Abstract
Hepatitis B virus (HBV) infection is still a public health problem worldwide, being endemic in some parts of the world. It can lead to serious liver diseases such as chronic hepatitis, cirrhosis, and hepatocellular cancer. The differences in host immune response can be one of the reasons for the various clinical presentations of HBV infection. Polymorphisms of genes encoding the proinflammatory and antiinflammatory cytokines, which are responsible for regulation of the immune response, can affect the clinical presentation of the infection. Particularly, the polymorphisms of the genes encoding cytokines such as interleukin (IL)-1, IL-6, IL-8, IL-10, IL-18, IL-28B, interferon-γ, tumor necrosis factor-α, tumor growth factor-β1, and regulatory molecules like vitamin D receptor and chemokine receptor 5 can be responsible for different clinical presentations of HBV infections. The genomic information about cytokines and other mediators can be important for determining high-risk people for developing chronic hepatitis or hepatocellular cancer and may be used to plan treatment and preventive approaches for these people. In this review, the current knowledge in the literature on the association between cytokine/regulatory molecule gene polymorphisms and clinical course of chronic HBV infection is summarized, and the clinical implementations and future prospects regarding this knowledge are discussed.
Keywords: Hepatitis B virus, Cytokine, Polymorphism, Chronic hepatitis
Core tip: The specific polymorphisms of genes encoding cytokines, such as interleukin (IL)-1, IL-8, IL-10, IL-18, IL-28B, tumor necrosis factor-α, interferon-γ, tumor growth factor-β1, and regulatory molecules such as vitamin D receptor and chemokine receptor 5 affect the clinical course of chronic hepatitis B virus (HBV) infection. This review aims to summarize the literature on cytokine gene polymorphisms and chronic HBV infection and discuss future prospects regarding the clinical implication of these polymorphisms.
INTRODUCTION
Hepatitis B virus (HBV) infection is a serious and common infectious disease of the liver, affecting 240 million people worldwide with an estimated 600000 deaths per year, and remains the major cause for chronic hepatitis, cirrhosis, and hepatocellular carcinoma[1-3]. HBV infection is endemic, particularly in developing countries, and is a serious public health problem[3].
Following acute HBV infection, 1%-5% of adults develop chronic infection[4] (Figure 1). Rate of chronicity is inversely proportional to age, being higher in newborns and children than in adults. The prevalence of chronic HBV infection is also higher (over 8%) in areas where the disease is highly endemic than in those with intermediate and low endemicity[4].
Figure 1.
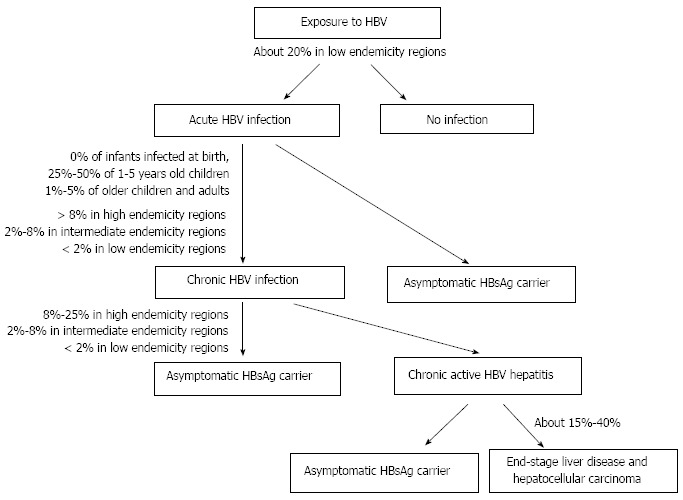
Clinical presentations of hepatitis B virus infections[4,100]. HBV: Hepatitis B virus; HBsAg: Hepatitis B surface antigen.
The chronic diseases caused by HBV are chronic hepatitis, cirrhosis, and primary hepatocellular carcinoma[5]. Chronic hepatitis can lead to end-stage liver disease in 15%-40% of patients[6]. A number of factors, including host-related factors (e.g., genetic and immunological background), pathogen-related factors (e.g., viral load, genotype), and environmental factors (e.g., hygiene, nutrition, treatment, vaccination)[7] affect the outcome of HBV infection.
It has been well known that the genetic background of the host and host-pathogen interactions influence the outcome of HBV infection[8-12]. Hepatitis B surface antigen positivity is more common in identical twins than in fraternal twins[13], which indicates that host-related genetic factors have an impact on the course of HBV infection.
Gene polymorphisms such as the single nucleotide polymorphism (SNP; replacement of a nucleotide with another one) may change the structure and biological function of the protein coded by that gene. A SNP in the promoter region of a gene may cause increased or decreased production of the relevant protein. The presence of these types of inherited gene polymorphisms may make a person more susceptible or resistant to a certain disease[14].
Cytokines and regulatory molecules play a fundamental role in the immunopathogenesis of HBV infection. The gene loci for cytokines are defined, and polymorphisms of these genes are suggested to influence the outcome of HBV infection[11]. Therefore, many recent studies have focused on the effect of gene polymorphisms of cytokines on disease outcome and response to vaccination and treatment[10]. Understanding the genetic background of this common public health problem may give rise to new strategies for prevention, treatment, and control of HBV infection.
In this systematic review of the literature, the impact of gene polymorphisms on the course of chronic HBV infection is evaluated and discussed with a focus on polymorphisms of genes encoding cytokines and regulatory proteins.
CYTOKINES
Cytokines represent a large family of molecules, including tumor necrosis factor-alpha (TNF-α), interleukin (IL)-1β, IL-4, IL-6, IL-8, IL-10, IL-18, IL-28, interferon (IFN)-γ, IFN-α, and tumor growth factor-beta (TGF-β). Cytokines play an important role in the initiation and regulation of immune responses and, therefore, might affect susceptibility to HBV and/or the natural course of the infection[9].
In addition to cytokines, antioxidant enzymes (e.g., nitric oxide synthase, manganese superoxide dismutase, glutathione S-transferase), and regulatory proteins (e.g., chemokine receptor 5 (CCR5), vitamin D receptor (VDR), estrogen receptor, mannose binding lectin) may also have a role in the course of HBV infection and polymorphisms of the genes encoding these proteins are evaluated in various studies[14].
The role of polymorphisms of genes encoding cytokines and some regulatory proteins in chronic HBV infection is summarized below (Table 1).
Table 1.
Role of polymorphisms of genes encoding cytokines and some regulatory proteins in chronic hepatitis B virus infection
| Cytokine | Allele/polymorphism | Effect | Ref. |
| IL-1 | |||
| IL-1α | |||
| IL-1β | -511C | Persistent infection | [20-22] |
| IL-1RN | 2 | Protective against HBV infection | [18] |
| IL-6 | -174 G/C | No effect | [26] |
| IL-8 | -251AA | Protective against HBV-related cirrhosis | [28] |
| IL-10 | -1082G | Virus clearance, lower HBV viral load, protective against HBV infection | [30,31] |
| Persistent HBV infection | [35] | ||
| -592CA | Virus clearance | [35] | |
| Persistent HBV infection | [36] | ||
| IL-10R K47E | Persistent, chronic infection | [38,99] | |
| IL-18 | -148C | Virus clearance | [40] |
| +8925G | Virus clearance | [40] | |
| +13925C | Virus clearance | [40] | |
| -137C | Protective against HBV | [43,44] | |
| -607AA | Inhibition of HBV DNA replication | [43,44] | |
| IL-28B | Virus clearance, prevent HBV progression | [46] | |
| No effect | [51] | ||
| TNF-α | -863A | Virus clearance, persistent infection | [68,71,77] |
| -238A | Persistent infection | [37,58-62,71] | |
| -308A | Progressive disease | [55,58,66,77] | |
| Protective against chronic HBV infection | [76] | ||
| -857CC | Persistent infection | [37,61,62,68] | |
| Protective against chronic HBV infection | [75] | ||
| IFN-γ | +874AA | Viral load, persistent infection | [54] |
| TGF-β1 | -509C | Development of cirrhosis | [80] |
| Codon 10T | Development of cirrhosis | [80,83] | |
| Progression to hepatocellular cancer | [81,82] |
HBV: Hepatitis B virus; IL: Interleukin; TNF: Tumor necrosis factor; IFN: Interferon; TGF: Tumor growth factor.
IL-1
IL-1 is a proinflammatory cytokine with various biological activities[15]. The IL-1 gene family encodes IL-1α, IL-1β, and their natural inhibitor, IL-1 receptor antagonist (IL-1RN)[15,16]. IL-1RN allele 2 polymorphism is associated with an increase in IL-1β production[17], which then increases the production of other cytokines (e.g., IL-2, IL-6, and TNF-α), and stimulates the clearance of HBV[18]. IL-1RN polymorphisms, thus, have a protective role against HBV infection[18].
In addition to its proinflammatory action, IL-1β has a role in tumor growth[19]. Polymorphism of IL-1β at -511C allele is associated with increased IL-1β level and is a genetic indicator of hepatocellular cancer development in chronic HBV-infected patients[20]. IL-1β and IL-1RN accessory protein gene polymorphisms are related to chronic and persistent HBV infection[18,21]. Fontanini et al[22] reported that IL-1β proinflammatory polymorphisms are associated with cirrhosis and end stage liver disease, which are more pronounced in males.
IL-6
IL-6 is an important cytokine that regulates the immune response to HBV infection[15]. IL-6 level is significantly increased in chronic HBV infection[23]. However, studies from Korea and Israel showed that there is no relation between IL-6 gene polymorphism and chronic HBV infection[24,25]. Similarly, studies from other populations indicate no significant effect of IL-6 polymorphism at -174G/C on chronic HBV infection[26].
IL-8
IL-8 has been associated with tumors and chronic inflammatory diseases through its mitogenic and angiogenic functions[27]. Qin et al[28] indicated that the polymorphism of the IL-8 gene at -251AA might be protective for HBV-related cirrhosis.
IL-10
IL-10 is secreted mainly from T cells and has an inhibitory action on both inflammatory and immunoproliferative responses. It stimulates the differentiation and proliferation of B cells producing immunglobulin M (IgM), IgG and IgA. Moreover, IL-10 inhibits secretion of various cytokines from T cells and monocytes/macrophages[29]. The polymorphism of IL-10 at -1082 region that results in increased production of G allele is correlated with virus clearance during intrauterine HBV infection. Moreover, increased IL-10 production has a protective effect against HBV infection[30]. The G/G genotype at -1082 is further associated with lower HBV viral load at the immune inflammatory phase in children with chronic HBV infection[31].
However, there are some conflicting results in the literature evaluating the effect of IL-10 gene polymorphism on HBV infection. Polymorphisms of genes encoding IL-10 are related to increased hepatocellular cancer risk in Korean, Taiwanese, and Chinese patients[32-34]. A meta-analysis of seven studies by Zhang et al[35] indicated that there is an association between the gene polymorphism IL-10 -1082GA and persistent HBV infection susceptibility. Moreover, this meta-analysis also showed that the gene polymorphism IL-10 -592CA and the clearance of HBV are associated[35]. The carriers of the -592A allele in the IL-10 promoter region are proposed to have a higher risk of persistent HBV infection[36]. However, according to some data in the literature, there is no association between IL-10 gene polymorphisms and chronic HBV infection[37].
IL-10RB is a subunit of receptor complexes for IFN-λ and IL-22, which have antiviral- and hepatocyte-protective activity, respectively. Polymorphism of IL-10RB codon 47 is related to chronic HBV infection in the Korean population[38].
IL-18
IL-18 is a potent proinflammatory cytokine and an immune activator. It is mainly produced in active macrophages and increases induction of IFN-γ and TNF-α, and cytotoxicity of natural killer cells[39].
IL-18 can promote hepatitis B virus clearance. Three polymorphic sites in the IL-18 gene at alleles -148C, +8925G, and +13925C are associated with HBV clearance in the Korean population[40]. A possible positive relationship between serum IL-18 level and disease severity of HBV infection has been indicated in clinical studies[41]. Three SNPs are defined in the promoter region of the IL-18 gene that can affect IL-18 production and in return IFN-γ expression[42]. In a study of a Chinese population, the polymorphism at -137 with C allele was associated with protection against HBV infection[43]. Moreover, AA genotype at -607 position causes an inhibition of HBV DNA replication[43]. Migita et al[44] studied 204 chronically HBV-infected patients; of these, 43 were inactive HBV carriers and 161 had chronic progressive liver disease including cirrhosis. The authors found that the AA genotype of IL-18 gene-promoter polymorphisms at position -607 and C allele at position -137 are significantly higher in inactive HBV carriers than in those with chronic progressive liver disease, suggesting that the polymorphisms of the IL-18 promoter regions (-607 and -137) can be associated with different outcomes of HBV infection[44].
IL-28B
IL-28B, which is also known as IFN-λ-3, is encoded by the IL-28B gene. IL-28B inhibits HBV replication in hepatocyte cell lines and has been considered as a potential new treatment for viral hepatitis[45]. The genetic polymorphisms near the IL-28B gene are strongly associated with sustained viral response and spontaneous viral clearance in patients with chronic HBV infection. Thus, genetic variation of IL-28B may prevent progression of HBV infection by reducing viral load and liver inflammation[46].
However, some conflicting results have been reported so far. IFN-λ-3 (IL-28B) polymorphism is a reliable predictor of IFN therapy outcome in patients with chronic HBV infection[47]. Moreover, it is a protective factor for HBV infection recurrence and hepatic dysfunction after liver transplantation[48,49]. However, IFN-λ-3 genotype was reported to have no role in the development of chronic HBV infection among HIV-infected patients[50]. Moreover, a study comparing patients with persistent infection with individuals recovered from HBV infection found that IL-28B polymorphism has no association with clearance of HBV and does not influence the outcomes of HBV infection[51].
IFN-γ
IFN-γ has a regulatory role in cellular immunity and functions of cytotoxic T lymphocytes. Antiviral, antiproliferative, immunoregulatory, and proinflammatory actions of IFN-γ play a key role in host defense mechanisms. IFN-γ is secreted from T cells and natural killer cells and regulates T cell response, activating monocytes and macrophages, which then produce an antiviral response by releasing free radicals and proinflammatory cytokines such as TNF-α[52]. The core response element of HBV is sensitive to TNF-α, IFN-γ and IFN-α. Increased levels of TNF-α and IFN-γ intensify the antiviral activity of T lymphocytes[53]. IFN-γ gene polymorphism at position +879 causing low IFN-γ level is reported to be higher in patients with chronic HBV infection compared to a control group[25]. Additionally, a negative correlation between necroinflammatory/fibrosis scores and genetic production of IFN-γ and TGF-β1 was reported in HBV-infected patients[25]. A recent study from Turkey revealed that IFN-γ gene polymorphism at position +874AA is correlated with viral load and chronic HBV infection[54].
Conde et al[55] showed in a recent study performed on 153 patients that higher serum levels of IFN-γ and TGF-β1 are associated with chronic HBV infection, and serum level of IL-10 is lower in patients with active disease[55]. Furthermore, the authors reported that the presence of allele A of the TNF-α -308 polymorphism is a risk factor for progressive disease.
TNF-α
TNF-α is a key cytokine that determines host immune response to HBV and viral clearance. Therefore, TNF-α gene polymorphism can have a role in the course of HBV infection. TNF-α level and TNF-α receptor expression are increased in HBV-infected patients[56,57]. The TNF-α gene is localized at MHC HLA region III and two polymorphisms at -308 G/A and -238 G/A positions of the promoter region may affect TNF-α expression[58,59]. Polymorphisms at these regions may cause an elevation of TNF-α transcriptional activity and increase TNF-α serum level[58]. In HBV-infected German patients, the promoter variant at -238A location is significantly correlated with chronicity of HBV infection[60]. Similarly, a study on Chinese patients showed that polymorphisms at the promoter region at -238GA and -857CC locations are associated with persistence of HBV infection[37,61,62]. Although TNF-α polymorphism is not a determinant of HBV clearance in the Italian population, it is suggested to play a role in the prognosis of patients with chronic HBV infection[63]. However, a study performed on Iranian patients reported that TNF-α polymorphism has no role in HBV pathogenesis[64]. Similarly, in HBV-infected Japanese patients, there is no association between TNF-α polymorphism and progression to hepatocellular carcinoma[65].
A genetic analysis of 956 Chinese Han subjects revealed an association between the polymorphism in the promoter region of TNF-α located at -308A and HBV disease progression[66]. A similar result was reported in a study of 27 Turkish patients[67]. TNF-α polymorphisms at position -857CC and -863AA are also associated with the development of persistent HBV infection in the Chinese Han population[68]. Another study from the South Indian population also showed that TNF-α promoter polymorphisms (at positions -238A, -308A, -857T, -863A and -1031C) are important host genetic factors that may determine the variable outcome of HBV infection[69].
Cytotoxic T lymphocyte-associated antigen 4 (CTLA-4) polymorphism may also affect the host immune response, including production of cytokines. Han et al[70] reported that CTLA4 +49GG genotype is associated with lower TNF-α and IFN-γ levels in patients with chronic HBV infection.
Overall results of a meta-analysis involving 19 studies (5245 chronic HBV infection cases and 3181 controls with G238A genotypes) and 11 studies (3576 cases and 2044 controls with C863A genotypes) suggested that there is no significant association between TNF-α -238 and TNF-α -863 gene promoter polymorphisms and chronic HBV infection[71]. When subgroups were analyzed by ethnicity in this study, no significant association was found in Asian populations, but the TNF-α -238A allele is still a risk for chronic HBV infection in European populations[71]. Moreover, carriers of -863A genotype were reported to have increased levels of TNF-α in the liver in response to HBV infection, and this induces hepatocyte damage that may lead to hepatocellular carcinoma[72]. Kao et al[73] reported that polymorphism at -863A locus of the promoter region of the TNF-α gene is associated with lower TNF-α production and persistence of HBV infection[74]. Furthermore, a meta-analysis including 14 studies (4929 chronic HBV infection cases and 2702 controls with -857 genotype) showed that the TNF-α -857T allele reduces the risk of chronic HBV infection in the Asian population[75]. Similarly, it was proposed that the TNF-α -308A allele is protective against chronic HBV infection in the Mongolian population[76].
A meta-analysis of 12 studies suggested that polymorphisms -863A and -308G in the TNF-α promoter region might be a risk factor for HBV persistence[77]. Since ethnicity plays an important role in HBV infection outcome, conflicting results are reported on the association between TNF-α promoter gene polymorphisms and HBV infection outcome.
TGF-β1
TGF-β1 shows an inhibitory effect in the early stages of tumor development, while it stimulates tumor growth, invasion, and metastasis in advanced stages[78]. TGF-β1 plays a critical role in the pathogenesis of liver fibrosis by stimulating extracellular matrix proteins and inhibiting their destruction[79]. Therefore, mechanisms increasing the level of biologically active TGF-β1 have a potential role in the development of liver fibrosis. A study of Chinese patients revealed that even though there is no association between TGF-β1 -509C polymorphism and cirrhosis, this polymorphism might affect TGF-β1 levels and development of cirrhosis[80]. However, in the very same study, codon 10T polymorphism is related to the development of cirrhosis, but not with progression of disease and plasma TGF-β1 levels[80]. Codon 10T polymorphism in the TGF-β1 gene was also reported to be associated with progression to hepatocellular cancer[81,82] and cirrhosis[83] in patients with chronic HBV infection.
REGULATORY PROTEINS
Vitamin D
The active metabolite of vitamin D, 1,25-dihydroxyvitamin-D, has immunomodulatory action in addition to its regulatory role in calcium metabolism. It activates monocytes, increases cell-regulated immunity, inhibits lymphocyte proliferation, immunoglobulin, and cytokine synthesis, and inhibits type 1 cytokine secreting T helper (Th1) response while activating Th2 response. Additionally, vitamin D plays a role in programmed cell death. Monocytes, macrophages, and active T lymphocytes carry VDR. While the stimulation of VDR on monocytes and macrophages increases production of TNF-α, IL-1, and prostaglandin E2, stimulation of VDR on lymphocytes inhibits T cell proliferation and production of IFN-γ, IL-2 and TNF-β[84]. Four polymorphisms of the VDR gene are associated with various immune diseases[84]. Furthermore, being homozygous for VDR gene polymorphism at codon 352 (genotype tt) is significantly less frequent in patients positive for hepatitis B surface antigen, and it was suggested that this genotype provides resistance to chronicity of HBV infection[85]. VDR a/a allele is also associated with severity of HBV-related liver disease and with higher viral load[86].
CCR5
An efficient immune response against viral hepatitis should promote inflammatory cells to be activated and to migrate to the liver. Chemokines have important functions during this process by means of their chemotactic and immunoregulatory actions. The CCR5 acts as a receptor for chemokines. Among the chemokines, regulated on activation normal T cell expressed and secreted (RANTES; CCL5), macrophage inhibitory protein-1α (MIP-1α; CCL3), and MIP-1β (CCL4) are natural ligands of CCR5. Both these chemokines and CCR5 regulate T cell functions by mediating polarization, activation, and differentiation of Th1 and cytotoxic T cells[87]. Besides, CCR5 has a regulatory function for the immunoregulatory action of vitamin D.
The frequency of heterozygosity of the CCR5-delta 32 gene is higher in chronic hepatitis B patients than in controls, which shows the relation of this polymorphism with susceptibility to HBV-related liver disease[86]. CCR5 59029A and 59029G alleles are associated with increased chronic HBV infection risk and spontaneous HBV clearance, respectively[88]. The frequency of CCR5 Wt/mt allele is higher in chronic HBV patients than in healthy subjects, while CCR5 Wt/Wt allele is more common in patients with severe liver disease than in mild cases[86].
CURRENT INTERESTS AND FUTURE PROSPECTS
Recent gene polymorphism studies have focused on the clinical implication of polymorphism-HBV infection associations such as gene therapy targets[46], prediction of infection risk, disease progression, chronicity, response to treatment[89] or vaccine[90-94], and susceptibility to mother-to-child transmission of HBV[95].
IL-28B genotyping is suggested to predict the response to pegylated interferon[96] and to provide a valuable gene therapy target due to its reducing effect on HBV viral load and hepatic inflammation[46].
Gene polymorphisms of IL-1B, IL-4, IL-4R, IL-13[90,93,94], IL12A and IL12B[92] are suggested to predict the immune response to HBV vaccination.
Since TNF-α and vitamin D pathways are involved in the susceptibility to, and the outcome of, HBV infection acquired early in life, they can be used clinically to determine the susceptibility to mother-to-child transmission of HBV[95].
Although studies on the clinical application of gene polymorphisms of cytokines have been increasing recently, further clinical studies are needed for widespread use of genotyping in the course of HBV infections.
CONCLUSION
Along with the establishment of the key role of endogenous mediators in the response to infection, effects of host-related factors on the course of chronic HBV infection have been investigated from different perspectives. Some of these studies have focused on the effect of diversity in genes encoding endogenous mediators of inflammatory response to HBV infections.
Inflammatory processes are mostly regulated by proinflammatory and antiinflammatory cytokines, and other mediators, which are determinative for the course of disease. Polymorphisms in genes encoding endogenous mediators may be the underlying cause of clinical differences between patients. Results of the studies summarized in this review suggest that cytokine gene polymorphisms affect the level of cytokines during the inflammatory response to HBV, and thus determine the clinical course of chronic HBV infection. Genomic information with regard to cytokines and other mediators can be used for identifying individuals who are at high risk of developing chronic hepatitis and hepatocellular carcinoma, and for the planning of preventive measures and treatment approaches.
As recent studies have indicated, gene polymorphisms of inflammatory mediators may be important in determining the response to both treatment and vaccine. For example, serum levels of TNF-α in patients who respond to treatment with interferon were found to be higher than those in nonresponders[97]. Granulocyte-macrophage colony-stimulating factor has been reported to increase the response rate to recombinant hepatitis B vaccine[98]. Additionally, genetic factors may play a role in the development of adverse reactions secondary to the vaccine such as arthritis, multiple sclerosis, and other autoimmune diseases. However, further studies are still needed to investigate in detail the effects of genetic polymorphisms and their clinical implications for the response to treatment and vaccine, and development of adverse events.
In conclusion, there is currently a vast amount of evidence on the association between polymorphisms of genes encoding cytokines/regulatory molecules and the clinical course of chronic HBV infection. Conflicting results on the role of specific polymorphisms are probably due to various ethnic groups studied. In the future, determining genetic polymorphisms of mediators that have a role in both the natural course of the infection and the response to treatment and vaccination will contribute significantly to the prevention and treatment of HBV infections by eliminating possible risk factors prior to disease and by development of new treatment approaches.
Footnotes
P- Reviewers: Cunha C, Koubaa M S- Editor: Wen LL L- Editor: Logan S E- Editor: Wu HL
References
- 1.Chisari FV, Ferrari C. Hepatitis B virus immunopathogenesis. Annu Rev Immunol. 1995;13:29–60. doi: 10.1146/annurev.iy.13.040195.000333. [DOI] [PubMed] [Google Scholar]
- 2.Lee WM. Hepatitis B virus infection. N Engl J Med. 1997;337:1733–1745. doi: 10.1056/NEJM199712113372406. [DOI] [PubMed] [Google Scholar]
- 3.World Health Organization. Hepatitis B. Fact Sheet No 204. July 2013. Available from: http://www.who.int/mediacentre/factsheets/fs204/en/ Accessed on September 23, 2013.
- 4.World Health Organization. Global Alert and Response (GAR). Hepatitis B. Available from: http://www.who.int/csr/disease/hepatitis/whocdscsrlyo20022/en/index1.html Accessed on September 23, 2013.
- 5.Wang FS. Current status and prospects of studies on human genetic alleles associated with hepatitis B virus infection. World J Gastroenterol. 2003;9:641–644. doi: 10.3748/wjg.v9.i4.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Park W, Keeffe EB. Diagnosis and treatment of chronic hepatitis B. Minerva Gastroenterol Dietol. 2004;50:289–303. [PubMed] [Google Scholar]
- 7.Thursz M. Genetic susceptibility in infectious diseases. Biotechnol Genet Eng Rev. 2000;17:253–264. [PubMed] [Google Scholar]
- 8.Thursz M, Yee L, Khakoo S. Understanding the host genetics of chronic hepatitis B and C. Semin Liver Dis. 2011;31:115–127. doi: 10.1055/s-0031-1276642. [DOI] [PubMed] [Google Scholar]
- 9.Grünhage F, Nattermann J. Viral hepatitis: human genes that limit infection. Best Pract Res Clin Gastroenterol. 2010;24:709–723. doi: 10.1016/j.bpg.2010.07.009. [DOI] [PubMed] [Google Scholar]
- 10.McNicholl JM, Downer MV, Udhayakumar V, Alper CA, Swerdlow DL. Host-pathogen interactions in emerging and re-emerging infectious diseases: a genomic perspective of tuberculosis, malaria, human immunodeficiency virus infection, hepatitis B, and cholera. Annu Rev Public Health. 2000;21:15–46. doi: 10.1146/annurev.publhealth.21.1.15. [DOI] [PubMed] [Google Scholar]
- 11.Thursz MR. Host genetic factors influencing the outcome of hepatitis. J Viral Hepat. 1997;4:215–220. doi: 10.1046/j.1365-2893.1997.00052.x. [DOI] [PubMed] [Google Scholar]
- 12.Mackay IR. Genetic susceptibility to chronic hepatitis B virus infection. J Gastroenterol Hepatol. 2006;21:1087–1088. doi: 10.1111/j.1440-1746.2006.04430.x. [DOI] [PubMed] [Google Scholar]
- 13.Lin TM, Chen CJ, Wu MM, Yang CS, Chen JS, Lin CC, Kwang TY, Hsu ST, Lin SY, Hsu LC. Hepatitis B virus markers in Chinese twins. Anticancer Res. 1989;9:737–741. [PubMed] [Google Scholar]
- 14.de Andrade DR, de Andrade DR. The influence of the human genome on chronic viral hepatitis outcome. Rev Inst Med Trop Sao Paulo. 2004;46:119–126. [PubMed] [Google Scholar]
- 15.Dinarello CA. Biologic basis for interleukin-1 in disease. Blood. 1996;87:2095–2147. [PubMed] [Google Scholar]
- 16.Nicklin MJ, Weith A, Duff GW. A physical map of the region encompassing the human interleukin-1 alpha, interleukin-1 beta, and interleukin-1 receptor antagonist genes. Genomics. 1994;19:382–384. doi: 10.1006/geno.1994.1076. [DOI] [PubMed] [Google Scholar]
- 17.Hwang IR, Kodama T, Kikuchi S, Sakai K, Peterson LE, Graham DY, Yamaoka Y. Effect of interleukin 1 polymorphisms on gastric mucosal interleukin 1beta production in Helicobacter pylori infection. Gastroenterology. 2002;123:1793–1803. doi: 10.1053/gast.2002.37043. [DOI] [PubMed] [Google Scholar]
- 18.Zhang PA, Li Y, Xu P, Wu JM. Polymorphisms of interleukin-1B and interleukin-1 receptor antagonist genes in patients with chronic hepatitis B. World J Gastroenterol. 2004;10:1826–1829. doi: 10.3748/wjg.v10.i12.1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rahman MA, Dhar DK, Yamaguchi E, Maruyama S, Sato T, Hayashi H, Ono T, Yamanoi A, Kohno H, Nagasue N. Coexpression of inducible nitric oxide synthase and COX-2 in hepatocellular carcinoma and surrounding liver: possible involvement of COX-2 in the angiogenesis of hepatitis C virus-positive cases. Clin Cancer Res. 2001;7:1325–1332. [PubMed] [Google Scholar]
- 20.Hirankarn N, Kimkong I, Kummee P, Tangkijvanich P, Poovorawan Y. Interleukin-1beta gene polymorphism associated with hepatocellular carcinoma in hepatitis B virus infection. World J Gastroenterol. 2006;12:776–779. doi: 10.3748/wjg.v12.i5.776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim SS, Cheong JY, Lee D, Lee SK, Kim MH, Kwack K, Yang SJ, Lee HY, Cho SW. Interleukin-1ß and interleukin-1 receptor accessory protein gene polymorphisms are associated with persistent hepatitis B virus infection. Hepatogastroenterology. 2012;59:190–197. doi: 10.5754/hge10375. [DOI] [PubMed] [Google Scholar]
- 22.Fontanini E, Cussigh A, Fabris C, Falleti E, Toniutto P, Bitetto D, Cmet S, Fumolo E, Fornasiere E, Bignulin S, et al. Gender-related distribution of the interleukin-1 beta and interleukin-1 receptor antagonist gene polymorphisms in patients with end-stage liver disease. Inflammation. 2010;33:251–258. doi: 10.1007/s10753-009-9179-2. [DOI] [PubMed] [Google Scholar]
- 23.Song W, Zhang F, Li Z. A quantitative analysis of IL-6 mRNA expression of peripheral blood monocyte cell in patients with chronic hepatitis B. Zhonghua Ganzangbing Zazhi. 2000;8:346–347. [PubMed] [Google Scholar]
- 24.Park BL, Lee HS, Kim YJ, Kim JY, Jung JH, Kim LH, Shin HD. Association between interleukin 6 promoter variants and chronic hepatitis B progression. Exp Mol Med. 2003;35:76–82. doi: 10.1038/emm.2003.11. [DOI] [PubMed] [Google Scholar]
- 25.Ben-Ari Z, Mor E, Papo O, Kfir B, Sulkes J, Tambur AR, Tur-Kaspa R, Klein T. Cytokine gene polymorphisms in patients infected with hepatitis B virus. Am J Gastroenterol. 2003;98:144–150. doi: 10.1111/j.1572-0241.2003.07179.x. [DOI] [PubMed] [Google Scholar]
- 26.Giannitrapani L, Soresi M, Balasus D, Licata A, Montalto G. Genetic association of interleukin-6 polymorphism (-174 G/C) with chronic liver diseases and hepatocellular carcinoma. World J Gastroenterol. 2013;19:2449–2455. doi: 10.3748/wjg.v19.i16.2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wei YS, Lan Y, Tang RG, Xu QQ, Huang Y, Nong HB, Huang WT. Single nucleotide polymorphism and haplotype association of the interleukin-8 gene with nasopharyngeal carcinoma. Clin Immunol. 2007;125:309–317. doi: 10.1016/j.clim.2007.07.010. [DOI] [PubMed] [Google Scholar]
- 28.Qin X, Deng Y, Liao XC, Mo CJ, Li X, Wu HL, He YN, Huang XM, Peng T, Chen ZP, et al. The IL-8 gene polymorphisms and the risk of the hepatitis B virus/infected patients. DNA Cell Biol. 2012;31:1125–1130. doi: 10.1089/dna.2011.1438. [DOI] [PubMed] [Google Scholar]
- 29.Levings MK, Sangregorio R, Galbiati F, Squadrone S, de Waal Malefyt R, Roncarolo MG. IFN-alpha and IL-10 induce the differentiation of human type 1 T regulatory cells. J Immunol. 2001;166:5530–5539. doi: 10.4049/jimmunol.166.9.5530. [DOI] [PubMed] [Google Scholar]
- 30.Zhu QR, Ge YL, Gu SQ, Yu H, Wang JS, Gu XH, Fei LE, Dong ZQ. Relationship between cytokines gene polymorphism and susceptibility to hepatitis B virus intrauterine infection. Chin Med J (Engl) 2005;118:1604–1609. [PubMed] [Google Scholar]
- 31.Wu JF, Ni YH, Lin YT, Lee TJ, Hsu SH, Chen HL, Tsuei DJ, Hsu HY, Chang MH. Human interleukin-10 genotypes are associated with different precore/core gene mutation patterns in children with chronic hepatitis B virus infection. J Pediatr. 2011;158:808–813. doi: 10.1016/j.jpeds.2010.11.015. [DOI] [PubMed] [Google Scholar]
- 32.Shin HD, Park BL, Kim LH, Jung JH, Kim JY, Yoon JH, Kim YJ, Lee HS. Interleukin 10 haplotype associated with increased risk of hepatocellular carcinoma. Hum Mol Genet. 2003;12:901–906. doi: 10.1093/hmg/ddg104. [DOI] [PubMed] [Google Scholar]
- 33.Tseng LH, Lin MT, Shau WY, Lin WC, Chang FY, Chien KL, Hansen JA, Chen DS, Chen PJ. Correlation of interleukin-10 gene haplotype with hepatocellular carcinoma in Taiwan. Tissue Antigens. 2006;67:127–133. doi: 10.1111/j.1399-0039.2006.00536.x. [DOI] [PubMed] [Google Scholar]
- 34.Wang J, Ni H, Chen L, Song WQ. Interleukin-10 promoter polymorphisms in patients with hepatitis B virus infection or hepatocellular carcinoma in Chinese Han ethnic population. Hepatobiliary Pancreat Dis Int. 2006;5:60–64. [PubMed] [Google Scholar]
- 35.Zhang TC, Pan FM, Zhang LZ, Gao YF, Zhang ZH, Gao J, Ge R, Mei Y, Shen BB, Duan ZH, et al. A meta-analysis of the relation of polymorphism at sites -1082 and -592 of the IL-10 gene promoter with susceptibility and clearance to persistent hepatitis B virus infection in the Chinese population. Infection. 2011;39:21–27. doi: 10.1007/s15010-010-0075-3. [DOI] [PubMed] [Google Scholar]
- 36.Cheong JY, Cho SW, Hwang IL, Yoon SK, Lee JH, Park CS, Lee JE, Hahm KB, Kim JH. Association between chronic hepatitis B virus infection and interleukin-10, tumor necrosis factor-alpha gene promoter polymorphisms. J Gastroenterol Hepatol. 2006;21:1163–1169. doi: 10.1111/j.1440-1746.2006.04304.x. [DOI] [PubMed] [Google Scholar]
- 37.Li C, Zhi-Xin C, Li-Juan Z, Chen P, Xiao-Zhong W. The association between cytokine gene polymorphisms and the outcomes of chronic HBV infection. Hepatol Res. 2006;36:158–166. doi: 10.1016/j.hepres.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 38.Cho O, Cheong JY, Jun KJ, Kim SS, Chwae YJ, Kim K, Park S, Cho SW. Relevance of interleukin-10RB to chronic hepatitis B virus infection and biological activities of interferon-λ and interleukin-22. Hepatol Int. 2013;7:111–118. doi: 10.1007/s12072-012-9361-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoshino T, Wiltrout RH, Young HA. IL-18 is a potent coinducer of IL-13 in NK and T cells: a new potential role for IL-18 in modulating the immune response. J Immunol. 1999;162:5070–5077. [PubMed] [Google Scholar]
- 40.Cheong JY, Cho SW, Oh B, Kimm K, Lee KM, Shin SJ, Lee JA, Park BL, Cheong HS, Shin HD, et al. Association of interleukin-18 gene polymorphisms with hepatitis B virus clearance. Dig Dis Sci. 2010;55:1113–1119. doi: 10.1007/s10620-009-0819-z. [DOI] [PubMed] [Google Scholar]
- 41.Sun Y, Chen HY, Wang F, Zhang X, Jiang HQ, Shao FJ, Zhu SH. [Effect of IL-18 on peripheral blood monocytes from chronic hepatitis B patients] Zhonghua Ganzangbing Zazhi. 2003;11:470–473. [PubMed] [Google Scholar]
- 42.Giedraitis V, He B, Huang WX, Hillert J. Cloning and mutation analysis of the human IL-18 promoter: a possible role of polymorphisms in expression regulation. J Neuroimmunol. 2001;112:146–152. doi: 10.1016/s0165-5728(00)00407-0. [DOI] [PubMed] [Google Scholar]
- 43.Zhang PA, Wu JM, Li Y, Yang XS. Association of polymorphisms of interleukin-18 gene promoter region with chronic hepatitis B in Chinese Han population. World J Gastroenterol. 2005;11:1594–1598. doi: 10.3748/wjg.v11.i11.1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Migita K, Sawakami-Kobayashi K, Maeda Y, Nakao K, Kondoh S, Sugiura M, Kawasumi R, Segawa O, Tajima H, Machida M, et al. Interleukin-18 promoter polymorphisms and the disease progression of Hepatitis B virus-related liver disease. Transl Res. 2009;153:91–96. doi: 10.1016/j.trsl.2008.11.008. [DOI] [PubMed] [Google Scholar]
- 45.Robek MD, Boyd BS, Chisari FV. Lambda interferon inhibits hepatitis B and C virus replication. J Virol. 2005;79:3851–3854. doi: 10.1128/JVI.79.6.3851-3854.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li W, Jiang Y, Jin Q, Shi X, Jin J, Gao Y, Pan Y, Zhang H, Jiang J, Niu J. Expression and gene polymorphisms of interleukin 28B and hepatitis B virus infection in a Chinese Han population. Liver Int. 2011;31:1118–1126. doi: 10.1111/j.1478-3231.2011.02507.x. [DOI] [PubMed] [Google Scholar]
- 47.Lampertico P, Viganò M, Cheroni C, Facchetti F, Invernizzi F, Valveri V, Soffredini R, Abrignani S, De Francesco R, Colombo M. IL28B polymorphisms predict interferon-related hepatitis B surface antigen seroclearance in genotype D hepatitis B e antigen-negative patients with chronic hepatitis B. Hepatology. 2013;57:890–896. doi: 10.1002/hep.25749. [DOI] [PubMed] [Google Scholar]
- 48.Li Y, Shi Y, Chen J, Cai B, Ying B, Wang L. Association of polymorphisms in interleukin-18 and interleukin-28B with hepatitis B recurrence after liver transplantation in Chinese Han population. Int J Immunogenet. 2012;39:346–352. doi: 10.1111/j.1744-313X.2012.01097.x. [DOI] [PubMed] [Google Scholar]
- 49.Chen J, Li Y, Wang L, Fu Y, Liao Y, Zhang J. Association of three SNPs in interleukin-28B with graft hepatic dysfunction after liver transplantation in Chinese Han population. Gene. 2012;508:121–124. doi: 10.1016/j.gene.2012.07.065. [DOI] [PubMed] [Google Scholar]
- 50.Martín-Carbonero L, Rallón NI, Benito JM, Poveda E, González-Lahoz J, Soriano V. Short communication: Does interleukin-28B single nucleotide polymorphisms influence the natural history of hepatitis B? AIDS Res Hum Retroviruses. 2012;28:1262–1264. doi: 10.1089/AID.2011.0365. [DOI] [PubMed] [Google Scholar]
- 51.Peng LJ, Guo JS, Zhang Z, Shi H, Wang J, Wang JY. IL28B rs12979860 polymorphism does not influence outcomes of hepatitis B virus infection. Tissue Antigens. 2012;79:302–305. doi: 10.1111/j.1399-0039.2011.01835.x. [DOI] [PubMed] [Google Scholar]
- 52.Biron CA, Nguyen KB, Pien GC, Cousens LP, Salazar-Mather TP. Natural killer cells in antiviral defense: function and regulation by innate cytokines. Annu Rev Immunol. 1999;17:189–220. doi: 10.1146/annurev.immunol.17.1.189. [DOI] [PubMed] [Google Scholar]
- 53.Romero R, Lavine JE. Cytokine inhibition of the hepatitis B virus core promoter. Hepatology. 1996;23:17–23. doi: 10.1002/hep.510230103. [DOI] [PubMed] [Google Scholar]
- 54.Korachi M, Ceran N, Adaleti R, Nigdelioglu A, Sökmen M. An association study of functional polymorphic genes IRF-1, IFNGR-1, and IFN-γ with disease progression, aspartate aminotransferase, alanine aminotransferase, and viral load in chronic hepatitis B and C. Int J Infect Dis. 2013;17:e44–e49. doi: 10.1016/j.ijid.2012.08.004. [DOI] [PubMed] [Google Scholar]
- 55.Conde SR, Feitosa RN, Freitas FB, Hermes RB, Demachki S, Araújo MT, Soares MC, Ishak R, Vallinoto AC. Association of cytokine gene polymorphisms and serum concentrations with the outcome of chronic hepatitis B. Cytokine. 2013;61:940–944. doi: 10.1016/j.cyto.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 56.Sheron N, Lau J, Daniels H, Goka J, Eddleston A, Alexander GJ, Williams R. Increased production of tumour necrosis factor alpha in chronic hepatitis B virus infection. J Hepatol. 1991;12:241–245. doi: 10.1016/0168-8278(91)90945-8. [DOI] [PubMed] [Google Scholar]
- 57.Zhang G, Li Z, Han Q, Li N, Zhu Q, Li F, Lv Y, Chen J, Lou S, Liu Z. Altered TNF-α and IFN-γ levels associated with PD1 but not TNFA polymorphisms in patients with chronic HBV infection. Infect Genet Evol. 2011;11:1624–1630. doi: 10.1016/j.meegid.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 58.Abraham LJ, Kroeger KM. Impact of the -308 TNF promoter polymorphism on the transcriptional regulation of the TNF gene: relevance to disease. J Leukoc Biol. 1999;66:562–566. doi: 10.1002/jlb.66.4.562. [DOI] [PubMed] [Google Scholar]
- 59.Wilson AG, de Vries N, Pociot F, di Giovine FS, van der Putte LB, Duff GW. An allelic polymorphism within the human tumor necrosis factor alpha promoter region is strongly associated with HLA A1, B8, and DR3 alleles. J Exp Med. 1993;177:557–560. doi: 10.1084/jem.177.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Höhler T, Kruger A, Gerken G, Schneider PM, Meyer zum Büschenefelde KH, Rittner C. A tumor necrosis factor-alpha (TNF-alpha) promoter polymorphism is associated with chronic hepatitis B infection. Clin Exp Immunol. 1998;111:579–582. doi: 10.1046/j.1365-2249.1998.00534.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li HQ, Li Z, Liu Y, Li JH, Dong JQ, Gao JR, Gou CY, Li H. Association of polymorphism of tumor necrosis factor-alpha gene promoter region with outcome of hepatitis B virus infection. World J Gastroenterol. 2005;11:5213–5217. doi: 10.3748/wjg.v11.i33.5213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xu XW, Lu MH, Tan DM. Association between tumour necrosis factor gene polymorphisms and the clinical types of patients with chronic hepatitis B virus infection. Clin Microbiol Infect. 2005;11:52–56. doi: 10.1111/j.1469-0691.2004.01029.x. [DOI] [PubMed] [Google Scholar]
- 63.Niro GA, Fontana R, Gioffreda D, Valvano MR, Lacobellis A, Facciorusso D, Andriulli A. Tumor necrosis factor gene polymorphisms and clearance or progression of hepatitis B virus infection. Liver Int. 2005;25:1175–1181. doi: 10.1111/j.1478-3231.2005.01166.x. [DOI] [PubMed] [Google Scholar]
- 64.Somi MH, Najafi L, Noori BN, Alizadeh AH, Aghah MR, Shavakhi A, Ehsani MJ, Aghazadeh R, Masoodi M, Amini S, et al. Tumor necrosis factor-alpha gene promoter polymorphism in Iranian patients with chronic hepatitis B. Indian J Gastroenterol. 2006;25:14–15. [PubMed] [Google Scholar]
- 65.Migita K, Miyazoe S, Maeda Y, Daikoku M, Abiru S, Ueki T, Yano K, Nagaoka S, Matsumoto T, Nakao K, et al. Cytokine gene polymorphisms in Japanese patients with hepatitis B virus infection--association between TGF-beta1 polymorphisms and hepatocellular carcinoma. J Hepatol. 2005;42:505–510. doi: 10.1016/j.jhep.2004.11.026. [DOI] [PubMed] [Google Scholar]
- 66.Wang B, Wang J, Zheng Y, Zhou S, Zheng J, Wang F, Ma X, Zeng Z. A study of TNF-alpha-238 and -308 polymorphisms with different outcomes of persistent hepatitis B virus infection in China. Pathology. 2010;42:674–680. doi: 10.3109/00313025.2010.523696. [DOI] [PubMed] [Google Scholar]
- 67.Basturk B, Karasu Z, Kilic M, Ulukaya S, Boyacioglu S, Oral B. Association of TNF-alpha -308 polymorphism with the outcome of hepatitis B virus infection in Turkey. Infect Genet Evol. 2008;8:20–25. doi: 10.1016/j.meegid.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 68.Qiu B, Wang X, Zhang P, Shi C, Zhang J, Qiu W, Wang W, Li D. Association of TNF-α promoter polymorphisms with the outcome of persistent HBV infection in a northeast Chinese Han population. Acta Biochim Biophys Sin (Shanghai) 2012;44:712–718. doi: 10.1093/abbs/gms046. [DOI] [PubMed] [Google Scholar]
- 69.Fletcher GJ, Samuel P, Christdas J, Gnanamony M, Ismail AM, Anantharam R, Eapen CE, Chacko MP, Daniel D, Kannangai R, et al. Association of HLA and TNF polymorphisms with the outcome of HBV infection in the South Indian population. Genes Immun. 2011;12:552–558. doi: 10.1038/gene.2011.32. [DOI] [PubMed] [Google Scholar]
- 70.Han Q, Duan S, Zhang G, Li Z, Li N, Zhu Q, Lv Y, Chen J, Liu Z. Associations between cytotoxic T lymphocyte-associated antigen-4 polymorphisms and serum tumor necrosis factor-α and interferon-γ levels in patients with chronic hepatitis B virus infection. Inflamm Res. 2011;60:1071–1078. doi: 10.1007/s00011-011-0368-8. [DOI] [PubMed] [Google Scholar]
- 71.Zheng MH, Xiao DD, Lin XF, Wu SJ, Peng MM, Yu XY, Liu WY, Li LF, Shi KQ, Fan YC, et al. The tumour necrosis factor-α-238A allele increases the risk of chronic HBV infection in European populations. J Viral Hepat. 2012;19:e11–e17. doi: 10.1111/j.1365-2893.2011.01491.x. [DOI] [PubMed] [Google Scholar]
- 72.Kummee P, Tangkijvanich P, Poovorawan Y, Hirankarn N. Association of HLA-DRB1*13 and TNF-alpha gene polymorphisms with clearance of chronic hepatitis B infection and risk of hepatocellular carcinoma in Thai population. J Viral Hepat. 2007;14:841–848. doi: 10.1111/j.1365-2893.2007.00880.x. [DOI] [PubMed] [Google Scholar]
- 73.Kao PC, Wu JF, Ni YH, Lin YT, Chen HL, Hsu SH, Hsu HY, Chang MH. Tumour necrosis factor-α promoter region polymorphisms affect the course of spontaneous HBsAg clearance. Liver Int. 2010;30:1448–1453. doi: 10.1111/j.1478-3231.2010.02340.x. [DOI] [PubMed] [Google Scholar]
- 74.Filik L. Tumour necrosis factor-α promoter region polymorphisms and spontaneous HBsAg clearance. Liver Int. 2011;31:1062; authors reply 1062. doi: 10.1111/j.1478-3231.2011.02456.x. [DOI] [PubMed] [Google Scholar]
- 75.Shi KQ, Cai XH, Xiao DD, Wu SJ, Peng MM, Lin XF, Liu WY, Fan YC, Chen YP, Zheng MH. Tumour necrosis factor-α-857T allele reduces the risk of hepatitis B virus infection in an Asian population. J Viral Hepat. 2012;19:e66–e72. doi: 10.1111/j.1365-2893.2011.01540.x. [DOI] [PubMed] [Google Scholar]
- 76.Zheng MH, Qiu LX, Xin YN, Pan HF, Shi KQ, Chen YP. Tumor necrosis factor-alpha-308A allele may have a protective effect for chronic hepatitis B virus infection in Mongoloid populations. Int J Infect Dis. 2010;14:e580–e585. doi: 10.1016/j.ijid.2009.08.010. [DOI] [PubMed] [Google Scholar]
- 77.Xia Q, Zhou L, Liu D, Chen Z, Chen F. Relationship between TNF-α lt; alpha& gt; gene promoter polymorphisms and outcomes of hepatitis B virus infections: a meta-analysis. PLoS One. 2011;6:e19606. doi: 10.1371/journal.pone.0019606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cui W, Fowlis DJ, Bryson S, Duffie E, Ireland H, Balmain A, Akhurst RJ. TGFbeta1 inhibits the formation of benign skin tumors, but enhances progression to invasive spindle carcinomas in transgenic mice. Cell. 1996;86:531–542. doi: 10.1016/s0092-8674(00)80127-0. [DOI] [PubMed] [Google Scholar]
- 79.Gressner AM, Weiskirchen R, Breitkopf K, Dooley S. Roles of TGF-beta in hepatic fibrosis. Front Biosci. 2002;7:d793–d807. doi: 10.2741/A812. [DOI] [PubMed] [Google Scholar]
- 80.Yang ZX, Wang H, Gao CF, Xu LL, Zhao WJ. [Effect of polymorphism of transforming growth factor beta1 gene on HBV-induced liver cirrhosis] Zhonghua Yixue Zazhi. 2005;85:1021–1026. [PubMed] [Google Scholar]
- 81.Gupta V, Arora R, Saha A, Dhir A, Kar P, Bamezai R. Novel variations in the signal peptide region of transforming growth factor beta1 gene in patients with hepatitis: a brief report from India. Int J Immunogenet. 2005;32:79–82. doi: 10.1111/j.1744-313X.2005.00499.x. [DOI] [PubMed] [Google Scholar]
- 82.Kim YJ, Lee HS, Im JP, Min BH, Kim HD, Jeong JB, Yoon JH, Kim CY, Kim MS, Kim JY, et al. Association of transforming growth factor-beta1 gene polymorphisms with a hepatocellular carcinoma risk in patients with chronic hepatitis B virus infection. Exp Mol Med. 2003;35:196–202. doi: 10.1038/emm.2003.27. [DOI] [PubMed] [Google Scholar]
- 83.Yu SK, Kwon OS, Jung HS, Bae KS, Kwon KA, Kim YK, Kim YS, Kim JH. Influence of transforming growth factor-beta1 gene polymorphism at codon 10 on the development of cirrhosis in chronic hepatitis B virus carriers. J Korean Med Sci. 2010;25:564–569. doi: 10.3346/jkms.2010.25.4.564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Long KZ, Santos JI. Vitamins and the regulation of the immune response. Pediatr Infect Dis J. 1999;18:283–290. doi: 10.1097/00006454-199903000-00018. [DOI] [PubMed] [Google Scholar]
- 85.Bellamy R, Ruwende C, Corrah T, McAdam KP, Thursz M, Whittle HC, Hill AV. Tuberculosis and chronic hepatitis B virus infection in Africans and variation in the vitamin D receptor gene. J Infect Dis. 1999;179:721–724. doi: 10.1086/314614. [DOI] [PubMed] [Google Scholar]
- 86.Suneetha PV, Sarin SK, Goyal A, Kumar GT, Shukla DK, Hissar S. Association between vitamin D receptor, CCR5, TNF-alpha and TNF-beta gene polymorphisms and HBV infection and severity of liver disease. J Hepatol. 2006;44:856–863. doi: 10.1016/j.jhep.2006.01.028. [DOI] [PubMed] [Google Scholar]
- 87.Apolinario A, Majano PL, Alvarez-Pérez E, Saez A, Lozano C, Vargas J, García-Monzón C. Increased expression of T cell chemokines and their receptors in chronic hepatitis C: relationship with the histological activity of liver disease. Am J Gastroenterol. 2002;97:2861–2870. doi: 10.1111/j.1572-0241.2002.07054.x. [DOI] [PubMed] [Google Scholar]
- 88.Chang HY, Ahn SH, Kim DY, Shin JS, Kim YS, Hong SP, Chung HJ, Kim SO, Yoo WD, Han KH. [Association between CCR5 promoter polymorphisms and hepatitis B virus infection] Korean J Hepatol. 2005;11:116–124. [PubMed] [Google Scholar]
- 89.Gong QM, Kong XF, Yang ZT, Xu J, Wang L, Li XH, Jin GD, Gao J, Zhang DH, Jiang JH, et al. Association study of IFNAR2 and IL10RB genes with the susceptibility and interferon response in HBV infection. J Viral Hepat. 2009;16:674–680. doi: 10.1111/j.1365-2893.2009.01130.x. [DOI] [PubMed] [Google Scholar]
- 90.Lin YJ, Lan YC, Huang YC, Lin TH, Huang SM, Lai CC, Liu CS, Lin CW, Chen SY, Tsai FJ. Effects of cytokine and cytokine receptor gene variation on high anti-HB titers: following up on Taiwan’s neonatal hepatitis B immunization program. Clin Chim Acta. 2012;413:1194–1198. doi: 10.1016/j.cca.2012.03.004. [DOI] [PubMed] [Google Scholar]
- 91.Macedo LC, Isolani AP, Visentainer JE, Moliterno RA. Association of cytokine genetic polymorphisms with the humoral immune response to recombinant vaccine against HBV in infants. J Med Virol. 2010;82:929–933. doi: 10.1002/jmv.21762. [DOI] [PubMed] [Google Scholar]
- 92.Pan L, Zhang W, Liang Z, Wu X, Zhu X, Li J, Li T, Wang L, Li H, Liu Y. Association between polymorphisms of the cytokine and cytokine receptor genes and immune response to hepatitis B vaccination in a Chinese Han population. J Med Virol. 2012;84:26–33. doi: 10.1002/jmv.22251. [DOI] [PubMed] [Google Scholar]
- 93.Chen J, Liang Z, Lu F, Fang X, Liu S, Zeng Y, Zhu F, Chen X, Shen T, Li J, et al. Toll-like receptors and cytokines/cytokine receptors polymorphisms associate with non-response to hepatitis B vaccine. Vaccine. 2011;29:706–711. doi: 10.1016/j.vaccine.2010.11.023. [DOI] [PubMed] [Google Scholar]
- 94.Wang Y, Xu P, Zhu D, Zhang S, Bi Y, Hu Y, Zhou YH. Association of polymorphisms of cytokine and TLR-2 genes with long-term immunity to hepatitis B in children vaccinated early in life. Vaccine. 2012;30:5708–5713. doi: 10.1016/j.vaccine.2012.07.010. [DOI] [PubMed] [Google Scholar]
- 95.Chatzidaki V, Choumerianou D, Dimitriou H, Kouroumalis E, Galanakis E. Genetic variants associated with susceptibility to mother-to-child transmission of hepatitis B virus. Eur J Gastroenterol Hepatol. 2012;24:1185–1190. doi: 10.1097/MEG.0b013e328356440f. [DOI] [PubMed] [Google Scholar]
- 96.Sonneveld MJ, Wong VW, Woltman AM, Wong GL, Cakaloglu Y, Zeuzem S, Buster EH, Uitterlinden AG, Hansen BE, Chan HL, et al. Polymorphisms near IL28B and serologic response to peginterferon in HBeAg-positive patients with chronic hepatitis B. Gastroenterology. 2012;142:513–520.e1. doi: 10.1053/j.gastro.2011.11.025. [DOI] [PubMed] [Google Scholar]
- 97.Daniels HM, Meager A, Eddleston AL, Alexander GJ, Williams R. Spontaneous production of tumour necrosis factor alpha and interleukin-1 beta during interferon-alpha treatment of chronic HBV infection. Lancet. 1990;335:875–877. doi: 10.1016/0140-6736(90)90475-k. [DOI] [PubMed] [Google Scholar]
- 98.Kapoor D, Aggarwal SR, Singh NP, Thakur V, Sarin SK. Granulocyte-macrophage colony-stimulating factor enhances the efficacy of hepatitis B virus vaccine in previously unvaccinated haemodialysis patients. J Viral Hepat. 1999;6:405–409. doi: 10.1046/j.1365-2893.1999.00180.x. [DOI] [PubMed] [Google Scholar]
- 99.Frodsham AJ, Zhang L, Dumpis U, Taib NA, Best S, Durham A, Hennig BJ, Hellier S, Knapp S, Wright M, et al. Class II cytokine receptor gene cluster is a major locus for hepatitis B persistence. Proc Natl Acad Sci USA. 2006;103:9148–9153. doi: 10.1073/pnas.0602800103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Center for Disease Control and Prevention. Hepatitis B, Epidemiology and Prevention of Vaccine-Preventable Diseases. The Pink Book: Course Textbook - 12th Edition Second Printing (May 2012) Available from: http://www.cdc.gov/vaccines/pubs/pinkbook/hepb.html Accessed on September 23, 2013.


