Abstract
Although the crystalline S-layer arrays that form the exoskeleton of many archaea and bacteria have been studied for decades, a long-awaited crystal structure coupled with a growing understanding of the S-layer assembly process are injecting new excitement in the field. The trend is amplified by computational strategies that allow for in silico design of protein building blocks capable of self-assembling into 2D lattices and other prescribed quaternary structures. We review these and other recent developments towards achieving unparalleled control over the geometry, chemistry and function of protein-based 2D objects from the nano- to the mesoscale.
Introduction
Biological building blocks that self-assemble into predetermined supramolecular structures are of considerable interest in bionanotechnology where an ability to control shape, size, geometry and surface chemistry is crucial to the production of advanced materials with tailored properties. Predictive control of shape has been particularly effective with nucleic acids where a variety of one, two and three-dimensional (3D) nanostructures have been produced via strand exchange and DNA Origami technologies [1]. Peptides and peptoids (polymers whose constituent monomers resemble amino acids but have side chains appended to the amide nitrogen rather than to the α carbon) have also been engineered to assemble into 2D structures [2–4]. Compared to these molecules, proteins offer a richer and more versatile structural, chemical and functional palette that can be further expanded through rational design, selection and directed evolution.
Two-dimensional (2D) protein arrays are of particular interest in bionanotechnology because they allow for the high-density display of peptides and proteins in sensor, diagnostic and vaccine applications. They also enable the periodic organization (or templating) of inorganic particles with nanoscale control of position for plasmonic, opto-electronic, magnetic and catalytic applications. In nature, 2D protein arrays are only found in the purple membrane patches of Halobacteria species [5], and the surface (S-) layer exoskeleton of nearly all archaea and many bacteria [6]. Here we will not discuss the purple membrane – a crystalline assembly consisting of trimers of bacteriorhodopsin tightly packed in a lipid-containing hexagonal array – because its structure and potential for optical applications have been reviewed elsewhere [7,8]. Instead, we focus this review on recent developments in our understanding of S-layer structure-function relationship and on progress in the computational design of entirely new kinds of protein arrays.
S-layer structure
S-layers are monomolecular lattices of (glyco)proteins that encapsulate certain bacteria and archaea and connect to the cell surface through one or several N-terminal glycan-binding domains. Their function ranges from protective coating, cell adhesion, surface recognition, molecular sieving and ion trapping, to scaffolding for enzymes and virulence factors [6,9]. Slayers are 5-to-20 nm thick in bacteria and up to 70-nm thick in archaea. They have a smooth, hydrophobic outer surface with net neutral charge and a corrugated inner surface that tends to be hydrophilic and carries either a net negative or positive charge [10]. Individual S-layer proteins have molecular masses between 40 and 200-kDa and form morphological units composed of one, two, three, four of six subunits which assemble with oblique (p1, p2), square (p4), or hexagonal (p3, p6) 2D rotational symmetries (Fig. 1A–B) [10]. Center-to-center unit spacing ranges between ≈ 5 and 30 nm and two or more classes of 2-to-6 nm pores typically perforate the array.
Fig. 1.
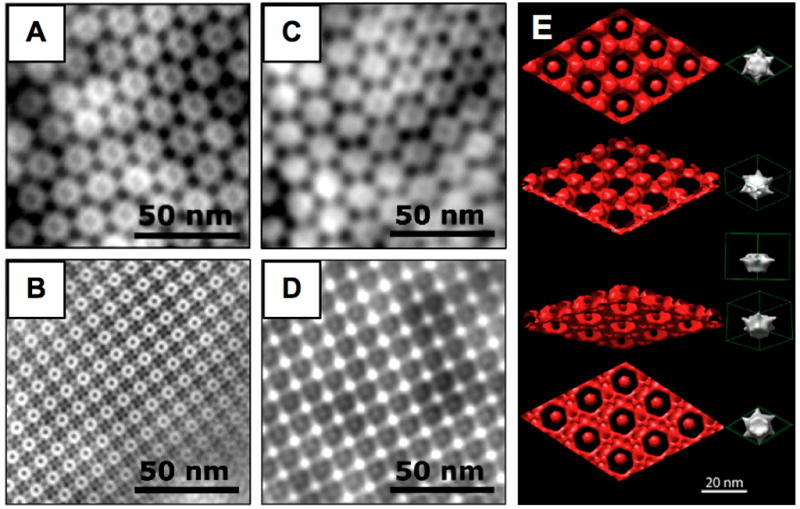
TEM images of negatively stained D. radiodurans (A) and S ureae (B) S-layers and of the corresponding electrodeposited Cu2O films (C,D). (E) TEM-based 3D reconstruction of nanostructured Cu2O (red). Four different angles are shown along with a protein unit cell (right). Panel E is reprinted with permission from reference [15].
Technological uses
Crystalline patches of S-layer proteins can be stripped from bacteria and archaea via detergent extraction, or by using other agents that disrupt their interaction with the cell wall, and directly used for practical applications [11]. In some cases, S-layer proteins can be expressed in heterologous hosts such as E. coli, unfolded by GuHCl or urea treatment, and re-assembled by dilution or dialysis [6]. Recrystallization from unfolded subunits is most reliably performed at the air-water interface using a Langmuir-Blodgett trough, but is also possible on the surface of zwitterionic lipids and certain technologically-relevant substrates such as silicon, carbon and metals. Reassembled S-layers are a mosaic of well-ordered domains that range in size from about 100 nm to 1–2 μm. However, interdomain dislocations and gaps are not uncommon. The reassembly process is influenced by protein concentration, buffer composition, identity of the surface or interface onto which the array is reassembled, and nature and concentration of added divalent cations, which can induce reassembly transitions from sheets, to cylinders, to morphologically poorly defined structures [12].
S-layers have been evaluated for a myriad of applications including: ultrafiltration membranes; drug delivery systems; scaffolds for immunogen displays; and substrates for the spatial organization of functional molecules, metals, and semiconducting nanoparticles (for recent reviews, see references [13•] [9,11]). Because they have a large void content (30–70% porosity), S-layers can also be used to template the synthesis of inorganic structures conformational to the geometry of the pores. Examples include the precipitation of CdS nanoparticles within the pores of the B. stearothermophilus S-layer via solution chemistry [14] and work from our own laboratories showing that the D. radiodurans and S. ureae S-layers can be used as a resist to template the electrodeposition of a broad range of materials including Pt, Ni, Co and Cu2O (Fig. 1C–E) [15,16].
Although such inorganic nanostructures hold promise for catalysis and opto-electronics applications, they are difficult to produce at large scales with the current S-layer “patch” technology. Furthermore, the presence of two or more types of pores in an array means that structures of distinct size and shape are also generated. This could be an enormous advantage if one could fill them with two separate materials. However, achieving this –let alone mineralizing crystalline materials within the confined environment provided by the pores– has so far proven elusive. Other S-layer idiosyncrasies, (e.g., the fact that inner and outer faces have different topography and chemistry) may complicate the control of surface interactions and interfere with in-registry stacking, thus making the production of 3D structures challenging.
Insights from the SbsB crystal structure
Although Baumeister and coworkers produced outstanding low-resolution TEM reconstructions of S-layers in the mid-eighties [17,18], X-ray crystallography attempts have long been thwarted by the difficulty of obtaining 3D crystals from proteins that evolved to self-assemble into 2D lattices [19••]. As a result, topological information has been obtained the hard way. For instance, in a bid to identify surface-exposed amino acids in the 98-kDa Geobacillus stearothermophilus SbsB protein, Howorka and coworkers created 75 cysteine substitution mutants and screened the solvent accessibility of these residues in both the monomeric, dimeric and assembled forms of the protein [20,21]. This heroic effort established that amongst 23 residues that were highly accessible in the monomer, 8 were interfacial, 10 were located on the inner face of the S-layer and 5 were on its outer face.
The long-awaited crystal structure of an S-layer protein (Geobacillus stearothermophilus SbsB) has validated this work and provided major insights on the mechanism by which S-layers form [19••]. SbsB consists of an unresolved cell-wall-attachment domain followed by six immunoglobulin-like domains organized into a disk shaped like the Greek letter ϕ [19••]. Four Ca2+ ions stabilize inter- and intra-domain contacts and are critical for quaternary structure acquisition (Fig. 2A). Indeed, unfolded SbsB only reassembles into an array if the buffer is supplemented with calcium and, while individual domains retain secondary structure upon EDTA treatment, all quaternary structure is lost [19••]. In short, Ca2+ binding induces a structural transition that alters the conformation of SbsB monomers from extended to a ϕ-shaped structure that is assembly-competent. The main contacts stabilizing the oblique (p1) lattice are between domains IV and VII and domains II and IV of neighboring subunits and they bury comparable surface area (450 Å2 and 490 Å2, respectively; Fig. 2A). Interestingly, domain II, which orients the cell wall-binding domain I towards the cell side, appears to be able to adopt multiple conformations in order to better handle topological defects on the cell surface. Overall, the SbsB structure provides a good explanation for the essential role of calcium in the S-layer assembly process and valuable insights for S-layer engineering.
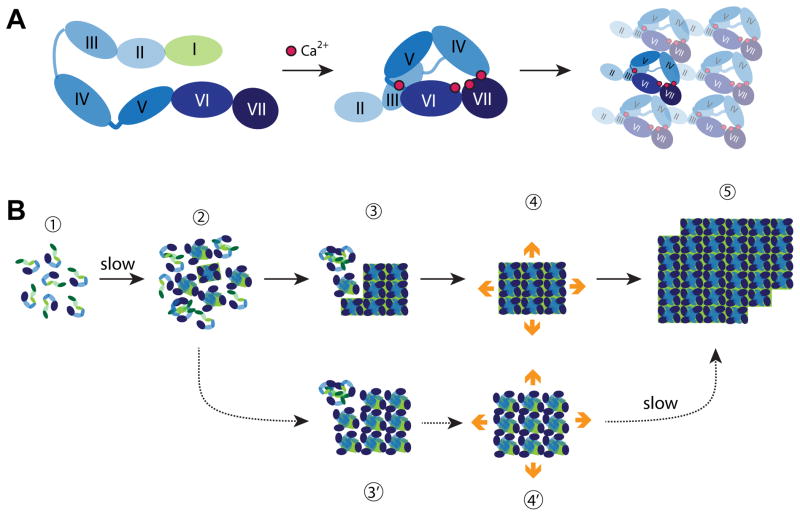
Fig. 2.
Building S-layers. (A) Schematic structure and assembly pathway of the G. stearothermophilus SbsB S-layer protein. The cell-wall attachment domain (light green) and six consecutive immunoglobulin-like domains (shades of blue) are schematically depicted in the extended monomer structure (left). The four calcium ions stabilizing intra- and inter-domain contacts are shown as red spheres in the assembly-competent monomer (middle). The cell-wall attachment domain is not resolved in the crystal structure and thus not shown. Structure of the oblique array formed upon assembly of compact monomers viewed from the outside of the cell (right). All drawings are based on reference [19••]. (B) Growth of L. sphaericus SbpA at interfaces. Monomers are believed to consist of three cell wall anchoring domains (green) followed by a calcium binding domain and three C-terminal immunoglobulin-like domains (shades of blue) [24]. Steps involved in S-layer crystallization on supported lipid bilayers are indicated with solid arrows and labeled 1 to 5. S-layers are viewed from their external side. Mica can stabilize a subpopulation of less compact but yet crystalline clusters (3′ and 4′) that eventually convert to the compact and lower energy form (step 5). All drawings based on references [22••] and [23].
Reassembly at surfaces
Recent in situ AFM imaging studies of the assembly of Lysinibacillus sphaericus SbpA at interfaces [22••] [23] have provided complementary insights on the mechanism by which Slayers nucleate and grow. SbpA is a 132-kDa protein that readily self-assembles into lattices exhibiting square (p4) symmetry and composed of tetrameric morphological units. As with SbsB, there is evidence that SbpA protomers have different conformations in monomeric and tetrameric states [24] and lattice self-assembly depends upon calcium addition. The assembly of SbpA on supported lipid bilayers is schematically depicted in Fig. 2B and involves the following steps: (i) adsorption of monomers in an extended conformation from the bulk solution and onto the surface (step 1); (ii) nucleation of amorphous clusters that are uniform in height in a process that is slow because it requires significant adsorbed protein surface coverage (step 2); (iii) comparably rapid (5–10 min) rearrangement into more compact crystalline clusters composed of SbsB tetramers (step 3); and (iv) extension of the clusters in all directions of the plane by addition of tetramers at crystalline cluster edges with a preference for sites that have a large number of nearest neighbors (steps 3 to 5) [22••]. Of particular interest, new tetramers were only detected at the perimeter of growing clusters. This suggests that interactions with the crystalline phase converts vicinal monomers and/or loosely folded oligomers (dimers and tetramers?) into compact tetramers that can be incorporated into the growing lattice. In a separate study of the reassembly of the same protein on mica [23], De Yoreo and coworkers were able to detect kinetically trapped clusters that are crystalline in nature but more loosely packed than the lower energy compact arrays that they eventually transform into (Fig. 1B, steps 3′ to 5) [23]. While these studies tell us little about the process of S-layer formation in vivo, they have strong practical implications. Indeed, understanding the mechanics of the crystallization process and how it is influenced by the interface will be key to the production of uniform structures exhibiting nanoscale periodicity and very long-range order.
S-layer protein engineering
Because no crystal structure of a S-layer protein was available until recently [19••], most protein engineering efforts have been conducted by trial and error and have focused on a limited number of candidates [13•]. A fragment identified by deletion analysis and spanning the first 1037 amino acids of mature L. sphaericus SbpA (SbpA31–1068) [25], has been especially useful as an N-terminal fusion partner because it remains capable of self-assembling in a crystalline lattice of p4 symmetry when various peptides and proteins are fused to it [13•]. This has been exploited to build immobilization platforms, sensor heads and immunogens in which the fused detection moiety, ligand, enzyme or antigen/allergen is displayed with high density and, in a repetitive geometry on the reconstituted crystal [13•]. Interestingly, a fusion protein between SbpA31–1068 and a short tag of sequence GSLCTPSRLEHHHHHH was recently reported to form bilayers of stacked monolayers [26]. When considering this result with the fact that truncation of 150 additional residues from the SbpA31–1068 C-terminus converts the lattice space group symmetry from square to oblique [25], it becomes clear that modifications at the C-terminus can have quite unexpected consequences on quaternary structure.
Beyond S-layers: artificial 2D protein arrays
Although the development of protein building blocks that self-assemble into well-defined architectures lags behind DNA nanotechnology, progress in the computational design of protein-protein interfaces has accelerated over the past few years [27,28]. For example, interfacial metal coordination [29,30], disulfide bonds [31], novel α-helical coiled-coil [32–34] and β-strands [35], and fusions between domains derived from distinct oligomeric proteins [36–38] have all been exploited to create new quaternary structures. Even more exciting is the recent success in using computational approaches to design entirely new interfaces and orthogonal protein pairs in which weak interactions collectively drive higher order structure formation with the possibility of exquisite control over position and orientation [39–41] [42••].
An attractive alternative to the engineering of natural S-layer proteins is, therefore, to computationally design protein building blocks that self-assemble into extended periodic arrays. One approach to do so is inspired builds on early work by the Yeates lab [37] and relies on the construction of genetic fusions between peptide chains derived from separate oligomeric proteins that exhibit rotational symmetry axes of equal order [43]. As an example, Sinclair et al. [38•] fused the Streptag octopeptide to the C-terminus of E. coli aminolevulinic acid dehydrogenase (ALAD; a tetrameric protein) and added Streptavidin to the purified protein to “stitch” a 2D lattice (Fig. 3A). The same group fused the Lac Repressor-derived Lac21E and Lac21K peptides [44] to ALAD and exploited the formation of coiled-coils to assemble a 2D lattice (Fig. 3B). These groundbreaking results were mitigated by the fact that the resulting structures lacked long-range order and were as small as a few hundred nanometers for the Lac21 fusions and at most a few micrometers for Strepavidin-assembled lattices. Furthermore, the material produced was reportedly sensitive to heat and small molecule inhibitors.
Fig. 3.
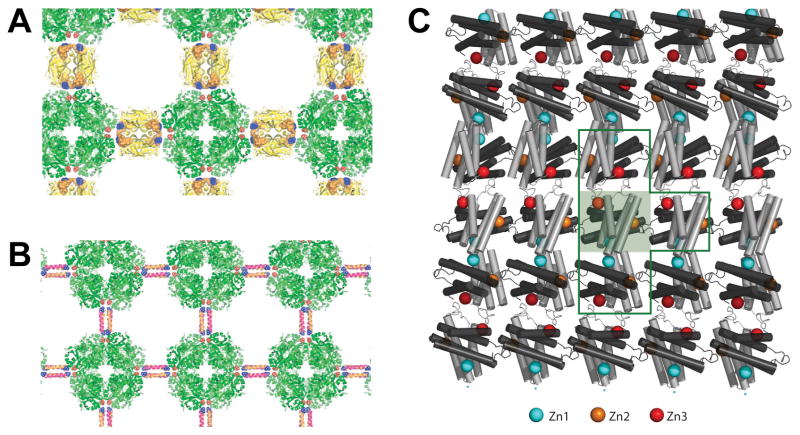
Schematic representation of 2D lattices generated by (A) fusing a Streptag to ALAD (green) and using Streptavidin (yellow) to stitch the structure; or (B) ALAD-Lac21E and ALAD-Lac21K fusions to assemble the array via coiled-coil interactions. Adapted from reference [38•] with permission. (C) Molecular arrangement of 2D sheets obtained with the RIDC3 derivative of cyt cb562 in the presence of zinc. The shaded green box contains a single C2-dimer. Three Zn coordination environments enable 2D self assembly. The Zn1 (blue) and Zn2 (orange) sites are formed by the high-affinity coordination motif and the Zn3 site (red) by the low-affinity coordination motif. Adapted from reference [29•] with permission.
An alternative pioneered by the Tezcan group relies on engineering metal coordination sites at the surface of proteins for cation-driven assembly of multidimensional structures [30]. Recently, Brodin and coworkers [29•] computationally redesigned the surface of cytochrome cb562 (cyt cb562, a four-helix bundle heme protein) to allow for the formation of C2-symmetric dimers through a high affinity Zn2+-binding site, while simultaneously allowing for dimer self-assembly in two perpendicular directions via a low affinity Zn2+-binding site (Fig. 3C). By controlling the Zn2+ to protein ratio and the protonation state of the zinc-coordinating histidine residues, the team showed that it was possible to form 2D nanoplates under conditions of slow nucleation (low pH and/or zinc) or helical nanotubes under conditions favoring fast nucleation events (high pH or low pH and high zinc concentration). Protein sheets were observed upon reordering of precipitates after 5 to 7 days of incubation. These sheets consisted of non-perforated squares or rectangles with characteristic dimensions varying between 5 and 10 μm. Whether this engineered protein (called RIDC3) can be further manipulated to incorporate guest peptides or fusion partners without losing its ability to assemble remains unclear, as is the robustness and scalability of the self-assembly process.
Conclusion and the road ahead
The availability of an S-layer protein crystal structure combined with recent advances in computational structural biology has opened the door to a two-pronged approach for the production of next generation 2D protein arrays. More than four decades of S-layer work by Uwe Sleytr and other laboratories all but guarantees that the SbsB structure (and hopefully that of other S-layer proteins) will be exploited to optimize the location of fusion joints and help fine-tune existing designs for robustness and usefulness in real world settings. However, an incomplete understanding of the protomer folding and assembly pathway might frustrate complex redesign attempts.
Computationally designed protein arrays remain in their infancy but they offer tremendous opportunities for engineering any and all desirable features on a nearly blank slate. These include lattice periodicity and symmetry; location of insertion and fusion joints; number, size, shape and chemistry of the pores; and corrugation, composition and electrostatics of “top” and “bottom” surfaces. Exciting short-term prospects include the redesign of protein interfacial contacts to produce building blocks that self-assemble via collective interactions and with atomic-level accuracy, the design of lattices that exhibit arbitrarily specified properties with nanoscale periodicity through the use of multiple such blocks, and the construction of 3D structures made up of stacked 2D arrays.
With artificial arrays, it should also be possible to take full advantage of the promise of solid binding peptides (SBPs), which are short sequences of amino acids that have been selected by combinatorial display for an ability to bind to inorganic or synthetic materials [45,46]. Insertion of SBPs at computationally defined, solvent-exposed locations should provide unparalleled control over array immobilization, orientation, and conceivably actuation, on virtually any surface(s) or interface(s) of technological interest. Because certain SBPs have the ability to promote inorganic mineralization [47], their incorporation within the architecture of custom-designed pores could be used to manufacture inorganic nanostructures of predictable size, shape, composition, and perhaps crystallinity over extended distances. Finally, it should be easier to access orthogonal chemistries for array derivatization by encoding non-natural amino acids in computationally designed protomers. (Although this could also be done with recombinant Slayer proteins, and especially when using new expression hosts [48], genetic instability and low-level expression issues might complicate the prospect).
Whether based on S-layers or computationally designed, 2D protein arrays face similar challenges. To make a technological difference, it will be necessary to reduce expression and purification costs and to develop robust technologies for the assembly and manipulation of very large lattices that retain order at the nanoscale. The size needed will of course depend on the application For instance, whereas mosaicity should be acceptable in the case of catalytic and structural nanoarrays, long-range order in the hundreds of micrometers to millimeters might be desirable for photonic, plasmonic and magnetic applications. The production of such defect-free crystals will likely require a detailed understanding of how environmental and process conditions influence assembly and growth. It will also be important to identify application spaces where protein-based 2D structures offer unique advantages over those fabricated by chemical self-assembly and top-down writing and patterning processes. It will be even more interesting to integrate these technologies in the pursuit of disruptive materials, systems and devices [49].
Highlights.
S-layer structure, engineering and technological use are briefly reviewed
New developments in S-layer protein structure and assembly are presented
Computational approaches to 2D protein array design are described
Forthcoming developments are discussed
Acknowledgments
JFM gratefully acknowledges financial support from NIH trough a Cancer Nanotechnology Training Grant (T32CA138312). This work was supported in part by the Office of Naval Research (BRC-11123566).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Seeman NC. Nanomaterials based on DNA. Annu Rev Biochem. 2010;79:65–87. doi: 10.1146/annurev-biochem-060308-102244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kudirka R, Tran H, Sanii B, Nam KT, Choi PH, Venkateswaran N, Chen R, Whitelam S, Zuckermann RN. Folding of a single-chain, information-rich polypeptoid sequence into a highly ordered nanosheet. Biopolymers. 2011;96:586–595. doi: 10.1002/bip.21590. [DOI] [PubMed] [Google Scholar]
- 3.Nam KT, Shelby SA, Choi PH, Marciel AB, Chen R, Tan L, Chu TK, Mesch RA, Lee BC, Connolly MD, et al. Free-floating ultrathin two-dimensional crystals from sequence-specific peptoid polymers. Nat Mater. 2010;9:454–460. doi: 10.1038/nmat2742. [DOI] [PubMed] [Google Scholar]
- 4.Woolfson DN, Mahmoud ZN. More than just bare scaffolds: towards multi-component and decorated fibrous biomaterials. Chem Soc Rev. 2010;39:3464–3479. doi: 10.1039/c0cs00032a. [DOI] [PubMed] [Google Scholar]
- 5.Blaurock AE, Stoeckenius W. Structure of the purple membrane. Nat New Biol. 1971;233:152–155. doi: 10.1038/newbio233152a0. [DOI] [PubMed] [Google Scholar]
- 6.Sara M, Sleytr UB. S-layer proteins. J Bacteriol. 2000;182:859–868. doi: 10.1128/jb.182.4.859-868.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cartailler JP, Luecke H. X-ray crystallographic analysis of lipid-protein interactions in the bacteriorhodopsin purple membrane. Annu Rev Biophys Biomol Struct. 2003;32 :285–310. doi: 10.1146/annurev.biophys.32.110601.142516. [DOI] [PubMed] [Google Scholar]
- 8.Hampp N. Bacteriorhodopsin as a photochromic retinal protein for optical memories. Chehm Rev. 2000;100:1755–1776. doi: 10.1021/cr980072x. [DOI] [PubMed] [Google Scholar]
- 9.Sleytr UB, Huber C, Ilk N, Pum D, Schuster B, Egelseer EM. S-layers as a tool kit for nanobiotechnological applications. FEMS Microbiol Lett. 2007;267:131–144. doi: 10.1111/j.1574-6968.2006.00573.x. [DOI] [PubMed] [Google Scholar]
- 10.Howorka S. Rationally engineering natural protein assemblies in nanobiotechnology. Curr Opin Biotechnol. 2011;22:485–491. doi: 10.1016/j.copbio.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 11.Pum D, Toca-Herrera JL, Sleytr UB. S-layer protein self-assembly. Int J Mol Sci. 2013;14 :2484–2501. doi: 10.3390/ijms14022484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Messner P, Pum D, Sleytr UB. Characterization of the ultrastructure and the self- assembly of the surface layer of Bacillus stearothermophilus strain NRS 2004/3a. J Ultrastruct Mol Struct Res. 1986;97:73–88. doi: 10.1016/s0889-1605(86)80008-8. [DOI] [PubMed] [Google Scholar]
- 13•.Ilk N, Egelseer EM, Sleytr UB. S-layer fusion proteins - construction principles and applications. Curr Opin Biotechnol. 2011;22:824–831. doi: 10.1016/j.copbio.2011.05.510. A thorough and informative review of the broad range of applications in which S-layer fusion proteins have been used. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shenton W, Pum D, Sleytr UB, Mann S. Synthesis of cadmium sulphide superlattices using self-assembled bacterial S-layers. Nature. 1997;389:585–587. [Google Scholar]
- 15.Allred DB, Cheng A, Sarikaya M, Baneyx F, Schwartz DT. 3D architecture of inorganic nanoarrays electrodeposited through an S-layer protein mask. Nano Lett. 2008;8 :1434–1438. doi: 10.1021/nl0803444. [DOI] [PubMed] [Google Scholar]
- 16.Allred DB, Sarikaya M, Baneyx F, Schwartz DT. Electrochemical nanofabrication using crystalline protein masks. Nano Lett. 2005;5:609–613. doi: 10.1021/nl047967b. [DOI] [PubMed] [Google Scholar]
- 17.Wildhaber I, Baumeister W. The cell envelope of Thermoproteus tenax: three-dimensional structure of the surface layer and its role in shape maintenance. EMBO J. 1987;6:1475–1480. doi: 10.1002/j.1460-2075.1987.tb02389.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wildhaber I, Santarius U, Baumeister W. Three-dimensional structure of the surface protein of Desulfurococcus mobilis. J Bacteriol. 1987;169:5563–5568. doi: 10.1128/jb.169.12.5563-5568.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19••.Baranova E, Fronzes R, Garcia-Pino A, Van Gerven N, Papapostolou D, Pehau-Arnaudet G, Pardon E, Steyaert J, Howorka S, Remaut H. SbsB structure and lattice reconstruction unveil Ca2+ triggered S-layer assembly. Nature. 2012;487:119–122. doi: 10.1038/nature11155. The first crystal structure of an S-layer protein was obtained using nanobodies (single domain antibodies) as crystallization chaperones to break the propensity of SbsB to polymerize and to promote the formation of 3D crystals. This strategy will likely prove useful to solve the structure of additional S-layer proteins. [DOI] [PubMed] [Google Scholar]
- 20.Howorka S, Sara M, Wang Y, Kuen B, Sleytr UB, Lubitz W, Bayley H. Surface-accessible residues in the monomeric and assembled forms of a bacterials surface layer protein. J Biol Chem. 2000;275:37876–37886. doi: 10.1074/jbc.M003838200. [DOI] [PubMed] [Google Scholar]
- 21.Kinns H, Howorka S. The surface location of individual residues in a bacterial S-layer protein. J Mol Biol. 2008;377:589–604. doi: 10.1016/j.jmb.2008.01.019. [DOI] [PubMed] [Google Scholar]
- 22••.Chung S, Shin SH, Bertozzi CR, De Yoreo JJ. Self-catalyzed growth of S-layers via an amorphous-to-crystalline transition limited by folding kinetics. Proc Natl Acad Sci U S A. 2010;107:16536–16541. doi: 10.1073/pnas.1008280107. This landmark study of the SbpA assembly process on supported lipid bilayers shows that S-layer growth is catalytic and challenges the assumption of rapid exchange of material between growing phase and bulk solution in classic nucleation theory. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shin SH, Chung S, Sanii B, Comolli LR, Bertozzi CR, De Yoreo JJ. Direct observation of kinetic traps associated with structural transformations leading to multiple pathways of S-layer assembly. Proc Natl Acad Sci U S A. 2012;109:12968–12973. doi: 10.1073/pnas.1201504109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horejs C, Gollner H, Pum D, Sleytr UB, Peterlik H, Jungbauer A, Tscheliessnig R. Atomistic structure of monomolecular surface layer self-assemblies: toward functionalized nanostructures. ACS Nano. 2011;5:2288–2297. doi: 10.1021/nn1035729. [DOI] [PubMed] [Google Scholar]
- 25.Huber C, Ilk N, Rünzler D, Egelseer EM, Weigert S, Sleytr UB, Sara M. The three S-layer-like homology motifs of the S-layer protein SbpA of Bacillus sphaericus CCM 2177 are not sufficient for binding to the pyruvylated secondary cell wall polymer. Mol Microbiol. 2005;55:197–205. doi: 10.1111/j.1365-2958.2004.04351.x. [DOI] [PubMed] [Google Scholar]
- 26.Shin SH, Comolli LR, Tscheliessnig R, Wang C, Nam KT, Hexemer A, Siegerist CE, De Yoreo JJ, Bertozzi CR. Self-assembly of “S-bilayers”, a step towards expanding the dimensionality of S-layer assemblies. ACS Nano. 2013;7:4946–4953. doi: 10.1021/nn400263j. [DOI] [PubMed] [Google Scholar]
- 27.Karanicolas J, Kuhlman B. Computational design of affinity and specificity at protein-protein interfaces. Curr Opin Struct Biol. 2009;19:458–463. doi: 10.1016/j.sbi.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mandell DJ, Kortemme T. Computer-aided design of functional protein interactions. Nat Chem Biol. 2009;5:797–807. doi: 10.1038/nchembio.251. [DOI] [PubMed] [Google Scholar]
- 29•.Brodin JD, Ambroggio XI, Tang C, Parent KN, Baker TS, Tezcan FA. Metal-directed, chemically tunable assembly of one-, two- and three-dimensional crystalline protein arrays. Nat Chem. 2012;4:375–382. doi: 10.1038/nchem.1290. A monomeric protein is redesigned with metal coordination motifs and interfacial mutations for Zn2+-controlled assembly of crystalline arrays. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Salgado EN, Ambroggio XI, Brodin JD, Lewis RA, Kuhlman B, Tezcan FA. Metal templated design of protein interfaces. Proc Natl Acad Sci U S A. 2010;107:1827–1832. doi: 10.1073/pnas.0906852107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Medina-Morales A, Perez A, Brodin JD, Tezcan FA. In vitro and cellular self-assembly of a Zn-binding protein cryptand via templated disulfide bonds. J Am Chem Soc. 2013;135:12013–12022. doi: 10.1021/ja405318d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lanci CJ, MacDermaid CM, Kang SG, Acharya R, North B, Yang X, Qiu XJ, DeGrado WF, Saven JG. Computational design of a protein crystal. Proc Natl Acad Sci U S A. 2012;109:7304–7309. doi: 10.1073/pnas.1112595109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McAllister KA, Zou H, Cochran FV, Bender GM, Senes A, Fry HC, Nanda V, Keenan PA, Lear JD, Saven JG, et al. Using alpha-helical coiled-coils to design nanostructured metalloporphyrin arrays. J Am Chem Soc. 2008;130:11921–11927. doi: 10.1021/ja800697g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zaccai NR, Chi B, Thomson AR, Boyle AL, Bartlett GJ, Bruning M, Linden N, Sessions RB, Booth PJ, Brady RL, et al. A de novo peptide hexamer with a mutable channel. Nat Chem Biol. 2011;7:935–941. doi: 10.1038/nchembio.692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stranges PB, Machius M, Miley MJ, Tripathy A, Kuhlman B. Computational design of a symmetric homodimer using beta-strand assembly. Proc Natl Acad Sci U S A. 2011;108:20562–20567. doi: 10.1073/pnas.1115124108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lai YT, Cascio D, Yeates TO. Structure of a 16-nm cage designed by using protein oligomers. Science. 2012;336:1129. doi: 10.1126/science.1219351. [DOI] [PubMed] [Google Scholar]
- 37.Padilla JE, Colovos C, Yeates TO. Nanohedra: using symmetry to design self assembling protein cages, layers, crystals, and filaments. Proc Natl Acad Sci U S A. 2001;98:2217–2221. doi: 10.1073/pnas.041614998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38•.Sinclair JC, Davies KM, Venien-Bryan C, Noble ME. Generation of protein lattices by fusing proteins with matching rotational symmetry. Nat Nanotechnol. 2011;6:558–562. doi: 10.1038/nnano.2011.122. This study articulates a set of symmetry-based design rules for the production of 1-, 2- and 3D protein arrays with practical demonstration of two approaches for the generation of 2D lattices. [DOI] [PubMed] [Google Scholar]
- 39.Grueninger D, Treiber N, Ziegler MO, Koetter JW, Schulze MS, Schulz GE. Designed protein-protein association. Science. 2008;319:206–209. doi: 10.1126/science.1150421. [DOI] [PubMed] [Google Scholar]
- 40.Karanicolas J, Corn JE, Chen I, Joachimiak LA, Dym O, Peck SH, Albeck S, Unger T, Hu W, Liu G, et al. A de novo protein binding pair by computational design and directed evolution. Mol Cell. 2011;42:250–260. doi: 10.1016/j.molcel.2011.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fleishman SJ, Whitehead TA, Ekiert DC, Dreyfus C, Corn JE, Strauch EM, Wilson IA, Baker D. Computational design of proteins targeting the conserved stem region of influenza hemagglutinin. Science. 2011;332:816–821. doi: 10.1126/science.1202617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42••.King NP, Sheffler W, Sawaya MR, Vollmar BS, Sumida JP, Andre I, Gonen T, Yeates TO, Baker D. Computational design of self-assembling protein nanomaterials with atomic level accuracy. Science. 2012;336:1171–1174. doi: 10.1126/science.1219364. A seminal paper describing a generic method for the design of self-assembling building blocks which involves symmetry docking and interface engineering. Cage-like structures are produced with atomic level accuracy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lai YT, King NP, Yeates TO. Principles for designing ordered protein assemblies. Trends Cell Biol. 2012;22:653–661. doi: 10.1016/j.tcb.2012.08.004. [DOI] [PubMed] [Google Scholar]
- 44.Fairman R, Chao HG, Lavoie TB, Villafranca JJ, Matsueda GR, Novotny J. Design of heterotetrameric coiled coils: evidence for increased stabilization by Glu(−)-Lys(+) ion pair interactions. Biochemistry. 1996;35:2824–2829. doi: 10.1021/bi952784c. [DOI] [PubMed] [Google Scholar]
- 45.Baneyx F, Schwartz DT. Selection and analysis of solid-binding peptides. Curr Opin Biotechnol. 2007;18:312–317. doi: 10.1016/j.copbio.2007.04.008. [DOI] [PubMed] [Google Scholar]
- 46.Sarikaya M, Tamerler C, Jen AK, Schulten K, Baneyx F. Molecular biomimetics: nanotechnology through biology. Nat Mater. 2003;2:577–585. doi: 10.1038/nmat964. [DOI] [PubMed] [Google Scholar]
- 47.Coyle BL, Zhou W, Baneyx F. Protein-aided mineralization of inorganic nanostructures. In: Rehm BHA, editor. Bionanotechnology: biological self-assembly and its applications. Caister Academic Press; 2013. [Google Scholar]
- 48.Knobloch D, Ostermann K, Rödel G. Production, secretion, and cell surface display of recombinant Sporosarcina ureae S-layer fusion proteins in Bacillus megaterium. Appl Env Microbiol. 2012;78:560–567. doi: 10.1128/AEM.06127-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rothbauer M, Küpcü S, Sticker D, Sleytr UB, Ertl P. Exploitation of S-layer anisotropy: pH-dependent nanolayer orientation for cellular nanopatterning. ACS Nano. 2013;9 :8020–8030. doi: 10.1021/nn403198a. [DOI] [PubMed] [Google Scholar]