Abstract
With the exception of high latitudes, life has evolved under bright days and dark nights. Most organisms have developed endogenously driven circadian rhythms which are synchronized to this daily light/dark cycle. In recent years, humans have shifted away from the naturally occurring solar light cycle in favor of artificial and sometimes irregular light schedules produced by electrical lighting. Exposure to unnatural light cycles is increasingly associated with obesity and metabolic syndrome; however the means by which environmental lighting alters metabolism are poorly understood. Thus, we exposed mice to nighttime light and investigated changes in the circadian system and body weight. Here we report that exposure to ecologically relevant levels of dim (5 lux) light at night attenuate core circadian clock rhythms in the SCN at both the gene and protein level. Moreover, circadian clock rhythms were perturbed in the liver by nighttime light exposure. Changes in the circadian clock were associated with temporal alterations in feeding behavior and increased weight gain. These results are significant because they provide mechanistic evidence for how mild changes in environmental lighting can alter circadian and metabolic function.
INTRODUCTION
For >3 billion years, life outside of the highest latitudes has evolved under bright days and dark nights. Many organisms have developed endogenous 24-h rhythms, called circadian rhythms, which are synchronized to this light/dark cycle. With the widespread adoption of electrical lighting ~150 years ago, humans began brightly illuminating their nocturnal environments. Exposure to light at night is now pervasive in modern society and typically considered a mild environmental perturbation. However, the use of light at night (LAN) began prior to a deep appreciation of the importance of circadian rhythms for normal biological functions (Fonken and Nelson, 2011; Gerstner, 2012).
Recent evidence suggests that exposure to unnatural light cycles increases the risk for cancer (Stevens, 2009), sleep disturbances (Kohyama, 2009), and mood disorders (Driesen et al., 2010). Furthermore, exposure to light at night is increasingly associated with changes in metabolism. Shift workers who experience sustained nighttime illumination are at increased risk for cardiovascular disease and elevated body mass index (Ha and Park, 2005; Knutsson, 2003; Parkes, 2002; van Amelsvoort et al., 1999). Increases in nighttime light exposure at home are associated with increased body mass, waist circumference and triglyceride levels, and poor cholesterol balance (Obayashi et al., 2013). Even brief exposure to altered light and food schedules can result in adverse metabolic and cardiovascular consequences (Scheer et al., 2009). Moreover, we have reported that mice chronically exposed to dimly illuminated, as opposed to dark, nights elevate body mass independently of changes in total daily activity or caloric intake (Fonken et al., 2012; Fonken et al., 2010). Mice exposed to dim nights shift the timing of food intake toward the light phase and restricting food access to the dark phase prevents dLAN-associate body mass gain. The mechanism by which light at night induces these changes is not fully understood.
Here we propose that light alters metabolic homeostasis in mammals by disrupting the circadian system. Light is the most potent synchronizing factor for the circadian system. Light information travels directly from intrinsically photosensitive ganglion cells in the retina to the master circadian clock located in the suprachiasmatic nuclei (SCN) of the hypothalamus (Chen et al., 2011). Pacemaker neurons within the SCN thereby drive the circadian clock with an autoregulatory transcriptional-translational feedback loop of transcription activators and repressors (Albrecht, 2002). Although the SCN are the primary pacemakers in mammals, most if not all central and peripheral tissues contain the molecular machinery necessary for self sustaining circadian oscillation (Mohawk et al., 2012). The master clock converts external light/dark information to neural and endocrine signals that synchronize peripheral clocks (Guo et al., 2005; McNamara et al., 2001).
Exposure to a pulse of light during the night can phase advance or delay the circadian clock depending on the strength and time of the light signal (Miyake et al., 2000). Exposure to constant light can alter activity rhythms and ablate circadian rhythms in glucocorticoids, two principle outputs of the circadian system (Coomans et al., 2013; Fonken et al., 2010). Moreover, specifically timed nighttime light pulses can be used to ablate the circadian system (Ruby et al., 2008). Because disruption in circadian clock genes are associated with significant changes in metabolism (Bechtold et al., 2008; Carvas et al., 2012; Delezie et al., 2012; Marcheva et al., 2010; Paschos et al., 2012), we hypothesized that exposure to light at night alters metabolism through disrupting the circadian system. To assess the effects of light at night on the circadian clock we exposed mice to either total darkness or dim light (5 lux) during the night and then characterized the expression of several circadian clock genes and proteins in the SCN, hippocampus, liver, and adipose tissue. Five lux of nighttime light exposure was selected because (1) it is approximately five times bright than maximal moonlight (2) it is comparable to levels of light pollution found in urban areas (Kloog et al., 2009) and sleeping environments (Obayashi et al., 2013), yet (3) it is highly distinct from daytime light levels.
METHODS
Animals
One hundred and twenty male Swiss Webster mice (~8 weeks of age) were obtained from Charles River for use in this study. Mice were individually housed in propylene cages (dimensions: 33 × 19 × 14cm) at an ambient temperature of 23 ±2°C and provided with Harlan Teklad 8640 food (Madison, WI) and filtered tap water ad libitum. All mice were maintained in a standard light dark cycle (LD; 14:10 light (~150 lux)/dark (0 lux)) for one week following arrival. After the 1 week acclimation period mice were randomly assigned a group and transferred to a cabinet with either LD or a light/dim light cycle (14:10 light (~150 lux)/dim light (5 lux)). Daytime lighting was provided with white LEDs on the walls of the cabinets and dim light was administered with a flexible strip of cool white LEDs wrapped around the rack on which the mouse cages were placed. The lighting intensity was measured inside the home cage and was highly consistent between cages. Mice were also assigned to 1 of 6 tissue collection time points (Zeitgeber Time (ZT) 2, 6, 10, 14, 18, 22). Mice were weighed at the start of the experimental light treatment and weekly throughout the study. After 3 weeks in lighting conditions food was weighed twice daily for four days to determine the timing of food intake. Four weeks after placement in light conditions blood and tissue were collected for either quantitative PCR (qPCR) or immunohistochemical analyses (n=5 per group/use/time point).
qPCR
Mice were anesthetized with isofluorane vapors and rapidly decapitated. Peripheral tissue was dissected out, immediately weighed on a fine balance, and flash frozen. Weighed tissues include liver, spleen, pancreas, white adipose tissue (epididymal and inguinal), brown adipose tissue, adrenals, heart, and skeletal muscle. Brains were collected, placed in RNAlater, and after > 24 hr the hypothalamus and hippocampus were dissected out for PCR. Total RNA was extracted from liver, white adipose, hippocampal, and hypothalamic tissues using a homogenizer (Ultra-Turrax T8, IKAWorks, Wilmington, NC) and an RNeasy Mini Kit following the manufacturers protocol (Qiagen, Austin, TX). For fat extractions an additional chloroform step was added following homogenization and prior to the use of the Mini Kit. RNA concentration and purity were measured on an ND-1000 spectrophotometer (Fischer Scientific, PLACE). RNA concentrations were equalized with sterile water and RNA was reverse transcribed into cDNA with M-MLV Reverse Transcriptase enzyme (Invitrogen, Carlsbad, CA) according to the manufacturer’s protocol. Gene expression for Clock, Bmal1, Per1, Per2, Cry1, Cry2 and Rev-erbα were determined using inventoried primer and probe assays (Applied Biosystems, Foster City, CA) on an ABI 7500 Fast Real Time PCR System using Taqman® Universal PCR Master Mix. The universal two-step RT-PCR cycling conditions used were: 50 °C for 2 min, 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. Relative gene expression of individual samples run in duplicate was calculated by comparison to a relative standard curve and standardized by comparison to 18S rRNA signal.
Immonohistochemistry
Mice were given a lethal dose of sodium pentobarbital and perfused transcardially with ice-cold 0.1M PBS followed by 4% paraformaldehyde. Brains were removed, post-fixed overnight, cyroprotected in 30% sucrose, and frozen with dry ice. Brains were serially sectioned at 40 µm into cryoprotectant and stored at −20°C. Sets of tissue collected at 240 µm intervals were used for immunohistochemical detection of BMAL, CLOCK, PER1, and PER2 using antibodies generously provided by David Weaver (LeSauter et al., 2012). Sections were rinsed in PBS, blocked for 1 hr in 4% bovine serum albumin in 0.1 M PBS + 0.3% TX, and then incubated overnight at room temperature with primary antibody at 1:5000. The following day sections were rinsed and incubated for 1 hr with biotinylated goat anti-rabbit at 1:1000 (Vector Laboratories, Burligame, CA). Endogenous peroxidase activity was quenched with 3% H2O2 in methanol for 20 min and then the signal was amplified with avidin-biotin complex (Vectastain Elite ABC kit, Vector Laboratories) and tissue was developed using DAB. Tissue was then mounted onto gel coated slides, dehydrated through a series of graded ethanol washes, cleared with xylene, and coverslipped using Permount. Images of sections containing the SCN were captured at 20X using a Nikon E800 microscope. The number of immonoreactive cells in the SCN was counted in ImageJ (NIH) by a condition blind observer and averaged across section and sides of the bilateral structure to obtain one value per each mouse.
Statistical analyses
Body mass was analyzed using a repeated-measures analysis of variance (ANOVA) with time as the within subject factor and lighting condition as the between subject factor. Comparisons between lighting conditions with respect to weight gain, tissue masses, and percentage of daytime food intake were conducted using a one-way ANOVA. Blood glucose concentrations, qPCR results, and IHC results were analyzed using a two-way ANOVA with lighting condition and time as the between subjects factors. Following a significant F score, multiple comparisons were conducted with Tukey’s HSD test. All statistical analyses were performed using StatView software (v.5.0.1, Cary, NC). In all cases, differences between group means were considered statistically significant if p ≤ 0.05.
Results
Exposure to Dim Light at Night Increases Body Mass
Body mass increased among both groups over the course of the study (F3,354 = 304.187; p < 0.0001); however, mice exposed to dim light at night significantly elevated body mass compared to mice housed in dark nights (F3,354 = 15.820; p < 0.0001; Fig 1A). Overall, mice exposed to dim light at night had a greater body mass gain at the conclusion of the study compared to mice exposed to dark nights (F1,118 = 15.476; p < 0.001; Fig 1B). There were no differences in spleen, liver, pancreas, BAT, adrenal, or heart masses between groups (p > 0.05 in all cases). Epididymal fat pad mass was elevated among mice exposed to dim light at night (F1,58 = 7.520; p < 0.01; Fig 1D) suggesting increases in body mass may reflect increases in white adipose tissue.
Figure 1.
Mice exposed to light at night increased body mass. (A) Body mass in mice exposed to dimly lit and dark nights. (B) At the conclusions of the study body mass gain was increased among mice exposed to dim light at night as compared to dark nights. (C) Epididymal fat pad mass, an index of overall adiposity, was also increased in mice exposed to light at night. (D) Mice exposed to dim light at night altered timing of food intake consuming more during the light phase than mice exposed to dark nights. (E) There were no differences in blood glucose rhythms between lighting conditions. All data are presented as (mean ± SEM). *indicates dim light differs from dark nights.
Light at Night Attenuates Nocturnal Feeding Behavior
Despite increases in body mass among mice exposed to dLAN, there were no differences in total daily food intake between groups (p > 0.05; Fig S1). In agreement with previous results (Fonken et al., 2012; Fonken et al., 2010), exposure to light at night increased the percentage of food consumed during the light phase (F1,118 = 16.595; p < 0.0001; Fig 1D). Mice displayed a diurnal variation in blood glucose levels (F5,106 = 14.023; p < 0.0001; Fig 1E) with no differences between lighting conditions (p > 0.05).
Clock Gene Expression is Disrupted by Nighttime Light Exposure
To test the hypothesis that nighttime light exposure affects core clock gene expression, we analyzed the diurnal expression of transcripts encoding Clock, Bmal1, Per1, Per2, Cry1, and Cry2 in the hypothalamus, hippocampus, fat, and liver every 4 hr after 4 weeks of exposure to dim or dark nights. All of the core clock genes assessed in the hypothalamus displayed diurnal variation (p < 0.05; Fig. 2A). Expression of Clock, Bmal, and Cry1 were unaffected by lighting conditions, however, rhythmic expression of Per1, Per2, and Cry2 were all attenuated by exposure to dim light as compared to dark nights (p < 0.05; Fig 2A). Specifically, gene expression of Per2 and Cry1 was significantly reduced at ZT18, 6 hours after lights off, and Per1 expression was reduced at both ZT6 and ZT14.
Figure 2.
Dim light at night attenuates the amplitude of clock gene expression in the hypothalamus and liver. Transcripts of the core circadian clock genes Clock, Bmal1, Per1, Per2, Cry1, and Cry2 were analyzed by quantitative PCR in the (A) hypothalamus (B) hippocampus, (C) white adipose tissue, and (D) liver. Tissues were collected every 4 hr from mice exposed to either dark (black lines) or dimly lit (dotted lines) nights for 4 weeks. Values are expressed as relative abundance (mean ± SEM) after normalization to 18S *indicates dim light differs from dark nights.
In order to determine whether the effects of nighttime light exposure in the brain are specific to the master circadian clock we examined core clock gene expression in the hippocampus, a brain region known to show robust circadian oscillations. There was clear cycling of Clock, Bmal1, Per1, Per2, Cry1, and Cry2 in the hippocampus of mice exposed to both dark and dim nights (p < 0.001; Fig 2B). Overall, lighting condition had no effect on hippocampal clock gene expression (p > 0.05). This suggests that within the brain, changes in clock gene expression provoked by exposure to light at night may be regionally specific.
Recent work has demonstrated the importance of tissue specific clocks in regulating metabolism (e.g., Marcheva et al., 2010). Thus, we investigated the effects of nighttime light exposure on core clock gene expression in peripheral white adipose and liver tissues. All of the clock genes analyzed except for Clock displayed rhythmic variation in white adipose tissue and there was no effect of nighttime light exposure on mRNA expression levels (p > 0.05; Fig 2C). In contrast, rhythmic expression of Bmal1, Per1, Per2, Cry1, and Cry2 were all attenuated in the liver of mice exposed to dim nights (p < 0.05; Fig 2D). Taken together, these results reveal that exposure to low levels of light at night produce both tissue- and gene-specific changes in the expression levels of several core circadian clock genes.
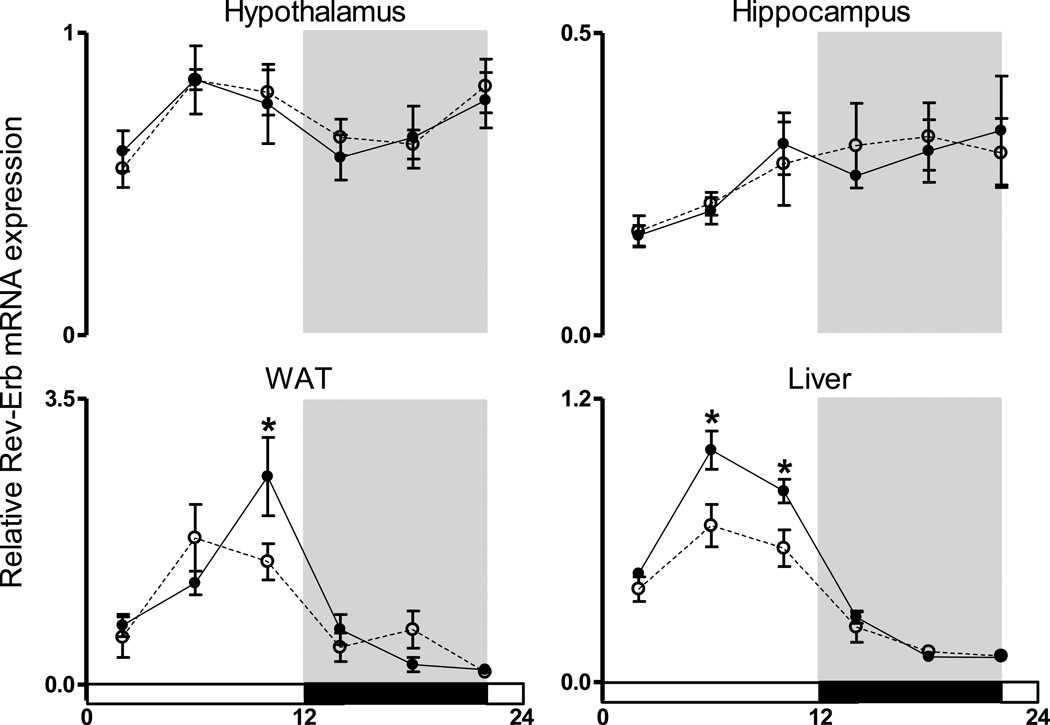
To further explore the effects of light at night on clock transcriptional networks we studied the 24 h pattern of expression of Rev-Erb mRNA in hypothalamic, hippocampal, WAT, and liver tissues. Rev-Erb is a nuclear receptor that has prominent functions for both circadian oscillations and metabolic homeostasis (reviewed in Ribberger and Albrecht, 2012). Diurnal rhythmicity in Rev-Erb expression was observed in all tissues evaluated (p < 0.05; Fig 3). However, exposure to dim light at night reduced Rev-Erb expression levels during the light phase in both the WAT and liver (p < 0.05). Decreased Rev-Erb expression was specific to the periphery as there were no changes in hypothalamic or hippocampal Rev-Erb mRNA levels.
Figure 3.
Dim light at night suppresses the amplitude of Rev-Erb mRNA expression in peripheral tissue. Rev-Erb gene expression was quantified in the (A) hypothalamus (B) hippocampus, (C) white adipose tissue, and (D) liver. Values are expressed as relative abundance (mean ± SEM) after normalization to 18S †indicates main effect of time, *indicates dim light differs from dark nights.
Core Clock Protein Expression in the Suprachiasmatic Nucleus
In order to fully characterize changes in circadian clock function within the SCN after nighttime light exposure we evaluated clock protein expression every 4 h in mice housed under dark or dimly lit nights for 4 weeks. Rhythmic CLOCK and BMAL protein expression was observed within the SCN with no effect of nighttime light exposure (Fig 4A/B). Expression levels of both PER1 and PER2 were altered by exposure to dim light as compared to dark nights (p < 0.05; Fig 4C/D). Whereas mice exposed to dark nights had rhythmic expression of PER1, mice exposed to dim light at night showed no diurnal variation in PER1. Specifically, the peak in PER1 expression at ZT10 was abolished by exposure to dim light at night. PER2 expression was also suppressed in mice exposed to dim light at night at ZT14. These results confirm gene expression findings demonstrating dim light at night specifically targets hypothalamic Per1 and Per2.
Figure 4.
PER1 and PER2 protein expression are reduced in the suprachiasmatic nucleus of the hypothalamus of mice exposed to dimly lit as compared to dark nights. CLOCK, BMAL, PER1 and PER2 immunoreactivity were analyzed in hypothalamic tissue collected every 4 hr from mice exposed to either dark (black lines) or dimly lit (dotted lines) nights for 4 weeks. Images of sections containing the SCN were captured at 20X and the number of immonoreactive cells was counted and averaged across section and sides of the bilateral structure. Data are presented as (mean ± SEM). *indicates dim light differs from dark nights.
Discussion
Exposure to electrical light at night can lead to disruptions in metabolic energy homeostasis in rodents and humans (Ha and Park, 2005; Knutsson, 2003; Obayashi et al., 2013; Parkes, 2002; van Amelsvoort et al., 1999). However, it remains unclear how environmental lighting affects metabolism. Thus, we exposed mice to dim light at night and investigated changes in body mass and the circadian system. Here we establish that exposure to ecologically relevant levels of dim light during the night attenuate circadian clock gene and protein rhythms, change feeding behavior, and lead to weight gain. These observations indicate that exposure to dim light at night, a commonplace and innocuous seeming environmental manipulation, can influence the circadian system and metabolism.
Mice exposed to dim light at night showed rapid and sustained body mass elevations. Increases in body mass may reflect increases in white adipose tissue as epididymal fat pad mass was elevated among mice exposed to dim nights. Although total daily caloric intake was comparable between groups, mice exposed to light at night consumed more food during the light period and less during the dark period than mice housed in dark nights. Disorganization in the feeding rhythm may contribute to increased body weight. Indeed, altered timing of food intake can cause weight gain and restricting feeding to specific hours prevents development of obesity (Arble et al., 2009; Bray et al., 2010; Hatori et al., 2012; Sherman et al., 2012). Furthermore, mice fed a high fat diet disrupt daily feeding patterns and attenuate circadian clock gene rhythms in peripheral tissue (Kohsaka et al., 2007).
Genetic models indicate a close association between the molecular events underlying metabolism and those involved in the generation of circadian rhythms. For example, Clock mutant mice become overweight on a high fat diet and develop symptoms characteristic of metabolic syndrome (Turek et al., 2005). Mice with mutations in other clock genes including Bmal1, Per1, Per2, Vipr2, and Rev-erbα display similar metabolic outcomes (Bechtold et al., 2008; Carvas et al., 2012; Delezie et al., 2012; Marcheva et al., 2010). These studies indicate alterations in core clock transcription factors within both the central clock and peripheral tissues alter metabolism. Importantly, our results suggest that changes in the circadian clock genes can be induced with exposure to ecologically relevant levels of dim light at night. Mice exposed to dim light at night suppress Per1 and Per2 expression at both the gene and protein level in the SCN. Importantly, there were no differences in clock gene expression in the hippocampus, suggesting changes in central clock gene expression provoked by exposure to light at night are regionally specific.
In addition to altering clock gene expression in the hypothalamus, exposure to dim as opposed to dark nights attenuated the rhythm in all but one of the core circadian clock genes assessed in the liver. Peripheral clocks are entrained by neural and endocrine signaling from the SCN (Guo et al., 2005; McNamara et al., 2001), as well as local factors such as nutritional signals (Vollmers et al., 2009). Recent work highlights the importance of peripheral clocks in regulating metabolism as single tissue clock gene deletions in the liver or fat can result in metabolic disturbances (Lamia et al., 2008; Marcheva et al., 2010; Paschos et al., 2012).
In addition to disruption in core clock mechanisms, mice exposed to dim light at night attenuated Rev-Erb expression in the liver and adipose tissue. Although previously considered an accessory feedback loop, REV-ERBs are increasingly demonstrated to be essential for circadian clock function and regulation of rhythmic metabolism. Mice deficient in both isoforms of REV-ERB show circadian rhythm adjustments and pronounced changes in metabolically related functions (Bugge et al., 2012; Cho et al., 2012).
Here we provide an extensive characterization of expression of circadian clock genes and proteins in both the master circadian oscillator in the brain and tissue specific clocks in mice exposed to light at night. Overall, our findings indicate that exposure to light at night attenuates core circadian clock mechanisms in the SCN at both the gene and protein level. Moreover, circadian clock function is disrupted in metabolically relevant peripheral tissue (i.e., white adipose tissue and liver) by nighttime light exposure. These changes in circadian clock function are associated with alterations in feeding behavior and increased weight gain. These finding are significant because they provide evidence for how mild changes in environmental lighting can alter circadian and metabolic function. Exposure to light at night is pervasive in modern society and typically considered a harmless environmental perturbation; however, our results demonstrate nighttime light exposure alters homeostatic functions. Detailed analysis of temporal changes induced by nighttime light exposure may provide insight into the onset and progression of obesity and metabolic syndrome and other disorders involving sleep and circadian disruption.
Acknowledgements
We thank Tracy Bedrosian, Joseph Ferraro, and Zachary McHenry for technical assistance. This research was supported by NSF grant IOS-11-18792 to RJN. LKF was supported by an American Heart Association Predoctoral Fellowship.
Footnotes
Disclosure. The authors declare no conflict of interest.
References
- Albrecht U. Invited review: regulation of mammalian circadian clock genes. J Appl Physiol. 2002;92:1348–1355. doi: 10.1152/japplphysiol.00759.2001. [DOI] [PubMed] [Google Scholar]
- Arble DM, Bass J, Laposky AD, Vitaterna MH, Turek FW. Circadian timing of food intake contributes to weight gain. Obesity (Silver Spring) 2009;17:2100–2102. doi: 10.1038/oby.2009.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold DA, Brown TM, Luckman SM, Piggins HD. Metabolic rhythm abnormalities in mice lacking VIP-VPAC2 signaling. Am J Physiol Regul Integr Comp Physiol. 2008;294:R344–R351. doi: 10.1152/ajpregu.00667.2007. [DOI] [PubMed] [Google Scholar]
- Bray MS, Tsai JY, Villegas-Montoya C, Boland BB, Blasier Z, Egbejimi O, Kueht M, Young ME. Time-of-day-dependent dietary fat consumption influences multiple cardiometabolic syndrome parameters in mice. Int J Obes (Lond) 2010;34:1589–1598. doi: 10.1038/ijo.2010.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugge A, Feng D, Everett LJ, Briggs ER, Mullican SE, Wang F, Jager J, Lazar MA. Rev-erbalpha and Rev-erbbeta coordinately protect the circadian clock and normal metabolic function. Genes Dev. 2012;26:657–667. doi: 10.1101/gad.186858.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvas JM, Vukolic A, Yepuri G, Xiong Y, Popp K, Schmutz I, Chappuis S, Albrecht U, Ming XF, Montani JP, Yang Z. Period2 gene mutant mice show compromised insulin-mediated endothelial nitric oxide release and altered glucose homeostasis. Front Physiol. 2012;3:337. doi: 10.3389/fphys.2012.00337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SK, Badea TC, Hattar S. Photoentrainment and pupillary light reflex are mediated by distinct populations of ipRGCs. Nature. 2011;476 doi: 10.1038/nature10206. 92-+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, Zhao X, Hatori M, Yu RT, Barish GD, Lam MT, Chong LW, DiTacchio L, Atkins AR, Glass CK, Liddle C, Auwerx J, Downes M, Panda S, Evans RM. Regulation of circadian behaviour and metabolism by REV-ERB-alpha and REV-ERB-beta. Nature. 2012;485:123–127. doi: 10.1038/nature11048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coomans CP, van den Berg SA, Houben T, van Klinken JB, van den Berg R, Pronk AC, Havekes LM, Romijn JA, van Dijk KW, Biermasz NR, Meijer JH. Detrimental effects of constant light exposure and high-fat diet on circadian energy metabolism and insulin sensitivity. Faseb J. 2013 doi: 10.1096/fj.12-210898. [DOI] [PubMed] [Google Scholar]
- Delezie J, Dumont S, Dardente H, Oudart H, Grechez-Cassiau A, Klosen P, Teboul M, Delaunay F, Pevet P, Challet E. The nuclear receptor REV-ERBalpha is required for the daily balance of carbohydrate and lipid metabolism. Faseb J. 2012;26:3321–3335. doi: 10.1096/fj.12-208751. [DOI] [PubMed] [Google Scholar]
- Driesen K, Jansen NW, Kant I, Mohren DC, van Amelsvoort LG. Depressed mood in the working population: associations with work schedules and working hours. Chronobiol Int. 2010;27:1062–1079. doi: 10.3109/07420528.2010.489877. [DOI] [PubMed] [Google Scholar]
- Fonken LK, Nelson RJ. Illuminating the deleterious effects of light at night. F1000 Med Rep. 2011;3:18. doi: 10.3410/M3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonken LK, Weil ZM, Nelson RJ. Dark nights reverse metabolic disruption caused by dim light at night. Obesity (Silver Spring) 2012 doi: 10.1002/oby.20108. In press. [DOI] [PubMed] [Google Scholar]
- Fonken LK, Workman JL, Walton JC, Weil ZM, Morris JS, Haim A, Nelson RJ. Light at night increases body mass by shifting the time of food intake. Proc Natl Acad Sci U S A. 2010;107:18664–18669. doi: 10.1073/pnas.1008734107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstner JR. On the evolution of memory: a time for clocks. Front Mol Neurosci. 2012;5:23. doi: 10.3389/fnmol.2012.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Brewer JM, Champhekar A, Harris RB, Bittman EL. Differential control of peripheral circadian rhythms by suprachiasmatic-dependent neural signals. Proc Natl Acad Sci U S A. 2005;102:3111–3116. doi: 10.1073/pnas.0409734102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha M, Park J. Shiftwork and metabolic risk factors of cardiovascular disease. J Occup Health. 2005;47:89–95. doi: 10.1539/joh.47.89. [DOI] [PubMed] [Google Scholar]
- Hatori M, Vollmers C, Zarrinpar A, Ditacchio L, Bushong EA, Gill S, Leblanc M, Chaix A, Joens M, Fitzpatrick JA, Ellisman MH, Panda S. Time-Restricted Feeding without Reducing Caloric Intake Prevents Metabolic Diseases in Mice Fed a High-Fat Diet. Cell Metab. 2012;15:848–860. doi: 10.1016/j.cmet.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloog I, Haim A, Portnov BA. Using kernel density function as an urban analysis tool: Investigating the association between nightlight exposure and the incidence of breast cancer in Haifa, Israel. Computers Environment and Urban Systems. 2009;33:55–63. [Google Scholar]
- Knutsson A. Health disorders of shift workers. Occup Med (Lond) 2003;53:103–108. doi: 10.1093/occmed/kqg048. [DOI] [PubMed] [Google Scholar]
- Kohsaka A, Laposky AD, Ramsey KM, Estrada C, Joshu C, Kobayashi Y, Turek FW, Bass J. High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab. 2007;6:414–421. doi: 10.1016/j.cmet.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Kohyama J. A newly proposed disease condition produced by light exposure during night: asynchronization. Brain Dev. 2009;31:255–273. doi: 10.1016/j.braindev.2008.07.006. [DOI] [PubMed] [Google Scholar]
- Lamia KA, Storch KF, Weitz CJ. Physiological significance of a peripheral tissue circadian clock. Proc Natl Acad Sci U S A. 2008;105:15172–15177. doi: 10.1073/pnas.0806717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeSauter J, Lambert CM, Robotham MR, Model Z, Silver R, Weaver DR. Antibodies for assessing circadian clock proteins in the rodent suprachiasmatic nucleus. PLoS One. 2012;7:e35938. doi: 10.1371/journal.pone.0035938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcheva B, Ramsey KM, Buhr ED, Kobayashi Y, Su H, Ko CH, Ivanova G, Omura C, Mo S, Vitaterna MH, Lopez JP, Philipson LH, Bradfield CA, Crosby SD, JeBailey L, Wang XZ, Takahashi JS, Bass J. Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature. 2010;466:627–631. doi: 10.1038/nature09253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamara P, Seo SB, Rudic RD, Sehgal A, Chakravarti D, FitzGerald GA. Regulation of CLOCK and MOP4 by nuclear hormone receptors in the vasculature: a humoral mechanism to reset a peripheral clock. Cell. 2001;105:877–889. doi: 10.1016/s0092-8674(01)00401-9. [DOI] [PubMed] [Google Scholar]
- Miyake S, Sumi Y, Yan L, Takekida S, Fukuyama T, Ishida Y, Yamaguchi S, Yagita K, Okamura H. Phase-dependent responses of Per1 and Per2 genes to a light-stimulus in the suprachiasmatic nucleus of the rat. Neurosci Lett. 2000;294:41–44. doi: 10.1016/s0304-3940(00)01545-7. [DOI] [PubMed] [Google Scholar]
- Mohawk JA, Green CB, Takahashi JS. Central and peripheral circadian clocks in mammals. Annu Rev Neurosci. 2012;35:445–462. doi: 10.1146/annurev-neuro-060909-153128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obayashi K, Saeki K, Iwamoto J, Okamoto N, Tomioka K, Nezu S, Ikada Y, Kurumatani N. Exposure to Light at Night, Nocturnal Urinary Melatonin Excretion, and Obesity/Dyslipidemia in the Elderly: A Cross-Sectional Analysis of the HEIJO-KYO Study. J Clin Endocrinol Metab. 2013;98:337–344. doi: 10.1210/jc.2012-2874. [DOI] [PubMed] [Google Scholar]
- Parkes KR. Shift work and age as interactive predictors of body mass index among offshore workers. Scand J Work Environ Health. 2002;28:64–71. doi: 10.5271/sjweh.648. [DOI] [PubMed] [Google Scholar]
- Paschos GK, Ibrahim S, Song WL, Kunieda T, Grant G, Reyes TM, Bradfield CA, Vaughan CH, Eiden M, Masoodi M, Griffin JL, Wang F, Lawson JA, Fitzgerald GA. Obesity in mice with adipocyte-specific deletion of clock component Arntl. Nat Med. 2012 doi: 10.1038/nm.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby NF, Hwang CE, Wessells C, Fernandez F, Zhang P, Sapolsky R, Heller HC. Hippocampal-dependent learning requires a functional circadian system. Proc Natl Acad Sci U S A. 2008;105:15593–15598. doi: 10.1073/pnas.0808259105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer FA, Hilton MF, Mantzoros CS, Shea SA. Adverse metabolic and cardiovascular consequences of circadian misalignment. Proc Natl Acad Sci U S A. 2009;106:4453–4458. doi: 10.1073/pnas.0808180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman H, Genzer Y, Cohen R, Chapnik N, Madar Z, Froy O. Timed high-fat diet resets circadian metabolism and prevents obesity. Faseb J. 2012 doi: 10.1096/fj.12-208868. [DOI] [PubMed] [Google Scholar]
- Stevens RG. Light-at-night, circadian disruption and breast cancer: assessment of existing evidence. Int J Epidemiol. 2009;38:963–970. doi: 10.1093/ije/dyp178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turek FW, Joshu C, Kohsaka A, Lin E, Ivanova G, McDearmon E, Laposky A, Losee-Olson S, Easton A, Jensen DR, Eckel RH, Takahashi JS, Bass J. Obesity and metabolic syndrome in circadian Clock mutant mice. Science. 2005;308:1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Amelsvoort LG, Schouten EG, Kok FJ. Duration of shiftwork related to body mass index and waist to hip ratio. Int J Obes Relat Metab Disord. 1999;23:973–978. doi: 10.1038/sj.ijo.0801028. [DOI] [PubMed] [Google Scholar]
- Vollmers C, Gill S, DiTacchio L, Pulivarthy SR, Le HD, Panda S. Time of feeding and the intrinsic circadian clock drive rhythms in hepatic gene expression. Proc Natl Acad Sci U S A. 2009;106:21453–21458. doi: 10.1073/pnas.0909591106. [DOI] [PMC free article] [PubMed] [Google Scholar]