Abstract
Lactobacillus vini was recently described as a contaminant in industrial ethanol fermentations and its co-occurrence with Dekkera bruxellensis was noted. We investigated the growth characteristics of L. vini in cocultivation together with either Saccharomyces cerevisiae or D. bruxellensis. Lower cell numbers of both the yeasts and L. vini as well as a decrease in ethanol and lactate formation in mixed batch cultures compared with pure cultures were noted. L. vini formed cell aggregates (flocs) in all cultivation media with different shapes in Man–Rogosa–Sharpe and yeast extract–peptone–dextrose media. Flocs’ size and proportion of cells bound to flocs increased with increasing ethanol concentration. In coculture, formation of lactic acid bacteria–yeast cell aggregates consisting of a bacterial core with an outer layer of yeast cells was observed. L. vini–D. bruxellensis flocs had a bigger surface, due to cells protruding from the pseudomycelium. The involvement of mannose residues in the flocculation between L. vini and yeasts was tested. The presence of mannose induced deflocculation in a concentration-dependent manner. Less mannose was required for the deflocculation of D. bruxellensis as compared with S. cerevisiae.
Keywords: lactic acid bacteria, yeast, flocculation, ethanol production, contamination
1. Introduction
Lactic acid bacteria (LAB) constitute one major group of contaminants in industrial yeast fermentations, and a variety of species have been isolated from those processes 1–3. Lactobacillus fermentum and Lactobacillus brevis were reported to be the most common contaminants in ethanol production 4. High growth rates, tolerance to low pH and high temperature, and resistance to high ethanol concentrations enable a variety of LAB to compete directly with yeast for the sugar substrate and may also inhibit the fermentation process by producing organic acids, mainly lactic acid. The species Lactobacillus vini was described a few years ago 5. The bacterium has been isolated from several ethanol production plants in Sweden and Brazil; therefore, there is a demand to understand the interaction of L. vini with yeasts 2,6. The involved ethanol plants run in a continuous mode with cell recirculation, at sugar limitation and ethanol concentrations up to 100 g/L. In the Swedish plant, low pH (3.6) and temperatures of 36 °C have been documented 6. In most documented cases, L. vini was associated with the presence of Dekkera bruxellensis. D. bruxellensis is a nonconventional yeast that outcompeted traditionally used Saccharomyces cerevisiae in several ethanol production plants running continuous fermentation with cell recirculation 2,6. Although D. bruxellensis is usually considered as a contaminant in ethanol production 7,8, it has been shown to have ethanol yields similar to or even higher than S. cerevisiae as well as low glycerol formation, high biomass yield, a high ethanol tolerance, and a broad range of temperature and pH tolerance 9–11. On the other hand, it has lower specific ethanol production and growth rates than S. cerevisiae and may therefore not be competitive in batch fermentations.
It is currently unclear why D. bruxellensis so frequently co-occurs with L. vini in industrial ethanol fermentation 12. To investigate LAB–yeast interactions, cocultivations under conditions that are conducive to both microorganisms are required.
This study aimed to test growth requirements of L. vini and to assess its growth dynamics in batch pure cultures and in cocultures with D. bruxellensis and S. cerevisiae.
2. Materials and Methods
2.1. Strains and culture media
The yeasts strains D. bruxellensis CBS 11270 and S. cerevisiae J672 9 and the LAB strain L. vini 2FLAB5 isolated from a Swedish ethanol plant 6 were used in this study.
L. vini was pregrown on Man–Rogosa–Sharpe (MRS) agar (Oxoid, Basingstoke, New Hampshire, England) in an anaerobic environment using a BD BBL™ GasPak™ Plus Anaerobic System (Becton, Dickinson and Company, Franklin Lakes, NJ, USA).
Yeast extract–peptone–dextrose medium (YPD) contained 10 g/L yeast extract, 20 g/L bacteriological peptone (both from Oxoid), and 20 g/L d(+) glucose (VWR International Ltd., Poole, England). Solid media also contained 16 g/L agar. For testing the influence of ethanol on flocculation, 10, 50, or 100 g/L ethanol were added to liquid YPD. Synthetic medium contained KH2PO4 18.75 g/L, (NH4)2HPO4 6 g/L, MgSO4·7H2O 1.13 g/L, yeast nitrogen base (Difco™; Becton Dickinson and Company) 6.5 g/L, and glucose 40 g/L. Several supplements of synthetic medium were tested: 5 g/L yeast extract or a mixture of ergosterol (18 µg/L) and Tween 80 (3 g/L) combined with 20 g/L bacteriological peptone or all 20 proteinogenic amino acids (100 mg/L each). The ergosterol/Tween 80 solution was prepared according to Ref. [13].
2.2. Precultures
Yeast precultures were incubated in 150 mL synthetic medium in 500 mL shake-flasks with shaking (200 rpm) at 30 °C for approximately 72 H for D. bruxellensis and for 24 H for S. cerevisiae.
Precultures of L. vini were grown in 200 mL MRS medium [MRS broth 52 g/L, (Oxoid)] in 250 mL flasks (Schott Duran, Wertheim/Main, Germany) with shaking (200 rpm) at 30 °C for approximately 100 H.
2.3. Growth tests of L. vini
L. vini was inoculated to an OD600 of about 1.0 (determination was not exact because flocs could not completely be resuspended by vigorous shaking) into 20 mL serum vials (PharmaPack Packaging System AB, Södertälje, Sweden) containing 10 mL of corresponding medium. The initial pH was adjusted to 5, using concentrated hydrochloric acid. Experiments were performed in triplicate. The vials were incubated at 30 °C and shaken at a speed of 200 rpm on a rotary table.
2.4. Batch cocultivation
For cocultures, every tested microorganism was inoculated to an OD600 of approximately 1 into 20 mL serum vials containing 15 mL of YPD, which were closed with a rubber stopper perforated with a syringe needle. The initial pH was adjusted to 5. Pure cultures inoculated to an OD600 of approximately 1 were run as controls. Experiments were performed in triplicate. The vials were incubated at 30 °C and shaken at a speed of 200 rpm on a rotary table.
2.5. Influence of mannose on flocculation
Precultured yeast and bacterial cells were washed with sterile saline (NaCl solution, 9 g/L). For testing the effect of mannose on LAB and LAB–yeast flocculations, every examined microorganism was inoculated to an OD600 of approximately 0.2 into 2 mL tubes containing 2 mL of 10−1, 10−2, 10−3, 10−4, or 10−5 M mannose in a buffer solution (9.35 g/L KH2PO4, 3 g/L (NH4)2PO4, 1 g/L MgSO4 × 7H20, pH 5). Experiments were performed in triplicate.
2.6. Analytical methods
Ethanol, acetate, and glycerol were measured by high-pressure liquid chromatography as described earlier 14.
Cell viability was determined by viable counts where serial dilutions were plated on YPD-agar plates. The plates were incubated at 30 °C for 24 H (S. cerevisiae) or 72 H (D. bruxellensis). L. vini plates were kept in anaerobic jars. Differentiations of the yeasts and bacteria species were based on the differences in colony morphology. S. cerevisiae colonies had a yellowish color and dull surface with deepening in the center. D. bruxellensis had smaller colonies of a whitish color with a glittering and protuberant surface. L. vini colonies were the smallest size with a swelling center and had a whitish translucent color.
Flocculation was studied at 400-fold magnification using an Olympus BH2 microscope.
The influence of mannose on flocculation was also monitored by optical density measurements immediately after suspending cells in the buffer and 7 Min after at a wavelength of 600 nm (OD600) by an Ultrospec 1100 pro spectrophotometer.
2.7. Statistical analysis
To test whether there were significant differences between the treatments, Student's t-test was performed with a significance level of 5% using the software MS Excel (Version 2007).
3. Results
3.1. Growth characteristics of L. vini in batch culture
We initially determined the growth of L. vini in different media that could potentially be used for cocultivation of LAB and yeasts. L. vini was able to grow in YPD medium, reached approximately 1.0 × 108 cells/mL (Fig. 1), and did not increase further even though there was still a significant amount of glucose present in the medium (8.4 ± 1.1 g/L).
Figure 1.
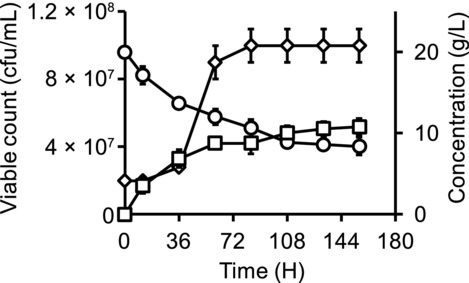
Lactate (squares) formation, glucose (circles) consumption, and growth of L. vini (diamonds) in YPD medium.
We tested whether it was possible to replace components in the rich medium with defined supplements. Yeast extract could be replaced by a mixture of ergosterol and Tween 80, which is also essential for anaerobic cultivation of yeasts 15. However, peptone could not be replaced by a mixture of all 20 amino acids. It is possible that L. vini requires oligopeptides for growth, which has been observed in other LAB species 16. Substantial flocculation was observed in all media tested in this study. Flocs in YPD medium were spherical, whereas flocs in MRS medium tended to be irregular clumps. When ethanol was added to the medium at concentrations of 10, 20, and 100 g/L, the degree of L. vini flocculation increased in a concentration-dependent manner. We observed both cell aggregates of different sizes as well as single cells in the absence of ethanol. The amount of single cells decreased when 10 g/L ethanol was added to a growing culture, and no individual cells could be detected at 100 g/L ethanol.
3.2. Batch cocultivation of L. vini with yeast
To investigate the impact of L. vini on yeast growth, it was cocultivated with D. bruxellensis and S. cerevisiae. In both cocultures with S. cerevisiae and D. bruxellensis, the cfu number of L. vini reached the same level of 7.1 × 107 ± 2.0 × 106 cfu/mL, which was only 70% of the corresponding cell number in pure culture (Figs. 2A and 2B). Lactate production was significantly reduced by 10%–20% in both cocultures. In coculture, D. bruxellensis viable counts were higher than those of S. cerevisiae (5.0 × 107 ± 1.5 × 106 cfu/mL compared with 4.5 × 107 ± 1.5 × 106 cfu/mL), respectively. However, the final ethanol concentration in L. vini–D. bruxellensis coculture was significantly (P = 0.016) lower (5.3 ± 0.2 g/L) than in the L. vini–S. cerevisiae coculture (6.0 ± 0.2 g/L). Final ethanol concentrations in the corresponding D. bruxellensis and S. cerevisiae pure cultures were 7.2 ± 0.1 and 7.3 ± 0.4 g/L, respectively (Fig. 2). Low amounts of glycerol and acetate were also formed in both cocultures. The maximum concentrations after glucose was finished for glycerol and acetate, respectively, were 0.04 ± 0.03 and 0.19 ± 0.08 g/L in the D. bruxellensis–L. vini coculture and 0.52 ± 0.02 and 0.19 ± 0.05 g/L in the S. cerevisiae–L. vini coculture.
Figure 2.
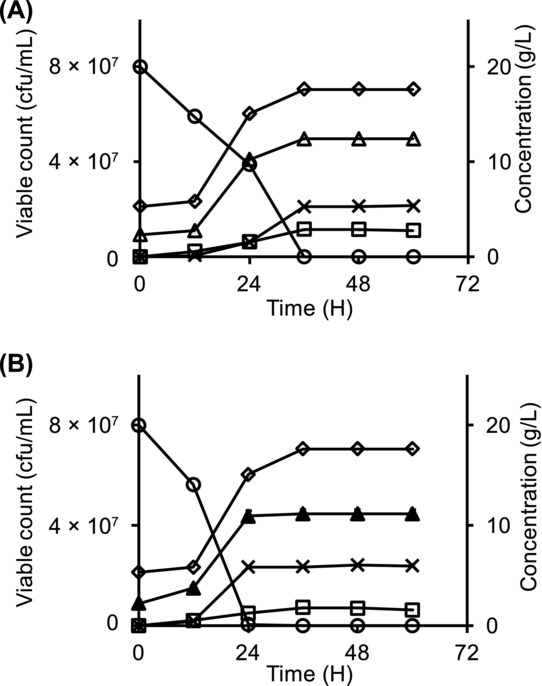
Lactate (squares) formation, ethanol (crosses) production, glucose (circles) consumption, and growth of corresponding microbes during batch cocultivation of L. vini (diamonds) with yeast. (A) Cocultivation of L. vini and D. bruxellensis (open triangles), (B) cocultivation of L. vini and S. cerevisiae (filled triangles).
3.3. Coflocculation of L. vini and yeast
Microscopic investigation of L. vini–yeast cocultures revealed that L. vini formed aggregates together with either yeast (Figs. 3A and 3B). Cell aggregation occurred immediately after addition of the L. vini cells to the yeast cultures. The flocs consisted of a bacterial core surrounded by yeast cells. Several aggregates were completely covered with yeast cells. Moreover, cells of D. bruxellensis formed pseudohyphae that protruded from the flocs of budding cells of S. cerevisiae mainly formed a monolayer around the bacterial core (Figs. 3A and 3B).
Figure 3.
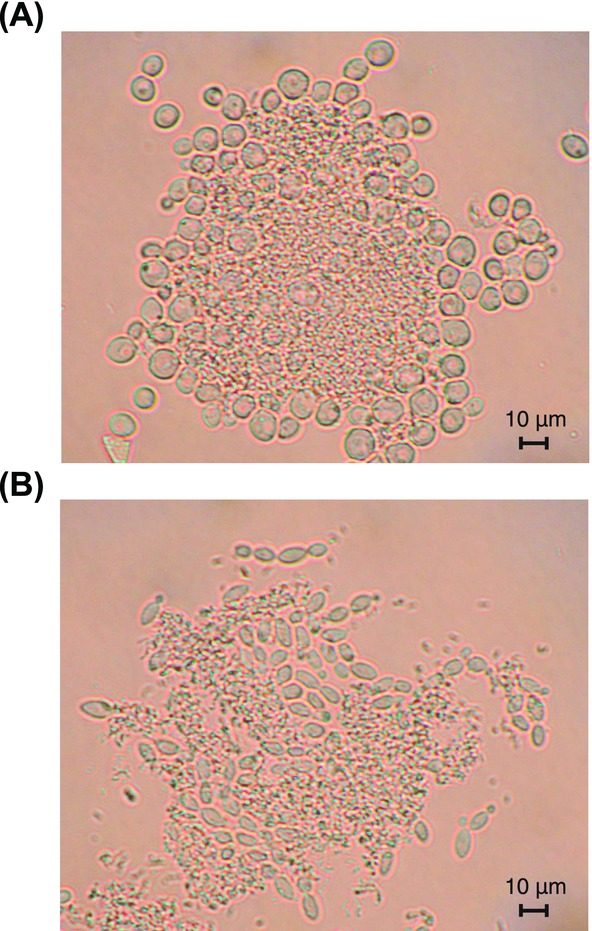
Coflocculation of L. vini with yeasts during batch cocultivation. (A) L. vini and S. cerevisiae, (B) L. vini and D. bruxellensis. (Magnification ×400.)
3.4. Influence of mannose on flocculation
The degree of L. vini, L. vini–D. bruxellensis, and L. vini–S. cerevisiae flocculation decreased when mannose was present in the suspension in a concentration-dependent manner. This effect was more pronounced 7 Min after suspending the cells in the buffer as compared with the time zero (Fig. 4). This was due to sedimentation of the flocs in samples with low mannose content. Microscopic investigation revealed a difference in the mannose effect on the L. vini–D. bruxellensis and L. vini–S. cerevisiae flocs. The concentration efficient for complete deflocculation of L. vini–D. bruxellensis was 10−2 M mannose, whereas it was 10−1 M for L. vini–S. cerevisiae. Mannose addition also affected floc formation by bacterial cells both in pure and in mixed culture. However, LAB flocs were never completely resolved within the tested mannose concentrations.
Figure 4.
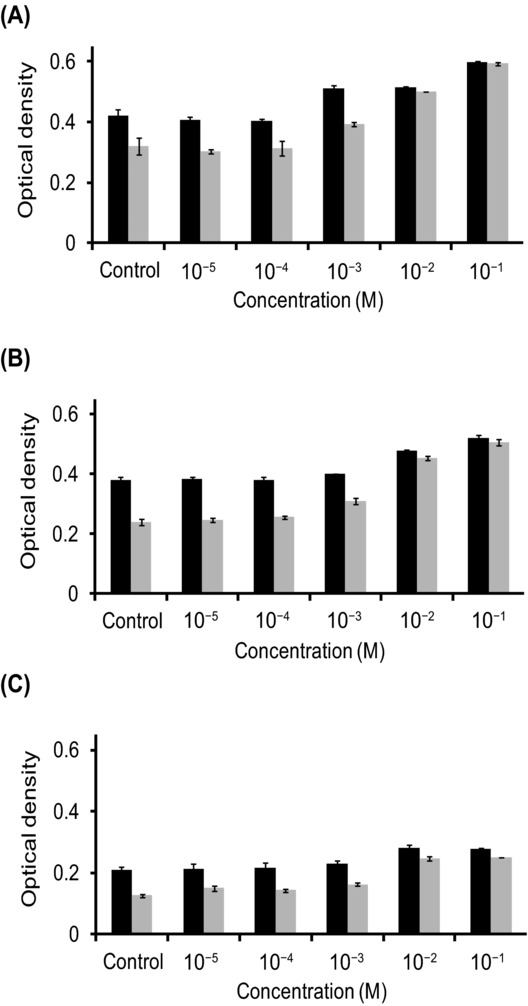
Influence of mannose concentration on the optical density of flocculating cultures immediately after suspending cells in the buffer (black) and 7 Min after (gray). (A) Coculture of L. vini and D. bruxellensis, (B) coculture of L. vini and S. cerevisiae, and (C) pure culture of L. vini.
4. Discussion
This study investigated the growth of the ethanol-tolerant LAB L. vini in liquid medium that can be used for cocultivation with yeasts. A high degree of flocculation was observed for this bacterium in all tested media. A similar phenomenon was shown for other LAB species 17,18. Possibly, the formation of flocs can reduce the surface exposed to ethanol and thus help the bacteria to survive the stressful conditions of the ethanol process. This conclusion is also supported by the observed higher flocculation activity of L. vini in the presence of ethanol. On the other hand, a smaller surface area also decreases sugar and nutrient uptake by the cells.
Interestingly, L. vini formed coaggregates with the yeast cells when cultivated in mixed cultures. Coflocculation between yeasts and LAB has rarely been investigated, but it has been shown that L. fermentum, which is also present in industrial ethanol processes, can also form aggregates with S. cerevisiae 19. The tendency of L. vini to form aggregates with the yeast makes it difficult to detect contamination by microscopic investigations when the bacteria are hidden in the flocs.
A growth cessation of L. vini in pure culture was observed while glucose was still present in the medium. The likely reason for this should be the relatively high concentration of lactate (10–12 g/L). The LAB L. fermentum and L. brevis also stopped growing in the presence of 10 g/L of lactate 4.
In industrial ethanol processes with cell recirculation, L. vini was at several occasions demonstrated to be associated with D. bruxellensis 2,6. The cocultivation experiments described in this study are a first attempt to understand the specific interaction between these two species; however, the results of our experiments did not provide an obvious explanation for this frequent coexistence. D. bruxellensis seemed to be more affected by L. vini than S. cerevisiae, because ethanol concentration decreased on the expanse of formed lactic acid in the corresponding coculture. However, this may apply only for a batch culture, where the conditions are not favorable for D. bruxellensis due to its lower specific glucose uptake and growth rates 9. The behavior during the stationary phase might closely resemble the conditions in the continuous ethanol process, but no impact on survival during this phase was seen in any of yeasts.
In the batch culture, L. vini apparently acts as a contaminant, as it channels sugar to lactate and bacterial biomass, at the expense of ethanol formation and yeast cell numbers. There is no proof yet about whether and how L. vini influences the competition between D. bruxellensis and S. cerevisiae. Attachment of LAB to yeast cells is attributed to mannose binding proteins of the bacteria 20. Observed differences in the mannose effect on the L. vini–D. bruxellensis and L. vini–S. cerevisiae flocs may be due to the higher mannose content of the cell wall of S. cerevisiae, compared with D. bruxellensis 21, which can lead to higher capacity for L. vini binding to S. cerevisiae. Cells incorporated within the flocs tend to sediment while single cells stay in suspension. Thus, frequent cooccurrence of D. bruxellensis and L. vini may be due to the fact that D. bruxellensis is less involved in flocculation and thus less impacted by L. vini.
It has been found that D. bruxellensis can outcompete S. cerevisiae in glucose limited cocultivations without the involvement of any bacteria 22. However, it is possible that the smaller flocs with the protruding cells formed by D. bruxellensis–L. vini represent an advantage for the cells because the surface is enlarged, enabling more efficient uptake of the limited nutrients while at the same time protecting the cells in the inner part of the aggregate from stressful conditions 23,24. Thus, our results indicate that LAB can affect ethanol production not only by competing for substrate or lactic acid formation but also by influencing spatial structure of the population in the fermentor.
Acknowledgments
This study was financed by the research program MicroDrivE (http://microdrive.slu.se) at the Swedish University of Agricultural Sciences, Uppsala and the Swedish Energy Agency (Energimyndigheten). The authors wish to thank Afsar Ali for technical assistance, Dr. Tomas Linder for critically reading the manuscript and support during the preparation of figures, and Dr. Hans Jonsson for valuable discussions.
Glossary
- CFU
colony-forming units
- LAB
lactic acid bacteria
- MRS
Man–Rogosa–Sharpe medium
- YPD
yeast extract–peptone–dextrose medium
6. References
- Beckner M, Ivey ML, Phister TG. Lett. Appl. Microbiol. 2011;53:387–394. doi: 10.1111/j.1472-765X.2011.03124.x. [DOI] [PubMed] [Google Scholar]
- Lucena BT, dos Santos BM, Moreira JL, Moreira AP, Nunes AC, Azevedo V, Miyoshi A, Thompson Fl, de Morais MA., Jr BMC Microbiol. 2010;10:298. doi: 10.1186/1471-2180-10-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner KA, Leathers TD. J. Ind. Microbiol. Biotechnol. 2004;31:401–408. doi: 10.1007/s10295-004-0159-0. [DOI] [PubMed] [Google Scholar]
- Watanabe I, Nakamura T, Shima J. J. Ind. Microbiol. Biotechnol. 2008;35:1117–1122. doi: 10.1007/s10295-008-0390-1. [DOI] [PubMed] [Google Scholar]
- Rodas AM, Chenoll E, Macian MC, Ferrer S, Pardo I, Aznar R. Int. J. Syst. Evol. Microbiol. 2006;56:513–517. doi: 10.1099/ijs.0.63877-0. [DOI] [PubMed] [Google Scholar]
- Passoth V, Blomqvist J, Schnürer J. Appl. Environ. Microbiol. 2007;73:4354–4356. doi: 10.1128/AEM.00437-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza Liberal AT, Basilio AC, do Monte Resende A, Brasileiro BT, da Silva-Filho EA, de Morais JO, Simoes DA, de Morais MA., Jr J. Appl. Microbiol. 2007;102:538–547. doi: 10.1111/j.1365-2672.2006.03082.x. [DOI] [PubMed] [Google Scholar]
- Loureiro V, Malfeito-Ferreira M. Int. J. Food Microbiol. 2003;86:23–50. doi: 10.1016/s0168-1605(03)00246-0. [DOI] [PubMed] [Google Scholar]
- Blomqvist J, Eberhard T, Schnürer J, Passoth V. Appl. Microbiol. Biotechnol. 2010;87:1487–1497. doi: 10.1007/s00253-010-2619-y. [DOI] [PubMed] [Google Scholar]
- Pereira LF, Bassi AP, Avansini SH, Neto AG, Brasileiro BT, Ceccato-Antonini SR, de Morais MA., Jr Antonie Van Leeuwenhoek. 2012;101:529–539. doi: 10.1007/s10482-011-9662-2. [DOI] [PubMed] [Google Scholar]
- Blomqvist J, South E, Tiukova I, Momeni MH, Hansson H, Ståhlberg J, Horn SJ, Schnürer J, Passoth V. Lett. Appl. Microbiol. 2011;53:73–78. doi: 10.1111/j.1472-765X.2011.03067.x. [DOI] [PubMed] [Google Scholar]
- de Souza RB, dos Santos BM, de Souza RDR, da Silva PKN, Lucena BTL, de Morais MA. J. Ind. Microbiol. Biotechnol. 2012;39:1645–1650. doi: 10.1007/s10295-012-1167-0. [DOI] [PubMed] [Google Scholar]
- Andreasen AA, Stier TJB. J. Cell. Compar. Physiol. 1954;43:271—281. doi: 10.1002/jcp.1030430303. [DOI] [PubMed] [Google Scholar]
- Fredlund E, Blank LM, Schnürer J, Sauer U, Passoth V. Appl. Environ. Microbiol. 2004;70:5905–5911. doi: 10.1128/AEM.70.10.5905-5911.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreasen AA, Stier TJB. J. Cell. Compar. Physiol. 1953;41:23–36. doi: 10.1002/jcp.1030410103. [DOI] [PubMed] [Google Scholar]
- Moretro T, Aasen IM, Storro I, Axelsson L. J. Appl. Microbiol. 2000;88:536–545. doi: 10.1046/j.1365-2672.2000.00994.x. [DOI] [PubMed] [Google Scholar]
- Liu H, Xu W, Luo Y, Tian H, Wang H, Guo X, Yuan Y, Huang K. Antonie Van Leeuwenhoek. 2011;99:579–589. doi: 10.1007/s10482-010-9529-y. [DOI] [PubMed] [Google Scholar]
- Roos S, Lindgren S, Jonsson H. Mol. Microbiol. 1999;32:427–436. doi: 10.1046/j.1365-2958.1999.01363.x. [DOI] [PubMed] [Google Scholar]
- Santos MT, Yokoya F. J. Ferment. Bioeng. 1993;75:151–154. [Google Scholar]
- Furukawa S, Nojima N, Yoshida K, Hirayama S, Ogihara H, Morinaga Y. Biosci. Biotechnol. Biochem. 2011;75:1430–1434. doi: 10.1271/bbb.100817. [DOI] [PubMed] [Google Scholar]
- Prillinger H, Dörfler C, Laaser G, Eckerlein B, Lehle L. Zeitschrift für Mykologie. 1990;56:219–250. [Google Scholar]
- Blomqvist J, Nogue VS, Gorwa-Grauslund M, Passoth V. Yeast. 2012;29:265–274. doi: 10.1002/yea.2904. [DOI] [PubMed] [Google Scholar]
- Bhinu VS. J. Mol. Microbiol. Biotechnol. 2005;10:15–21. doi: 10.1159/000090344. [DOI] [PubMed] [Google Scholar]
- Wuertz S, Okabe S, Hausner M. Water Sci. Technol. 2004;49:327–336. [PubMed] [Google Scholar]


