Abstract
Innate memory phenotype (IMP) CD8+ T cells are non-conventional αβ T cells exhibiting features of innate immune cells, and are significantly increased in the absence of ITK. Their developmental path and function are not clear. Here we show hematopoietic MHCI dependent generation of antigen specific IMP CD8+ T cells using bone marrow chimeras. WT bone marrow gives rise to IMP CD8+ T cells in MHCI−/− recipients, resembling those in Itk−/− mice, but distinct from those derived via homeostatic proliferation (HP), and independent of recipient thymus. By contrast, MHCI−/− bone marrow does not lead to IMP CD8+ T cells in WT recipients. OTI IMP CD8+ T cells generated via this method exhibited enhanced early response to antigen without prior primary stimulation. Our findings suggest a method to generate antigen specific “naïve” CD8+ IMP T cells, demonstrate that they are not HP cells and can promptly respond in an antigen specific fashion.
Keywords: Antigen specific response, Innate memory phenotype, Homeostatic expansion, ITK, Non-conventional T cells
Introduction
Conventional T cells arise as naïve T cells through thymic selection, and require further exposure to antigens to expand and acquire effector/memory function (1). While conventional T cells are selected on thymic stroma by peptide-MHC complexes, non-conventional innate T cells have distinct selection pathways, spontaneously express markers typically found on memory T cells, and have the ability to rapidly respond upon stimulation (1–7). Non-conventional CD8+ T cells are distinguished by their memory-like attributes of the effector program such as secretion of IFN-γ and expression of CD44 and CD122 (IL-2Rβ) (8, 9), and are thus termed “Innate Memory” or “Innate Memory Phenotype (IMP)” CD8+ T cells (1, 10–14). IMP CD8+ T cells may be selected independent of thymic MHCIa, with a dominant role played by hematopoietic MHCI (11, 13). However, it was also suggested recently that innate memory-like CD8+ thymocytes are mainly selected by nonhematopoietic MHCIa (15), strongly alluding to alternative pathways for IMP CD8+ T cell development.
IMP CD8+ exist in specific-pathogen-free as well as in germ-free mice (16), independent of immunization and infection, and are significantly increased in the absence of Tec kinase ITK, and the transcriptional regulators CBP, KLF2 and Id3, or mice carrying an ITK binding mutant of Slp-76 (1, 10, 11, 13, 17–20). IL-4 has been shown to be involved in the generation of these cells in cell extrinsic manner in the absence of ITK (12). Although it is known that IMP CD8+ are distinct from antigen-induced memory CD8+ T cells, they share markers of T cells that have undergone homeostatic expansion (21), and so it is not clear whether IMP CD8+ T cells arise through homeostatic proliferation. Functionally, IMP CD8+ T cells can facilitate the clearance of Listeria monocytogenes infection (10), however, it has been difficult to directly examine whether IMP CD8+ T cells can respond to antigen during an immune response due to the difficulty discriminating IMP CD8+ T cells from conventional memory CD8+ T cells.
Materials and Methods
Mice
WT, MHCI−/− (B2m−/−: B6.129P2-B2mtm1Unc/J), and nude (B6.Cg-Foxn1nu/J) mice were from the Jackson Laboratory (Bar Harbor, ME), on a a C57BL/6 background. All experiments were approved by the Office of Research Protection’s IACUC at Pennsylvania State and Cornell Universities.
Generation of bone marrow chimeras
Bone marrow from donor mice was injected i.v. into lethally irradiated recipients (6–9 weeks old). T cell depleted bone marrow was obtained by negative selection using biotinylated-anti-CD3ε antibody (eBioscience, San Diego, CA) and magnetic column separation (Miltenyi Biotec, Cambridge, MA). Mice were kept in specific pathogen free environment, and fed acid water containing antibiotics (1 mg/ml Gentamycin Sulfate) for 8–9 weeks post BMT prior to analysis.
Cell sorting, microarray and quantitative real-time PCR
Cell sorting was performed on a FACSAria Cell Sorter (BD Biosciences, San Jose, CA). IMP CD8+ T cells (TCRβ+CD8α+CD44hi) were sorted from splenocytes of WT→MHCI−/− chimeras 8 weeks post transplantation and 8-week old Itk−/− mice. HP cells were generated by i.v. injecting naïve CD8+ T cells (TCRβ+CD8α+CD44lo) into Rag1−/− recipients (0.5×106/mouse), followed by sorting of TCRβ+CD8α+CD44hi cells 8 weeks post transfer. mRNA from sorted cells was extracted, amplified and used for microarray (GeneChip Mouse Genome 430 2.0 Array, Affymetrix, Santa Clara, CA) at the Cornell University Life Sciences Core Laboratories Center. Microarray data were analyzed using GeneSpring GX software (Agilent Technologies, Clara, CA). RMA-normalized probe values were used to generate correlation coefficient matrix, further converted to gene expression values with Quantile normalization, followed by analysis of gene differential expression. Data have been deposited into the NCBI GEO repository (http://www.ncbi.nlm.nih.gov/gds; accession # GSE41482). Quantitative real-time PCR was carried out using Taqman probe sets (Applied Biosystems, Foster City, CA).
Data analysis
Two-tailed Student’s t test was performed using GraphPad Prism v5.00 (GraphPad, San Diego, CA), with p < 0.05 considered statistically significant.
Results and Discussion
Development of IMP CD8+ T cells via hematopoietic MHCI selection independent of the thymus
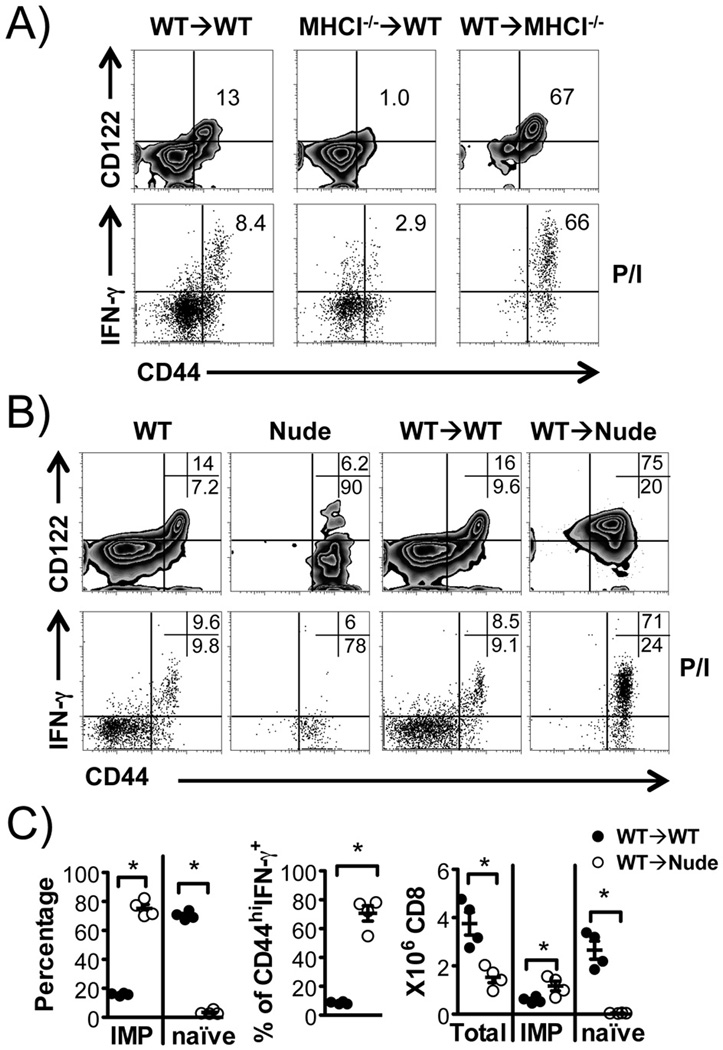
IMP CD8+ T cells have been reported to be able to develop in the thymus independent of MHCI molecules on thymic stroma (11). We took advantage of this to generate mice carrying predominantly IMP or naïve CD8+ T cells through BMTs utilizing WT and MHCI−/− (B2m−/−) mice. The spleens of WT→MHCI−/− mice had predominantly CD44hiCD122+ IMP CD8+ T cells that rapidly produced IFN-γ upon P/I stimulation, as has been observed (1, 10–14). By contrast, MHCI−/−→WT mice had predominantly naïve CD8+ T cells (Fig. 1A). As expected, hematopoietic MHCI expression is necessary and sufficient for development of IMP CD8+ T cells.
FIGURE 1. Development of IMP CD8+ T cells via hematopoietic MHCI selection independent of the thymus.
(A) Representative flow cytometric analysis of IMP CD8+ T cells (upper panel, CD44hiCD122+) and P/I induced IFN-γ production (lower panel). Donor TCRβ+CD8α+ cells from WT→WT (MHCI+CD45.2+), MHCI−/−→WT (MHCI−CD45.2+) and WT→MHCI−/− (MHCI+CD45.2−) chimeras are shown. (B&C) The thymus is not required for development of IMP CD8+ T cells. (B) Bone marrow chimeras were generated as indicated and donor TCRβ+CD8α+ cells analyzed by flow cytometry for CD44 and CD122, and IFN-γ producing capacity induced by P/I, compared to those from WT and Nude mice. (C) Percentages of IMP, naïve and P/I induced CD44hi IFN-γ producing CD8+ T cells, along with numbers of total, IMP and naïve CD8+ T cells, in “WT→WT” and “WT→Nude” chimeras. n = 4 in each group. *p < 0.05 by Student’s t test.
Further examination revealed that the thymus is dispensable for IMP CD8+ T cell development, since transplant of WT bone marrow into athymic Nude mice resulted in the development of similar cells to that seen in the WT→MHCI−/− transplants (Fig. 1B&C). Of note, while endogenous CD8+ T cells in nude mice are CD44hi, they do not express CD122 and fail to rapidly produce IFN-γ in response to P/I (Fig. 1B).
IMP CD8+ T cells are distinct from homeostatic expanded CD8+ T cells
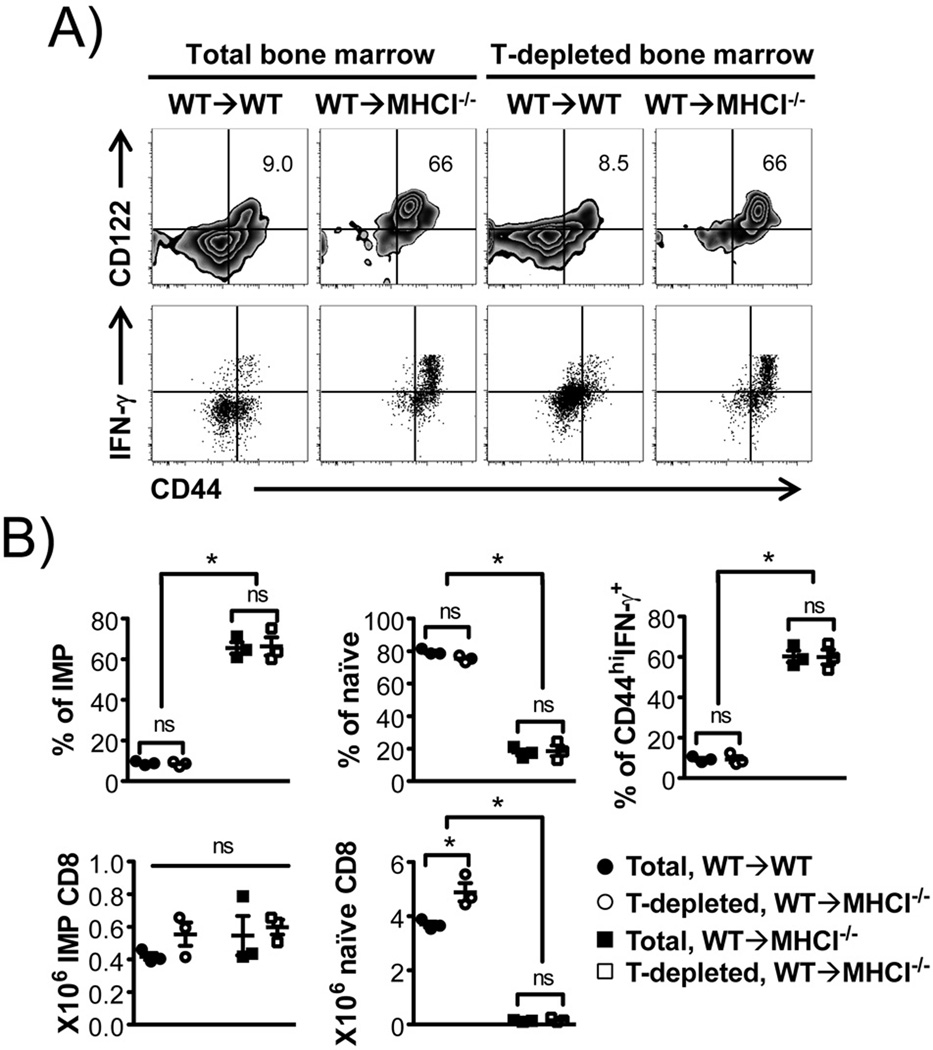
When transferred into lymphopenic environments, naïve CD8+ T cells undergo significant proliferation in an attempt to restore “normal” levels of T cells. Along with this proliferation, naïve CD44loCD8+ T cells acquire a phenotype resembling memory-like CD8+ T cells, a process termed “homeostatic expansion” (23, 24). Naïve CD8+ T cells also proliferate and differentiate upon antigen recognition and activation, converting into long-lived antigen specific memory CD8+ T cells that reside in part in bone marrow (25). It is therefore possible that the cells we observed are due to homeostatic expansion of a small number of naïve T cells and/or proliferation of memory T cells in the transferred donor bone marrow. However, we found that depletion of T cells prior to BMT did not affect the development of IMP CD8+ T cells (Fig. 2). This suggests that IMP CD8+ T cells observed in WT→MHCI−/− chimeric model are the result of hematopoietic MHCI selection and development rather than homeostatic expansion of T cells from donor bone marrow.
FIGURE 2. IMP CD8+ T cells in WT→MHCI−/− chimeras develop despite depletion of T cells from donor bone marrow.
WT bone marrow was either left intact or depleted of T-cells, and used as donors to generate “WT→WT” and “WT→MHCI−/−” chimeras. Donor TCRβ+CD8α+ cells were analyzed. (A) Flow cytometric analysis for CD44 and CD122, and percentage of CD44hi IFN-γ producing CD8+ T cells in response to P/I. (B) Percentages and numbers of IMP and naïve CD8+ T cells, and percentage of P/I induced CD44hi IFN-γ+ CD8+ T cells. n = 3 in each group. *p < 0.05, ns = not significant, by Student’s t test.
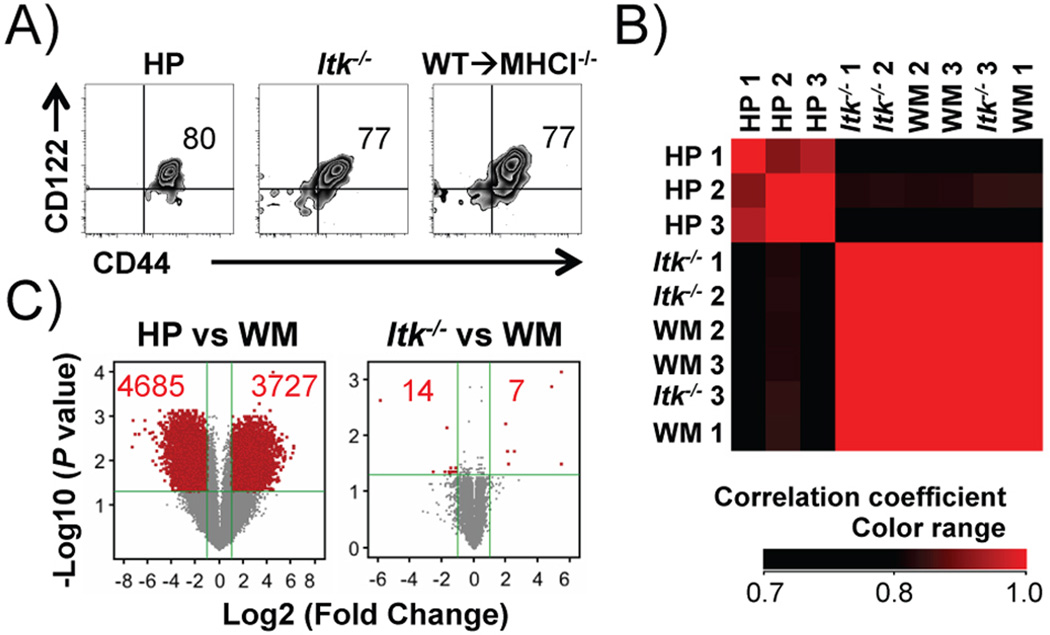
To confirm that the IMP CD8+ T cells in the WT→MHCI−/− chimeras are not the result of homeostatic expansion, we analyzed whole genome gene expression profiles of sorted IMP CD8+ T cells from WT→MHCI−/− chimeras (WM), Itk−/− mice, and CD8+ T cells derived from homeostatic expansion of naïve CD8+ T cells in Rag1−/− recipients (Fig. 3A). Correlation coefficient matrix shows that gene expression between WM IMP CD8+ T cells and Itk−/− IMP CD8+ T cells are highly correlated (Correlation coefficient > 0.98), while both are less correlated with HP CD8+ T cells (Correlation coefficient: 0.66 ~ 0.87) (Fig. 3B). Among 27800 genes, compared to WM CD8+ T cells, there are ~ 4000 up regulated and > 4000 down regulated by more than 2 fold in HP CD8+ T cells (P < 0.05, P values were generated by asymptotic computation with Benjamini-Hochberg false discovery rate correction), while there are only 21 of these in Itk−/− CD8+ T cells (Fig. 3C). We examined a few meaningful genes that were changed between these cells by q-RT-PCR and consistent with the differential expression profile identified by microarray (Supplemental Fig. 1A), we found up-regulation of TNFR (Tnfarsf1a) and NFκB2 (Nfkb2), and down-regulation of Eomesodermin (Eomes) and Bim (Bcl2l11) (Supplemental Fig. 1B), in HP CD8+ T cells but not Itk−/− CD8+ T cells compared to WM IMP CD8+ T cells. This whole genome expression analysis strongly suggests that IMP CD8+ cells generated in WT→MHCI−/− chimeras highly resemble IMP CD8+ T cells in Itk−/− mice, and are distinct from those derived from homeostatic expansion.
FIGURE 3. Hematopoietic MHCI dependent CD8+ T cells resemble innate memory CD8+ T cells in Itk−/− mice, but are distinct from HP cells.
Comparison of HP, Itk−/− and WM (WT→MHCI−/−) CD8+ T cells. (A) HP, Itk−/− and WM CD8+ T cells share expression of CD44 and CD122. (B) Itk−/− and WM CD8+ T cells show extremely high correlation in whole genome gene expression, which is distinct from HP CD8+ T cells. Samples were clustered based on the hierarchy of correlation. (C) HP but not Itk−/− CD8+ T cells, exhibit significantly higher number of differentially expressed genes compared to WM CD8+ T cells. Genes with significant change (fold change > 2, corrected P < 0.05) are shown in red. Numbers in red indicate numbers of gene that significantly up- or down- regulated.
Antigen-naïve OVA specific IMP CD8+ T cells promptly respond to antigen in the absence of primary antigen exposure
The BMT approach suggests a method to generate antigen-naïve, antigen specific IMP CD8+ T cells. To study the primary response of antigen specific IMP CD8+ T cells, we utilized OTI mice (expressing OVA specific T cell receptor on CD8+ T cells) as bone marrow donors, to generate antigen-naïve OVA specific IMP CD8+ T cells in MHCI−/− recipients. Reciprocally, OTI transgenic mice that lack MHCI−/− (MHCI−/−OTI) were used as donors to generate OVA specific naïve CD8+ T cells in WT recipients, with OTI→WT chimeras as controls. Similar to the results of BMTs in non-transgenic backgrounds, OTI→MHCI−/− chimeras had predominantly TCR transgene positive OVA specific CD44hiCD122+ IMP CD8+ T cells that rapidly produced IFN-γ. Analogously, MHCI−/−OTI→WT chimeras had predominantly TCR transgene positive OVA specific CD44loCD122− naïve CD8+ T cells (Fig. 4A&B). Using this model, we examined whether antigen-naïve, antigen specific IMP CD8+ T cells can rapidly respond to antigen in the absence of prior antigenic exposure. OTI transgenic IMP and naïve CD8+ T cells were stimulated in vitro with OVA or OVA257–264 epitope of OVA recognized by the OTI TCR, without prior primary antigen exposure in vivo. We found that OVA/OVA257–264 specific IFN-γ and TNF-α production by CD44hi CD8+ T cells peaked on the third day post stimulation in OTI IMP CD8+ T cells (OTI→MHCI−/−) (Fig. 4C), suggesting that these cells were highly functional IFN-γ/TNF-α double producers (Fig. 4C), and indicating that antigen naïve OVA specific IMP CD8+ T cells can rapidly respond in a highly functional manner to specific antigen in the absence of primary antigenic exposure. By contrast, naïve OTI cells (MHCI−/−OTI→WT) did not show significant cytokine response until > 6 days of stimulation as would be expected in vivo.
FIGURE 4. Hematopoietic MHCI dependent OTI IMP CD8+ T cells exhibit prompt and potent antigen specific response in vitro without primary antigen exposure.
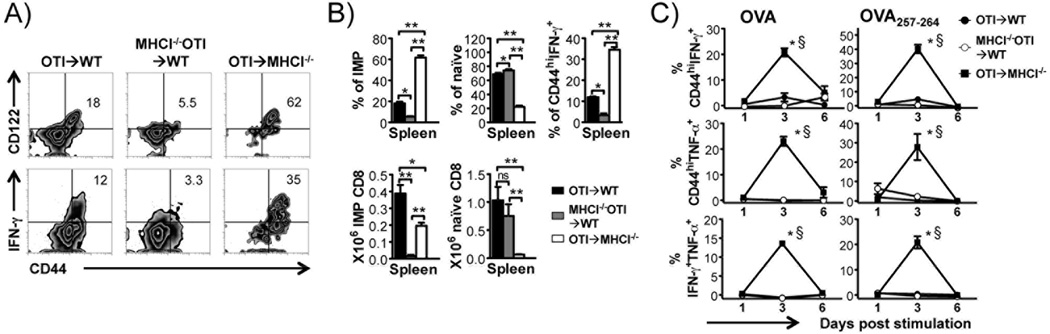
BMTs were done as indicated and donor TCR transgene positive CD8α+ cells from OTI→WT (CD45.2+MHCI+), MHCI−/−OTI→WT (CD45.2+MHCI−) and OTI→MHCI−/− (CD45.2+MHCI+) chimeras analyzed. (A) Flow cytometric analysis of CD8+ T cells for CD44 and CD122 and P/I induced IFN-γ production in chimeric spleens. (B) Percentages and numbers of CD44hiCD122+ IMP CD8+ T cells, CD44loCD122− naïve CD8+ T cells, and proportion of CD44hi IFN-γ producing cells in response to P/I. *p < 0.05, ns = not significant, by Student’s t test. Data represent results from three independent experiments. (C) Percentages of CD44hi IFN-γ+, CD44hi TNF-α+ and IFN-γ+/TNF-α+ double positive donor CD8+ T cells in response to OVA and OVA257–264 peptide along the time course. All values were corrected for the response of non-stimulated controls. *p < 0.05, compared to “OTI→WT” chimeras, §p < 0.05, compared to “MHCI−/−OTI→WT” chimeras, by two-way ANOVA. Data represent results from two independent experiments.
The function of IMP CD8+ T cells, and the mechanism behind their development, has elicited significant interest. Recently, Rafei et. al. showed that antigen-naïve OTI transgenic CD44hiCD8+ thymocytes selected on thymic MHCIa and that have never left thymus, can promptly respond to OVA-peptide with cytokine secretion (15). In addition, Haluszczak et. al. showed that a proportion of functional antigen-specific peripheral CD8+ T cells in germ-free mice have a memory-like phenotype, although one cannot rule out that these cells are antigen-experienced (21). Using bone marrow chimeras and transgenic approaches, we show for the first time that we can develop significant numbers of antigen-naïve, antigen specific IMP CD8+ T cells that are similar to those that develop in the absence of ITK, but different from those that develop due to homeostatic expansion, although they have similar phenotypes. Our work provides a model, and opens the door for more detailed analysis of the development and more importantly, function of such cells.
Our findings that endogenous CD8+ T cells in nude mice have a partial innate memory phenotype suggests that perhaps other signals may be required for the complete development and/or maturation of these cells, which may be indirectly dependent on the thymic structure or the Foxn1 gene. This could involve a precursor that traffics through the thymus prior to our BMT. In the absence of ITK or the transcription factor KLF2, the increase in innate memory-like CD8+ T cells have been suggested to be due to the influence of IL-4 produced by NKT-like cells in these mice (12). What role IL-4 signaling plays in the development of these hematopoietic MHCI-dependent IMP CD8+ T cells remains to be determined. Our preliminary analysis indicates that IMP CD8+ T cells differentially express the TNFR (lower than HP cells) and Bim and the transcription factor Eomesodermin (higher than HP cells), the latter previously observed in IMP CD8+ T cells (29). Future experiments will distinguish the role of such factors in the development of these cells.
In the immune system, innate immune cells initiate a relatively antigen non-specific response to primary infection, allowing cells of the adaptive immune system to develop more specific and exquisite antigen responses, along with the generation of immune memory (30, 31). In particular, antigen specific memory T cell responses are thought to be generated only after antigenic exposure (32). However, our work suggests that antigen naïve innate memory CD8+ T cells can generate potent memory-like antigen specific responses. We suggest that such IMP CD8+ T cells evolved to rapidly respond early in infections in an antigen specific fashion, assisting the innate immune response and allowing priming of the antigen specific adaptive immune response. These cells may occupy an essential niche in the early immune response to primary infection.
Supplementary Material
Acknowledgement
We thank Meg Potter and Amie Wood for animal care, Shailaja Hegde and Gabriel Balmus for assistance with mouse irradiation, Nicole Bem and Rod Getchell for assistance with flow cytometry, Lavanya Sayam for assistance in cell sorting and Jennifer D. Mosher for assistance with microarray.
Abbreviations Used
- BMT
bone marrow transplantation
- HP
Homeostatic proliferation
- IMP
innate memory phenotype
- PMA
phorbol 12-myristate 13-acetate
- P/I
phorbol 12-myristate 13-acetate and Ionomycin
Footnotes
This work was supported by grants from the National Institutes of Health (AI051626 and AI065566) to A.A..
The authors have no financial conflicts of interest.
References
- 1.Berg LJ. Signalling through TEC kinases regulates conventional versus innate CD8(+) T-cell development. Nat Rev Immunol. 2007;7:479–485. doi: 10.1038/nri2091. [DOI] [PubMed] [Google Scholar]
- 2.Behar SM, Porcelli SA. CD1-restricted T cells in host defense to infectious diseases. Curr Top Microbiol Immunol. 2007;314:215–250. doi: 10.1007/978-3-540-69511-0_9. [DOI] [PubMed] [Google Scholar]
- 3.Born WK, Reardon CL, O'Brien RL. The function of gammadelta T cells in innate immunity. Curr Opin Immunol. 2006;18:31–38. doi: 10.1016/j.coi.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 4.Oliveira CC, van Veelen PA, Querido B, de Ru A, Sluijter M, Laban S, Drijfhout JW, van der Burg SH, Offringa R, van Hall T. The nonpolymorphic MHC Qa-1b mediates CD8+ T cell surveillance of antigen-processing defects. J Exp Med. 2009;207:207–221. doi: 10.1084/jem.20091429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cho H, Bediako Y, Xu H, Choi HJ, Wang CR. Positive selecting cell type determines the phenotype of MHC class Ib-restricted CD8+ T cells. Proc Natl Acad Sci U S A. 2011;108:13241–13246. doi: 10.1073/pnas.1105118108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Urdahl KB, Sun JC, Bevan MJ. Positive selection of MHC class Ib-restricted CD8(+) T cells on hematopoietic cells. Nat Immunol. 2002;3:772–779. doi: 10.1038/ni814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anderson G, Owen JJ, Moore NC, Jenkinson EJ. Thymic epithelial cells provide unique signals for positive selection of CD4+CD8+ thymocytes in vitro. J Exp Med. 1994;179:2027–2031. doi: 10.1084/jem.179.6.2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Intlekofer AM, Takemoto N, Wherry EJ, Longworth SA, Northrup JT, Palanivel VR, Mullen AC, Gasink CR, Kaech SM, Miller JD, Gapin L, Ryan K, Russ AP, Lindsten T, Orange JS, Goldrath AW, Ahmed R, Reiner SL. Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nat Immunol. 2005;6:1236–1244. doi: 10.1038/ni1268. [DOI] [PubMed] [Google Scholar]
- 9.Pearce EL, Mullen AC, Martins GA, Krawczyk CM, Hutchins AS, Zediak VP, Banica M, DiCioccio CB, Gross DA, Mao CA, Shen H, Cereb N, Yang SY, Lindsten T, Rossant J, Hunter CA, Reiner SL. Control of effector CD8+ T cell function by the transcription factor Eomesodermin. Science. 2003;302:1041–1043. doi: 10.1126/science.1090148. [DOI] [PubMed] [Google Scholar]
- 10.Hu J, Sahu N, Walsh E, August A. Memory phenotype CD8+ T cells with innate function selectively develop in the absence of active Itk. Eur J Immunol. 2007;37:2892–2899. doi: 10.1002/eji.200737311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Broussard C, Fleischacker C, Horai R, Chetana M, Venegas AM, Sharp LL, Hedrick SM, Fowlkes BJ, Schwartzberg PL. Altered development of CD8+ T cell lineages in mice deficient for the Tec kinases Itk and Rlk. Immunity. 2006;25:93–104. doi: 10.1016/j.immuni.2006.05.011. [DOI] [PubMed] [Google Scholar]
- 12.Weinreich MA, Odumade OA, Jameson SC, Hogquist KA. T cells expressing the transcription factor PLZF regulate the development of memory-like CD8+ T cells. Nat Immunol. 2010;11:709–716. doi: 10.1038/ni.1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horai R, Mueller KL, Handon RA, Cannons JL, Anderson SM, Kirby MR, Schwartzberg PL. Requirements for selection of conventional and innate T lymphocyte lineages. Immunity. 2007;27:775–785. doi: 10.1016/j.immuni.2007.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lai D, Zhu J, Wang T, Hu-Li J, Terabe M, Berzofsky JA, Clayberger C, Krensky AM. KLF13 sustains thymic memory-like CD8(+) T cells in BALB/c mice by regulating IL-4-generating invariant natural killer T cells. J Exp Med. 2011;208:1093–1103. doi: 10.1084/jem.20101527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rafei M, Hardy MP, Williams P, Vanegas JR, Forner KA, Dulude G, Labrecque N, Galipeau J, Perreault C. Development and Function of Innate Polyclonal TCR{alpha}{beta}+ CD8+ Thymocytes. J Immunol. 2011 doi: 10.4049/jimmunol.1101097. [DOI] [PubMed] [Google Scholar]
- 16.Huang T, Wei B, Velazquez P, Borneman J, Braun J. Commensal microbiota alter the abundance and TCR responsiveness of splenic naive CD4+ T lymphocytes. Clin Immunol. 2005;117:221–230. doi: 10.1016/j.clim.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 17.Weinreich MA, Takada K, Skon C, Reiner SL, Jameson SC, Hogquist KA. KLF2 transcription-factor deficiency in T cells results in unrestrained cytokine production and upregulation of bystander chemokine receptors. Immunity. 2009;31:122–130. doi: 10.1016/j.immuni.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fukuyama T, Kasper LH, Boussouar F, Jeevan T, van Deursen J, Brindle PK. Histone acetyltransferase CBP is vital to demarcate conventional and innate CD8+ T-cell development. Mol Cell Biol. 2009;29:3894–3904. doi: 10.1128/MCB.01598-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Verykokakis M, Boos MD, Bendelac A, Kee BL. SAP protein-dependent natural killer T-like cells regulate the development of CD8(+) T cells with innate lymphocyte characteristics. Immunity. 2010;33:203–215. doi: 10.1016/j.immuni.2010.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jordan MS, Smith JE, Burns JC, Austin JE, Nichols KE, Aschenbrenner AC, Koretzky GA. Complementation in trans of altered thymocyte development in mice expressing mutant forms of the adaptor molecule SLP76. Immunity. 2008;28:359–369. doi: 10.1016/j.immuni.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haluszczak C, Akue AD, Hamilton SE, Johnson LD, Pujanauski L, Teodorovic L, Jameson SC, Kedl RM. The antigen-specific CD8+ T cell repertoire in unimmunized mice includes memory phenotype cells bearing markers of homeostatic expansion. J Exp Med. 2009;206:435–448. doi: 10.1084/jem.20081829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bruce D, Whitcomb JP, August A, McDowell MA, Cantorna MT. Elevated non-specific immunity and normal Listeria clearance in young and old vitamin D receptor knockout mice. Int Immunol. 2009;21:113–122. doi: 10.1093/intimm/dxn129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cho BK, Rao VP, Ge Q, Eisen HN, Chen J. Homeostasis-stimulated proliferation drives naive T cells to differentiate directly into memory T cells. J Exp Med. 2000;192:549–556. doi: 10.1084/jem.192.4.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sprent J, Cho JH, Boyman O, Surh CD. T cell homeostasis. Immunol Cell Biol. 2008;86:312–319. doi: 10.1038/icb.2008.12. [DOI] [PubMed] [Google Scholar]
- 25.Becker TC, Coley SM, Wherry EJ, Ahmed R. Bone marrow is a preferred site for homeostatic proliferation of memory CD8 T cells. J Immunol. 2005;174:1269–1273. doi: 10.4049/jimmunol.174.3.1269. [DOI] [PubMed] [Google Scholar]
- 26.Rodgers JR, Cook RG. MHC class Ib molecules bridge innate and acquired immunity. Nat Rev Immunol. 2005;5:459–471. doi: 10.1038/nri1635. [DOI] [PubMed] [Google Scholar]
- 27.Cho H, Choi HJ, Xu H, Felio K, Wang CR. Nonconventional CD8+ T cell responses to Listeria infection in mice lacking MHC class Ia and H2-M3. J Immunol. 2011;186:489–498. doi: 10.4049/jimmunol.1002639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sullivan BA, Reed-Loisel LM, Kersh GJ, Jensen PE. Homeostatic proliferation of a Qa-1b-restricted T cell: a distinction between the ligands required for positive selection and for proliferation in lymphopenic hosts. J Immunol. 2004;173:6065–6071. doi: 10.4049/jimmunol.173.10.6065. [DOI] [PubMed] [Google Scholar]
- 29.Lee Y, Jameson S, Hogquist K. Alternative memory in the CD8 T cell lineage. Trends Immunol. 2011;32:50–56. doi: 10.1016/j.it.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zinkernagel RM. Uncertainties - discrepancies in immunology. Immunol Rev. 2002;185:103–125. doi: 10.1034/j.1600-065x.2002.18511.x. [DOI] [PubMed] [Google Scholar]
- 31.Iwasaki A, Medzhitov R. Regulation of adaptive immunity by the innate immune system. Science. 2010;327:291–295. doi: 10.1126/science.1183021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Janeway CA, J, Travers P, Walport M, Schlomchik MJ. Garland Sciences. 6th ed. 2005. Immunology: The immune system in health and disease; p. 12. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.