Abstract
A polyspecific amber suppressor aminoacyl-tRNA synthetase/tRNA pair was evolved that genetically encodes a series of histidine analogues in both Escherichia coli and mammalian cells. In combination with tRNACUAPyl, a pyrrolysyl-tRNA synthetase mutant was able to site-specifically incorporate 3-methyl-histidine, 3-pyridyl-alanine, 2-furyl-alanine, and 3-(2-thienyl)-alanine into proteins in response to an amber codon. Substitution of His66 in the blue fluorescent protein (BFP) with these histidine analogues created mutant proteins with distinct spectral properties. This work further expands the structural and chemical diversity of unnatural amino acids (UAAs) that can be genetically encoded in prokaryotic and eukaryotic organisms and affords new probes of protein structure and function.
The ability to genetically incorporate unnatural amino acids (UAAs) into proteins in living cells provides an important tool to both probe and manipulate protein structure and function.1 In the past decade a large number of UAAs with diverse structures and properties have been successfully introduced into proteins in E. coli, S. cerevisiae, C. elegans, plants, and mammalian cells.1−10 These UAAs include photo-crosslinkers and biophysical probes, metal ion-binding and redox-active amino acids, and amino acids with bioorthogonal chemical reactivities. The UAA of interest is incorporated into proteins in response to a reassigned nonsense or frameshift codon by an aminoacyl-tRNA synthetase (aaRS)/tRNA pair that is orthogonal to the endogenous translational machinery of the host. In an effort to further expand the repertoire of genetically encoded UAAs, here we report an evolved orthogonal aaRS/tRNA pair that is specific for a number of histidine analogues. Histidine plays a functional role in many enzymes, where it can serve as a metal chelator, nucleophile, or general acid or base. Consequently, the ability to substitute histidine isosteres and analogues with altered properties would provide useful probes of the various functions of this canonical amino acid. To genetically encode histidine analogues, the pyrrolysyl-tRNA synthetase (PylRS)/tRNAPyl pair, which is characterized by high translational efficiency and orthogonality in both E. coli and mammalian cells, was subjected to rounds of positive and negative selection.11−14 The resulting PylRS/tRNAPyl pair was able to incorporate 3-methyl-histidine, 3-pyridyl-alanine, 2-furyl-alanine, 3-(2-thienyl)-alanine, and 2-(5-bromo-thienyl)-alanine site-specifically into proteins with high fidelity and efficiency. To demonstrate the utility of these histidine analogues, we substituted His66 in the blue fluorescent protein (BFP) with these UAAs to generate variant BFPs with altered spectral properties.
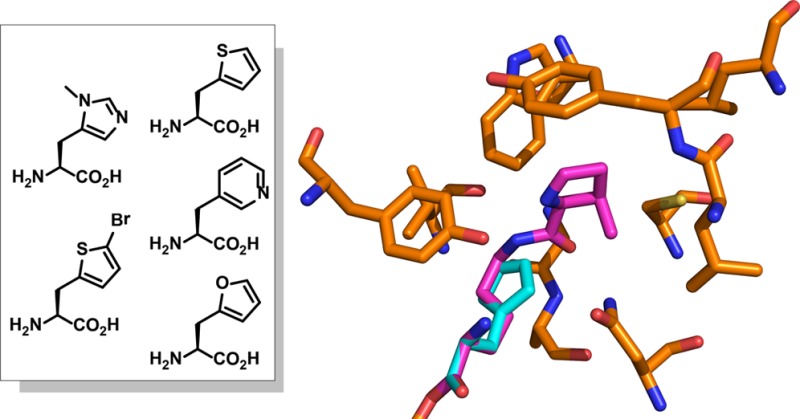
To explore the possibility of using the PylRS/tRNACUAPyl pair to encode new histidine analogues in E. coli, a previously reported Methanosarcina barkeri PylRS library was employed.13 On the basis of the crystal structure of the PylRS·pyrrolysyl-AMP complex, four amino acid residues involved in recognition of pyrrolysine (Leu270, Tyr271, Leu274, and Cys313; Figure 1B) were randomized by site-saturation mutagenesis (NNK randomization; N = any nucleotide, K = G or T), and Tyr349 was fixed as Phe by site-directed mutagenesis, since this substitution was previously shown to increase aminoacylation efficiency.15 More than 109 transformants were generated for the library, and no significant sequence bias was observed by sequence analysis of individual clones.
Figure 1.

(A) Structures of pyrrolysine (Pyl) and 3-methyl-histidine (3-Me-His). (B) X-ray crystal structure of the MmPylRS complex with pyrrolysyl-AMP.16 Histidine-AMP is overlaid with pyrrolysyl-AMP. The structure is from PDB id 2ZIM.
This library was then subjected to a double-sieve selection to identify PylRS variants that specifically utilize 3-methyl-histidine (3-Me-His, Figure 1A) as a substrate.1 Methylation of histidine at the 3 position is a naturally occurring modification that has been found in anserine in the skeletal muscle and brain of mammals and birds.17 PylRS library variants were first introduced into E. coli DH10B cells containing a pRep plasmid encoding tRNACUAPyl and a chloramphenicol acetyltransferase (CAT) gene with an Asp112TAG mutation. These cells were positively selected for chloramphenicol resistance in the presence of 3-Me-His (1 mM). The negative round of selection was carried out with a plasmid encoding a toxic barnase gene with amber mutations at three permissive sites (Gln2TAG, Asp44TAG, and Gly65TAG) in the absence of 3-Me-His. After two rounds of positive selection and one round of negative selection, 24 individual colonies that survived chloramphenicol (110 μg mL–1) in the positive selection only in the presence of 3-Me-His were characterized. Sequencing of these individual clones revealed a unique sequence (PylHRS; L270I, Y271F, L274G, C313F, and Y349F). The mutation of Cys313 to Phe likely diminishes the depth of the active site to better pack the smaller histidine analogue, as compared to the elongated native pyrrolysine substrate.
To test the fidelity and efficiency of the incorporation of 3-Me-His into proteins in E. coli, we expressed mutants of green fluorescent protein (GFP) using the evolved PylHRS/tRNACUAPyl pair. pLeiG-GFP-Asp134TAG containing a GFP variant with a C-terminal His6 tag and an amber codon at Asp134 was co-transformed with the PylHRS/tRNA pair in the pBK selection vector into the E. coli DH10B cell line.18 Protein expression was carried out in either minimal media or rich media, in the presence or the absence of 3-Me-His (1 mM). SDS-PAGE analysis of the GFP mutants purified by Ni2+-NTA affinity chromatography revealed that full-length GFP was expressed only in the presence of 1 mM 3-Me-His. ESI-MS analysis confirmed site-specific incorporation of 3-Me-His into GFP (Supplementary Figure S1). The yields of the purified GFP mutant containing 3-Me-His were 9.3 mg L–1 in 2x YT media and 7.1 mg L–1 in glycerol minimal media supplemented with leucine (GMML).
We have previously observed substrate polyspecificity for a subset of evolved aaRSs, in which the aaRS aminoacylates various UAAs, but not any of the canonical 20 amino acids.19 To determine if PylHRS can charge other histidine analogues, we grew DH10B cells containing pBK-PylHRS and pLeiG-GFP-Asp134TAG in GMML media supplemented with 1 mM concentration of various His analogues and structurally related UAAs (3-methyl-histidine, 1-methyl-histidine, 2-furyl-alanine, 2-thienyl-alanine, 4-thiazoly alanine, 2-thiazoly alanine, cyclopentyl-alanine, 2-pyridyl-alanine, 3-pyridyl-alanine, 4-pyridyl-alanine, 2-(5-bromo-thienyl)-alanine, 2,5-diiodo-histidine, 2-fluoro-phenylalanine, 1,2,4-triazol-alanine and tetrazolyl-alanine), and the relative fluorescent intensities of these cell cultures were then measured (Supplementary Figure S2). Higher GFP expression levels were observed in the presence of 3-pyridyl-alanine (3-Py-Ala, 2), 2-furyl-alanine (Fury-Ala, 3), 2-thienyl-alanine (Th-Ala, 4), and 2-(5-bromo-thienyl)-alanine (5-Br-Th, 5) (Figure 2A and Supplementary Figure S2). Full-length GFP mutants containing these UAAs were purified by Ni2+-NTA affinity chromatography, and the incorporation of these UAAs was further confirmed by ESI-MS (Supplementary Figure S1). Yields of GFP mutants expressed in GMML substituted with 3-Py-Ala, Fury-Ala, Th-Ala, and 5-Br-Th were 4.0, 3.0, 4.2, and 12.0 mg L–1, respectively. Th-Ala and 5-Br-Th also showed high incorporation fidelity and efficiency when protein expressions were carried out in 2x YT, with yields of 5.1 and 16.8 mg L–1, respectively.
Figure 2.

Evolution of MbPylRS to genetically encode histidine analogues. (A) Structures of UAAs 1–5. (B) Expression of GFP-Asp134TAG variants analyzed by SDS-PAGE (stained with Coomassie blue).
A significant advantage of using the PylRS/tRNAPyl pair is that it can be efficiently evolved in bacteria and then directly imported into mammalian cells to genetically encode the UAA. Therefore, we tested the ability of the PylHRS to incorporate histidine derivatives in HEK293T cells in combination with tRNAPyl. A plasmid, pAcBac2·tR2-PylHRS, encoding two tandem copies of the Methanosarcina mazei pyrrolysyl tRNA (MmtRNAPyl) cassette, as well as the PylHRS gene expressed under a CMV promoter, was constructed based on a previously reported mammalian suppression system.8,9 This plasmid was co-transfected into HEK293T cells with pAcBac2·tR2-EGFP, and the expression levels of full-length EGFP were monitored by fluorescence microscopy in the absence of UAA or the presence of 1 mM histidine analogues.9 Robust expression of EGFP was observed in the presence of 3-Me-His and Th-Ala (Supplementary Figure S3). Full-length EGFP mutants containing these histidine derivatives were isolated from cell cultures by Ni2+-NTA affinity chromatography. SDS-PAGE, ESI-MS, and MS/MS analysis confirmed the incorporation of histidine analogues into EGFPs in mammalian cells (Supplementary Figure S4). Yields of EGFP mutants substituted with 3-Me-His and Th-Ala were 4 μg/107 cells and 3 μg/107 cells, respectively. EGFPs containing 3-Py-Ala, Fury-Ala, and 5-Br-Th were not isolated, likely due to either the low suppression efficiency or toxicity of the UAAs in mammalian cells.
Blue fluorescent proteins (BFPs) provide a powerful tool to visualize protein expression and localization and probe protein function.21 The chromophores of BFPs result from the self-catalyzed post-translational cyclization of three internal amino acids (Ser65-His66-Gly67 in BFP) followed by dehydration of the His66 imidazole side chain (Figure 3A).22 Using canonical amino acid mutagenesis, several BFPs have been reported with increased brightness, improved photostability, and enhanced quantum yield.23−25 Thus substitution of His66 in BFP with the above histidine analogues may result in fluorescent proteins with distinct spectral properties. To express the BFP mutants, DH10B cells harboring pBK-PylHRS and pLeiG-BFP-His66TAG were grown in GMML media supplemented with the histidine analogues (1 mM). The full-length BFP mutants as well as wild-type BFP were further purified by Ni2+-NTA affinity chromatography and analyzed by SDS-PAGE and ESI-MS analyses, confirming specific incorporation of the UAAs (Figure 3B and Supplementary Figure S5).
Figure 3.
UAA mutagenesis of the BFP-chromophore. (A) Structure of the of BFP-chromophore.22 (B) Incorporation of various UAAs into BFP-His66TAG, analyzed by SDS-PAGE stained with Coomassie blue. His: wild-type BFP; (1): 3-Me-His mutant; (2): 3-Py-Ala mutant; (3): Fury-Ala mutant; (4): Th-Ala mutant; (5): 5-Br-Th mutant. (C) Absorption spectra for wild-type BFP and mutants. (D) Emission spectra for wild-type BFP and mutants.
The absorbance and fluorescence spectra of wild-type and mutant BFPs were measured in 20 mM Tris·HCl buffer, pH 8.0 (Figure 3C, D), and the spectral properties of these BFP mutants are summarized in Table 1. Wild-type BFP has an absorption maximum at 384 nm and emission maximum at 446 nm. Substitution of His66 with 3-Me-His, Fury-Ala, and Th-Ala resulted in similar absorption profiles. The absorption maxima of the BFP mutants containing 3-Py-Ala and 5-Br-Th exhibit a 33 nm blue-shift and a 20 nm red-shift, respectively. The emission maxima of the BFP mutants span the region from violet, blue to cyan. The BFPs containing Fury-Ala, Th-Ala and 5-Br-Th have a 34, 38, and 40 nm red-shift in the emission maxima and exhibit unusually large Stokes shifts (93, 96, and 82 nm, respectively). The X-ray crystal structure of BFP has revealed that in the more stable cis form of the chromophore of BFP, the His66 appears to hydrogen-bond with the nitrogen atom of His148.22 Replacement of His66 with 3-Me-His resulted in a 26-fold decrease in quantum yield suggesting that this mutation may increase the population of the trans isomer of the chromophore.25 Substitution of His66 with Fury-Ala and Th-Ala similarly resulted in decreased quantum yields.
Table 1. Spectral Properties of Wild Type and Mutant BFPs.
| amino acid at position 66 | absorbance max (nm)a | emission max (nm) | quantum yield |
|---|---|---|---|
| His | 384 | 446 | 0.157 ± 0.001 |
| 3-Me-His | 388 | 468 | 0.006 ± 0.001 |
| 3-Py-Ala | 351 | 429 | 0.075 ± 0.001 |
| Fury-Ala | 387 | 480 | 0.019 ± 0.001 |
| Th-Ala | 388 | 484 | 0.027 ± 0.001 |
| 5-Br-Th | 404 | 486 | 0.103 ± 0.008 |
BFP mutants are in 20 mM Tris buffer (pH = 8.0).
In conclusion, we have developed a novel PylRS/tRNACUAPyl pair to genetically encode histidine analogues in E. coli with good yield and high fidelity. Using this system, 3-methyl-histidine, 3-pyridyl-alanine, 2-furyl-alanine, 3-(2-thienyl)-alanine, and 2-(5-bromo-thienyl)-alanine were successfully incorporated into proteins in both E. coli and mammalian cells. The UAAs were then used to replace His66 in the chromophore to modulate the spectral properties of BFP. These histidine analogues should prove useful as probes of structure and function within proteins.
Methods
Expression and Purification of GFP
The pLeiG-GFP-Asp134TAG and pBK-PylHRS plasmids were transformed into E. coli DH10B strain. Cells were grown in GMML or 2x YT media, supplemented with chloramphenicol (25 μg/mL), kanamycin (25 μg/mL), and 1 mM unnatural amino acid at 37 °C to an OD600 of 0.6, at which point IPTG was added to a final concentration of 1 mM. After 16 h of incubation at 30 °C, the cells were harvested by centrifugation at 4,700 × g for 10 min. To purify the protein, cell pellets were resuspended in BugBuster protein extraction reagent and lysed at 30 °C. The resulting cell lysate was clarified by centrifugation at 18,000 × g for 30 min, and the proteins were purified on Ni2+-NTA resin following the manufacturer’s (Qiagen) instructions.
Determination of aaRS Polyspecificity
The pLeiG-GFP-Asp134TAG and pBK-PylHRS plasmids were transformed into E. coli DH10B. Cells were grown in GMML or 2x YT media, supplemented with chloramphenicol (25 μg/mL), kanamycin (25 μg/mL), and 1 mM concentration of various unnatural amino acids at 37 °C to an OD600 of 0.6, at which point IPTG was added to a final concentration of 1 mM. After 16 h at 30 °C, the cells were harvested by centrifugation at 4,700 × g for 10 min and washed three times with PBS. Cells were resuspended in PBS and transferred to a clear bottom 96-well plate, and GFP fluorescence was measured using a plate reader (485 nm excitation and 515 nm emission).
Expression and Purification of BFP
The pLeiG-BFP-His66TAG and pBK-PylHRS plasmids were transformed into E. coli DH10B. Cells were grown in minimal (GMML) media, supplemented with chloramphenicol (25 μg/mL), kanamycin (25 μg/mL), and 1 mM UAA, at 37 °C to an OD600 of 0.6, at which point IPTG was added to a final concentration of 1 mM. Protein was expressed and purified using the protocol described above.
Spectroscopic Measurements
Fractions containing purified proteins were dialyzed against 20 mM Tris buffer, pH 8. Protein concentration was adjusted such that the absorbance was within the range 0.5–1.0 (arbitrary unit). Fluorescence emission spectra of protein samples were measured on a Varian Cary Eclipse fluorescence spectrophotometer. Fluorescence quantum yields were calculated on protein samples with an OD not higher than 0.05, using 9-aminoacridine as a standard (quantum yield = 0.97 in water). For wild-type BFP, the 385–600 nm region of the fluorescence emission spectra was integrated with excitation at 382 nm. The quantum yield of wild-type BFP was calculated by comparing of the integrated area of the emission spectra between the protein sample and 9-aminoacridine. The quantum yields of other BFP mutants were similarly measured: 3-Me-His mutant: excitation at 388 nm, integration from 405 to 600 nm; 3-Py-Ala mutant: excitation at 352 nm, integration from 370 to 600 nm; Fury-Ala mutant: excitation at 387 nm, integration from 404 to 600 nm; Th-Ala mutant: excitation at 388 nm, integration from 404 to 600 nm; 5-Br-Th mutant: excitation at 404 nm, integration from 405 to 600 nm.
Acknowledgments
We would like to acknowledge V. Seely for her assistance in manuscript preparation and X. Zhang for his help in spectral analysis. We thank A. Chatterjee and J. Furman for helpful discussions. This is paper no. 26068 from The Scripps Research Institute. This work was funded by the NIH grant GM097206 (PGS).
Supporting Information Available
This material is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions
§ These authors contributed equally to this work.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Liu C. C.; Schultz P. G. (2010) Adding new chemistries to the genetic code. Annu. Rev. Biochem. 79, 413–444. [DOI] [PubMed] [Google Scholar]
- Chin J. W.; Cropp T. A.; Anderson J. C.; Mukherji M.; Zhang Z.; Schultz P. G. (2003) An expanded eukaryotic genetic code. Science 301, 964–967. [DOI] [PubMed] [Google Scholar]
- Liu W. S.; Brock A.; Chen S.; Chen S. B.; Schultz P. G. (2007) Genetic incorporation of unnatural amino acids into proteins in mammalian cells. Nat. Methods 4, 239–244. [DOI] [PubMed] [Google Scholar]
- Li F.; Zhang H.; Sun Y.; Pan Y.; Zhou J.; Wang J. (2013) Expanding the genetic code for photoclick chemistry in E. coli, mammalian cells, and A. thaliana. Angew. Chem., Int. Ed. 52, 9700–9704. [DOI] [PubMed] [Google Scholar]
- Greiss S.; Chin J. W. (2011) Expanding the genetic code of an animal. J. Am. Chem. Soc. 133, 14196–14199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco A.; Townsley F. M.; Greiss S.; Lang K.; Chin J. W. (2012) Expanding the genetic code of Drosophila melanogaster. Nat. Chem. Biol. 8, 748–750. [DOI] [PubMed] [Google Scholar]
- Chatterjee A.; Sun S. B.; Furman J. L.; Xiao H.; Schultz P. G. (2013) A versatile platform for single- and multiple-unnatural amino acid mutagenesis in Escherichia coli. Biochemistry 52, 1828–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H.; Chatterjee A.; Choi S. H.; Bajjuri K. M.; Sinha S. C.; Schultz P. G. (2013) Genetic incorporation of multiple unnatural amino acids into proteins in mammalian cells. Angew. Chem., Int. Ed. 52, 14080–14083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee A.; Xiao H.; Bollong M.; Ai H. W.; Schultz P. G. (2013) Efficient viral delivery system for unnatural amino acid mutagenesis in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 110, 11803–11808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C. H.; Axup J. Y.; Schultz P. G. (2013) Protein conjugation with genetically encoded unnatural amino acids. Curr. Opin. Chem. Biol. 17, 412–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takimoto J. K.; Dellas N.; Noel J. P.; Wang L. (2011) Stereochemical basis for engineered pyrrolysyl-tRNA synthetase and the efficient in vivo incorporation of structurally divergent non-native amino acids. ACS Chem. Biol. 6, 733–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. S.; Fang X.; Wallace A. L.; Wu B.; Liu W. R. (2012) A rationally designed pyrrolysyl-tRNA synthetase mutant with a broad substrate spectrum. J. Am. Chem. Soc. 134, 2950–2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groff D.; Chen P. R.; Peters F. B.; Schultz P. G. (2010) A genetically encoded epsilon-N-methyl lysine in mammalian cells. ChemBioChem 11, 1066–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann H.; Peak-Chew S. Y.; Chin J. W. (2008) Genetically encoding N(epsilon)-acetyllysine in recombinant proteins. Nat. Chem. Biol. 4, 232–234. [DOI] [PubMed] [Google Scholar]
- Yanagisawa T.; Ishii R.; Fukunaga R.; Kobayashi T.; Sakamoto K.; Yokoyama S. (2008) Multistep engineering of pyrrolysyl-tRNA synthetase to genetically encode N(epsilon)-(o-azidobenzyloxycarbonyl) lysine for site-specific protein modification. Chem. Biol. 15, 1187–1197. [DOI] [PubMed] [Google Scholar]
- Kavran J. M.; Gundliapalli S.; O’Donoghue P.; Englert M.; Soell D.; Steitz T. A. (2007) Structure of pyrrolysyl-tRNA synthetase, an archaeal enzyme for genetic code innovation. Proc. Natl. Acad. Sci. U.S.A. 104, 11268–11273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapp J. A.; Wilson D. W. (1938) Quantitative studies of carnosine and anserine in mammalian muscle: II. The distribution of carnosine and anserine in various muscles of different species. J. Biol. Chem. 126, 19–27. [Google Scholar]
- Kim C. H.; Kang M.; Kim H. J.; Chatterjee A.; Schultz P. G. (2012) Site-specific incorporation of epsilon-N-crotonyllysine into histones. Angew. Chem., Int. Ed. 51, 7246–7249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young D. D.; Young T. S.; Jahnz M.; Ahmad I.; Spraggon G.; Schultz P. G. (2011) An evolved aminoacyl-tRNA synthetase with atypical polysubstrate specificity. Biochemistry 50, 1894–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chudakov D. M.; Matz M. V.; Lukyanov S.; Lukyanov K. A. (2010) Fluorescent proteins and their applications in imaging living cells and tissues. Physiol. Rev. 90, 1103–1163. [DOI] [PubMed] [Google Scholar]
- Wachter R. M.; King B. A.; Heim R.; Kallio K.; Tsien R. Y.; Boxer S. G.; Remington S. J. (1997) Crystal structure and photodynamic behavior of the blue emission variant Y66H/Y145F of green fluorescent protein. Biochemistry 36, 9759–9765. [DOI] [PubMed] [Google Scholar]
- Heim R.; Tsien R. Y. (1996) Engineering green fluorescent protein for improved brightness, longer wavelengths and fluorescence resonance energy transfer. Curr. Biol. 6, 178–182. [DOI] [PubMed] [Google Scholar]
- Ai H. W.; Shaner N. C.; Cheng Z.; Tsien R. Y.; Campbell R. E. (2007) Exploration of new chromophore structures leads to the identification of improved blue fluorescent proteins. Biochemistry 46, 5904–5910. [DOI] [PubMed] [Google Scholar]
- Jablonski A. E.; Vegh R. B.; Hsiang J. C.; Bommarius B.; Chen Y. C.; Solntsev K. M.; Bommarius A. S.; Tolbert L. M.; Dickson R. M. (2013) Optically Modulatable Blue Fluorescent Proteins. J. Am. Chem. Soc. 135, 16410–16417. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



