Abstract
MicroRNAs (miRNAs) regulate many aspects of human biology. They target mRNAs for translational repression or degradation through base-pairing with 3’ UTRs, primarily via seed sequences (nucleotides 2-8 in the mature miRNA sequence). A number of individual miRNAs and miRNA families share seed sequences and targets, but differ in the sequences outside of the seed. miRNAs have been implicated in the etiology of a wide variety of human diseases and therefore represent promising therapeutic targets. However, potential redundancy and compensatory action of different miRNAs sharing the same seed sequence, and the challenge of simultaneously targeting miRNAs that differ significantly in non-seed sequences complicates therapeutic targeting approaches. We recently demonstrated effective inhibition of entire miRNA families using seed-targeting 8-mer locked nucleic acid (LNA)-modified antimiRs in short-term experiments in mammalian cells and in mice. However, the long-term efficacy and safety of this approach in higher organisms, such as humans and non-human primates, has not been determined. Here, we show that pharmacological inhibition of the miR-33 family, key regulators of cholesterol/lipid homeostasis, by a subcutaneously delivered 8-mer LNA-modified antimiR in obese and insulin-resistant non-human primates results in de-repression of miR-33 targets, such as ABCA1, increases circulating high-density lipoprotein-cholesterol (HDL-C), and is well tolerated over 108 days of treatment. These findings demonstrate the efficacy and safety of an 8-mer LNA-antimiR against a miRNA family in a non-human primate metabolic disease model, suggesting that this could be a feasible approach for therapeutic targeting of miRNA families sharing the same seed sequence in human diseases.
Introduction
MiRNAs are short (~22 nucleotide) non-coding RNAs with diverse functions in development, metabolism and disease (1). They regulate gene expression by base-pairing with partially complementary sequences in the 3’UTRs of their target mRNAs and thereby promote mRNA degradation or translational repression (2-5). Each miRNA has the potential to target a large number of mRNAs as predicted by complementarity to the miRNA seed region and non-seed sequences (3). Aberrant expression or function of miRNAs has been linked to a number of diseases, and inhibition of several disease-associated miRNAs with antimiRs has recently been explored as a therapeutic intervention (6). Indeed, inhibition of miR-122 for the treatment of hepatitis C virus infection (7) by the LNA-modified antimiR termed miravirsen recently completed a human Phase II trial (8). A potential hurdle in the development of miRNA inhibitors as therapeutics is the redundancy and compensatory action among miRNA family members and other miRNAs sharing the same seed sequence. In humans, 47% (41/87) of the highly conserved miRNA families (as defined by TargetScan 6.2) contain two or more family members that exhibit the same seed sequence (9). For example, the highly conserved let-7/miR-98 family contains nine members in humans and mice. The let-7/miR-98 family also shares its seed sequence with other miRNAs, such as miR-4458 and miR-4500, but differs from them in the sequences outside the seed. This highlights the challenge of sequence-specific antisense targeting of potentially redundant miRNAs. We recently examined the possibility of using “tiny” seed-targeting 8-mer LNA-antimiRs to target miRNAs sharing the same seed sequence, and demonstrated that this approach can be used to effectively inhibit all members of a given miRNA family, without loss of specificity or display of observable toxicity, in mammalian cells and in short-term studies in mice (10). The potency as well as lack of apparent off-target effects is quite remarkable given the short sequence used, and the mechanism underlying this specificity remains to be determined. Nevertheless, several additional studies have now successfully used 8-mer antimiRs to target miRNA families in mice, suggesting that this approach might have broader applicability (11-13). However, the long-term efficacy and safety of 8-mer antimiRs has not yet been explored in humans or non-human primates.
Human miR-33a and miR-33b, which differ by two nucleotides outside the seed sequence, were recently identified in introns of the SREBF2 and SREBF1 genes, respectively, and control cholesterol/lipid homeostasis in concert with their host gene products, the Sterol Regulatory Element-Binding Protein (SREBP) transcription factors (14-21). In contrast to humans and other mammals, mice and other rodents only have miR-33a, present in the SREBF2 gene, while the miR-33b sequence is missing in the rodent SREBF1 gene. One of the best characterized targets of miR-33a/b is the ATP-binding cassette transporter ABCA1, which is critical for cholesterol efflux from peripheral cells and tissues, including atherogenic macrophages/foam cells, and for generation of nascent, lipid-poor high density lipoprotein (HDL) from liver and other tissues (22). Low concentrations of circulating HDL-cholesterol (HDL-C) are associated with metabolic syndrome and atherosclerosis, whereas blood HDL-C concentrations above a threshold are linked to protection against cardiovascular disease. Intensive efforts have thus focused on raising circulating HDL-C in high-risk groups. However, it is unclear whether current therapeutic approaches to elevate HDL-C, such as treatment with niacin or cholesteryl ester transfer protein (CETP) inhibitors, are effective in promoting the entire path of reverse cholesterol transport (RCT) from macrophages to the liver and biliary excretion, a crucial mechanism to decrease atherosclerosis (23). Hence, therapeutic avenues directly aimed at increasing RCT from atherogenic macrophages are needed. We and others have recently shown that antisense-based antagonism or knockout of miR-33a and/or b promotes ABCA1-mediated cholesterol efflux from macrophages and stimulates the production of nascent HDL that is able to accept excess cholesterol from atherogenic macrophages, thereby ameliorating atherosclerosis (14-16, 18, 20, 21, 24). This demonstrates the potential of therapeutic targeting of the miR-33 family members as an alternative approach to combat cardiovascular disease.
In addition to their effect on cholesterol efflux and HDL-C synthesis, miR-33a/b also inhibit the expression of genes involved in other aspects of cholesterol/lipid homeostasis, such as fatty acid β-oxidation, regulation of energy homeostasis by AMP kinase and insulin signaling, as well as hepatic cholesterol/bile acid clearance (18, 19, 25, 26). These miRNAs could therefore also potentially represent attractive targets for the treatment of other aspects of metabolic syndrome, such as insulin resistance, hypertriglyceridemia, and non-alcoholic fatty liver disease (NAFLD) (19, 26). However, because mice lack the miR-33b homolog, studies in other mammals, such as non-human primates that harbor both miR-33a and miR-33b are necessary to determine their relative functional importance and potential redundancy in cholesterol/lipid regulation, and contribution to human physiology and disease.
In this study, we show that the two miR-33 family members (miR-33a/b) are acting in a redundant manner to control hepatic ABCA1 expression and HDL-cholesterol homeostasis in obese and insulin-resistant non-human primates. Importantly, we demonstrate marked pharmacological inhibition of the miR-33 family by a once-weekly subcutaneously delivered seed-targeting 8-mer LNA-antimiR, which results in elevation of HDL-C by up to ~40% in an African green monkey metabolic disease model. Furthermore, we show that long-term treatment with the 8-mer LNA-antimiR was well tolerated with no evidence of off-target effects or toxicity. These findings demonstrate for the first time that a seed-targeting 8-mer LNA-antimiR can effectively inhibit a miRNA family in non-human primates, with a clean long-term safety profile.
Results
Selected miR-33-targeting antimiRs are effective in vitro and in mice
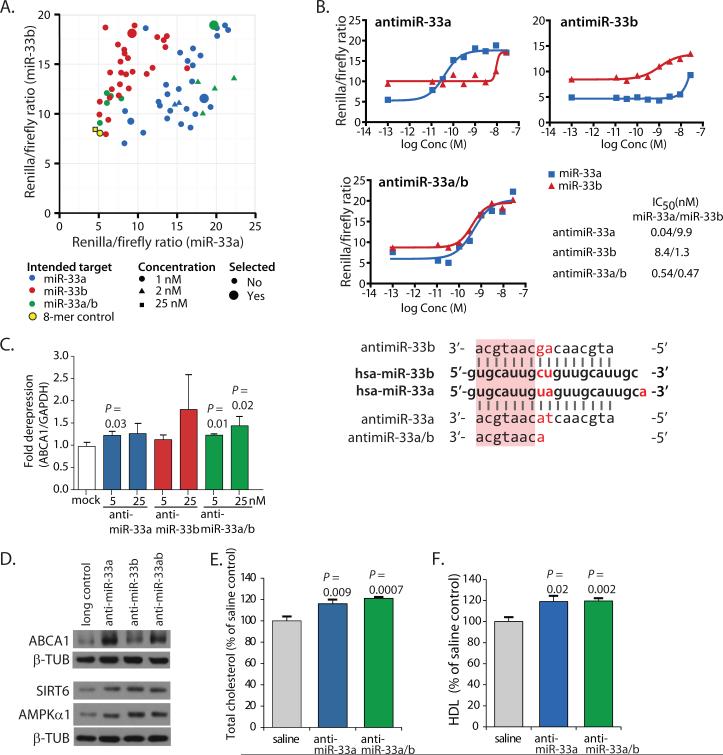
We set out to explore the efficacy and safety of miR-33a/b-targeting LNA-antimiRs for potential therapeutic purposes as well as to assess the importance of targeting both miRNAs simultaneously in a non-human primate model of metabolic disease. Initially, we screened a library of 66 LNA-modified antimiRs to identify compounds that selectively inhibit miR-33a or miR-33b, or inhibit both equally, using 3’UTR luciferase reporter assays in the human hepatoma cell line HepG2 (Fig. 1A). From these assays, we selected the most effective LNA-antimiRs for targeting miR-33a, miR-33b or both (miR-33a/b) and established duplex melting temperatures (Tm) and IC50 values (table S1) to assess their specificity (Fig. 1B). We also evaluated their efficacy in increasing the expression of ABCA1 in HepG2 cells (Fig. 1C, table S2). Finally, we compared their in vivo effects on hepatic ABCA1 expression in mice, as well as total serum cholesterol and circulating HDL-C. Based on results from these studies, we selected three LNA-modified antimiRs; (i) antimiR-33a, (ii) antimiR-33b and (iii) an 8-mer seed-targeting antimiR inhibiting both miR-33a and b (antimiR-33a/b) for our subsequent studies (Fig. 1B, table S1). The 16-mer LNA/DNA-antimiRs showed selective inhibition of miR-33a and miR-33b, respectively, while the 8-mer antimiR-33a/b inhibited both miR-33a and miR-33b (Fig. 1B). Effects on ABCA1 mRNA levels were modest but significant for LNA-antimiRs targeting miR-33a and miR-33a/b (P = 0.03; P = 0.01 and 0.02) (Fig. 1C, table S2). All three LNA-antimiRs substantially de-repressed the miR-33 targets ABCA1, SIRT6 and AMPKα1 (also known as PRKAA1) at the protein level in human HepG2 hepatoma cells (Fig. 1D) (19, 26). Moreover, total cholesterol and HDL-C were also increased significantly in mice by antimiR-33a and antimiR-33a/b (P = 0.009, 0.0007; P = 0.02, 0.002) (Fig. 1, E and F, table S2). In contrast, the long LNA/DNA and 8-mer scrambled control LNA oligonucleotides did not show significant effects in any assay (P = 0.37, 0.75, 0.19, 0.33; P = 0.18, 0.06) (fig. S1, table S2).
Fig. 1. Inhibition of miR-33 function in HepG2 cells and mice using LNA-modified antimiR oligonucleotides.
(A) Relative luciferase activity of the miR-33 reporters containing a perfect match target site for miR-33a and miR-33b, respectively, co-transfected into HepG2 cells with different LNA-modified antimiRs or the 8-mer scrambled control LNA oligonucleotide. The antimiR-33 oligonucleotides selected for the non-human primate study are marked as large circles. (B) Specificity of the three selected antimiR-33 oligonucleotides targeting miR-33a, miR-33b, or miR-33a/b, respectively, was assessed using the miR-33a and miR-33b luciferase reporters co-transfected into HepG2 cells with the selected antimiRs at varying concentrations. Luciferase activity was measured after 24 hours, and the half-maximal inhibitory concentration (IC50) values were determined. Alignment of miR-33a and miR-33b with the selected antimiR oligonucleotide sequences is shown below the graphs: differences between miR-33a and b are marked in red and the shared seed sequence is marked with a pink rectangle. (C) Transfection with antimiR-33 oligonucleotides (5 and 25 nM) resulted in de-repression of ABCA1 mRNA expression in HepG2 human hepatoma cells. (D) Transfection with antimiR-33 oligonucleotides (5 nM) de-repressed ABCA1, SIRT6, and AMPKα1 targets at the protein level in HepG2 cells. β-TUB denotes β-Tubulin, which was used as a loading control. (E) Treatment with antimiR oligonucleotides targeting miR-33a or miR-33a/b increased total plasma cholesterol in mice. (F) Treatment with antimiR oligonucleotides targeting miR-33a or miR-33a/b increased circulating HDL-C in mice. Error bars for (C) represent standard deviation (SD), and for panels (E) and (F) standard error of the mean (SEM). Statistical analysis was carried out with Student's t-test. A scrambled 8-mer LNA oligonucleotide (8-mer control) and a scrambled 16-mer LNA/DNA oligonucleotide (long control) were used as controls (13).
Pharmacological inhibition of the miR-33a/b family elevates HDL-C in non-human primates
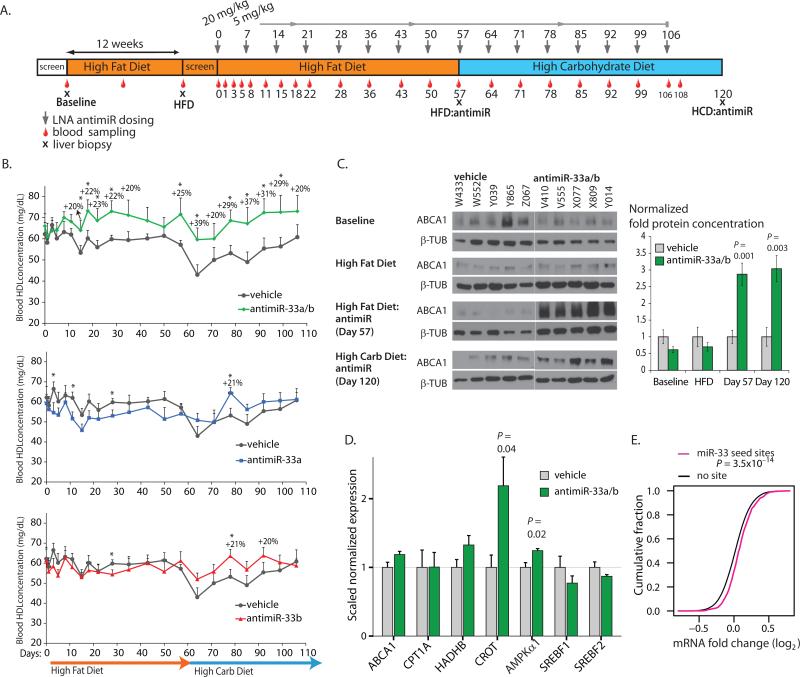
To assess the efficacy and safety of the antimiRs targeting miR-33a, miR-33b and miR-33a/b in a non-human primate model of metabolic disease, we selected a population of 20 obese African green monkeys (weight averaged 59% over normal weight (27)) (table S3). To further induce metabolic abnormalities seen in humans with metabolic syndrome (insulin resistance/hyperglycemia, abnormal LDL/HDL-cholesterol ratios, and hypertriglyceridemia), all animals were fed a high-fat diet (HFD) with excess cholesterol for 12 weeks. Subsequently, 4 groups of 5 animals each were subcutaneously injected with vehicle, antimiR-33a, antimiR-33b, or antimiR-33a/b compound, respectively, with an initial 20 mg/kg loading dose followed by weekly maintenance doses of 5 mg/kg (Fig. 2A). To determine the effects of a high-carbohydrate diet (HCD), we shifted the animals to a diet high in fructose after 8 weeks of treatment. To evaluate potential therapeutic benefits of the antimiR treatments, we measured HDL-C concentration in plasma weekly, as well as determined total cholesterol, LDL-cholesterol, and triglycerides (Fig. 2A). We found that treatment with the 8-mer antimiR-33a/b increased HDL-C by up to 39% as compared to vehicle-treated animals at the same time point (Fig. 2B, table S2). Treatment with the 16-mer antimiRs selectively targeting either miR-33a or miR-33b did not show significant overall effects on circulating HDL-C (Fig. 2B, table S2). These results illustrate the apparent redundancy of the miR-33 family members, and highlight the importance of targeting both miR-33a and b simultaneously. We also analyzed the effects of antimiR treatment in the liver by taking biopsies at the start of the study (Baseline), after 12 weeks of HFD (HFD), after 8 weeks of LNA-antimiR treatment (HFD:antimiR), and after 7 additional weeks of antimiR treatment on the HCD (HCD:antimiR) (Fig. 2A). We found that the ABCA1 target was strongly de-repressed at the protein level by the 8-mer antimiR-33a/b treatment (Fig. 2C, table S2). In addition, quantitative RT-PCR analysis revealed a modest increase in the hepatic expression of selected miR-33 targets, such as ABCA1, CROT, and AMPKα1 (14, 15, 18), after 8-mer antimiR-33a/b treatment (Day 57) (Fig. 2D, table S2). The antimiRs selectively targeting miR-33a or miR-33b were less efficacious, and inconsistent in their effects on direct miR-33 targets (fig. S2, table S2), again emphasizing the redundancy of miR-33a and b. We also performed DNA microarray-based genome-wide expression analysis on liver samples taken before switching to the high-carbohydrate diet to assess overall gene expression changes. We found significant de-repression of TargetScan 6.2-predicted miR-33 targets in antimiR-33a/b treated animals compared to vehicle-treated animals (Fig. 2E). A list of confirmed and predicted miR-33 targets found to be de-repressed is shown in table S4. These findings indicate that treatment with the 8-mer antimiR-33a/b in vivo results in the expected changes in a non-human primate model of metabolic disease.
Fig. 2. Inhibition of the miR-33 family in obese African green monkeys by a seed-targeting 8-mer antimiR oligonucleotide.
(A) Design and timeline of the non-human primate study. (B) Plasma HDL-C concentrations in antimiR-33a/b-, antimiR-33a- and antimiR-33b-treated African green monkeys compared to vehicle control-treated animals. * represents P <0.05. P-values are given in table S2. (C) Western blot analysis of hepatic ABCA1 protein in vehicle- and antimiR-33a/b-treated African green monkeys. (D) De-repression of miR-33 target mRNAs in antimiR-33a/b-treated African green monkeys. Error bars represent standard error of the mean (SEM). Statistical analysis was performed with Student's t-test. (E) De-repression of liver mRNAs with predicted miR-33 seed match sites in antimiR-33a/b-treated monkeys compared to vehicle-treated control animals. Cumulative distributions of mRNA changes between antimiR-33a/b- and vehicle-treated animals are shown for TargetScan 6.2-predicted miR-33 target mRNAs (magenta) and mRNAs without miR-33 seed match sites (black), demonstrating that miR-33 target mRNAs are de-repressed in monkey livers after antimiR-33a/b treatment (P = 3.5x10-14, one sided Kolmogorov-Smirnov test).
To investigate the effects of miR-33 inhibition on insulin resistance, we performed an intravenous glucose tolerance test at the end of the study, expecting the vehicle-treated monkeys to suffer from hyperglycemia and elevated circulating insulin as a result of the obese phenotype and the challenging diet. However, although we found that all animals in our study displayed elevated fasting glucose levels and poor glucose regulation, baseline insulin levels were extremely low (fig. S3, table S2). This suggests progression toward a diabetic phenotype with chronic low pancreatic insulin production. There was no apparent difference in glucose homeostasis or insulin sensitivity between vehicle and antimiR-33a/b-treated animals (fig. S3, table S2).
A recent study showed that a high-carbohydrate diet produced an increase in hepatic miR-33b expression in normal male African green monkeys (21). However, we found that hepatic miR-33b expression was similar to miR-33a expression and essentially unchanged in our cohort of obese, insulin-resistant female monkeys after the introduction of the high-carbohydrate diet (fig. S4). These findings are consistent with the low insulin levels observed in this severe metabolic disease model. Elevated circulating insulin associated with obesity-related insulin resistance in humans has been linked to excess circulating triglycerides (28). In agreement with the low insulin and lack of HCD-dependent changes in miR-33b expression in our monkeys, we did not detect a reduction in triglyceride concentrations in response to antimiR-33a/b treatment, as compared to vehicle-treated animals (fig. S5, table S2).
Long-term inhibition of the miR-33 family does not cause adverse effects
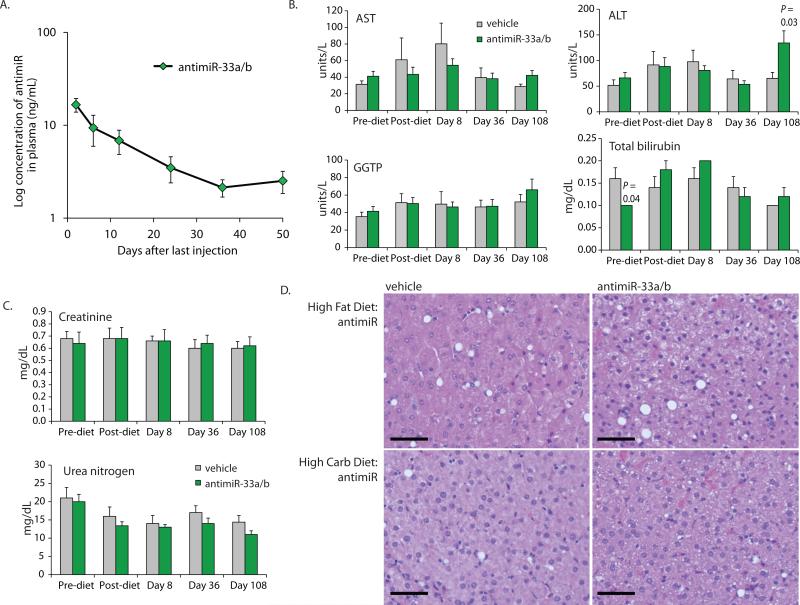
To assess the pharmacokinetics and safety profile of the 8-mer antimiR-33a/b treatment, we analyzed the plasma stability of antimiR-33a/b after the last dose. The terminal plasma half-life of antimiR-33a/b was 17.5 days, which is comparable to the half-life previously reported for other LNA-modified antisense oligonucleotides ranging from 12-16 nucleotides in length (Fig. 3A, table S5) (7, 29). No evidence of injection site reactions was observed after loading dose or maintenance dose administration. In addition, no adverse reactions were noted during routine cage-side observations, indicating a lack of local toxicity. Next, we performed clinical chemistry analysis on selected blood samples (Fig. 2A). Markers of liver function, such as alanine aminotransferase (ALT), aspartate aminotransferase (AST), bilirubin and gamma glutamyl transpeptidase (GGTP) were not markedly changed after treatment with the 8-mer antimiR-33a/b, suggesting the absence of hepatic toxicity (Fig. 3B, table S2). Kidney toxicity parameters, such as circulating creatinine and blood urea nitrogen (BUN) also remained within the normal range (Fig. 3C, table S2). Additional clinical chemistry data are included in the supplemental data (fig. S6, table S2). To further evaluate potential pathological effects of the 8-mer antimiR-33a/b treatment, representative sections of liver biopsies from each animal were stained with H&E, and evaluated for histologic changes, including hepatic steatosis, inflammation, and fibrosis. Hepatic steatosis was not a prominent feature in either antimiR-33a/b treated or vehicle groups at baseline or after treatment (Fig. 3D), and ranged from 0-4% (table S6). Moreover, no significant inflammation or fibrosis was visible. Additionally, we examined the differentially expressed genes from the microarray analysis and did not find statistically significant enrichment of genes with gene ontology (DAVID) terms associated with toxicity or other potential adverse effects, such as “xenobiotic detoxification”, “inflammation”, “immune response”, and “cancer” in any of the treatment groups (table S7). These results indicate that treatment with the seed- targeting 8-mer antimiR-33a/b compound for over 100 days was well tolerated in non-human primates.
Fig. 3. Safety of antimiR-33a/b treatment in obese African green monkeys.
(A) Plasma concentrations of subcutaneously administered antimiR-33a/b compound in obese African green monkeys are shown after the last dose (Day 106, shown on the graph as day 0). (B) Liver function was monitored by assessing plasma concentrations of Aspartate Aminotransferase (AST), Alanine Aminotransferase (ALT), Gamma Glutamyl Transpeptidase (GGTP) and Total Bilirubin in vehicle-and antimiR-33a/b-treated African green monkeys during the study. (C) Assessment of renal function by measuring plasma Creatinine and Blood Urea Nitrogen (BUN) in the antimiR-33a/b-treated monkeys compared to vehicle-treated control animals. (D) Assessment of hepatic steatosis in antimiR-33a/b treated African green monkeys. Representative hematoxylin & eosin (H&E)-stained liver sections of vehicle and antimiR-33a/b-treated animals on high-fat and high-carbohydrate diets, respectively. Scale Bars are 500 µm. Error bars represent standard error of the mean (SEM). Statistical analysis for panels B-C was carried out with Student's t-test.
Discussion
Pharmacological inhibition of disease-associated miRNAs holds the promise of a potential therapeutic approach for a number of diseases. However, many therapeutically interesting miRNAs belong to families with two or more family members exhibiting differences in sequences outside of the seed, and unrelated miRNAs may also share the same seed, rendering therapeutic targeting efforts based on sequence-specific antisense inhibition challenging. In a previous study, we showed that miRNA families could be effectively inhibited in mice using 8-mer LNA-antimiRs targeting the seed sequence shared among the miRNA family members (10). In this report, we demonstrate pharmacological inhibition of a family of miRNAs, miR-33a/b, by an 8-mer seed-targeting LNA-antimiR in non-human primates. Our results show that such targeting is efficacious and without detectable adverse effects, indicating that a seed-targeting approach could potentially be explored for the inhibition of miR-33 and other disease-associated miRNA families in humans.
The miR-33 family of miRNAs is a regulator of circulating HDL-C and reverse cholesterol transport from cholesterol-loaded macrophages (14, 15, 20). Antisense inhibition or knockout of the single miR-33 species in mice has been shown to reduce arterial plaque burden, suggesting a therapeutic strategy for the treatment of atherosclerosis (20, 24). With this study, we confirm that inhibition of both miR-33a and b in female obese and insulin-resistant non-human primates increases HDL-C by up to ~40% compared with vehicle-treated controls at the same time point. Accordingly, we find hepatic up-regulation of the ABCA1 cholesterol transporter as well as prominent de-repression of other confirmed targets, such as CROT and AMPKα1. We found no indication of injection site reactions or other adverse effects of treatment for over 100 days with the 8-mer antimiR-33a/b targeting both miR-33a and b. The clinical chemistry and pathologic analyses demonstrated no evidence of toxicity, inflammation, or other deleterious changes associated with the 8-mer antimiR-33a/b treatment, indicating its apparent safety.
Using a longer 21-mer 2’-F/MOE-modified antimiR with a single mismatch to both miR-33a and b, Rayner et al. recently showed that inhibition of miR-33a and b in healthy male non-human primates increased circulating HDL-C concentration (21), but they did not examine the possibility that miR-33a and b might act in a redundant fashion. Our data demonstrate that miR-33 family members indeed function redundantly to control HDL-C, highlighting the importance of targeting both family members simultaneously. In addition to effects on HDL-C, the Rayner et al. study showed that miR-33 antagonism also reduced VLDL-associated triglycerides in their cohort of normal male African green monkeys (21). We did not observe significant changes in triglyceride concentrations upon miR-33a/b-targeting LNA-antimiR treatment, but the obese female non-human primates in our study displayed elevated fasting glucose, poor glucose regulation in intravenous glucose tolerance tests, and extremely low fasting insulin as compared to previously published data with normal animals (30), consistent with insulin resistance progressing towards a diabetic phenotype. Because insulin activates hepatic SREBP-1c expression and promotes lipogenesis and hepatic triglyceride synthesis (31), the low insulin concentrations observed in our obese African green monkey cohort could result in low SREBP-1c expression with concomitant low miR-33b expression, and thus milder effects of miR-33 antagonism on triglycerides. Accordingly, in contrast with the findings of Rayner et al., the high-carbohydrate diet in our study did not result in miR-33b up-regulation. Nevertheless, it is important to note that even under conditions of obesity, hyperglycemia, and low insulin responsiveness in this severe metabolic disease primate model, pharmacological inhibition of both miR-33a and b by an 8-mer antimiR oligonucleotide results in increased circulating HDL-C. In contrast to our study as well as other studies (21, 24, 32), a recent report by Marquart et al. showed that LDLr null mice on a very high (1.25%) cholesterol/high-fat diet treated with an antimiR-33 oligonucleotide for 12 weeks did not exhibit elevated HDL-C, had no improvement in atherosclerosis, and also displayed elevated triglycerides (33). The different outcomes of these studies illustrate that inhibition of miR-33 might have distinct effects under different experimental circumstances (such as large excess versus moderate dietary cholesterol), and further underlines the importance of testing the potential therapeutic use of miR-33 inhibition in an animal model as close to the clinic as possible, as in our current primate study.
Taken together, our results show that inhibition of a miRNA family by an 8-mer antimiR in non-human primates is effective and well tolerated. This opens the path for clinical studies of seed-targeting antimiR-33a/b inhibitors for the treatment of cardiovascular disease in humans, as well as the potential use of seed-targeting 8-mer LNA-modified antimiRs for the inhibition of other miRNA families in human diseases.
Materials and Methods
Study design
We and others have recently shown that miRNA families can be targeted by seed-targeting 8-mer LNA-antimiR oligonucleotides in vitro and in mice, however this approach has not been evaluated in non-human primates or humans. In this study, we employed LNA-antimiR oligonucleotides targeting selectively miR-33a (antimiR-33a) or miR-33b (antimiR-33b) or targeting both miR-33a and b (antimiR-33a/b) to assess the potential redundancy of the miR-33a/b family members in regulating circulating HDL-C as well as the efficacy and safety of the seed-targeting 8-mer antimiR-33a/b in obese African green monkeys.
We first verified the efficacy of the LNA-modified antimiRs in mice (6 mice per group, saline as vehicle control). For the non-human primate study, 20 obese female African green monkeys were fed a high fat diet for 12 weeks, followed by a high carbohydrate diet. Animals were assigned randomly to experimental groups based on ranks of levels of HDL, triglyceride and LDL after 12 weeks on the high fat diet feeding, and the animals were treated with antimiR-33a, antimir-33b or antimiR-33a/b or with saline vehicle (5 monkeys per study group) for a total of 108 days.
Oligonucleotides
The LNA-antimiR and control oligonucleotides were synthesized with a complete phosphorothioate backbone, and duplex melting temperatures were determined as previously described (34) (table S1).
Cell Culture Experiments
HepG2 cells (ECACC #85011430) were cultured in EMEM medium (with 10% FBS, 2 mM Glutamax and 25 µg/ml gentamicin) for the qRT-PCR experiments. For the western blot analysis, HepG2 cells were cultured in MEM (with 2 mM L-Glutamine, 50 units/ml penicillin, 50 µg/ml streptomycin and 1mM Sodium Pyruvate). Transfections for the luciferase assays and qRT-PCR analyses were performed using Lipofectamine 2000 (Invitrogen). In brief, 6.5x105 HepG2 cells were seeded per well in a 6-well plate. Cells were transfected with 0.6 µg miR-33a or miR-33b luciferase reporter plasmid (psiCHECK2; Promega) together with varying concentrations of the LNA-antimiR oligonucleotides and 2.55 µl Lipofectamine 2000 per well according to the manufacturer’s instructions. After 24 hours, the cells were harvested and luciferase activity was measured.
Transfections for western blot analysis were done using the Amaxa system (Lonza). Briefly, before transfection, 6-well plates were treated with 10 µg/ml poly-D-lysine hydrobromide (Sigma P7405) solution for 10 min. and thoroughly rinsed with PBS. LNA-antimiR or control oligonucleotide was added to 2x106 HepG2 cells and transfected using Amaxa according to the manufacturer's instructions at a final concentration of 5 nM. At indicated time points, the cells were harvested and subjected to immunoblotting.
Luciferase Reporter Assays
The miR-33 luciferase reporters were generated by cloning annealed 5’ phosphorylated oligonucleotides corresponding to single perfect match target sites for human miR-33a and miR-33b, respectively, into the 3’UTR of the Renilla luciferase gene in the psiCHECK2 plasmid (Promega) (XhoI/NotI sites). In addition to the Renilla luciferase gene, the psiCHECK-2 dual-luciferase plasmid also contains a constitutively expressed firefly luciferase gene for normalization of Renilla luciferase expression. Cell transfection and luciferase activity measurements were carried out according to the manufacturer's instructions (Invitrogen Lipofectamine 2000 and Promega Dual-luciferase kits).
RNA Isolation and Real-Time Quantitative RT-PCR
RNA from cells and mouse tissues was isolated using the Trizol reagent according to the manufacturer's instructions (Invitrogen). RNA from the non-human primate liver biopsies was isolated using the mirVana miRNA Isolation Kit protocol (Life Technologies). mRNA quantification was carried out using TaqMan assays and an Applied Biosystems 7500 real-time PCR instrument (Applied Biosystems), and miR-33a and miR-33b quantification was performed using the Exiqon miRCURY LNA™ Universal RT microRNA PCR assays according to the manufacturers’ protocols.
Mouse Studies
C57BL/6J female mice (Taconic) with an average body weight of 25 g at first injection received regular chow diet (Altromin #1324, Brogaarden). The animal housing rooms were maintained on a 12:12-h light:dark cycle with relative humidity of 55 ± 10% and a temperature of 21°C ± 2°C. The experiments were performed according to the principles stated in the Danish law on animal experiments and were approved by the Danish National Committee for Animal Experiments, Ministry of Justice, Denmark. All LNA-antimiR compounds were formulated in physiological saline (0.9% NaCl) to a final concentration allowing the mice to receive a tail-vein injection volume of 10 ml/kg. C57BL/6J mice were dosed for three consecutive days with daily doses of 25 mg/kg of the LNA-antimiR compounds or saline, and were sacrificed 24 hours after the last dose. At sacrifice, serum samples were taken and the mouse livers were snap-frozen at -80°C.
Non-Human Primate Studies
All African green monkey experiments were performed at the RxGen primate facility on the St. Kitts Biomedical Research Foundation campus, St. Kitts, in accordance with National Institutes of Health guide for the Care and Use of Laboratory Animals and were approved by the institutional Animal Care and Use Committee. Twenty moderately obese (table S3) adult female African green monkeys were selected for the study and fed a high-fat diet for 12 weeks. High-fat diet chow was formulated by adding 1 L of melted lard (Crisco, J.M. Smucker Company) containing 10 g of cholesterol (Sigma #C8503) to 10 kg of standard monkey chow (Teklad diet #2055). Before the administration of the LNA-antimiR compounds the animals were randomly divided into 4 groups of 5 animals. The LNA-modified antimiR-33a, antimiR-33b and antimiR-33a/b compounds formulated in a 0.9% NaCl solution were subcutaneously delivered at a dose of 20 mg/kg on day 0, and a 5 mg/kg maintenance dose at weekly intervals until day 106. Vehicle controls received a 0.9% NaCl solution. At day 57 (after 8 weeks of treatment), the animals were shifted to a high-carbohydrate diet. Standard monkey chow (10 kg, TekLad diet #2055) was mixed with 1 L of a 3 kg/L consumer-grade granulated fructose (Mysage.com) solution resulting in 0.23 g of fructose per gram of modified chow and an estimated maximal available daily intake of 28 g of fructose in 120 g of modified diet. A percutaneous liver biopsy was performed on all monkeys at four time points during the course of the study (pre-high fat diet (Baseline), 12 weeks after initiating the high-fat diet (HFD), at day 57 and day 120). For liver biopsies animals were initially sedated by intramuscular injection of ketamine (10 mg/kg) and anesthesia subsequently maintained using 2% isoflurane.
Blood Chemistry
Glucose, blood urea nitrogen (BUN), creatinine, total protein, albumin, total bilirubin, alkaline phosphatase, alanine aminotransferase (ALT), aspartate aminotransferase (AST), cholesterol, calcium, phosphorus, sodium, potassium, chloride, albumin/globulin ratio, BUN/creatinine (calculated), globulins (calculated), lipase, amylase, triglycerides, creatinine phosphokinase (CPK), lactate dehydrogenase, gamma glutamyl transpeptidase (GGTP), and magnesium were measured by Antech Diagnostics on an Olympus AU5400 fully automated analyzer. Analysis was conducted on plasma samples collected during prescreening and at pre-diet baseline, 12 weeks after initiating high-fat diet feeding and on study days 1, 8, 36 and 108.
Lipid Chemistry
For the mouse studies, serum was analyzed for total cholesterol using ABX Pentra Cholesterol CP (Horiba ABX Diagnostics) according to the manufacturer’s instructions. The different serum lipoprotein classes were separated by FPLC (fast protein liquid chromatography) using a Shimadzu FPLC system with a knitted teflon coil (Supelco) as reaction chamber and a 25 ml Superose 6 column (GE Healthcare). In brief, 10-15 µl aliquots of serum were injected by a Shimadzu autosampler and separated in lipoprotein fractions by size exclusion chromatography before entering the reaction chamber where it was mixed with colorimetric cholesterol reagent (Infinity reagent, TR13521, Thermo Trace). The reaction proceeded at 37°C for 5 min. resulting in full color development followed by detection (480 nM) and quantification by LC solution software (Shimadzu). For the non-human primate studies, total cholesterol, HDL-cholesterol, and triglycerides were measured in plasma samples collected at pre-diet baseline, 12 weeks after initiating high-fat diet feeding and on study days 0, 1, 3, 5, 8, 11, 15, 18, 22, 28, 36, 43, 50, 57, 71, 78, 85, 92, 99 and 106 by Antech GLP on an Olympus AU640 fully automated analysis system using reagent sets #OSR6116 (total cholesterol), #OSR6133 (triglycerides), #OSR6156 (HDL) and #OSR6196 (LDL).
Insulin and Glucose Measurements
Plasma insulin was measured with an ultrasensitive human insulin ELISA (Mercodia) according to the manufacturer's protocol. Plasma glucose was measured with a colorimetric assay kit (Cayman Chemical).
Western Blot Analysis
Liver biopsies were taken at indicated time points, and placed in RNAlater (Qiagen). A small piece of liver tissue was cut off and put into 500 µl of high-salt (450 mM NaCl) RIPA buffer and homogenized on ice. Samples were briefly sonicated, spun at high speed for 10 min. at 4°C, and the supernatant was aliquoted and flash frozen in liquid nitrogen. Western blotting was performed according to standard procedures. Briefly, samples with added sample buffer were run on 4-12% Bis-Tris NuPage gels (Life Sciences). Proteins were then transferred onto nitrocellulose membranes. Blots were incubated with antibodies (ABCA1, Abcam #ab18180; SIRT6, Abcam #ab62739; AMPKα1, #ab32047; β-tubulin, Sigma #T7816) overnight at 4°C. Blots were then washed in TBST, incubated with secondary antibody for one hour at room temperature, washed briefly and then exposed (Immobilon Western reagent, Millipore). Band intensity of ABCA1 was normalized to β-tubulin using ImageJ (NIH).
Hematoxylin & Eosin Staining
The liver biopsies, performed at each study interval, were assessed for histological changes after antimiR treatment and dietary interventions. Tissue samples were placed in paraformaldehyde and paraffin embedded. Representative sections were stained with hematoxylin and eosin and assessed for hepatic steatosis. Metamorph® NX software was used to calculate percentage of hepatic steatosis from digital images obtained at 20x magnification.
Pharmacokinetics Analysis
Blood samples were collected at different time points after the last dose of antimiR-33a/b into tubes containing K2-EDTA. Plasma was subsequently prepared by centrifugation for 10 min. at 1,300xg and 4°C. The plasma samples and plasma standards (~10 mg/L) were diluted 10 times into 150 µl of a 35 nM solution of a biotinylated and digoxigenin-modified capture and detection probe and mixed for an hour. Streptavidin-coated plates (Nunc Immobilizer Streptavidin F96 CLEAR module plate Nunc #436014) were washed three times. The plasma samples (100 µL) were transferred to the streptavidin-coated plates and incubated for one hour. The wells were aspirated and washed three times with 300 μl of 2x SSCT buffer. 100 units of S1 nuclease (Invitrogen 18001-016) in nuclease buffer (0.28 M NaCl, 50 mM NaAc, 4.5 mM ZnSO4, pH 4.6) were added per well and the plates were incubated at 35°C for 1 hour. The wells were aspirated and washed three times with 300 μl of 2x SSCT buffer. One hundred microliters of anti-Dig-AP Fab fragments (Roche Applied Science, #11 093 274 910) diluted 1:2,000 in PBST (Phosphate buffered saline, pH 7.2) were added to the wells and incubated for 1 hour at room temperature. The wells were aspirated and washed three times with 300 μl of 2x SSCT buffer. One hundred microliters of substrate solution (KPL BluePhos Microwell Phosphatase substrate system 50-88- 00) were added to each well. The intensity of the color development was measured spectrophotometrically at 615 nm every 5 min. after shaking. The test samples were referenced against the standard samples. The determined oligonucleotide concentrations in plasma samples were used in the PK analysis. The mean values of the individual oligonucleotide plasma concentrations at the different collection times (after the last dose) were subjected to noncompartmental PK evaluation using the PC-based software WinNonlin, version 5.3. (Pharsight Corporation).
DNA Microarray Analysis
DNA microarray analysis was performed on RNA extracted from the liver biopsies of African green monkeys taken at the HFD:antimiR time point (Day 57) during the study. A 12x135K human expression array from NimbleGen (Roche) was used for the RNA expression analysis. In brief, double-stranded cDNA (ds cDNA) was prepared from 1 µg of total RNA using the cDNA Synthesis System (Roche). DsDNA was labeled with Cy3 using the Roche One Color DNA Labeling kit following the manufacturer's protocol. Labeled cDNA (6 µg) was hybridized to the array for 20 hours following the manufacturer's protocol. Arrays were washed using the NimbleGen wash buffer kit following the manufacturer's protocol and scanned on the ms200 scanner. Images were aligned and features were extracted using Nimblescan 2.6 software. Data were analyzed with the Agilent GeneSpring GX 11.0 using the Percentile Shift Normalization algorithm. Fold changes and GO term analysis were also performed in Agilent GeneSpring GX 11.0. The array data are deposited in the Gene Expression Omnibus repository (accession numbers: GSE50116, and GSM1214398 to GSM1214407).
Statistical analysis
Microsoft Excel was used for data analysis, except for the microarray data. Testing of statistical significance was performed using Student's t-test (two-tailed, homoscedastic). Error bars in the figures denote standard deviations for all cell culture experiments and standard error of the mean for animal experiments. Microarray data were normalized using the Percentile Shift Normalization algorithm in Agilent GeneSpring GX 11.0. Calculations of cumulative distributions and the Kolmogorov-Smirnov tests were performed in R, an environment for statistical computing and graphics.
Supplementary Material
One Sentence Summary.
Long-term treatment of obese, insulin-resistant non-human primates with a subcutaneously delivered 8-mer seed-targeting antimiR oligonucleotide against the microRNA-33 family effectively de-represses hepatic expression of miR-33 targets, increases circulating HDL-cholesterol, and is associated with a clean safety profile.
Acknowledgments
The authors thank Dianna Elwood, Ernell Nisbett, Mike Struharik and Michelle Ocana for technical assistance and Robert Persson for input on the pharmacokinetics analysis. We also thank the MGH Neurobiology Department and the Neurobiology Imaging Facility for consultation and instrument availability that supported this work. Funding: This work was supported by the following grants to A.M.N.: US National Institutes of Health (NIH) grants R21DK084459 and R01DK094184, and the Partners Innovation Fund. R.T.C. was supported by NIH DK078772 (mentorship grant), and A.S.D. by NIH DK093216-01A1 (F32 Grant). JRW was supported by NIH R01GM097360, CA059267and RSG-13-115-01-CCG.The MGH Neurobiology Imaging facility is supported in part by the Neural Imaging Center as part of an NINDS P30 Core Center grant #NS072030.
Footnotes
Author contributions: A.M.N., S.K., and S.H.N., conceptualized the study, V.R., S.O. and M.W.L. designed and performed the experiments and carried out data analysis, A.P. performed data analysis, R.M., S.S. and R.E.G. designed and performed the insulin and glucose measurements, J.C.B. and J.R.W. designed and performed the DNA microarray experiments, A.S.D. and R.T.C. performed the pathology analysis on the liver biopsy samples, H.F.H. performed the pharmacokinetics analysis, R.G., S.W., S.H. and R.B. designed and performed the non- human primate experiments and sample collection, A.M.N. and S.K. supervised the study, V.R. and A.M.N. wrote the manuscript with input from all authors.
Competing interests: S.O., M.W.L. and H.F.H. are employees of Santaris Pharma A/S, a clinical tage biopharmaceutical company that develops RNA-based therapeutics. S.K. and A.P. are former employees of Santaris Pharma A/S; S.K. is now Professor and A.P. is Bioinformatician at the Department of Haematology, Aalborg University Hospital. A.M.N. has patents pending on miR-33a and miR-33b antisense targeting for the treatment of cardiometabolic disorders.
Data and materials availability: The DNA microarray data for this study have been deposited in the Gene Expression Omnibus repository (accession numbers: GSE50116, GSM1214398-GSM1214407).
References
- 1.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hertel J, Lindemeyer M, Missal K, Fried C, Tanzer A, Flamm C, Hofacker IL, Stadler PF. The expansion of the metazoan microRNA repertoire. BMC Genomics. 2006;7:25. doi: 10.1186/1471-2164-7-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 6.Mendell JT, Olson EN. MicroRNAs in stress signaling and human disease. Cell. 2012;148:1172–1187. doi: 10.1016/j.cell.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk ME, Kauppinen S, Orum H. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Janssen HL, Reesink HW, Lawitz EJ, Zeuzem S, Rodriguez-Torres M, Patel K, van der Meer AJ, Patick AK, Chen A, Zhou Y, Persson R, King BD, Kauppinen S, Levin AA, Hodges MR. Treatment of HCV infection by targeting microRNA. N Engl J Med. 2013;368:1685–1694. doi: 10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]
- 9.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Molecular cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Obad S, dos Santos CO, Petri A, Heidenblad M, Broom O, Ruse C, Fu C, Lindow M, Stenvang J, Straarup EM, Hansen HF, Koch T, Pappin D, Hannon GJ, Kauppinen S. Silencing of microRNA families by seed-targeting tiny LNAs. Nat Genet. 2011;43:371–378. doi: 10.1038/ng.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y, Roccaro AM, Rombaoa C, Flores L, Obad S, Fernandes SM, Sacco A, Liu Y, Ngo H, Quang P, Azab AK, Azab F, Maiso P, Reagan M, Brown JR, Thai TH, Kauppinen S, Ghobrial IM. LNA-mediated anti-miR-155 silencing in low-grade B-cell lymphomas. Blood. 2012;120:1678–1686. doi: 10.1182/blood-2012-02-410647. [DOI] [PubMed] [Google Scholar]
- 12.Leucci E, Zriwil A, Gregersen LH, Jensen KT, Obad S, Bellan C, Leoncini L, Kauppinen S, Lund AH. Inhibition of miR-9 de-represses HuR and DICER1 and impairs Hodgkin lymphoma tumour outgrowth in vivo. Oncogene. 2012 doi: 10.1038/onc.2012.15. [DOI] [PubMed] [Google Scholar]
- 13.Bernardo BC, Gao XM, Winbanks CE, Boey EJ, Tham YK, Kiriazis H, Gregorevic P, Obad S, Kauppinen S, Du XJ, Lin RC, McMullen JR. Therapeutic inhibition of the miR-34 family attenuates pathological cardiac remodeling and improves heart function. Proc Natl Acad Sci U S A. 2012;109:17615–17620. doi: 10.1073/pnas.1206432109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Najafi-Shoushtari SH, Kristo F, Li Y, Shioda T, Cohen DE, Gerszten RE, Naar AM. MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science. 2010;328:1566–1569. doi: 10.1126/science.1189123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rayner KJ, Suarez Y, Davalos A, Parathath S, Fitzgerald ML, Tamehiro N, Fisher EA, Moore KJ, Fernandez-Hernando C. MiR-33 contributes to the regulation of cholesterol homeostasis. Science. 2010;328:1570–1573. doi: 10.1126/science.1189862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marquart TJ, Allen RM, Ory DS, Baldan A. miR-33 links SREBP-2 induction to repression of sterol transporters. Proc Natl Acad Sci U S A. 2010;107:12228–12232. doi: 10.1073/pnas.1005191107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horie T, Ono K, Horiguchi M, Nishi H, Nakamura T, Nagao K, Kinoshita M, Kuwabara Y, Marusawa H, Iwanaga Y, Hasegawa K, Yokode M, Kimura T, Kita T. MicroRNA-33 encoded by an intron of sterol regulatory element-binding protein 2 (Srebp2) regulates HDL in vivo. Proc Natl Acad Sci U S A. 2010;107:17321–17326. doi: 10.1073/pnas.1008499107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gerin I, Clerbaux LA, Haumont O, Lanthier N, Das AK, Burant CF, Leclercq IA, MacDougald OA, Bommer GT. Expression of miR-33 from an SREBP2 intron inhibits cholesterol export and fatty acid oxidation. The Journal of biological chemistry. 2010;285:33652–33661. doi: 10.1074/jbc.M110.152090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davalos A, Goedeke L, Smibert P, Ramirez CM, Warrier NP, Andreo U, Cirera-Salinas D, Rayner K, Suresh U, Pastor-Pareja JC, Esplugues E, Fisher EA, Penalva LO, Moore KJ, Suarez Y, Lai EC, Fernandez-Hernando C. miR-33a/b contribute to the regulation of fatty acid metabolism and insulin signaling. Proc Natl Acad Sci U S A. 2011;108:9232–9237. doi: 10.1073/pnas.1102281108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rayner KJ, Sheedy FJ, Esau CC, Hussain FN, Temel RE, Parathath S, van Gils JM, Rayner AJ, Chang AN, Suarez Y, Fernandez-Hernando C, Fisher EA, Moore KJ. Antagonism of miR-33 in mice promotes reverse cholesterol transport and regression of atherosclerosis. J Clin Invest. 2011 doi: 10.1172/JCI57275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rayner KJ, Esau CC, Hussain FN, McDaniel AL, Marshall SM, van Gils JM, Ray TD, Sheedy FJ, Goedeke L, Liu X, Khatsenko OG, Kaimal V, Lees CJ, Fernandez-Hernando C, Fisher EA, Temel RE, Moore KJ. Inhibition of miR-33a/b in non-human primates raises plasma HDL and lowers VLDL triglycerides. Nature. 2011;478:404–407. doi: 10.1038/nature10486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Attie AD. ABCA1: at the nexus of cholesterol, HDL and atherosclerosis. Trends Biochem Sci. 2007;32:172–179. doi: 10.1016/j.tibs.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 23.Rader DJ, Tall AR. The not-so-simple HDL story: Is it time to revise the HDL cholesterol hypothesis? Nat Med. 2012;18:1344–1346. doi: 10.1038/nm.2937. [DOI] [PubMed] [Google Scholar]
- 24.Horie T, Osamu B, Kuwabara Y, Chujo Y, Watanabe S, Kinoshita M, Horiguchi M, Nakamura T, Chonabayashi K, Hishizawa M, Hasegawa K, Kume N, Yokode M, Kita T, Kimura T, Ono K. MicroRNA-33 Deficiency Reduces the Progression of Atherosclerotic Plaque in ApoE−/− Mice. JAMA. 2012 doi: 10.1161/JAHA.112.003376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Allen RM, Marquart TJ, Albert CJ, Suchy FJ, Wang DQ, Ananthanarayanan M, Ford DA, Baldan A. miR-33 controls the expression of biliary transporters, and mediates statin- and diet-induced hepatotoxicity. EMBO Mol Med. 2012 doi: 10.1002/emmm.201201228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rottiers V, Najafi-Shoushtari SH, Kristo F, Gurumurthy S, Zhong L, Li Y, Cohen DE, Gerszten RE, Bardeesy N, Mostoslavsky R, Naar AM. MicroRNAs in Metabolism and Metabolic Diseases. Cold Spring Harb Symp Quant Biol. 2011 doi: 10.1101/sqb.2011.76.011049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liddie S, Goody RJ, Valles R, Lawrence MS. Clinical chemistry and hematology values in a Caribbean population of African green monkeys. J Med Primatol. 2010;39:389–398. doi: 10.1111/j.1600-0684.2010.00422.x. [DOI] [PubMed] [Google Scholar]
- 28.Avramoglu RK, Basciano H, Adeli K. Lipid and lipoprotein dysregulation in insulin resistant states. Clin Chim Acta. 2006;368:1–19. doi: 10.1016/j.cca.2005.12.026. [DOI] [PubMed] [Google Scholar]
- 29.Straarup EM, Fisker N, Hedtjarn M, Lindholm MW, Rosenbohm C, Aarup V, Hansen HF, Orum H, Hansen JB, Koch T. Short locked nucleic acid antisense oligonucleotides potently reduce apolipoprotein B mRNA and serum cholesterol in mice and non-human primates. Nucleic Acids Res. 2010;38:7100–7111. doi: 10.1093/nar/gkq457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cann JA, Kavanagh K, Jorgensen MJ, Mohanan S, Howard TD, Gray SB, Hawkins GA, Fairbanks LA, Wagner JD. Clinicopathologic characterization of naturally occurring diabetes mellitus in vervet monkeys. Vet Pathol. 2010;47:713–718. doi: 10.1177/0300985810370011. [DOI] [PubMed] [Google Scholar]
- 31.Chen G, Liang G, Ou J, Goldstein JL, Brown MS. Central role for liver X receptor in insulin-mediated activation of Srebp-1c transcription and stimulation of fatty acid synthesis in liver. Proc Natl Acad Sci U S A. 2004;101:11245–11250. doi: 10.1073/pnas.0404297101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rotllan N, Ramirez CM, Aryal B, Esau CC, Fernandez-Hernando C. Therapeutic Silencing of MicroRNA-33 Inhibits the Progression of Atherosclerosis in Ldlr-/- Mice. Arterioscler Thromb Vasc Biol. 2013 doi: 10.1161/ATVBAHA.113.301732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marquart TJ, Wu J, Lusis AJ, Baldan A. Anti-miR-33 therapy does not alter the progression of atherosclerosis in low-density lipoprotein receptor-deficient mice. Arterioscler Thromb Vasc Biol. 2013;33:455–458. doi: 10.1161/ATVBAHA.112.300639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Elmen J, Lindow M, Silahtaroglu A, Bak M, Christensen M, Lind-Thomsen A, Hedtjarn M, Hansen JB, Hansen HF, Straarup EM, McCullagh K, Kearney P, Kauppinen S. Antagonism of microRNA-122 in mice by systemically administered LNA-antimiR leads to up-regulation of a large set of predicted target mRNAs in the liver. Nucleic Acids Res. 2008;36:1153–1162. doi: 10.1093/nar/gkm1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.