table 1). We examined the same models reported in Table 1 excluding individuals with low values (<60 for LUMA, <74 for LINE-1) and found very similar results and no overall change in inferences (data not shown). In addition, we examined the models reported in Table 1 excluding the 15 individuals with a prior cancer; the strong inverse statistically significant associations with baseline values for each methylation assay remained with standardized β values of −0.71, −0.50, and −0.28 for LUMA, LINE-1, and Sat2, respectively.
Table 1.
Multivariable linear regression models of absolute change in genomic methylation markers by participant characteristics and baseline DNA methylation level
| Variable | Change in LUMA (R2 = 0.58)a
|
Change in LINE-1 (R2 = 0.35)a
|
Change in Sat2 (R2 = 0.23)a
|
|||
|---|---|---|---|---|---|---|
| Standardized β | P | Standardized β | P | Standardized β | P | |
| Gender (male vs. female) | −0.05 | 0.52 | 0.03 | 0.78 | 0.08 | 0.49 |
| Age at baseline | −0.04 | 0.61 | 0.22 | 0.04 | −0.09 | 0.43 |
| Time between blood draws | −0.16 | 0.05 | 0.13 | 0.22 | 0.14 | 0.20 |
| Prior cancer (yes vs. no)b | −0.003 | 0.99 | −0.34 | 0.001 | −0.14 | 0.18 |
| BRCA1/2 mutation (yes vs. no) | −0.06 | 0.45 | 0.04 | 0.68 | −0.17 | 0.11 |
| BMI at baseline | 0.04 | 0.59 | −0.10 | 0.31 | −0.02 | 0.86 |
| Smoking status at baseline (ever vs. never) | −0.02 | 0.79 | 0.02 | 0.82 | 0.01 | 0.90 |
| Baseline methylation | −0.78 | <0.0001 | −0.41 | <0.0001 | −0.43 | 0.0002 |
R2 from multivariable regression model for absolute change in each marker simultaneously adjusted for all variables in the table.
Fifteen individuals had a prior cancer reported at either time 1 or time 2, when we excluded these 15 subjects, the overall inferences were the same; baseline methylation levels remained highly associated with absolute change (see Results).
Discussion
Overall, we observed a large percent of decreases and increases in DNA methylation levels over time. Consistent with another within-individual study reporting on the LUMA assay over time (12), we observed large differences in LUMA levels over time with roughly the same number of participants decreasing as increasing over time. Thus, intraindividual change might be missed if measuring the population average DNA methylation through cross-sectional sampling. We also observed that the number of individuals that had large changes in genomic DNA methylation over time (≥10% changes) differed by assay type with approximately 1/3, 2/3, and only 4% experiencing large changes in LUMA, Sat2, and LINE-1, respectively, over time.
Genomic DNA methylation levels measured in blood DNA have been associated with white blood cell counts and some risk factors of cancer (9, 14). Few studies, however, have examined whether risk factors are related to changes in methylation over time. Comparing within-individual changes over time, we did not observe a clear pattern between age and genomic DNA methylation levels for any of the 3 assays we used. To date, most studies of age and genomic DNA methylation have investigated LINE-1 methylation in cross-sectional studies (4, 8, 9, 14). We found that changes in LINE-1 methylation in PBMC DNA differed between individuals with and without a history of cancer. This is consistent with the lower levels of blood LINE-1 methylation in bladder (3, 4) and head and neck squamous cell (8) but not breast (7) cancer patients. We also found that subjects with a BRCA1 mutation had lower Sat2 methylation levels, although this association did not remain statistically significant in multivariable models adjusting for other factors.
The strongest predictors of absolute change, however, were baseline values of each respective assay. After excluding observations with very low baseline values of LUMA and LINE-1, the overall associations observed in the regression models were similar and the overall inferences the same as before the exclusions (data not shown). Overall the CV for each assay was quite small suggesting that the lack of reliability did not drive our overall findings. For example, the inter-CV for the lowest LUMA value was 2.2. Although we did not repeat the LINE-1 analysis for the samples with low values, the interassay CV for the 5% of samples that were repeated was 0.9, again suggesting that unreliable measures were not a likely explanation for our findings. Storage issues can sometimes explain differences in laboratory assays over time, but we did adjust for time between the blood draws in our analyses. As we observed increases and decreases in methylation levels for each assay over time, it is unclear how storage time may help explain these opposing patterns.
These data, if replicated, suggest that changes in DNA methylation over time vary by assay type and are most associated with baseline values of the assay. We observed greater differences over time in Sat2 measures, a marker that we have found to be associated with breast cancer within families (20), than we did with LUMA and LINE-1, 2 markers that we did not find to be associated with breast cancer within the same families (20, 21). Thus, these findings suggest that assays that change more over time may warrant consideration for studies that measure later life exposures. These findings have implications when designing studies to examine the associations between exposures and blood DNA methylation levels and in examining the association between DNA methylation levels and disease.
Acknowledgments
Grant Support
This work was supported by an award from the Breast Cancer Research Foundation and NIH grants U01 CA69398, 1R01 CA138822, P30 CA13696, and P30 ES009089, by the National Cancer Institute, NIH under RFA # CA-06-503, and through cooperative agreements with members of the Breast Cancer Family Registry and P.I.s.
Footnotes
Disclosure of Potential Conflicts of Interest
The content of this article does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating centers in the CFR, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government or the CFR.
Authors’ Contributions
Conception and design: H.-C. Wu, R.M. Santella, M.B. Terry
Development of methodology: L. Delgado-Cruzata, R.M. Santella, M.B. Terry
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): Q. Wang, R.M. Santella, M.B. Terry
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): H.-C. Wu, R.M. Santella, M.B. Terry
Writing, review, and/or revision of the manuscript: H.-C. Wu, R.M. Santella, M.B. Terry
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): M.B. Terry
References
- 1.Lim U, Flood A, Choi S, Albanes D, Cross A, Schatzkin A, et al. Genomic methylation of leukocyte DNA in relation to colorectal adenoma among asymptomatic women. Gastroenterology. 2008;134:47–55. doi: 10.1053/j.gastro.2007.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pufulete M, Al-Ghnaniem R, Leather A, Appleby P, Gout S, Terry C, et al. Folate status, genomic DNA hypomethylation, and risk of colorectal adenoma and cancer: a case control study. Gastroenterology. 2003;124:1240–8. doi: 10.1016/s0016-5085(03)00279-8. [DOI] [PubMed] [Google Scholar]
- 3.Cash H, Tao L, Yuan J, Marsit C, Houseman E, Xiang Y, et al. LINE-1 hypomethylation is associated with bladder cancer risk among non-smoking Chinese. Int J Cancer. 2012;130:1151–9. doi: 10.1002/ijc.26098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilhelm C, Kelsey K, Butler R, Plaza S, Gagne L, Zens MS, et al. Implications of LINE1 methylation for bladder cancer risk in women. Clin Cancer Res. 2010;16:1682–9. doi: 10.1158/1078-0432.CCR-09-2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moore LE, Pfeiffer RM, Poscablo C, Real FX, Kogevinas M, Silverman D, et al. Genomic DNA hypomethylation as a biomarker for bladder cancer susceptibility in the Spanish Bladder Cancer Study: a case–control study. Lancet Oncol. 2008;9:359–66. doi: 10.1016/S1470-2045(08)70038-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hou L, Wang H, Sartori S, Gawron A, Lissowska J, Bollati V, et al. Blood leukocyte DNA hypomethylation and gastric cancer risk in a high-risk Polish population. Int J Cancer. 2010;127:1866–74. doi: 10.1002/ijc.25190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choi J, James S, Link P, McCann S, Hong C, Davis W, et al. Association between global DNA hypomethylation in leukocytes and risk of breast cancer. Carcinogenesis. 2009;30:1889–1897. doi: 10.1093/carcin/bgp143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ting Hsiung D, Marsit CJ, Houseman EA, Eddy K, Furniss CS, McClean MD, et al. Global DNA methylation level in whole blood as a biomarker in head and neck squamous cell carcinoma. Cancer Epidemiol Biomarkers Prevent. 2007;16:108–114. doi: 10.1158/1055-9965.EPI-06-0636. [DOI] [PubMed] [Google Scholar]
- 9.Terry MB, Delgado-Cruzata L, Vin-Raviv N, Wu H, Santella R. DNA methylation in white blood cells: Association with risk factors in epidemiologic studies. Epigenetics. 2011;6:828–37. doi: 10.4161/epi.6.7.16500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fuke C, Shimabukuro M, Petronis A, Sugimoto J, Oda T, Miura K, et al. Age related changes in 5-methylcytosine content in human peripheral leukocytes and placentas: an HPLC-based study. Ann Hum Genet. 2004;68:196–204. doi: 10.1046/j.1529-8817.2004.00081.x. [DOI] [PubMed] [Google Scholar]
- 11.Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, Ballestar ML, et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci U S Am. 2005;102:10604–9. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bjornsson HT, Sigurdsson MI, Fallin MD, Irizarry RA, Aspelund T, Cui H, et al. Intra-individual change over time in DNA methylation with familial clustering. JAMA. 2008;299:2877–83. doi: 10.1001/jama.299.24.2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bollati V, Schwartz J, Wright R, Litonjua A, Tarantini L, Suh H, et al. Decline in genomic DNA methylation through aging in a cohort of elderly subjects. Mechan Ageing Dev. 2009;130:234–9. doi: 10.1016/j.mad.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu Z-Z, Hou L, Bollati V, Tarantini L, Marinelli B, Cantone L, et al. Predictors of global methylation levels in blood DNA of healthy subjects: a combined analysis. Int J Epidemiol. 2010;41:126–139. doi: 10.1093/ije/dyq154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bollati V, Baccarelli A, Hou L, Bonzini M, Fustinoni S, Cavallo D, et al. Changes in DNA methylation patterns in subjects exposed to low-dose benzene. Cancer Res. 2007;67:876–80. doi: 10.1158/0008-5472.CAN-06-2995. [DOI] [PubMed] [Google Scholar]
- 16.Pilsner J, Liu X, Ahsan H, IIievski V, Slavkovich V, Levy D, et al. Genomic methylation of peripheral blood leukocyte DNA influences of arsenic and folate in Bangladeshi adults. Am J Clin Nutr. 2007;86:1179–86. doi: 10.1093/ajcn/86.4.1179. [DOI] [PubMed] [Google Scholar]
- 17.John E, Hopper J, Beck J, Knight J, Neuhausen S, Senie R, et al. The Breast Cancer Family Registry: an infrastructure for cooperative multinational, interdisciplinary and translational studies of the genetic epidemiology of breast cancer. Breast Cancer Res. 2004;6:R375–89. doi: 10.1186/bcr801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu H, Delgado-Cruzata L, Flom J, Kappil M, Ferris J, Liao Y, et al. Global methylation profiles in DNA from different blood cell types. Epigenetics. 2011;6:76–85. doi: 10.4161/epi.6.1.13391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weisenberger D, Campan M, Long T, Kim M, Woods C, Fiala E, et al. Analysis of repetitive element DNA methylation by MethyLight. Nucleic Acids Res. 2005;33:6823–6836. doi: 10.1093/nar/gki987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu HC, Wang Q, Delgado-Cruzata L, Flom JD, Perrin M, Liao Y, et al. Repetitive element DNA methylation levels in white blood cell DNA from sisters discordant for breast cancer from the New York site of the Breast Cancer Family Registry. Carcinogenesis. 2012 doi: 10.1093/carcin/bgs201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Delgado-Cruzata L, Wu HC, Wang Q, Perrin M, Liao Y, Kappil M, et al. Global DNA methylation levels in white blood cell DNA from sisters discordant for breast cancer from the New York site of the Breast Cancer Family Registry. Epigenetics. 2012;7(8) doi: 10.4161/epi.20830. [DOI] [PMC free article] [PubMed] [Google Scholar]