Abstract
The role of positive selection in human evolution remains controversial. On the one hand, scans for positive selection have identified hundreds of candidate loci, and the genome-wide patterns of polymorphism show signatures consistent with frequent positive selection. On the other hand, recent studies have argued that many of the candidate loci are false positives and that most genome-wide signatures of adaptation are in fact due to reduction of neutral diversity by linked deleterious mutations, known as background selection. Here we analyze human polymorphism data from the 1000 Genomes Project and detect signatures of positive selection once we correct for the effects of background selection. We show that levels of neutral polymorphism are lower near amino acid substitutions, with the strongest reduction observed specifically near functionally consequential amino acid substitutions. Furthermore, amino acid substitutions are associated with signatures of recent adaptation that should not be generated by background selection, such as unusually long and frequent haplotypes and specific distortions in the site frequency spectrum. We use forward simulations to argue that the observed signatures require a high rate of strongly adaptive substitutions near amino acid changes. We further demonstrate that the observed signatures of positive selection correlate better with the presence of regulatory sequences, as predicted by the ENCODE Project Consortium, than with the positions of amino acid substitutions. Our results suggest that adaptation was frequent in human evolution and provide support for the hypothesis of King and Wilson that adaptive divergence is primarily driven by regulatory changes.
The rate and patterns of positive selection are of fundamental interest for the study of human evolution. Population genomic studies should, in principle, allow us to quantify positive selection from its expected signatures in sequence polymorphism and divergence data. Surprisingly, despite the sequencing of thousands of human genomes (The 1000 Genomes Project Consortium 2012) and the availability of whole-genome sequences of closely related species, the extent to which adaptation has left identifiable signatures in the patterns of polymorphism in the human genome remains highly controversial (Akey 2009; Hernandez et al. 2011).
On the one hand, recent studies have identified a large number of loci showing signatures of recent selective sweeps (Voight et al. 2006; Sabeti et al. 2007; Williamson et al. 2007; Pickrell et al. 2009; Grossman et al. 2013), and McDonald-Kreitman (MK) analyses inferred that ∼10%–20% of amino acid changes have been adaptive in human evolution (Boyko et al. 2008; Messer and Petrov 2013). Consistently, regions of high functional density, high rate of amino acid substitutions, and low recombination all show reduced levels of neutral diversity (Cai et al. 2009; Lohmueller et al. 2011), as expected under recurrent selective sweeps in functional regions.
On the other hand, there are reasons to question the notion that adaptation left clear signatures in the human genome. First, different scans for positive selection have identified largely nonoverlapping sets of candidates (Akey 2009), which could be due to a high rate of false positives. Second, MK analyses can be confounded by a number of factors, such as perturbations left by demographic events and by the presence of slightly deleterious mutations (Eyre-Walker and Keightley 2009; Messer and Petrov 2013), and some MK analyses have failed to find evidence for adaptation in the human lineage (Eyre-Walker and Keightley 2009). Finally, it has been shown that background selection (BGS) (Charlesworth et al. 1993), a process in which deleterious mutations remove linked neutral variation from the population, reduces levels of polymorphism in regions of higher functional density and low recombination, providing an alternative explanation for the observation of these correlations in the human genome.
One signature of positive selection—lower levels of neutral variation near functional substitutions (Andolfatto 2007; Macpherson et al. 2007; Cai et al. 2009)—is not generally expected under BGS and should therefore provide the clearest genomic evidence for the action of positive selection. While this signature was found in the human genome by Cai et al. (2009), it could not be detected by two recent studies using the newest large-scale data sets of human diversity (Hernandez et al. 2011; Lohmueller et al. 2011). In particular, Hernandez et al. (2011) searched for lower levels of neutral diversity near functional substitutions by contrasting levels of neutral diversity near nonsynonymous compared with synonymous substitutions. They did not find this signature in the human genome and, moreover, found that diversity might in fact be marginally higher near nonsynonymous substitutions. Simulations showed that this puts sharp limits on the amount of adaptation by classic selective sweeps in recent human evolution (Hernandez et al. 2011).
However, it is likely that the study design of Hernandez et al. (2011) (also implemented in Drosophila by Sattath et al. 2011) is strongly biased against finding signatures of positive selection in the human genome and all other genomes with sharply variable levels of genomic constraint. This is because, as we show in the Results, nonsynonymous substitutions in the human genome tend to be located in regions of weaker constraint and thus weaker BGS compared with synonymous substitutions. These differences in levels of BGS should elevate neutral diversity near nonsynonymous compared with synonymous substitutions. The approach of Hernandez et al. (2011) would thus detect positive selection only if the reduction of diversity due to positive selection near nonsynonymous substitutions happens to be greater than the initial difference in the opposite direction due to BGS.
Here we utilize a number of more sensitive approaches in the search for signatures of positive selection while attempting to reduce the confounding effects of BGS to the greatest extent possible. Our results suggest that positive selection was frequent in human history and might have involved adaptive mutations of substantial selective effect. We estimate that a few hundred strong adaptive events are likely to be detectable in the human genome, consistent with the latest scan for positive selection (Grossman et al. 2013). Moreover, we provide evidence that the majority of adaptive substitutions were due to cis-regulatory rather than protein-coding changes, consistent with the King and Wilson (1975) hypothesis that adaptive divergence is primarily driven by regulatory changes.
Results
The search for signals of positive selection in the human genome is complicated by highly variable levels of functional constraint, and thus BGS, along the genome. To be able to detect positive selection, the reduction of neutral polymorphism near nonsynonymous substitutions needs to be stronger than the elevation of polymorphism due to weaker BGS in the same regions. This bias against detecting evidence of positive selection should be particularly strong when levels of neutral polymorphism near synonymous and nonsynonymous substitutions are contrasted, as in the approach pioneered by Hernandez et al. (2011) and Sattath et al. (2011). First, these approaches have limited power in the human genome as the majority (∼65%) of all synonymous substitutions are located extremely close (<0.02 cM) to nonsynonymous ones (Methods). What is more troubling is that the synonymous substitutions that are located far from nonsynonymous substitutions mark regions of particularly strong selective constraint and thus particularly strong BGS. Specifically, we find that 82% of synonymous substitutions are found within conserved segments of the genome predicted by phastCons (Methods; Siepel et al. 2005), versus only 56% of nonsynonymous ones.
The reason for this difference between synonymous and nonsynonymous substitutions is that selectively constrained regions, by definition, lack nonsynonymous substitutions but still allow changes at less constrained synonymous sites. Because constrained regions have stronger BGS, this means that BGS could in principle reduce diversity more drastically near synonymous than near nonsynonymous substitutions to an extent sufficient to mask positive selection. To test whether this could happen in the human genome, we simulate BGS in regions with varying levels of constraint under currently accepted population size, recombination, and selection parameters (Supplemental Material). The simulated regions contain 10% of potentially functional sites, and we vary the proportion of functional sites that are under constraint from zero to 100%. Note that even in simulations with 100% functional sites under constraint, we use a distribution of fitness effects (DFE) with many mutations being virtually neutral, making our analysis overall conservative (Supplemental Material). These simulations show that diversity in the strongly constrained regions can be reduced by as much as 10% on average (Supplemental Fig. 1). This decrease is similar or even greater than what is expected to be generated by positive selection (Hernandez et al. 2011) and thus could have been sufficient to hamper previous attempts to detect positive selection near nonsynonymous changes. We therefore believe that using the synonymous substitutions as the control for detecting reduction of neutral polymorphism near nonsynonymous substitutions in the functionally heterogeneous human genome might be unduly conservative.
Below we devise sensitive methods for the detection of positive selection in the human genome. We first search for reduction of neutral polymorphism near all and near functionally important nonsynonymous substitutions. We match regions near and far from nonsynonymous substitutions by levels of BGS measured using a variety of correlates of BGS, such as levels of functional constraint and recombination. We focus specifically on regions of low BGS, because in these regions the bias against finding positive selection should be the weakest. In addition to level of diversity, we also use haplotype-based statistics iHS and XPEHH. Unlike the overall level of neutral polymorphism used in the first set of tests, we demonstrate that these haplotype statistics are virtually insensitive to BGS and, as a result, that their extreme deviations are predictive of recent and strong selective sweeps.
All data analyses are carried out with the 1000 Genomes Phase 1 data 20100804 release (http://www.1000genomes.org). Levels of neutral diversity are calculated as the average pairwise heterozygosity at putatively neutral sites, scaled by divergence between human and macaque. Human-specific substitutions at synonymous and nonsynonymous sites are inferred from human–chimpanzee–orangutan alignments (Methods).
Choosing analysis windows
BGS is expected to be stronger in regions of low recombination: Consistent with this, the correlation between neutral diversity and recombination rate measured in 500-kb windows sliding every 1000 kb is strong and positive (n = 2,247, Spearman’s ρ = 0.45, P < 2 × 10−16). Note that all correlations reported in this section use 500-kb windows that are at least 500 kb from each other to ensure independence of the estimates to the greatest extent possible. BGS should also be stronger in regions of high functional constraint. We can measure functional constraint using multiple variables, all of which show strong negative correlations with levels of neutral diversity in 500-kb windows, including (1) density of coding sequences (CDS) (n = 2247, ρ = −0.22, P < 2 × 10−16), (2) density of conserved coding sequences (CCDS) and noncoding sequences in all mammals or just in primates according to phastCons (ρ = −0.22, P < 2 × 10−16 and ρ = −0.25, P < 2 × 10−16, respectively), and (3) the density of UTRs (ρ = −0.23, P < 2 × 10−16). All of these correlations were computed as partial correlations controlling for recombination rate. In addition, we also find a strong negative partial correlation between diversity and GC content, controlling for recombination (ρ = −0.2, P < 2 × 10−16), which might be related to high GC content of coding regions (Lander et al. 2001) or some other property correlated with the GC content.
The segments of conserved DNA identified by phastCons are shared by mammals and/or primates and represent averaged constraint over long evolutionary periods of time. The density of mammalian and primate conserved sequences being equal, large regions that are particularly devoid of human-specific nonsynonymous substitutions may still be under stronger constraint. To detect such regions of unusually strong constraint, and thus BGS, we plot the distribution of distances to the nearest amino acid substitution in the human genome in all regions that have CCDS density >0.1% (to make sure there are CCDS in the windows); 67.4% of the windows are located <0.1 cM away from a human-specific amino acid substitution, 30% are located 0.1–1 cM, and 2.6% are located farther than 1 cM. These latter windows may represent regions of unusually strong constraint.
We quantify whether the regions of moderate to high functional density (CCDS density >0.5%) located far (>1 cM) from any amino acid substitution are indeed subject to stronger BGS by conducting a bootstrap procedure (Methods). For each window located between 0.1 and 1 cM away from an amino acid change, we match a randomly sampled window located 1 cM or farther whose functional density, GC content, and recombination do not differ by more than empirically fixed thresholds compared with the 0.1- to 1-cM window. Windows <0.1 cM away are excluded from this comparison since they are the ones most likely to be affected by positive selection, and we want to focus only on BGS as a function of distance to the nearest amino acid change. Thresholds of the bootstrap are adjusted such that the 0.1- to 1-cM windows and >1-cM windows have similar average functional density, GC content, and recombination rates. Windows for which no good match can be found are excluded (the detailed bootstrap procedure is described in Methods).
Neutral diversity is indeed substantially reduced in regions that are located >1 cM away from any amino acid substitution, controlling for functional density, GC content, and recombination (Methods). Overall, the reduction is 7% (randomization test, P = 8.6 × 10−3) and becomes even stronger (∼15%; randomization test, P = 2.4 × 10−2) in regions where recombination rates do not exceed 1 cM/Mb. This is consistent with our interpretation that regions of substantial functional density that are located very far from any amino acid change are more constrained in a way that cannot be accounted for using average conservation in more distant mammalian and primate species. Below we exclude the 2.6% of the windows that are located >1 cM from an amino acid substitution unless stated otherwise.
The near-vs-far test
One key expectation under positive selection is a reduction of neutral diversity near functional substitutions. We first test this prediction by contrasting neutral diversity in 500-kb windows near (<0.1 cM) compared to far (>0.5 cM) from any of the 21,278 amino acid substitutions we identified in the human lineage (Methods). Importantly, the windows are matched by all parameters associated with BGS that we described above.
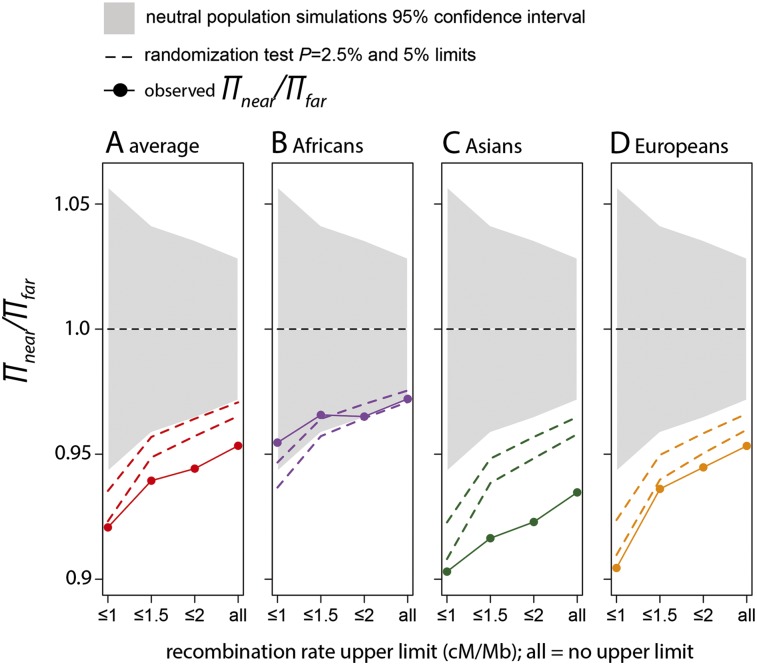
We first carry out this test in the regions with low density of conserved coding sequences (CCDS density<0.5%) and thus weak effects of BGS. This analysis reveals a substantial 5% decrease of neutral diversity near amino acid changes (Fig. 1A; randomization test P = 6 × 10−3; Methods). As expected under frequent positive selection, the decrease is more pronounced in low recombination regions (<1 cM/Mb), where the decrease of diversity is 8% on average (P = 1.5 × 10−2). The decrease is stronger in the Asian (9.5%, P = 1.2 × 10−2) and European (9.5%, P = 1.2 × 10−2) populations than in Africa (5%, P = 7.5 × 10−2) (Fig. 1B–D).
Figure 1.
Lower diversity near versus far from amino acid substitutions. Each panel shows the level of synonymous heterozygosity near amino acid changes (πnear) compared with that far from amino acid changes (πfar) for the particular subpopulation. πnear is the average over all near windows (<0.1 cM) from the bootstrap procedure (Methods). πfar is the average over all far windows (>0.5 cM but <1 cM). The gray area depicts the 95% confidence intervals based on neutral simulations (Methods). The dashed lines show 95% and 97.5% confidence intervals based on a randomization test (Methods). Randomization tests result in at most 35% greater variance than the neutral simulations. This is expected given that neutral simulations do not account for complex demography and other sources of noise in the data. On the x-axis, ≤1, ≤1.5, etc., means that we use only windows with recombination rates ≤1 cM/Mb, 1.5 cM/Mb, etc., to compare diversity near and far from amino acid substitutions. (All) All windows are used independently of their recombination rates.
When we include regions of higher conserved coding density (>0.5%), as well as those located >1 cM away from any amino acid substitution, we fail to detect any decrease in neutral diversity near amino acid substitutions. In fact, we find the opposite pattern of, on average, 4% higher diversity near amino acid substitutions (P = 4.6 × 10−2), reminiscent of the results of Hernandez et al. (2011). This suggests that BGS can indeed obscure signatures of positive selection in the human genome, making it essential to control for BGS and reduce its effects as much as possible when searching for positive selection.
The functional-vs-nonfunctional test
The regions of low BGS in the near-vs-far test above correspond to ∼30% of all the regions in which the test can be applied in principle, and ∼17% of the genome in total (290 Mb of “near” and 236 Mb of “far” windows; Supplemental Table 1). We are thus unable to apply this test to the majority of the genome. In addition, the choice of the threshold of CCDS <0.5% is somewhat ad hoc and was driven by the need to have enough windows for the bootstrap procedure while reducing the effect of BGS as much as possible.
In order to find additional signatures of positive selection that are less sensitive to BGS and can be applied to more of the genome, we modify the near-vs-far test to compare windows that have the same overall number of amino acid substitutions (i.e., all the windows are “near”), and then contrast the windows that differ by the presence or absence of predicted functionally consequential substitutions, as defined by PolyPhen-2 (Methods; Adzhubei et al. 2010). We reason that predicted functionally consequential substitutions are more likely to be adaptive than predicted neutral ones and should be associated with a more pronounced reduction of neutral polymorphism in their vicinity. At the same time, controlling for the total overall number of nonsynonymous substitutions in a window naturally controls for the variation in BGS.
We compare neutral diversity in 500-kb windows either near predicted functional amino acid substitutions (<0.1 cM) or near predicted neutral amino acid substitutions (<0.1 cM from a neutral substitution and >0.5 cM from a functional one). The matching windows must have the same (plus/minus one) total number of amino acid substitutions. In addition, we again control for the key genomic variables (densities of coding, conserved coding and noncoding sequences, recombination rate, and GC content; Methods). In total, this functional-vs-nonfunctional test includes 823 Mb near functional substitutions and 768 Mb near nonfunctional ones (∼50% of the genome in total; Supplemental Table 1) and therefore greatly extends the span of the human genome we are able to analyze.
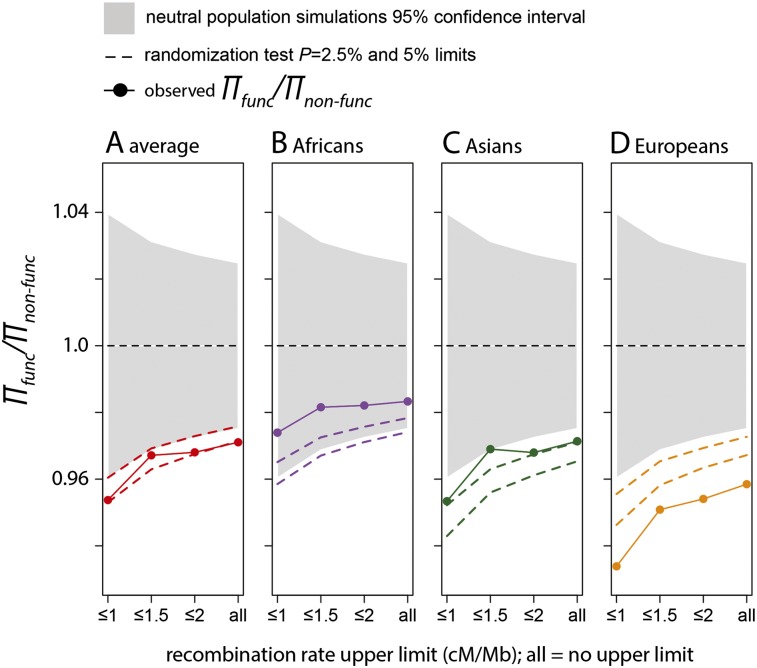
Using this test, we find that neutral diversity is decreased by ∼3% on average near functional compared to nonfunctional amino acid substitutions (Fig. 2A). This decrease is statistically significant but marginally so (randomization test P = 4.8 × 10−2). As expected, the decrease of diversity is more pronounced in regions with low rates of recombination (5% on average, P = 2 × 10−2; <1 cM/Mb) (Fig. 2A). The decrease is again weaker in the African population (3%, P = 0.1) compared with the Asian (5%, P = 4.5 × 10−2) and European populations (7%, P = 5 × 10−3) (Fig. 2B–D). Together with the near vs far test, this test further suggests a detectable effect of recent positive selection on human genetic diversity.
Figure 2.
Lower diversity near functional amino acid substitutions. We compared heterozygosity near functional amino acid changes, πfunc, with heterozygosity near nonfunctional amino acid changes, πnon-func. πfunc is the average over all functional windows from the bootstrap procedure (Methods). πnon-func is the average over all nonfunctional windows. The gray area depicts the 95% confidence intervals based on the neutral simulations (Methods). The dashed lines show 95% and 97.5% confidence intervals established based on a randomization test (Methods). On the x-axis, ≤1, ≤1.5, etc., means that we use only windows with recombination rates ≤1 cM/Mb, 1.5 cM/Mb, etc., to compare diversity near and far from functional amino acid substitutions. (All) All windows are used independently of their recombination rates.
The omnibus near-vs-far and functional-vs-nonfunctional test
The near-vs-far test (Fig. 1) and the functional-vs-nonfunctional test (Fig. 2) search for different signals in the data and should be independent of one another. Indeed, all regions in the functional-vs-nonfunctional test are located near an amino acid substitution, and thus they are all in the “near” category in the near-vs-far test. The fact that the “near” regions have lower diversity than the “far” regions should not affect the results of a test that looks only within the “near” regions. In addition, we confirm by simulation that the finite number of regions used in the bootstrap procedure does not generate spurious correlations between the two tests (Supplemental Material).
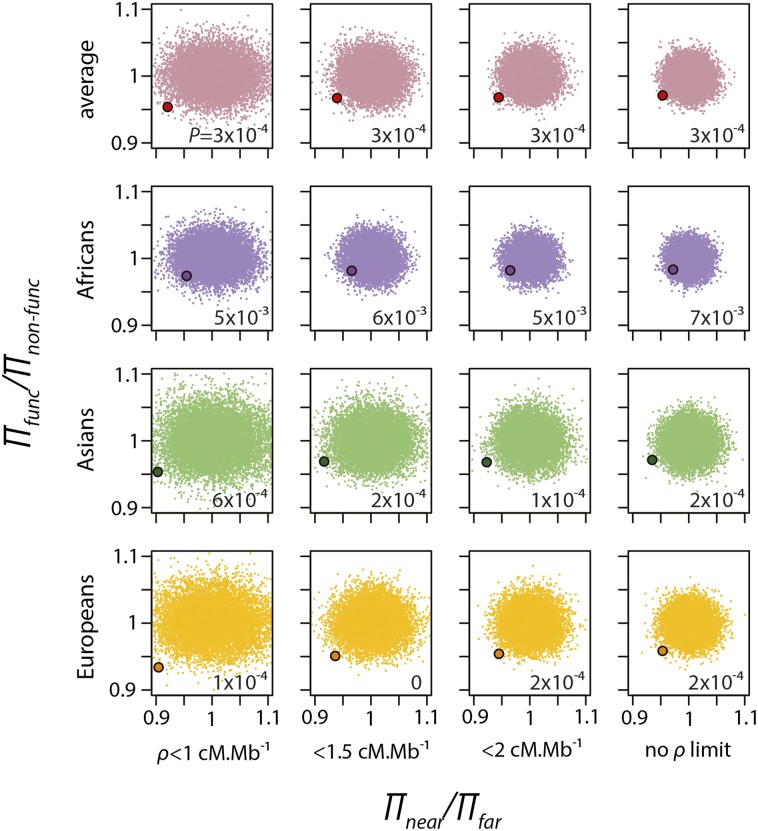
The independence of the two tests allows us to combine them into a single, omnibus test and calculate a joint P-value (Fig. 3). In all human populations, the observed combined decreases are highly statistically significant, as shown by the P-values of the combined randomization test in Figure 3 (all populations combined, P = 3 × 10−4; Asian, P = 2 × 10−4; African, P = 7 × 10−3; European, P = 2 × 10−4). Even in Africa, where the signal of positive selection is consistently weaker in both the near-vs-far and the functional-vs-nonfunctional tests, the probability of both observed decreasing by chance is <1%. In European and Asian populations, the same probability is <0.1%. Taken together, these results strongly suggest that positive selection has significantly decreased neutral diversity in the human genome.
Figure 3.
Combined near-vs-far and functional-vs-nonfunctional tests. Clouds of small dots represent the ratios πnear/πfar and πfunc/πnon-func obtained with the randomization test. The larger dot in each graph represents the observed πnear/πfar and πfunc/πnon-func. The numerical values at the lower right of each graph are the P-values obtained after 10,000 iterations of the randomization test. The P-values are estimated as the proportion of the randomizations that give values below the observed value in both tests.
Extreme values of XPEHH and iHS near and far from nonsynonymous substitutions
Both positive selection and BGS are expected to reduce the overall level of neutral polymorphism. Yet only positive selection, and not BGS, is expected to drive individual haplotypes to unusually high frequencies. Tests based on the presence of unusually frequent and long haplotypes, such as iHS (Sabeti et al. 2002; Voight et al. 2006) and XPEHH (Sabeti et al. 2007), should therefore be insensitive to BGS and provide a less confounded approach for the systematic detection of positive selection in the genome.
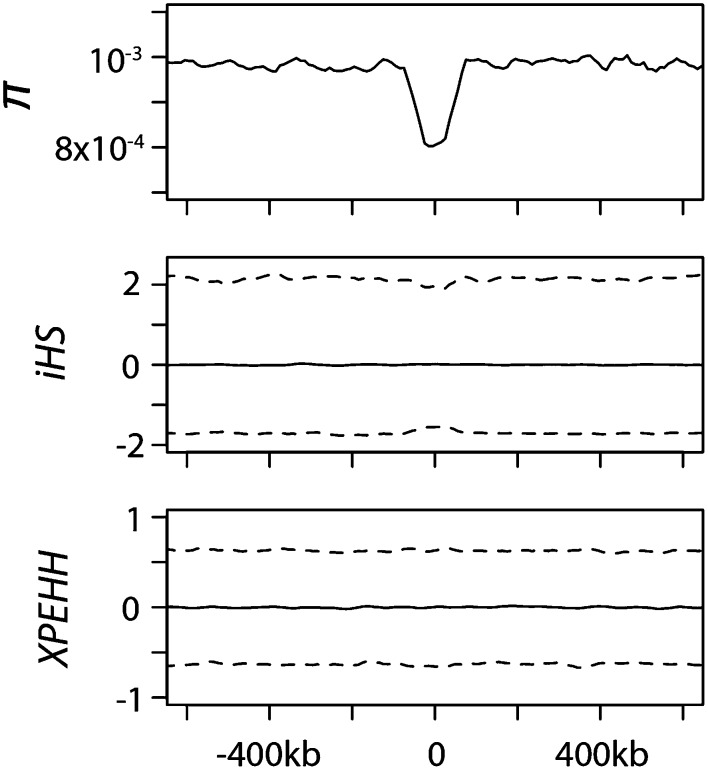
We first use extensive forward simulations to confirm this intuition. We use SLiM (Messer 2013) to simulate 4-Mb regions that include a 100-kb central region where deleterious mutations occur with a predefined strength of selection and rate (Supplemental Material). We analyze a range of distributions of selective effects of deleterious mutations (Supplemental Fig. 2), including a gamma distribution that matches our best current estimate of the DFE of functional mutations in the human genome (Keightley and Eyre-Walker 2007). As expected, BGS has a strong effect on the levels of diversity (Fig. 4) but has no detectable effect on XPEHH and only a marginal effect on iHS. BGS slightly decreases the variance of iHS values, thereby making scans for positive selection using extreme values of iHS conservative in the presence of BGS.
Figure 4.
Robustness of iHS and XPEHH to BGS. We tested the effect of BGS on iHS and XPEHH (Results; Supplemental Material). (Top) Average heterozygosity; (middle) iHS; (bottom) XPEHH. The full lines represent average iHS or XPEHH along the simulated region. The dashed lines represent the limits of iHS or XPEHH 95% confidence intervals.
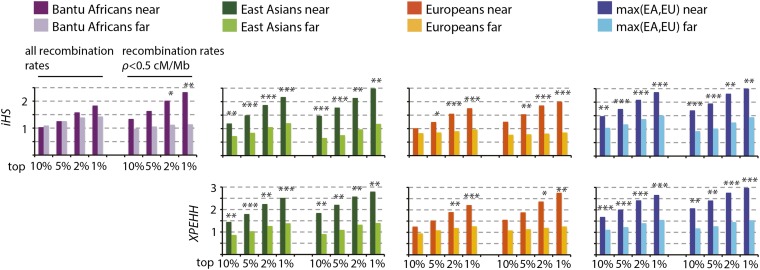
We modify the near-vs-far test by using extreme values of iHS and XPEHH instead of overall levels of neutral diversity near and far from amino acid substitutions as a measure of positive selection. For iHS, we consider the distribution of absolute values to capture adaptation driven by both ancestral and derived alleles and to avoid issues due to potential mispolarization of ancestral states. Specifically, we compare the most extreme values of XPEHH and iHS near and far from amino acid substitutions (Fig. 5). For example, we use the 10% of windows with the highest iHS values near amino acid changes and use the average iHS for these windows. We then compare this value with the average iHS of the 10% most extreme windows far from amino acid changes (Fig. 5). This comparison is repeated using the 5%, 2%, or 1% most extreme windows (Fig. 5). We use values of iHS and XPEHH calculated for the HGDP panel by Pickrell et al. (2009) for the Bantu, East Asian, and European populations. As before, the “near” windows are <0.1 cM and the “far” windows are >0.5 cM from any amino acid substitution. We control for levels of recombination and coding density in the bootstrap procedure. The significance of the differences between the near and far windows is again calculated using the randomization test (Methods).
Figure 5.
Stronger haplotype- and SFS-based signals of positive selection near amino acid changes. This figure shows the comparison of the top 10%, 5%, 2%, and 1% XPEHH and iHS windows near and far from amino acid changes. Randomization test: *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001. (Left side of histograms) All regions irrespective of recombination rates. (Right side) Only regions with recombination rates <0.5 cM/Mb. The max(EA,EU) histograms show the results obtained when retaining for each window the maximum signal of the East Asian and European populations.
Figure 5 shows clear signatures of positive selection in the iHS and XPEHH modification of the near-vs-far test. iHS shows significantly more extreme values near amino acid changes in all three tested populations (Fig. 5, upper row). In line with our prediction, this pattern is more pronounced in low recombination regions (<0.5 cM/Mbp) (Fig. 5, right side of histograms), especially in the African population. In order to increase the statistical power, we also compare the maximum values of iHS in East Asians and Europeans in each window near and far from amino acid changes and find that this test yields even more significant results.
Results are essentially the same when using the XPEHH modification of the near-vs-far test (Fig. 5, second row). We choose the African population as the reference population and apply the XPEHH modification of the near-vs-far test in the two remaining populations. The results are significant in both populations and again become more pronounced in the low recombination regions (<0.5 cM/Mbp) and when the maximum value of XPEHH in the two populations is used as a test statistic. Similar results are also obtained using the CLR test (Supplemental Material; Supplemental Fig. 3; Williamson et al. 2007).
Because iHS and XPEHH are insensitive to BGS, we were able to carry out these tests even in regions that have high coding density and in which the tests that rely on the overall level of polymorphism are too biased by BGS against the detection of positive selection. Specifically, the “near” windows in the iHS and XPEHH tests represent a total of 1.56 Gb, and “far” windows represent a total of 618 Mb, extending the analysis to ∼70% of the human genome (Supplemental Table 1).
Forward simulations of positive selection
We run forward simulations of positive selection using SLiM (Methods; Messer 2013) in order to determine the frequency and strength of selective sweeps required to decrease neutral diversity near amino acid substitutions between 2% and 9.5% in 500-kb windows, as observed in the data (Figs. 1, 2). In particular, we focus on regions of low recombination (<1 cM/Mb) and simulate adaptation with three different rates of adaptive amino acid substitutions (proportion of substitutions that are adaptive α = 10%, 20%, and 40%) and two different selection regimes (selection coefficient s = 0.01 and 0.05). Surprisingly, the amount of strong positive selection required to explain the observed reduction in diversity is very high (Fig. 6). The observed 9.5% reduction in Europe and Asia is similar to the average reduction expected if 40% of amino acid substitutions were adaptive with a selection coefficient of s = 0.05; they are in the high range if 10% or 20% of amino acid changes were adaptive with s = 0.01.
Figure 6.
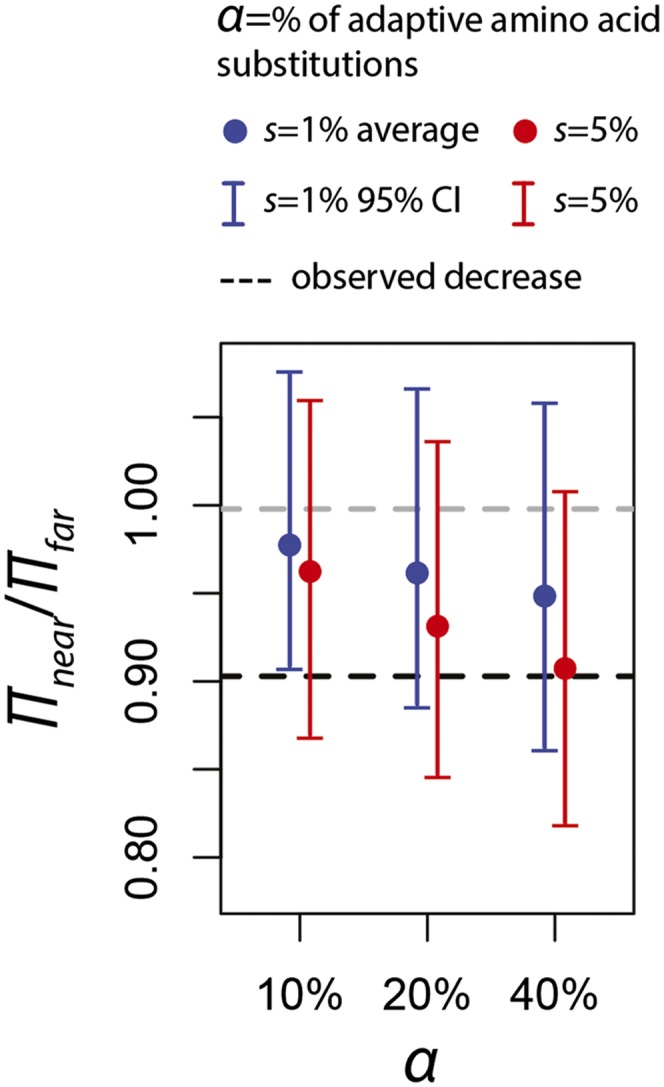
Simulated decreases of diversity for different rates and strengths of positive selection. We ran 100 forward simulations (Methods) to estimate the average and 95% confidence intervals (CI) for the decrease of diversity near amino acid changes under different rates and strengths of positive selection. To be conservative, we extended the confidence intervals from simulations by 35% given that neutral simulations underestimate the variance by ∼35% in the simulated regions, as shown in Figure 1.
This rate appears higher than that estimated with MK approaches, which predict that 10%–20% or fewer amino acid changes (Boyko et al. 2008; Messer and Petrov 2013) were adaptive in the human lineage. While MK estimates include both strongly (s > 0.01) and weakly (s < 0.001) selected substitutions, our simulations suggest that at least 10% of strongly selected (s = 0.01) amino acid substitutions would be required to obtain the observed decrease in diversity. This implies either that at least half of the adaptive amino acid substitutions were driven by strong selection or, alternatively, that the majority of adaptive changes are not amino acid substitutions themselves but instead are adaptations at nearby, possibly regulatory, sites.
Adaptation is centered at the ENCODE-defined regulatory elements
The above simulations suggest that adaptation by amino acid substitutions is unlikely to generate all of the observed signatures of adaptation. We therefore search for adaptation at regulatory regions by focusing on the regulatory elements defined by the ENCODE Project Consortium (ENCODE-defined regulatory elements, or EREs) (Gerstein et al. 2012). ERE density in our analysis is the density of elements predicted as DNase I hypersensitive sites and also as transcription factor binding sites identified via ChIP-seq by the ENCODE Project Consortium (Gerstein et al. 2012). Note that our strict definition of ERE elements leaves us with only 4% of the positions in the analyzed windows. Moreover, the ERE content correlates strongly with coding density (n = 2189, Spearman’s ρ = 0.73, P < 2 × 10−16), suggesting that EREs are indeed often functional.
We examine the correlation between the density of ERE and iHS in three populations of the 1000 Genomes Phase 1 project (Supplemental Material). In Europe and Asia, absolute values of iHS correlate positively with ERE density (Fig. 7B,C), but the correlation is more subtle in Africa, where it becomes positive only in low recombination regions (Fig. 7A). The correlation is notably stronger in regions with low recombination rates, as expected under frequent positive selection.
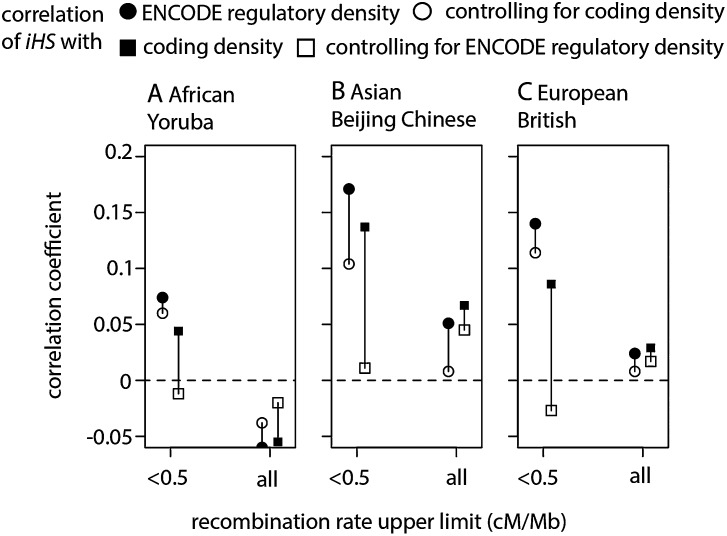
Figure 7.
Most human recent positive selection occurs in regulatory sequences. The filled circles and squares show the correlation coefficients of the absolute values of iHS with the density of regulatory and coding sequence density, respectively, controlling for recombination and average pairwise diversity (Methods). The open circles and squares show partial correlations. For instance, an open circle shows partial correlation between absolute values of iHS and regulatory density controlling for coding density (also controlling for recombination and average pairwise diversity). All correlations are Spearman’s rank correlations or partial correlations. Correlation coefficients >0.05 are all highly significant (P < 2 × 10−16).
Because ERE density correlates with the coding density and the coding density correlates with iHS (Fig. 7), it is important to disentangle the respective contributions of coding and regulatory sequences to the observed signals of recent positive selection. In order to do so, we calculate the reciprocal partial correlations between (1) iHS and ERE density controlling for coding density and (2) iHS and coding density controlling for ERE density. When using the whole genome regardless of recombination, partial correlations between iHS and ERE or coding density are weak and inconsistent between different human populations, being either positive in Asia or negative in Africa (Fig. 7A–C). In low recombination regions (<0.5 cM/Mb), where the effects are expected to be the strongest and clearest, the results are striking (Fig. 7A–C; Supplemental Table 2): While the partial correlation between iHS and ERE density appears virtually independent of coding density, the correlation between iHS and coding density disappears entirely when controlling for ERE density. These results are robust to the amount of overlap between the windows used to measure the correlations (Supplemental Table 2). In particular, we measure significant correlations with windows that are located at least 1 Mbp from each other and thus expected to provide largely independent values of all statistics (Supplemental Table 2). These results provide evidence that many signals of positive selection in the human genome may indeed be due to adaptation centered in regulatory rather than in coding sequences.
Discussion
In this study, we have used a number of independent approaches to search for signatures of positive selection in the patterns of variation in the human genome. Our results show that BGS can inhibit the detection of even frequent positive selection in humans. The key prediction of recurrent positive selection is that neutral polymorphism should be lower in regions with more functional substitutions, for instance, nonsynonymous substitutions. Perhaps counterintuitively, BGS is expected to generate the opposite signature: Regions of the genome with coding sequences but very few nonsynonymous substitutions are likely to exhibit stronger BGS and lower levels of neutral polymorphism. This means that the standard approaches that search for adaptation using the signature of low levels of polymorphism next to nonsynonymous substitutions—such as those of Andolfatto (2007), Macpherson et al. (2007), Cai et al. (2009), Hernandez et al. (2011), and Sattath et al. (2011)—are likely to underestimate the effect of positive selection. This underestimation is likely to be marginal in small and functionally dense genomes, such as that of Drosophila, where levels of BGS are expected to be homogeneous along the genome. However, in larger genomes with heterogeneous distribution of functional sequences, such as that of humans, the levels of BGS vary sharply along the genome, and this bias against finding signatures of positive selection can become profound. We confirmed this assertion using forward simulations of BGS in the human genome.
We were able to detect lower levels of polymorphism near nonsynonymous substitutions. Surprisingly, our results indicate that carefully matching windows near and far from nonsynonymous substitutions for a number of factors known to correlate with selective constraint and diversity in general is essential but not sufficient to fully control for BGS. Indeed, decreased diversity near nonsynonymous substitutions is apparent only when genomic regions of low functional density, and hence weak BGS, are analyzed. The opposite pattern of higher diversity near nonsynonymous substitutions is seen in regions of high functional density and hence strong BGS. Our interpretation of this pattern is that the very fact of observing a nonsynonymous substitution carries substantial information not only about rates of adaptation but also about the lower level of constraint and thus BGS. The latter effect becomes dominant in regions with strong BGS in general. Note, that although on balance we believe this is the most parsimonious explanation of the data and is consistent with the results based on haplotype statistics that are insensitive to BGS, we cannot exclude that some unknown variable correlating with average pairwise diversity might affect our results or that windows in regions of low functional density exhibit drastically different DFE of deleterious mutations.
Although unlikely, it is therefore still in principle possible to imagine scenarios where BGS alone could explain our results with average pairwise diversity. Thus it is essential that we were able to detect positive selection using the presence of long and frequent haplotypes that are unlikely to be mimicked by BGS. Indeed, we conducted extensive simulations of BGS under varying rates and patterns of deleterious mutation and showed that tests of selection based on the presence of long and frequent haplotypes (iHS and XPEHH) are insensitive to BGS. As expected under positive selection, we detected significantly more extreme values of iHS and XPEHH near amino acid substitutions. Because these statistics are insensitive to BGS, we were able to carry out this analysis systematically on a genome-wide scale, without being restricted to only the regions with low functional density, as in the case of the near-vs-far test using average pairwise diversity.
All the evidence together suggests that positive selection left detectable effects on patterns of variation in the human genome. However, it is also clear that these patterns are challenging to detect and quantify due to a number of factors in addition to BGS. First, demographic perturbations such as bottlenecks and admixture can generate variability in the levels of polymorphism and haplotype structure. Yet it is hard to imagine a scenario in which these demographic perturbations would affect windows near amino acid substitutions differently from those that are far from amino acid substitutions in the long history of evolution since divergence of humans and chimpanzees. Indeed, the vast majority of the amino acid substitutions happened long ago, prior to any demographic event in question. In addition, the windows near and far from amino acid substitutions that are used in the comparisons have had exactly the same demographic history. Thus the main effect of demography is to increase variance in levels of polymorphism in windows both near and far from amino acid substitutions, but it is unlikely to generate false positives by itself. Both the forward simulations and permutation tests carried out here highlight substantial variance in levels of polymorphism due to drift, which strongly limits our ability to obtain precise estimates of the rate and strength of recent positive selection. Second, it is very important to use windows with a large physical size (500 kb in our case) to correctly control for BGS (Methods). This means, however, that our analysis is likely limited to detecting only the effects of strong recent positive selection, given that only selected mutations with selection coefficients on the order of ≥1% can affect diversity in regions of several hundreds of kilobases (Sabeti et al. 2006). This bias against weaker positive selection further complicates attempts to precisely quantify the rate and strength of positive selection. Therefore our results suggest that BGS may make it far more difficult to distinguish and quantify the rates of weakly and strongly advantageous mutations, as could be done in Drosophila (Macpherson et al. 2007; Sattath et al. 2011). Third, patterns of recombination may also make it more difficult to detect positive selection in human compared with Drosophila (Myers et al. 2005). Indeed, recombination rates are known to be more heterogeneous in human than in Drosophila, and this may add even more variance in our analyses.
In our simulations of positive selection, we estimate that the observed decrease of diversity of ∼10% near amino acid substitutions would correspond to roughly 100 strong sweeps (s = 0.05) in the past 100,000 yr. However, confidence intervals in our simulations (Fig. 6) and the lack of knowledge of the DFE of advantageous mutations (Fig. 6) make it very hard to estimate the rate of adaptation precisely. While our results do exclude the possibility that adaptation had no effect on neutral diversity, they provide only a very rough order of magnitude estimate for the rate of recent positive selection.
It is also worth noting that although signals of positive selection are detectable in all tested populations, these signals are systematically stronger in the out-of-Africa populations. This pattern will require further investigation, as it could be due to many different and nonmutually exclusive reasons such as variation in demography, differences in patterns of linkage disequilibrium, different rates of adaptation, different patterns of monogenic and polygenic adaptation in different populations, and differences in the proportion of adaptation from de novo mutations versus from standing genetic variation.
In this study, we detect and quantify a number of signatures left by apparently abundant recent positive selection in the genome-wide signals in diversity in humans. We also provide suggestive evidence that positive selection may have been driven largely by regulatory rather than coding sequences. The primary evidence for this comes from two observations. First, haplotype signatures of positive selection captured by extreme values of iHS correlate better with the density of ENCODE regulatory elements (ERE) than with the density of coding sequences (Fig. 7). Second, admittedly simplified simulations suggest that without a substantial number of strongly advantageous regulatory substitutions taking place in the same regions as the amino acid substitutions, the total impact of positive selection on diversity would require a seemingly unreasonable number of strongly advantageous amino acid substitutions. Because windows near amino acid substitutions have 21% more human-specific fixed regulatory (ERE) substitutions compared with the windows far away from amino acid substitutions (as defined in the near-vs-far test, Fig. 1) and because ERE substitutions are ∼30 times more common than amino acid substitutions, even a modest difference in the rate of adaptation within EREs between near and far windows could potentially explain our results for the near vs far test.
Although our results support that most adaptation is regulatory, the possibility also remains that rates of coding adaptation obtained by MK approaches may be strong underestimates, as previously discussed by Eyre-Walker and Keightley (2009). This said, our conjecture is consistent with recent results showing that adaptation between different human populations may have been driven primarily by regulatory rather than coding differences as well (Fraser 2013). The challenge for the future is to better quantify and identify the nature of recent human-specific adaptations. This will likely require improved modeling of BGS in the human genome, based on a deeper knowledge of the DFE and its variability along the human genome.
Methods
Human-specific nonsynonymous and synonymous fixed substitutions
Human-specific nonsynonymous and synonymous substitutions were obtained using human–chimpanzee–orangutan coding DNA sequence (CDS) alignments. Human CDS are first extracted from the Ensembl v64 database (Flicek et al. 2012; http://www.ensembl.org/). For each gene, only the longest CDS is retained. The longest human CDS are then mapped onto the chimpanzee and orangutan genomes using BLAT (protein–protein BLAT, 60% minimum identity) (Kent 2002). The best, highest identity chimpanzee and orangutan BLAT hit sequences are then mapped back on the human genome. Only those human–chimpanzee and human–orangutan best reciprocal hits are retained for further analysis. Extracting chimpanzee and orangutan CDSs from their respective genomes using BLAT instead of directly using Ensembl annotations ensures that the sequences used during subsequent global alignment steps have good local similarity. The analysis is further restricted to those best BLAT reciprocal hits that coincide with Ensembl v64 one-to-one orthologs. A total of 17,237 CDS multiple alignments are finally obtained using PRANK (Löytynoja and Goldman 2008) under the codon evolution model settings. PRANK used with its codon evolution model was previously shown to be the most accurate solution to align CDS (Fletcher and Yang 2010). From these alignments, a total of 27,538 and 40,709 nonsynonymous and synonymous human-specific substitutions are identified, respectively. This includes only those cases where chimpanzee and orangutan both exhibit the same nucleotide at the orthologous position. Of the 27,538 nonsynonymous substitutions, a total of 21,278 are fixed in all African, Asian, and European populations. Of the 40,709 synonymous substitutions, 32,666 are fixed. The ratio of the number of fixed nonsynonymous to fixed synonymous substitutions is 65.1%, which is in very good agreement with the previous result of 64% obtained by Boyko et al. (2008). Only diversity patterns close to fixed substitutions are analyzed in the near-vs-far and the functional-vs-nonfunctional tests. Focusing on fixed substitutions is therefore intended to make results easier to interpret. This is also expected to be conservative when searching for sweeps, because we exclude fixations that occurred after the split of African and non-African populations.
PolyPhen-2 analysis
We use PolyPhen-2 (Adzhubei et al. 2010) to identify which human-specific amino acid substitutions are more likely to be functionally consequential. PolyPhen-2 annotates SNPs but can also be used to annotate fixed amino acid changes by using the REVERSE option. Of the 21,278 fixed amino acid changes specific to the human lineage, 18,924 (89%) can be annotated. Of these, 15,488 are annotated as benign, 1874 as possibly damaging, and 1562 as probably damaging by PolyPhen-2. The possibly damaging and probably damaging amino acid changes (18% of the total) are more likely to be functionally consequential than the benign ones. Thus, in the functional-vs-nonfunctional test (main text and section “Bootstrap Procedure” below), functional windows are those close to a possibly or probably damaging amino acid change, and the nonfunctional windows are those close to a benign amino acid change, but far from any possibly or probably damaging one.
Neutral diversity
Neutral diversity is measured using average heterozygosity π, measured as 2f(1 − f)n/(n − 1), where f is the frequency of the nonreference allele in the 1000 Genomes Phase 1 20100804 release (December 2010 update), and n is the number of chromosomes in 500-kb windows (see below for an in-depth discussion on window size). More specifically, average heterozygosity is calculated separately for the three African, Asian, and European populations. We use only positions outside of CDS, UTRs (from Ensembl v64) and phastCons CNEs (from the UCSC Genome Browser), simple repeats, and transposable elements identified by RepeatMasker (http://genome.ucsc.edu/). Excluding functional elements, repeats, and positions not aligned with a nucleotide in macaque, approximately a third of the positions within the windows can be used on average to measure neutral diversity. We also exclude all windows closer than 5 Mb to centromeres or telomeres from our analysis. Diversity is further scaled by the number of positions found to be divergent between human and macaque in human–macaque BLASTZ (Schwartz et al. 2003) alignments retrieved from the UCSC Genome Browser (http://genome.ucsc.edu/). This is done to eliminate the effect of local variations in mutation rate or remaining strong selective constraint. Because local changes in mutation rate and strong selective constraint affect both diversity and divergence equally, using the ratio of diversity on divergence removes at least partially the effects of heterogeneous mutation rates and selective constraint. Using scaled diversity implies that only those positions where a nucleotide (non-N or any other undefined position) is aligned with a nucleotide in macaque are used. Scaled neutral diversity is calculated within the 500-kb windows sliding every 5 kb in the genome. For a complete explanation on why we choose large windows with a fixed physical rather than genetic size, see Supplemental Material.
Bootstrap procedure
In humans, local functional density is very heterogeneous and is a main determinant of neutral diversity. Regions of high functional density have higher levels of BGS and hence lower levels of neutral diversity (McVicker et al. 2009; Lohmueller et al. 2011). GC content and recombination also have a strong influence on levels of neutral diversity (Results). In our study we want to characterize the effect of positive selection on neutral diversity. This is done by comparing neutral diversity in regions of the genome where the rate of positive selection is expected to be higher with neutral diversity in regions where the rate of positive selection is expected to be lower. Genomic windows with potentially higher rates of positive selection are called tested windows, and genomic windows with potentially lower rates of positive selection are called control windows. In the near-vs-far test, tested windows are the windows near amino acid changes (nearest amino acid change at <0.1 cM from the center of the window), and the control windows are windows far from any amino acid change (>0.5 cM). In the functional-vs-nonfunctional test, tested windows are the windows near functional amino acid changes according to PolyPhen-2 (<0.1 cM), and the control windows are windows near nonfunctional amino acid changes (<0.1 cM) but far from any functional amino acid change (>0.5 cM). In addition to positive selection, we also tested whether windows very far from any amino acid change (>1 cM) experience more BGS than windows moderately far from amino acid changes (between 0.1 cM and 1 cM). In this case tested windows are the windows between 0.1 cM and 1 cM, and control windows are the windows >1 cM from any amino acid change.
The major challenge when testing positive selection by comparing tested and control windows is to make sure that both kinds of windows are as similar as possible. One may think of an example where in tested windows the percentage of positions within CDS is 2% on average and only 0.5% in control windows. In this case, there are four times more CDS in the tested windows than in the control windows. BGS is thus stronger in the tested windows. In such an example, neutral diversity is lower in tested windows than in control windows not because of positive selection but because of stronger BGS, and it is impossible to conclude anything about positive selection. This example shows that in order to be conclusive about positive selection, we need to compare windows with levels of BGS as similar as possible. This means that the tested and control windows need to have, on average, similar functional densities, in addition to similar recombination rates and GC content. This is achieved by using a simple bootstrap procedure. For each tested window, we match a control window whose characteristics are not more different than fixed thresholds compared with the tested window. These characteristics are the average recombination rate in the window obtained from the most recent deCode 2010 genetic map (Kong et al. 2010), GC content, CDS density (Ensembl v64), conserved coding sequences (CCDS) density (Ensembl v64), UTR density (Ensembl v64), and total functional density (TFD). CCDS are the 83% of coding sequences that overlap conserved segments (mammal-wide and/or primate-wide) predicted by phastCons (Siepel et al. 2005) and available at the UCSC Genome Browser (phastCons applied to a genome alignment of 44 mammals). TFD is the percentage of positions in a window that are in at least one of these different types of functional elements: CDSs, CCDSs, UTRs, and phastCons conserved noncoding elements (CNEs). In addition, we also control for the amount of surrounding CDS, which is the number of positions within a CDS up to 0.1 cM upstream of and 0.1 cM downstream from a window.
For each tested window, we find a matching control window whose recombination, GC content, CDS, CCDS, UTR, TFD, and surrounding CDS are comprised of between x% and y% of their values in the tested window. The values of x and y are specific to each of the controlled factors, and x is smaller than one while y is greater than one. For example, we could ask control windows to have a CDS density comprising between x = 80% and y = 120% of the tested window CDS density. In practice, we adjust the thresholds so that when the bootstrap is complete, tested windows and control windows have very similar average recombination rates, average GC content, and average CDS, CCDS, UTR, TFD, and surrounding CDS. In addition, we also make sure that they have very similar phastCons CNE density.
Although we cannot avoid slight differences, we make sure they are in the conservative direction. For example, the average CDS density in the control windows may be 3% higher than in the tested windows, and the average recombination rate may be 5% lower. When no matching control window is found in the genome, the tested window is excluded from the analysis. The same control window can be used several times as a match for several tested windows. The different amounts of sequences that could be used for each test are shown in Supplemental Table 1. The x% and y% thresholds used for the different tests conducted in this analysis are provided in Supplemental Table 3. Note that the thresholds were adjusted so that they could be used for all the repetitions of a given test in various conditions. For example, in the near-vs-far test, we used thresholds that can be applied whether or not we use only windows below a fixed recombination threshold and whether or not we use only low CCDS windows (Results). This is to ensure that the results obtained under these different conditions can be fairly compared between each other. For the near-vs-far test, low CCDS, and recombination rate <1 cM/Mb (Results), we further controlled that the observed decrease of diversity is robust to changing x and y for several factors, while still having conservative comparisons between near and far windows (Supplemental Table 4).
For each test, the bootstrap procedure is conducted 10 times, independently. Each time, we calculate the average neutral diversity in tested windows Πtested, the average neutral diversity in control windows Πcontrol, and the ratio Πtested/Πcontrol. Different realizations of the bootstrap procedure give very similar Πtested/Πcontrol ratios. For all tests and for each realization, the ratio Πtested/Πcontrol never differs by >10% of its average over the 10 realizations. The observed ratios Πtested/Πcontrol shown in Figures 1 and 2 represent the average over the 10 realizations of the bootstrap procedure. Because there is so little variation between the different realizations of the bootstrap procedure, we always use the first realization for running populations simulations (see section “Population Simulations” below) and for calculating P-values of the randomization test (see section “Randomization Test” below). Note also that we do not include average sequencing depth in the windows as one of the controlled variables, although it is well known to have an effect on the estimation of neutral diversity. This is because we found this is not necessary since, on average, the tested and control windows retained by the bootstrap procedure have extremely similar average sequencing depths that never vary by >0.5% from each other.
Randomization test
We use a randomization test to estimate the significance of the differences of neutral diversity we observe between tested and control windows used in the bootstrap procedure. In order to obtain a random distribution of Πtested/Πcontrol for a given realization of the bootstrap procedure, we need to shuffle tested and control windows while accounting for a number of features of the analysis. First, the tested and the control windows are often clustered together, much like the windows represented along a chromosome in Supplemental Figure 5. Πtested and Πcontrol are calculated from groups of neighboring, overlapping windows that have correlated neutral diversity values. Compared to a situation where we would have the same number of windows but all independent from each other, this grouping substantially increases the variance of Πtested, Πcontrol and thus of the ratio Πtested/Πcontrol. Shuffling individual windows independently from each other is therefore very likely to greatly underestimate the true variance of the ratio. Second, during the bootstrap procedure, the same control window can be matched with several tested windows, which should also be taken into account during the randomization process. In order to maintain the structure of the sampling scheme used in the bootstrap procedure, we shuffle blocks of neighboring windows (Supplemental Fig. 4). Windows used in the bootstrap procedure are first ordered according to their genomic positions. We then cut 20 segments of equal size (Supplemental Fig. 4 represents a situation with only three segments). This is done to maintain the grouping of windows. The 20 segments are then shuffled to obtain a new random ordering of windows. In addition, a segment can be flipped with a probability of 50%. The same sampling scheme that was used during the bootstrap procedure is finally applied to the randomized windows. For example, in the genome, the positions 19, 20, and 21 are occupied by tested windows tested_19, tested_20, and _tested_21, which are all matched to the same control window, control_29 at position 29 (Supplemental Fig. 2). After the randomization, positions 19, 20, and 21 are now occupied by the tested windows tested_8, tested_9, and tested_10 that are now all matched to window tested_18 at position 29. This way, the neighboring windows, tested_19, tested_20, and tested_21 have been replaced by three other neighboring windows, and window tested_18 matches three times as window control_29. The randomization process is repeated 10,000 times to obtain the P-value for the test. P-values are calculated as the proportion of randomizations where random Πtested/Πcontrol is lower or higher than the observed Πtested/Πcontrol depending on the case studied. This means that the randomization test is a one-sided test.
Population simulations
In our study we use forward simulations to estimate the ranges of the ratios of ∏near/∏far and ∏func/∏non-func under both a demographic scenario of panmixia with no advantageous mutation and under a scenario of panmixia with different rates and strengths of positive selection. Simulations were conducted using SLiM (Messer 2013). We simulate segments of the human genome where windows were sampled by the bootstrap procedure. Supplemental Figure 3 shows how those segments are defined based on where the sampled windows are in the genome and how far they are from each other. In Supplemental Figure 5, 500-kb sampled windows define three nonoverlapping groups along a chromosome. The first and second groups (starting from the left) are at a distance of 0.23 cM from each other. These two groups are fused together to form a genomic segment that includes them both. The segment is further extended 0.1 cM upstream and 0.1 cM downstream to avoid edge effects and to include the effect of eventual neighboring advantageous mutations not included in, but close to, the sampled windows (Supplemental Fig. 5). The third group is at 0.84 cM and is treated as an independent segment. Overall, groups of windows closer than 0.5 cM from each other are fused together, while groups >0.5 cM from each other are treated as independent simulated segments.
All the segments in the genome are simulated independently, and the simulated ratios Πtested/Πcontrol are calculated exactly as they are when using the bootstrapping procedure. This means that the same 500-kb windows are used and that within each window, variants whose coordinates fall within a functional element or a repeated element or do not align with macaque in the real genome are excluded from the calculation of simulated diversity. The whole operation is repeated 100 times for the estimation of confidence intervals of Πtested/Πcontrol.
The recombination maps used in each segment match the deCode 2010 recombination map (Kong et al. 2010). The simulations were conducted using a population of 500 individuals, and the recombination and mutation rates were rescaled accordingly to match the average recombination rate (1.16 cM/Mb) and the average heterozygosity (0.001) observed in the human genome. After a burn-in of 5000 generations, the neutral simulations are continued for 1000 additional generations (this is equivalent to 20,000 generations in a nonrescaled 10,000 individuals human population). Simulations with positive selection are continued for 2500 generations after the burn-in to ensure that all advantageous mutations introduced after the burn-in are given a fair amount of time to fix.
For the simulations with positive selection, we introduce advantageous mutations at random generation times with a fixed rescaled selection coefficient at positions where amino acid changes are found in the human genome. As an example, we can simulate a scenario where 10% of the amino acid changes were adaptive with s = 1%. The selection coefficient of 1% in a population of 10,000 individuals is rescaled to 20% in our 500 individuals simulated population to maintain the same intensity of selection. In order to obtain 10% of fixed adaptive mutations given the probability of fixation (2s = 40%), we need 25% of the introduced mutations with s = 20%. These advantageous mutations are introduced randomly among all the locations with an amino acid change. For the sake of speed in our simulations with positive selection, we use 2500 generations after burn-in, although in our rescaled population the number of generations to the human–chimpanzee most recent ancestor is 10,000 generations (rescaled from 200,000 generations assuming a TMRCA of 5 Myr and a generation time of 25 yr). Advantageous mutations were thus attributed an introduction time between 1 and 10,000 generations after burn-in, but only those mutations having a random introduction generation between 1 and 2500 were actually introduced in the population.
Acknowledgments
We thank Hugues Roest Crollius (ENS Paris) for sharing his computational resources, Pardis Sabeti, Ryan Hernandez, Kirk Loehmueller, Hunter Fraser, Noah Rosenberg, Daniel Weissman, and members of the Petrov laboratory, especially Pleuni Pennings, Fabian Staubach, Diamantis Sellis, Rajiv McCoy, Anna-Sophie Fiston-Lavier, and Nandita Garud for helpful comments on the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.164822.113.
References
- The 1000 Genomes Project Consortium. 2012. An integrated map of genetic variation from 1,092 human genomes. Nature 491: 56–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR 2010. A method and server for predicting damaging missense mutations. Nat Methods 7: 248–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akey JM 2009. Constructing genomic maps of positive selection in humans: Where do we go from here? Genome Res 19: 711–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andolfatto P 2007. Hitchhiking effects of recurrent beneficial amino acid substitutions in the Drosophila melanogaster genome. Genome Res 17: 1755–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyko AR, Williamson SH, Indap AR, Degenhardt JD, Hernandez RD, Lohmueller KE, Adams MD, Schmidt S, Sninsky JJ, Sunyaev SR, et al. 2008. Assessing the evolutionary impact of amino acid mutations in the human genome. PLoS Genet 4: e1000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai JJ, Macpherson JM, Sella G, Petrov DA 2009. Pervasive hitchhiking at coding and regulatory sites in humans. PLoS Genet 5: e1000336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B, Morgan MT, Charlesworth D 1993. The effect of deleterious mutations on neutral molecular variation. Genetics 134: 1289–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre-Walker A, Keightley PD 2009. Estimating the rate of adaptive molecular evolution in the presence of slightly deleterious mutations and population size change. Mol Biol Evol 26: 2097–2108 [DOI] [PubMed] [Google Scholar]
- Fletcher W, Yang Z 2010. The effect of insertions, deletions, and alignment errors on the branch-site test of positive selection. Mol Biol Evol 27: 2257–2267 [DOI] [PubMed] [Google Scholar]
- Flicek P, Amode MR, Barrell D, Beal K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fairley S, Fitzgerald S, et al. 2012. Ensembl 2012. Nucleic Acids Res 40: D84–D90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser HB 2013. Gene expression drives local adaptation in humans. Genome Res 23: 1089–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein MB, Kundaje A, Hariharan M, Landt SG, Yan KK, Cheng C, Mu XJ, Khurana E, Rozowsky J, Alexander R, et al. 2012. Architecture of the human regulatory network derived from ENCODE data. Nature 489: 91–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman SR, Andersen KG, Shlyakhter I, Tabrizi S, Winnicki S, Yen A, Park DJ, Griesemer D, Karlsson EK, Wong SH, et al. 2013. Identifying recent adaptations in large-scale genomic data. Cell 152: 703–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez RD, Kelley JL, Elyashiv E, Melton SC, Auton A, McVean G, Sella G, Przeworski M 2011. Classic selective sweeps were rare in recent human evolution. Science 331: 920–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley PD, Eyre-Walker A 2007. Joint inference of the distribution of fitness effects of deleterious mutations and population demography based on nucleotide polymorphism frequencies. Genetics 177: 2251–2261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ 2002. BLAT: the BLAST-like alignment tool. Genome Res 12: 656–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King MC, Wilson AC 1975. Evolution at two levels in humans and chimpanzees. Science 188: 107–116 [DOI] [PubMed] [Google Scholar]
- Kong A, Thorleifsson G, Gudbjartsson DF, Masson G, Sigurdsson A, Jonasdottir A, Walters GB, Gylfason A, Kristinsson KT, Gudjonsson SA, et al. 2010. Fine-scale recombination rate differences between sexes, populations and individuals. Nature 467: 1099–1103 [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921 [DOI] [PubMed] [Google Scholar]
- Lohmueller KE, Albrechtsen A, Li Y, Kim SY, Korneliussen T, Vinckenbosch N, Tian G, Huerta-Sanchez E, Feder AF, Grarup N, et al. 2011. Natural selection affects multiple aspects of genetic variation at putatively neutral sites across the human genome. PLoS Genet 7: e1002326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löytynoja A, Goldman N 2008. Phylogeny-aware gap placement prevents errors in sequence alignment and evolutionary analysis. Science 320: 1632–1635 [DOI] [PubMed] [Google Scholar]
- Macpherson JM, Sella G, Davis JC, Petrov DA 2007. Genomewide spatial correspondence between nonsynonymous divergence and neutral polymorphism reveals extensive adaptation in Drosophila. Genetics 177: 2083–2099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVicker G, Gordon D, Davis C, Green P 2009. Widespread genomic signatures of natural selection in hominid evolution. PLoS Genet 5: e1000471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messer PW 2013. SLiM: simulating evolution with selection and linkage. Genetics 194: 1037–1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messer PW, Petrov DA 2013. Frequent adaptation and the McDonald-Kreitman test. Proc Natl Acad Sci 110: 8615–8620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers S, Bottolo L, Freeman C, McVean G, Donnelly P 2005. A fine-scale map of recombination rates and hotspots across the human genome. Science 310: 321–324 [DOI] [PubMed] [Google Scholar]
- Pickrell JK, Coop G, Novembre J, Kudaravalli S, Li JZ, Absher D, Srinivasan BS, Barsh GS, Myers RM, Feldman MW, et al. 2009. Signals of recent positive selection in a worldwide sample of human populations. Genome Res 19: 826–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabeti PC, Reich DE, Higgins JM, Levine HZ, Richter DJ, Schaffner SF, Gabriel SB, Platko JV, Patterson NJ, McDonald GJ, et al. 2002. Detecting recent positive selection in the human genome from haplotype structure. Nature 419: 832–837 [DOI] [PubMed] [Google Scholar]
- Sabeti PC, Schaffner SF, Fry B, Lohmueller J, Varilly P, Shamovsky O, Palma A, Mikkelsen TS, Altshuler D, Lander ES 2006. Positive natural selection in the human lineage. Science 312: 1614–1620 [DOI] [PubMed] [Google Scholar]
- Sabeti PC, Varilly P, Fry B, Lohmueller J, Hostetter E, Cotsapas C, Xie X, Byrne EH, McCarroll SA, Gaudet R, et al. 2007. Genome-wide detection and characterization of positive selection in human populations. Nature 449: 913–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattath S, Elyashiv E, Kolodny O, Rinott Y, Sella G 2011. Pervasive adaptive protein evolution apparent in diversity patterns around amino acid substitutions in Drosophila simulans. PLoS Genet 7: e1001302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Kent WJ, Smit A, Zhang Z, Baertsch R, Hardison RC, Haussler D, Miller W 2003. Human-mouse alignments with BLASTZ. Genome Res 13: 103–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al. 2005. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res 15: 1034–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voight BF, Kudaravalli S, Wen X, Pritchard JK 2006. A map of recent positive selection in the human genome. PLoS Biol 4: e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson SH, Hubisz MJ, Clark AG, Payseur BA, Bustamante CD, Nielsen R 2007. Localizing recent adaptive evolution in the human genome. PLoS Genet 3: e90. [DOI] [PMC free article] [PubMed] [Google Scholar]