Abstract
The significance of horizontal gene transfer (HGT) in eukaryotic evolution remains controversial. Although many eukaryotic genes are of bacterial origin, they are often interpreted as being derived from mitochondria or plastids. Because of their fixed gene pool and gene loss, however, mitochondria and plastids alone cannot adequately explain the presence of all, or even the majority, of bacterial genes in eukaryotes. Available data indicate that no insurmountable barrier to HGT exists, even in complex multicellular eukaryotes. In addition, the discovery of both recent and ancient HGT events in all major eukaryotic groups suggests that HGT has been a regular occurrence throughout the history of eukaryotic evolution. A model of HGT is proposed that suggests both unicellular and early developmental stages as likely entry points for foreign genes into multicellular eukaryotes.
Keywords: endosymbiosis, eukaryotic evolution, gene acquisition, genome evolution, organellar gene transfer
Introduction
About a decade ago, Doolittle et al. raised a question about the number of bacterial genes in protists, speculating that many bacterial genes should have accumulated in genomes of protists through feeding activities 1,2. Back then, horizontal gene transfer (HGT) had been documented widely as a mechanism to gain foreign genetic materials in prokaryotes, but remained largely an exotic concept in eukaryotes, with little substantial evidence. It is now clear that HGT has occurred in all major eukaryotic lineages. Horizontally acquired genes are not only frequent in unicellular eukaryotes 3–5, but also found in various multicellular eukaryotes, including cnidarians 6,7, mites 8, insects 9–12, nematodes 13–15, fish 16, and land plants 17–22. Although reports of HGT in eukaryotes are still frequently met with skepticism, evidence for HGT throughout eukaryotic evolution is abundant and increasing.
In this paper, I discuss issues related to HGT in eukaryotes. Because most foreign genes reported in eukaryotes thus far are from bacteria, I will focus on bacterial genes. I argue that many bacterial genes in eukaryotes cannot be explained simply as gene transfers from mitochondria or plastids; rather, HGT in eukaryotes should be widespread and expected. Further, I propose a mechanism for integration of foreign DNA into eukaryotic genomes during unicellular or early developmental stages, when their nuclear DNA is relatively exposed to potential sources of HGT.
No barrier to HGT in eukaryotes is insurmountable
To understand the scale of HGT in eukaryotes, it is important to consider the presumed barriers to gene acquisition. Historically, protists were thought to be more likely to acquire genes than multicellular eukaryotes 1,3. Many protists are intimately associated with bacteria or microbial eukaryotes, which serve as food sources, pathogens, or symbionts. Certain species (e.g. Acanthamoeba sp.) can harbor a wide range of bacterial endosymbionts; in effect, they are training camps for bacterial adaptation to the intracellular environments of eukaryotic hosts 23,24. Although mitochondria and plastids – derived from α-proteobacterial and cyanobacterial endosymbionts, respectively – often receive the most prominent recognition, their bacterial ancestors likely represented only a portion of microbial diversity present within their ancient host cells. Temporal, transient, or obligate endosymbionts occur in many organisms 25, and differences between obligate endosymbionts and organelles can be marginal. For example, there has been heated debate over whether Prochlorococcus-related cyanobacterial endosymbionts (chromatophores) in Paulinella should be considered organelles 26–28. Although not to the same extent as in mitochondria and plastids, chromatophore genomes are highly reduced 29, with some essential genes transferred to the nucleus 30. Translocation machinery also has evolved to re-import the protein products of transferred genes back into chromatophores 31,32.
Relative to protists, HGT is often assumed to be rare in complex multicellular eukaryotes because the physical isolation of germ cells from somatic cells may prevent foreign genes from being transmitted to offspring 33,34. Despite this assumption, recent HGT events have been frequently reported in both animals and plants 8,11,19–21,35,36. Some of these horizontally acquired genes in animals and plants are derived from symbionts, either inside or outside germ cells 7,10,36, whereas others are from free-living organisms 16. The magnitude of HGT from obligate bacterial endosymbionts to their insect hosts can be staggering 10. In some other cases, genes even were acquired from other sources to help maintain obligate endosymbionts 37. HGT appears to be particularly frequent in plant mitochondria 38–40, which often harbor mitochondrial genes from distantly related plants. It has been speculated that the acquisition of foreign genes in plant mitochondria was mediated through parasitism, transfer agents (e.g. insects, pathogens, or viruses), illegitimate pollination or other mechanisms 38,41. In any case, these foreign genes must ultimately pass through germ cells to be transmitted to the mitochondria of offspring. These data suggest that, although isolated germ cells may indeed be barriers to HGT in animals and plants, they are not insurmountable.
Bacterial genes in eukaryotes: How many are of organellar origin?
Given that barriers to HGT clearly are not insurmountable, we can examine available data to contemplate the number of HGT-derived genes in eukaryotes. It is well known that eukaryotic genomes contain many bacterial genes 42. Because of the α-proteobacterial and cyanobacterial origins of mitochondria and plastids, respectively, these genes are often presumed to predominantly be mitochondrial or plastid derivations 43–45. Indeed, a supertree analysis of 185 genomes from all three domains of life revealed that the strongest phylogenetic signals among eukaryotic genes come from cyanobacteria, α-proteobacteria, and archaebacteria. Presumably these are attributable to plastids, mitochondria and an archaeon that was likely involved in the origin of eukaryotic cells 46. However, strong signals also exist for different proteobacterial groups (16.7%) as well as for various other bacteria (13.8%), raising the question: how many of these remaining bacterial genes are indeed of mitochondrial or plastid origin?
It can be argued that all these bacterial genes are possibly derived from mitochondria or plastids. Prokaryotic genomes are fluid and shaped by constant gene acquisition and gene loss 47,48. Over time, such fluidity could erase the α-proteobacterial or cyanobacterial signal of an organellar gene 45,49,50. This scenario is not unlikely, particularly if the ancient progenitor of mitochondria or plastids was phylogenetically basal to extant α-proteobacteria or cyanobacteria; a single homologous replacement in either the endosymbiotic ancestor or its sister taxon could completely obliterate the true phylogenetic identity of an organelle-derived gene. Fluid prokaryotic genomes, however, appear not to be a significant issue for identifying plastid-derived genes. The majority of functional genes in plastid genomes are either closely related or highly similar to cyanobacterial sequences 51, despite evidence for HGT to the cyanobacterial progenitor of plastids 52. Similarly, cyanobacterial signal remains the strongest for all bacterial genes in eukaryotes 46, even though some plastid-derived genes might have evolved rapidly as a result of functional decoupling from plastids 53. Therefore, it is doubtful that the fluidity of prokaryotic genomes could account for all or the majority of genes of other bacterial origins.
The argument that all or most bacterial genes in eukaryotes are of organellar origin is inherently linked to the dynamic process of DNA transfer from organelles to the nucleus 43,54,55. During the evolution of mitochondria and plastids, many organellar genes were either gradually lost or transferred to the nucleus. I here call the latter process organellar gene transfer (OGT) to distinguish it from endosymbiotic gene transfer (EGT), which also frequently involves other endosymbionts. Indeed, OGT has led to the incorporation of numerous genes of organellar origin into the nuclear genome. However, it is rarely mentioned that the transfer of functional genes from mitochondria or plastids to the nucleus is constrained by the gene pool of these organelles. Genes acquired from mitochondria and plastids in any eukaryotic genome must be fewer in number than the original gene contents of the α-proteobacterial and cyanobacterial progenitors of two organelles. Although ongoing DNA transfer from organelles to the nucleus has been demonstrated experimentally 56,57, functional genes are rarely involved. Conversely, frequent gene loss events during organellar evolution, occurring independently or as a result of OGT, led to increasingly reduced transferrable gene pool in organellar genomes 54,58. Major reductions of genomes are common features of many endosymbionts, bacterial, or eukaryotic 58,59. Although the scale of gene loss versus transfer to the nucleus is not entirely clear, the loss of organellar genes can be significant in some organisms 60,61. For instance, the tremendous loss of organellar genes is particularly evident in apicomplexans, both the plastid and mitochondrial genomes of which are highly reduced or completely lost 62,63; their nuclear genomes, with genes presumably derived from five genomes (including plastid, mitochondrial, and nuclear genomes of an algal endosymbiont), sometimes contain less than 4,000 genes 62,64,65. Once a gene has been lost from the organellar genome, it is likely gone forever unless a homolog is acquired again from another source, a very rare phenomenon. Over time, the gene pool available for OGT becomes increasingly smaller. In this sense, the process of OGT represents a closed system; it will eventually approach a dead end when the transferable gene pool is depleted. While this does not diminish the significance of OGT in eukaryotic genome evolution, it also suggests that mitochondria and plastids should not be granted unbound power to explain all bacterial genes in eukaryotes.
Compared to OGT, HGT represents an open system, one theoretically allowing genes to be acquired from virtually unlimited sources. Given the ultimate constraint on OGT, many genes of other bacterial origins may at least be equally explained by HGT. Importantly, the large numbers of proteobacterial and cyanobacterial genes in eukaryotes are also consistent with the observation that proteobacteria and cyanobacteria are the most common endosymbionts in many eukaryotic groups (e.g. amoebae 23,66, fungi, 67, plants 68, insects, nematodes, and other animals 69,70). In particular, Wolbachia and Rickettsia are not only common endosymbionts, but also potentially closely related to the bacterial ancestor of mitochondria 43,71. In many respects, they are similar to mitochondria in having reduced genomes resulting from both gene loss and transfer to the nucleus 72,73. In other cases, they might have been secondarily lost, leaving only their genes in the host nucleus 74. If similar proteobacterial endosymbionts existed during early eukaryotic evolution, it would be nearly impossible to distinguish their phylogenetic signal from that of mitochondria.
HGT occurs continually in major eukaryotic lineages
There are many straightforward cases of HGT in eukaryotes that involve recently acquired genes 10,18,36,75,76. Because phylogenetic signal from their donors remains clear, these recently acquired genes can be readily identified. There are also cases of convergent gene acquisitions or recurrent transfers of the same genes. For instance, acquisition of genes encoding enzymes for plant cell wall degradation occurred independently in multiple major lineages 15,77,78. Recurrent HGT events involving bacteria as ultimate donors have been observed in plants, choanoflagellates, amoebae, and others 17,78,79. Not only can HGT lead to acquisition of individual genes, but also entire metabolic pathways 11,14,76,80,81. The proficiency of some eukaryotes in acquiring foreign genes is further evidenced by their stunning ability to recycle plastids 82,83. In some cases, protein products of horizontally acquired genes function in permanently established plastids 84,85; in others, transient plastids were stolen from algal prey and genes were acquired for plastid maintenance 86. These observations showcase HGT as a dynamic process in eukaryotes, and one that is usually under-appreciated.
The dynamic nature of HGT is also reflected in its continual occurrence over time. Although complex multicellular eukaryotes appear to have fewer recently acquired genes than protists, this does not diminish the possibility that they acquired many genes more anciently. Unicellularity is the most common form of eukaryotic life, and it is known that unicellular eukaryotes are prone to HGT 3,4. In fact, acquired genes can be found in numerous unicellular eukaryotes including many obligate intracellular parasites 76,87–89, which often have streamlined genomes and retain fewer foreign genes. The fact that all multicellular eukaryotes descend from unicellular ancestors points to potentially more frequent ancient HGT 20,90. Indeed, foreign genes were introduced regularly at major historical stages during the evolution of primary photosynthetic eukaryotes 17,20,91–96. In some unicellular eukaryotes such as rumen ciliates and red alga Galdieria sulphuraria, acquired genes account for 4–5% of the total genome 77,96; similarly, HGT has contributed to over 3% of the nematode genome 97 and at least 8–9% of the gene content in bdelloid rotifers 98. These numbers may still be underestimations because of the loss of phylogenetic signal in many anciently acquired genes.
Tracing this dynamic process back to the inception of eukaryotic evolution leads to the following conjecture. If the ancestral eukaryote was indeed chimeric and derived from a symbiosis of bacteria and archaea as often suggested 99,100, then we should expect that the earliest eukaryotes were similar to extant prokaryotes in proficiency of DNA uptake. Thus, they probably acquired many genes from external sources. Similarly, if the host cell or the ancestral eukaryote was able to engulf the proteobacterial progenitor of mitochondria, it might engulf other bacteria or bacterial DNA as well. Assuming the nucleus or other unique eukaryotic features did not appear in a sudden event, specific barriers to HGT now present in eukaryotic cells would have evolved gradually. Therefore, HGT during early eukaryotic evolution might occur as frequently as in modern bacteria and archaea, allowing foreign genes to trickle into early eukaryotes continually. Even with a low fixation rate, many foreign genes could have accumulated in extant eukaryotes over a long evolutionary time period 1,2. This process theoretically is open-ended and could have introduced miscellaneous genes independently in different lineages through time. Conceivably, genes acquired later from other sources might have replaced mitochondria- or plastids-derived homologs in the nucleus, explaining in part the observation that many proteins of non-organellar origin function in organelles in various lineages 84,85,94.
The assumption that organelles are the sole or primary source of bacterial genes not only contradicts many apparent cases of HGT in eukaryotes 3–5,35, but also provides little explanation for eukaryotic adaptation to diverse habitats. Adaptation to shifting environments is often accompanied by acquisition of new genes and loss of others 47,101. Bacteria and archaea are able to adapt to their environments by sampling from a large global gene pool and maintaining fluid genomes 47,48. If novel genetic information cannot be achieved, the adaptability of any eukaryote with a limited gene pool will be hampered. Particularly for early eukaryotes, the task for surviving in various niches and further diversifying into major lineages was probably daunting. Given the fact that point mutation, recombination, gene duplication, and genome rearrangement only operate on pre-existing genes, it would have been disadvantageous for early eukaryotes to completely abandon their ability to acquire ready-to-use genes from other sources.
The weak-link model explains frequent HGT in eukaryotes
One of the most popular models for HGT in eukaryotes is the gene ratchet mechanism proposed by Doolittle 1,2. Under this model, bacteria phagocytized by protists as food sources are lysed within host cells, allowing their DNA to be incorporated into host genomes. Though elegant, this model does not explain the widespread existence of foreign genes in eukaryotes that do not engage in phagocytosis. Given the occurrence of HGT in eukaryotes with miscellaneous lifestyles 3–5,35, I here offer a different perspective on HGT mechanisms in eukaryotes.
For any foreign gene to be acquired and stably inherited by a recipient organism, it must (i) enter recipient cells, (ii) be integrated into the recipient genome, and (iii) be transmitted to offspring. Foreign genes could enter cells of the recipient organism at any weakly protected stage of the lifecycle in natural environments. This process could be facilitated if the recipient and donor organisms maintain an intimate physical association through symbiosis, parasitism, infection, or other known forms of contact 18,35. For unicellular eukaryotes, the HGT process could be similar to the gene ratchet mechanism proposed by Doolittle 1, but does not specifically require feeding activities for foreign genes to enter recipient cells. Once inside the cell, integration of foreign genes into recipient genomes does not appear to be particularly difficult, given the still frequent movement of organellar DNA fragments into the nucleus in different organisms 56,57. Even if the initial attempt to integrate into the nuclear genome fails, as long as foreign genes can enter recipient cells without significant difficulties, successive attempts may eventually lead to successful integration of foreign genes into the recipient genome 1. Subsequently, transmission of integrated foreign genes to offspring can be accomplished simply through mitosis in these unicellular organisms (Fig. 1).
Figure 1.
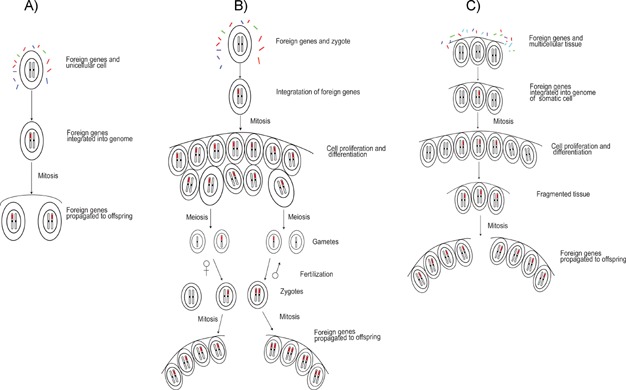
Illustration of the weak-link model of HGT. The unicellular or early developmental stages (spore, zygotes, embryos, etc.) are exposed to foreign genes. These weakly protected stages allow the entry and integration of foreign genes. A: In unicellular eukaryotes, foreign genes may be directly transmitted to offspring through mitosis. B: In multicellular eukaryotes with sexual reproduction, cell proliferation and differentiation spread the foreign genes to all cells including germ cells, which then give rise to male and female gametes. Subsequent fertilization allows foreign genes to be transmitted to offspring. C: In asexual multicellular eukaryotes, propagation of cells carrying foreign genes allows the foreign genes to be transmitted to offspring.
For complex multicellular eukaryotes such as plants and animals, foreign genes must be passed through specialized reproductive cells (germ lines) to be transmitted to offspring. Therefore, the isolation of germ cells from somatic cells is often considered to be the major barrier to HGT in animals and, to a lesser extent, higher plants 33,34. Overcoming this barrier is certainly not easy, but possible. For example, the close association of Wolbachia with germ cells of arthropods or acquisition of bacterial endosymbionts during embryonic development could promote stable HGT from these endosymbionts to their hosts 9,10. Similarly, if a plant or an animal is exposed to and readily incorporates foreign DNA during its very early developmental stages, subsequent cell proliferation and differentiation will spread these foreign genes to other tissues, including germ cells (Fig. 1). For instance, in nonvascular and seedless vascular plants, female gametes are weakly protected in archegonia and exposed to external environments during fertilization, and male gametes generally are exposed completely prior to reaching an oocyte. External fertilization occurs in animals inhabiting aquatic environments, meaning gametes and zygotes are, likewise, freely exposed to foreign sources of DNA. Structurally internalized gametes in seed plants and animals in terrestrial environments may be protected from mechanical damages, but not necessarily foreign DNA from symbiotic bacteria, pathogens, or other microbes 8,10,19,38. Foreign genes introduced during zygotic or embryonic development will be propagated through mitosis into germ cells and, therefore, next generation. Propagation of foreign genes also is possible through gene transfer among neighboring cells, as demonstrated in natural plant grafts 102,103. In these respects, the entry points in early developmental stages represent the weak link in recipient organisms for initiating foreign gene transfer; as such, they ultimately control the transmission of foreign genes to offspring. Once foreign genes are passed onto offspring, they can be fixed in the population through drift or positive selection on newly acquired functions.
This model of gene acquisition critically depends on the presence of weak or unprotected points for foreign genes to enter recipient cells. Other than early developmental stages (e.g. zygotes, embryos, or spores) of multicellular eukaryotes, weak-link entry points for foreign genes include all lifecycle stages in unicellular eukaryotes. In multicellular eukaryotes with complex sexual reproduction, the propagation of foreign genes through cell proliferation and differentiation does not require direct contact between the donor and differentiated reproductive tissues of the recipient (Fig. 1). In multicellular eukaryotes with asexual reproduction, this process allows foreign genes to be transmitted directly to offspring by mitotic propagation of cells carrying these genes (Fig. 1). Because foreign genes are expected to decay into pseudogenes if not selectively advantageous to the recipient organism, the actual number of acquired genes should vary among organisms of different lifestyles. However, given the potential role of HGT in allowing organisms to explore new resources and niches 47,48,101,104, foreign genes with novel functions could be fixed more frequently in recipients under resource limitation or in shifting environments.
This model also makes the following specific predictions regarding the occurrence or overall frequency of HGT in eukaryotes of different lifestyles:
Frequent HGT in unicellular eukaryotes. Since all developmental stages of unicellular eukaryotes represent weak-link entry points, there are ample opportunities for foreign genes to be integrated and, therefore, transmitted to offspring.
Occurrence of foreign genes in multicellular eukaryotes with fully exposed unicellular or early developmental stages (e.g. spores, zygotes, or embryos) in their lifecycles (see above).
Frequent HGT in asexual multicellular eukaryotes. The absence of specific germ cells means that any cell carrying foreign genes may propagate them into offspring. The frequency of HGT should be even higher if bacterial endosymbionts exist in asexual structures, such as spores and hyphae in fungi 67.
Existence of many anciently acquired genes in multicellular eukaryotes. Because multicellular eukaryotes are ultimately derived from unicellular ancestors, it is expected that many foreign genes acquired by their unicellular ancestors remain in the genomes of their multicellular descendants.
HGT may still be underestimated in eukaryotes
Despite potentially frequent HGT in many eukaryotes, identification of acquired genes often is complicated. Although foreign genes may gradually accumulate in recipient genomes, their phylogenetic signal tying them to specific source taxa may be muted or completely erased by substitutions over time. Additionally, HGT from uncultivated or extinct bacterial lineages may not be properly identified 105. Even if phylogenetic signal is retained, recovery of accurate phylogenies can be complicated. In particular, many gene families are patchily distributed in prokaryotes and eukaryotes, and explanations of such patchiness can be controversial 106–108.
Interpretations of patchy distributions hinge on underlying assumptions 2. For researchers who view vertical inheritance as the sole or dominant genetic paradigm, HGT rarely offers a satisfying explanation. In such cases, a patchy distribution is best explained by differential gene losses, misidentification of genes, or simply phylogenetic artifacts. Although these factors can create patchy distributions, indiscriminately resorting to them as the chief explanation not only discounts the obvious existence of HGT in many eukaryotes, but also ignores the gene pool constraints from the common ancestor of eukaryotes and progenitors of organelles. Clearly, some reported cases of HGT turn out to be artifacts 4,35, but the existence of some established artifacts does not discount the likelihood of HGT in many other cases.
On the other hand, patchy distributions are easily explained based on current knowledge of HGT. For examples, HGT from prokaryotes, sometimes involving the same genes independently and recurrently 78,109,110, can spread prokaryotic genes among unrelated eukaryotes. Further, the bacterial ancestry of mitochondria and plastids, the widespread distribution of secondary, tertiary, or transient plastids, and the presence of bacterial endosymbionts (e.g. Wolbachia and Rickettsia in animals) in many eukaryotes, are all known to lead to gene transfer and, therefore, bacterial genes in eukaryotic genomes. In such cases, patchy distributions not only are expected, but also clearly reflect the very nature of HGT in eukaryotes 111.
Given the difficulties and complications discussed above, it is important that putative cases of HGT in eukaryotes be investigated carefully. To do so, independent lines of evidence and alternative scenarios should be considered. Many cases of patchy distribution probably reflect combined effects of duplication, gene loss, HGT and other processes 80,112,113. Nevertheless, as long as vertical inheritance remains the null hypothesis, HGT in eukaryotes will likely be underestimated. Therefore, it is useful to bear in mind that HGT, although difficult to “prove” in every individual case, offers a valid explanation for many of the atypical gene distributions in eukaryotes.
Conclusions and outlook
A large percentage of eukaryotic genes are unquestionably of bacterial origin. Because mitochondria and plastids represent fixed gene pools, from which many genes have been lost completely during their evolution, OGT alone cannot adequately explain the large number of bacterial genes in eukaryotic genomes. The occurrence of recent HGT events in all major eukaryotic groups indicates that there are no insurmountable barriers to HGT, even in complex multicellular forms. Additionally, the finding of many anciently acquired genes in eukaryotes suggests that HGT is a dynamic process that has operated continually throughout the history of eukaryotic evolution. The weak-link model of HGT hypothesizes that unicellular and early developmental stages are the most likely entry points for foreign genes into recipient cells. Given the universal existence of these weak-link entry points, HGT is expected to occur frequently, on an evolutionary time scale, in all groups of eukaryotes. The weak-link hypothesis makes several explicit predictions that can be tested either by genome analyses or by experiments under controlled conditions. Future work is critically needed to understand the overall scale of HGT, but also the contribution of HGT, compared to other genetic mechanisms such as de novo gene generation and duplication, to the expansion of gene pool in different eukaryotic lineages throughout evolutionary time. Such work can be accomplished through careful evolutionary genomic analyses and will benefit our understanding of the role of HGT in the innovation and evolution of eukaryotes.
Acknowledgments
I am grateful to John Stiller, Peter Gogarten, and Trip Lamb for their comments and editing the manuscript. I also thank Jipei Yue for discussions and assistance in drawing the diagram. This work is supported in part by an NSF Assembling the Tree of Life (ATOL) grant (DEB 0830024), the CAS/SAFEA International Partnership Program for Creative Research Teams, and an internal grant from the Kunming Institute of Botany, the Chinese Academy of Sciences.
Glossary
- HGT
horizontal gene transfer
- OGT
organellar gene transfer
References
- Doolittle WF. You are what you eat: a gene transfer ratchet could account for bacterial genes in eukaryotic nuclear genomes. Trends Genet. 1998;14:307–11. doi: 10.1016/s0168-9525(98)01494-2. [DOI] [PubMed] [Google Scholar]
- Doolittle WF, Boucher Y, Nesbo CL, Douady CJ, et al. How big is the iceberg of which organellar genes in nuclear genomes are but the tip. Philos Trans R Soc Lond B Biol Sci. 2003;358:39–57;. doi: 10.1098/rstb.2002.1185. discussion 57–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson JO. Lateral gene transfer in eukaryotes. Cell Mol Life Sci. 2005;62:1182–97. doi: 10.1007/s00018-005-4539-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling PJ, Palmer JD. Horizontal gene transfer in eukaryotic evolution. Nat Rev Genet. 2008;9:605–18. doi: 10.1038/nrg2386. [DOI] [PubMed] [Google Scholar]
- Andersson JO. Gene transfer and diversification of microbial eukaryotes. Annu Rev Microbiol. 2009;63:177–93. doi: 10.1146/annurev.micro.091208.073203. [DOI] [PubMed] [Google Scholar]
- Denker E, Bapteste E, Le Guyader H, Manuel M, et al. Horizontal gene transfer and the evolution of cnidarian stinging cells. Curr Biol. 2008;18:R858–9. doi: 10.1016/j.cub.2008.07.031. [DOI] [PubMed] [Google Scholar]
- Chapman JA, Kirkness EF, Simakov O, Hampson SE, et al. The dynamic genome of Hydra. Nature. 2010;464:592–6. doi: 10.1038/nature08830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grbic M, Van Leeuwen T, Clark RM, Rombauts S, et al. The genome of Tetranychus urticae reveals herbivorous pest adaptations. Nature. 2011;479:487–92. doi: 10.1038/nature10640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo N, Nikoh N, Ijichi N, Shimada M, et al. Genome fragment of Wolbachia endosymbiont transferred to X chromosome of host insect. Proc Natl Acad Sci USA. 2002;99:14280–5. doi: 10.1073/pnas.222228199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotopp JC, Clark ME, Oliveira DC, Foster JM, et al. Widespread lateral gene transfer from intracellular bacteria to multicellular eukaryotes. Science. 2007;317:1753–6. doi: 10.1126/science.1142490. [DOI] [PubMed] [Google Scholar]
- Moran NA, Jarvik T. Lateral transfer of genes from fungi underlies carotenoid production in aphids. Science. 2010;328:624–7. doi: 10.1126/science.1187113. [DOI] [PubMed] [Google Scholar]
- Nikoh N, McCutcheon JP, Kudo T, Miyagishima SY, et al. Bacterial genes in the aphid genome: absence of functional gene transfer from Buchnera to its host. PLoS Genet. 2010;6:e1000827. doi: 10.1371/journal.pgen.1000827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholl EH, Thorne JL, McCarter JP, Bird DM. Horizontally transferred genes in plant-parasitic nematodes: a high-throughput genomic approach. Genome Biol. 2003;4:R39. doi: 10.1186/gb-2003-4-6-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig JP, Bekal S, Hudson M, Domier L, et al. Analysis of a horizontally transferred pathway involved in vitamin B6 biosynthesis from the soybean cyst nematode Heterodera glycines. Mol Biol Evol. 2008;25:2085–98. doi: 10.1093/molbev/msn141. [DOI] [PubMed] [Google Scholar]
- Danchin EG, Rosso MN, Vieira P, de Almeida-Engler J, et al. Multiple lateral gene transfers and duplications have promoted plant parasitism ability in nematodes. Proc Natl Acad Sci USA. 2010;107:17651–6. doi: 10.1073/pnas.1008486107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham LA, Lougheed SC, Ewart KV, Davies PL. Lateral transfer of a lectin-like antifreeze protein gene in fishes. PLoS One. 2008;3:e2616. doi: 10.1371/journal.pone.0002616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards TA, Soanes DM, Foster PG, Leonard G, et al. Phylogenomic analysis demonstrates a pattern of rare and ancient horizontal gene transfer between plants and fungi. Plant Cell. 2009;21:1897–911. doi: 10.1105/tpc.109.065805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S, Maruyama S, Nozaki H, Shirasu K. Horizontal gene transfer by the parasitic plant Striga hermonthica. Science. 2010;328:1128. doi: 10.1126/science.1187145. [DOI] [PubMed] [Google Scholar]
- Christin PA, Edwards EJ, Besnard G, Boxall SF, et al. Adaptive evolution of C(4) photosynthesis through recurrent lateral gene transfer. Curr Biol. 2012;22:445–9. doi: 10.1016/j.cub.2012.01.054. [DOI] [PubMed] [Google Scholar]
- Yue J, Hu X, Sun H, Yang Y, et al. Widespread impact of horizontal gene transfer on plant colonization of land. Nat Commun. 2012;3:1152. doi: 10.1038/ncomms2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Z, Bradley RK, Wurdack KJ, Wong K, et al. Horizontal transfer of expressed genes in a parasitic flowering plant. BMC Genomics. 2012;13:227. doi: 10.1186/1471-2164-13-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Fernandez-Aparicio M, Wafula EK, Das M, et al. Evolution of a horizontally acquired legume gene, albumin 1, in the parasitic plant Phelipanche aegyptiaca and related species. BMC Evol Biol. 2013;13:48. doi: 10.1186/1471-2148-13-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn M, Wagner M. Bacterial endosymbionts of free-living amoebae. J Eukaryot Microbiol. 2004;51:509–14. doi: 10.1111/j.1550-7408.2004.tb00278.x. [DOI] [PubMed] [Google Scholar]
- Schmitz-Esser S, Toenshoff ER, Haider S, Heinz E, et al. Diversity of bacterial endosymbionts of environmental acanthamoeba isolates. Appl Environ Microbiol. 2008;74:5822–31. doi: 10.1128/AEM.01093-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowack EC, Melkonian M. Endosymbiotic associations within protists. Philos Trans R Soc Lond B Biol Sci. 2010;365:699–712. doi: 10.1098/rstb.2009.0188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theissen U, Martin W. The difference between organelles and endosymbionts. Curr Biol. 2006;16:R1016–7. doi: 10.1016/j.cub.2006.11.020. author reply R7-8. [DOI] [PubMed] [Google Scholar]
- Bodyl A, Mackiewicz P, Stiller JW. The intracellular cyanobacteria of Paulinella chromatophora: endosymbionts or organelles. Trends Microbiol. 2007;15:295–6. doi: 10.1016/j.tim.2007.05.002. [DOI] [PubMed] [Google Scholar]
- Keeling PJ, Archibald JM. Organelle evolution: what's in a name. Curr Biol. 2008;18:R345–7. doi: 10.1016/j.cub.2008.02.065. [DOI] [PubMed] [Google Scholar]
- Nowack EC, Melkonian M, Glockner G. Chromatophore genome sequence of Paulinella sheds light on acquisition of photosynthesis by eukaryotes. Curr Biol. 2008;18:410–8. doi: 10.1016/j.cub.2008.02.051. [DOI] [PubMed] [Google Scholar]
- Nowack EC, Vogel H, Groth M, Grossman AR, et al. Endosymbiotic gene transfer and transcriptional regulation of transferred genes in Paulinella chromatophora. Mol Biol Evol. 2011;28:407–22. doi: 10.1093/molbev/msq209. [DOI] [PubMed] [Google Scholar]
- Mackiewicz P, Bodyl A, Gagat P. Possible import routes of proteins into the cyanobacterial endosymbionts/plastids of Paulinella chromatophora. Theory Biosci. 2012;131:1–18. doi: 10.1007/s12064-011-0147-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowack EC, Grossman AR. Trafficking of protein into the recently established photosynthetic organelles of Paulinella chromatophora. Proc Natl Acad Sci USA. 2012;109:5340–5. doi: 10.1073/pnas.1118800109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurland CG, Canback B, Berg OG. Horizontal gene transfer: a critical view. Proc Natl Acad Sci USA. 2003;100:9658–62. doi: 10.1073/pnas.1632870100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock R. The give-and-take of DNA: horizontal gene transfer in plants. Trends Plant Sci. 2010;15:11–22. doi: 10.1016/j.tplants.2009.10.001. [DOI] [PubMed] [Google Scholar]
- Dunning Hotopp JC. Horizontal gene transfer between bacteria and animals. Trends Genet. 2011;27:157–63. doi: 10.1016/j.tig.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acuna R, Padilla BE, Florez-Ramos CP, Rubio JD, et al. Adaptive horizontal transfer of a bacterial gene to an invasive insect pest of coffee. Proc Natl Acad Sci USA. 2012;109:4197–202. doi: 10.1073/pnas.1121190109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikoh N, Nakabachi A. Aphids acquired symbiotic genes via lateral gene transfer. BMC Biol. 2009;7:12. doi: 10.1186/1741-7007-7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergthorsson U, Richardson AO, Young GJ, Goertzen LR, et al. Massive horizontal transfer of mitochondrial genes from diverse land plant donors to the basal angiosperm Amborella. Proc Natl Acad Sci USA. 2004;101:17747–52. doi: 10.1073/pnas.0408336102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Z, Wang Y, Bradley RK, Sugumaran M, et al. Massive mitochondrial gene transfer in a parasitic flowering plant clade. PLoS Genet. 2013;9:e1003265. doi: 10.1371/journal.pgen.1003265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mower JP, Stefanovic S, Hao W, Gummow JS, et al. Horizontal acquisition of multiple mitochondrial genes from a parasitic plant followed by gene conversion with host mitochondrial genes. BMC Biol. 2010;8:150. doi: 10.1186/1741-7007-8-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson AO, Palmer JD. Horizontal gene transfer in plants. J Exp Bot. 2007;58:1–9. doi: 10.1093/jxb/erl148. [DOI] [PubMed] [Google Scholar]
- Koonin EV, Fedorova ND, Jackson JD, Jacobs AR, et al. A comprehensive evolutionary classification of proteins encoded in complete eukaryotic genomes. Genome Biol. 2004;5:R7. doi: 10.1186/gb-2004-5-2-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esser C, Ahmadinejad N, Wiegand C, Rotte C, et al. A genome phylogeny for mitochondria among alpha-proteobacteria and a predominantly eubacterial ancestry of yeast nuclear genes. Mol Biol Evol. 2004;21:1643–60. doi: 10.1093/molbev/msh160. [DOI] [PubMed] [Google Scholar]
- Koonin EV. Darwinian evolution in the light of genomics. Nucleic Acids Res. 2009;37:1011–34. doi: 10.1093/nar/gkp089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiergart T, Landan G, Schenk M, Dagan T, et al. An evolutionary network of genes present in the eukaryote common ancestor polls genomes on eukaryotic and mitochondrial origin. Genome Biol Evol. 2012;4:466–85. doi: 10.1093/gbe/evs018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisani D, Cotton JA, McInerney JO. Supertrees disentangle the chimerical origin of eukaryotic genomes. Mol Biol Evol. 2007;24:1752–60. doi: 10.1093/molbev/msm095. [DOI] [PubMed] [Google Scholar]
- Ochman H, Lawrence JG, Groisman EA. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405:299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- Gogarten JP, Doolittle WF, Lawrence JG. Prokaryotic evolution in light of gene transfer. Mol Biol Evol. 2002;19:2226–38. doi: 10.1093/oxfordjournals.molbev.a004046. [DOI] [PubMed] [Google Scholar]
- Esser C, Martin W, Dagan T. The origin of mitochondria in light of a fluid prokaryotic chromosome model. Biol Lett. 2007;3:180–4. doi: 10.1098/rsbl.2006.0582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards TA, Archibald JM. Cell evolution: gene transfer agents and the origin of mitochondria. Curr Biol. 2011;21:R112–4. doi: 10.1016/j.cub.2010.12.036. [DOI] [PubMed] [Google Scholar]
- Rice DW, Palmer JD. An exceptional horizontal gene transfer in plastids: gene replacement by a distant bacterial paralog and evidence that haptophyte and cryptophyte plastids are sisters. BMC Biol. 2006;4:31. doi: 10.1186/1741-7007-4-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross J, Meurer J, Bhattacharya D. Evidence of a chimeric genome in the cyanobacterial ancestor of plastids. BMC Evol Biol. 2008;8:117. doi: 10.1186/1471-2148-8-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin W, Hazkani-Covo E, Shavit-Greivink L, Schmitt V, et al. Gene transfers from organelles to the nucleus: How much, what happens, and why none in Elysia. J Endocytobiosis Cell Res. 2012;23:16–20. [Google Scholar]
- Martin W, Herrmann RG. Gene transfer from organelles to the nucleus: how much, what happens, and Why. Plant Physiol. 1998;118:9–17. doi: 10.1104/pp.118.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noutsos C, Richly E, Leister D. Generation and evolutionary fate of insertions of organelle DNA in the nuclear genomes of flowering plants. Genome Res. 2005;15:616–28. doi: 10.1101/gr.3788705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CY, Ayliffe MA, Timmis JN. Direct measurement of the transfer rate of chloroplast DNA into the nucleus. Nature. 2003;422:72–6. doi: 10.1038/nature01435. [DOI] [PubMed] [Google Scholar]
- Stegemann S, Hartmann S, Ruf S, Bock R. High-frequency gene transfer from the chloroplast genome to the nucleus. Proc Natl Acad Sci USA. 2003;100:8828–33. doi: 10.1073/pnas.1430924100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutcheon JP, Moran NA. Extreme genome reduction in symbiotic bacteria. Nat Rev Microbiol. 2012;10:13–26. doi: 10.1038/nrmicro2670. [DOI] [PubMed] [Google Scholar]
- Archibald JM, Lane CE. Going, going, not quite gone: nucleomorphs as a case study in nuclear genome reduction. J Hered. 2009;100:582–90. doi: 10.1093/jhered/esp055. [DOI] [PubMed] [Google Scholar]
- Huang J, Mullapudi N, Lancto CA, Scott M, et al. Phylogenomic evidence supports past endosymbiosis, intracellular and horizontal gene transfer in Cryptosporidium parvum. Genome Biol. 2004;5:R88. doi: 10.1186/gb-2004-5-11-r88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison HG, McArthur AG, Gillin FD, Aley SB, et al. Genomic minimalism in the early diverging intestinal parasite Giardia lamblia. Science. 2007;317:1921–6. doi: 10.1126/science.1143837. [DOI] [PubMed] [Google Scholar]
- Brayton KA, Lau AO, Herndon DR, Hannick L, et al. Genome sequence of Babesia bovis and comparative analysis of apicomplexan hemoprotozoa. PLoS Pathog. 2007;3:1401–13. doi: 10.1371/journal.ppat.0030148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissinger JC, DeBarry J. Genome cartography: charting the apicomplexan genome. Trends Parasitol. 2011;27:345–54. doi: 10.1016/j.pt.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pain A, Renauld H, Berriman M, Murphy L, et al. Genome of the host-cell transforming parasite Theileria annulata compared with T. parva. Science. 2005;309:131–3. doi: 10.1126/science.1110418. [DOI] [PubMed] [Google Scholar]
- Abrahamsen MS, Templeton TJ, Enomoto S, Abrahante JE, et al. Complete genome sequence of the apicomplexan, Cryptosporidium parvum. Science. 2004;304:441–5. doi: 10.1126/science.1094786. [DOI] [PubMed] [Google Scholar]
- Taylor M, Mediannikov O, Raoult D, Greub G. Endosymbiotic bacteria associated with nematodes, ticks and amoebae. FEMS Immunol Med Microbiol. 2012;64:21–31. doi: 10.1111/j.1574-695X.2011.00916.x. [DOI] [PubMed] [Google Scholar]
- Kobayashi DY, Crouch JA. Bacterial/fungal interactions: from pathogens to mutualistic endosymbionts. Annu Rev Phytopathol. 2009;47:63–82. doi: 10.1146/annurev-phyto-080508-081729. [DOI] [PubMed] [Google Scholar]
- Reinhold-Hurek B, Hurek T. Living inside plants: bacterial endophytes. Curr Opin Plant Biol. 2011;14:435–43. doi: 10.1016/j.pbi.2011.04.004. [DOI] [PubMed] [Google Scholar]
- Erwin PM, Lopez-Legentil S, Gonzalez-Pech R, Turon X. A specific mix of generalists: bacterial symbionts in Mediterranean Ircinia spp. FEMS Microbiol Ecol. 2012;79:619–37. doi: 10.1111/j.1574-6941.2011.01243.x. [DOI] [PubMed] [Google Scholar]
- Moran NA, Baumann P. Bacterial endosymbionts in animals. Curr Opin Microbiol. 2000;3:270–5. doi: 10.1016/s1369-5274(00)00088-6. [DOI] [PubMed] [Google Scholar]
- Gray MW, Burger G, Lang BF. The origin and early evolution of mitochondria. Genome Biol. 2001;2:REVIEWS1018. doi: 10.1186/gb-2001-2-6-reviews1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Sun LV, Vamathevan J, Riegler M, et al. Phylogenomics of the reproductive parasite Wolbachia pipientis wMel: a streamlined genome overrun by mobile genetic elements. PLoS Biol. 2004;2:E69. doi: 10.1371/journal.pbio.0020069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster J, Ganatra M, Kamal I, Ware J, et al. The Wolbachia genome of Brugia malayi: endosymbiont evolution within a human pathogenic nematode. PLoS Biol. 2005;3:e121. doi: 10.1371/journal.pbio.0030121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNulty SN, Foster JM, Mitreva M, Dunning Hotopp JC, et al. Endosymbiont DNA in endobacteria-free filarial nematodes indicates ancient horizontal genetic transfer. PLoS One. 2010;5:e11029. doi: 10.1371/journal.pone.0011029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Koning AP, Brinkman FS, Jones SJ, Keeling PJ. Lateral gene transfer and metabolic adaptation in the human parasite Trichomonas vaginalis. Mol Biol Evol. 2000;17:1769–73. doi: 10.1093/oxfordjournals.molbev.a026275. [DOI] [PubMed] [Google Scholar]
- Striepen B, Pruijssers AJ, Huang J, Li C, et al. Gene transfer in the evolution of parasite nucleotide biosynthesis. Proc Natl Acad Sci USA. 2004;101:3154–9. doi: 10.1073/pnas.0304686101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricard G, McEwan NR, Dutilh BE, Jouany JP, et al. Horizontal gene transfer from bacteria to rumen ciliates indicates adaptation to their anaerobic, carbohydrates-rich environment. BMC Genomics. 2006;7:22. doi: 10.1186/1471-2164-7-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun G, Yang Z, Ishwar A, Huang J. Algal genes in the closest relatives of animals. Mol Biol Evol. 2010;27:2879–89. doi: 10.1093/molbev/msq175. [DOI] [PubMed] [Google Scholar]
- Andersson JO. Evolution of patchily distributed proteins shared between eukaryotes and prokaryotes: Dictyostelium as a case study. J Mol Microbiol Biotechnol. 2011;20:83–95. doi: 10.1159/000324505. [DOI] [PubMed] [Google Scholar]
- Sun G, Huang J. Horizontally acquired DAP pathway as a unit of self-regulation. J Evol Biol. 2011;24:587–95. doi: 10.1111/j.1420-9101.2010.02192.x. [DOI] [PubMed] [Google Scholar]
- Monier A, Pagarete A, de Vargas C, Allen MJ, et al. Horizontal gene transfer of an entire metabolic pathway between a eukaryotic alga and its DNA virus. Genome Res. 2009;19:1441–9. doi: 10.1101/gr.091686.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archibald JM, Keeling PJ. Recycled plastids: a ‘green movement’ in eukaryotic evolution. Trends Genet. 2002;18:577–84. doi: 10.1016/s0168-9525(02)02777-4. [DOI] [PubMed] [Google Scholar]
- Bhattacharya D, Yoon HS, Hackett JD. Photosynthetic eukaryotes unite: endosymbiosis connects the dots. BioEssays. 2004;26:50–60. doi: 10.1002/bies.10376. [DOI] [PubMed] [Google Scholar]
- Archibald JM, Rogers MB, Toop M, Ishida K, et al. Lateral gene transfer and the evolution of plastid-targeted proteins in the secondary plastid-containing alga Bigelowiella natans. Proc Natl Acad Sci USA. 2003;100:7678–83. doi: 10.1073/pnas.1230951100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosenko T, Lidie KL, Van Dolah FM, Lindquist E, et al. Chimeric plastid proteome in the Florida “red tide” dinoflagellate Karenia brevis. Mol Biol Evol. 2006;23:2026–38. doi: 10.1093/molbev/msl074. [DOI] [PubMed] [Google Scholar]
- Wisecaver JH, Hackett JD. Transcriptome analysis reveals nuclear-encoded proteins for the maintenance of temporary plastids in the dinoflagellate Dinophysis acuminata. BMC Genomics. 2010;11:366. doi: 10.1186/1471-2164-11-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson JO, Sjogren AM, Davis LA, Embley TM, et al. Phylogenetic analyses of diplomonad genes reveal frequent lateral gene transfers affecting eukaryotes. Curr Biol. 2003;13:94–104. doi: 10.1016/s0960-9822(03)00003-4. [DOI] [PubMed] [Google Scholar]
- Huang J, Mullapudi N, Sicheritz-Ponten T, Kissinger JC. A first glimpse into the pattern and scale of gene transfer in Apicomplexa. Int J Parasitol. 2004;34:265–74. doi: 10.1016/j.ijpara.2003.11.025. [DOI] [PubMed] [Google Scholar]
- Pombert JF, Selman M, Burki F, Bardell FT, et al. Gain and loss of multiple functionally related, horizontally transferred genes in the reduced genomes of two microsporidian parasites. Proc Natl Acad Sci USA. 2012;109:12638–43. doi: 10.1073/pnas.1205020109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Gogarten JP. Ancient horizontal gene transfer can benefit phylogenetic reconstruction. Trends Genet. 2006;22:361–6. doi: 10.1016/j.tig.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Huang J, Gogarten P. Did an ancient chlamydial endosymbiosis facilitate the establishment of primary plastids. Genome Biol. 2007;8:R99. doi: 10.1186/gb-2007-8-6-r99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyra HM, Linka M, Weber AP, Bhattacharya D. Host origin of plastid solute transporters in the first photosynthetic eukaryotes. Genome Biol. 2007;8:R212. doi: 10.1186/gb-2007-8-10-r212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Gogarten JP. Concerted gene recruitment in early plant evolution. Genome Biol. 2008;9:R109. doi: 10.1186/gb-2008-9-7-r109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Miyagishima SY. Eukaryotic and eubacterial contributions to the establishment of plastid proteome estimated by large-scale phylogenetic analyses. Mol Biol Evol. 2010;27:581–90. doi: 10.1093/molbev/msp273. [DOI] [PubMed] [Google Scholar]
- Price DC, Chan CX, Yoon HS, Yang EC, et al. Cyanophora paradoxa genome elucidates origin of photosynthesis in algae and plants. Science. 2012;335:843–7. doi: 10.1126/science.1213561. [DOI] [PubMed] [Google Scholar]
- Schonknecht G, Chen WH, Ternes CM, Barbier GG, et al. Gene transfer from bacteria and archaea facilitated evolution of an extremophilic eukaryote. Science. 2013;339:1207–10. doi: 10.1126/science.1231707. [DOI] [PubMed] [Google Scholar]
- Paganini J, Campan-Fournier A, Da Rocha M, Gouret P, et al. Contribution of lateral gene transfers to the genome composition and parasitic ability of root-knot nematodes. PLoS One. 2012;7:e50875. doi: 10.1371/journal.pone.0050875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boschetti C, Carr A, Crisp A, Eyres I, et al. Biochemical diversification through foreign gene expression in bdelloid rotifers. PLoS Genet. 2012;8:e1003035. doi: 10.1371/journal.pgen.1003035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV. The origin and early evolution of eukaryotes in the light of phylogenomics. Genome Biol. 2010;11:209. doi: 10.1186/gb-2010-11-5-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin WF. Early evolution without a tree of life. Biol Direct. 2011;6:36. doi: 10.1186/1745-6150-6-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling PJ. Functional and ecological impacts of horizontal gene transfer in eukaryotes. Curr Opin Genet Dev. 2009;19:613–9. doi: 10.1016/j.gde.2009.10.001. [DOI] [PubMed] [Google Scholar]
- Stegemann S, Bock R. Exchange of genetic material between cells in plant tissue grafts. Science. 2009;324:649–51. doi: 10.1126/science.1170397. [DOI] [PubMed] [Google Scholar]
- Stegemann S, Keuthe M, Greiner S, Bock R. Horizontal transfer of chloroplast genomes between plant species. Proc Natl Acad Sci USA. 2012;109:2434–8. doi: 10.1073/pnas.1114076109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haegeman A, Jones JT, Danchin EG. Horizontal gene transfer in nematodes: a catalyst for plant parasitism. Mol Plant Microbe Interact. 2011;24:879–87. doi: 10.1094/MPMI-03-11-0055. [DOI] [PubMed] [Google Scholar]
- Fournier GP, Huang J, Gogarten JP. Horizontal gene transfer from extinct and extant lineages: biological innovation and the coral of life. Philos Trans R Soc Lond B Biol Sci. 2009;364:2229–39. doi: 10.1098/rstb.2009.0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble GP, Rogers MB, Keeling PJ. Complex distribution of EFL and EF-1alpha proteins in the green algal lineage. BMC Evol Biol. 2007;7:82. doi: 10.1186/1471-2148-7-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocquyt E, Verbruggen H, Leliaert F, Zechman FW, et al. Gain and loss of elongation factor genes in green algae. BMC Evol Biol. 2009;9:39. doi: 10.1186/1471-2148-9-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabova J, Ruzicka P, Verner Z, Hampl V, et al. Experimental examination of EFL and MATX eukaryotic horizontal gene transfers: coexistence of mutually exclusive transcripts predates functional rescue. Mol Biol Evol. 2011;28:2371–8. doi: 10.1093/molbev/msr060. [DOI] [PubMed] [Google Scholar]
- Loftus B, Anderson I, Davies R, Alsmark UC, et al. The genome of the protist parasite Entamoeba histolytica. Nature. 2005;433:865–8. doi: 10.1038/nature03291. [DOI] [PubMed] [Google Scholar]
- Moran Y, Fredman D, Szczesny P, Grynberg M, et al. Recurrent horizontal transfer of bacterial toxin genes to eukaryotes. Mol Biol Evol. 2012;29:2223–30. doi: 10.1093/molbev/mss089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson JO. Phylogenomic approaches underestimate eukaryotic gene transfer. Mob Genet Elements. 2012;2:59–62. doi: 10.4161/mge.19668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson JO, Hirt RP, Foster PG, Roger AJ. Evolution of four gene families with patchy phylogenetic distributions: influx of genes into protist genomes. BMC Evol Biol. 2006;6:27. doi: 10.1186/1471-2148-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama S, Misawa K, Iseki M, Watanabe M, et al. Origins of a cyanobacterial 6-phosphogluconate dehydrogenase in plastid-lacking eukaryotes. BMC Evol Biol. 2008;8:151. doi: 10.1186/1471-2148-8-151. [DOI] [PMC free article] [PubMed] [Google Scholar]


