Abstract
Compact viral genomes such as those found in noroviruses, which cause significant enteric disease in humans, often encode only a few proteins, but affect a wide range of processes in their hosts and ensure efficient propagation of the virus. Both human and mouse noroviruses persistently replicate and are shed in stool, a highly effective strategy for spreading between hosts. For mouse norovirus (MNV), the presence of a glutamate rather than an aspartate at position 94 of the NS1/2 protein was previously shown to be essential for persistent replication and shedding. Here, we analyze these critical sequences of NS1/2 at the structural level. Using solution NMR methods we determined folded NS1/2 domain structures from a nonpersistent murine norovirus strain CW3, a persistent strain CR6, and a persistent mutant strain CW3D94E. We found an unstructured PEST-like domain followed by a novel folded domain in the N-terminus of NS1/2. All three forms of the domain are stable and monomeric in solution. Residue 94, critical for determining persistence, is located in a reverse turn following an α-helix in the folded domain. The longer sidechain of glutamate, but not aspartate, allows interaction with the indole group of the nearby tryptophan, reshaping the surface of the domain. The discrimination between glutamyl and aspartyl residue is imposed by the stable tertiary conformation. These structural requirements correlate with the in vivo function of NS1/2 in persistence, a key element of norovirus biology and infection.
Keywords: polyprotein, MNV, protein NMR, sidechain rotamers, PEST motif, helix-strand-helix
Introduction
Human noroviruses cause non-bacterial gastroenteritis and are a significant public health problem worldwide, causing both outbreaks and sporadic cases1,2. Asymptomatic shedding within the human population likely leads to symptomatic infection, as no evidence for an animal reservoir of human noroviruses exists3,4. There are no effective anti-noroviral drugs, and there is no vaccine. The persistence of noroviruses in human and animal populations leads to recurrence of viral infections, outbreaks, and epidemics; however, our understanding of viral and host factors determining the duration of persistence and, consequently, the risk posed by a previously infected asymptomatic host is limited.
A recent analysis of viral persistence in the intestinal tract revealed genetic elements in the NS1/2 protein that are responsible for persistence of the virus in the mouse4,5. The murine norovirus RNA genome contains four open reading frames6. ORF-1, encoded at the 5′ end of the viral genome, is translated into a non-structural polyprotein that is then cleaved into 6 mature products designated NS1/2, and NS3 through NS7, with NS6 being the protease processing the polyprotein7,8. NS1/2 localizes to organelles in the secretory pathway, and human norovirus NS1/2 has been reported to disrupt cellular protein secretion9. While murine norovirus NS1/2 is not recognized to be cleaved by the viral protease, it does contain sequences that can be cleaved by caspase 3 near the junction between the divergent N-terminal 120 residues and the conserved remainder of its sequence10. Interestingly, homologs of NS1/2 in other RNA viruses are cleaved into two proteins10. The membrane localization of NS1/2 has been attributed to the last 40–80 residues in its sequence, which are well conserved and contain a hydrophobic region11–13. The middle domain, residues 120-240, is also well conserved at the amino acid sequence level among noroviruses and has been annotated into the NlpC/P60 family14. The possible enzymatic activity of this domain has not been identified yet. The N-terminal 120 residues of NS1/2 show little conservation among noroviral genetic groups; however, this domain is where a single mutation, D94E, was found to be sufficient to impart a persistent phenotype on the virus4. Herein, we have extended this analysis to the structural level by elucidating the differences between tertiary structures of proteins found in persistent and nonpersistent viruses.
Materials and Methods
Protein Expression and Purification
Sequences corresponding to residues 1-341 of NS1/2 encoded by the MNV strains CR6 and CW3 were PCR amplified from a plasmid containing the MNV genome using the following oligonucleotide primers with BamH I and Kpn I restriction endonuclease sites: forward-5′-GACGGATCCAGGATGGCAACGCCATCTTCTGC-3′, CW3 reverse-5′-GACGGTACCTTATTCGGCCTGCCATTCCCCGAAG-3′, CR6 reverse-5′-GACGGTACCTTATTCGGCCTGCCATTCCCCGAAG-3′. The DNA coding for NS1/2 CW3 residues 1-133 was generated by reverse amplification from the full length NS1/2 encoding sequence using a stop codon-containing forward primer 5′-GCGGAGGACGCTATGGATGCCAAGtagCCTGTGATCGCTCTATCTTG-3′ and reverse primer 5′-CAAGATAGAGCCGATCACAGGctaCTTGGCATCCATAGCGTCCTCCGC-3′. The reaction mixture was digested with the methylation specific restriction enzyme Dpn I for 2 h at 37 °C to remove the original methylated template, and then transformed into E. coli Scarabxpress™ T7 lac cells (ScarabGenomics, Madison, WI). Shortened constructs of NS1/2 from strains CW3, CR6, and CW3D94E, consisting of residues 28-114 and 58-114 were PCR amplified using the forward primers 5′-GACGGATCCGTCTCCTTTGGAGCACCTAGCCCCCTCTCT-3′ and 5′-GGCGCCCTTGCGGCCCTTCATGCGG-3′ respectively, and the same reverse primer 5′-GACGGTACCCTACCGCAGATGGGGGTAACGGCGGAAGATA-3′. The PCR products were cloned into a modified pET vector (pET-BNK) that introduced an N-terminal, 12-residue His6 tag (MRGSHHHHHHGS). For all protein constructs cloned in the pET-BNK vector, transformed E. coli cells were grown to OD600 = 1.2 in LB or M9 media, supplemented with 0.1% glucose, 100 μg/mL ampicillin. Cells were induced with 1 mM IPTG for 4 h, harvested by centrifugation (8 min., 11,000 x g), and lysed by sonication in binding buffer (50 mM NaH2PO4, 20 mM imidazole, 0.5 M NaCl, pH 7.5, 1 mM PMSF). Cell extract was clarified to remove insoluble debris and loaded onto HisTalon™ Superflow cartridges (5 mL, Clontech). A gradient of increasing imidazole concentration over 20 column volumes ended with 50 mM NaH2PO4, 30 mM imidazole, 0.5 M NaCl, pH 7.5, and was followed by a gradient elution over 10 column volumes, reaching 0.5 M imidazole. Protein-containing fractions were pooled and dialyzed to the size-exclusion chromatography buffer, 50 mM NaH2PO4, 0.3 M NaCl, and centrifugally concentrated to 5 mL using the Vivaspin 10K MWCO (Sartorius), before loading on the 26/60 column of the Superdex™ 200 (GE Healthcare). The SDS-PAGE analyzed pure fractions of eluted protein were concentrated to 1 mM. 15N- and 15N, 13C -labeled samples were prepared by growing cells in M9 minimal media with 1 g/L 15NH4Cl and/or 4 g/L 13C-u-glucose (CIL, Andover, MA) as sole sources of nitrogen and/or carbon.
NMR Experiments and Structure Determination
NMR experiments were performed on Bruker Avance 600, 800 and 900 spectrometers equipped with cryoprobes. Samples were prepared at approximately 0.5 mM protein concentration in 50 mM NaH2PO4, 0.3 M NaCl, 0.001% DSS, pH 7.5 and placed in elliptical NMR tubes (Bruker Biospin) to optimize cryoprobe sensitivity and shorten the pulse width. All experiments used for resonance assignments were performed at 23 °C. 15N-labeled samples were used to acquire 2D HSQC experiments. 15N, 13C-labeled samples of N-terminally His6-tagged CW3D94E 28-114 were used to acquire 3D HNCO, HNCACO, CBCANH, CBCACONH, CCCONH, and HCCCONH experiments15 used for backbone and sidechain assignments. Homonuclear 1H, 2D NOESY, and 3D 15N- and 13C-edited NOESY-HSQC experiments15 were used for assigning NOESY crosspeaks used in structure calculations of CW3D94E. Distance restraints for calculations of CW3 58-114 and CR6 58-114 structures were derived from homonuclear 1H-1H NOESY experiments in 95% H2O/5% D2O, and 100% D2O recorded at 800 and 900 MHz. The high resolution of these spectra allowed assignments of almost the same number of NOE peaks as did the combination of heteronuclear-edited NOESY experiments for CW3D94E.
A heteronuclear 1H-15N NOE experiment was recorded to estimate the backbone dynamics for both CW3 58-114 and CR6 58-114. The ratio of the intensities of the peak in the NOE and no-NOE spectra was taken as a measure of the steady-state heteronuclear NOE. Residues with peaks that overlapped in the spectrum were excluded.
NMR data were processed using TOPSPIN (Bruker Biospin) and NMRPipe16 and analyzed using SPARKY (T. D. Goddard and D. G. Kneller, SPARKY 3, University of California, San Francisco). Structural ensembles were calculated using CYANA17 with 30,000 steps for each structure. Distance restraints were calibrated automatically using CYANA routines. Hydrogen bond restraints were included only in later stages of calculations when they could be identified in a majority of structures. A total of 90 backbone φ and ψ dihedral angle restraints calculated using TALOS18 were employed in the calculations. For the final round of calculations, 900 structures were calculated in CYANA, and the 50 with the lowest target function were energy-minimized using AMBER 11 with 3,500 steps of steepest-descent energy minimization. Energy-minimized structures were analyzed with PROCHECK-NMR19. The final ensemble consists of the 20 structures with the lowest energies and the best non-bonded backbone geometry. Chimera20 was used for visualization of structures and for producing figures.
Determination of the Stability of the Secondary Structure by Circular Dichroism
Samples of CR6, CW3 and CW3D94E 58-114 were prepared at 50 μM in 0.3 M NaCl, 25 mM NaH2PO4, pH 7.5. Thermal melting experiments were performed on an Applied Photophysics Chirascan CD spectrometer in a 1 mm cuvette. The temperature was incremented by 1 °C every 2 minutes with 280 to 200 nm wavelength scan at each temperature point between 5 and 90 °C. The data were fitted to 6 parameter equations21 using XLSTAT nonlinear regression routine.
Results
Structured Domain within MNV NS1/2 Protein
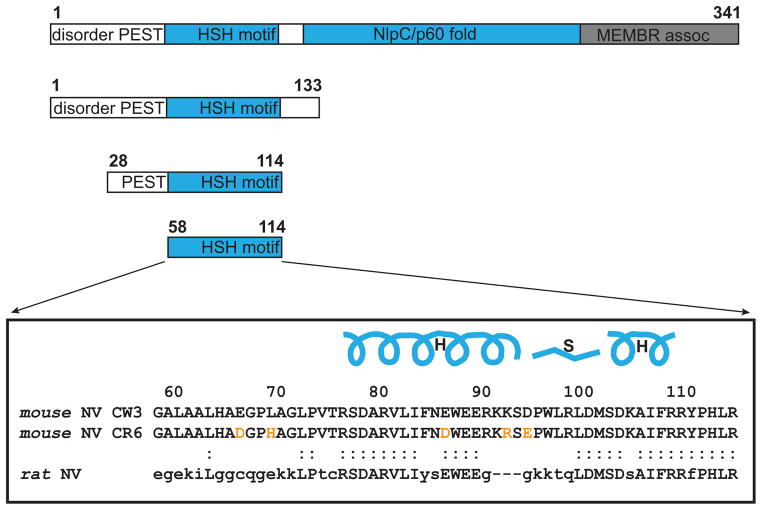
The heterologously expressed full-length 341-residue polypeptide of MNV NS1/2 was rapidly degraded during purification from E. coli cells. Because the sites of functional significance for persistent infection mapped to the N-terminal portion of the protein, we produced several different N-terminal constructs using DNA from CW3 and CR6 strains (Fig. 1). These proteins were expressed in E. coli cells and they were stable during purification. First, we purified the N-terminal fragment of NS1/2 CW3 protein including residues 1 to 133 with the N-terminal His6 tag. The sample was used to record 2D NOESY spectra, which showed a number of cross peaks indicative of a stable tertiary structure. The 15N-1H HSQC spectra recorded with 15N-labeled sample confirmed this observation and also suggested that about half of the 133 residues behave as an unstructured polypeptide. The NMR spectra of a smaller fragment encompassing residues 28-114 retained all signals of the structured domain showing significant dispersion of signals. Following the resonance assignments and heteronuclear NOE analysis, the structured domain was finally defined to residues 58-114 in the murine NS1/2 sequence. The corresponding CW3D94E and CR6 variants showed similar behavior. All NS1/2 protein constructs of 133 residues or fewer showed NMR linewidths, and Superdex 200 (separation range 10 to 600 kDa) size-exclusion chromatography (SEC) behavior consistent with monomeric species (Fig. S1). The SEC-estimated molecular masses of N-terminally His6-tagged 28-114 and 58-114 fragments of NS1/2 were 12.3 and 5.1 kDa, respectively. The theoretical calculated masses for these proteins are 11.1 and 7.9 kDa in good agreement with SEC estimates and consistent with a monomeric state in solution (Fig. S1).
Fig. 1.
Murine Norovirus NS1/2 protein constructs. Domains identified in NS1/2 are labeled. Disordered segments are shown as white bars, including the PEST motif. Soluble, stably-structured domains are shown in blue and the membrane associated domain is shown in gray. Aligned sequences of 58-114 domains of CW3 and CR6 strains are shown with amino acid differences highlighted in orange and secondary structure elements of helix-strand-helix (HSH) motif drawn above the sequence. Rat norovirus sequence corresponding to mouse NS1/2 58-114 is also shown with residues identical to strains CW3 and/or CR6 shown in upper case. A colon indicates identity between any currently known rat and mouse norovirus.
Circular Dichroism Analysis of NS1/2 N-terminal Domain 58-114
We tested the stability of NS1/2 domains 58-114 using temperature-dependent circular dichroism (CD) spectrometry (Fig. 2). The CD spectra at 24 °C showed strong helical signatures (Fig. 2A), which were completely absent at 66 °C (Fig. 2B). The calculated midpoints (Tm) of unfolding transitions for CR6, CW3, and CW3D94E (69 residues including purification tag) in 25 mM NaH2PO4, 0.3 M NaCl, pH 7.5, were 47, 53, and 54 °C (Fig. 2C), and the calculated enthalpies were −55, −48, and −51 kcal/mol, respectively. Considering the small size of the structured domains (57 residues), all three variants are very stably folded.
Fig. 2.
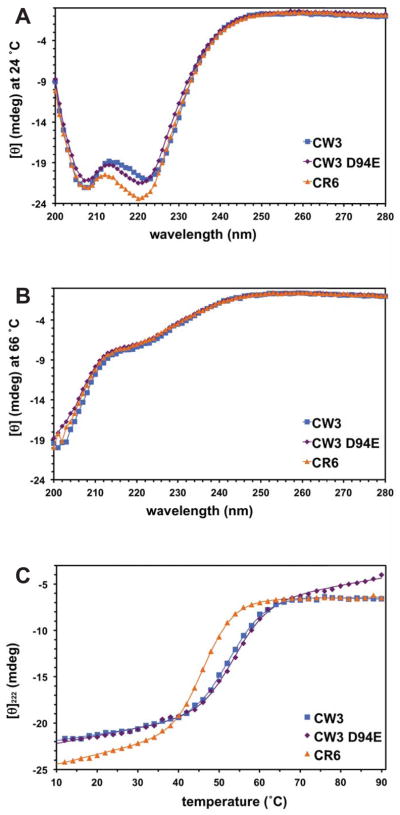
Circular dichroism spectra on NS1/2 58-114 domains. A. Spectra at 24 °C. B. Spectra at 66 °C. C. Thermal denaturation followed by circular dichroism at 222 nm for the NS1 58-114 variants, CR6 (orange triangles), CW3 (blue squares) and CW3D94E (purple diamonds). Protein samples were 50 μM in 25 mM NaH2PO4, 0.3 M NaCl, pH 7.5. The calculated Tm values were 47, 53, 54 °C, respectively. In A and B, solid lines show nonlinear fits to the data points.
NMR Assignments and Structure Calculations
The first NMR assignments were carried out on a 98-residue N-terminally His6-tagged NS1/2 CW3D94E 28-114. We followed a standard triple resonance approach, relying on a collection of several experiments acquired on a 15N,13C-labeled sample. Distance restraints were based on NOE cross peaks observed in 3D 15N-edited, 3D 13C-edited and 2D homonuclear NOESY spectra. Following restraint assignments, the determination of the tertiary structure and measurements of the backbone dynamics defined the structured domain to residues 58-114. The tertiary structures of these domains were very well-determined for all three variants (Table I).
Table I.
Structural statistics for ensembles of 20 structures
| Norovirus strain/PDB code | CW3/2mch | CW3D94E/2mcd | CR6/2mck |
|---|---|---|---|
| Nonredundant NOE restraints | 729 | 785 | 662 |
| Intraresidue | 203 | 251 | 190 |
| Short | 172 | 225 | 153 |
| Medium | 128 | 117 | 124 |
| Long | 226 | 192 | 195 |
| Hydrogen bond restraints | 50 | 48 | 40 |
| TALOS dihedral angle restraints | 94 | 90 | 94 |
| Average CYANA target function | 1.28 | 1.53 | 1.17 |
| Number of violations > 0.2 Å | 0 | 5 | 3 |
| Average AMBER energies (± standard deviation) | |||
| Input structures | −1815 | −2292 | −2096 |
| Energy minimized structures | −2720 | −3381 | −2879 |
| Avg. Ramachandran stats from PROCHECK (res. 60–113) | |||
| Most favored (%) | 93.5 | 92.4 | 93.6 |
| Additionally allowed (%) | 6.5 | 7.6 | 6.4 |
| Generously allowed (%) | 0 | 0 | 0 |
| Disallowed (%) | 0 | 0 | 0 |
| Average RMSD from mean structure (Å, res. 60-113) | |||
| Backbone (N,Cα,C′,O) | .22 | .28 | .21 |
| Heavy atoms | .88 | .82 | .84 |
The 30-residue sequence between positions 28 and 57, SVSFGAPSPLSSESEDEINYMTPPEQEAQP, contains 5 Pro, 5 Glu, 6 Ser and one Thr, making it a good candidate for a PEST-like motif22,23. Although well-conserved between CW3 and CR6 strains (only change I/V45), this segment showed no observable through-space NOE interactions and the chemical shifts of its resonances were characteristic of unstructured segments in proteins.
Similarities and Differences at the Spectral Level in HSQC and 2D 1H-1H NOESY
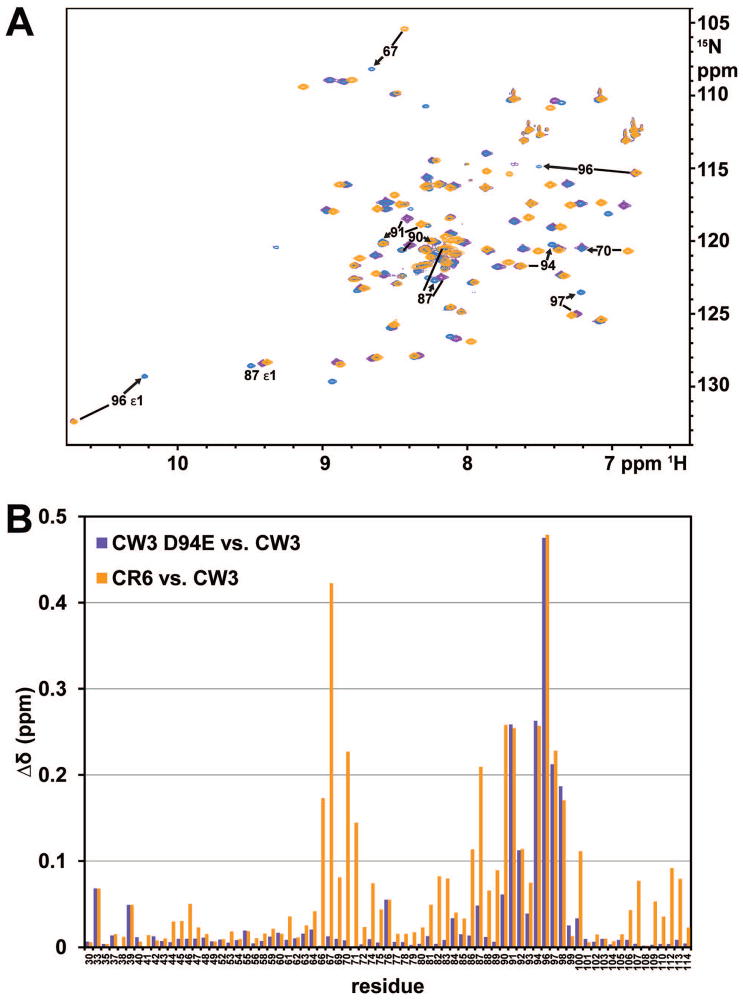
The sequences of NS1/2 28-114 domains from strains CW3 and CR6 are 92% identical, and their NMR spectra are generally similar (Fig. 3A). The most significant differences are visible for backbone and sidechain resonances of W96 and other residues near the mutation site at position 94 (Fig. 3B). There are also significant shifts at positions 66–71, where CW3 residues E66 and L69 are replaced by CR6 D66 and H69, however the overall pattern of cross peaks in their NOESY spectra remains very similar. These differences are consistent with residue substitutions that do not remodel the 3D structure of the protein.
Fig. 3.
Amide 15N and 1H chemical shift differences among NS1/2 domains from different strains. A. Overlaid 15N-1H HSQC spectra of NS1/2 CW3 (blue, middle), CW3D94E (purple, bottom) and CR6 (orange, top) domains 28-114, at 24 °C. Many peaks are visible only in the color of the top CR6 orange layer. Peaks with Δδ above 0.2 among 3 forms are labeled with a corresponding residue number. Arrows always point to the position of the peak in the CW3 form; in some cases these peaks were covered by upper layers. B. Plot of combined 1H and 15N amide chemical shift differences for non-proline residues. Differences between CW3D94E and CW3 are shown as purple bars, and between CR6 and CW3 as orange bars. Combined difference values were calculated as .
In the CW3D94E mutant, a single, conservative substitution of D94 to E94 results in large shifts of amide resonances of W96 and residues K91, K92, L97, and R98 (Fig. 3B). The positions of these shifted resonances coincide with positions observed in the CR6 form. The extent of these changes can be rationalized only in the context of the 3D structure of this domain.
Conformation of the MNV NS1/2 N-terminal Domain 58-114
The domain has an L-shaped structure with the tip of the long stem formed by residues 93-95, and the tip of the short stem formed by residues 65-68, (Fig. 4). The first well-structured residues are A61-L63, which extend into a somewhat irregular, hairpin-shaped loop ending with T75. Within this loop, residues E66-L69 adopt a 310-helical conformation and reverse the direction of the chain. Next, the following α-helix-1 extends from T75 to E94 ending with a reverse turn at P95-W96. Residues L97 to M101 extend along helix-1, with sidechains of L97, L99 and M101 making hydrophobic contacts with R90, W87, F84 and I83, respectively. This extended segment continues to α-helix-2, between S102 and Y110, followed by a reverse turn of P111-H112, which is stabilized by sidechain interactions of L113 with I106 and Y110. The small hydrophobic core consists of residues P73, A79, I83, I106, Y110, and L113. The CW3 domain has a theoretical pI of 9.5 with a rather polar surface dominated by positively charged side chains of 7 arginyl and 3 lysyl residues. Of those, sidechains of R76, R80, K92, R98 and R114 appear highly dynamic with no observable NOE interactions. Among 4 aspartyl and 4 glutamyl residues, E66 and E86 are also highly mobile. The carboxylate-bearing residue at position 94 is a well-defined rotamer with χ1 and χ2 falling into −60°, 180° minima for E94 and with χ1 for D94 into the −60° minimum. The rest of the charged sidechains show ordering to a variable extent. In particular, both H64 and H112 are well-ordered and interact with one another as well as with L69, or H69 in the case of the CR6 variant. The overall conformations are very similar in all three forms of the domain, with rmsd of backbone atoms below 1 Å (Table I, Fig. 5).
Fig. 4.
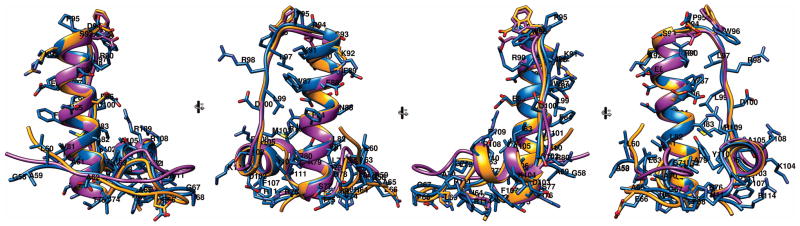
Four 90° rotated views of superimposed ribbon diagrams of NS1/2 58-114. CW3 in blue, is shown with residue numbers and sidechains, CR6 in orange, and CW3D94E in purple. The sidechains of CR6 and CW3D94E different from CW3 are also shown.
Fig. 5.
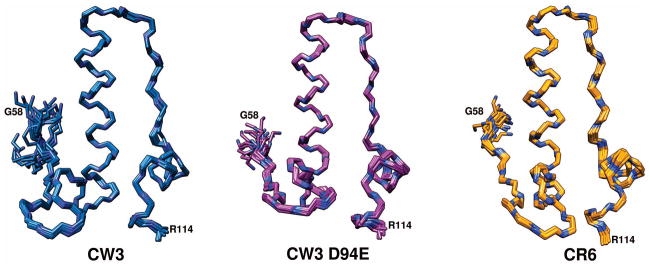
The backbone atoms of superimposed ensembles of 20 calculated structures for NS1/2 58-114. CW3 in blue, CW3D94E in purple and CR6 in orange. Backbone nitrogen atoms are shown in dark blue.
Impact of W96 Sidechain on Structure, Dynamics and Molecular Surface of NS1/2 58-114
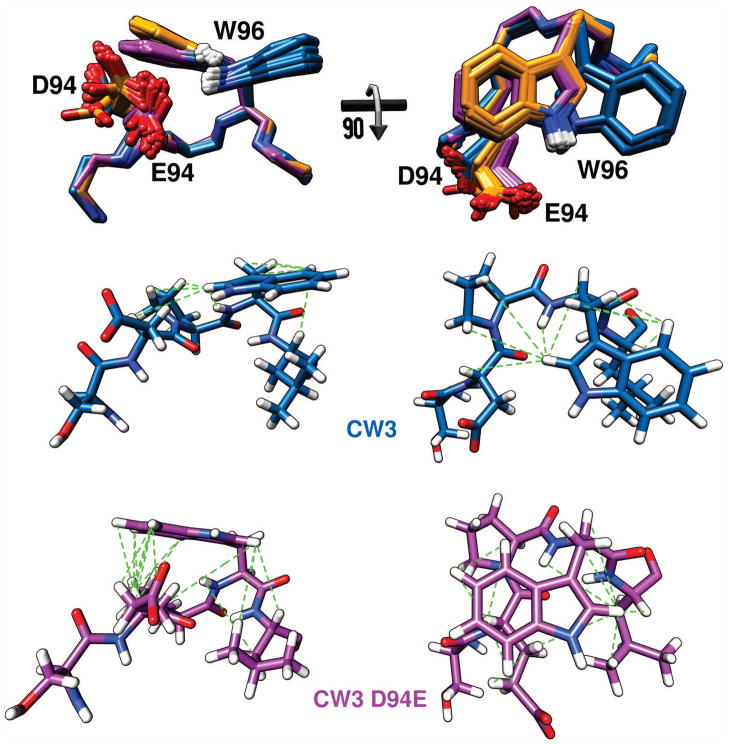
The sidechain of W96 appears as a different rotamer depending on the presence of a glutamyl or an aspartyl group in position 94. Residues 94-97 produce a type I reverse β-turn. With an aspartyl sidechain at position 94, the torsion angles of W96 take on χ1 near +60° and χ2 near −90° minima24. The presence of a glutamyl sidechain does not change the preference of the χ1 angle of W96, but χ2 rotates 180° into the +90° minimum. The proximity of the E94 carboxylate oxygen to the indole NH group of W96, as well as downfield chemical shift changes of 3.4 ppm for 15Nε1 and 0.5 ppm for 1Hε1 compared to D94, strongly suggest a hydrogen-bonding interaction between these sidechains (Fig. 6). We never included this interaction as a restraint in our calculations. In the CW3D94E ensemble, the average W96 Hε1-E94 Oε and W96 Nε1-E94 Oε distances were 3.15 Å and 3.79 Å, respectively. The average distances for those same atom pairs were 3.83 Å and 4.40 Å in CR6. The slightly longer distances for CR6 were a consequence of using only homonuclear NOESY derived distance restraints, while the CW3D94E ensemble calculations included 15N- and 13C-edited NOE experiments. For CW3, also calculated with homonuclear NOESY data only, the W96 Hε1-D94 Oδ and W96 Nε1-D94 Oδ distances were 4.98 Å and 5.58 Å, well beyond acceptable hydrogen bond distances. The shorter sidechain of D94 makes such interactions sterically impossible.
Fig. 6.
Side and top view of backbone atoms of residues 93-97 along with sidechains of D/E94 and W96. Ensembles of 10 superimposed structures for CW3 (blue), CR6 (orange) and CW3D94E (purple) are shown. For clarity only the Hε1 of W96 indole is shown (white). Below, in the same orientations, one structure of CW3 (blue) and CW3D94E (purple) are shown with all atoms (hydrogens in white). Green broken lines represent NOE distance restraints to the W96 sidechain observed for the respective structure.
For CW3 and CR6 domains 58-114, we compared the dynamics of backbone and tryptophan side chains using 1H-15N heteronuclear NOE experiments (Fig. S2). The region of the P95-W96 β-turn showed only a small increase in mobility compared to the majority of the backbone. For residues 94, 96, 97 of CW3 we found an average NOE of 0.69 versus an average of 0.78 for residues 78-93, and the corresponding values for CR6 were 0.74 and 0.78. The measurement errors were below 0.015. This difference points to a small increase in ordering of the backbone of residues 94-97 upon Asp to Glu substitution at position 94. Similarly, the CW3 W96 indole NH group NOE was 0.45 versus CR6 NOE of 0.54, with measurement errors below 0.1. For a reference and comparison, the more buried W87 indole NH had NOE values of 0.73 and 0.74, respectively in CW3 and CR6 forms (error below 0.01).
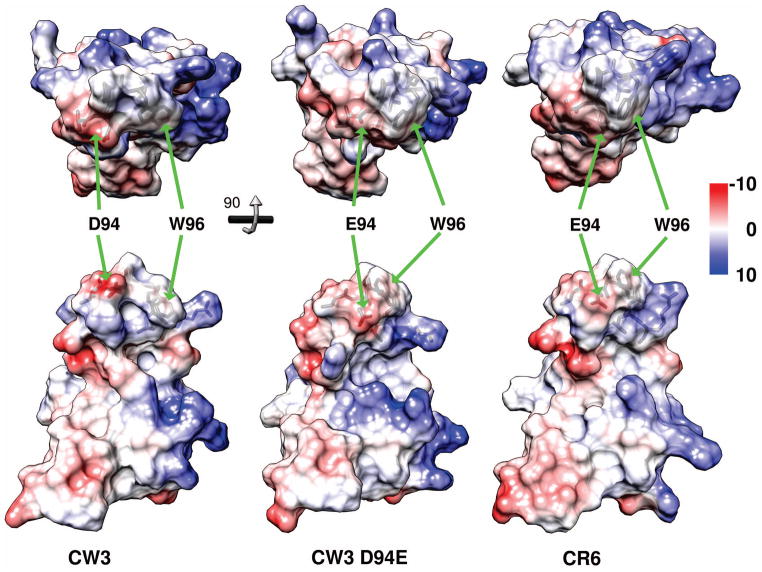
The rotation of the indole ring of W96 changes the shape and charge distribution of the molecular surface (Fig. 7). These changes are localized to the top of the long stem of the L-shaped domain, which is very distant from both termini of the polypeptide chain. Consequently, similar changes are very likely to occur in the full-length native structures of NS1/2 functioning in vivo. The presence of W96 χ2 in the +90° minimum (as in CR6 and CW3D94E) brings the indole ring close to P95, producing a rather pointed hydrophobic tip surrounded by mostly positively charged sidechains of R90, K91, R/K92, and R98. The opposite orientation of χ2 in the −90° minimum produces a wider surface in this location with a more exposed P95-W96 peptide bond and the indole ring in closer proximity to the L97 sidechain. However local, such a difference in the molecular surface can be sufficient to determine the outcome of molecular recognition events, for example binding to a complementarily-shaped hydrophobic pocket surrounded by negative charges.
Fig. 7.
Top and side views of molecular surfaces of the 95-96 region for NS1/2 58-114. Domains from strains CW3, CW3D94E, and CR6 are left, middle, and right, respectively. Sidechains of residues 94 and 96 are indicated with green arrows. Coulombic surface coloring, with blue indicating positive and red negative charge. Lower raw models were rotated 90° around x-axis, relative to the top raw models. Heavy atoms of residues 93-97 are visible through the semitransparent surface in each model.
Discussion
The N-terminal sequences of NS1/2 proteins of several strains of murine norovirus were found to harbor functionally essential residues for viral persistence in the intestine, which genetic analyses defined to very specific positions and residue types4. Also recently, NS1/2 sequences from multiple noroviral genetic groups have been investigated, primarily using bioinformatics techniques, and the N-terminal 120 residues were suggested to be intrinsically disordered13. As the identification of novel structural domains remains challenging for bioinformatics, we have taken an experimental approach. We began with the NMR analysis of purified fragments of NS1/2, and found a stable tertiary structure for residues 58-114 in the N-terminal portion of this polypeptide. A more conserved (among all noroviral genetic groups) NS1/2 C-terminal domain (residues 134-265, annotated as a member NlpC/P60 family) also contains stable tertiary structure, suggested by bioinformatics, and confirmed by our preliminary experiments (data not shown).
When isolated, the first 133 residues of MNV NS1/2 showed both intrinsically-disordered and stably-folded components. In particular, the first 57 residues of NS1/2 showed no stable secondary and/or tertiary structure. Additionally, residues 26-57 show sequence composition typical for PEST-like motifs with a PEST score of 7.52 (EMBOSS 6.3.1:epestfind25). PEST motifs in most described examples serve as proteolytic signals23,26,27, however the functional importance of the predicted PEST motif in NS1/2 remains to be determined.
Unlike residues 1-57, the following 57-residue fragment of NS1/2 (58-114) produced well-resolved NMR spectra, with multiple through-space interactions indicative of a stable tertiary structure. The presence of stable, largely helical secondary structure was also apparent in CD measurements, with mid-points of melting transitions close to 50° C for both CW3 and CR6 strains. We have produced this domain and found it to recapitulate all well-dispersed NMR signals we observed for samples of NS1/2 1-133 and NS1/2 28-114 fragments. When analyzed by a number of sequence predicting algorithms, this structured domain scores positively, albeit weakly, as a disordered or unstructured segment. The lack of detectable sequence conservation among noroviral genetic groups within this region of NS1/2 makes predicting the existence of this domain even more difficult. Nevertheless, this portion of the N terminus of NS1/2 folds into a stable structure. To understand the role of its critical residues, we determined the 3D structures of this domain from strains CW3 and CR6, as well as the D94E mutant form of CW3.
The recently published genome of the rat norovirus is the closest evolutionary relative of the mouse norovirus28. These two viruses show 48% identity in the first 120 residues of NS1/2, and 67% identity between residues 72 and 114 (Fig. 1). The prominent difference is a three-residue deletion at the end of helix-1. The result of this change is very likely a helix that is one turn shorter, followed by a reverse turn including a glycyl residue. The surface of this area in the rat norovirus NS1/2 domain is going to be much more charged as both prolyl and tryptophanyl sidechains are missing. This suggests that elements of the secondary and tertiary structure have been preserved over evolutionary time while the species-specific interaction surfaces and disordered segments have diverged between mouse and rat noroviruses.
The small size and monomeric state in solution made the NS1/2 58-114 domain an ideal candidate for solution structure determination by NMR techniques. The strain variants showed very similar patterns in NMR spectra, and only a few very distinct differences, pointing to a common conserved fold with a few local differences. Approximately the C-terminal half of this fold forms a helix-strand-helix motif. It consists of a 20-residue helix-1 followed by an 8-residue extended strand running proximal and antiparallel to helix-1, followed then by a shorter 7-residue helix packing perpendicularly to helix-1 (Fig. 4). This 35-residue motif can be discerned in the context of a small number of larger folds. DALI searches29 in the Protein Data Bank find only three structures that gave significant structural similarity Z scores above 2. The PDB codes and Z scores were: 1hlv, 2.8; 1jra, 2.3; 2qkm, 2.3. Only residues 140-175 of 1jra, the FAD-binding yeast thiol oxidase Erv2p30, constitute a structurally independent C-terminal element that resembles closely residues 75-110 of NS1/2. However, in Erv2p, these residues form one side of the FAD binding pocket, and only 4 of them (7%) are identical to those in NS1/2. Additionally, in Erv2p, the end of the extended strand is disulfide-bridged to the longer helix, and this helix packs against a preceding helix, unlike the exposed NS1/2 helix-1 (Fig. 8).
Fig. 8.
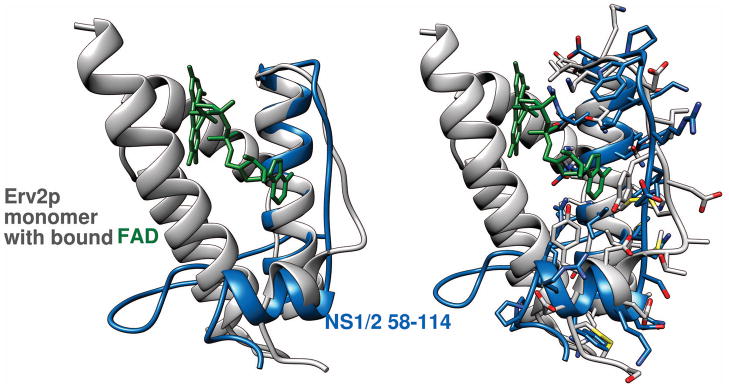
The helix-strand-helix motif of NS1/2 58-114 CW3 (blue) superimposed on a monomer of yeast Erv2p thiol oxidase (PDB code 1jra, gray). The FAD bound by Erv2p is also shown (green). Ribbon diagrams only are shown in the left side image. Sidechains for corresponding superimposed sequences in both proteins are shown in the right side image.
The determinant sufficient for persistence of MNV was mapped to NS1/2 residue 94, where a conservative change of Asp to Glu residue produced a dramatic change in the biological behavior of the virus4. Because of its placement within a well-structured portion of the N-terminus of NS1/2, we were able to examine the D94E mutation at the structural level. The immediately apparent changes in NMR spectra of the D94E mutant were not exactly in residue 94 or its sequential neighbors. These spectral differences are actually caused by the W96 sidechain indole ring covering the peptide bond of the β-turn at the end of helix-1. The longer sidechain of E94 allows direct interaction between its carboxylate and the indole HN of W96. On the structural level, the change of Asp to Glu flips the χ2 of W96 from −90° to +90° resulting in numerous changes in chemical shifts as the ring currents of W96 affect different residues depending on its proximity. The observed rotamers of W96 differ also in the details of their molecular surfaces, where the E94 associated rotamer shows a more pointed hydrophobic tip made of sidechains of P95 and W96. In contrast, the D94-associated rotamer has the 95-96 peptide bond more exposed, with the indole ring covering primarily the L97 sidechain. Although very local, these differences may be sufficient to change the affinity of interactions of this domain with other proteins.
Between D94 and E94 forms of NS1/2 58-114, there is a small but measurable difference in dynamic properties. The E94 stabilizes not only the movements of the W96 indole ring but also the backbone of residues 96-99 (Fig. S2). The effect is local and the overall stability of CW3 versus CW3D94E remains practically unchanged, as measured by a very small difference in CD thermal unfolding experiments (Fig. 2).
The observed phenotypic characteristics of the viral life cycle and the atomic-level protein structural changes are phenomena at very different levels of biological organization. How the biochemical characteristics of this novel fold fit into cellular pathways engaged during viral infection, resulting in evasion of immunity and consequently viral persistence, are still open questions, and we and others are actively pursuing them.
Supplementary Material
Acknowledgments
Grant support: NIH RO1 AI 054483 to HWV.
Footnotes
Institution at which the work was performed:
Washington University School of Medicine, St. Louis, MO 63110
Data deposition: The atomic coordinates and NMR assignments have been deposited in the Protein Data Bank and BMRB-BioMagResBank.
PDB code for NS1/2 58-114 strain CW3: 2mch, BMRB accession no. 19439
PDB code for NS1/2 58-114 strain CW3D94E: 2mcd, BMRB accession no. 19436
PDB code for NS1/2 58-114 strain CR6: 2mck, BMRB accession no. 19444
Contributor Information
Brendan N. Borin, NMR Accelerator, San Diego, CA
Wei Tang, Department of Biochemistry & Molecular Biophysics, Washington University School of Medicine, St. Louis, MO 63110.
Timothy J. Nice, Department of Pathology & Immunology, Washington University School of Medicine, St. Louis, MO 63110
Broc T. McCune, Department of Pathology & Immunology, Washington University School of Medicine, St. Louis, MO 63110
Herbert W. Virgin, Department of Pathology & Immunology, Washington University School of Medicine, St. Louis, MO 63110
Andrzej M. Krezel, Departments of Biochemistry & Molecular Biophysics and Pathology & Immunology, Washington University School of Medicine, St. Louis, MO 63110
References
- 1.Glass RI, Parashar UD, Estes MK. Norovirus gastroenteritis. N Engl J Med. 2009;361(18):1776–1785. doi: 10.1056/NEJMra0804575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koo HL, Ajami N, Atmar RL, DuPont HL. Noroviruses: The leading cause of gastroenteritis worldwide. Discov Med. 2010;10(50):61–70. [PMC free article] [PubMed] [Google Scholar]
- 3.Sukhrie FH, Siebenga JJ, Beersma MF, Koopmans M. Chronic shedders as reservoir for nosocomial transmission of norovirus. J Clin Microbiol. 2010;48(11):4303–4305. doi: 10.1128/JCM.01308-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nice TJ, Strong DW, McCune BT, Pohl CS, Virgin HW. A single-amino-acid change in murine norovirus NS1/2 is sufficient for colonic tropism and persistence. Journal of virology. 2013;87(1):327–334. doi: 10.1128/JVI.01864-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karst SM, Wobus CE, Lay M, Davidson J, Virgin HW. STAT1-dependent innate immunity to a Norwalk-like virus. Science. 2003;299(5612):1575–1578. doi: 10.1126/science.1077905. [DOI] [PubMed] [Google Scholar]
- 6.Wobus CE, Thackray LB, Virgin HW. Murine norovirus: a model system to study norovirus biology and pathogenesis. Journal of virology. 2006;80(11):5104–5112. doi: 10.1128/JVI.02346-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang X, Wang M, Wang K, Estes MK. Sequence and genomic organization of Norwalk virus. Virology. 1993;195(1):51–61. doi: 10.1006/viro.1993.1345. [DOI] [PubMed] [Google Scholar]
- 8.Lambden PR, Caul EO, Ashley CR, Clarke IN. Sequence and genome organization of a human small round-structured (Norwalk-like) virus. Science. 1993;259(5094):516–519. doi: 10.1126/science.8380940. [DOI] [PubMed] [Google Scholar]
- 9.Hyde JL, Mackenzie JM. Subcellular localization of the MNV-1 ORF1 proteins and their potential roles in the formation of the MNV-1 replication complex. Virology. 2010;406(1):138–148. doi: 10.1016/j.virol.2010.06.047. [DOI] [PubMed] [Google Scholar]
- 10.Sosnovtsev SV, Belliot G, Chang KO, Prikhodko VG, Thackray LB, Wobus CE, Karst SM, Virgin HW, Green KY. Cleavage map and proteolytic processing of the murine norovirus nonstructural polyprotein in infected cells. Journal of virology. 2006;80(16):7816–7831. doi: 10.1128/JVI.00532-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ettayebi K, Hardy ME. Norwalk virus nonstructural protein p48 forms a complex with the SNARE regulator VAP-A and prevents cell surface expression of vesicular stomatitis virus G protein. Journal of virology. 2003;77(21):11790–11797. doi: 10.1128/JVI.77.21.11790-11797.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fernandez-Vega V, Sosnovtsev SV, Belliot G, King AD, Mitra T, Gorbalenya A, Green KY. Norwalk virus N-terminal nonstructural protein is associated with disassembly of the Golgi complex in transfected cells. Journal of virology. 2004;78(9):4827–4837. doi: 10.1128/JVI.78.9.4827-4837.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baker ES, Luckner SR, Krause KL, Lambden PR, Clarke IN, Ward VK. Inherent structural disorder and dimerisation of murine norovirus NS1-2 protein. PloS one. 2012;7(2):e30534. doi: 10.1371/journal.pone.0030534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anantharaman V, Aravind L. Evolutionary history, structural features and biochemical diversity of the NlpC/P60 superfamily of enzymes. Genome Biol. 2003;4(2):R11. doi: 10.1186/gb-2003-4-2-r11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sattler M, Schleucher J, Griesinger C. Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Progress in Nuclear Magnetic Resonance Spectroscopy. 1999;34:93–158. [Google Scholar]
- 16.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. Journal of biomolecular NMR. 1995;6(3):277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 17.Guntert P, Mumenthaler C, Wuthrich K. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J Mol Biol. 1997;273(1):283–298. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- 18.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. Journal of biomolecular NMR. 1999;13(3):289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 19.Laskowski RA, Rullmann JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8(4):477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 20.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. Journal of computational chemistry. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 21.Greenfield NJ. Using circular dichroism collected as a function of temperature to determine the thermodynamics of protein unfolding and binding interactions. Nat Protoc. 2006;1(6):2527–2535. doi: 10.1038/nprot.2006.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rogers S, Wells R, Rechsteiner M. Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science. 1986;234(4774):364–368. doi: 10.1126/science.2876518. [DOI] [PubMed] [Google Scholar]
- 23.Rechsteiner M, Rogers SW. PEST sequences and regulation by proteolysis. Trends Biochem Sci. 1996;21(7):267–271. [PubMed] [Google Scholar]
- 24.Miyanoiri Y, Takeda M, Jee J, Ono AM, Okuma K, Terauchi T, Kainosho M. Alternative SAIL-Trp for robust aromatic signal assignment and determination of the chi(2) conformation by intra-residue NOEs. Journal of biomolecular NMR. 2011;51(4):425–435. doi: 10.1007/s10858-011-9568-3. [DOI] [PubMed] [Google Scholar]
- 25.Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16(6):276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 26.Antonsson A, Payne E, Hengst K, McMillan NA. The human papillomavirus type 16 E7 protein binds human interferon regulatory factor-9 via a novel PEST domain required for transformation. J Interferon Cytokine Res. 2006;26(7):455–461. doi: 10.1089/jir.2006.26.455. [DOI] [PubMed] [Google Scholar]
- 27.Lepere-Douard C, Trotard M, Le Seyec J, Gripon P. The first transmembrane domain of the hepatitis B virus large envelope protein is crucial for infectivity. Journal of virology. 2009;83(22):11819–11829. doi: 10.1128/JVI.01026-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tse H, Chan WM, Lam CS, Lau SK, Woo PC, Yuen KY. Complete genome sequences of novel rat noroviruses in Hong Kong. Journal of virology. 2012;86(22):12435–12436. doi: 10.1128/JVI.01976-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38(Web Server issue):W545–549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gross E, Sevier CS, Vala A, Kaiser CA, Fass D. A new FAD-binding fold and intersubunit disulfide shuttle in the thiol oxidase Erv2p. Nat Struct Biol. 2002;9(1):61–67. doi: 10.1038/nsb740. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.