Abstract
The metal hyperaccumulator Noccaea caerulescens is an established model to study the adaptation of plants to metalliferous soils. Various comparators have been used in these studies. The choice of suitable comparators is important and depends on the hypothesis to be tested and methods to be used. In high-throughput analyses such as microarray, N. caerulescens has been compared to non-tolerant, non-accumulator plants like Arabidopsis thaliana or Thlaspi arvense rather than to the related hypertolerant or hyperaccumulator plants. An underutilized source is N. caerulescens populations with considerable variation in their capacity to accumulate and tolerate metals. Whole transcriptome sequencing (RNA-Seq) is revealing interesting variation in their gene expression profiles. Combining physiological characteristics of N. caerulescens accessions with their RNA-Seq has a great potential to provide detailed insight into the underlying molecular mechanisms, including entirely new gene products. In this review we will critically consider comparative transcriptome analyses carried out to explore metal hyperaccumulation and hypertolerance of N. caerulescens, and demonstrate the potential of RNA-Seq analysis as a tool in evolutionary genomics.
Keywords: RNA-Seq, deep sequencing, NGS, Noccaea caerulescens, Thlaspi caerulescens, hyperaccumulation, metal tolerance, Brassicaceae
INTRODUCTION
The Alpine pennycress Noccaea caerulescens (previously Thlaspi caerulescens) from the Brassicaceae family has been extensively studied at the physiological level for its ability to hyperaccumulate and hypertolerate metals such as Zn, Cd, and Ni (Meerts and Van Isacker, 1997; Lombi et al., 2000; Reeves et al., 2001; Assunção et al., 2003c; Roosens et al., 2003; Escarré et al., 2013). To explore the basis for these traits, a number of plants with differences in hyperaccumulation and hypertolerance have been compared. This includes cross- and intra-species comparisons, and comparisons between accumulators and non-accumulators. The choice of comparators is important and depends on the hypothesis to be tested and methodology used. While deep sequencing techniques have the potential to greatly increase our understanding about the mechanisms of plant metal-related traits and their evolution during the adaptation to different environments, a careful consideration of appropriate comparators becomes increasingly important. A major challenge is to find the genes of interest among those differentially expressed between the plants. In this review we consider the choice of comparators in exploring metal hyperaccumulation and hypertolerance characteristics of N. caerulescens, and discuss the pros and cons of the approaches in relation to RNA-Seq analysis. Applying RNA-Seq analysis to N. caerulescens populations with contrasting metal tolerance or hyperaccumulation capacities is a superior tool to analyze the evolutionary genomics of plant adaptation strategies to metalliferous soils.
TRANSCRIPTOME COMPARISON BETWEEN N. caerulescens AND NON-TOLERANT NON-ACCUMULATOR PLANTS
The non-accumulator, non-tolerant Arabidopsis thaliana, a model plant from the Brassicaceae family, is considered as a good comparator to N. caerulescens because of considerable sequence similarity: 87-88% identity in intergenic transcribed spacer regions (Peer et al., 2003), and ca. 88.5% nucleotide identity within transcribed regions (Rigola et al., 2006). Furthermore, genetic resources available for A. thaliana are superior compared to any other plant.
One of the first efforts to characterize the N. caerulescens transcriptome was made by Rigola et al. (2006), comparing ESTs (expressed sequence tags) of A. thaliana and N. caerulescens accession La Calamine, a Zn-tolerant Zn hyperaccumulator (Assunção et al., 2003c). Besides two species, the complex comparison thus included Zn hyperaccumulator/non-accumulator pair, Zn tolerant/non-tolerant pair as well as different Zn concentrations. The assembled partial cDNA sequences (unigenes) represented only 13% of the root and shoot transcriptomes, but some interesting findings emerged. Several unigenes showed similarity to genes for which a role in metal hyperaccumulation, tolerance or homeostasis was previously implicated. Three percent of the unigenes corresponded to A. thaliana orthologues not known to be expressed, and ca. 8% were considered N. caerulescens – specific. An effort was made to estimate transcriptional activity of highly expressed genes from the frequency of their detection but, unlike RNA-Seq, EST analysis provides limited information about transcript abundances.
To identify genes involved in Zn tolerance and/or accumulation, van de Mortel et al. (2006) compared root gene expression profiles of A. thaliana and N. caerulescens (La Calamine) under Zn excess and deficiency. For both species, a 60-mer oligo DNA microarray covering nearly complete A. thaliana transcriptome was used. An uncertainty in this approach was that the probes generally did not hybridize to N. caerulescens cDNA as efficiently as to A. thaliana cDNA, partly because they were designed to fit less conserved regions of A. thaliana transcripts. Furthermore, in the absence of N. caerulescens genome data, a marked effort was needed to develop primers to verify the results by PCR, as microarray does not provide accurate information about sequences. Over 2000 genes were significantly differentially expressed between A. thaliana and N. caerulescens. A challenge remained that most of the differentially expressed genes would not be directly linked to metal homeostasis. Furthermore, considering Zn hyperaccumulation as a constitutive species-level trait in N. caerulescens (Rascio and Navari-Izzo, 2011), many metal homeostasis genes being constitutively highly expressed in hyperaccumulators (Hammond et al., 2006; van de Mortel et al., 2006, 2008), Zn-dependence of the expression would not be expected for these genes.
In order to identify genes primarily involved in Cd tolerance, van de Mortel et al. (2008) investigated the root gene expression profiles of A. thaliana and N. caerulescens (La Calamine) under different Cd and Zn exposures, using the same A. thaliana array platform as van de Mortel et al. (2006). The authors concluded that these two species have specific responses to Cd, and emphasized the role of lignin, glutathione and sulfate metabolism. Only 93 out of 409 N. caerulescens genes differentially expressed in response to Zn exposures were identical to those reported by van de Mortel et al. (2006), highlighting the fact that differences in plant growth conditions, sampling, but also probe hybridization conditions lead to major differences in the outcomes. Interestingly, although similarities were found in gene expression profiles between N. caerulescens and Arabidopsis halleri, another metal hyperaccumulator species, compared to A. thaliana, there were also clear differences (Weber et al., 2004; Talke et al., 2006), which implied either species-specific differences in the mechanisms of metal hyperaccumulation or methodological differences.
Another non-accumulator plant, Thlaspi arvense, has been used as a comparator to N. caerulescens. The average similarity between the coding regions of A. thaliana and N. caerulescens or T. arvense genes was calculated by Hammond et al. (2006) as 81.5%. To increase the understanding about Zn hyperaccumulation mechanisms, Hammond et al. (2006) compared shoot transcriptomes of agar- and compost-grown N. caerulescens (Viviez population) with T. arvense by using an A. thaliana array, in which each gene was represented by a set of oligo probes. The array was extensively validated in order to overcome the potential limitations arising from sequence divergence between orthologous genes in A. thaliana, N. caerulescens and T. arvense, as the probe selection was demonstrated to dramatically affect the estimates of differential gene expression. Approximately 5000 differentially expressed genes were found, including several genes previously implicated in Zn homeostasis. Literature comparison indicated that very few of those genes were common to the genes identified when A. halleri and A. thaliana were compared which, according to Hammond et al. (2006), might have been a consequence of not accounting for sequence divergences between A. thaliana and A. halleri. Furthermore, although Viviez population shows constitutive Zn hyperaccumulation capacity, it is primarily a Cd hyperaccumulator and thus perhaps not the best comparator to study Zn hyperaccumulation (Schwartz et al., 2003).
Until now, no transcriptome comparisons between N. caerulescens and related metal hyperaccumulators are available. In principle, such comparisons might be useful if the plants provide unique accumulation/tolerance profiles. Even with similar profiles, these comparisons might help to reject or support a hypothesis and could tell whether the genetic basis of any specific tolerance or accumulation mechanism is species-specific or more universal. Examples of such plants are the Ni accumulator N. goesingense (Lombi et al., 2000) and the Zn/Cd hyperaccumulator N. praecox, which accumulates Cd similarly to N. caerulescens Ganges accession but is less Cd-tolerant (Vogel-Mikuš et al., 2005). Another comparator of interest is the more distantly related A. halleri, which accumulates Cd and Zn but not Ni, is not Ni tolerant, shows different Zn compartmentation, and has differences in the regulation of Cd uptake compared to N. caerulescens (Cosio et al., 2004; Marquès et al., 2004; Broadley et al., 2007). Comparing N. caerulescens and A. halleri at specific tissue- or even single cell level might help determining the specific mechanisms responsible for metal accumulation (e.g., different Zn compartmentation).
MICROARRAY ANALYSIS VERSUS RNA-SEQ
While cross-species microarray analyses have provided valuable information about N. caerulescens and its responses to environment compared to the related non-accumulators A. thaliana and T. arvense, the approaches have limitations, which may lead to a conclusion that there is a link between metal hyperaccumulation and a specific gene expression when in fact there is not, and vice versa. All microarray-based methods have limitations in terms of sensitivity and dynamic range, but a major challenge in cross-species comparisons is the sequence divergence of orthologous genes and thus the probe design. This can lead to inaccuracies, e.g., in estimating relative transcript abundances and to the lack of identification of novel transcripts.
RNA-Seq analysis is rapidly becoming the method of choice in the cross- and intra-species comparisons. Some advantages are its superior sensitivity and dynamic range. An example of the sensitivity is the MYB72 gene, which is of interest because the A. thaliana myb72 loss-of-function mutant exhibits decreased tolerance to Zn and Fe (van de Mortel et al., 2008). No expression of an orthologue could be established in N. caerulescens by microarray or low-stringency semi-quantitative RT-PCR (van de Mortel et al., 2008). Low-level expression was, however, found by RNA-Seq analysis, which opens the possibility that this transcription factor is involved in Zn tolerance of N. caerulescens (Halimaa et al., 2014). It should be noted that the detection of lowly expressed genes in RNA-Seq analysis is particularly sensitive to sequencing depth and coverage (Rapaport et al., 2013).
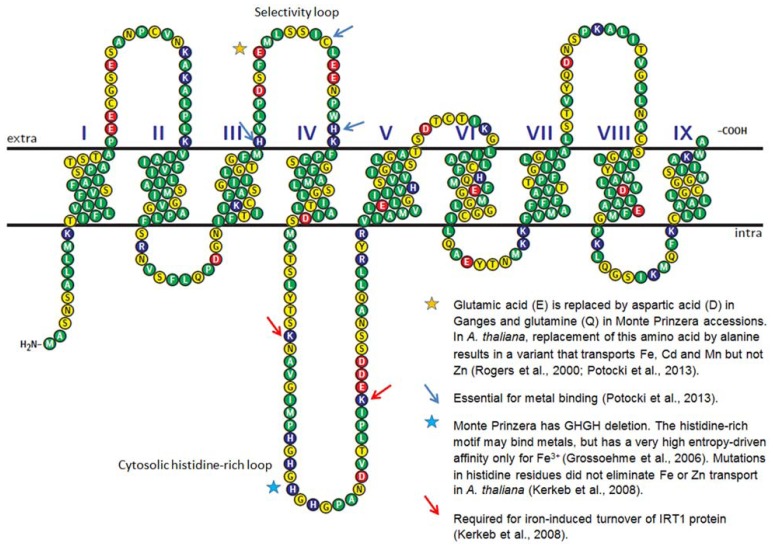
Since the output is made available as sequences and read counts rather than signal intensities like in microarrays, RNA-Seq is particularly attractive for species that have not been fully sequenced, such as N. caerulescens. Apart from being able to reveal novel transcripts, RNA-Seq can provide detailed information about single nucleotide polymorphisms (SNPs), splice variants and small RNAs. An example of nucleotide polymorphism leading to differences in amino acid sequences is shown in Figure 1 for IRT1 transcript assembled from Illumina RNA-Seq data in three N. caerulescens accessions (Halimaa et al., unpublished). Some of the differences in the corresponding amino acid sequences of this metal transporter might contribute to differences in metal specificity among the accessions, e.g., by influencing the affinity to different metals. In Arabidopsis, IRT1 has broad metal specificity (Eide et al., 1996; Korshunova et al., 1999).
FIGURE 1.
Predicted secondary structure of A. thaliana IRT1 metal transporter. The amino acid sequence was retrieved from TAIR10 database. The secondary structure of the protein was predicted using HMMTOP 2.0 (Tusnády and Simon, 1998, 2001). The diagram was generated with TEXtopo package (Beitz, 2000). Amino acids known to be important for the function of IRT1 are labeled with stars and arrows. Stars also indicate the locations where N. caerulescens accessions differ from A. thaliana. The N. caerulescens amino acid sequences were derived from Illumina RNA-Seq analysis of three accessions: Lellingen, Ganges, and Monte Prinzera.
The RNA-Seq technology does not fully solve the problems faced with cross-species comparisons. When comparing, e.g., N. caerulescens and A. thaliana to a common reference genome, the two species will have different mapping properties, complicating the calculation of transcript abundances (Derrien et al., 2012). In cross-species comparisons, the safest approach is thus to compare the profiles of sets of genes rather than transcript levels of individual genes. This problem can be partly overcome by de novo assembling the N. caerulescens transcriptome. However, this introduces a new challenge of identifying orthologues between the two species, particularly for gene families with many similar sequences (Romero et al., 2012). The reliability of comparisons increases with closer phylogenetic relationship between the plants to be compared (Koch and German, 2013).
Noccaea Caerulescens ACCESSIONS AS THE SOLUTION FOR THE COMPARATIVE RNA-SEQ ANALYSIS
Whereas caution has to be exercised when comparing different species, different N. caerulescens accessions and crosses could be almost ideal comparators in RNA-Seq, particularly if the accessions have been self-pollinated for several generations and are physiologically well-characterized. Comparison of IRT1 sequences showed that, whereas differences were found on average in 12.4% of the nucleotides compared to A. thaliana, the polymorphism among three N. caerulescens accessions was only 1.7%, apart from a deletion in the histidine motif in Monte Prinzera (Figure 1; Halimaa et al., unpublished). Furthermore, when comparing transcript abundances between accessions by mapping the RNA-Seq reads to a common reference genome, the read counts can simply be normalized by the sequencing depth of the library (for a review of normalization methods see Dillies et al., 2012).
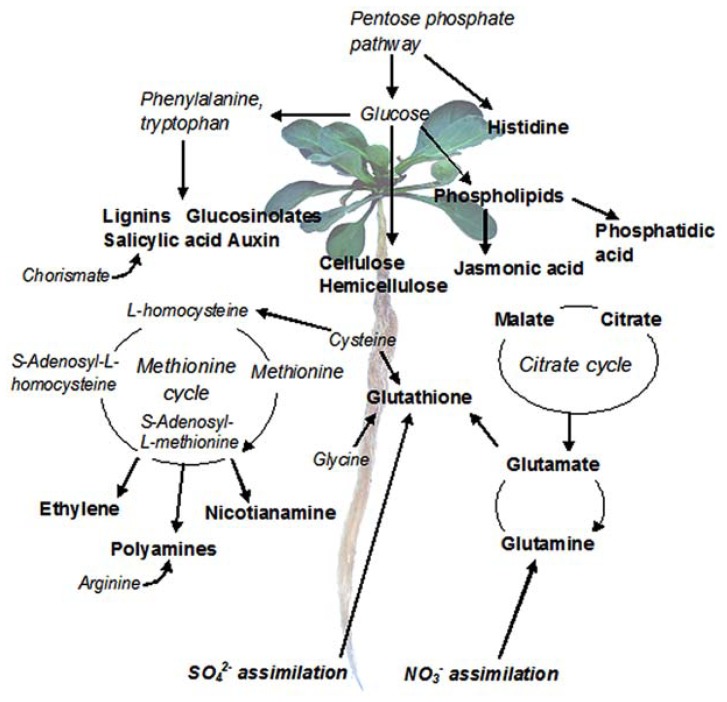
Another advantage of RNA-Seq is that different transcripts within one sample can be compared using FPKM normalization, i.e., adjustment by the length of each complete transcript. This can be used, e.g., to answer the question which pathways dominate in a specific tissue. As an example, we have used SOLiD RNA-Seq system to analyze the pathways that dominate in the roots of three N. caerulescens accessions (Figure 2; Halimaa et al., 2014). The biosynthesis of lignin, glucosinolates, and auxin were among the most highly expressed pathways in all three accessions. Lignin and auxin biosynthesis are related to general root development, while glucosinolate biosynthesis is associated with the Brassicaceae family. Genes related to sulfate and nitrate assimilation were also highly expressed, providing essential raw material to metal ligand synthesis. Nitrate enhances Zn hyperaccumulation in the roots and shoots of N. caerulescens (White-Monsant and Tang, 2013). Methionine cycle that generates intermediates to nicotianamine, ethylene, and polyamine synthesis was very highly expressed. Additionally, genes encoding the synthesis of metal ligands (e.g., glutathione, histidine, glutamine, metallothioneins, nicotianamine, and citrate) were abundantly expressed. Furthermore, our data indicated high expression of many genes linked to scavenging of reactive oxygen species (ROS) and related signaling networks. This is in line with previous findings linking salicylate and jasmonate biosynthesis to signaling of glutathione-mediated Ni tolerance (Freeman et al., 2005), and to Cd tolerance (Tolrà et al., 2006), respectively. As all known N. caerulescens populations have basal levels of metal tolerance and hyperaccumulation that exceed those found in non-accumulator plants (Assunção et al., 2003b), some of the highly expressed pathways probably contribute to these traits.
FIGURE 2.
The most active metabolic pathways in N. caerulescens roots. The data are based on SOLiD RNA-Seq analysis of three biological replicates of three N. caerulescens accessions: La Calamine, Ganges, and Monte Prinzera. The read counts were normalized by the size of the libraries and the length of the genes, assuming an equal length for orthologous genes in A. thaliana and N. caerulescens, and the most highly expressed genes (expression value > 1000) were subjected to a more detailed analysis. The genes were manually linked to metabolic pathways in KEGG (Kyoto Encyclopedia of Genes and Genomes).
Apart from the more or less constitutive high expression of the above-mentioned pathways contributing to the basal level of tolerance and accumulation, metabolic differences can be expected between N. caerulescens populations adapted to environments with different metal complements. The different populations are potentially a rich source of variation in the uptake, translocation, accumulation of and tolerance to several metals, and could thus serve as useful platforms in comparative analyses to explore the underlying molecular and evolutionary mechanisms. For example, one of the most studied N. caerulescens accessions found near a Zn/Cd smelter in Prayon, Belgium (Vázquez et al., 1992), accumulates 3.4% Zn in shoot dry weight, with shoot to root ratio of ca. six (Monsant et al., 2011), but accumulates much less Cd than some populations from Southern France (Robinson et al., 1998; Lombi et al., 2000; Reeves et al., 2001; Roosens et al., 2003). Assunção et al. (2003c) studied the physiological differences among four N. caerulescens accessions: Ganges from Southern France is the best Cd accumulator; La Calamine from a calamine ore waste in Belgium enriched with Zn, Cd, and Pb has the lowest level of accumulation of all the metals tested, and accumulates much less Cd and Ni than the non-accumulator T. arvense; Monte Prinzera from a Ni-enriched serpentine soil in Italy is characterized by its superior Ni tolerance and accumulation but is very sensitive to Cd; Lellingen from a non-metalliferous soil in Luxembourg is relatively sensitive to all three metals. Ganges and La Calamine have similar tolerance profiles, being the most tolerant to Zn and Cd, but least tolerant to Ni. Xing et al. (2008) demonstrated large variation between ten N. caerulescens accessions in Zn and Cd uptake and translocation. Escarré et al. (2013) found large heterogeneity in Zn, Cd, and Ni concentrations among eighteen metallicolous, non-metallicolous and serpentine N. caerulescens populations in response to different cultivation conditions. These examples demonstrate highly contrasting metal tolerance and accumulation characteristics among N. caerulescens populations, and points to their potential for comparative RNA-Seq combined with association analyses.
Comparing just two N. caerulescens accessions might not allow the conclusion why one population accumulates Ni and another one Cd or Zn while sharing the same genes. For example, comparison of N. caerulescens accessions Prayon and Ganges suggested that HMA4 expression does not correlate with Cd translocation efficiency (Xing et al., 2008), whereas opposite conclusion was drawn from the expression patterns of three different accessions, i.e., St-Félix-de-Pallières, Puente Basadre, and Prayon (Craciun et al., 2012), supported by our RNA-Seq comparison of Ganges, La Calamine, and Monte Prinzera accessions (Halimaa et al., 2014). However, both studies are consistent with initial findings of constitutive high expression of HMA4 in hyperaccumulators (Bernard et al., 2004; Talke et al., 2006; van de Mortel et al., 2006; Hammond et al., 2006), which is largely due to gene duplication (Hanikenne et al., 2008; Lochlainn et al., 2011; Craciun et al., 2012). A careful combination of accessions can thus significantly improve the predictability of comparisons. It is, however, important to recognize that in RNA-Seq analysis the number of replicated samples is the most predominant factor in providing detection power for differential expression (Rapaport et al., 2013).
The application of RNA-Seq and multiple predictive tools to the comparison of several N. caerulescens accessions is a powerful way to provide insights into the evolution of the populations in different environments. As an example, Gene Ontology (GO) enrichment analysis indicated that the most significant differences between N. caerulescens accessions La Calamine, Ganges and Monte Prinzera are related to metal ion (di-, tri-valent inorganic cation) transmembrane transporter activity, iron and calcium ion binding (inorganic) anion transmembrane transporter activity, and antioxidant activity (Halimaa et al., 2014).
Co-segregation, genetic linkage mapping and quantitative trait loci (QTL) analyses make use of crosses between genotypes with contrasting phenotypes. Several crosses have been made between N. caerulescens accessions (Assunção et al., 2003a, 2006; Deniau et al., 2006; Richau and Schat, 2009). However, expression QTL (eQTL) has not been applied so far to address metal hyperaccumulation or tolerance. Candidate gene identification can be further improved by applying RNA-Seq to recombinant inbred lines selected for particular tolerance or accumulation capacities or, preferably, their associated QTL markers.
PROSPECTS
Deep sequencing technologies with unparalleled accuracy, resolution and throughput have revolutionized transcriptomic and genomic research. So far full advantage of these techniques has not been taken to study the regulatory mechanisms underlying metal hyperaccumulation and tolerance. Environmental stress factors can cause changes in chromatin properties and in the production of small RNAs that contribute to regulation of gene expression. Deep sequencing has the potential to detect the small RNAs with low abundance. For example, several micro-RNAs (miRNAs) regulated by drought, cold and salt in rice, and responding to aluminum treatment in Medicago truncatula, were identified with deep sequencing (Barrera-Figueroa et al., 2012; Chen et al., 2012). Metals cause epigenetic changes, and epigenetics may have a role in plant adaptation to metalliferous environments, but there are only few studies of metal effect at DNA methylation level (Aina et al., 2004; Wada et al., 2004; Choi and Sano, 2007; Ou et al., 2012).
Having populations with a wide range of metal hyperaccumulation and hypertolerance capacities, N. caerulescens shows a great potential to serve as an excellent model in plant evolutionary genomics. Combined with high-throughput sequencing, a significant leap forward is expected in the understanding of the metabolic adaptation of this plant to different metal environments. However, even with its increasing power, RNA-Seq still remains a screening method that provides candidate genes, a more detailed analysis being needed to prove the true significance of the findings.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was financially supported by the Academy of Finland (project No. 260552). We acknowledge CSC – IT Center for Science Ltd. for the allocation of computational resources.
REFERENCES
- Aina R., Sgorbati S., Santagostino A., Labra M., Ghiani A., Citterio S. (2004). Specific hypomethylation of DNA is induced by heavy metals in white clover and industrial hemp. Physiol. Plantarum 121 472–480 10.1111/j.1399-3054.2004.00343.x [DOI] [Google Scholar]
- Assunção A. G. L., Ten Bookum W. M., Nelissen H. J. M., Vooijs R., Schat H, Ernst W. H. O. (2003a). A cosegregation analysis of zinc (Zn) accumulation and Zn tolerance in the Zn hyperaccumulator Thlaspi caerulescens. New Phytol. 159 383–390 10.1046/j.1469-8137.2003.00758.x [DOI] [PubMed] [Google Scholar]
- Assunção A. G. L., Schat H, Aarts M. G. M. (2003b). Thlaspi caerulescens, an attractive model species to study heavy metal hyperaccumulation in plants. New Phytol. 159 351–360 10.1046/j.1469-8137.2003.00820.x [DOI] [PubMed] [Google Scholar]
- Assunção A. G. L., Ten Bookum W. M., Nelissen H. J. M., Vooijs R., Schat H, Ernst W. H. O. (2003c). Differential metal tolerance and accumulation patterns among Thlaspi caerulescens populations originating from different soil types. New Phytol. 159 411–419 10.1046/j.1469-8137.2003.00819.x [DOI] [PubMed] [Google Scholar]
- Assunção A. G. L., Pieper B., Vromans J., Lindhout P., Aarts M. G. M., Schat H. (2006). Construction of a genetic linkage map of Thlaspi caerulescens and quantitative trait loci analysis of zinc accumulation. New Phytol. 170 21–32 10.1111/j.1469-8137.2005.01631.x [DOI] [PubMed] [Google Scholar]
- Barrera-Figueroa B. E., Gao L., Wu Z., Zhou X., Zhu J., Jin H., et al. (2012). High throughput sequencing reveals novel and abiotic stress-regulated microRNAs in the inflorescences of rice. BMC Plant Biol. 12:132 10.1186/1471-2229-12-132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beitz E. (2000). TEXtopo: shaded membrane protein topology plots in LATEX 2ε. Bioinformatics 16 1050–1051 10.1093/bioinformatics/16.11.1050 [DOI] [PubMed] [Google Scholar]
- Bernard C., Roosens N., Czernic P., Lebrun M., Verbruggen N. (2004). A novel CPx-ATPase from the cadmium hyperaccumulator Thlaspi caerulescens. FEBS Lett. 569 140–148 10.1016/j.febslet.2004.05.036 [DOI] [PubMed] [Google Scholar]
- Broadley M. R., White P. J., Hammond J. P., Zelko I., Lux A. (2007). Zinc in plants. New Phytol. 173 677–702 10.1111/j.1469-8137.2007.01996.x [DOI] [PubMed] [Google Scholar]
- Chen L., Wang T., Zhao M., Tian Q., Zhang W. H. (2012). Identification of aluminum-responsive microRNAs in Medicago truncatula by genome-wide high-throughput sequencing. Planta 235 375–386 10.1007/s00425-011-1514-9 [DOI] [PubMed] [Google Scholar]
- Choi C. S., Sano H. (2007). Abiotic-stress induces demethylation and transcriptional activation of a gene encoding a glycerophosphodiesterase-like protein in tobacco plants. Mol. Genet. Genomics 277 589–600 10.1007/s00438-007-0209-1 [DOI] [PubMed] [Google Scholar]
- Cosio C., Martinoia E., Keller C. (2004). Hyperaccumulation of cadmium and zinc in Thlaspi caerulescens and Arabidopsis halleri at the leaf cellular level. Plant Physiol. 134 716–725 10.1104/pp.103.031948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craciun A. R., Meyer C.-L., Chen J., Roosens N., De Groodt R., Hilson P., et al. (2012). Variation in HMA4 gene copy number and expression among Noccaea caerulescens populations presenting different levels of Cd tolerance and accumulation. J. Exp. Bot. 63 4179–4189 10.1093/jxb/ers104 [DOI] [PubMed] [Google Scholar]
- Deniau A. X., Pieper B., Ten Bookum W. M., Lindhout P., Aarts M. G. M., Schat H. (2006). QTL analysis of cadmium and zinc accumulation in the heavy metal hyperaccumulator Thlaspi caerulescens. Theor. Appl. Genet. 113 907–920 10.1007/s00122-006-0350-y [DOI] [PubMed] [Google Scholar]
- Derrien T., Estellé J., Sola S. M., Knowles D. G., Raineri E., Guigó R., et al. (2012). Fast computation and applications of genome mappability. PLoS ONE 7:e30377 10.1371/journal.pone.0030377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillies M.-A., Rau A., Aubert J., Hennequet-Antier C., Jeanmougin M., Servant N., et al. (2012). A comprehensive evaluation of normalization methods for Illumina high-throughput RNA sequencing data analysis. Brief Bioinform. 14 671-683 10.1093/bib/bbs046 [DOI] [PubMed] [Google Scholar]
- Eide D., Broderius M., Fett J., Guerinot M. L. (1996). A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proc. Natl. Acad. Sci. U.S.A. 93 5624–5628 10.1073/pnas.93.11.5624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escarré J., Lefèbvre C., Frérot H., Mahieu S., Noret N. (2013). Metal concentration and metal mass of metallicolous, non metallicolous and serpentine Noccaea caerulescens populations, cultivated in different growth media. Plant Soil 370 197–221 10.1007/s11104-013-1618-z [DOI] [Google Scholar]
- Freeman J. L., Garcia D., Kim D., Hopf A., Salt D. E. (2005). Constitutively elevated salicylic acid signals glutathione-mediated nickel tolerance in Thlaspi nickel hyperaccumulators. Plant Physiol. 137 1082–1091 10.1104/pp.104.055293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossoehme N. E., Akilesh S., Guerinot M. L., Wilcox D. E. (2006). Metal-binding thermodynamics of the histidine-rich sequence from the metal-transport protein IRT1 of Arabidopsis thaliana. Inorg. Chem. 45 8500–8508 10.1021/ic0606431 [DOI] [PubMed] [Google Scholar]
- Halimaa P., Lin Y.-F., Ahonen V., Blande D., Clemens S., Gyenesei A., et al. (2014). Gene expression differences between Noccaea caerulescens ecotypes help to identify candidate genes for metal phytoremediation. Environ. Sci. Technol. 48 3344–3353 10.1021/es4042995 [DOI] [PubMed] [Google Scholar]
- Hammond J. P., Bowen H. C., White P. J., Mills V., Pyke K. A., Baker A. J. M., et al. (2006). A comparison of the Thlaspi caerulescens and Thlaspi arvense shoot transcriptomes. New Phytol. 170 239–260 10.1111/j.1469-8137.2006.01662.x [DOI] [PubMed] [Google Scholar]
- Hanikenne M., Talke I. N., Haydon M. J., Lanz C., Nolte A., Motte P., et al. (2008). Evolution of metal hyperaccumulation required cis-regulatory changes and triplication of HMA4. Nature 453 391–395 10.1038/nature06877 [DOI] [PubMed] [Google Scholar]
- Kerkeb L., Mukherjee I., Chatterjee I., Lahner B., Salt D. E., Connolly E. L. (2008). Iron-induced turnover of the Arabidopsis Iron-Regulated Transporter1 metal transporter requires lysine residues. Plant Physiol. 146 1964–1973 10.1104/pp.107.113282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch M. A., German D. A. (2013). Taxonomy and systematics are key to biological information: Arabidopsis, Eutrema (Thellungiella), Noccaea and Schrenkiella (Brassicaceae) as examples. Front. Plant Sci. 4:267 10.3389/fpls.2013.00267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korshunova Y., Eide D., Clark G., Guerinot M., Pakrasi H. (1999). The IRT1 protein from Arabidopsis thaliana is a metal transporter with a broad substrate range. Plant Mol. Biol. 40 37–44 10.1023/A:1026438615520 [DOI] [PubMed] [Google Scholar]
- Lochlainn S. Ó., Bowen H. C., Fray R. G., Hammond J. P., King G. J., White P. J., et al. (2011). Tandem quadruplication of HMA4 in the zinc (Zn) and cadmium (Cd) hyperaccumulator Noccaea caerulescens. PLoS ONE 6:e17814 10.1371/journal.pone.0017814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombi E., Zhao F. J., Dunham S. J., McGrath S. P. (2000). Cadmium accumulation in populations of Thlaspi caerulescens and Thlaspi goesingense. New Phytol. 145 11–20 10.1046/j.1469-8137.2000.00560.x [DOI] [Google Scholar]
- Marquès L., Cossegal M., Bodin S., Czernic P., Lebrun M. (2004). Heavy metal specificity of cellular tolerance in two hyperaccumulating plants, Arabidopsis halleri and Thlaspi caerulescens. New Phytol. 164 289–295 10.1111/j.1469-8137.2004.01178.x [DOI] [PubMed] [Google Scholar]
- Meerts P, Van Isacker N. (1997). Heavy metal tolerance and accumulation in metallicolous and non-metallicolous populations of Thlaspi caerulescens from continental Europe. Plant Ecol. 133 221–231 10.1023/A:1009717619579 [DOI] [Google Scholar]
- Monsant A. C., Kappen P., Wang Y., Pigram P. J., Baker A. J. M., Tang C. (2011). In vivo speciation of zinc in Noccaea caerulescens in response to nitrogen form and zinc exposure. Plant Soil 348 167–183 10.1007/s11104-011-0887-7 [DOI] [Google Scholar]
- Ou X., Zhang Y., Xu C., Lin X., Zang Q., Zhuang T., et al. (2012). Transgenerational inheritance of modified DNA methylation patterns and enhanced tolerance induced by heavy metal stress in rice (Oryza sativa L.). PLoS ONE 7:e41143 10.1371/journal.pone.0041143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peer W. A., Mamoudian M., Lahner B., Reeves R. D., Murphy A. S., Salt D. E. (2003). Identifying model metal hyperaccumulating plants: germplasm analysis of 20 Brassicaceae accessions from a wide geographical area. New Phytol. 159 421–430 10.1046/j.1469-8137.2003.00822.x [DOI] [PubMed] [Google Scholar]
- Potocki S., Valensin D., Camponeschi F., Kozlowski H. (2013). The extracellular loop of IRT1 ZIP protein - the chosen one for zinc? J. Inorg. Biochem. 127 246–252 10.1016/j.jinorgbio.2013.05.003 [DOI] [PubMed] [Google Scholar]
- Rapaport F., Khanin R., Liang Y., Krek A., Zumbo P., Mason C. E., et al. (2013). Comprehensive evaluation of differential expression analysis methods for RNA-seq data. Genome Biol. 14:R95 10.1186/gb-2013-14-9-r95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rascio N., Navari-Izzo F. (2011). Heavy metal hyperaccumulating plants: how and why do they do it? And what makes them so interesting? Plant Sci. 180 169–181 10.1016/j.plantsci.2010.08.016 [DOI] [PubMed] [Google Scholar]
- Reeves R. D., Schwartz C., Morel J. L., Edmondson J. (2001). Distribution and metal-accumulating behavior of Thlaspi caerulescens and associated metallophytes in France. Int. J. Phytoremediat. 3 145–172 10.1080/15226510108500054 [DOI] [Google Scholar]
- Richau K. H., Schat H. (2009). Intraspecific variation of nickeland zinc accumulation and tolerance in the hyperaccumulatoro Thlaspi caerulescens. Plant Soil 314 253–262 10.1007/s11104-008-9724-z [DOI] [Google Scholar]
- Rigola D., Fiers M., Vurro E, Aarts M. G. M. (2006). The heavy metal hyperaccumulator Thlaspi caerulescens expresses many species-specific genes, as identified by comparative expressed sequence tag analysis. New Phytol. 170 753–765 10.1111/j.1469-8137.2006.01714.x [DOI] [PubMed] [Google Scholar]
- Robinson B. H., Leblanc M., Petit D., Brooks R. R., Kirkman J. H, Gregg P. E. H. (1998). The potential of Thlaspi caerulescens for phytoremediation of contaminated soils. Plant Soil 203 47–56 10.1023/A:1004328816645 [DOI] [Google Scholar]
- Rogers E. E., Eide D. J., Guerinot M. L. (2000). Altered selectivity in an Arabidopsis metal transporter. Proc. Natl. Acad. Sci. U.S.A. 97 12356–12360 10.1073/pnas.210214197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero I. G., Ruvinsky I., Gilad Y. (2012). Comparative studies of gene expression and the evolution of gene regulation. Nat. Rev. Genet. 13 505–516 10.1038/nrg3229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roosens N., Verbruggen N., Meerts P., Ximénez-Embún P, Smith J. A. C. (2003). Natural variation in cadmium tolerance and its relationship to metal hyperaccumulation for sevel populations of Thlaspi caerulescens from Western Europe. Plant Cell Environ. 26 1657–1672 10.1046/j.1365-3040.2003.01084.x [DOI] [Google Scholar]
- Schwartz C., Echevarria G., Morel J. L. (2003). Phytoextraction of cadmium with Thlaspi caerulescens. Plant Soil 249 27–35 10.1023/A:1022584220411 [DOI] [Google Scholar]
- Talke I. N., Hanikenne M, Krämer U. (2006). Zinc-dependent global transcriptional control, transcriptional deregulation, and higher gene copy number for genes in metal homeostasis of the hyperaccumulator Arabidopsis halleri. Plant Physiol. 142 148–167 10.1104/pp.105.076232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolrà R., Pongrac P., Poschenrieder C., Vogel-Mikuš K., Regvar M, Barceló J. (2006). Distinctive effects of cadmium on glucosinolate profiles in Cd hyperaccumulator Thlaspi praecox and non-hyperaccumulator Thlaspi arvense. Plant Soil 288 333–341 10.1007/s11104-006-9124-1 [DOI] [Google Scholar]
- Tusnády G. E., Simon I. (1998). Principles governing amino acid composition of integral membrane proteins: applications to topology prediction. J. Mol. Biol. 283 489–506 10.1006/jmbi.1998.2107 [DOI] [PubMed] [Google Scholar]
- Tusnády G. E., Simon I. (2001). The HMMTOP transmembrane topology prediction server. Bioinformatics 17 849–850 10.1093/bioinformatics/17.9.849 [DOI] [PubMed] [Google Scholar]
- van de Mortel J. E., Schat H., Moerland P. D., Ver Looren van Themaat E., van der Ent S., Blankestijn H., et al. (2008). Expression differences for genes involved in lignin, glutathione and sulfate metabolism in response to cadmium in Arabidopsis thaliana and the related Zn/Cd-hyperaccumulator Thlaspi caerulescens. Plant Cell Environ. 31 301–324 10.1111/j.1365-3040.2007.01764.x [DOI] [PubMed] [Google Scholar]
- van de Mortel J. E., Villanueva L. A., Schat H., Kwekkeboom J., Coughlan S., Moerland P. D., et al. (2006). Large expression differences in genes for iron and zinc homeostasis, stress response, and lignin biosynthesis distinguish roots of Arabidopsis thaliana and the related metal hyperaccumulator Thlaspi caerulescens. Plant Physiol. 142 1127–1147 10.1104/pp.106.082073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vázquez M. D., Barceló J., Poschenrieder C., Mádico J., Hatton P., Baker A. J. M., et al. (1992). Localization of zinc and cadmium in Thlaspi caerulescens (Brassicaceae), a metallophyte that can hyperaccumulate both metals. J. Plant Physiol. 140 350–355 10.1016/S0176-1617(11)81091-6 [DOI] [Google Scholar]
- Vogel-Mikuš K., Drobne D., Regvar M. (2005). Zn, Cd, and Pb accumulation and arbuscular mycorrhizal colonization of pennycress Thlaspi praecox Wulf. (Brassicaceae) from the vicinity of a lead mine and smelter in Slovenia. Environ. Pollut. 133 233–242 10.1016/j.envpol.2004.06.021 [DOI] [PubMed] [Google Scholar]
- Wada Y., Miyamoto K., Kusano T., Sano H. (2004). Association between up-regulation of stress-responsive genes and hypomethylation of genomic DNA in tobacco plants. Mol. Genet. Genom. 271 658–666 10.1007/s00438-004-1018-4 [DOI] [PubMed] [Google Scholar]
- Weber M., Harada E., Vess C., von Roepenack-Lahaye E., Clemens S. (2004). Comparative microarray analysis of Arabidopsis thaliana and Arabidopsis halleri roots identifies nicotianamine synthase, a ZIP transporter and other genes as potential metal hyperaccumulation factors. Plant J. 37 269–281 10.1046/j.1365-313X.2003.01960.x [DOI] [PubMed] [Google Scholar]
- White-Monsant A. C., Tang C. (2013). Organic acids are not specifically involved in the nitrate-enhanced Zn hyperaccumulation mechanism in Noccaea caerulescens. Environ. Exp. Bot. 91 12–21 10.1016/j.envexpbot.2013.02.006 [DOI] [Google Scholar]
- Xing J. P., Jiang R. F., Ueno D., Ma J. F., Schat H., McGrath S. P., et al. (2008). Variation in root-to-shoot translocation of cadmium and zinc among different accessions of the hyperaccumulators Thlaspi caerulescens and Thlaspi praecox. New Phytol. 178 315–325 10.1111/j.1469-8137.2008.02376.x [DOI] [PubMed] [Google Scholar]