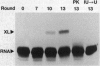
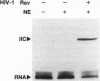
Abstract
We have used an in vitro selection procedure called crosslinking SELEX (SELEX = systematic evolution of ligands by exponential enrichment) to identify RNA sequences that bind with high affinity and crosslink to the Rev protein from human immunodeficiency virus type 1 (HIV-1). A randomized RNA library substituted with the photoreactive chromophore 5-iodouracil was irradiated with monochromatic UV light in the presence of Rev. Those sequences with the ability to photocrosslink to Rev were partitioned from the rest of the RNA pool, amplified, and used for the next round of selection. Rounds of photocrosslinking selection were alternated with rounds of selection for RNA sequences with high affinity to Rev. This iterative, dual-selection method yielded RNA molecules with subnanomolar dissociation constants and high efficiency photocrosslinking to Rev. Some of the RNA molecules isolated by this procedure form a stable complex with Rev that is resistant to denaturing gel electrophoresis in the absence of UV irradiation. In vitro selection of nucleic acids by using modified nucleotides allows the isolation of nucleic acid molecules with potentially limitless chemical capacities to covalently attack a target molecule.
Full text
PDF

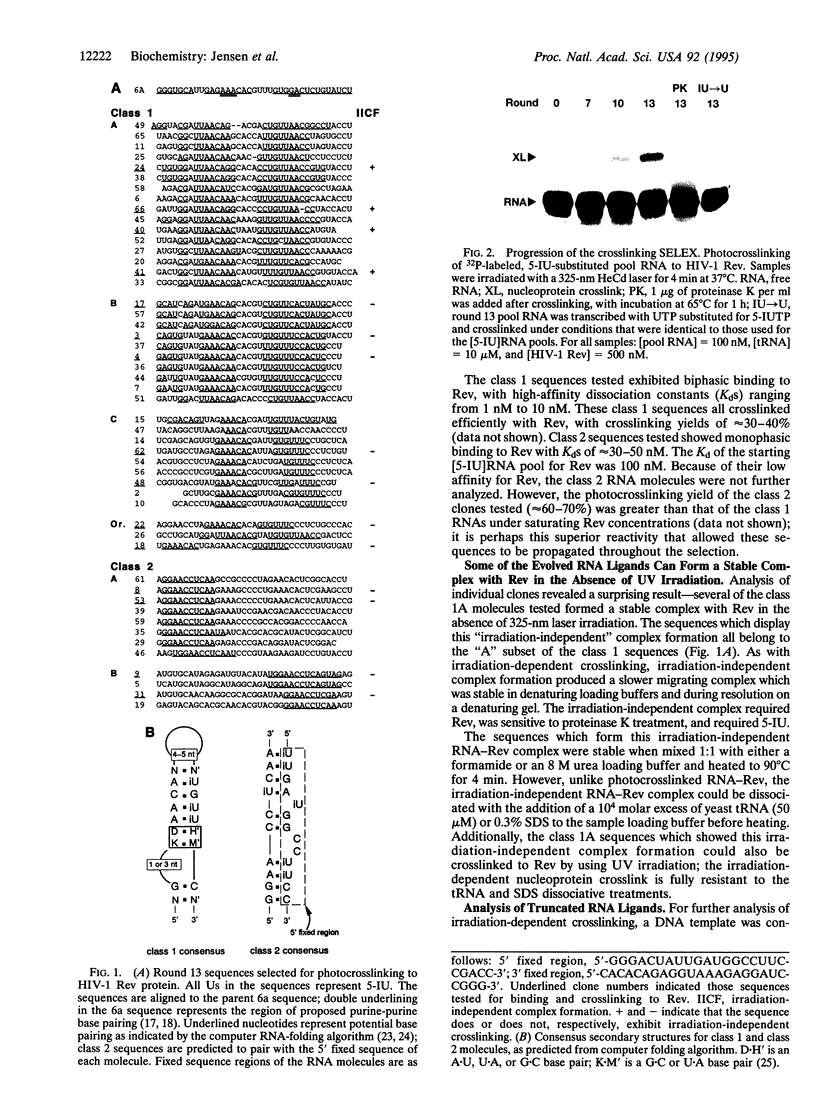
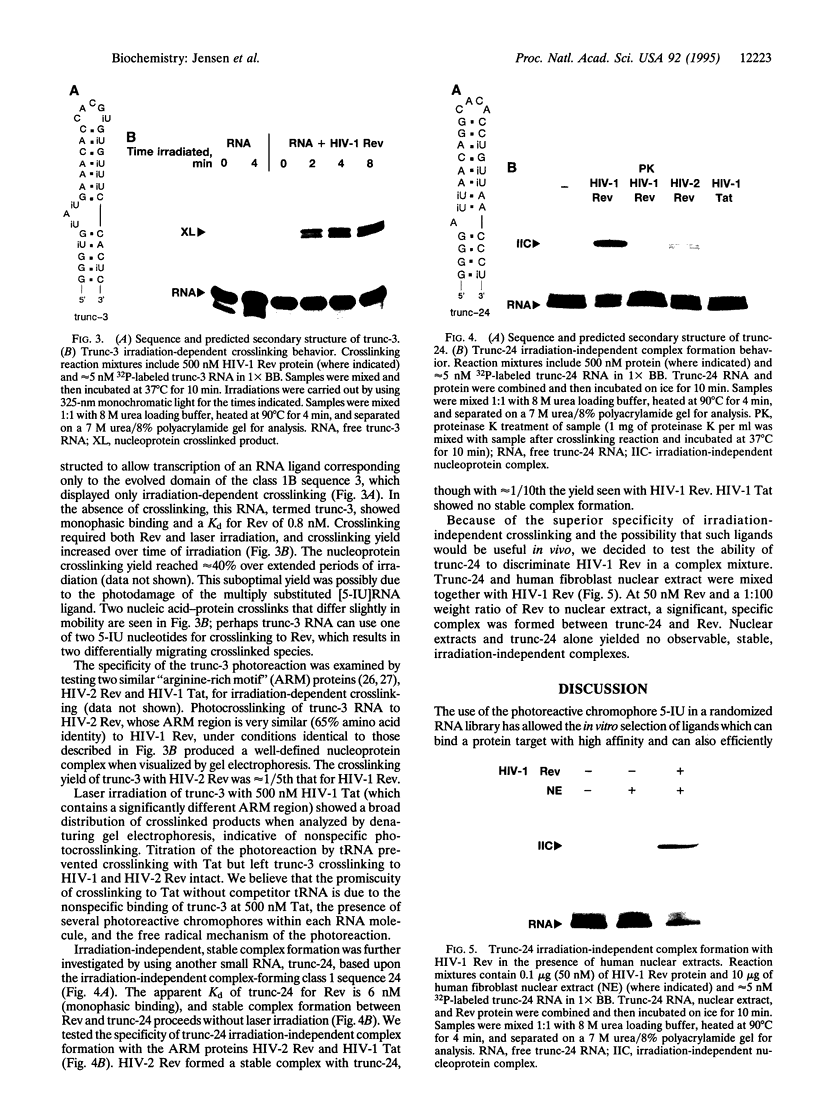

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bartel D. P., Zapp M. L., Green M. R., Szostak J. W. HIV-1 Rev regulation involves recognition of non-Watson-Crick base pairs in viral RNA. Cell. 1991 Nov 1;67(3):529–536. doi: 10.1016/0092-8674(91)90527-6. [DOI] [PubMed] [Google Scholar]
- Battiste J. L., Tan R., Frankel A. D., Williamson J. R. Binding of an HIV Rev peptide to Rev responsive element RNA induces formation of purine-purine base pairs. Biochemistry. 1994 Mar 15;33(10):2741–2747. doi: 10.1021/bi00176a001. [DOI] [PubMed] [Google Scholar]
- Burd C. G., Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994 Jul 29;265(5172):615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- Carey J., Cameron V., de Haseth P. L., Uhlenbeck O. C. Sequence-specific interaction of R17 coat protein with its ribonucleic acid binding site. Biochemistry. 1983 May 24;22(11):2601–2610. doi: 10.1021/bi00280a002. [DOI] [PubMed] [Google Scholar]
- Cook K. S., Fisk G. J., Hauber J., Usman N., Daly T. J., Rusche J. R. Characterization of HIV-1 REV protein: binding stoichiometry and minimal RNA substrate. Nucleic Acids Res. 1991 Apr 11;19(7):1577–1583. doi: 10.1093/nar/19.7.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly T. J., Cook K. S., Gray G. S., Maione T. E., Rusche J. R. Specific binding of HIV-1 recombinant Rev protein to the Rev-responsive element in vitro. Nature. 1989 Dec 14;342(6251):816–819. doi: 10.1038/342816a0. [DOI] [PubMed] [Google Scholar]
- Eaton B. E., Pieken W. A. Ribonucleosides and RNA. Annu Rev Biochem. 1995;64:837–863. doi: 10.1146/annurev.bi.64.070195.004201. [DOI] [PubMed] [Google Scholar]
- Foote J., Eisen H. N. Kinetic and affinity limits on antibodies produced during immune responses. Proc Natl Acad Sci U S A. 1995 Feb 28;92(5):1254–1256. doi: 10.1073/pnas.92.5.1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett C., Wataya Y., Santi D. V. Thymidylate synthetase. Catalysis of dehalogenation of 5-bromo- and 5-iodo-2'-deoxyuridylate. Biochemistry. 1979 Jun 26;18(13):2798–2804. doi: 10.1021/bi00580a017. [DOI] [PubMed] [Google Scholar]
- Giver L., Bartel D., Zapp M., Pawul A., Green M., Ellington A. D. Selective optimization of the Rev-binding element of HIV-1. Nucleic Acids Res. 1993 Nov 25;21(23):5509–5516. doi: 10.1093/nar/21.23.5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold L. Oligonucleotides as research, diagnostic, and therapeutic agents. J Biol Chem. 1995 Jun 9;270(23):13581–13584. doi: 10.1074/jbc.270.23.13581. [DOI] [PubMed] [Google Scholar]
- Gold L., Polisky B., Uhlenbeck O., Yarus M. Diversity of oligonucleotide functions. Annu Rev Biochem. 1995;64:763–797. doi: 10.1146/annurev.bi.64.070195.003555. [DOI] [PubMed] [Google Scholar]
- Heaphy S., Dingwall C., Ernberg I., Gait M. J., Green S. M., Karn J., Lowe A. D., Singh M., Skinner M. A. HIV-1 regulator of virion expression (Rev) protein binds to an RNA stem-loop structure located within the Rev response element region. Cell. 1990 Feb 23;60(4):685–693. doi: 10.1016/0092-8674(90)90671-z. [DOI] [PubMed] [Google Scholar]
- Heaphy S., Finch J. T., Gait M. J., Karn J., Singh M. Human immunodeficiency virus type 1 regulator of virion expression, rev, forms nucleoprotein filaments after binding to a purine-rich "bubble" located within the rev-responsive region of viral mRNAs. Proc Natl Acad Sci U S A. 1991 Aug 15;88(16):7366–7370. doi: 10.1073/pnas.88.16.7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanetich K. M., Santi D. V. 5,6-dihydropyrimidine adducts in the reactions and interactions of pyrimidines with proteins. Prog Nucleic Acid Res Mol Biol. 1992;42:127–156. doi: 10.1016/s0079-6603(08)60575-9. [DOI] [PubMed] [Google Scholar]
- Jaeger J. A., Turner D. H., Zuker M. Improved predictions of secondary structures for RNA. Proc Natl Acad Sci U S A. 1989 Oct;86(20):7706–7710. doi: 10.1073/pnas.86.20.7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jellinek D., Green L. S., Bell C., Janjić N. Inhibition of receptor binding by high-affinity RNA ligands to vascular endothelial growth factor. Biochemistry. 1994 Aug 30;33(34):10450–10456. doi: 10.1021/bi00200a028. [DOI] [PubMed] [Google Scholar]
- Jensen K. B., Green L., MacDougal-Waugh S., Tuerk C. Characterization of an in vitro-selected RNA ligand to the HIV-1 Rev protein. J Mol Biol. 1994 Jan 7;235(1):237–247. doi: 10.1016/s0022-2836(05)80030-0. [DOI] [PubMed] [Google Scholar]
- Karn J., Dingwall C., Finch J. T., Heaphy S., Gait M. J. RNA binding by the tat and rev proteins of HIV-1. Biochimie. 1991 Jan;73(1):9–16. doi: 10.1016/0300-9084(91)90068-c. [DOI] [PubMed] [Google Scholar]
- Lazinski D., Grzadzielska E., Das A. Sequence-specific recognition of RNA hairpins by bacteriophage antiterminators requires a conserved arginine-rich motif. Cell. 1989 Oct 6;59(1):207–218. doi: 10.1016/0092-8674(89)90882-9. [DOI] [PubMed] [Google Scholar]
- Najita L., Sarnow P. Oxidation-reduction sensitive interaction of a cellular 50-kDa protein with an RNA hairpin in the 5' noncoding region of the poliovirus genome. Proc Natl Acad Sci U S A. 1990 Aug;87(15):5846–5850. doi: 10.1073/pnas.87.15.5846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson R. D., Bartel D. P., Szostak J. W., Horvath S. J., Feigon J. 1H NMR studies of the high-affinity Rev binding site of the Rev responsive element of HIV-1 mRNA: base pairing in the core binding element. Biochemistry. 1994 May 10;33(18):5357–5366. doi: 10.1021/bi00184a001. [DOI] [PubMed] [Google Scholar]
- Pritchard C. E., Grasby J. A., Hamy F., Zacharek A. M., Singh M., Karn J., Gait M. J. Methylphosphonate mapping of phosphate contacts critical for RNA recognition by the human immunodeficiency virus tat and rev proteins. Nucleic Acids Res. 1994 Jul 11;22(13):2592–2600. doi: 10.1093/nar/22.13.2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romaniuk P. J., Uhlenbeck O. C. Nucleoside and nucleotide inactivation of R17 coat protein: evidence for a transient covalent RNA-protein bond. Biochemistry. 1985 Jul 16;24(15):4239–4244. doi: 10.1021/bi00336a064. [DOI] [PubMed] [Google Scholar]
- Schneider D., Tuerk C., Gold L. Selection of high affinity RNA ligands to the bacteriophage R17 coat protein. J Mol Biol. 1992 Dec 5;228(3):862–869. doi: 10.1016/0022-2836(92)90870-p. [DOI] [PubMed] [Google Scholar]
- Starzyk R. M., Koontz S. W., Schimmel P. A covalent adduct between the uracil ring and the active site of an aminoacyl tRNA synthetase. Nature. 1982 Jul 8;298(5870):136–140. doi: 10.1038/298136a0. [DOI] [PubMed] [Google Scholar]
- Stump W. T., Hall K. B. Crosslinking of an iodo-uridine-RNA hairpin to a single site on the human U1A N-terminal RNA binding domain. RNA. 1995 Mar;1(1):55–63. [PMC free article] [PubMed] [Google Scholar]
- Tuerk C., Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990 Aug 3;249(4968):505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- Tuerk C., MacDougal-Waugh S. In vitro evolution of functional nucleic acids: high-affinity RNA ligands of HIV-1 proteins. Gene. 1993 Dec 27;137(1):33–39. doi: 10.1016/0378-1119(93)90248-2. [DOI] [PubMed] [Google Scholar]
- Willis M. C., Hicke B. J., Uhlenbeck O. C., Cech T. R., Koch T. H. Photocrosslinking of 5-iodouracil-substituted RNA and DNA to proteins. Science. 1993 Nov 19;262(5137):1255–1257. doi: 10.1126/science.7694369. [DOI] [PubMed] [Google Scholar]
- Zapp M. L., Green M. R. Sequence-specific RNA binding by the HIV-1 Rev protein. Nature. 1989 Dec 7;342(6250):714–716. doi: 10.1038/342714a0. [DOI] [PubMed] [Google Scholar]
- Zuker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989 Apr 7;244(4900):48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]