Abstract
Cytoplasmic dynein is a homodimeric AAA+ motor that transports a multitude of cargos toward the microtubule minus end. How the two catalytic head domains interact and move relative to each other during processive movement is unclear. Here, we tracked the relative positions of both heads with nanometer precision and directly observed the heads moving independently along the microtubule. The heads remained widely separated, and their stepping behavior varied as a function of interhead separation. One active head was sufficient for processive movement, and an active head could drag an inactive partner head forward. Thus, dynein moves processively without interhead coordination, a mechanism fundamentally distinct from the hand-over-hand stepping of kinesin and myosin.
Cytoplasmic dynein forms a 1.2-MD complex that uses adenosine triphosphate (ATP) hydrolysis to power minus-end– directed motility along microtubules. The catalytic head domain is composed of six AAA+ [adenosine triphosphatases (ATPases) associated with diverse cellular activities] modules arranged in a ring (1).ATPase activity at AAA1 is essential for dynein motility (2). The two rings are connected by the dimerization of N-terminal tail domains and bind to the microtubule through a ~15-nm coiled-coil stalk bearing a small microtubule-binding domain (MTBD). Dynein is required for many cellular processes, including organelle transport and cell division, and dynein malfunction can lead to motor neuron degeneration (3).
Despite recent advances in understanding dynein’s structure (4) and mechanism (5, 6), the stepping mechanism of dimeric dynein during processive motion remains unclear. Studies of kinesin and myosin motors have shown that their heads take alternating steps in a hand-over-hand (HoH) fashion (7–9). Dynein motility also requires two heads (6), but it is unclear whether they coordinate with each other to achieve processive motion (10). Dynein’s structure and origin are distinct from kinesin and myosin, suggesting that it may use a different mechanism.
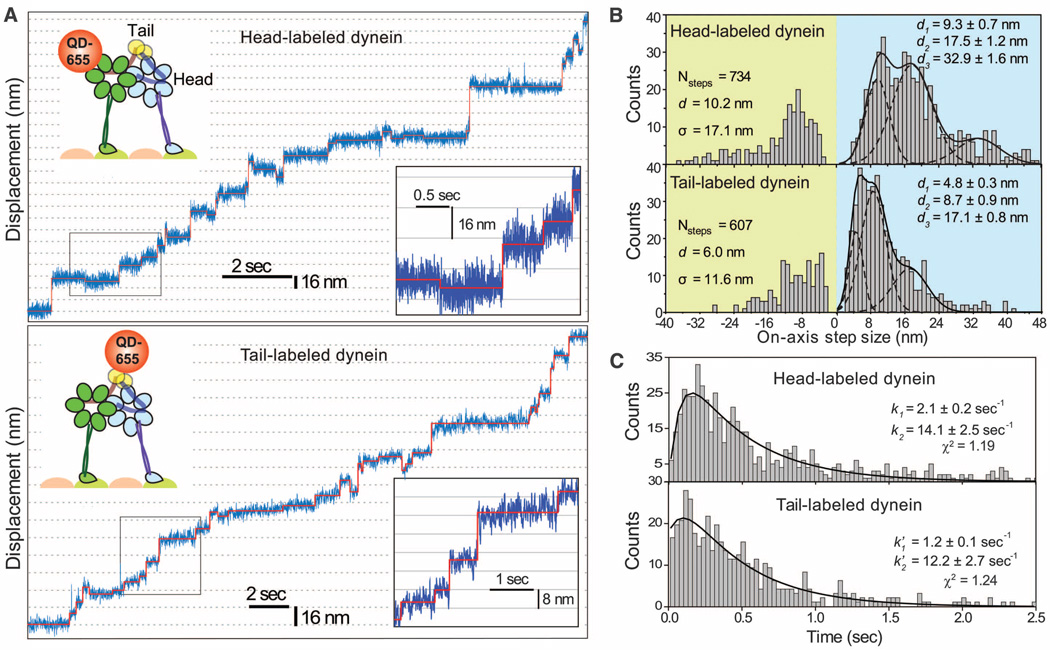
To investigate dynein’s stepping mechanism, we tracked the movement of an artificially dimerized, tail-truncated yeast cytoplasmic dynein [glutathione S-transferase–Dyn331kD (GST-Dyn331kD)], which has similar stepping properties to native dynein (6). First, we tracked the movement of dynein labeled with a single quantum dot (QD) at 2 msec temporal resolution in the presence of rate-limiting ATP (Fig. 1A and fig. S1). As reported previously (5, 6), the distribution of step sizes was multimodal. The step-size histogram of head-labeled dynein revealed two major peaks at 9.3 ±± 0.7 and 17.5 ± 1.2 nm that were nearly twice as large as the peaks observed in the tail histogram (4.8 ± 0.3 and 8.7 ± 0.9 nm) (Fig. 1B). The probability of backward stepping (pBW) was 0.2. The dwell-time histogram for head-labeled dynein at 12 µM ATP (Fig. 1C) was best fit by a convolution of one slow (k1=2.1 ± 0.2 sec−1) and one fast (k2 = 14.1 ± 2.5 sec−1) exponential rate constant. The product of k2 and the average head step size (dhead = 10.2 nm) agrees well with the average velocity of dynein at saturating [ATP] (124 nm/sec). This data excludes the symmetric HoH model, which predicts the stepping kinetics of a head to be a convolution of two equal rate constants (fig. S2). Instead, the dwell-time histogram of tail-labeled dynein was well described by a kinetic model in which the heads can step independently and the tail moves each time either head takes a step (k′1=1.2 ± 0.1 sec−1; k′2=12.2 ± 2.7 sec−1) (fig. S2).
Fig. 1.
(A) Step-size measurements of tail- and head-labeled GST-Dyn331kD with a single QD-655 at 2-msec temporal resolution. The QD position (blue traces) was fit by a step-finding algorithm (solid red lines). (B) Multiple Gaussian fits to step-size histograms reveal two major peaks at 9.3 and 17.5 nm for the head and 4.8 and 8.7 nm for the tail. (C) Dwell-time histogram of head-stepping fitted to a convolution of two unequal rate constants and that of tail-stepping fitted to a model assuming uncoordinated stepping between two heads, each with two unequal rate constants.
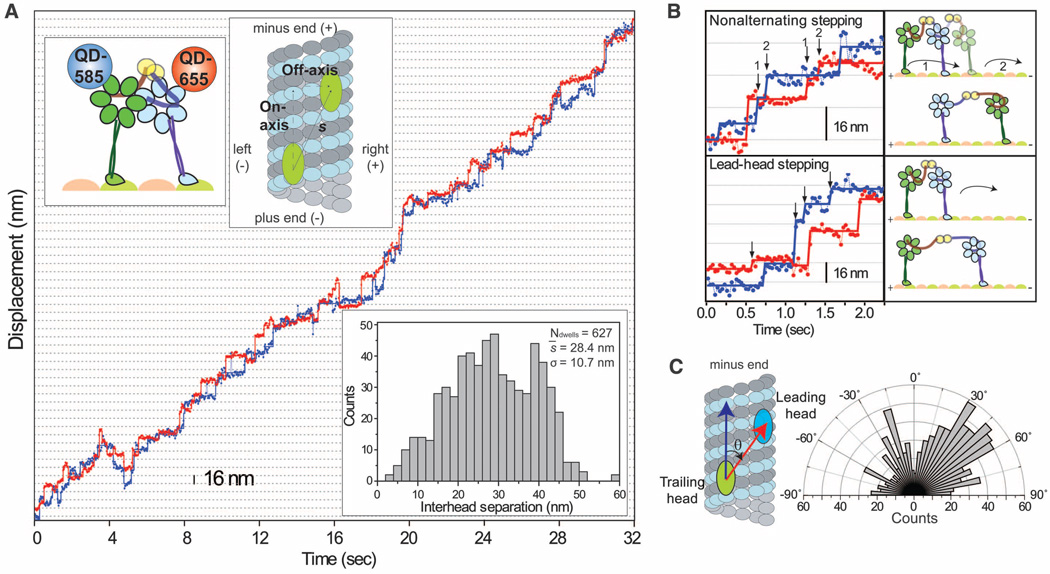
To directly address how the two heads interact and move relative to each other, we labeled GST-Dyn331kD with different-colored QDs (17 to 22 nm in size) (table S1) at the C termini and simultaneously tracked the positions of the heads during processive movement (11). The fluorescent signal was split into two channels and registered to 3-nm precision (figs. S3 to S5). Representative traces (Fig. 2A, fig. S6, and movies S1 and S2) clearly showed that either head could take a step regardless of which head was leading, a mechanism distinct from HoH. Although most steps showed a canonical alternating pattern, one head could take multiple consecutive (nonalternating) steps before the other head moved (Fig. 2B). Nonalternating stepping occurred about half as often as alternating stepping (32% of steps, fig. S7). This may partly be due to the time needed for a head to complete its ATPase cycle before it can take another step.
Fig. 2.
(A) Stepping trace of GST-Dyn331kD labeled with QD-585 (blue) and QD-655 (red) shows that the heads move independently of each other during processive runs. The heads are separated by 28.4 ± 10.7 nm (bottom inset). (B) Examples of nonalternating and lead-head stepping (arrows) show that dynein stepping deviates from the HoH mechanism. (C) Histogram of the angles between the interhead separation vector (red arrow) and the microtubule long axis (blue arrow).
The heads often walked along different protofilaments (fig. S6), with the leading head preferentially located to the right of the trailing head (Fig. 2C and fig. S8). Measurements using organic dyes showed that the heads were separated by 23.0 ± 13.2 nm (mean ± SD) (fig. S9), excluding the possibility of a stacking interaction between the AAA+ rings (12).
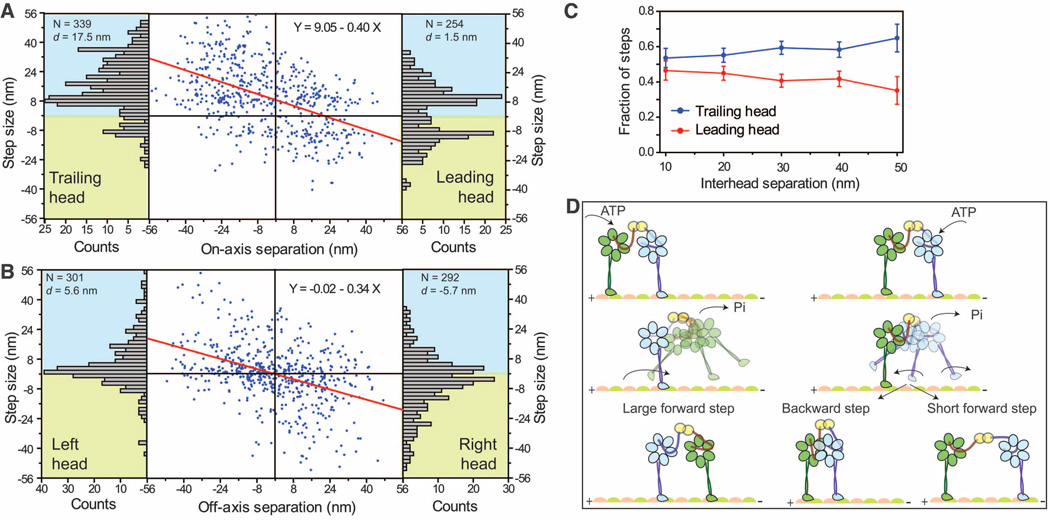
The heads frequently swapped the lead, but not strictly after every step of the motor. The stepping characteristics of a head in the leading and trailing positions differed substantially depending on interhead separation, forming the basis of the high variability in dynein’s step size (6). The on-axis (parallel to the microtubule long axis) step size decreased by ~0.4 nm per nanometer increase in interhead separation (Fig. 3A). The trailing head took larger (d = 17.5 nm) forward and fewer (pBW= 0.14) backward steps. In contrast, the leading head took significantly shorter (d = 1.5 nm) forward and more frequent (pBW = 0.45) backward steps. The off-axis (perpendicular to the microtubule long axis) step size also decreased with interhead separation but without a net bias toward left or right (Fig. 3B). The stepping probabilities of the leading and trailing heads were nearly identical when they were positioned close (10 to 20 nm) to each other. At larger separations, 65% of the steps were taken by the trailing head (Fig. 3C).
Fig. 3.
(A) The on-axis step size (blue dots) of GST-Dyn331kD decreases linearly (red line) as a function of interhead separation and is biased forward by 9.1 ± 0.6 nm. The leading head takes shorter (d = 1.5 nm) steps with more frequent (pBW = 0.45) backward steps, compared with the trailing head (d = 17.5 nm; pBW = 0.14) (bar graphs). (B) Off-axis step sizes show a linear dependence on interhead separation without a bias to move toward the right (positive) or left (negative). (C) Fraction of the steps taken by the leading and trailing heads at different interhead separations (mean ± SEM). (D) Tethered excursion model for the dynein stepping mechanism. Either the leading or the trailing head can hydrolyze ATP and release from the microtubule. A diffusional search of the trailing head (green) is biased forward by interhead tension and the linker swing, resulting in a large forward step. In contrast, linker swing and tension bias the diffusion of the leading head (blue) in opposing directions, resulting in either a backward step or a short forward step.
The heads of native dynein also moved independently, and their stepping behavior varied as a function of interhead separation, similar to GST-Dyn331kD (figs. S10 and S11). The results show that the tail domain is not involved in interhead coordination.
Our results show that dynein’s stepping mechanism is different from processive kinesins and myosins. The stepping motion of these motors is driven by the power stroke of the bound head, which moves the trailing head forward. For dynein, we propose a tethered excursion model (Fig. 3D), in which a conformational change that produces the minus-end bias occurs in the unbound head. Either head can bind ATP at the AAA1 site, followed by its release from the microtubule (13). The linker undergoes a minus-end–directed priming stroke upon ATP hydrolysis (14), which moves its MTBD forward. The large and flexible linker allows the released head to diffuse over a wide area, resulting in both large interhead separations and variable step sizes.
The heads experience intramolecular tension at large separations, and the power stroke of the bound head may occasionally trigger the release of the other head. In fact, we observed that the stepping probability of the trailing head increased at large separations (Fig. 3C). This result is consistent with force-induced movement of dynein, which requires less force when it is pulled toward the minus end (15). In addition, tension can bias the diffusional search of the tethered head toward the bound head, preventing further extension of the dimer (Fig. 3D).
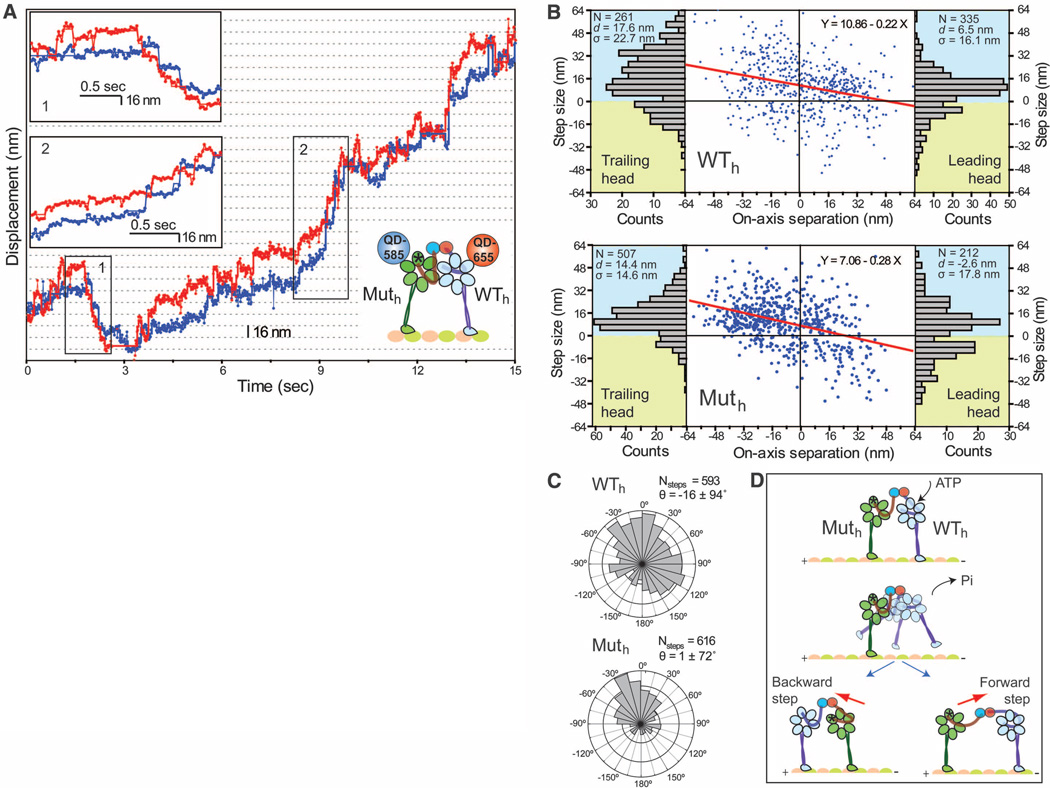
The tethered excursion model predicts that a single force-generating head in a dynein dimer can both take a step and drag an inactive head forward. We tested the motility of a heterodimeric dynein with one wild-type head (WTh) and one mutant head lacking the ability to hydrolyze ATP at the AAA1 domain (Muth) (6, 16). Muth cannot undergo a power stroke (17) and is weakly associated with the microtubule (13). The WTh\Muth heterodimer moved processively toward the microtubule minus end (movies S3 and S4, Fig. 4A, and figs. S12 and S13).WTh was found in the lead 66% of the time (fig. S14). Similarly, during occasional short (4 to 5 steps) backward runs, WTh remained in the leading position toward the plus end.
Fig. 4.
(A) Stepping trace of the WTh\Muth heterodimer. Muth and WTh were labeled with QD-585 (blue trace) and QD-655 (red trace), respectively. WTh mostly remains in the lead during a short backward run (insert 1) and a long forward run (insert 2). (B) Plot of on-axis step size (blue dots) of WTh and Muth versus interhead separation. The step size decreases linearly (red line) with increased interhead separation and is biased forward by 10.9 ± 0.8 nm for WTh and 7.1 ± 0.7 nm for Muth. Step-size histograms (bar graphs) show that dWT = 6.5 nm whereas dMut = −2.6 nm in the lead. (C) Histogram of the angles between interhead separation and stepping vectors of Muth and WTh. (D) Model for WTh\Muth processivity. ATP binding to WTh triggers its release from the microtubule and the subsequent diffusional search is biased forward by the power stroke of the WTh linker. The weakly attached Muth moves toward WTh under linker tension.
Muth exhibited different stepping characteristics in both the leading and trailing positions (Fig. 4B) than WTh. The on-axis step size of Muth had reduced (7.1 ± 0.7 nm) minus-end– directed bias, compared with WTh (10.9 ± 0.8 nm). The probability of Muth being in the lead decreased as interhead separation increased (<25% at 30+ nm separations) (fig. S15), and Muth was more likely to step backward from the lead (d = –2.6 nm; pBW = 0.44) (Fig. 4B). The average step size and stepping rate of Muth were similar to those of WTh (figs. S14 and S15). Muth stepping was mostly directed toward WTh, whereas the direction of WTh stepping was largely independent of the position of Muth (Fig. 4C).
Thus, dynein motility does not require allosteric communication between the AAA1 sites, and only one force-generating head is sufficient for processive movement. WTh usually remains in the lead and drives forward movement. The detachment of Muth from the microtubule can be facilitated by ATP binding to its AAA1 site (13). Alternatively, Muth can release under tension generated through the power stroke of WTh. Because Muth lacks the ability to generate a power stroke, its step size is mainly biased toward the WTh under tension (Fig. 4D).
Our results challenge established views of motor processivity that require coordination between the mechanochemical cycles of the heads. Kinesin (18, 19) and myosin (20) motility rely on mechanical and chemical gating mechanisms that allow the leading head to stay bound to its track while the trailing head moves forward. Dynein clearly moves by a different mechanism. We see no evidence of strict gating that keeps the heads out of phase. Processivity requires two heads to be physically connected (6) to prevent detachment of the motor when one of the heads steps forward. It is possible that simultaneous detachment of both heads is stochastic. Dynein’s high duty ratio (21) may allow the motor to take many steps before dissociation. Processivity in the absence of gating is also achieved by multiple monomeric kinesins (22) and engineered dimeric motors that have poor mechanochemical communication between the heads (18, 23). Further understanding of the molecular basis of dynein processivity will require simultaneous monitoring of the linker conformations and stepping motion of the leading and trailing head domains.
Supplementary Material
Acknowledgments
We thank R. D. Vale and A. P. Carter for providing yeast strains; T. Bilyard, F. B. Cleary, and S. Shih for critical evaluation of the manuscript; and K. Schimert for technical assisstance. This work is supported by NIH [GM094522 (A.Y.), GM08295 (M.A.D. and P.A.C.)], NSF [MCB-1055017 (A.Y.)], and Burroughs Wellcome Foundation (A.Y.).
Footnotes
Supporting Online Material
www.sciencemag.org/cgi/content/full/science.1215804/DC1
Materials and Methods
Figs. S1 to S15
Table S1
References (24–29)
Movies S1 to S4
References and Notes
- 1.Burgess SA, Walker ML, Sakakibara H, Knight PJ, Oiwa K. Nature. 2003;421:715. doi: 10.1038/nature01377. [DOI] [PubMed] [Google Scholar]
- 2.Kon T, Nishiura M, Ohkura R, Toyoshima YY, Sutoh K. Biochemistry. 2004;43:11266. doi: 10.1021/bi048985a. [DOI] [PubMed] [Google Scholar]
- 3.Ori-McKenney KM, Xu J, Gross SP, Vallee RB. Nat. Cell Biol. 2010;12:1228. doi: 10.1038/ncb2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carter AP, Cho C, Jin L, Vale RD. Science. 2011;331:1159. doi: 10.1126/science.1202393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mallik R, Carter BC, Lex SA, King SJ, Gross SP. Nature. 2004;427:649. doi: 10.1038/nature02293. [DOI] [PubMed] [Google Scholar]
- 6.Reck-Peterson SL, et al. Cell. 2006;126:335. doi: 10.1016/j.cell.2006.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yildiz A, et al. Science. 2003;300:2061. doi: 10.1126/science.1084398. [DOI] [PubMed] [Google Scholar]
- 8.Asbury CL, Fehr AN, Block SM. Science. 2003;302:2130. doi: 10.1126/science.1092985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yildiz A, Tomishige M, Vale RD, Selvin PR. Science. 2004;303:676. doi: 10.1126/science.1093753. [DOI] [PubMed] [Google Scholar]
- 10.Tsygankov D, Serohijos AW, Dokholyan NV, Elston TC. J. Chem. Phys. 2009;130 doi: 10.1063/1.3050098. 025101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Churchman LS, Okten Z, Rock RS, Dawson JF, Spudich JA. Proc. Natl. Acad. Sci. U.S.A. 2005;102:1419. doi: 10.1073/pnas.0409487102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nicastro D, et al. Science. 2006;313:944. doi: 10.1126/science.1128618. [DOI] [PubMed] [Google Scholar]
- 13.Imamula K, Kon T, Ohkura R, Sutoh K. Proc. Natl. Acad. Sci. U.S.A. 2007;104:16134. doi: 10.1073/pnas.0702370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roberts AJ, et al. Cell. 2009;136:485. doi: 10.1016/j.cell.2008.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gennerich A, Carter AP, Reck-Peterson SL, Vale RD. Cell. 2007;131:952. doi: 10.1016/j.cell.2007.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cho C, Reck-Peterson SL, Vale RD. J. Biol. Chem. 2008;283:25839. doi: 10.1074/jbc.M802951200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kon T, Mogami T, Ohkura R, Nishiura M, Sutoh K. Nat. Struct. Mol. Biol. 2005;12:513. doi: 10.1038/nsmb930. [DOI] [PubMed] [Google Scholar]
- 18.Yildiz A, Tomishige M, Gennerich A, Vale RD. Cell. 2008;134:1030. doi: 10.1016/j.cell.2008.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clancy BE, Behnke-Parks WM, Andreasson JO, Rosenfeld SS, Block SM. Nat. Struct. Mol. Biol. 2011;18:1020. doi: 10.1038/nsmb.2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nishikawa S, et al. Cell. 2010;142:879. doi: 10.1016/j.cell.2010.08.033. [DOI] [PubMed] [Google Scholar]
- 21.Shima T, Imamula K, Kon T, Ohkura R, Sutoh K. J. Struct. Biol. 2006;156:182. doi: 10.1016/j.jsb.2006.03.014. [DOI] [PubMed] [Google Scholar]
- 22.Kamei T, Kakuta S, Higuchi H. Biophys. J. 2005;88:2068. doi: 10.1529/biophysj.104.049759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liao JC, Elting MW, Delp SL, Spudich JA, Bryant Z. J. Mol. Biol. 2009;392:862. doi: 10.1016/j.jmb.2009.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.