Abstract
AIM: To investigate the feasibility of detecting aberrantly hypermethylated Wnt-antagonist gene promoters (SFRP2 and WIF-1) in fecal DNA as non-invasive biomarkers for early colorectal cancer (CRC).
METHODS: The methylation-specific polymerase chain reaction assay was performed to blindly analyze the methylation status of SFRP2 and WIF-1 gene promoters in fecal samples from 48 subjects with CRC, 35 with adenomas, 32 with hyperplastic polyps and 30 endoscopically normal subjects. Additionally, we compared the diagnostic efficiency of measuring the hypermethylated SFRP2 and WIF-1 genes in the feces to the fecal occult blood test (FOBT) for the early detection of CRC.
RESULTS: Hypermethylated SFRP2 was detected in the feces of 56.3% (27/48) of CRC cases, 51.4% (18/35) of adenoma cases and 12.5% (4/32) of patients with hyperplastic polyps. The hypermethylation of WIF-1 was detected in 60.4% (29/48), 45.7% (16/35) and 18.7% (6/32) of fecal samples from CRC, adenoma and hyperplastic polyp patients, respectively. At least one hypermethylated gene was detected in 81.3% (39/48) of CRC and 65.7% (23/35) of adenoma samples. In contrast, only a hypermethylated WIF-1 gene was detected in one case of normal fecal samples. Moreover, no significant associations were observed between SFPR2 and WIF-1 hypermethylation and clinicopathological features. Additionally, 81.8% of CRC cases diagnosed as Dukes A stage or advanced adenomas had at least one hypermethylated gene detected, while the detection rate with the FOBT was only 31.8% (P < 0.001).
CONCLUSION: Hypermethylated SFRP2 and WIF-1 genes in fecal DNA are novel and promising molecular biomarkers that have great diagnostic potential for early CRC.
Keywords: Colorectal carcinoma, Secreted frizzled-related protein 2, Wnt inhibitory factor-1, Stool, Methylation
Core tip: Currently available approaches for the early detection of colorectal carcinoma (CRC) are suboptimal. The analysis of methylation markers in the stool as a non-invasive test is important for the early diagnosis of CRC. The epigenetic silencing of Wnt antagonist genes occurs in the early stages of CRC, and up to 90% of colorectal cancers result in the aberrant activation of Wnt signaling. In our study, we found that hypermethylated SFRP2 and WIF-1 genes in fecal DNA are novel and promising molecular biomarkers that have a great potential for the diagnosis of early CRC.
INTRODUCTION
Colorectal cancer (CRC) is a leading cause of cancer-related death in Western countries, and the incidence rate has rapidly increased in Eastern countries in the past decade, especially in China[1,2]. The survival is closely related to CRC stage at the time of diagnosis. The 5-year survival rate for stage I CRC has reached 90%, but less than 10% for CRC cases who have distant metastase[3,4]. However, most CRC patients are diagnosed in the middle or late stages because no typical symptoms for the early stage of CRC exist[5]. Therefore, the diagnosis of CRC in early stage has great importance for reducing CRC mortality.
The critical target lesions have included the early stages of CRC and the highest risk of precancerous lesions[6]. Historically, colorectal adenoma has been recognized as the most important precancerous lesion of CRC. Early detection of CRC or the precancerous lesions is a major challenge to improve patient’s survival and widen the window of therapeutic intervention.
Although the incidence of CRC in the United States has decreased by 2%-3% every year over the past 15 years, the detection rate of early CRC is still less than 5%[6]. Currently available approaches to early detection of colorectal tumors are suboptimal. With evolving insights into the molecular changes during CRC development, assays to detect tumor-derived DNA alterations in the feces are a promising approach that may improve screening effectiveness and user friendliness for the screening and early diagnosis of colorectal neoplasia.
Genetic alterations are believed to drive the histological progression of CRC in the colorectal adenoma-carcinoma sequence. The aberrant methylation of genes appears to act in concert with other genetic alterations and has been recognized as one of the most common molecular alterations in CRC. The epigenetic silencing of Wnt antagonist genes occurs in the early stages of CRC, and up to 90% of colorectal cancers result in the aberrant activation of Wnt signaling[7,8]. The Wnt antagonist genes, secreted frizzled related protein gene family (SFRPs) and Wnt inhibitory factor-1 (WIF-1), have been found to be hypermethylated and are frequently not expressed in CRC cell lines and primary CRC samples[7,9]. Our previous study demonstrated that Wnt antagonist genes with promoter hypermethylation were frequently found in early CRC[9].
In this study, we extended the previous work and analyzed the methylation status of SFRP2 and WIF-1 genes in stools taken from CRC patients, from patients with benign colorectal diseases and from normal controls and then assessed the potential of fecal SFRP2 and WIF-1 gene hypermethylation as a non-invasive screening tool for CRC. Additionally, we compared the early CRC diagnostic efficiencies of the detection of hypermethylated SFRP2 and WIF-1 genes in the stool to the fecal occult blood test (FOBT).
MATERIALS AND METHODS
Stool sample collection
Stool samples were collected from 48 patients with sporadic CRC, 67 patients with benign colorectal diseases (35 colorectal adenomas and 32 hyperplastic polyps) and 30 endoscopically normal patients undergoing surgery and endoscopy at the Zhongnan Hospital of Wuhan University between February 2012 and March 2013. The patients’ clinicopathological features are listed in Table 1. All samples were stored at -80 °C until processing and all patient diagnoses were confirmed by pathology after surgery or colonoscopy. None of the patients had received chemotherapy or radiation therapy prior to surgery. All matched samples were also analyzed with a FOBT (Fecal Occult Blood Gold Gel Stripe, Van Ed Chapman Biological Engineering Company), which was performed independently at the laboratory of the Zhongnan Hospital of Wuhan University. This study was approved by the Ethical Committee of our university, and all patients gave informed consent for their participation.
Table 1.
Correlation of the methylation status of fecal SFRP2 and WIF-1 genes with patients’ clinicopathological parameters n (%)
| Parameter | Case |
WIF-1 |
SFRP2 |
WIF-1+
SFRP2 |
Sensitivity | Specificity | |||
| (n) | Methylation | P value | Methylation | P value | Methylation | P value | |||
| CRC | 48 | 27 (56.3) | 0.0001 | 29 (60.4) | 0.0001 | 39 (81.3) | 0.0001 | 81.3% | |
| Gender | |||||||||
| Male | 24 | 17 (70.8) | 0.140 | 14 (58.3) | 0.771 | ||||
| Female | 24 | 12 (29.2) | 13 (54.2) | ||||||
| Age (yr) | |||||||||
| > 55 | 32 | 21 (65.6) | 0.297 | 18 (56.3) | 1.000 | ||||
| ≤ 55 | 16 | 8 (50) | 9 (56.3) | ||||||
| Tumor location | |||||||||
| Left colon | 32 | 21 (65.6) | 0.267 | 18 (56.3) | 1.000 | ||||
| Right colon | 16 | 8 (50) | 9 (56.3) | ||||||
| TNM stage | |||||||||
| A-B | 27 | 18 (66.7) | 0.315 | 15 (55.6) | 0.912 | ||||
| C-D | 21 | 11 (52.4) | 12 (57.1) | ||||||
| Lymph node metastasis | |||||||||
| Positive | 21 | 11 (52.4) | 0.315 | 12 (57.1) | 0.912 | ||||
| Negative | 27 | 18 (66.7) | 15 (55.6) | ||||||
| Distant metastasis | |||||||||
| Positive | 7 | 4 (57.1) | 0.848 | 4 (57.1) | 0.959 | ||||
| Negative | 41 | 25 (61.0) | 23 (56.1) | ||||||
| Adenoma | 35 | 18 (51.4) | 0.0001 | 16 (45.7) | 0.0001 | 23 (65.7) | 0.0001 | 65.7% | |
| Advanced | 15 | ||||||||
| Non-advanced | 20 | ||||||||
| Hyperplastic polyp | 32 | 4 (12.5) | 0.0451 | 6 (18.7) | 0.0551 | 8 (25.0) | 0.0161 | 25.0% | 96.7% |
| Normal control | 30 | 0 (0) | 1 (3.3) | 1 (3.3) | |||||
Represents the P value vs normal subjects. Sensitivity represents the ability of the DNA methylation test to detect the presence of at least one hypermethylated gene. CRC: Colorectal cancer; SFRP: Secreted frizzled related protein; WIF-1: Wnt inhibitory factor-1.
Carcinomas were classified according to the Union for International Cancer Control TNM classification system. Advanced adenoma was defined as an adenoma of least 10 mm in diameter, with high-grade dysplasia, villous or tubulovillous histological characteristics, or any combination of the three[10]. The left colon was defined as the region from the rectum to the splenic flexure, and the right colon was defined as being above the transverse colon.
DNA extraction
All stool samples were coded before being randomized and processed. Genomic DNA was extracted from clinical fecal samples (200 mg) with the QIAamp DNA Stool Mini Kit (Qiagen, Germany) according to the manufacturer’s instructions.
Bisulfite modification
Bisulfite genomic DNA modification, which converts all unmethylated cytosines to uracils, was performed as reported previously[11].For verification of the quality of the extracted DNA, the human β-actin gene must be amplified by polymerase chain reaction (PCR). Each PCR reaction mix consisted of a total volume of 25 μL containing 10 × PCR buffer (Thermo, United States), 12 μL PCR-Mix (Topsun Group, China), 1 μmol/L of each of the primers β-actin-F (TGGTGATGGAGGAGGCTCAGCAAGT ) and β-actin-R (AGCCAATGGGACCTGCTCCTCCCTTGA), and 5-20 ng fecal DNA. The PCR samples were subjected to 94 °C for 3 min, followed by 35 cycles at 94 °C for 30 s, 60 °C for 30 s, and 72 °C for 1 min, and finally a 10 min extension at 72 °C. PCR products were analyzed by 3% agarose gel electrophoresis.
Methylation-specific PCR analysis
After the bisulfite treatment, the target genes were amplified with primers for the methylated and unmethylated genes.The primer sequences are listed in Table 2.
Table 2.
SFRP2 and WIF-1 gene primer sequences for methylation-specific polymerase chain reaction assays
| Gene | Primer (5’-3’) | Product size (bp) | Annealing temperature (°C) |
| M: SFRP2 | GGGTCGGAGTTTTTCGGAGTTGCGC | 138 | 62 |
| CCGCTCTCTTCGCTAAATACGACTCG | (38) | ||
| U: SFRP2 | TTTTGGGTTGGAGTTTTTTGGAGTTGTGT | 145 | 52 |
| AACCCACTCTCTTCACTAAATACAACTCA | (38) | ||
| M: WIF-1 | GGGCGTTTTATTGGGCGTAT | 197 | 58 |
| AAACCAACAATCAACGAAC | (38) | ||
| U: WIF-1 | GGGTGTTTTATTGGGTGTAT | 198 | 55 |
| AAACCAACAATCAACAAAAC | (38) |
SFRP: Secreted frizzled related protein; WIF-1: Wnt inhibitory factor-1.
The procedures of methylation-specific PCR are as follows: 2 μL modified DNA was amplified in a total volume of 25 μL containing 2.5 μL of 10 × PCR buffer, 1.5 mmol/L MgCl2, 200 μmol/L of each dNTP, 0.5 μmol/L of each primer, and 1.0 unit of Taq polymerase. Amplification conditions included a hot start at 95 °C for 5 min, 40 cycles at 95 °C for 45 s, annealing temperature of each gene for 45 s, 72 °C for 45 s, and a final 8 min extension step at 72 °C. Products were visualized by 3% agarose gel electrophoresis with ultraviolet illumination using a gel imaging analyzing system (Genesnap Syngene, China). Methylation pattern analysis was based on the distribution of visible bands.
Statistical analysis
Statistical analysis was performed using the SPSS statistical software version 17.0. Associations between the discrete variables were assessed using the two-sided Fisher’s exact test or Pearson’s χ2 tests. A P value less than 0.05 was regarded as statistically significant.
RESULTS
Hypermethylation of SFRP2 and WIF-1 genes in fecal DNA
Hypermethylation of SFRP2 and WIF-1 in fecal samples was detected in 56.3% (27/48) and 60.4% (29/48) of 48 CRC samples, respectively, and 81.3% (39/48) of CRC samples had at least one hypermethylated gene detected. The frequencies of aberrant methylation of SFRP2 and WIF-1 were 51.4% (18/35) and 45.7% (16/35) in colorectal adenoma samples, and 65.7% (23/35) of adenoma samples had at least one hypermethylated gene. Hypermethylation of SFRP2 and WIF-1 in fecal samples was detected in 12.5% (4/32) and 18.7% (6/32) of 32 hyperplastic polyp samples, respectively, and 25% (8/32) of the hyperplastic polyp samples had at least one hypermethylated gene. In 30 normal fecal samples, one case exhibited a band representative of methylated WIF-1 (Figure 1, Table 3).
Figure 1.
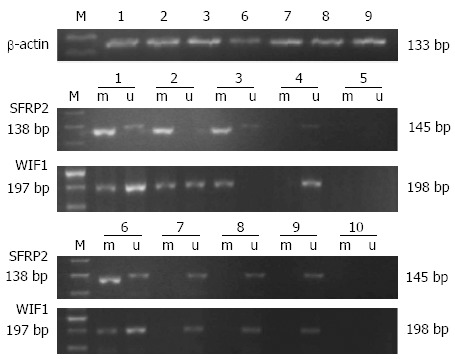
Methylation status of SFRP2 and WIF-1 genes in clinical stool samples. 1-3: Colorectal cancers; 6-8: Polyps (colorectal adenomas or hyperplastic polyps); 4, 9: Normal controls; 5, 10: Blank controls; M: 50 bp DNA marker; m: Methylated bands; u: Unmethylated bands; SFRP: Secreted frizzled related protein; WIF-1: Wnt inhibitory factor-1.
Table 3.
Hypermethylation status of SFRP2 and WIF-1 in fecal DNA samples n (%)
| Group | Case |
Hypermethylated genes |
||
| SFRP2 | WIF-1 | SFRP2 + WIF-1 | ||
| CRC | 48 | 27 (56.3) | 29 (60.4) | 39 (81.3) |
| Dukes A-B | 27 | 15 (55.6) | 18 (66.7) | 22 (81.5) |
| Dukes A | 7 | 5 (71.4) | 4 (57.1) | 6 (85.7) |
| Dukes B | 20 | 10 (50.0) | 14 (70.0) | 16 (80.0) |
| Dukes C-D | 21 | 12 (57.1) | 11 (52.4) | 17 (81.0) |
| Dukes C | 14 | 8 (57.1) | 7 (50.0) | 10 (71.4) |
| Dukes D | 7 | 4 (57.1) | 4 (57.1) | 7 (100) |
| Adenoma | 35 | 18 (51.4) | 16 (45.7) | 23 (65.7) |
| Advanced | 15 | 9 (60.0) | 8 (53.3) | 12 (80.0) |
| Non-advanced | 20 | 8 (40.0) | 7 (35.0) | 11 (55.0) |
| Hyperplastic polyp | 32 | 4 (12.5) | 6 (18.7) | 8 (25.0) |
| Normal control | 30 | 0 (0) | 1 (3.3) | 1 (3.3) |
CRC: Colorectal cancer; SFRP: Secreted frizzled related protein; WIF-1: Wnt inhibitory factor-1.
The rates of hypermethylation of SFRP2 and WIF-1 genes were significantly different between the carcinoma and normal control groups (P < 0.001 for both), between the carcinoma and hyperplastic polyp samples (P < 0.001 for both), between the adenoma group and normal controls (P < 0.001 for both), between the adenoma and hyperplastic polyp groups (P < 0.001 for SFRP2, P = 0.005 for WIF-1), and between the hyperplastic polyp group and normal controls (P = 0.045 for SFRP2, but P = 0.055 for WIF-1). No significant difference was found between the carcinoma and adenoma groups (P = 0.663 for SFRP2, and P = 0.184 for WIF-1).
Hypermethylated SFRP2 and WIF-1 genes were detected in 71.4% (5/7) and 57.1% (4/7) of fecal samples from patients with Dukes A stage of CRC, and 60.0% (9/15) and 53.3% (8/15) from those with advanced adenoma, respectively. The frequency of combined SFRP2 and WIF-1 hypermethylation for detection of CRC was higher than that of single gene. The frequency of hypermethylated SFRP2 and WIF-1 in fecal samples reached 80.0% for advanced adenoma, which is higher than that for non-advanced adenoma (55%). Additionally, 81.8% (18/22) of Dukes A stage of CRC and advanced adenoma patients had at least one hypermethylated gene detected in the fecal samples.
Moreover, no significant associations were observed between the hypermethylation of SFRP2 and WIF-1 genes and other clinicopathological features, including gender, age, tumor location, TNM stage, lymph node status and distant metastasis, etc. (Table 1).
Hypermethylated genes in fecal DNA and FOBT
There were 83 colorectal tumors (48 CRCs and 35 adenomas) and there was a distinct difference (P < 0.001) in the efficiency of diagnosing colorectal tumors when using the fecal hypermethylated SFRP2 and WIF-1 genes (74.7%, 62/83) vs FOBT (43.4%, 36/83), as well as in tumors classified as Dukes A of CRC (P = 0.031) and adenoma (P < 0.001). There was no significant difference when comparing diagnosis by hypermethylation or FOBT in Dukes B-D of CRC (P > 0.05), samples of hyperplastic polyps and normal subjects (P > 0.05) (Table 4).
Table 4.
Hypermethylated SFRP2 and WIF-1 genes and fecal occult blood test for the detection of colorectal tumors n (%)
| Group | n | Hypermethylated SFRP2 and WIF-1 | Fecal occult blood test | P value |
| CRC | 48 | 39 (81.3) | 27 (56.3) | 0.008 |
| Dukes A-B | 27 | 22 (81.5) | 14 (51.9) | 0.021 |
| Dukes A | 7 | 6 (85.7) | 2 (28.5) | 0.031 |
| Dukes B | 20 | 16 (80.0) | 12 (60.0) | 0.168 |
| Dukes C-D | 21 | 17 (81.0) | 13 (61.9) | 0.170 |
| Dukes C | 14 | 10 (71.4) | 8 (57.1) | 0.430 |
| Dukes D | 7 | 7 (100) | 5 (71.4) | 0.127 |
| Adenoma | 35 | 23 (65.7) | 9 (25.7) | < 0.001 |
| Advanced | 15 | 12 (80.0) | 5 (33.3) | 0.010 |
| Non-advanced | 20 | 11 (55.0) | 4 (20.0) | 0.022 |
| Hyperplastic polyp | 32 | 8 (25.0) | 5 (15.6) | 0.351 |
| Normal control | 30 | 1 (3.3) | 1 (3.3) | 1.000 |
CRC: Colorectal cancer; SFRP: Secreted frizzled related protein; WIF-1: Wnt inhibitory factor-1.
DISCUSSION
Sporadic CRC development is a multistep process with the accumulation of genetic and epigenetic alterations that transform colonic epithelial cells into colon adenocarcinoma cells[12]. Recently, the epigenetic silencing of tumor suppressor genes has been increasingly recognized as an important mechanism in tumorigenesis, and the detection of hypermethylated genes in patient stools has emerged as a biologically rational and user-friendly strategy for early CRC[13-15].
Aberrant activation of the Wnt signaling pathway is associated with a variety of human cancers, particularly colorectal cancer, as well as the physiological function of adult organisms. Aberrant promoter hypermethylation of Wnt antagonist genes frequently occurs in the early stages of the pathogenesis of human cancers, and up to 90% of colorectal cancers result in aberrant activation of Wnt signaling[8]. Previous studies have identified that SFRP2 and WIF-1, both secreted Wnt antagonists, are silenced epigenetically[8,16].
Our results here and in our previous study have shown that the promoters of SFRP2 and WIF-1 genes are frequently hypermethylated in patients with colorectal tumors and can be detected in their fecal samples[9]. The results showed that SFRP2 and WIF-1 were hypermethylated in 56.3% and 60.4% of CRC, 51.4% and 45.7% of adenoma, and 12.5% and 18.7% of hyperplastic polyp cases, respectively, thereby indicating that the hypermethylation of SFRP2 and WIF-1 occurs in hyperplastic polyps and becomes more frequent in adenomas and then finally in CRCs. Our results showed that the rate of hypermethylation of SFRP2 and WIF-1 genes in fecal samples had reached 81.8% (18/22) in advanced adenoma and Dukes A of CRC patients. Therefore, examination of SFRP2 and WIF-1 hypermethylation status may provide useful information for detecting the early onset of CRC, indicating that hypermethylation of SFRP2 and WIF-1 is closely linked to the initiation and progression of CRC carcinogenesis.
Historically, colorectal adenoma has been recognized as the most important precancerous lesion occurring prior to fulminant CRC. It is estimated that 50% of individuals will develop adenomas in their lifetime, but only 6% will progress into CRC[5,17]. Therefore, most adenomas do not progress to cancer. We showed that the frequency of hypermethylated SFRP2 was higher in advanced adenomas than in non-advanced adenomas. Moreover, the difference between the methylation states of genes in CRC and advanced adenoma was not significant (P > 0.05), while the difference between epigenetic changes in CRC and non-advanced adenoma was significant (P < 0.05). This suggests that advanced adenomas rather than non-advanced ones are important in the screening for early CRC. Additionally, screening out advanced adenomas could be used complementary with colonoscopy to reduce the high cost of surveillance endoscopy and could improve patient compliance to surveillance, which is currently poor, even among high-risk patients[18-20].
The diagnostic sensitivity attained by screening for combined SFPR2 and WIF-1 hypermethylation is higher than screening for a single hypermethylated gene. The clinical sensitivities for detecting CRC, advanced adenoma and hyperplastic polyps reached 81.3%, 80.0% and 25.0%, respectively, and the specificity was 96.7% for detection of CRC and benign colorectal diseases. In addition, there was a trend, although not statistically significant, of more frequent SFRP2 and WIF-1 hypermethylation among the adenomas accompanied with epithelial dysplasia. Hypermethylation of the WIF-1 gene was found in one endoscopically normal subject, which was likely attributed to the presence of premalignant lesions, such as aberrant crypt foci (ACF).
To further address the clinical relevance of hypermethylated SFRP2 and WIF-1 in CRC, our data suggested that assaying for hypermethylated genes works equally well for detecting proximal and distal colorectal tumors, in agreement with a previous study illustrating that the detection of hypermethylated genes was not influenced by the site of the neoplasm[21]. Considering the proportional rise in incidence of CRC and precancerous lesions in the proximal colon, measuring DNA hypermethylation in fecal samples plays an important role in the detection of proximal neoplasms[22,23]. Although determinate studies are needed, our results suggest the potential usefulness of assaying the hypermethylation of Wnt antagonist genes in fecal samples for the detection of early CRC.
It is known that the majority of sporadic colorectal dysplasias and malignancies require the inactivation of the APC gene, which promotes β-catenin mediated transcription. Therefore, in the absence of epigenetic silencing, APC mutation and the consequent β-catenin activity would lead to Wnt antagonist induction in a negative feedback loop, starving the tumor cell of Wnt ligand[24]. Because the epigenetic silencing of Wnt antagonist genes occurs in early sporadic colorectal tumorigenesis, the activation of Wnt signaling may be necessary for survival of the early tumor[16]. In addition, the restoration of SFRP and WIF-1 function in CRC cells attenuates Wnt/β-catenin signaling, resulting in a significant inhibition of colony formation and cell proliferation even in the presence of downstream mutations in APC or β-catenin. The hypermethylation of SFRP2 and WIF-1 has been detected in more than 90% and 80%, respectively, of primary CRC cases[25]. As the activation of Wnt pathway in colorectal tumors has been observed at high frequencies, and considering the important role it plays in both initiation and progression of CRC, the targeted inhibition of the Wnt signaling pathway provides a rational and promising approach for the screening of early CRC.
Colonocytes from the surface of the colonic mucosa are continuously shed into the lumen of the colon and excreted as a normal component of stool. These cells are shed from the lower crypts at a rate of at least 1010 cells per day, each with a lifespan of 3-4 d, and within four days, the entire colonic mucosa is renewed[26,27]. Current advances in colon cytobiology have led to the understanding that exfoliation is triggered by apoptosis in the normal colon, while in malignant lesions, genetic and/or epigenetic changes prevent this destruction of colonocytes, allowing them to survive within the stool[28]. Additionally, the number of colonocytes exfoliated from malignant lesions is 4- to 5-fold greater than from normal tissue[27], so it could be simple and effective to collect fecal samples containing colonic DNA for the detection of early CRC.
FOBT, especially immunochemical FOBT, is the most widely utilized procedure for non-invasive CRC screening. Although FOBT is confirmed to reduce the mortality of CRC, the test has little impact on the incidence of CRC because of the low level of sensitivity to detect precursor lesions[29]. Stool DNA testing has emerged as a biologically rational and user-friendly strategy for the non-invasive detection of both CRC and precursor lesions[10].
In our study, the sensitivity of detection of hypermethylated Wnt antagonist genes represents an apparent advantage over FOBT in the detection of CRC and colorectal adenoma, especially in the early cancer and advanced adenoma stages. This shows that measuring the hypermethylation of Wnt antagonist genes in fecal samples was superior to FOBT for the detection of colorectal tumors. This detection advantage may be attributed to the apparently continuous exfoliation of cells from colorectal neoplasms, while occult bleeding is intermittent[30,31].
In summary, the detection of tumor-derived DNA alterations in the stool is an intriguing new approach with a high potential for the non-invasive detection of CRC. Our results demonstrate that the hypermethylation of SFRP2 and WIF-1 in fecal samples shows promise for the accurate detection of CRC. However, a large number of studies are required to further accurately confirm the role of hypermethylation of SFRP2 and WIF-1 in the early CRC diagnosis.
COMMENTS
Background
Colorectal cancer (CRC) is the third most common malignancy and the second leading cause of cancer-related deaths in Western countries. The diagnosis of CRC in early stages has great importance for reducing CRC mortality. Although significant advances have been achieved in diagnostic technologies, the currently available modalities for diagnosing CRC remain suboptimal.
Research frontiers
DNA hypermethylation can result in the transcriptional silencing of tumor suppressor genes and plays an important role in the initiation and progression of sporadic CRC. Detection of hypermethylation of Wnt antagonist genes in human stool DNA might provide a novel and promising strategy for early CRC diagnosis.
Innovations and breakthroughs
Methylation-specific polymerase chain reaction was performed to analyze the promoter hypermethylation status of the SFRP2 and WIF-1 genes in a blinded fashion in stool samples from patients with CRC and colorectal benign diseases. Furthermore, for the diagnosis of early CRC, the authors compared the sensitivity of assaying for the hypermethylated SFRP2 and WIF-1 genes to the standard fecal occult blood test in matched stool samples from the same patients.
Applications
This study demonstrated initially that hypermethylated SFRP2 and WIF-1 genes in stool are promising and non-invasive sensitive markers for screening early CRC.
Terminology
Wnt signaling pathway: Signals are transduced through binding of Wnt ligands to the Frizzled (Fz) family of seven-pass transmembrane receptors by competition for binding to Wnt ligands or by the direct formation of non-functional complexes with Fz receptors. Wnt-antagonists: Wnt antagonists regulate the upstream signaling of the Wnt pathway outside of the cell, which are divided into two functional classes. The first functional class is the SFRPs (secreted Frizzled-related proteins), which include the SFRP family (SFRP1-5), wnt inhibitory factor-1 (WIF-1) and Cerberus, which bind directly to Wnts. The second functional class is the Dickkopf (Dkk) class that binds to Wnts indirectly.
Peer review
This paper studied the use of hypermethylation of SFRP2 and WIF-1 promoters as a novel and promising approach for early CRC diagnosis. The authors showed that there exists a high prevalence of SFRP2 and WIF-1 hypermethylation in the stool DNA of colorectal neoplasms. They conclude that the hypermethylation of SFRP2 and WIF-1 may be a sensitive diagnostic test for colorectal neoplasms.
Footnotes
Supported by The National Natural Science Foundation of China, No. 81101868; The Natural Science Foundation of Hubei Province of China, No. 2011CDB505
P- Reviewers: Mueller WC, Roncucci L, Sporea I S- Editor: Zhai HH L- Editor: Wang TQ E- Editor: Wang CH
References
- 1.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.The gastroenterology of the Chinese Medical Association. A consensus of the early detection, treatment, and the comprehensive prevention of colorectal cancer in China. Zhonghua Xiaohua Zazhi. 2012;32:1–10. [Google Scholar]
- 3.Chen WD, Han ZJ, Skoletsky J, Olson J, Sah J, Myeroff L, Platzer P, Lu S, Dawson D, Willis J, et al. Detection in fecal DNA of colon cancer-specific methylation of the nonexpressed vimentin gene. J Natl Cancer Inst. 2005;97:1124–1132. doi: 10.1093/jnci/dji204. [DOI] [PubMed] [Google Scholar]
- 4.Bosch LJ, Carvalho B, Fijneman RJ, Jimenez CR, Pinedo HM, van Engeland M, Meijer GA. Molecular tests for colorectal cancer screening. Clin Colorectal Cancer. 2011;10:8–23. doi: 10.3816/CCC.2011.n.002. [DOI] [PubMed] [Google Scholar]
- 5.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, Thun MJ. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 6.Kohler BA, Ward E, McCarthy BJ, Schymura MJ, Ries LA, Eheman C, Jemal A, Anderson RN, Ajani UA, Edwards BK. Annual report to the nation on the status of cancer, 1975-2007, featuring tumors of the brain and other nervous system. J Natl Cancer Inst. 2011;103:714–736. doi: 10.1093/jnci/djr077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suzuki H, Gabrielson E, Chen W, Anbazhagan R, van Engeland M, Weijenberg MP, Herman JG, Baylin SB. A genomic screen for genes upregulated by demethylation and histone deacetylase inhibition in human colorectal cancer. Nat Genet. 2002;31:141–149. doi: 10.1038/ng892. [DOI] [PubMed] [Google Scholar]
- 8.Suzuki H, Watkins DN, Jair KW, Schuebel KE, Markowitz SD, Chen WD, Pretlow TP, Yang B, Akiyama Y, Van Engeland M, et al. Epigenetic inactivation of SFRP genes allows constitutive WNT signaling in colorectal cancer. Nat Genet. 2004;36:417–422. doi: 10.1038/ng1330. [DOI] [PubMed] [Google Scholar]
- 9.Qi J, Zhu YQ, Luo J, Tao WH. Hypermethylation and expression regulation of secreted frizzled-related protein genes in colorectal tumor. World J Gastroenterol. 2006;12:7113–7117. doi: 10.3748/wjg.v12.i44.7113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qi J, Zhu YQ. Targeting the most upstream site of Wnt signaling pathway provides a strategic advantage for therapy in colorectal cancer. Curr Drug Targets. 2008;9:548–557. doi: 10.2174/138945008784911769. [DOI] [PubMed] [Google Scholar]
- 11.Ahlquist DA. Molecular detection of colorectal neoplasia. Gastroenterology. 2010;138:2127–2139. doi: 10.1053/j.gastro.2010.01.055. [DOI] [PubMed] [Google Scholar]
- 12.Herman JG, Graff JR, Myöhänen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grady WM, Carethers JM. Genomic and epigenetic instability in colorectal cancer pathogenesis. Gastroenterology. 2008;135:1079–1099. doi: 10.1053/j.gastro.2008.07.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lao VV, Grady WM. Epigenetics and colorectal cancer. Nat Rev Gastroenterol Hepatol. 2011;8:686–700. doi: 10.1038/nrgastro.2011.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berger BM, Ahlquist DA. Stool DNA screening for colorectal neoplasia: biological and technical basis for high detection rates. Pathology. 2012;44:80–88. doi: 10.1097/PAT.0b013e3283502fdf. [DOI] [PubMed] [Google Scholar]
- 16.Zou H, Taylor WR, Harrington JJ, Hussain FT, Cao X, Loprinzi CL, Levine TR, Rex DK, Ahnen D, Knigge KL, et al. High detection rates of colorectal neoplasia by stool DNA testing with a novel digital melt curve assay. Gastroenterology. 2009;136:459–470. doi: 10.1053/j.gastro.2008.10.023. [DOI] [PubMed] [Google Scholar]
- 17.He B, Reguart N, You L, Mazieres J, Xu Z, Lee AY, Mikami I, McCormick F, Jablons DM. Blockade of Wnt-1 signaling induces apoptosis in human colorectal cancer cells containing downstream mutations. Oncogene. 2005;24:3054–3058. doi: 10.1038/sj.onc.1208511. [DOI] [PubMed] [Google Scholar]
- 18.Strul H, Kariv R, Leshno M, Halak A, Jakubowicz M, Santo M, Umansky M, Shirin H, Degani Y, Revivo M, et al. The prevalence rate and anatomic location of colorectal adenoma and cancer detected by colonoscopy in average-risk individuals aged 40-80 years. Am J Gastroenterol. 2006;101:255–262. doi: 10.1111/j.1572-0241.2006.00430.x. [DOI] [PubMed] [Google Scholar]
- 19.Rubenstein JH, Waljee AK, Jeter JM, Velayos FS, Ladabaum U, Higgins PD. Cost effectiveness of ulcerative colitis surveillance in the setting of 5-aminosalicylates. Am J Gastroenterol. 2009;104:2222–2232. doi: 10.1038/ajg.2009.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Velayos FS, Liu L, Lewis JD, Allison JE, Flowers N, Hutfless S, Abramson O, Perry GS, Herrinton LJ. Prevalence of colorectal cancer surveillance for ulcerative colitis in an integrated health care delivery system. Gastroenterology. 2010;139:1511–1518. doi: 10.1053/j.gastro.2010.07.039. [DOI] [PubMed] [Google Scholar]
- 21.Vienne A, Simon T, Cosnes J, Baudry C, Bouhnik Y, Soulé JC, Chaussade S, Marteau P, Jian R, Delchier JC, et al. Low prevalence of colonoscopic surveillance of inflammatory bowel disease patients with longstanding extensive colitis: a clinical practice survey nested in the CESAME cohort. Aliment Pharmacol Ther. 2011;34:188–195. doi: 10.1111/j.1365-2036.2011.04711.x. [DOI] [PubMed] [Google Scholar]
- 22.Ahlquist DA, Zou H, Domanico M, Mahoney DW, Yab TC, Taylor WR, Butz ML, Thibodeau SN, Rabeneck L, Paszat LF, et al. Next-generation stool DNA test accurately detects colorectal cancer and large adenomas. Gastroenterology. 2012;142:248–256; quiz e25-26. doi: 10.1053/j.gastro.2011.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gupta AK, Melton LJ, Petersen GM, Timmons LJ, Vege SS, Harmsen WS, Diehl NN, Zinsmeister AR, Ahlquist DA. Changing trends in the incidence, stage, survival, and screen-detection of colorectal cancer: a population-based study. Clin Gastroenterol Hepatol. 2005;3:150–158. doi: 10.1016/s1542-3565(04)00664-0. [DOI] [PubMed] [Google Scholar]
- 24.Park SY, Kim BC, Shin SJ, Lee SK, Kim TI, Kim WH. Proximal shift in the distribution of adenomatous polyps in Korea over the past ten years. Hepatogastroenterology. 2009;56:677–681. [PubMed] [Google Scholar]
- 25.Caldwell GM, Jones CE, Soon Y, Warrack R, Morton DG, Matthews GM. Reorganisation of Wnt-response pathways in colorectal tumorigenesis. Br J Cancer. 2008;98:1437–1442. doi: 10.1038/sj.bjc.6604327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Itzkowitz S, Brand R, Jandorf L, Durkee K, Millholland J, Rabeneck L, Schroy PC, Sontag S, Johnson D, Markowitz S, et al. A simplified, noninvasive stool DNA test for colorectal cancer detection. Am J Gastroenterol. 2008;103:2862–2870. doi: 10.1111/j.1572-0241.2008.02088.x. [DOI] [PubMed] [Google Scholar]
- 27.Davies RJ, Miller R, Coleman N. Colorectal cancer screening: prospects for molecular stool analysis. Nat Rev Cancer. 2005;5:199–209. doi: 10.1038/nrc1569. [DOI] [PubMed] [Google Scholar]
- 28.Nair P, Lagerholm S, Dutta S, Shami S, Davis K, Ma S, Malayeri M. Coprocytobiology: on the nature of cellular elements from stools in the pathophysiology of colonic disease. J Clin Gastroenterol. 2003;36:S84–S93; discussion S94-S96. doi: 10.1097/00004836-200305001-00015. [DOI] [PubMed] [Google Scholar]
- 29.Heresbach D, Manfredi S, D’halluin PN, Bretagne JF, Branger B. Review in depth and meta-analysis of controlled trials on colorectal cancer screening by faecal occult blood test. Eur J Gastroenterol Hepatol. 2006;18:427–433. doi: 10.1097/00042737-200604000-00018. [DOI] [PubMed] [Google Scholar]
- 30.Ahlquist DA, Harrington JJ, Burgart LJ, Roche PC. Morphometric analysis of the „mucocellular layer“ overlying colorectal cancer and normal mucosa: relevance to exfoliation and stool screening. Hum Pathol. 2000;31:51–57. doi: 10.1016/s0046-8177(00)80198-7. [DOI] [PubMed] [Google Scholar]
- 31.Ahlquist DA, Sargent DJ, Loprinzi CL, Levin TR, Rex DK, Ahnen DJ, Knigge K, Lance MP, Burgart LJ, Hamilton SR, et al. Stool DNA and occult blood testing for screen detection of colorectal neoplasia. Ann Intern Med. 2008;149:441–450, W81. doi: 10.7326/0003-4819-149-7-200810070-00004. [DOI] [PMC free article] [PubMed] [Google Scholar]


