Abstract
AIM: To analyze the expression profiles of long non-coding RNAs (lncRNAs) in hepatocellular carcinoma.
METHODS: Hepatocellular carcinoma (HCC) tissues and matched adjacent non-tumor (NT) liver tissues were collected from 29 patients with HCC, immediately after liver resection, between March 2011 and July 2013. The diagnosis of HCC was made based on histological examination. Differentially expressed lncRNAs between HCC and NT tissues were revealed through microarray-based lncRNAs expression profiling. Further, quantification of selected lncRNAs was performed using quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR).
RESULTS: Six hundred and fifty-nine lncRNAs were differentially expressed between HCC and NT tissues, of which five [TCONS_00018278, AK093543, D16366, ENST00000501583, NR_002819 (MALAT1)] were selected for validation. Four of them were significantly downregulated in HCC tissues compared with NT tissues (P = 0.012, 0.045, 0.000 and 0.000, respectively), and the expression level of MALAT1 showed no significant difference (P = 0.114).
CONCLUSION: This study identified a set of lncRNAs differentially expressed in HCC tissues and provided useful information for exploring potential therapeutic targets and diagnostic biomarkers of this cancer.
Keywords: Long non-coding RNA, Hepatocellular carcinoma, MALAT1, Expression signature, Microarray
Core tip: Long non-coding RNAs (lncRNAs) possess prominent and diverse regulatory functions in cancer processes. In this study, the expression profiles of lncRNAs in hepatocellular carcinoma (HCC) were analyzed using a microarray-based method. One hundred and seventy-one lncRNAs were significantly downregulated and 488 were upregulated in HCC tissues. Validation was performed in 29 subjects with HCC using quantitative real-time polymerase chain reaction and the expressions of four lncRNAs were significantly downregulated in HCC, whereas the expression of lncRNA MALAT1 showed no significant difference. The same result was obtained in a subset of patients with hepatitis B virus-related HCC.
INTRODUCTION
Hepatocellular carcinoma (HCC) is the sixth most prevalent neoplasm and the third leading cause of cancer-related death[1-3]. In particular, new cases of HCC in China account for almost 55% of all patients worldwide[4]. In the United States, the incidence of HCC is increasing faster than that of any other cancer[5]. HCC is strongly associated with chronic hepatitis B virus (HBV) and 50% of HCC cases are attributable to chronic infection with HBV[1,3,6]. With a dismal 5-year survival rate of approximately 5%-6%, HCC has one the worst prognoses among cancers and causes enormous social cost and burden, despite noteworthy advances in surgical techniques and medical treatment[5,7]. There is a great need to determine the relationships between clinicopathological features and molecular changes in HCC to develop new diagnosis and treatment strategies, and to improve the prognosis of diagnosed patients[8,9].
The human genome comprises not only protein-coding genes, but also encodes a far larger set of non-protein-coding transcripts that have crucial functions for diseases[10,11]. Though much attention has focused on small non-coding RNAs [18-200 nucleotides (nt)] in recent years, long non-coding RNAs (lncRNAs), a newly discovered class of non-coding RNAs greater than 200 nt in length, have been shown to possess prominent and diverse regulatory functions in a wide range of biological processes[12-15]. Studies indicate that lncRNAs are associated with various human diseases, including cancer[16-19]. Altered lncRNAs expression levels correlate with patient clinicopathological features and prognosis. For example, Gupta et al[20] found that the expression of lncRNA HOX antisense intergenic RNA (HOTAIR) was increased in primary breast tumors and metastases, and the HOTAIR expression level in primary tumors was a powerful predictor of eventual metastasis and death. More recently, Yang et al[21] found that the expression level of lncRNA high expression in HCC (lncRNA-HEIH) in HBV-related HCC could effectively predict recurrence and was an independent prognostic factor for survival. Furthermore, using loss-of-function and gain-of-function approaches, they identified that lncRNA-HEIH plays a key role in cell-cycle regulation. In spite of this progress, studies on the roles of lncRNAs in HCC are still at a preliminary stage and their clinical significance remains largely unknown.
In this study, the lncRNA expression signatures from four subjects with HCC, were compared with matched adjacent non-tumor (NT) liver tissues. Five of these lncRNAs were evaluated by quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR) in twenty-nine HCC tissues and NT tissues. Our results demonstrated that lncRNAs profiles may provide new molecular biomarkers or therapeutic targets for HCC.
MATERIALS AND METHODS
Patients
In this prospective study, twenty-nine patients aged 30-73 years were enrolled from March 2011 to July 2013. The human ethics committees of the Peking Union Medical College Hospital (PUMCH) approved this study. Before surgery, written informed consent was obtained from all patients. All these patients received liver resection at PUMCH and were diagnosed as HCC histopathologically after surgery. There was no radiotherapy or chemotherapy before surgery. Four of these patients were used for microarray analysis of lncRNAs and twenty-five were used for an extra evaluation. Demographic and clinical characterizations of the study population are summarized in Table 1.
Table 1.
Demographic and clinical characteristics of patients with hepatocellular carcinoma for the study of tissue lncRNAs expression n (%)
| Parameters | Description | Minimum | Maximum |
| Age (yr) | 56.59 ± 10.97 | 30 | 73 |
| Gender | |||
| Male | 26 (89.70) | ||
| Female | 3 (10.30) | ||
| Hepatitis virus infection | |||
| HBV | 22 (75.90) | ||
| HCV | 3 (10.30) | ||
| Negative | 4 (13.80) | ||
| Tumor size (cm) | 6.44 ± 3.65 | 2 | 19 |
| Tumor differentiation | |||
| Well | 6 (20.70) | ||
| Moderate | 16 (55.20) | ||
| Poor | 7 (24.10) | ||
| TNM stage | |||
| I | 10 (34.50) | ||
| II | 11 (37.90) | ||
| III | 7 (14.0) | ||
| IV | 1 (3.40) | ||
| BCLC stage | |||
| A | 21 (72.40) | ||
| B | 1 (3.40) | ||
| C | 7 (24.10) | ||
| AFP (ng/mL) | |||
| ≤ 400 | 17 (58.60) | ||
| > 400 | 12 (41.40) | ||
| PVTT | |||
| Present | 23 (79.30) | ||
| Absent | 6 (20.70) | ||
| Tumor number | |||
| Single | 27 (93.10) | ||
| Multiple | 2 (6.90) | ||
| Capsule invasive | |||
| Present | 6 (20.70) | ||
| Absent | 23 (79.30) | ||
| ALP (U/L) | 42.90 ± 32.24 | 11 | 159 |
| ALB (g/L) | 39.86 ± 3.72 | 28 | 47 |
| TBIL (μmol/L) | 15.36 ± 7.41 | 7.1 | 42.6 |
| DBIL (μmol/L) | 6.10 ± 3.08 | 3 | 16.5 |
Data are expressed as mean ± SD or n (%). TNM: Tumor-node-metastasis staging system; BCLC: The Barcelona Clinic Liver Cancer staging system; HBV: Hepatitis B virus; HCV: Hepatitis C virus; AFP: Alpha fetoprotein; PVTT: Portal vein tumor thrombus; ALP: Alkaline phosphatase; ALB: Albumin; TBIL: Total bilirubin; DBIL: Direct bilirubin.
Tissue collection and total RNA extraction
Paired HCC tissues and adjacent NT liver tissues from every subject were snap-frozen in liquid nitrogen immediately after resection and stored at -80 °C until use. The mirVanaTM RNA Isolation Kit (Applied Biosystems, Foster City, CA, United States) was used to extract total RNA from frozen samples, according to the manufacturer’s protocols, which was then eluted with 100 mL of nuclease-free water. A NanoDrop 2000 spectrophotometer (Thermo Scientific, Wilmington, DE, United States) determined the RNA yield, and agarose gel electrophoresis with ethidium bromide staining evaluated the RNA integrity.
Microarray and computational analysis
Four HCC tissues and corresponding non-tumor liver tissues were used for lncRNA microarray profiling. 200 ng of total RNA was labeled using the LowInput Quick-Amp Labeling Kit, One-Color (24*) (Agilent, p/n 5190-2305, Santa Clara, CA, United States) and hybridized on the Gene Expression Hybridization Kit (Agilent, p/n 5188-4242). The Agilent human lncRNA array contains 46506 human lncRNAs probes and 30656 human mRNAs probes. Hybridization signals were detected using The Microarray Scanner (Agilent, p/n G2505C) detected the hybridization signals and the Feature Extraction Software (Agilent) analyzed the scanned images. OE Biotech, Shanghai, China performed the microarray work.
Real-time quantitative reverse transcription-PCR
A two-step reaction process was used for quantification: reverse transcription (RT) and PCR. Each RT reaction consisted of 0.5 μg RNA, 2 μL of PrimerScript Buffer, 0.5 μL of oligo dT, 0.5 μL of random 6 mers and 0.5 μL of PrimerScript RT Enzyme Mix I (TaKaRa, Japan), in a total volume of 10 μL. Reactions were performed in a GeneAmp® PCR System 9700 (Applied Biosystems) for 15 min at 37 °C, followed by heat inactivation of RT for 5 s at 85 °C. The 10 μL RT reaction mix was then diluted × 10 in nuclease-free water and held at -20 °C.
Real-time PCR was performed using LightCycler® 480 II Real-time PCR Instrument (Roche, Basel, Switzerland) with 10 μL PCR reaction mixture that included 1 μL of cDNA, 5 μL of 2 × LightCycler® 480 SYBR Green I Master (Roche), 0.2 μL of forward primer, 0.2 μL of reverse primer and 3.6 μL of nuclease-free water. Reactions were incubated in a 384-well optical plate (Roche) at 95 °C for 10 min; followed by 40 cycles of 95 °C for 10 s, 60 °C for 30 s. Each sample was run in triplicate for analysis. At the end of the PCR cycles, melting curve analysis was performed to validate the specific generation of the expected PCR product. All experiments were done in triplicate. The expression levels of lncRNAs were normalized to glyceraldehyde-3-phosphate dehydrogenase and were calculated using the 2-ΔΔCt method[22]. The primer sequences were designed in the laboratory and synthesized by Generay Biotech (Generay, Shanghai, PRC), based on the mRNA sequences obtained from the NCBI database and are shown in Table 2.
Table 2.
The primer sequences used in this study
| Target ID | Forward primer | Reverse primer |
| TCONS_00018278 | ATGAGGAACTGAGGTCTTTG | GATTGAAGTTAGAGGGTCCAT |
| AK093543 | GCAGATTGATAAACCTACGGA | ACTTTCTCAGCAGTTTGTTATG |
| D16366 | TGACAATTCTGGGTGCGA | AGAGAACCACAAGTACAGC |
| ENST00000501583 | TCACACTCTCACACTCGCA | TCGTCTGACTCTGGGTTC |
| NR_002819 | CAGCAGCAGACAGGATTC | TTCCTTCACCAAATCGCAC |
| GAPDH | TGTTGCCATCAATGACCCCTT | CTCCACGACGTACTCAGCG |
GAPDH: Glyceraldehyde-3-phosphate dehydrogenase.
Statistical analysis
The Statistical Program for Social Sciences (SPSS) 18.0 software (SPSS, Chicago, IL, United States) performed all the statistical analyses. All data were expressed as the mean ± SD or proportions where appropriate. For comparisons, paired t-tests and unpaired t-tests were performed where appropriate. GraphPad Prism 5.01 for Microsoft Windows (GraphPad Software, San Diego, CA, United States) was used to plot all graphs. P values of 0.05 (two-tailed) were considered statistically significant.
RESULTS
LncRNAs expression profiles in HCC
LncRNA profiling detected 659 lncRNAs with significant differential expression levels in HCC tissues compared with paired NT tissues (> 2-fold change; P < 0.05): 488 were significantly upregulated and 171 were significantly downregulated in HCC tissues. The list of the top twenty differentially expressed lncRNAs identified by microarray analysis is shown in Table 3. Unsupervised hierarchical clustering analysis showed systematic variations in the expression of the 659 lncRNAs and 690 mRNAs between paired HCC and NT tissues. The samples were self-segregated into the HCC and NT clusters. These normalized microarray expression data were used to generate heat maps (Figure 1).
Table 3.
The top twenty differentially expressed lncRNAs identified by the microarray in four hepatocellular carcinoma patients
| lncRNAs | Fold change (Ca/Nt) | Regulation direction |
| BC015396 | 23.924885 | Up |
| NR_046337 | 15.497891 | Up |
| ENST00000542490 | 11.481871 | Up |
| Hs.568152 | 10.981759 | Up |
| ENST00000548057 | 10.762795 | Up |
| D16366 | 10.744075 | Down |
| ENST00000548036 | 10.218715 | Up |
| uc003bho.3 | 10.091635 | Up |
| TCONS_l2_00006115 | 9.758178 | Up |
| TCONS_l2_00006118 | 8.632748 | Up |
| AK021443 | 7.991496 | Up |
| AY027770 | 7.947163 | Up |
| TCONS_00007547 | 7.920387 | Up |
| Hs.171192 | 7.900335 | Up |
| ENST00000568976 | 7.852221 | Up |
| TCONS_l2_00006113 | 7.546310 | Up |
| XR_110225 | 7.462819 | Down |
| ENST00000500112 | 6.609479 | Up |
| ENST00000548475 | 6.573556 | Up |
| ENST00000527297 | 6.532015 | Up |
Hepatocellular carcinoma samples (Ca) vs matched non-tumor (Nt).
Figure 1.
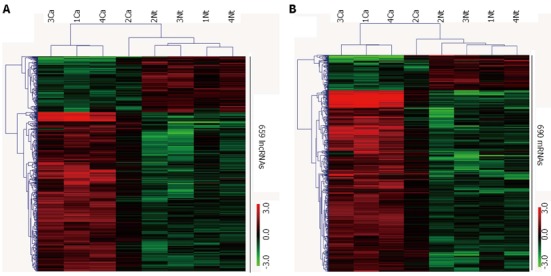
Heat map showing differentially expressed (fold change > 2) 659 lncRNAs (A) and 690 mRNAs (B) between hepatocellular carcinoma samples and non-tumor samples (greater than 2.0-fold change, P < 0.05). Each row represents one non-coding/coding RNA, and each column represents a tissue sample. The relative expression values are depicted according to the color scale. Red indicates upregulation; green indicates downregulation. Ca: Cancer samples; Nt: Matched non-tumor samples.
Overview of the mRNA profile in HCC
Through 30656 coding transcripts probes, up to 13664 mRNAs could be detected in four pairs of tissue samples. The expression levels of 577 mRNAs were significantly upregulated in HCC tissues compared with the matched NT tissues, while the expression levels of 113 mRNAs were significantly downregulated (> 2-fold change; P < 0.05) among all four pairs of samples. Gene ontology (GO) and pathway analysis showed that the differentially expressed mRNAs might be involved in regulation of transcription, and cell cycle and cell division-associated signal pathways. These results support the view that carcinogenesis of HCC is a complex multistep process involving many altered signaling cascades that lead to a heterogeneous molecular profile[2].
Validation of profiling data using quantitative real-time polymerase chain reaction
To validate the microarray analysis results, five lncRNAs were randomly selected from the differential lncRNAs and their expressions were analyzed using qRT-PCR in 29 pairs of HCC and matched NT tissues (Figure 2A). Our data indicated that the expressions of TCONS_00018278, AK093543, D16366 and ENST00000501583 were significantly downregulated in HCC (P = 0.012, 0.045, 0.000 and 0.000, respectively), whereas the expression of NR_002819 showed no significant difference (P = 0.114). We also performed a subgroup analysis in twenty-two pairs of HBV-related HCC and matched NT tissues. Except NR_002819 (P = 0.141), the remaining four lncRNAs demonstrated a statistically significant difference in expression between two groups of tissues (P = 0.029, 0.024, 0.005 and 0.008, respectively) (Figure 2B). These data confirm that a collection of lncRNAs is often dysregulated in HCC tissues.
Figure 2.
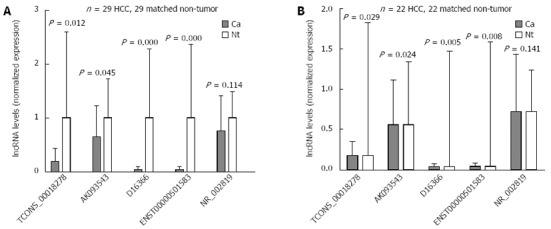
Validation of the differential expression of five lncRNAs using quantitative real-time reverse transcription polymerase chain reaction. A: In 29 paired hepatocellular carcinoma and non-tumor samples; B: In 22 paired hepatitis B virus-related hepatocellular carcinoma and non-tumor samples. Ca: Cancer samples; Nt: Matched non-tumor samples; HCC: Hepatocellular carcinoma; lncRNAs: Long non-coding RNAs.
DISCUSSION
To date, there have been few studies focusing on lncRNAs expression profiles in HCC or illustrating the association of lncRNA expression with clinicopathological features and outcomes in HCC. The carcinogenesis of HCC involves synergy of multiple environmental factors and genes, needs multiple pathological stages and involves various molecular events[2]. Technological improvements and thorough genetic characterization will help to find new diagnostic or prognostic markers, identify potential therapeutic targets, and develop new therapeutic strategies[23].
LncRNAs, which are non-coding RNAs greater than 200 nt in length, were first described by Brockdorff et al[24]. The authors described a 19 kb non-protein-coding transcript, X-inactive specific transcript (XIST), which is responsible for the inactivation of one of the two X chromosome in placental females through DNA methylation. Since then, especially in recent years, an increasing number of studies have reported that lncRNAs are involved in the progression of diverse diseases, especially cancers[25-28]. A great number of lncRNAs can be commonly detected in various tumors, such as colon cancer-associated transcript (CCAT1), metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), HOTAIR and H19[15,29-31]. Accumulating evidence suggests that unique lncRNAs are associated with the development and progression of cancer through diverse pathways, including regulation of the cell cycle[21], apoptosis[32], metastasis[33], autophagy[34], chemotherapy resistance[35], and epigenetic regulation[36] in tumor tissues or malignant cell lines. Thus, lncRNAs have opened a new field of cancer genomics. Although there are no drugs that act against lncRNAs presently, it will be fascinating to observe whether drugs could be developed that specifically target lncRNAs. Notably, lncRNAs can be detected in human body fluids and hold great promise as biomarkers[30,37,38].
In this study, we focused on exploration and description of the microarray-based lncRNAs expression signatures of HCC. With abundant and varied probes amounted to 46506 lncRNAs and 30656 mRNAs in the microarray, a large number of lncRNAs could be determined quantitatively. Comprehensive in-depth analysis of the expression profiles of lncRNAs in HCC was executed to understand the role of lncRNAs in HCC. On the basis of the lncRNA array data, we confirmed that tens of thousands of lncRNAs were expressed; however, only a few hundred of them were differentially expressed in each tissue sample. In all four HCC tissues, 171 lncRNAs downregulated and 488 were upregulated compared with adjacent non-tumor tissues. Most of these differentially expressed lncRNAs have not been characterized functionally. They might be involved in the regulation of the development and progression of HCC and might be helpful in exploring the molecular basis of HCC.
To validate the microarray analysis findings, five lncRNAs (TCONS_00018278, AK093543, D16366, ENST00000501583, NR_002819) were subjected to qRT-PCR in twenty-nine pairs of samples. The results showed significantly lower expression of TCONS_00018278, AK093543, D16366 and ENST00000501583 in HCC tissues compared with matched NT tissues. So far, little is known about the roles of these four lncRNAs in diseases. However, based on their significantly downregulated expression levels in HCC tissues, we assumed that some of them may be associated with the pathogenesis or progression of HCC, which should be explored in future studies.
Our data indicated that the expression of NR_002819 showed no significant differences between tumor tissues and adjacent non-tumor tissues either in all twenty-nine HCC patients (P = 0.114) or in the subgroup of twenty-two HBV-related HCC patients (P = 0.141).
NR_002819, generally known as MALAT1 or nuclear-enriched abundant transcript 2, is a highly abundant nucleus-restricted RNA localized to nuclear speckles, which influences alternative splicing of pre-mRNAs by modulating the cellular distribution and activity of SR splicing factors[26,39,40]. Overexpression of MALAT1 was first found in non-small cell lung cancers and then in many other cancer types, such as breast, pancreas, colon, prostate, implying key roles in carcinogenesis and tumor metastasis[23,26,41].
Currently, little is known about the function and mechanism of MALAT1 in HCC. Luo JH and his colleagues found that the expression level of MALAT1 was significantly higher in hepatoblastoma (HPBL) tissues than HCC tissues[42]. Lin et al[43] found that MALAT1 expression in human carcinomas vs the corresponding non-tumor tissues was markedly elevated in four patients with HCC. Lai et al[44] found that MALAT1 was upregulated in HCC tissues compared with adjacent non-tumor tissues in sixty patients, which correlated with HCC prognosis. Liu et al[45] failed to find a significant association between single nucleotide polymorphisms (SNPs) in MALAT1 and decreased susceptibility to HCC in HBV persistent carriers in a case-control study involving over 3000 subjects. In vitro studies have demonstrated that MALAT-1 interacts with hnRNP C and regulates the progress of the cell cycle in HepG2 and HeLa cell lines[46].
The present study, and the studies[43,44] are the only three papers focusing on the quantitative assessment of MALAT1 in HCC tissues. The discrepancy between our findings and those of Lin et al[43] and Lai et al[44] suggested that further validation is necessary. It should be noted that the sample sizes of all of these three studies are comparatively small. For instance, project of Lai et al[44] included 60 patients and only four subjects were involved in study[43]. Follow-up studies in larger cohorts of patients should be conducted at a later date.
The results of qRT-PCR were consistent with the data from the microarray and revealed that there were unique lncRNAs expression signatures in these tissues. These differentially expressed lncRNAs represent a potential novel way to detect HCC. Some studies revealed the existence of lncRNAs in body fluids, such as serum and plasma[30,38,47]. However, it is too early to utilize this collection of lncRNAs as biomarkers of HCC based on the present data. Further studies to investigate their functions and structures are necessary. The data obtained from this present research will benefit the exploration of novel diagnostic markers and therapeutic targets in HCC. The sample size of our pilot study was small and only five lncRNAs were selected for validation, so further studies in larger cohorts of patients are necessary.
This study described the lncRNAs expression signatures of HCC based on a microarray, which provided a promising new opportunity to detect HCC. Also, this study identified a four dysregulated lncRNAs in HCC. Our data indicated that the expression of MALAT1 showed no significant differences in HCC tissues. These results warrant further analysis of the relationship between lncRNAs and clinicopathological features of HCC, which will help our understanding of its carcinogenic mechanisms, and permit us to explore new early diagnostic markers and novel target genes to prevent HCC development.
COMMENTS
Background
Hepatocellular carcinoma (HCC) is the sixth most prevalent neoplasm and the third leading cause of cancer-related death. In particular, new cases of HCC in China account for almost 55% of all patients worldwide. With a dismal 5-year survival rate of approximately 5%-6%, HCC has one of the worst prognoses among cancers and causes enormous social cost and burden, despite notable advances in surgical techniques and medical treatment. The relationships between clinicopathological features and molecular changes in HCC must be determined to develop new diagnosis and treatment strategies and improve the prognosis of diagnosed patients.
Research frontiers
Long non-coding RNSs (lncRNAs) possess diverse regulatory functions in a broad range of biological processes. Studies indicate that lncRNAs are associated with various human diseases, including cancer. Altered lncRNA expression levels correlate with patient clinicopathological features and prognosis. However, the roles of lncRNAs in HCC and their clinical significance remain largely unknown.
Innovations and breakthroughs
In this study, the authors presented the lncRNAs expression profiles in four subjects with HCC, compared with matched adjacent non-tumor liver tissues. Five of these lncRNAs were evaluated by quantitative real-time polymerase chain reaction (qRT-PCR) in a total of twenty-nine pairs of tissues. The results demonstrated that lncRNA profiles may provide new molecular biomarkers or the basis for the diagnosis of HCC.
Applications
This study identified a set of lncRNAs differentially expressed in HCC tissues and provided useful information for exploring potential therapeutic targets and diagnostic biomarkers of this malignancy.
Terminology
lncRNAs: a newly discovered class of non-coding RNAs greater than 200 nt in length.
Peer review
The authors of this manuscript analyze expression profiles of lncRNAs in HCC using a microarray and found that 659 lncRNAs were differentially expressed between HCC and non-tumor tissues. Moreover, they selected five lncRNAs for validation using qRT-PCR. This explorative work is suitable for publication. The English is excellent and it contains new data.
Footnotes
Supported by Grants from the China Medical Board of New York, No. 11-045; and National Key Technology Research and Development Program of China, No. BAI06B01
P- Reviewer: Shih B S- Editor: Song XX L- Editor: Stewart G E- Editor: Zhang DN
References
- 1.Bruix J, Llovet JM. Prognostic prediction and treatment strategy in hepatocellular carcinoma. Hepatology. 2002;35:519–524. doi: 10.1053/jhep.2002.32089. [DOI] [PubMed] [Google Scholar]
- 2.Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet. 2012;379:1245–1255. doi: 10.1016/S0140-6736(11)61347-0. [DOI] [PubMed] [Google Scholar]
- 3.Nguyen VT, Law MG, Dore GJ. Hepatitis B-related hepatocellular carcinoma: epidemiological characteristics and disease burden. J Viral Hepat. 2009;16:453–463. doi: 10.1111/j.1365-2893.2009.01117.x. [DOI] [PubMed] [Google Scholar]
- 4.Shariff MI, Cox IJ, Gomaa AI, Khan SA, Gedroyc W, Taylor-Robinson SD. Hepatocellular carcinoma: current trends in worldwide epidemiology, risk factors, diagnosis and therapeutics. Expert Rev Gastroenterol Hepatol. 2009;3:353–367. doi: 10.1586/egh.09.35. [DOI] [PubMed] [Google Scholar]
- 5.Hao K, Luk JM, Lee NP, Mao M, Zhang C, Ferguson MD, Lamb J, Dai H, Ng IO, Sham PC, et al. Predicting prognosis in hepatocellular carcinoma after curative surgery with common clinicopathologic parameters. BMC Cancer. 2009;9:389. doi: 10.1186/1471-2407-9-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.But DY, Lai CL, Yuen MF. Natural history of hepatitis-related hepatocellular carcinoma. World J Gastroenterol. 2008;14:1652–1656. doi: 10.3748/wjg.14.1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Siegel R, Ward E, Brawley O, Jemal A. Cancer statistics, 2011: the impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J Clin. 2011;61:212–236. doi: 10.3322/caac.20121. [DOI] [PubMed] [Google Scholar]
- 8.Willyard C. Researchers look for ‘sweet’ method to diagnose cancer. Nat Med. 2007;13:1267. doi: 10.1038/nm1107-1267. [DOI] [PubMed] [Google Scholar]
- 9.Imbeaud S, Ladeiro Y, Zucman-Rossi J. Identification of novel oncogenes and tumor suppressors in hepatocellular carcinoma. Semin Liver Dis. 2010;30:75–86. doi: 10.1055/s-0030-1247134. [DOI] [PubMed] [Google Scholar]
- 10.Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou H, Xiao B, Zhou F, Deng H, Zhang X, Lou Y, Gong Z, Du C, Guo J. MiR-421 is a functional marker of circulating tumor cells in gastric cancer patients. Biomarkers. 2012;17:104–110. doi: 10.3109/1354750X.2011.614961. [DOI] [PubMed] [Google Scholar]
- 12.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 15.Yang F, Bi J, Xue X, Zheng L, Zhi K, Hua J, Fang G. Up-regulated long non-coding RNA H19 contributes to proliferation of gastric cancer cells. FEBS J. 2012;279:3159–3165. doi: 10.1111/j.1742-4658.2012.08694.x. [DOI] [PubMed] [Google Scholar]
- 16.Tahira AC, Kubrusly MS, Faria MF, Dazzani B, Fonseca RS, Maracaja-Coutinho V, Verjovski-Almeida S, Machado MC, Reis EM. Long noncoding intronic RNAs are differentially expressed in primary and metastatic pancreatic cancer. Mol Cancer. 2011;10:141. doi: 10.1186/1476-4598-10-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qureshi IA, Mattick JS, Mehler MF. Long non-coding RNAs in nervous system function and disease. Brain Res. 2010;1338:20–35. doi: 10.1016/j.brainres.2010.03.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 20.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang F, Zhang L, Huo XS, Yuan JH, Xu D, Yuan SX, Zhu N, Zhou WP, Yang GS, Wang YZ, et al. Long noncoding RNA high expression in hepatocellular carcinoma facilitates tumor growth through enhancer of zeste homolog 2 in humans. Hepatology. 2011;54:1679–1689. doi: 10.1002/hep.24563. [DOI] [PubMed] [Google Scholar]
- 22.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 23.Shi X, Sun M, Liu H, Yao Y, Song Y. Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett. 2013;339:159–166. doi: 10.1016/j.canlet.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 24.Brockdorff N, Ashworth A, Kay GF, McCabe VM, Norris DP, Cooper PJ, Swift S, Rastan S. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell. 1992;71:515–526. doi: 10.1016/0092-8674(92)90519-i. [DOI] [PubMed] [Google Scholar]
- 25.Aguilo F, Zhou MM, Walsh MJ. Long noncoding RNA, polycomb, and the ghosts haunting INK4b-ARF-INK4a expression. Cancer Res. 2011;71:5365–5369. doi: 10.1158/0008-5472.CAN-10-4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ji P, Diederichs S, Wang W, Böing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 27.Wang J, Liu X, Wu H, Ni P, Gu Z, Qiao Y, Chen N, Sun F, Fan Q. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res. 2010;38:5366–5383. doi: 10.1093/nar/gkq285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 29.He S, Liu S, Zhu H. The sequence, structure and evolutionary features of HOTAIR in mammals. BMC Evol Biol. 2011;11:102. doi: 10.1186/1471-2148-11-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ren S, Wang F, Shen J, Sun Y, Xu W, Lu J, Wei M, Xu C, Wu C, Zhang Z, et al. Long non-coding RNA metastasis associated in lung adenocarcinoma transcript 1 derived miniRNA as a novel plasma-based biomarker for diagnosing prostate cancer. Eur J Cancer. 2013;49:2949–2959. doi: 10.1016/j.ejca.2013.04.026. [DOI] [PubMed] [Google Scholar]
- 31.Yang F, Xue X, Bi J, Zheng L, Zhi K, Gu Y, Fang G. Long noncoding RNA CCAT1, which could be activated by c-Myc, promotes the progression of gastric carcinoma. J Cancer Res Clin Oncol. 2013;139:437–445. doi: 10.1007/s00432-012-1324-x. [DOI] [PubMed] [Google Scholar]
- 32.Khaitan D, Dinger ME, Mazar J, Crawford J, Smith MA, Mattick JS, Perera RJ. The melanoma-upregulated long noncoding RNA SPRY4-IT1 modulates apoptosis and invasion. Cancer Res. 2011;71:3852–3862. doi: 10.1158/0008-5472.CAN-10-4460. [DOI] [PubMed] [Google Scholar]
- 33.Yang F, Huo XS, Yuan SX, Zhang L, Zhou WP, Wang F, Sun SH. Repression of the long noncoding RNA-LET by histone deacetylase 3 contributes to hypoxia-mediated metastasis. Mol Cell. 2013;49:1083–1096. doi: 10.1016/j.molcel.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 34.Ying L, Huang Y, Chen H, Wang Y, Xia L, Chen Y, Liu Y, Qiu F. Downregulated MEG3 activates autophagy and increases cell proliferation in bladder cancer. Mol Biosyst. 2013;9:407–411. doi: 10.1039/c2mb25386k. [DOI] [PubMed] [Google Scholar]
- 35.Yang Y, Li H, Hou S, Hu B, Liu J, Wang J. The noncoding RNA expression profile and the effect of lncRNA AK126698 on cisplatin resistance in non-small-cell lung cancer cell. PLoS One. 2013;8:e65309. doi: 10.1371/journal.pone.0065309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Augoff K, McCue B, Plow EF, Sossey-Alaoui K. miR-31 and its host gene lncRNA LOC554202 are regulated by promoter hypermethylation in triple-negative breast cancer. Mol Cancer. 2012;11:5. doi: 10.1186/1476-4598-11-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gaughwin PM, Ciesla M, Lahiri N, Tabrizi SJ, Brundin P, Björkqvist M. Hsa-miR-34b is a plasma-stable microRNA that is elevated in pre-manifest Huntington’s disease. Hum Mol Genet. 2011;20:2225–2237. doi: 10.1093/hmg/ddr111. [DOI] [PubMed] [Google Scholar]
- 38.Xie H, Ma H, Zhou D. Plasma HULC as a promising novel biomarker for the detection of hepatocellular carcinoma. Biomed Res Int. 2013;2013:136106. doi: 10.1155/2013/136106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hutchinson JN, Ensminger AW, Clemson CM, Lynch CR, Lawrence JB, Chess A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics. 2007;8:39. doi: 10.1186/1471-2164-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Änkö ML, Müller-McNicoll M, Brandl H, Curk T, Gorup C, Henry I, Ule J, Neugebauer KM. The RNA-binding landscapes of two SR proteins reveal unique functions and binding to diverse RNA classes. Genome Biol. 2012;13:R17. doi: 10.1186/gb-2012-13-3-r17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tano K, Akimitsu N. Long non-coding RNAs in cancer progression. Front Genet. 2012;3:219. doi: 10.3389/fgene.2012.00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luo JH, Ren B, Keryanov S, Tseng GC, Rao UN, Monga SP, Strom S, Demetris AJ, Nalesnik M, Yu YP, et al. Transcriptomic and genomic analysis of human hepatocellular carcinomas and hepatoblastomas. Hepatology. 2006;44:1012–1024. doi: 10.1002/hep.21328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin R, Maeda S, Liu C, Karin M, Edgington TS. A large noncoding RNA is a marker for murine hepatocellular carcinomas and a spectrum of human carcinomas. Oncogene. 2007;26:851–858. doi: 10.1038/sj.onc.1209846. [DOI] [PubMed] [Google Scholar]
- 44.Lai MC, Yang Z, Zhou L, Zhu QQ, Xie HY, Zhang F, Wu LM, Chen LM, Zheng SS. Long non-coding RNA MALAT-1 overexpression predicts tumor recurrence of hepatocellular carcinoma after liver transplantation. Med Oncol. 2012;29:1810–1816. doi: 10.1007/s12032-011-0004-z. [DOI] [PubMed] [Google Scholar]
- 45.Liu Y, Pan S, Liu L, Zhai X, Liu J, Wen J, Zhang Y, Chen J, Shen H, Hu Z. A genetic variant in long non-coding RNA HULC contributes to risk of HBV-related hepatocellular carcinoma in a Chinese population. PLoS One. 2012;7:e35145. doi: 10.1371/journal.pone.0035145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang F, Yi F, Han X, Du Q, Liang Z. MALAT-1 interacts with hnRNP C in cell cycle regulation. FEBS Lett. 2013;587:3175–3181. doi: 10.1016/j.febslet.2013.07.048. [DOI] [PubMed] [Google Scholar]
- 47.Reis EM, Verjovski-Almeida S. Perspectives of Long Non-Coding RNAs in Cancer Diagnostics. Front Genet. 2012;3:32. doi: 10.3389/fgene.2012.00032. [DOI] [PMC free article] [PubMed] [Google Scholar]


