Abstract
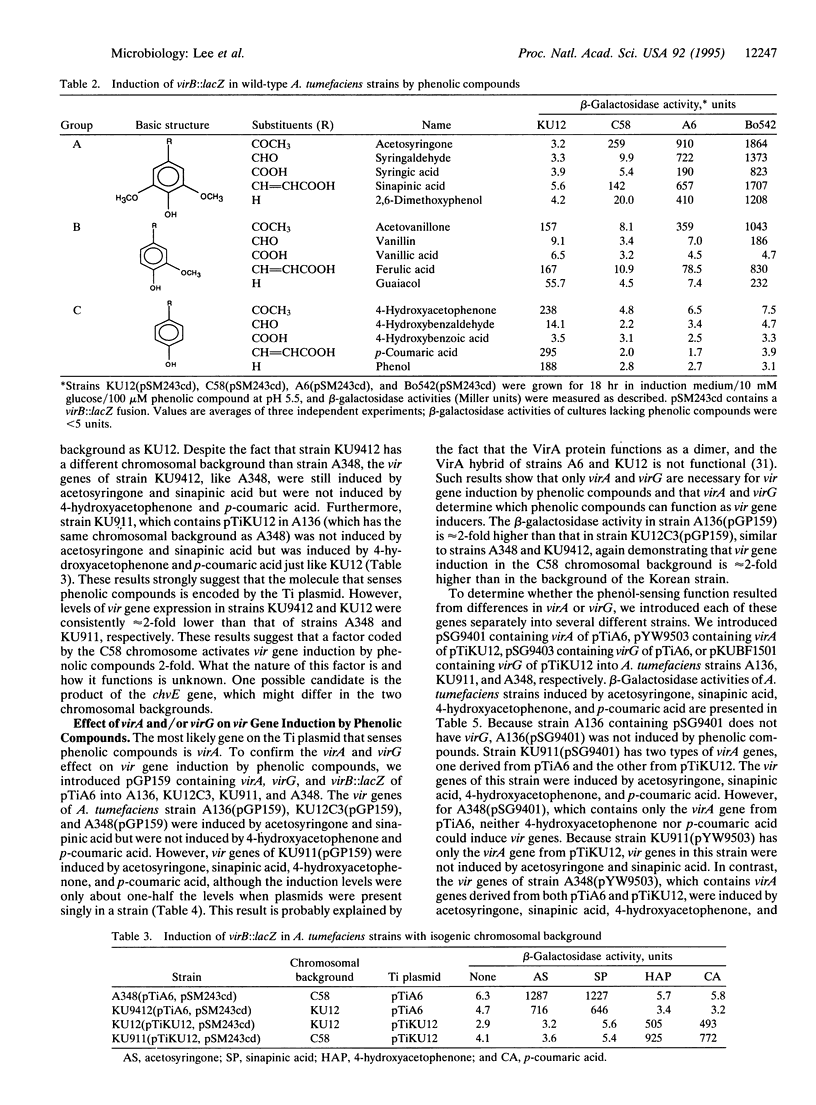
The virulence (vir) genes of Agrobacterium tumefaciens are induced by low-molecular-weight phenolic compounds and monosaccharides through a two-component regulatory system consisting of the VirA and VirG proteins. However, it is not clear how the phenolic compounds are sensed by the VirA/VirG system. We tested the vir-inducing abilities of 15 different phenolic compounds using four wild-type strains of A. tumefaciens--KU12, C58, A6, and Bo542. We analyzed the relationship between structures of the phenolic compounds and levels of vir gene expression in these strains. In strain KU12, vir genes were not induced by phenolic compounds containing 4'-hydroxy, 3'-methoxy, and 5'-methoxy groups, such as acetosyringone, which strongly induced vir genes of the other three strains. On the other hand, vir genes of strain KU12 were induced by phenolic compounds containing only a 4'-hydroxy group, such as 4-hydroxyacetophenone, which did not induce vir genes of the other three strains. The vir genes of strains KU12, A6, and Bo542 were all induced by phenolic compounds containing 4'-hydroxy and 3'-methoxy groups, such as acetovanillone. By transferring different Ti plasmids into isogenic chromosomal backgrounds, we showed that the phenolic-sensing determinant is associated with Ti plasmid. Subcloning of Ti plasmid indicates that the virA locus determines which phenolic compounds can function as vir gene inducers. These results suggest that the VirA protein directly senses the phenolic compounds for vir gene activation.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ankenbauer R. G., Best E. A., Palanca C. A., Nester E. W. Mutants of the Agrobacterium tumefaciens virA gene exhibiting acetosyringone-independent expression of the vir regulon. Mol Plant Microbe Interact. 1991 Jul-Aug;4(4):400–406. doi: 10.1094/mpmi-4-400. [DOI] [PubMed] [Google Scholar]
- Burn J. E., Hamilton W. D., Wootton J. C., Johnston A. W. Single and multiple mutations affecting properties of the regulatory gene nodD of Rhizobium. Mol Microbiol. 1989 Nov;3(11):1567–1577. doi: 10.1111/j.1365-2958.1989.tb00142.x. [DOI] [PubMed] [Google Scholar]
- Cangelosi G. A., Best E. A., Martinetti G., Nester E. W. Genetic analysis of Agrobacterium. Methods Enzymol. 1991;204:384–397. doi: 10.1016/0076-6879(91)04020-o. [DOI] [PubMed] [Google Scholar]
- Chang C. H., Winans S. C. Functional roles assigned to the periplasmic, linker, and receiver domains of the Agrobacterium tumefaciens VirA protein. J Bacteriol. 1992 Nov;174(21):7033–7039. doi: 10.1128/jb.174.21.7033-7039.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chilton M. D., Currier T. C., Farrand S. K., Bendich A. J., Gordon M. P., Nester E. W. Agrobacterium tumefaciens DNA and PS8 bacteriophage DNA not detected in crown gall tumors. Proc Natl Acad Sci U S A. 1974 Sep;71(9):3672–3676. doi: 10.1073/pnas.71.9.3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Close T. J., Zaitlin D., Kado C. I. Design and development of amplifiable broad-host-range cloning vectors: analysis of the vir region of Agrobacterium tumefaciens plasmid pTiC58. Plasmid. 1984 Sep;12(2):111–118. doi: 10.1016/0147-619x(84)90057-x. [DOI] [PubMed] [Google Scholar]
- Das A., Pazour G. J. Delineation of the regulatory region sequences of Agrobacterium tumefaciens virB operon. Nucleic Acids Res. 1989 Jun 26;17(12):4541–4550. doi: 10.1093/nar/17.12.4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Feyter R., Yang Y., Gabriel D. W. Gene-for-genes interactions between cotton R genes and Xanthomonas campestris pv. malvacearum avr genes. Mol Plant Microbe Interact. 1993 Mar-Apr;6(2):225–237. doi: 10.1094/mpmi-6-225. [DOI] [PubMed] [Google Scholar]
- Duban M. E., Lee K., Lynn D. G. Strategies in pathogenesis: mechanistic specificity in the detection of generic signals. Mol Microbiol. 1993 Mar;7(5):637–645. doi: 10.1111/j.1365-2958.1993.tb01155.x. [DOI] [PubMed] [Google Scholar]
- Garfinkel D. J., Nester E. W. Agrobacterium tumefaciens mutants affected in crown gall tumorigenesis and octopine catabolism. J Bacteriol. 1980 Nov;144(2):732–743. doi: 10.1128/jb.144.2.732-743.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath B., Bachem C. W., Schell J., Kondorosi A. Host-specific regulation of nodulation genes in Rhizobium is mediated by a plant-signal, interacting with the nodD gene product. EMBO J. 1987 Apr;6(4):841–848. doi: 10.1002/j.1460-2075.1987.tb04829.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S. G., Roitsch T., Christie P. J., Nester E. W. The regulatory VirG protein specifically binds to a cis-acting regulatory sequence involved in transcriptional activation of Agrobacterium tumefaciens virulence genes. J Bacteriol. 1990 Feb;172(2):531–537. doi: 10.1128/jb.172.2.531-537.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S., Song Y., Pan S. Q., Nester E. W. Characterization of a virG mutation that confers constitutive virulence gene expression in Agrobacterium. Mol Microbiol. 1993 Feb;7(4):555–562. doi: 10.1111/j.1365-2958.1993.tb01146.x. [DOI] [PubMed] [Google Scholar]
- Lee K., Dudley M. W., Hess K. M., Lynn D. G., Joerger R. D., Binns A. N. Mechanism of activation of Agrobacterium virulence genes: identification of phenol-binding proteins. Proc Natl Acad Sci U S A. 1992 Sep 15;89(18):8666–8670. doi: 10.1073/pnas.89.18.8666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melchers L. S., Regensburg-Tuïnk A. J., Schilperoort R. A., Hooykaas P. J. Specificity of signal molecules in the activation of Agrobacterium virulence gene expression. Mol Microbiol. 1989 Jul;3(7):969–977. doi: 10.1111/j.1365-2958.1989.tb00246.x. [DOI] [PubMed] [Google Scholar]
- Messens E., Dekeyser R., Stachel S. E. A nontransformable Triticum monococcum monocotyledonous culture produces the potent Agrobacterium vir-inducing compound ethyl ferulate. Proc Natl Acad Sci U S A. 1990 Jun;87(11):4368–4372. doi: 10.1073/pnas.87.11.4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris J. W., Morris R. O. Identification of an Agrobacterium tumefaciens virulence gene inducer from the pinaceous gymnosperm Pseudotsuga menziesii. Proc Natl Acad Sci U S A. 1990 May;87(9):3614–3618. doi: 10.1073/pnas.87.9.3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan S. Q., Charles T., Jin S., Wu Z. L., Nester E. W. Preformed dimeric state of the sensor protein VirA is involved in plant--Agrobacterium signal transduction. Proc Natl Acad Sci U S A. 1993 Nov 1;90(21):9939–9943. doi: 10.1073/pnas.90.21.9939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sciaky D., Montoya A. L., Chilton M. D. Fingerprints of Agrobacterium Ti plasmids. Plasmid. 1978 Feb;1(2):238–253. doi: 10.1016/0147-619x(78)90042-2. [DOI] [PubMed] [Google Scholar]
- Stachel S. E., An G., Flores C., Nester E. W. A Tn3 lacZ transposon for the random generation of beta-galactosidase gene fusions: application to the analysis of gene expression in Agrobacterium. EMBO J. 1985 Apr;4(4):891–898. doi: 10.1002/j.1460-2075.1985.tb03715.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachel S. E., Zambryski P. C. virA and virG control the plant-induced activation of the T-DNA transfer process of A. tumefaciens. Cell. 1986 Aug 1;46(3):325–333. doi: 10.1016/0092-8674(86)90653-7. [DOI] [PubMed] [Google Scholar]
- Tanaka K., Fujiwara T., Kumatori A., Shin S., Yoshimura T., Ichihara A., Tokunaga F., Aruga R., Iwanaga S., Kakizuka A. Molecular cloning of cDNA for proteasomes from rat liver: primary structure of component C3 with a possible tyrosine phosphorylation site. Biochemistry. 1990 Apr 17;29(15):3777–3785. doi: 10.1021/bi00467a026. [DOI] [PubMed] [Google Scholar]
- Turk S. C., van Lange R. P., Regensburg-Tuïnk T. J., Hooykaas P. J. Localization of the VirA domain involved in acetosyringone-mediated vir gene induction in Agrobacterium tumefaciens. Plant Mol Biol. 1994 Aug;25(5):899–907. doi: 10.1007/BF00028884. [DOI] [PubMed] [Google Scholar]
- Winans S. C., Ebert P. R., Stachel S. E., Gordon M. P., Nester E. W. A gene essential for Agrobacterium virulence is homologous to a family of positive regulatory loci. Proc Natl Acad Sci U S A. 1986 Nov;83(21):8278–8282. doi: 10.1073/pnas.83.21.8278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winans S. C., Kerstetter R. A., Nester E. W. Transcriptional regulation of the virA and virG genes of Agrobacterium tumefaciens. J Bacteriol. 1988 Sep;170(9):4047–4054. doi: 10.1128/jb.170.9.4047-4054.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winans S. C., Kerstetter R. A., Ward J. E., Nester E. W. A protein required for transcriptional regulation of Agrobacterium virulence genes spans the cytoplasmic membrane. J Bacteriol. 1989 Mar;171(3):1616–1622. doi: 10.1128/jb.171.3.1616-1622.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winans S. C. Two-way chemical signaling in Agrobacterium-plant interactions. Microbiol Rev. 1992 Mar;56(1):12–31. doi: 10.1128/mr.56.1.12-31.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rhijn P., Vanderleyden J. The Rhizobium-plant symbiosis. Microbiol Rev. 1995 Mar;59(1):124–142. doi: 10.1128/mr.59.1.124-142.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]



