Abstract
TRiC is an important group II chaperonin that facilitates the folding of many eukaryotic proteins. The TRiC complex consists of two stacked rings, each comprised of eight paralogous subunits with a mutual sequence identity of 30–35%. Each subunit has unique functional roles that are manifested by corresponding sequence conservation. It is generally assumed that the subunit order within each ring is fixed, but this order is still uncertain. Here we address the problem of the intra-ring subunit order by combining two sources of information: evolutionary conservation and a structural hypothesis. Specifically, we identify residues in the TRiC subunits that are likely to be part of the intra-unit interface, based on homology modeling to the solved thermosome structure. Within this set of residues, we search for a subset that shows an evolutionary conservation pattern that is indicative of the subunit order key. This pattern shows considerable conservation across species, but large variation across the eight subunits. By this approach we were able to locate two parts of the interface where complementary interactions seem to favor certain pairing of subunits. This knowledge leads to restrictions on the 5,040 (=7!) possible subunits arrangements in the ring, and limits them to just 72. Although our findings give only partial understanding of the inter-subunit interactions that determine their order, we conclude that they are comprised of complementary charged, polar and hydrophobic interactions that occur in both the equatorial and middle domains of each subunit.
1. Introduction
Chaperonins are large protein complexes that assist in the folding of nascent and misfolded polypeptide chains. The group II chaperonins are found only in eukaryotes and archaea and share a similar overall structure [1]. They consist of two stacked rings with flexible tops that can open and close through an ATP binding and hydrolysis cycle. Upon closure, an isolated cavity is formed within each ring, where refolding of polypeptide chains can occur.
TRiC is an important member of the group II chaperonins, and is highly conserved in most eukaryote families. It has been implicated in the folding pathways of many proteins, most notably actins and tubulins [2,3,4]. Each ring in TRiC consists of eight paralogous subunits: A, B, G, D, E, H, Q and Z. The specific differentiation of the eight TRiC subunits is highly conserved. For example, the sequence identity between the α subunit of yeast and the α subunit of bovine is greater than 60%, yet the sequence identity between different subunits in bovine is only ~30%. Clearly, this remarkable conservation is likely to ensure a constant order of the subunits within the ring in addition to its other functional roles in TRiC [5,6].
Because of their allosteric nature, the lid closure and the ATP cycle are highly influenced by the order of subunits within the ring [7]. Yet, despite its functional role, the order of subunits in the ring is still debated. Liou and Willison [16] proposed an arrangement of subunits in the ring that is based on a very difficult experimental protocol that analyzes micro-complexes, i.e. very partial fragments of TRiC rings found in cell extracts. This arrangement is used to this day [7,8], but has not been validated by a different methodology. Unfortunately, structural data that could unambiguously resolve the order of the subunits is not available. TRiC is very refractory to crystallization, and cryo-EM reconstructions of TRiC are currently at insufficient resolution to differentiate the subunits [8].
In order to make progress in the elucidation of ring order, we tried a computational approach. The subunit order in TRiC is a challenging modeling problem because of the large number of possible arrangements for this complex. In addition, the sequence identity between the subunits is high as well (~30%) making the molecular key to the subunit order more subtle. To overcome these difficulties we combined two sources of information: evolutionary conservation and a structural hypothesis. The structural model is based on the crystal structures for a close orthologue of TRiC, the archaeal thermosome [9]. The thermosome is also a group II chaperonin that consists of only two subunit types, which are not very differentiated (60% sequence identity). We analyzed the model by looking for residues that were part of the subunit interface and showed a conservation pattern that may indicate of the subunit order. Specifically, we looked for patterns that showed considerable conservation across species, but large variability across the eight subunits. We identified several areas of the interface where complementary interactions could putatively favor certain subunit pairings. From this analysis we derived a set of constraints that limit the number of possible subunits arrangements from 5,040 to 72.
2. Methods
The evolutionary profile in each position is based on sequence alignments from 13 species (number in parenthesis is sequence identity to the bovine gene): Bovine (100%), Human (97%), Zebrafish (86%), Ciona (74%), Drosophila (73%), C. Elegans (68%), Arabidopsis (66%), Yeast (63%), Neospora (61%), Candida (61%), Dictyostelium (61%), Plasmodium (58%) and Paramecium (58%). This order of decreasing sequence identity to the bovine genes is kept (in a left to right manner) throughout the presentations in the tables. The main guideline in choosing these specific organisms was to form a diverse set of family representatives that spans the eukaryotic evolutionary tree. A secondary guideline was to use only fully annotated genomes that had clear annotation of the eight different subunits. The eight sequences from each organism were aligned to each other and to the thermosome sequences using the COBALT multiple sequence alignment tool from NCBI [10].
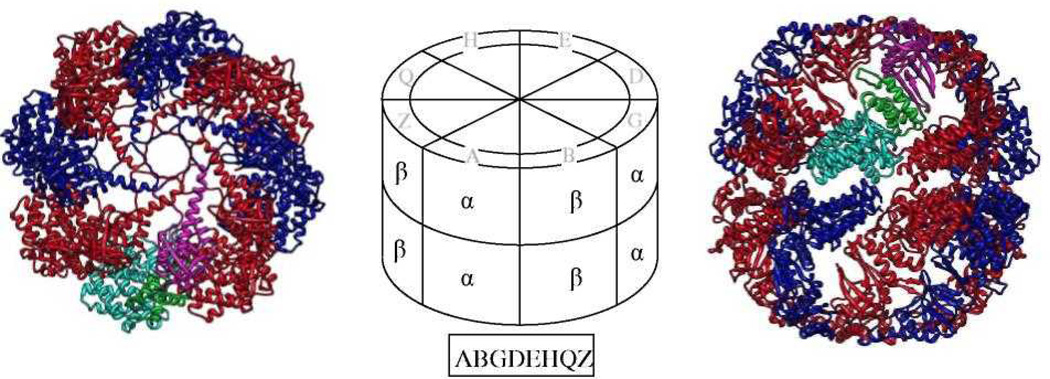
The structural model (Fig. 1) was based on the crystallographic structure of the archaeal thermosome from T. acidiphilum (PDB 1A6D). The sequence identity between TRiC and the thermosome is ~40%, indicating a highly reliable model. Unlike TRiC, which has eight different subunits, each ring in the thermosome is composed of four repeats of two different subunits (α and β). This arrangement leads to only two different interfaces, and we focused on the α-β interface. We exclude the apical domain from the structural analysis because it is unclear whether TRiC assembles in a close or open conformation. We also exclude the loops spanning residues 53–57 and 163–166 since they are structurally variable between the two thermosome subunits.
Figure 1.
Our structural model is based on the known structure of the thermosome (PDB file 1A6D), which has α (blue) and β (red) subunits that alternate around each of the two rings. The structure is viewed from the top (left) and side (right). The three domains of one of the α subunits in the top ring are colored cyan, green & magenta for the equatorial, middle & apical domains, respectively. The central schematic shows how to map the 8 subunits - A, B, G, D, E, H, Q & Z - of TRiC in a proposed arrangement (boxed) onto the top ring of the thermosome. This arrangement is brought as an example only, and is not part of our suggested set of 72.
Throughout the text we present possible subunit arrangements in the ring as a string of eight letters. These refer to the subunit order in a counterclockwise direction in the top ring when viewed from the top. Wherever we refer to the left and right subunits of an interface, we mean the respective interacting subunits in the top ring when viewed from the side (Figure 1, right)1.
A position in a certain subunit was considered conserved across species if it showed consistent physio-chemical properties in 11 or more of the species (i.e. mostly hydrophobic, negatively-charged, long-polar etc.). A position was considered as a possible candidate for the molecular key of the subunit order if at least two subunits were conserved across species, but showed remarkably different physio-chemical properties (i.e subunit Z and H in position 1 in Table 1). The definition of the interaction centers was more subjective and was based on manual inspection of clusters of positions that were characteristic of the order key.
Table 1.
The residue types observed in TRiC in the two homologous positions to the two marked thermosome residues in figure 2a.
| A | Position 1: Unit α D48 | Position 2: Unit β R515 | A | |||
| DDDDDDDDDDDDD | D | RRRRRRRRRRRRR | R | |||
| B | DDDDDDDDDDDDD | D | RRRRRRRRRRRRR | R | B | |
| G | MMMLLLLLLLLLL | hpb | RRRRRRRRRRRRR | R | G | |
| D | DDDDDDDDDDDDD | D | KKKKKKKRRRKKK | [KR] | D | |
| E | DDDDDDDDDDDDD | D | KKKKKKKKKKKKK | K | E | |
| H | DDDDDDDDDDDDD | D | SSSSSSSSGSSSS | [SG] | H | |
| Q | NNNNNNNNNNNNK | [NK] | RRRSKKRSSSRKR | [RSK] | Q | |
| Z | MMMMVLILILLYL | hpb | LLLLLLLLLLLLL | L | Z | |
For each subunit and position the data from 13 different organisms are shown in the order: Bovine, Human, Zebrafish, Ciona, Drosophila, C. Elegans, Arabidopsis, Yeast, Neospora, Candida, Dictyostelium, Plasmodium and Paramecium. The amino acids that occur in each row are shown to the right of that row.
“hp” is a conserved hydrophobic position.
3. Results
This study presents a practical approach towards elucidation of the intra-ring subunit order of TRiC by combining two sources of information: evolutionary conservation and a structural hypothesis. Three assumptions are made:
The subunit order within the ring is fixed, and each of the eight subunits appears once in the ring. This assumption greatly limits the possible number of arrangements in the ring to 5,040 (=7!). In particular, it removes the burden of analyzing the interactions of any subunit with a same copy of itself.
Structural hypothesis. The structural interface between subunits in the ring resembles the interface observed in the solved crystallographic structure of the archaeal thermosome [9]. This assumption is justified by the high sequence identity (~34%) between TRiC and the thermosome, as well as by supporting data from EM structural studies [8]. The structural analysis is therefore based on a straightforward alignment of the bovine TRiC sequences onto the solved thermosome structure.
Evolutionary information. The subunit order, as well as the molecular key that determines it, is conserved across different eukaryote species ranging from unicellular organisms to mammals. Strong support for this assumption comes from the much higher sequence identity observed between subunits of the same type across species relative to the identity between different subunit types in the same organism. This assumption is a valuable source of evolutionary information, as it implies that the residues participating in the order key are under selective evolutionary pressure.
Guided by these assumptions, analysis progressed in three steps. In the first step, all the residues in the thermosome structure that participate in an inter-subunit contact (within van der Waals contact distance) were pooled into a putative interface set. In the second step, we checked the evolutionary pattern of each residue in the interface set, and looked for conservation pattern that may confer order information. Specifically, we looked for residue positions that showed considerable conservation across species, but large variation across subunits (see for example Table 1). If several such positions were spatially clustered in the thermosome structure, we pooled them together into a putative interaction center. Five putative interaction centers were identified in this manner, however only two of them are amenable to reliable structural interpretation at this stage. In the last step, we tried to infer a number of structural restrictions from the two final interaction centers. We used these restrictions to discard the majority of the 5,040 subunit arrangements. This step is especially sensitive to the subtle structural differences between TRiC and the thermosome. We, therefore, were very conservative in our arguments, preferring to retain more possible arrangements. By this analysis, we were able to limit the number of possible subunit arrangements by 70 fold to just 72 (Table 3).
Table 3.
The 72 possible subunit arrangements in the ring sorted alphabetically.
| ABDEQGZH | ABDEQHGZ | ABDQGZHE | ABDQHGZE | ABEDQGZH | ABEDQHGZ |
| ABEQGZHD | ABEQHGZD | ABGZDEQH | ABGZDQHE | ABGZEDQH | ABGZEQHD |
| ABGZHDEQ | ABGZHDQE | ABGZHEDQ | ABGZHEQD | ADBEQGZH | ADBEQHGZ |
| ADBGZEQH | ADBGZHEQ | ADEQBGZH | ADEQGZHB | ADEQHBGZ | ADEQHGZB |
| ADQBGZHE | ADQEBGZH | ADQGZHBE | ADQGZHEB | ADQHBGZE | ADQHEBGZ |
| ADQHGZBE | ADQHGZEB | AEBDQGZH | AEBDQHGZ | AEBGZDQH | AEBGZHDQ |
| AEDQBGZH | AEDQGZHB | AEDQHBGZ | AEDQHGZB | AEQBGZHD | AEQDBGZH |
| AEQGZHBD | AEQGZHDB | AEQHBGZD | AEQHDBGZ | AEQHGZBD | AEQHGZDB |
| AQBGZHDE | AQBGZHED | AQDBGZHE | AQDEBGZH | AQEBGZHD | AQEDBGZH |
| AQGZHBDE | AQGZHBED | AQGZHDBE | AQGZHDEB | AQGZHEBD | AQGZHEDB |
| AQHBGZDE | AQHBGZED | AQHDBGZE | AQHDEBGZ | AQHEBGZD | AQHEDBGZ |
| AQHGZBDE | AQHGZBED | AQHGZDBE | AQHGZDEB | AQHGZEBD | AQHGZEDB |
3.1. Interaction Center 1
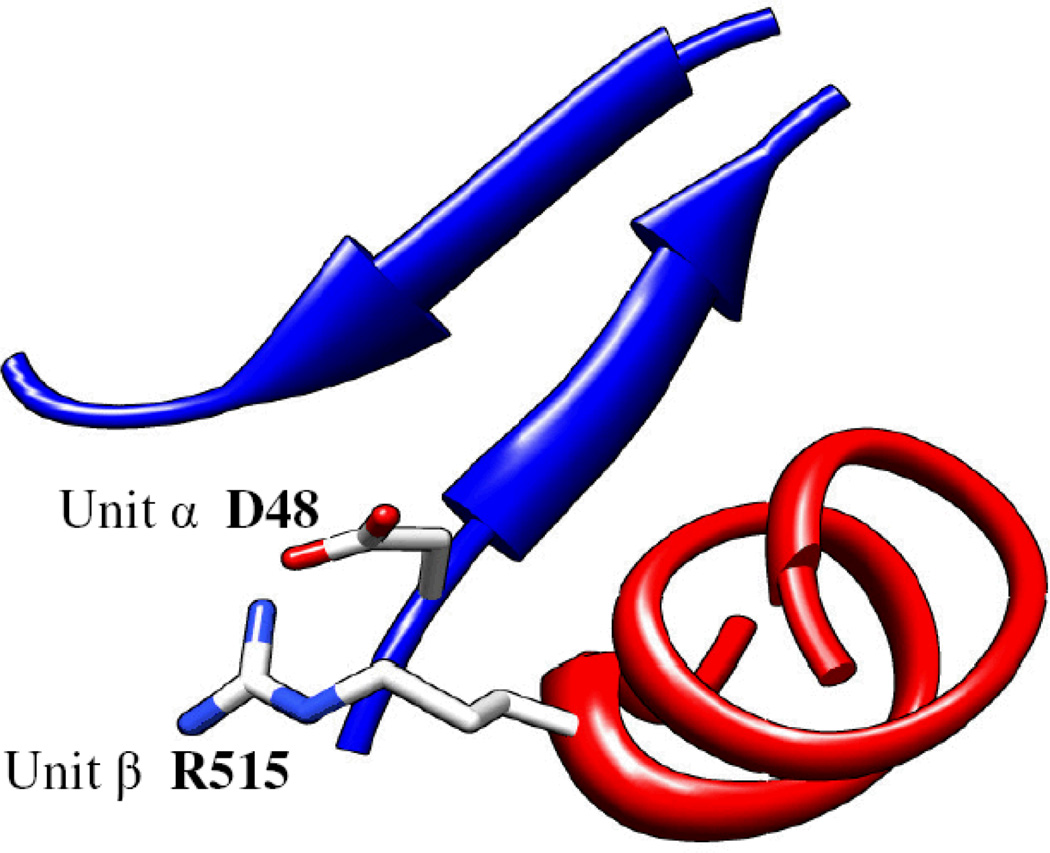
In the thermosome, residue D48 from the α subunit makes a partially buried salt bridge with residue R515 from the β subunit (Fig. 2). In this arrangement, the aspartic side-chain is quite buried within the interface, while the arginine side-chain is relatively free to move into the solvent. In the thermosome, this salt bridge is conserved in both the α-β and the β-α interfaces. Table 1 shows the residue types that are observed in these two positions across 13 species (rows) and across the different subunit types (columns). The 13 letters in each subunit and position are arranged in decreasing sequence similarity to the bovine genes, and range from bovine (left) to the unicellular paramecium (right). The table shows a consistent conservation pattern: each position conserves the residue property (charge, hydrophobicity etc.) across species, but large variation occurs between subunit types.
Figure 2.
Two complementary residues at the interface of two thermosome subunits. Unit α (blue) and subunit β (red) are the left and right subunits, respectively, in the top ring when viewed from the side of the particle. In TRiC this salt bridge is not conserved between all subunits. The amino acids at these two positions in different organisms are shown in Table 1.
The data suggest that the observed salt bridge from the thermosome is conserved between some of the subunits in TRiC, but is absent in others. Because of the partial burial of the aspartic side-chain, there will be a considerable energetic advantage if a positively-charged side-chain from the next subunit can form a salt bridge with it. Following this reasoning, the possible subunit arrangements are restricted to those where any of the {A,B,D,E,H} subunits is followed by any one of the {A,B,G,D,E,Q} subunits. The Q subunit is included in the latter group, even though it does not always have a positive charge in that position, so as to make our restrictions as conservative as possible. Applying this first restriction lowers the number of possible subunit arrangements from 5,040 to 480.
A conserved hydrophobic pattern is observed for subunits G and Z in Position 1 and for subunit Z alone in Position 2. Disregarding the Z-Z interface, the G-Z interface forms a very favorable hydrophobic interaction. Furthermore, a hydrophobic residue in Position 2 will cause unfavorable burial of any polar residue in Position 1. This reasoning restricts the subunit arrangements to those with a G-Z interface. Applying this restriction further lowers the number of possible subunit arrangements to 240. The G-Z restriction is validated independently by interaction center 2 (see below).
3.2. Interaction Center 21
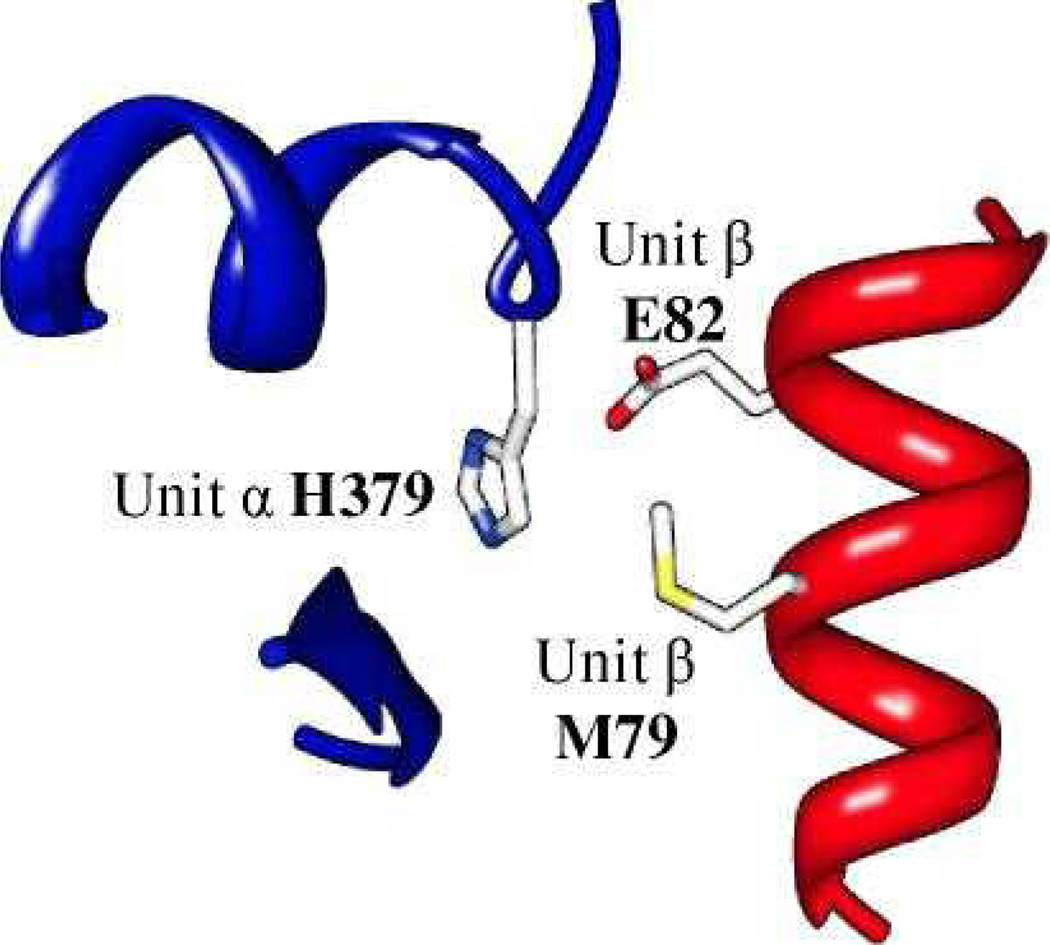
In the thermosome, the N-terminal part of a helix from the middle domain is interacting with the equatorial domain of the next subunit in the ring (Fig. 3). The interaction involves a glutamic acid side-chain (Position 3) that makes electrostatic interactions with both the positive histidine ring (Position 1) and the dipole-related charged N-terminal of the helix. In addition, the histidine side-chain makes a hydrophobic interaction with the methionine side-chain (Position 2). In TRiC, these positions are altered, but show conservation patterns that are consistent with the order key (Table 2). As with the previous center, the table refers to adjacent subunits in the ring and the positions are as marked in Figure 3.
Figure 3.
Three complementary residues at the interface between two thermosome subunits. Subunit α (blue) and subunit β (red) are the left and right subunits, respectively, in the top ring when viewed from the side of the particle. The amino acids at these three positions in different organisms are shown in Table 2.
Table 2.
The residue types observed in TRiC for the three homologous positions to the three marked thermosome residues in figure 3a.
| A | Position 1: Unit α H379 | Position 2: Unit β M79 | Position 3: Unit β E82 | A | |||
| FFFIFVYYFYLFL | hp | VVVVVVVIIIIIV | hp | EEEEEEEEDEQNE | [EDQN] | ||
| B | QQQQQQHQQQHHH | [QH] | VVVVIVVVVVIII | hp | DDDEDDDNNNDDE | [DNE] | B |
| G | EEEDDDDDDDDDD | [DE] | SSSSSSSSSSSSS | S | EEEEEEEEEEEEE | E | G |
| D | LLLLLLLMLMLLL | hp | MMMMMMMMMMMMM | M | EEEEEEEEQDEEE | [EQD] | D |
| E | MMMMMMMMMMMMM | M | LLLLLLLLLLLLL | L | EEEEQEEQEQQEE | [EQ] | E |
| H | QQQQQQQQQQQQQ | Q | TTTTTTITITTIV | [TIV] | DDDDDDDDDDDDD | D | H |
| Q | NNNNNSSNNNNNN | [NS] | MMMMLILVLVMIM | hp | MMMIMMLMMMMKM | [MILK] | Q |
| Z | HHHHHHHYHYHYH | [HY] | LLLLMMMLMMLMM | hp | KKKKRKRRRRRRR | [RK] | Z |
See footnotes to Table 2.
Position 3 is central in this network because it interacts with the helix terminal and possibly with all other positions. Most subunits in this position have a conserved a negatively-charged residue, which is compatible with the positive charge of the helix terminal as seen in the thermosome structure. In striking contrast, the Z subunit displays a conservation pattern of positively-charged amino acids. The expected electrostatic repulsion between the helix terminal and the Z subunit can be facilitated if any of the other positions provides a mediating negative charge. The only possibility is Position 1 in the G subunit that conserves negative charge. This match revalidates the G-Z condition we derived previously from interaction center 1. It also suggests that the helix terminal of the G subunit is protruding from the Z subunit, because the interaction is mediated by an additional side-chain.
Another unusual subunit in Position 3 is subunit Q, which shows a unique hydrophobic conservation. Together with Position 2, subunit Q displays a significant hydrophobic patch at this interaction center. A favorable hydrophobic interaction can form in the interface between this patch and one of the hydrophobic side-chains from Position 1. This reasoning restricts the subunit arrangements in the ring to those where the Q subunit is preceded by one of the subunits from the following group {A,D,E}. Applying this restriction further lowers the number of possible arrangements of subunits in a ring to 144. This interaction is also likely to lead to bulging at the helix terminal, since it does not form an interaction of the helix terminal with a negative side-chain in Position 3.
Finally, Position 2 shows a general hydrophobic conservation pattern. A strong exception is subunit G, which shows a conserved polar pattern (subunit H is similar, but less conserved). A hydrophobic side-chain from Position 1 is likely to bury this serine upon subunit binding. We can therefore forbid subunit arrangements in which the G subunit is preceded by one of the subunits from the group {A,D,E}. Applying this restriction further lowers the number of possible subunit arrangements to in the ring 72. Of all the restrictions discussed so far, this is the one we are least confident with, because of the weaker nature of polar burial as compared to charge burial. However, the two-fold reduction in possible subunits arrangements makes this restriction valuable.
The 72 possible arrangements of subunits we find here only sample 33 of the 56 different possible subunit pairings (see Table 4), and not in equal measure. This distribution could be used in experiment planning that can determine subunit pairing (such as the double immuno-labeling and cryo-EM reconstruction used by Martin-Benito et al. [12]). For example, an experiment of double labeling with antibodies against the Q and H subunits is guaranteed to lower the number of possible arrangements to 36. If the particle reconstruction reveals the extra density due to the bound antibodies on successive subunits, then the only possible pairing is QH as pairing HQ has already been eliminated. On the other hand, if the extra densities are further apart around the ring, then the only possible pairing for subunit H is ZH as there are only two allowed pairings with H to the right, QH and ZH.
Table 4.
Number of occurrences of allowed subunits pairing in the 72 possible ring arrangements
| Subunit | A | B | G | D | E | H | Q | Z | #Partners |
|---|---|---|---|---|---|---|---|---|---|
| A | - | 16 | - | 16 | 16 | - | 24 | - | 4 |
| B | 12 | - | 36 | 12 | 12 | - | - | - | 4 |
| G | - | - | - | - | - | - | - | 72 | 1 |
| D | 16 | 16 | - | - | 16 | - | 24 | - | 4 |
| E | 16 | 16 | - | 16 | - | - | 24 | - | 4 |
| H | 14 | 12 | 18 | 14 | 14 | - | - | - | 5 |
| Q | 4 | 6 | 18 | 4 | 4 | 36 | - | - | 6 |
| Z | 10 | 6 | - | 10 | 10 | 36 | - | - | 5 |
| #Partners | 6 | 6 | 3 | 6 | 6 | 2 | 3 | 1 |
4. Discussion
In this study we used evolutionary information and a structural model to narrow the number of possible subunit arrangements in the ring of TRiC from 5,040 to 72 (Table 3). This specific type of data analysis has already been used effectively in many other studies [13–15], but its application to TRiC is novel both in terms of the magnitude of the problem as well as in the level of detail demanded. Two sources of errors might jeopardize the validity of the results: wrong restrictions due to misinterpretation of the structural model or, more likely, because the model itself contains errors that reflect the difference between TRiC and the thermosome. These concerns are always inherent to this type of methodology, and we tried to alleviate them by adherence to the most conservative possible assumptions and views of the data. Somewhat paradoxically, the lack of great detail in our assumptions actually makes them more robust to errors in the model: minor changes in backbone or side-chain conformations are unlikely to break them.
We emphasize that the five positions described in the tables are not the entire molecular key for the subunit order. At least three other putative interaction centers show conservation patterns that are likely to be important for the interface specificity. These centers were not used here because we are currently unsure of their interpretations for two main reasons. First, some of these centers are more complex and involve three interactions or more. The analysis of so many interactions requires a more rigorous modeling of the side-chain positions. Second, in some areas a significant backbone change is likely as large variations are already evident between the two thermosome subunits (loop 53–57 for example). Analysis of these areas requires consideration for backbone flexibility.
Our results start to reveal the molecular basis for the subunit order within the ring. It appears that a hydrophobic core, which is conserved across all subunits, provides a general scaffold for the interface between two successive subunits in the ring. This scaffold is bordered by interaction centers that are subunit-pair specific. The subunit order results from complementary charged, polar, and (to lesser extent) hydrophobic interactions that make the correct pairing more favorable than the alternatives. The subunit similarity excludes the possibility of significant steric clashes as a source of order information. Only the equatorial domains are part of the general hydrophobic interface. Major ordering information is encoded in Interaction Center 2 from the middle domain. Since the middle domain mediates between the ATP site and the lid, it is likely that the subunit specificity in interaction center 2 has functional relevance.
In order to completely determine the subunit arrangement in the ring, we see two possible courses of action. Perhaps most intuitively, structural data from other studies could be used to further validate and limit the set of subunit arrangements presented here. We believe that EM structural studies will be extremely valuable in that context. Alternatively, more detailed modeling could lead to additional structural insights from the unexplored interaction centers, as well as to tighter restrictions from the two interaction centers already described. That level of detail requires the use of more computer intensive approach with explicit modeling of side-chain conformations and backbone flexibility. We are currently testing various computational tools for that purpose and hope to make further progress in the future.
4.1. Inconsistencies with the Liou and Willison model
Liou and Willison [16] suggested an arrangement of subunits in the ring that is still in use. They inferred this arrangement from 6 association patterns of TRiC micro-complexes, i.e. very partial fragments of TRiC rings found in cell extracts. Unfortunately, none of the 72 possibilities that we summarize in Table 3 is consistent with the Liou and Willison arrangement. The G-Z interaction is among the first restrictions to prohibit their arrangement, which does not include it. However, we see a very strong signal for its occurrence in this work. Furthermore, out of the 72 possibilities we suggest, only one is consistent with 4 out of the 6 association patterns from the Liou and Willison work, and the rest are consistent with 3 or less. The clear inconsistency between the two studies suggests that the micro-complexes they identified are not always genuine parts of the intact complex.
Acknowledgements
This study was supported by the BioX program as well as NIH Nanomedicine award 2PN2EY016525-02 (Wah Chiu PI).
Footnotes
Molecular graphics images were produced using the UCSF Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIH P41 RR-01081) [11].
Contributor Information
Nir Kalisman, Department of Structural biology, School of Medicine, Stanford University Stanford, California 94305, USA.
Michael Levitt, Department of Structural biology, School of Medicine, Stanford University Stanford, California 94305, USA.
References
- 1.Spiess C, Meyer AS, Reissmann S, Frydman J. Trends Cell Biol. 2004;14:598. doi: 10.1016/j.tcb.2004.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gao Y, Thomas JO, Chow RL, Lee GH, Cowan NJ. Cell. 1992;69:1043. doi: 10.1016/0092-8674(92)90622-j. [DOI] [PubMed] [Google Scholar]
- 3.Gao Y, Vainberg IE, Chow RL, Cowan NJ. Mol Cell Biol. 1993;13:2478. doi: 10.1128/mcb.13.4.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frydman J, Nimmesgern E, Erdjument-Bromage H, Wall JS, Tempst P, Hartl FU. EMBO J. 1992;11:4767. doi: 10.1002/j.1460-2075.1992.tb05582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spiess C, Miller EJ, McClellan AJ, Frydman J. Mol Cell. 2006;24:25. doi: 10.1016/j.molcel.2006.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tam S, Geller R, Spiess C, Frydman J. Nat Cell Biol. 2006;8:1155. doi: 10.1038/ncb1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rivenzon-Segal D, Wolf SG, Shimon L, Willison KR, Horovitz A. Nat Struct Mol Biol. 2005;12:233. doi: 10.1038/nsmb901. [DOI] [PubMed] [Google Scholar]
- 8.Booth CR, Meyer AS, Cong Y, Topf M, Sali A, Ludtke SJ, Chiu W, Frydman J. Nat Struct Mol Biol. 2008;15:746. doi: 10.1038/nsmb.1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ditzel L, Löwe J, Stock D, Stetter KO, Huber H, Huber R, Steinbacher S. Cell. 1998;93:125. doi: 10.1016/s0092-8674(00)81152-6. [DOI] [PubMed] [Google Scholar]
- 10.Papadopoulos JS, Agarwala R. Bioinformatics. 2007;23:1073. doi: 10.1093/bioinformatics/btm076. [DOI] [PubMed] [Google Scholar]
- 11.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. J Comput Chem. 2004;25:1605. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 12.Martín-Benito J, Grantham J, Boskovic J, Brackley KI, Carrascosa JL, Willison KR, Valpuesta JM. EMBO Rep. 2007;8:252. doi: 10.1038/sj.embor.7400894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Armon A, Graur D, Ben-Tal N. J Mol Biol. 2001;307:447. doi: 10.1006/jmbi.2000.4474. [DOI] [PubMed] [Google Scholar]
- 14.Ahmad S, Keskin O, Sarai A, Nussinov R. Nucleic Acids Res. 2008;36:5922. doi: 10.1093/nar/gkn573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Engelen S, Trojan LA, Sacquin-Mora S, Lavery R, Carbone A. PLoS Comput Biol. 2009;5:e1000267. doi: 10.1371/journal.pcbi.1000267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liou AK, Willison KR. EMBO J. 1997;16:4311. doi: 10.1093/emboj/16.14.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]