Abstract

Tyrosyl-DNA phosphodiesterase I (TDP1) repairs stalled topoisomerase I (Top1)–DNA covalent complexes and has been proposed to be a promising and attractive target for cancer treatment. Inhibitors of TDP1 could conceivably act synergistically with Top1 inhibitors and thereby potentiate the effects of Top1 poisons. This study describes the successful design and synthesis of 2-position-modified indenoisoquinolines as dual Top1–TDP1 inhibitors using a structure-based drug design approach. Enzyme inhibition studies indicate that indenoisoquinolines modified at the 2-position with three-carbon side chains ending with amino substituents show both promising Top1 and TDP1 inhibitory activity. Molecular modeling of selected target compounds bound to Top1 and TDP1 was used to rationalize the enzyme inhibition results and structure–activity relationship analysis.
Introduction
The topoisomerase I (Top1) family of eukaryotic enzymes is required to relax DNA supercoiling generated by replication, transcription, and chromatin remodeling.1−4 Human Top1 acts through a nucleophilic tyrosine residue (Tyr723), which nicks the phosphodiester backbone of double-stranded, supercoiled DNA and forms a transient cleavage complex in which the 3′ end of the broken DNA strand is covalently linked to the enzyme (Scheme 1).5−8 Camptothecin (1, Figure 1) is a natural product for which Top1 is its only cellular target.9 Two camptothecin derivatives, irinotecan (2) and topotecan (3), are the only current Top1 inhibitors approved by the U.S. Food and Drug Administration for the treatment of cancer.10,11 However, these camptothecin derivatives have several major drawbacks. First, camptothecins are compromised by the reversibility of the Top1–DNA cleavage complex, which necessitates long infusion times for maximum activity.12−14 Second, the lactone ring is inherently unstable and hydrolyzes to form an inactive hydroxy acid.10,15,16 In addition, the anti-cancer activities of the camptothecins are compromised by R364H17 and N722S18 mutations as well as by induction of the ABCG219−21 and MXR21 ATP-binding cassette drug efflux transporters. Myelosuppression is dose-limiting with topotecan (2),22 whereas the major dose-limiting toxicities of irinotecan (3) are neutropenia and diarrhea.23 These limitations have stimulated the search for non-camptothecin Top1 inhibitors as anti-cancer agents.
Scheme 1.
Figure 1.
Representative Top1 inhibitors.
Indenoisoquinoline NSC314622 (4) was found to be a Top1 inhibitor with anti-cancer activity after a COMPARE analysis of its cytotoxicity profile in the National Cancer Institute’s 60 (NCI60)-cell screen indicated a high degree of correlation with camptothecin. Subsequent studies confirmed that the mechanism of action of the indenoisoquinolines is identical to that of the camptothecins.24,25 Specifically, they stabilize the ternary cleavage complex by intercalation between the DNA base pairs at the cleavage site after single-strand cleavage by Top1, thus preventing religation of the broken phosphodiester backbone. These inhibitors are therefore classified as Top1 poisons as opposed to Top1 suppressors, which inhibit the DNA cleavage reaction.6,7,26−28
The indenoisoquinolines have several advantages over the camptothecins. First, the cleavage complexes induced by the indenoisoquinolines are more stable than those formed in the presence of camptothecin derivatives.25 Second, in contrast to the camptothecin derivatives, which have an unstable lactone ring, the indenoisoquinolines are chemically stable. Moreover, the DNA cleavage site specificity of indenoisoquinolines is different from that of camptothecin, so they target the genome differently, and the indenoisoquinolines are less affected by the R364H and N722N Top1-resistance mutations than is camptothecin.25,29,30 After lead optimization and SAR studies, two indenoisoquinoline topoisomerase I inhibitors [indotecan (6, also known as LMP400 or NSC 724998) and indimitecan (7, also known as LMP776 or NSC 725776)] have entered phase I clinical trials for the treatment of cancer patients at the National Cancer Institute, and definite plans are being formulated to commence phase II clinical trials.31−33 Indotecan (6) is not a substrate for the ABCG2 and MDR-1 efflux pumps, whereas indimitecan (7) is less affected than the camptothecins.29
Tyrosyl-DNA phosphodiesterase I (TDP1) is a member of the phospholipase D superfamily of enzymes that catalyzes the hydrolysis of 3′ phosphotyrosyl linkers and other 3′-end blocking lesions.34−38 When Top1 nicks double-stranded DNA, a covalent cleavage complex is formed that can be repaired by TDP1.39 The enzyme mechanism involves the following steps (Scheme 2): First, as Lys265 and Lys495 residues in the catalytic site coordinate the oxygen atoms of the phosphate group, His263 attacks the phosphorus atom linked to the oxygen of the Top1 catalytic residue, Tyr723, at the 3′ end of DNA.40,41 Second, the TDP1–DNA covalent intermediate formed is hydrolyzed by a His493-activated water molecule, leading to the generation of a DNA product with a 3′ phosphate.42 Lastly, further repair is finished by polynucleotide kinase phosphatase (PNKP), a bifunctional enzyme with 5′ kinase and 3′ phosphatase activities that catalyzes both the hydrolysis of the 3′ phosphate and the phosphorylation of the 5′ end of DNA to enable their rejoining.43,44
Scheme 2.
Prior studies demonstrated that TDP1 plays a critical role in the cellular repair of Top1-mediated DNA damage: hypersensitivity to camptothecin occurs when the TDP1 gene is silenced in yeast.45,46 Moreover, TDP1-defective spinocerebellar ataxia with axonal neuropathy-1 (SCAN1) cells, which have a TDP1 mutation, N493R, and accumulate the normally transient TDP1–DNA repair complex, are highly sensitive to camptothecin and accumulate DNA strand breaks upon treatment with camptothecin.47,48 Similarly, knocking out TDP1 in vertebrate cells renders them hypersensitive to Top1-targeted drugs.37,49 These observations suggest that inhibitors of TDP1 could act synergistically with Top1 inhibitors and could potentiate the effects of Top1 poisons.
Although TDP1 is a promising and attractive target for cancer treatment, only a limited number of inhibitors have been identified (Figure 2). Vanadate and tungstate can mimic the phosphate in the transition state but cannot serve as pharmacologically useful inhibitors because of their poor specificity and hypersensitivity to all phosphoryl transfer reactions.50,51 Furamidine (8) was identified as a micromolar-range TDP1 inhibitor, but it also has additional targets because of its DNA-binding activities.52 Steroid derivative 9 was found to be a potent TDP1 inhibitor with an IC50 of 7.7 μM through a high-throughput screening assay. However, it suffers common pharmacokinetic problems in cellular systems.53 Later, Wang et al. reported that arylidene thioxothiazolidinones can inhibit TDP1 and identified compound 10 as a submicromolar inhibitor of TDP1.54 Recently, indenoisoquinoline 11 and bis(indenoisoquinoline) 12 were found to be dual Top1–TDP1 inhibitors.55,56
Figure 2.
Representative TDP1 inhibitors.
After indenoisoquinoline 4 was found to be a Top1 inhibitor with anti-cancer activity, a number of structure–activity relationship studies were performed on the indenoisoquinolines, including modifications on the A ring,57 B ring (side chain on the lactam nitrogen),58−62 C ring,63,64 and D ring.57,61−63,65−68 However, there is very limited study on the modification of the A ring of indenoisoquinoline, especially on the 2-position. To date, the methoxy group is the only substituent that has been placed at the 2-position. The crystal structure of an indenoisoquinoline in a ternary complex with DNA and Top1 (PDB ID: 1SC7) indicates that the A ring is next to the cleaved DNA strand, where there is more room to accommodate substituents on the drug.69 Recent studies of the metabolism of indotecan (LMP400, 6) and indimitecan (LMP776, 7) involved the synthesis of 2-hydroxylated indenoisoquinolines, thus providing a strategically placed handle for the attachment of a variety of side chains.68 At the same time, prior molecular modeling studies based on the crystal structure of TDP1 (PDB ID: 1RFF)70 provide a foundation for the structure-based design of indenoisoquinoline Top1 inhibitors that also inhibit TDP1.55,56
In the present study, attention is focused on the modification of O-2-position of indenoisoquinoline using a structure-based drug design approach. At the same time, considering the limited number of TDP1 inhibitors reported in the literature and the critical role of TDP1 in cleaving stalled Top1–DNA covalent complexes, there is interest in the identification of potent small molecule TDP1 inhibitors for further therapeutic development. On the basis of the indenoisoquinoline skeleton, the present investigation was undertaken to define the structural parameters associated with dual Top1–TDP1 inhibitory activity. This report details the structure-based design, synthesis, and biological evaluation of O-2-derivatized indenoisoquinolines as dual Top1–TDP1 inhibitors.
Results and Discussion
A molecular docking study was performed to guide the structural modification of the indenoisoquinolines and to help understand the Top1 inhibition results. The energy-minimized structure of morpholine derivative 13(58) (Scheme 3) was docked into the crystal structure (PDB ID: 1SC7) of a Top1−DNA cleavage site with GOLD using the centroid coordinates of the indenoisoquinoline ligand. The energy-minimized, top-ranked GOLD pose of compound 13 in a ternary complex with DNA and Top1 is displayed in Figure 3. Compound 13 intercalates readily at the DNA cleavage site, between the +1 and −1 base pairs. Rings A and B stack with the scissile strand bases, whereas rings C and D stack with the noncleaved strand bases. The carbonyl group on the C ring forms a hydrogen bond to a nitrogen of the Arg364 side chain with an N–O distance of 2.5 Å, which is an important contact for the Top1 inhibitory activity.68 It is worth mentioning that Asp533 is also an important residue that is known to be required for enzyme sensitivity to camptothecin.71 The X-ray crystal structure of the ternary campthothecin–Top1–DNA complex indicates that camptothecin intercalates at the site of DNA cleavage and forms two hydrogen bonds with the active site. One hydrogen bond in the camptothecin ternary complex is from a nitrogen atom of Arg364 to a free electron pair of the B ring N-1 of camptothecin (N–N distance 2.9 Å); the other interaction is a hydrogen bond between C-20 hydroxyl and the oxygen atom of Asp533 (O–O distance 3.4 Å).69 In the present molecular docking study (Figure 3), the calculated distance between the 2-position oxygen atom of compound 13 and the carbonyl group of Asp533 is 4.7 Å. The docking pose suggests that aminoalkyl substituents attached to O-2, next to the cleaved DNA strand, could be used to target the carboxylate of Asp533. Therefore, a series of O-2 indenoisoquinoline derivatives was designed and synthesized with 2-OH indenoisoquinoline 13 as the starting material.
Scheme 3.
Figure 3.

Hypothetical binding mode of compound 13 in a ternary complex with DNA and Top1. All distances are measured from heavy atom to heavy atom. The diagram is programmed for wall-eyed (relaxed) viewing. Compound 13 is shown in yellow sticks, and the base pairs are displayed in lines.
The synthesis of indenoisoquinoline 13 was performed according to a previously reported method with some modifications.68 With 2-hydroxylated indenoisoquinoline 13 in hand, amine compound 17, which has a three-carbon side chain, was first prepared using the synthetic route shown in Scheme 3. Treatment of 13 with 1,3-dibromopropane in DMF in the presence of sodium hydride provided alkylation product 14, which was accompanied by smaller amounts of allyl compound 15 as a side product. Displacement of the bromide of compound 14 by sodium azide yielded intermediate 16, which was converted to amine 17 by Staudinger reduction.
Subsequently, as shown in Scheme 4, dimethylamino analogue 18 was synthesized by treatment of compound 14 with dimethylamine in the presence of sodium iodide. Similarly, compound 14 reacted with ethylamine in refluxing dioxane to afford compound 19. Treatment of compound 14 with morpholine and sodium iodide gave compound 20. In addition, compound 21 was prepared by treatment of bromide compound 14 with N-methylpiperazine in the presence of sodium iodide. It was assumed that the C-2 terminal amine appendages would be protonated at physiological pH and that the ammonium cations would form a salt bridge with the Asp533 carboxylate anion.
Scheme 4.
A different synthetic route was employed for the synthesis of compounds 22 and 23. 2-Hydroxylated indenoisoquinoline 13 reacted with 1-(3-chloropropyl)piperidine hydrochloride in the presence of potassium carbonate to provide compound 22 directly. Similarly, compound 23 was made by treatment of indenoisoquinoline 13 with 1-(3-chloropropyl)pyrrolidine hydrochloride in the presence of potassium carbonate (Scheme 5).
Scheme 5.
Ester intermediate 24 was prepared by treatment of compound 13 with methyl bromoacetate in the presence of sodium hydride. Treatment of compound 24 with hydrazine did not afford expected compound 25 but unexpectedly yielded the reduced 11-hydroxyl compound 26 instead (Scheme 6).
Scheme 6.
All of the new indenoisoquinoline derivatives with C-2 side chains were tested in Top1-mediated DNA cleavage assays. For this purpose, a 32P 3′-end-labeled 117 bp DNA fragment was incubated with Top1 and four 10-fold dilutions starting from 100 μM of a test compound. The DNA fragments were separated on 20% PAGE denaturing gels. The Top1 inhibitory activities were assigned on the basis of the visual inspection of the number and intensity of the gel bands corresponding to Top1-mediated DNA cleavage fragments. The results of this assay are designated relative to the Top1 inhibitory activity of compounds 1 and 5 and are expressed in semiquantitative fashion: 0, no detectable activity; +, weak activity; ++, similar activity to that of compound 5; +++, greater activity than that of 5; ++++, equipotent to 1. Ambiguous scores (e.g., between two values) are designated with parentheses (e.g., ++(+) would be between ++ and +++). As shown in Table 1, compound 17, which has an aminopropyl side chain, expressed low Top1 inhibitory activity at the 0/+ level. Interestingly, after conversion of the primary amine to a dimethylamine, the observed Top1 inhibitory activity increased from 0/+ for 17 to +++ for 18. A similar change was observed with the ethylaminopropyl compound 19, which displayed improved Top1 inhibitory activity relative to 17 at the +++ level. Introduction of a morpholine at the end of the propyl chain yielded compound 20, which was also found to be a promising Top1 inhibitor with activity at the +++ level. Subsequently, N-methylpiperazine, piperidine, and pyrrolidine were also introduced to the end of the propyl chain, and the corresponding compounds, 21–23, displayed good Top1 inhibitory activity at the +++, +++, and ++ levels, respectively. Compounds 15, 24, and 26, which lack aminopropyl side chain structures, were, in general, found to be more moderate Top1 inhibitors, with + or ++ activity. The Top1 inhibitory activity of primary amine 17 is low, but the activity was improved after conversion of the primary amine to a dimethylamine or to other cyclic amines. The extra steric bulk around the nitrogen may help to position the protonated nitrogen for binding to the Asp533 carboxylate. The Top1-mediated DNA fragmentation patterns produced by camptothecin, indenoisoquinoline 5, and compounds 18–23 are presented in Figure 4. The sequence preferences for trapping the Top1–DNA cleavage complexes by these indenoisoquinolines are similar to each other, but the pattern is different from camptothecin, indicating that the indenoisoquinolines target the genome differently from camptothecin. Interestingly, as is evident from the gel, these indenoisoquinolines suppress DNA cleavage at a high concentration of 100 μM. According to the DNA unwinding studies,66 this result can be attributed to the ability of these indenoisoquinolines to intercalate into free DNA at high drug concentrations, thus suppressing DNA cleavage by Top1 by making the DNA a poorer Top1 substrate. To rationalize the effect of the introduction of aminopropyl side chains on the O-2 position of indenoisoquinolines on their ability to improve the Top1 inhibitory activity, compound 20 was selected for a molecular docking study. As shown in Figure 5, compound 20 hypothetically intercalates at the site of DNA cleavage, between the +1 and −1 base pairs. Rings A and B stack with the scissile strand bases, whereas rings C and D stack with the noncleaved strand bases, which is consistent with the calculated binding mode of compound 13. The carbonyl group on the C ring in the minor DNA groove forms a hydrogen bond with a nitrogen of the Arg364 side chain, with an O–N distance of 2.5 Å, and as expected, there is also a hydrogen bond between the N atom in the morpholine ring in the major DNA groove and the Asp533 side chain, with a distance of 3.1 Å, which may contribute to the slightly improved Top1 inhibitory activity of these O-2-modified indenoisoquinolines relative to that of phenol 13.
Table 1. Top1 and TDP1 Activity of O-2-Modified Indenoisoquinolines.
| compd | Top1a | TDP1b |
|---|---|---|
| 13 | ++(+) | NT |
| 15 | + | 0 |
| 17 | 0/+ | + |
| 18 | +++ | ++ |
| 19 | +++ | ++ |
| 20 | +++ | 0 |
| 21 | +++ | 0 |
| 22 | +++ | ++ |
| 23 | ++ | ++(+) |
| 24 | ++ | 0 |
| 26 | + | 0 |
Compound-induced DNA cleavage resulting from Top1 inhibition is graded by the following semiquantitative scale relative to 1 μM camptothecin (1) or MJ-III-65 (5): 0, no detectable activity; +, weak activity; ++, similar activity to that of compound 5; +++, greater activity than that of 5; ++++, equipotent to 1. The (+) ranking indicates the activity lies between two given values. NT: not tested.
TDP1 IC50 was determined in duplicate using a semiquantitative scale: 0, IC50 > 111 μM; +, IC50 between 37 and 111 μM; ++, IC50 between 12 and 37 μM; +++, IC50 between 1 and 12 μM; ++++, IC50 < 1 μM.
Figure 4.

Top1-mediated DNA cleavage induced by indenoisoquinolines 18, 19, and 21–23: lane 1, DNA alone; lane 2, Top1 + DNA; lane 3, 1 (1 μM); lane 4:, 5 (1 μM); lanes 5–24, 18, 19, 21, 22, and 23 (each at 0.1, 1, 10, and 100 μM from left to right). Numbers and arrows on the left indicate cleavage site positions.
Figure 5.

Hypothetical binding mode of compound 20 in a ternary complex with DNA and Top1. All distances are measured from heavy atom to heavy atom. The diagram is programmed for wall-eyed (relaxed) viewing. Compound 20 is shown in pink sticks, and the base pairs are displayed in lines.
The TDP1 inhibitory activities of the O-2-substituented indenoisoquinolines were measured by determining their ability to inhibit the hydrolysis of the phosphodiester linkage between tyrosine and the 3′ end of a DNA oligonucleotide substrate, thus preventing the generation of an oligonucleotide with a free 3′ phosphate (N14P, Scheme 7).53 Therefore, the disappearance of the gel band for N14P indicates TDP1 inhibition. The TDP1 inhibitory activities of O-2-modified indenoisoquinolines are displayed in Table 1, and a representative gel demonstrating dose-dependent TDP1 inhibition is depicted in Figure 6. TDP1 IC50 was determined in duplicate using a semiquantitative scale: 0, IC50 > 111 μM; +, IC50 between 37 and 111 μM; ++, IC50 between 12 and 37 μM; +++, IC50 between 1 and 12 μM; and ++++, IC50 < 1 μM. From Table 1 and Figure 6, it is clear that compounds 18 and 19, which have dimethylamine or ethylamine at the end of the propyl side chain, display good TDP1 inhibitory activity with ++ potency. Compounds 22 and 23, with six- or five-membered rings on the end of the propyl side chain, also exhibit good inhibition of TDP1 with ++ and ++(+) activity, respectively. However, when the 4-position of the six-membered ring was substituted with a heteroatom (oxygen for compound 20 and nitrogen for compound 21), no TDP1 inhibitory activity was observed. The structure–activity relationships correlate well with the molecular docking studies. According to a previous report on TDP1,40 two specific regions of the enzyme are important for substrate binding, which have been termed the catalytic region and the hydrophobic region. The TDP1 catalytic region possesses two lysine (265 and 495) and two histidine (263 and 493) residues, which are responsible for the stabilization of the negatively charged phosphate backbone of the DNA, whereas the hydrophobic region consists of several residues (Ala520, Ala456, Phe259, Met491, Gly260, Tyr261, etc.) located at the top of the substrate channel. Compound 22, which has a propyl side chain attached at the terminal end to a piperidine, was docked into the active site of the TDP1 crystal structure (PDB ID: 1RFF) using GOLD. The energy-minimized, top-ranked GOLD pose of compound 22 in the TDP1 active site is displayed in Figure 7. Compound 22 fits well in the catalytic and hydrophobic regions with four hydrogen bonds. The 2-ether oxygen on the A ring and the 11-carbonyl group on the C ring of compound 22 are calculated to form hydrogen bonds with catalytic histidine residues 493 and 263, respectively. There are also hypothetical hydrogen bonds calculated between the 11-carbonyl group on the C ring and Asn516 as well as between the N atom of the piperidine in the side chain and Ser459. The propyl side chain ending with piperidine in compound 22 occupies the hydrophobic region, as expected, which provides a reason for the greater TDP1 activity of compounds with three-carbon side chains connected to amines with more hydrophobic substituents. For example, compounds 18 and 19, which have dimethylamino and ethylamino groups on the end of the propyl side chain, exhibit excellent TDP1 activity, and compounds 22 and 23, with piperidine and pyrrolidine on the end of the propyl side chain, also show promising TDP1 activity. In contrast, compounds 20 and 21, which have the less hydrophobic amines N-methylpiperazine and morpholine, display no TDP1 inhibitory activity.
Scheme 7. Schematic Representation of the TDP1 Gel-Based Assays Using Recombinant TDP1.
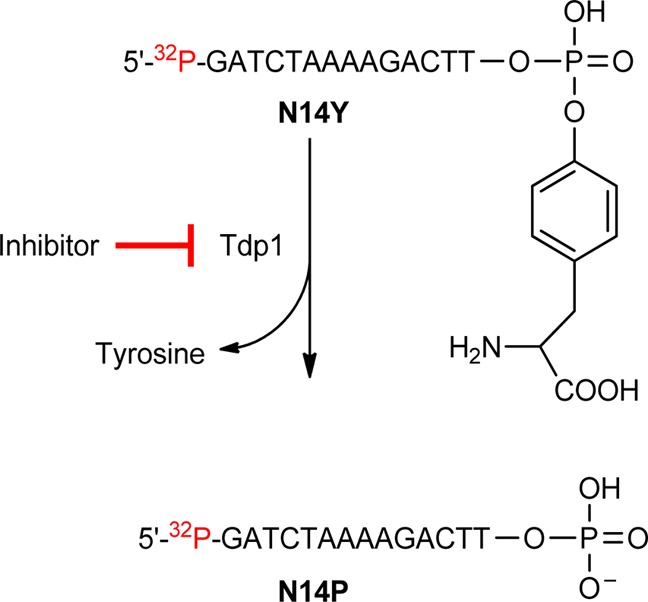
Figure 6.

Representative gels showing concentration-dependent TDP1 inhibition by O-2-modified indenoisoquinolines 21, 18, 19, and 22: lane 1, DNA alone; lane 2, TDP1 + DNA; and lanes 3–22, 21, 18, 19, and 22 (each at 1.4, 4.1, 12.3, 37, and 111 μM from left to right).
Figure 7.

Hypothetical binding mode of compound 22 in the active site of TDP1 (PDB ID: 1RFF). All distances are measured from heavy atom to heavy atom. The diagram is programmed for wall-eyed (relaxed) viewing. Compound 22 is shown in pink sticks.
Although it has been challenging to optimize the compounds for inhibition of two enzymes (Top1 and TDP1) simultaneously, molecular docking studies led to the hypothesis that the indenoisoquinoline platform present in Top1 inhibitors could be accommodated within the catalytic region in the TDP1 active site and that a three-carbon side chain at O-2 containing terminal amines bearing hydrophobic substituents could bind in a hydrophobic region of TDP1. This approach was validated by enzyme inhibition assays that demonstrated significant inhibition of both enzymes by compounds 18, 19, 22, and 23.
Selected compounds were tested for anti-proliferative activity in the National Cancer Institute’s developmental therapeutics assay 60-cell line screen (NCI60). The cells were incubated with the tested compounds at 100, 10, 1, 0.1, and 0.01 μM concentrations for 48 h before treatment with sulforhodamine B dye. Optical densities were recorded, and their ratios relative to that of the control were plotted as the percentage growth against the log of the tested compound concentrations. The concentration that corresponds to 50% growth inhibition (GI50) was calculated by interpolation between the points located above and below the 50% percentage growth inhibition. The results are listed in Table 2. Many of the new O-2-modified indenoisoquinolines display significant potency against various cell lines with GI50’s in the low micromolar (compounds 22 and 24) or submicromolar range (compounds 15, 17, and 20). Compounds 20, 22, and 23, which have potent inhibitory against Top1, also have cytotoxicities with mean graph midpoint (MGM) values ranging from 1.575 ± 0.233 to 0.24 ± 0.028 μM. Although the MGM values for compounds 15, 17, 20, 22, 23, and 24 do not differ greatly, in general, there is an intriguing lack of correlation between the rank order of observed cytotoxicities and inhibition of the two enzymes studied. For example, indenoisoquinoline 15 is the most cytotoxic compound, but it has low Top1 inhibitory activity and no detectable TDP1 inhibitory activity. In contrast, the cytotoxicity of 22 is comparatively low, but it has relatively high activity versus both enzymes. The GI50 values in individual cell lines vary more widely than the MGM values, and more significant differences are observed. For example, compound 24 is the most cytotoxic of the indenoisoquinolines versus the lung HOP-62 cell line, but it has the lowest overall cytotoxicity as indicated by the MGM value. Therefore, the lack of a strong correlation between enzyme inhibition and cytotoxicity is a complicated matter that may be influenced by the particular cell line under investigation as well as by differences in cellular penetration, distribution within the cell, metabolism, ejection from the cell, and possible off-target effects.
Table 2. Antiproliferative Potencies of Selected O-2-Modified Indenoisoquinolines.
| cytotoxicity (GI50; μM)a |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| lung | colon | CNS | melanoma | ovarian | renal | prostate | breast | ||
| compd | HOP-62 | HCT-116 | SF-539 | UACC-62 | OVCAR-3 | SN12C | DU-145 | MCF-7 | MGMb |
| 1(51) | 0.01 | 0.03 | 0.01 | 0.01 | 0.22 | 0.02 | 0.01 | 0.01 | 0.04 ± 0.018c |
| 4(51) | 1.3 | 35 | 41 | 4.2 | 73 | 68 | 37 | 1.58 | 20.0 ± 14 |
| 5(51) | 0.02 | 0.10 | 0.04 | 0.03 | 0.5 | <0.01 | <0.01 | <0.01 | 0.2 ± 0.19 |
| 13(58) | 0.3 | 0.39 | 0.3 | 0.3 | 0.9 | 0.2 | 0.3 | 0.1 | 0.4 ± 0.005 |
| 15 | <0.01 | 0.01 | 0.01 | <0.01 | 0.3 | <0.01 | <0.01 | <0.01 | 0.1 ± 0.035 |
| 17 | 0.3 | 0.2 | 0.2 | 0.3 | 0.4 | 0.3 | 0.3 | 0.2 | 0.5 ± 0.092 |
| 20 | 0.03 | 0.1 | 0.1 | 0.2 | 0.7 | 0.1 | 0.02 | 0.02 | 0.2 ± 0.028 |
| 22 | 0.3 | 1.1 | 0.03 | 9.6 | 5.4 | 0.4 | 0.2 | 0.04 | 1.6 ± 0.23 |
| 23 | 0.2 | 0.2 | 0.2 | 0.3 | 1.2 | 0.2 | 0.04 | 0.1 | 0.6 ± 0.37 |
| 24 | 0.1 | 1.1 | 0.1 | 1.0 | 5.8 | 0.8 | 0.9 | 0.4 | 3.1 ± 1.2 |
The cytotoxicity GI50 values are the concentrations corresponding to 50% growth inhibition.
Mean graph midpoint for growth inhibition of all human cancer cell lines successfully tested, ranging from 10–8 to 10–4 molar.
For MGM GI50 values in which a standard error appears, the GI50 values for individual cell lines are the average of two determinations.
Conclusions
A series of 2-position-substituted indenoisoquinolines with a three-carbon side chain linked at the end to amines was designed and synthesized for the development of dual Top1 and TDP1 inhibitors based on the hypotheses that (a) 2-OH indenoisoquinolines substituted with a three-carbon side chain ending with amino substitutions could bind to Asp533 in the Top1 active site, thus improving the Top1 inhibitory activity, and (b) the indenoisoquinoline core could be accommodated within the catalytic region in the TDP1 active site while a three-carbon side chain attached to amines with hydrophobic substitutions could bind to the hydrophobic region of TDP1. Top1 inhibition results reveal that the attachment of aminopropyl side chains targeting Asp533 results in a slight but consistent improvement in activity in the cases of 18, 19, 20, 21, and 22, but in the cases of primary amine 17 and pyrrolidine derivative 23, there was an unexpected drop in Top1 inhibitory activity. Enzyme inhibition results with both Top1 and TDP1 indicate that compounds 18, 19, 22, and 23 have good inhibitory activity against Top1 and also show promising TDP1 inhibitory activity.
Experimental Section
General
Solvents and reagents were purchased from commercial vendors and were used without any further purification. Melting points were determined using capillary tubes with a Mel-Temp apparatus and are uncorrected. Infrared spectra were obtained using KBr pellets. IR spectra were recorded using a PerkinElmer 1600 series or Spectrum One FTIR spectrometer. 1H NMR spectra were recorded at 300 MHz using a Bruker ARX300 spectrometer with a QNP probe. Mass spectral analyses were performed at the Purdue University Campus-Wide Mass Spectrometry Center. ESI–MS studies were performed using a FinniganMAT LCQ Classic mass spectrometer. EI/CI–MS studies were performed using a Hewlett-Packard Engine or GCQ FinniganMAT mass spectrometer. APCI–MS studies were carried out using an Agilent 6320 ion trap mass spectrometer. Analytical thin-layer chromatography was carried out on Baker-flex silica gel IB2-F plates, and compounds were visualized with short-wavelength UV light and ninhydrin staining. Silica gel flash chromatography was performed using 230–400 mesh silica gel. HPLC analyses were performed on a Waters 1525 binary HPLC pump/Waters 2487 dual λ absorbance detector system using a 5 μM C18 reverse-phase column. Compound purities were estimated by reversed-phase C18 HPLC with a UV detector at 254 nm, and the major peak area of each tested compound was ≥95% of the combined total peak area. All yields refer to isolated compounds.
2-(3-Bromopropoxy)-3-methoxy-6-(3-morpholinopropyl)-5H-[1,3]dioxolo[4′,5′:5,6]indeno[1,2-c]isoquinoline-5,12(6H)-dione (14)
A solution of compound 13(58) (0.100 g, 0.216 mmol) in DMF (6 mL) was treated with sodium hydride (0.011 g, 2.16 mmol). After 10 min, 1,3-dibromopropane was added. The mixture was stirred at room temperature for 3 h. The mixture was diluted to a volume of 200 mL with CHCl3, washed with H2O (2 × 50 mL) and saturated aqueous NaCl (50 mL), dried over anhydrous sodium sulfate, and concentrated. The residue was purified by flash column chromatography (SiO2, ∼40 g), eluting with 1% MeOH in CHCl3 to yield compound 14 as a solid (0.071 g, 56%). mp 197–199 °C. IR (film) 3434, 2102, 1638, 1498, 1304, 1115, 1032 cm–1. 1H NMR (CDCl3) δ 8.02 (s, 1H), 7.62 (s, 1H), 7.38 (s, 1H), 7.04 (s, 1H), 6.15 (s, 2H), 5.58–5.37 (m, 2H), 4.79–4.77 (d, J = 5.1 Hz, 2H), 4.69–4.52 (t, J = 8.4 Hz, 2H), 3.97 (s, 3H), 3.77 (s, 4H), 2.55 (s, 6H), 2.02 (s, 2H), 1.33 (s, 2H). ESI–MS m/z 585/587 (MH+), 505 (MH+ – HBr). HRESI–MS m/z 585.1236 (MH+); calcd for C28H30BrN2O7, 585.1243.
2-(Allyloxy)-3-methoxy-6-(3-morpholinopropyl)-5H-[1,3]dioxolo[4′,5′:5,6]indeno [1,2-c]isoquinoline-5,12(6H)-dione (15)
Column chromatography of the mixture described above also yielded side-product 15 as a solid (0.013 g, 10%). mp 199–200 °C (dec). IR (film) 2936, 1749, 1698, 1651, 1304, 1034, 786 cm–1. 1H NMR (CDCl3) δ 8.01 (s, 1H), 7.61 (s, 1H), 7.37 (s, 1H), 7.03 (s, 1H), 6.08 (s, 3H), 5.53–5.40 (m, 2H), 4.79–4.77 (d, J = 5.7 Hz, 2H), 4.51–4.45 (t, J = 7.5 Hz, 2H), 3.97 (s, 3H), 3.77 (s, 4H), 2.54 (s, 6H), 2.01 (s, 2H). ESI–MS m/z 505 (MH+). HRESI–MS m/z 505.1967 (MH+); calcd for C28H29N2O7, 505.1975. HPLC purity: 97.57% (C18 reverse phase, MeOH/H2O, 90:10).
2-(3-Azidopropoxy)-3-methoxy-6-(3-morpholinopropyl)-5H-[1,3]dioxolo[4′,5′:5,6]indeno[1,2-c]isoquinoline-5,12(6H)-dione (16)
Sodium azide (0.021 g, 0.22 mmol) and compound 14 (0.128 g, 0.22 mmol) were diluted with DMSO (4 mL), and the mixture was heated at 100 °C for 2 h. The mixture was diluted to a volume of 200 mL with CHCl3, washed with H2O (2 × 60 mL) and saturated aqueous NaCl (50 mL), dried over anhydrous sodium sulfate, and concentrated. The resulting residue was purified by flash column chromatography (SiO2, ∼40 g), eluting with 0.5% MeOH in CHCl3 to yield product 16 as a solid (0.036 g, 60%). The solid was used for the next step without further purification. ESI–MS m/z 548 (MH+). HRESI–MS m/z 548.2141 (MH+); calcd for C28H30N5O7, 548.2145.
2-(3-Aminopropoxy)-3-methoxy-6-(3-morpholinopropyl)-5H-[1,3]dioxolo[4′,5′:5,6]indeno[1,2-c]isoquinoline-5,12(6H)-dione (17)
Triethyl phosphite (0.022 mL, 0.183 mmol) was added to a solution of compound 16 (0.040 g, 0.073 mmol) in benzene (4 mL), and the mixture was heated at reflux for 24 h. The mixture was diluted to a volume of 200 mL with CHCl3, washed with H2O (2 × 50 mL) and saturated aqueous NaCl (50 mL), dried over anhydrous sodium sulfate, and concentrated. The resulting residue was purified by flash column chromatography (SiO2, ∼40 g), eluting with 0.5% MeOH in CHCl3 to yield the title compound as a solid. The solid (0.015 g, 0.023 mmol) was diluted with benzene (4 mL), and 2 M HCl in methanol (6 mL) was added to the solution. The mixture was heated at reflux for 3 h. The reaction mixture was allowed to cool to room temperature, and the precipitate was filtered to provide desired compound 17 as a solid (0.007 g, 58%). mp > 350 °C. IR (film) 3413, 2346, 1751, 1651, 1559, 1437, 1309, 737 cm–1. 1H NMR (D2O) δ 6.77 (s, 1H), 6.62 (s, 1H), 6.56 (s, 1H), 6.16 (s, 1H), 5.96 (s, 2H), 4.86–4.82 (m, 2H), 4.01 (s, 6H), 3.78 (s, 2H), 3.67 (s, 3H), 3.32 (s, 4H), 3.24–3.19 (t, J = 7.5 Hz, 2H), 2.19–2.17 (m, 4H). ESI–MS m/z 522 (MH+). HRESI–MS m/z 522.2249 (MH+); calcd for C28H32N3O7, 522.2246. HPLC purity: 95.23% (C18 reverse phase, MeOH/H2O, 85:15).
2-(3-(Dimethylamino)propoxy)-3-methoxy-6-(3-morpholinopropyl)-5H-[1,3]dioxolo[4′,5′:5,6]indeno[1,2-c]isoquinoline-5,12(6H)-dione (18)
Sodium iodide (0.061 g, 0.408 mmol) and compound 14 (0.020 g, 0.034 mmol) were diluted with dioxane (10 mL), and dimethylamine (0.023 mL, 0.408 mmol) was added dropwise. The mixture was stirred at reflux for 52 h. The mixture was diluted to a volume of 250 mL with CHCl3, washed with H2O (2 × 50 mL) and saturated aqueous NaCl (50 mL), dried over anhydrous sodium sulfate, and concentrated. The residue was purified by flash column chromatography (SiO2, ∼40 g), eluting first with 0.5% MeOH in CHCl3 and then with 1% MeOH in CHCl3 to yield product 18 as a solid (0.009 g, 47%). mp 176–178 °C (dec). IR (film) 2922, 1698, 1650, 1483, 1306, 1032, 786 cm–1. 1H NMR (CDCl3) δ 8.02 (s, 1H), 7.63 (s, 1H), 7.43 (s, 1H), 7.08 (s, 1H), 6.09 (s, 2H), 4.54–4.48 (t, J = 8.4 Hz, 2H), 4.29–4.25 (t, J = 6.3 Hz, 2H), 3.95 (s, 3H), 3.78–3.74 (t, J = 4.8 Hz, 2H), 2.57–2.53 (m, 14H), 2.27 (s, 2H), 2.03–2.02 (m, 2H). ESI–MS m/z 550 (MH+). HRESI–MS m/z 550.2558 (MH+); calcd for C30H36N3O7, 550.2553. HPLC purity: 97.57% (C18 reverse phase, MeOH/H2O, 85:15).
2-(3-(Ethylamino)propoxy)-3-methoxy-6-(3-morpholinopropyl)-5H-[1,3]dioxolo[4′,5′:5,6]indeno[1,2-c]isoquinoline-5,12(6H)-dione (19)
Sodium iodide (0.060 g, 0.408 mmol) and compound 14 (0.020 g, 0.034 mmol) were diluted with dioxane (8 mL), and ethylamine (0.020 mL, 0.408 mmol, 70 wt % solution in water) was added dropwise. The mixture was stirred at reflux for 26 h. The mixture was diluted to a volume of 200 mL with CHCl3, washed with H2O (2 × 50 mL) and saturated aqueous NaCl (50 mL), dried over anhydrous sodium sulfate, and concentrated. The residue was purified by flash column chromatography (SiO2, ∼40 g), eluting with 0.5% MeOH in CHCl3 to yield product 19 as a solid (0.009 g, 46%). mp 187–188 °C (dec). IR (film) 1648, 1553, 1392, 1254, 1116, 1033, 785 cm–1. 1H NMR (CDCl3) δ 7.53 (s, 1H), 7.24 (s, 1H), 6.90 (s, 1H), 6.60 (s, 1H), 5.89 (s, 2H), 4.06 (s, 2H), 3.85 (s, 6H), 3.72 (s, 3H), 3.49–3.38 (m, 4H), 3.19–3.12 (m, 9H), 2.25–2.19 (m, 4H). ESI–MS m/z 550 (MH+). HRESI–MS m/z 550.2553 (MH+); calcd for C30H36N3O7, 550.2553. HPLC purity: 98.36% (C18 reverse phase, MeOH/H2O, 80:20).
3-Methoxy-2-(3-morpholinopropoxy)-6-(3-morpholinopropyl)-5H-[1,3]dioxolo[4′,5′:5,6]indeno[1,2-c]isoquinoline-5,12(6H)-dione (20)
Sodium iodide (0.139 g, 0.924 mmol) and compound 14 (0.045 g, 0.077 mmol) were diluted with dioxane (10 mL), and morpholine (0.08 mL, 0.924 mmol) was added dropwise. The mixture was stirred at reflux for 24 h. The mixture was diluted to a volume of 200 mL with CHCl3, washed with H2O (2 × 60 mL) and saturated aqueous NaCl (50 mL), dried over anhydrous sodium sulfate, and concentrated. The residue was purified by flash column chromatography (SiO2, ∼40 g), eluting first with 0.5% MeOH in CHCl3 and then with 1% MeOH in CHCl3 to yield product 20 as a solid (0.018 g, 41%). mp 188–189 °C (dec). IR (film) 2956, 1869, 1749, 1650, 1508, 1307, 1032, 865 cm–1. 1H NMR (CDCl3) δ 7.99 (s, 1H), 7.61 (s, 1H), 7.39 (s, 1H), 7.04 (s, 1H), 6.08 (s, 2H), 4.64–4.51 (t, J = 7.5 Hz, 2H), 4.28–4.24 (t, J = 6.6 Hz, 2H), 3.95 (s, 3H), 3.76 (s, 8H), 2.53 (s, 12H), 2.15–2.11 (t, J = 6.3 Hz, 2H), 2.01 (s, 2H). ESI–MS m/z 592 (MH+). HRESI–MS m/z 592.2664 (MH+); calcd for C32H38N3O8, 592.2659. HPLC purity: 95.38% (C18 reverse phase, MeOH/H2O, 85:15).
3-Methoxy-2-(3-(4-methylpiperazin-1-yl)propoxy)-6-(3-morpholinopropyl)-5H-[1,3]dioxolo[4′,5′:5,6]indeno[1,2-c]isoquinoline-5,12(6H)-dione (21)
Sodium iodide (0.061 g, 0.408 mmol) and compound 14 (0.020 g, 0.034 mmol) were diluted with dioxane (5 mL), and N-methyl piperazine (0.041 mL, 0.408 mmol) was added dropwise. The mixture was stirred at reflux for 24 h. The mixture was diluted to a volume of 200 mL with CHCl3, washed with H2O (2 × 50 mL) and saturated aqueous NaCl (50 mL), dried over anhydrous sodium sulfate, and concentrated. The residue was purified by flash column chromatography (SiO2, ∼40 g), eluting first with 0.5% MeOH in CHCl3 and then with 3% MeOH in CHCl3 to yield product 21 as a solid (0.010 g, 49%). mp 181–183 °C (dec). IR (film) 2924, 1870, 1650, 1508, 1307, 1032, 868 cm–1. 1H NMR (CDCl3) δ 8.01 (s, 1H), 7.62 (s, 1H), 7.42 (s, 1H), 7.06 (s, 1H), 6.09 (s, 2H), 4.53–4.47 (t, J = 8.4 Hz, 2H), 4.28–4.23 (t, J = 6.6 Hz, 2H), 3.95 (s, 3H), 3.77–3.74 (t, J = 4.5 Hz, 2H), 2.62–2.53 (m, 16H), 2.35 (s, 3H), 2.13–2.06 (m, 2H), 2.05–2.00 (m, 2H). ESI–MS m/z 605 (MH+). HRESI–MS m/z 605.2986 (MH+); calcd for C33H41N4O7, 605.2975. HPLC purity: 100% (C18 reverse phase, MeOH/H2O, 90:10).
3-Methoxy-6-(3-morpholinopropyl)-2-(3-(piperidin-1-yl)propoxy)-5H-[1,3]dioxolo[4′,5′:5,6]indeno[1,2-c]isoquinoline-5,12(6H)-dione (22)
1-(3-Chloropropyl)piperidine hydrochloride (214 mg, 1.08 mmol) and K2CO3 (298 mg, 2.16 mmol) were added to a DMF (5 mL) solution of compound 13 (0.100 g, 0.216 mmol). The mixture was heated at 90 °C for 23 h. The mixture was diluted to a volume of 300 mL with CHCl3, washed with H2O (2 × 80 mL) and saturated aqueous NaCl (80 mL), dried over anhydrous sodium sulfate, and concentrated. The residue was purified by flash column chromatography (SiO2, ∼40 g), eluting with 0.25% MeOH in CHCl3 to yield product 22 as a solid (0.072 g, 58%). mp 156–157 °C (dec). IR (film) 3399, 2091, 1645, 1392, 1305 cm–1. 1H NMR (CDCl3) δ 7.99 (s, 1H), 7.60 (s, 1H), 7.39 (s, 1H), 7.04 (s, 1H), 6.08 (s, 2H), 4.51–4.46 (t, J = 7.8 Hz, 2H), 4.25–4.21 (t, J = 6.6 Hz, 2H), 3.95 (s, 3H), 3.77–3.74 (m, 4H), 2.58–2.46 (m, 12H), 2.18–2.09 (m, 2H), 2.04–1.95 (m, 2H), 1.61–1.58 (m, 4H), 1.46–1.44 (m, 2H). ESI–MS m/z 590 (MH+). HRESI–MS m/z 590.2859 (MH+); calcd for C33H40N3O7, 590.2866. HPLC purity: 95.88% (C18 reverse phase, MeOH/H2O, 95:05).
3-Methoxy-6-(3-morpholinopropyl)-2-(3-(pyrrolidin-1-yl)propoxy)-5H-[1,3]dioxolo[4′,5′:5,6]indeno[1,2-c]isoquinoline-5,12(6H)-dione (23)
1-(3-Chloropropyl)piperidine hydrochloride (158 mg, 0.86 mmol) and K2CO3 (237 mg, 1.72 mmol) were added to a DMF (5 mL) solution of compound 13 (0.80 g, 0.172 mmol). The mixture was heated at 90 °C for 19 h. The mixture was diluted to a volume of 300 mL with CHCl3, washed with H2O (2 × 80 mL) and saturated aqueous NaCl (80 mL), dried over anhydrous sodium sulfate, and concentrated. The residue was purified by flash column chromatography (SiO2, ∼40 g), eluting with 0.25% MeOH in CHCl3 to yield product 23 as a solid (0.051 g, 55%). mp 151–152 °C (dec). IR (film) 3418, 2936, 2119, 1660, 1392, 1225, 1105, 1063 cm–1. 1H NMR (CDCl3) δ 8.02 (s, 1H), 7.63 (s, 1H), 7.42 (s, 1H), 7.07 (s, 1H), 6.09 (s, 2H), 4.52–4.48 (t, J = 8.1 Hz, 2H), 4.29–4.24 (t, J = 6.6 Hz, 2H), 3.95 (s, 3H), 3.77–3.74 (t, J = 4.5 Hz, 2H), 2.80–2.77 (m, 2H), 2.70 (s, 2H), 2.57–2.53 (m, 6H), 2.25–2.20 (m, 2H), 2.03–1.98 (m, 2H), 1.87 (s, 6H). ESI–MS m/z 576 (MH+). HRESI–MS m/z 576.2705 (MH+); calcd for C32H38N3O7, 576.2710; HPLC purity: 96.29% (C18 reverse phase, MeOH/H2O, 80:20).
Methyl 2-((3-Methoxy-6-(3-morpholinopropyl)-5,12-dioxo-6,12-dihydro-5H-[1,3]dioxolo[4′,5′:5,6]indeno[1,2-c]isoquinolin-2-yl)oxy)acetate (24)
Sodium hydride (0.067 g, 2.8 mmol) and compound 13 (0.130 g, 0.28 mmol) were diluted with DMF (8 mL), and methyl bromoacetate (0.106 mL, 1.12 mmol) was added dropwise. The mixture was stirred at room temperature for 7 h. The mixture was diluted to a volume of 250 mL with CHCl3, washed with H2O (2 × 60 mL) and saturated aqueous NaCl (50 mL), dried over anhydrous sodium sulfate, and concentrated. The residue was purified by flash column chromatography (SiO2, ∼40 g), eluting with 0.5% MeOH in CHCl3 to yield product 24 as a solid (0.077 g, 51%). mp 226–227 °C. IR (film) 2345, 1869, 1749, 1650, 1508, 1031, 737 cm–1. 1H NMR (CDCl3) δ 7.91 (s, 1H), 7.65 (s, 1H), 7.40 (s, 1H), 7.04 (s, 1H), 6.09 (s, 2H), 4.87 (s, 2H), 4.52–4.47 (t, J = 7.2 Hz, 2H), 3.98 (s, 3H), 3.86 (s, 3H), 3.78 (s, 4H), 2.54 (s, 6H), 2.01 (s, 2H). ESI–MS m/z 537 (MH+). HRESI–MS m/z 537.1875 (MH+); calcd for C28H29N2O9, 537.1873. HPLC purity: 96.60% (C18 reverse phase, MeOH/H2O, 90:10).
2-((12-Hydroxy-3-methoxy-6-(3-morpholinopropyl)-5-oxo-6,12-dihydro-5H-[1,3]dioxolo[4′,5′:5,6]indeno[1,2-c]isoquinolin-2-yl)oxy)acetohydrazide (26)
Hydrazine (0.028 mL, 0.056 mmol) and compound 13 (0.015 g, 0.028 mmol) were diluted with EtOH (10 mL), and the mixture was heated at reflux for 16 h. The precipitate obtained was washed with hexane (10 mL) and ether (10 mL) to yield product 26 as a light yellow solid (0.006 g, 40%). mp 266–268 °C. IR (film) 2365, 1869, 1773, 1648, 1508, 1032, 738 cm–1. 1H NMR (CDCl3) δ 9.30 (s, 1H), 7.62 (s, 1H), 7.45 (s, 1H), 7.36 (s, 1H), 7.21 (s, 1H), 6.10 (s, 2H), 5.34 (s, 2H), 4.61 (s, 2H), 4.48 (s, 2H), 3.88 (s, 3H), 3.61 (s, 4H), 2.49 (m, 6H), 1.96 (m, 2H). ESI–MS m/z 539 (MH+). HRESI–MS m/z 539.2146 (MH+); calcd for C27H30N4O8, 539.2142. HPLC purity: 95.19% (C18 reverse phase, MeOH/H2O, 85:15).
Topoisomerase I-Mediated DNA Cleavage Reactions
Human recombinant Top1 was purified from baculovirus as previously described.26 DNA cleavage reactions were prepared as previously reported with the exception of the DNA substrate.23 Briefly, a 117 bp DNA oligonucleotide (Integrated DNA Technologies) encompassing the previously identified Top1 cleavage sites in the 161 bp fragment from pBluescript SK(−) phagemid DNA was employed. This 117 bp oligonucleotide contains a single 5′ cytosine overhang, which was 3′-end-labeled by a fill-in reaction with [α-32P]dGTP in React 2 buffer (50 mM Tris-HCl, pH 8.0, 100 mM MgCl2, and 50 mM NaCl) and 0.5 units of DNA polymerase I (Klenow fragment, New England Biolabs). Unincorporated [32P]dGTP was removed using mini Quick Spin DNA columns (Roche, Indianapolis, IN), and the eluate containing the 3′-end-labeled DNA substrate was collected. Approximately 2 nM radiolabeled DNA substrate was incubated with recombinant Top1 in 20 μL of reaction buffer [10 mM Tris-HCl, pH 7.5, 50 mM KCl, 5 mM MgCl2, 0.1 mM EDTA, and 15 μg/mL BSA] at 25 °C for 20 min in the presence of various concentrations of compounds. The reactions were terminated by adding SDS (0.5% final concentration) followed by the addition of two volumes of loading dye (80% formamide, 10 mM sodium hydroxide, 1 mM sodium EDTA, 0.1% xylene cyanol, and 0.1% bromphenol blue). Aliquots of each reaction mixture were subjected to 20% denaturing PAGE. Gels were dried and visualized using a phosphoimager and ImageQuant software (Molecular Dynamics). For simplicity, cleavage sites were numbered as previously described in the 161 bp fragment.
Gel-Based Assay Measuring the Inhibition of Recombinant TDP1
A 5′-[32P]-labeled single-stranded DNA oligonucleotide containing a 3′ phosphotyrosine (N14Y) was generated as described by Dexheimer et al.53 The DNA substrate was then incubated with 5 pM recombinant TDP1 in the absence or presence of inhibitor for 15 min at room temperature in a buffer containing 50 mM Tris HCl, pH 7.5, 80 mM KCl, 2 mM EDTA, 1 mM DTT, 40 μg/mL BSA, and 0.01% Tween-20. Reactions were terminated by the addition of one volume of gel loading buffer [99.5% (v/v) formamide, 5 mM EDTA, 0.01% (w/v) xylene cyanol, and 0.01% (w/v) bromophenol blue]. Samples were subjected to 16% denaturing PAGE, and gels were exposed after drying to a PhosphorImager screen (GE Healthcare). Gel images were scanned using a Typhoon 8600 (GE Healthcare), and densitometric analyses were performed using ImageQuant software (GE Healthcare).
Molecular Modeling
The Top1 crystal structure for docking was prepared, and the docking protocol was validated as previously described.58 The ternary complex ligand centroid coordinates for docking were defined using the ligand in the Top1–DNA–MJ238 crystal structure (PDB ID: 1SC7) as the center of the binding pocket (x = 21.3419, y = −3.9888, and z = 28.2163). The ligand was then deleted. Indenoisoquinolines to be modeled were constructed in SYBYL. Atom types were assigned using SYBYL atom typing. Hydrogens were added, and the ligands were minimized by the conjugate gradient method using the MMFF94s force field with MMFF94 charges, a distance-dependent dielectric function, and a 0.01 kcal mol–1 Å–1 energy gradient convergence criterion. Each ligand was docked into the mutant crystal structure using GOLD 3.2 with default parameters, and the coordinates were defined by the crystal structure as described above. The top four poses for each ligand were examined. The highest-ranked poses for these ligands were merged into the crystal structure, and the entire complex was subsequently subjected to minimization using a standard Powell method, the MMFF94s force field and MMFF94 charges, a distance-dependent dielectric function, and a 0.05 kcal mol–1 Å–1 energy gradient convergence criterion. During the energy minimization, the ligand and a 7 Å sphere surrounding the ligand were allowed to move while the structures outside this sphere were frozen in an aggregate.
The TDP1 crystal structure (PDB ID: 1RFF) was prepared by removing one of the monomers along with all crystallized waters, the polydeoxyribonucleotide 5′-D-(*AP*GP*TP*T)-3′, the Top1-derived peptide residues 720–727 (mutation L724Y), and all metal ions. The Lys265, Lys495, and His493 residues were protonated. Missing hydrogens were added as needed. GOLD docking was performed using centroid coordinates x = 7.194, y = 52.407, and z = 0.704. The hydrogen-bond length was set to 4 Å, and the van der Waals parameter was set to 10 Å. The top ligand-binding pose (highest GOLD score) was selected and merged with the prepared protein. The ligand was surrounded by a sphere with a 12 Å radius and energy-minimized by the conjugate gradient method using the MMFF94s force field and MMFF94 charges with SYBYL software. The calculation was terminated when the gradient reached a value of 0.05 kcal mol–1 Å–1.
Acknowledgments
This work was made possible by the National Institutes of Health (NIH) through support from research grant UO1 CA89566 and was supported by the Intramural Research Program, Center for Cancer Research, National Cancer Institute, NIH (Z01 BC 006161 and Z01 BC 006150). In vitro cytotoxicity testing was performed by the Developmental Therapeutics Program at the National Cancer Institute under contract NO1-CO-56000.
Glossary
Abbreviations Used
- APCI–MS
atmospheric-pressure chemical ionization mass spectrometry
- CI/EI–MS
chemical ionization/electron impact mass spectrometry
- CPT
camptothecin
- DMAP
4-dimethylaminopyridine
- DMSO-d6
dimethyl sulfoxide-d6
- ESI–MS
electrospray ionization mass spectrometry
- HRMS
high-resolution mass spectrometry
- SCAN1
spinocerebellar ataxia with axonal neuropathy
- TDP1
tyrosyl-DNA phosphodiesterase I
- TFA
trifluoroacetic acid
- Top1
topoisomerase type I
Supporting Information Available
1H NMR spectra, HPLC traces, and SMILES molecular formula strings. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Pommier Y. DNA Topoisomerase I Inhibitors: Chemistry, Biology, and Interfacial Inhibition. Chem. Rev. 2009, 109, 2894–2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pommier Y. Topoisomerase I Inhibitors: Camptothecins and Beyond. Nat. Rev. Cancer 2006, 6, 789–802. [DOI] [PubMed] [Google Scholar]
- Champoux J. J. DNA Topoisomerases: Structure, Function, and Mechanism. Annu. Rev. Biochem. 2001, 70, 369–413. [DOI] [PubMed] [Google Scholar]
- Stewart L.; Redinbo M. R.; Qiu X.; Hol W. G. J.; Champoux J. J. A Model for the Mechanism of Human Topoisomerase I. Science 1998, 279, 1534–1541. [DOI] [PubMed] [Google Scholar]
- Pommier Y.; Barcelo J. A.; Rao V. A.; Sordet O.; Jobson A. G.; Thibaut L.; Miao Z.-H.; Seiler J. A.; Zhang H.; Marchand C.; Agama K.; Redon C. Repair of Topoisomerase I-Mediated DNA Damage. Prog. Nucleic Acid Res. Mol. Biol. 2006, 81, 179–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sordet O.; Khan Q. A.; Kohn K. W.; Pommier Y. Apoptosis Induced by Topoisomerase Inhibitors. Curr. Med. Chem.: Anti-Cancer Agents 2003, 3, 271–290. [DOI] [PubMed] [Google Scholar]
- Staker B. L.; Hjerrild K.; Feese M. D.; Behnke C. A.; Burgin A. B.; Stewart L. The Mechanism of Topoisomerase I Poisoning by a Camptothecin Analogue. Proc. Natl. Acad. Sci. U.S.A. 2002, 99, 15387–15392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. C. Cellular Roles of DNA Topoisomerases: A Molecular Perspective. Nat. Rev. Mol. Cell Biol. 2002, 3, 430–440. [DOI] [PubMed] [Google Scholar]
- Wall M. E.; Wani M. C.; Cook C. E.; Palmer K. H.; McPhail A. T.; Sim G. A. Plant Antitumor Agents. I. The Isolation and Structure of Camptothecin, a Novel Alkaloidal Leukemia and Tumor Inhibitor from Camptotheca acuminata. J. Am. Chem. Soc. 1966, 88, 3888–3890. [Google Scholar]
- Teicher B. Next Generation of Topoisomerase I Inhibitors: Rationale and Biomarker Strategies. Biochem. Pharmacol. 2008, 75, 1262–1271. [DOI] [PubMed] [Google Scholar]
- Thomas C. J.; Rahier N. J.; Hecht S. M. Camptothecin: Current Perspectives. Bioorg. Med. Chem. 2004, 12, 1585–1604. [DOI] [PubMed] [Google Scholar]
- Li C. J.; Averboukh L.; Pardee A. B. Beta-Lapachone, a Novel DNA Topoisomerase I Inhibitor with a Mode of Action Different from Camptothecin. J. Biol. Chem. 1993, 268, 22463–22468. [PubMed] [Google Scholar]
- Jaxel C.; Kohn K. W.; Wani M. C.; Pommier Y. Structure–Activity Study of the Actions of Camptothecin Derivatives on Mammalian Topoisomerase I: Evidence for a Specific Receptor Site and a Relation to Antitumor Activity. Cancer Res. 1989, 49, 1465–1469. [PubMed] [Google Scholar]
- Minami H.; Beijen J. H.; Verwij J.; Ratain M. J. Limited Sampling Model for Area under the Concentration Time Curve of Total Topotecan. Clin. Cancer Res. 1996, 2, 43–46. [PubMed] [Google Scholar]
- Luzzio M. J.; Besterman J. M.; Emerson D. L.; Evans M. G.; Lackey K.; Leitner P. L.; McIntyre G.; Morton B.; Myers P. L.; Peel M.; Sisco J. M.; Sternbach D. D.; Tong W. Q.; Truesdale A.; Uehling D. E.; Vuong A.; Yates J. Synthesis and Antitumor-Activity of Novel Water-Soluble Derivatives of Camptothecin as Specific Inhibitors of Topoisomerase-I. J. Med. Chem. 1995, 38, 395–401. [DOI] [PubMed] [Google Scholar]
- Mi Z.; Burke T. G. Differential Interactions of Camptothecin Lactone and Carboxylate Forms with Human Blood Components. Biochemistry 1994, 33, 10325–10336. [DOI] [PubMed] [Google Scholar]
- Urasaki Y.; Laco G. S.; Pourquier P.; Takebayashi Y.; Kohlhagen G.; Gioffre C.; Zhang H. L.; Chatterjee D.; Pantazis P.; Pommier Y. Characterization of a Novel Topoisomerase I Mutation from a Camptothecin-Resistant Human Prostate Cancer Cell Line. Cancer Res. 2001, 61, 1964–1969. [PubMed] [Google Scholar]
- Fujimori A.; Harker W. G.; Kohlhagen G.; Hoki Y.; Pommier Y. Mutation at the Catalytic Site of Topoisomerase I in CEM/C2, a Human Leukemia Cell Resistant to Camptothecin. Cancer Res. 1995, 55, 1339–1346. [PubMed] [Google Scholar]
- Bates S. E.; Medina-Perez W. Y.; Kohlhagen G.; Antony S.; Nadjem T.; Robey R. W.; Pommier Y. ABCG2 Mediates Differential Resistance to SN-38 (7-Ethyl-10-hydroxycamptothecin) and Homocamptothecins. J. Pharmacol. Exp. Ther. 2004, 310, 836–842. [DOI] [PubMed] [Google Scholar]
- Hoki Y.; Fujimori A.; Pommier Y. Differential Cytotoxicity of Clinically Important Camptothecin Derivatives in P-Glycoprotein Overexpressing Cell Lines. Cancer Chemother. Pharmacol. 1997, 40, 433–438. [DOI] [PubMed] [Google Scholar]
- Brangi M.; Litman T.; Ciotti M.; Nishiyama K.; Kohlhagen G.; Takimoto C.; Robey R.; Pommier Y.; Fojo T.; Bates S. E. Camptothecin Resistance: Role of the ATP-Binding Cassette (ABC), Mitoxantrone-Resistance Half-Transporter (MXR), and Potential for Glucuronidation in MXR-Expressing Cells. Cancer Res. 1999, 59, 5938–5946. [PubMed] [Google Scholar]
- Armstrong D. K. Topotecan Dosing Guidelines in Ovarian Cancer: Reduction and Management of Hematologic Toxicity. Oncologist 2004, 9, 33–42. [DOI] [PubMed] [Google Scholar]
- Saltz L. B. Clinical Use of Irinotecan: Current Status and Future Considerations. Oncologist 1997, 2, 402–409. [PubMed] [Google Scholar]
- Paull K. D.; Shoemaker R. H.; Hodes L.; Monks A.; Scudiero D. A.; Rubinstein L.; Plowman J.; Boyd M. R. Display and Analysis of Patterns of Differential Activity of Drugs Against Human Tumor Cell Lines: Development of Mean Graph and COMPARE Algorithm. J. Natl. Cancer Inst. 1989, 81, 1088–1092. [DOI] [PubMed] [Google Scholar]
- Kohlhagen G.; Paull K.; Cushman M.; Nagafuji P.; Pommier Y. Protein-Linked DNA Strand Breaks Induced by NSC 314622, a Novel Noncamptothecin Topoisomerase I Poison. Mol. Pharmacol. 1998, 54, 50–58. [DOI] [PubMed] [Google Scholar]
- Bailly C. Topoisomerase I Poisons and Suppressors as Anticancer Drugs. Curr. Med. Chem. 2000, 7, 39–58. [DOI] [PubMed] [Google Scholar]
- Hsiang Y. H.; Hertzberg R.; Hecht S.; Liu L. F. Camptothecin Induces Protein-linked DNA Breaks via Mammalian DNA Topoisomerase I. J. Biol. Chem. 1985, 260, 14873–14878. [PubMed] [Google Scholar]
- Pommier Y.; Marchand C. Interfacial Inhibitors: Targeting Macromolecular Complexes. Nat. Rev. Drug Discovery 2012, 11, 25–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antony S.; Agama K. K.; Miao Z. H.; Takagi K.; Wright M. H.; Robles A. I.; Varticovski L.; Nagarajan M.; Morrell A.; Cushman M.; Pommier Y. Novel Indenoisoquinolines NSC 725776 and NSC 724998 Produce Persistent Topolsomerase I Cleavage Complexes and Overcome Multidrug Resistance. Cancer Res. 2007, 67, 10397–10405. [DOI] [PubMed] [Google Scholar]
- Chrencik J. E.; Staker B. L.; Burgin A. B.; Pourquier P.; Pommier Y.; Stewart L.; Redinbo M. R. Mechanisms of Camptothecin Resistance by Human Topoisomerase I Mutations. J. Mol. Biol. 2004, 339, 773–784. [DOI] [PubMed] [Google Scholar]
- Nagarajan M.; Morrell A.; Ioanoviciu A.; Antony S.; Kohlhagen G.; Agama K.; Hollingshead M.; Pommier Y.; Cushman M. Synthesis and Evaluation of Indenoisoquinoline Topoisomerase I Inhibitors Substituted with Nitrogen Heterocycles. J. Med. Chem. 2006, 49, 6283–6289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourquier P.; Ueng L.-M.; Fertala J.; Wang D.; Park H.-J.; Essigmann J. M.; Bjornsti M.-A.; Pommier Y. Induction of Reversible Complexes between Eukaryotic DNA Topoisomerase I and DNA-Containing Oxidative Base Damages. 7,8-Dihydro-8-oxoguanine and 5-Hydroxycytosine. J. Biol. Chem. 1999, 274, 8516–8523. [DOI] [PubMed] [Google Scholar]
- ClinicalTrials.gov. A Phase I Study of Indenoisoquinolines LMP400 and LMP776 in Adults With Relapsed Solid Tumors and Lymphomas. http://clinicaltrials.gov/ct2/show/study/NCT01051635 (accessed Feb 22, 2014).
- Interthal H.; Pouliott J. J.; Champoux J. J. The Tyrosyl-DNA Phosphodiesterase Tdp1 Is a Member of the Phospholipase D Superfamily. Proc. Natl. Acad. Sci. U.S.A. 2001, 98, 12009–12014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waite M. The PLD Superfamily: Insights into Catalysis. Biochim. Biophys. Acta, Mol. Cell Biol. Lipids 1999, 1439, 187–197. [DOI] [PubMed] [Google Scholar]
- Raymond A. C.; Rideout M. C.; Staker B.; Hjerrild K.; Burgin A. B. Analysis of Human Tyrosyl-DNA Phosphodiesterase I Catalytic Residues. J. Mol. Biol. 2004, 338, 895–906. [DOI] [PubMed] [Google Scholar]
- Murai J.; Huang S. Y. N.; Das B. B.; Dexheimer T. S.; Takeda S.; Pommier Y. Tyrosyl-DNA Phosphodiesterase 1 (TDP1) Repairs DNA Damage Induced by Topoisomerases I and II and Base Alkylation in Vertebrate Cells. J. Biol. Chem. 2012, 287, 12848–12857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S. Y. N.; Murai J.; Dalla Rosa I.; Dexheimer T. S.; Naumova A.; Gmeiner W. H.; Pommier Y. TDP1 Repairs Nuclear and Mitochondrial DNA Damage Induced by Chain-Terminating Anticancer and Antiviral Nucleoside Analogs. Nucleic Acids Res. 2013, 41, 7793–7803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pommier Y.; Leo E.; Zhang H. L.; Marchand C. DNA Topoisomerases and Their Poisoning by Anticancer and Antibacterial Drugs. Chem. Biol. 2010, 17, 421–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dexheimer T. S.; Antony S.; Marchand C.; Pommier Y. Tyrosyl-DNA Phosphodiesterase as a Target for Anticancer Therapy. Anti-Cancer Agents Med. Chem. 2008, 8, 381–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies D. R.; Interthal H.; Champoux J. J.; Hol W. G. J. Insights into Substrate Binding and Catalytic Mechanism of Human Tyrosyl-DNA Phosphodiesterase (Tdp1) from Vanadate and Tungstate-Inhibited Structures. J. Mol. Biol. 2002, 324, 917–932. [DOI] [PubMed] [Google Scholar]
- Strumberg D.; Pilon A. A.; Smith M.; Hickey R.; Malkas L.; Pommier Y. Conversion of Topoisomerase 1 Cleavage Complexes on the Leading Strand of Ribosomal DNA into 5′-Phosphorylated DNA Double-Strand Breaks by Replication Runoff. Mol. Cell. Biol. 2000, 20, 3977–3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlin E. B.; Rudolph A. E.; Zhao Y.; Matthews H. R.; Dixon J. E. Catalytic Mechanism of the Phospholipase D Superfamily Proceeds via a Covalent Phosphohistidine Intermediate. Proc. Natl. Acad. Sci. U.S.A. 1998, 95, 9202–9207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S. W.; Burgin A. B.; Huizenga B. N.; Robertson C. A.; Yao K. C.; Nash H. A. A Eukaryotic Enzyme that Can Disjoin Dead-End Covalent Complexes between DNA and Type I Topoisomerases. Proc. Natl. Acad. Sci. U.S.A. 1996, 93, 11534–11539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouliot J. J.; Robertson C. A.; Nash H. A. Pathways for Repair of Topoisomerase I Covalent Complexes in Saccharomyces cerevisiae. Genes Cells 2001, 6, 677–687. [DOI] [PubMed] [Google Scholar]
- Liu C. Y.; Pouliot J. J.; Nash H. A. The Role of TDP1 from Budding Yeast in the Repair of DNA Damage. DNA Repair 2004, 3, 593–601. [DOI] [PubMed] [Google Scholar]
- Miao Z. H.; Agama K.; Sordet O.; Povirk L.; Kohn K. W.; Pommier Y. Hereditary Ataxia SCAN1 Cells Are Defective for the Repair of Transcription-Dependent Topoisomerase I Cleavage Complexes. DNA Repair 2006, 5, 1489–1494. [DOI] [PubMed] [Google Scholar]
- Interthal H.; Chen H. J.; Kehl-Fie T. E.; Zotzmann J.; Leppard J. B.; Champoux J. J. SCAN1 Mutant Tdp1 Accumulates the Enzyme-DNA Intermediate and Causes Camptothecin Hypersensitivity. EMBO J. 2005, 24, 2224–2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maede Y.; Shimizu H.; Fukushima T.; Kogame T.; Nakamura T.; Miki T.; Takeda S.; Pommier Y.; Murai J. Differential and Common DNA Repair Pathways for Topoisomerase I- and II-Targeted Drugs in a Genetic DT40 Repair Cell Screen Panel. Mol. Cancer Ther. 2014, 13, 214–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies D. R.; Interthal H.; Champoux J. J.; Hol W. G. J. The Crystal Structure of Human Tyrosyl-DNA Phosphodiesterase, Tdp1. Structure 2002, 10, 237–248. [DOI] [PubMed] [Google Scholar]
- Davies D. R.; Interthal H.; Champoux J. J.; Hol W. G. J. Crystal Structure of a Transition State Mimic for Tdp1 Assembled from Vanadate, DNA, and a Topoisomerase I-Derived Peptide. Chem. Biol. 2003, 10, 139–147. [DOI] [PubMed] [Google Scholar]
- Antony S.; Marchand C.; Stephen A. G.; Thibaut L.; Agama K. K.; Fisher R. J.; Pommier Y. Novel High-Throughput Electrochemiluminescent Assay for Identification of Human Tyrosyl-DNA Phosphodiesterase (Tdp1) Inhibitors and Characterization of Furamidine (NSC 305831) as an Inhibitor of Tdp1. Nucleic Acids Res. 2007, 35, 4474–4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dexheimer T. S.; Gediya L. K.; Stephen A. G.; Weidlich I.; Antony S.; Marchand C.; Interthal H.; Nicklaus M.; Fisher R. J.; Njar V. C.; Pommier Y. 4-Pregnen-21-ol-3,20-dione-21-(4-bromobenzenesulfonate) (NSC 88915) and Related Novel Steroid Derivatives as Tyrosyl-DNA Phosphodiesterase (Tdp1) Inhibitors. J. Med. Chem. 2009, 52, 7122–7131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirivolu V. R.; Vernekar S. K. V.; Marchand C.; Naumova A.; Chergui A.; Renaud A.; Stephen A. G.; Chen F.; Sham Y. Y.; Pommier Y.; Wang Z. 5-Arylidenethioxothiazolidinones as Inhibitors of Tyrosyl-DNA Phosphodiesterase I. J. Med. Chem. 2012, 55, 8671–8684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T. X.; Morrell A.; Conda-Sheridan M.; Marchand C.; Agama K.; Bermingam A.; Stephen A. G.; Chergui A.; Naumova A.; Fisher R.; O’Keefe B. R.; Pommier Y.; Cushman M. Synthesis and Biological Evaluation of the First Dual Tyrosyl-DNA Phosphodiesterase I (Tdp1)–Topoisomerase I (Top1) Inhibitors. J. Med. Chem. 2012, 55, 4457–4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conda-Sheridan M.; Reddy P. V. N.; Morrell A.; Cobb B. T.; Marchand C.; Agama K.; Chergui A.; Renaud A.; Stephen A. G.; Bindu L. K.; Pommier Y.; Cushman M. Synthesis and Biological Evaluation of Indenoisoquinolines That Inhibit Both Tyrosyl-DNA Phosphodiesterase I (Tdp1) and Topoisomerase I (Top1). J. Med. Chem. 2013, 56, 182–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cushman M.; Jayaraman M.; Vroman J. A.; Fukunaga A. K.; Fox B. M.; Kohlhagen G.; Strumberg D.; Pommier Y. Synthesis of New Indeno[1,2-c]isoquinolines: Cytotoxic Non-Camptothecin Topoisomerase I Inhibitors. J. Med. Chem. 2000, 43, 3688–3698. [DOI] [PubMed] [Google Scholar]
- Nagarajan M.; Xiao X.; Antony S.; Kohlhagen G.; Pommier Y.; Cushman M. Design, Synthesis, and Biological Evaluation of Indenoisoquinoline Topoisomerase I Inhibitors Featuring Polyamine Side Chains on the Lactam Nitrogen. J. Med. Chem. 2003, 46, 5712–5724. [DOI] [PubMed] [Google Scholar]
- Nagarajan M.; Morrell A.; Fort B. C.; Meckley M. R.; Antony S.; Kohlhagen G.; Pommier Y.; Cushman M. Synthesis and Anticancer Activity of Simplified Indenoisoquinoline Topoisomerase I Inhibitors Lacking Substituents on the Aromatic Rings. J. Med. Chem. 2004, 47, 5651–5661. [DOI] [PubMed] [Google Scholar]
- Morrell A.; Placzek M. S.; Steffen J. D.; Antony S.; Agama K.; Pommier Y.; Cushman M. Investigation of the Lactam Side Chain Length Necessary for Optimal Indenoisoquinoline Topoisomerase I Inhibition and Cytotoxicity in Human Cancer Cell Cultures. J. Med. Chem. 2007, 50, 2040–2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrell A.; Placzek M.; Parmley S.; Antony S.; Dexheimer T. S.; Pommier Y.; Cushman M. Nitrated Indenoisoquinolines as Topoisomerase I Inhibitors: A Systematic Study and Optimization. J. Med. Chem. 2007, 50, 4419–4430. [DOI] [PubMed] [Google Scholar]
- Strumberg D.; Pommier Y.; Paull K.; Jayaraman M.; Nagafuji P.; Cushman M. Synthesis of Cytotoxic Indenoisoquinoline Topoisomerase I Poisons. J. Med. Chem. 1999, 42, 446–457. [DOI] [PubMed] [Google Scholar]
- Fox B. M.; Xiao X.; Antony S.; Kohlhagen G.; Pommier Y.; Staker B. L.; Stewart L.; Cushman M. Design, Synthesis, and Biological Evaluation of Cytotoxic 11-Alkenylindenoisoquinoline Topoisomerase I Inhibitors and Indenoisoquinoline–Camptothecin Hybrids. J. Med. Chem. 2003, 46, 3275–3282. [DOI] [PubMed] [Google Scholar]
- Kiselev E.; Dexheimer T. S.; Pommier Y.; Cushman M. Design, Synthesis, and Evaluation of Dibenzo[c,h][1,6]naphthyridines as Topoisomerase I Inhibitors and Potential Anticancer Agents. J. Med. Chem. 2010, 53, 8716–8726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrell A.; Placzek M.; Parmley S.; Grella B.; Antony S.; Pommier Y.; Cushman M. Optimization of the Indenone Ring of Indenoisoquinoline Topoisomerase I Inhibitors. J. Med. Chem. 2007, 50, 4388–4404. [DOI] [PubMed] [Google Scholar]
- Kiselev E.; DeGuire S.; Morrell A.; Agama K.; Dexheimer T. S.; Pommier Y.; Cushman M. 7-Azaindenoisoquinolines as Topoisomerase I Inhibitors and Potential Anticancer Agents. J. Med. Chem. 2011, 54, 6106–6116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiselev E.; Agama K.; Pommier Y.; Cushman M. Azaindenoisoquinolines as Topoisomerase I Inhibitors and Potential Anticancer Agents: A Systematic Study of Structure–Activity Relationships. J. Med. Chem. 2012, 55, 1682–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cinelli M. A.; Reddy P. V. N.; Lv P. C.; Liang J. H.; Chen L.; Agama K.; Pommier Y.; van Breemen R. B.; Cushman M. Identification, Synthesis, and Biological Evaluation of Metabolites of the Experimental Cancer Treatment Drugs Indotecan (LMP400) and Indimitecan (LMP776) and Investigation of Isomerically Hydroxylated Indenoisoquinoline Analogues as Topoisomerase I Poisons. J. Med. Chem. 2012, 55, 10844–10862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staker B. L.; Feese M. D.; Cushman M.; Pommier Y.; Zembower D.; Stewart L.; Burgin A. B. Structures of Three Classes of Anticancer Agents Bound to the Human Topoisomerase I–DNA Covalent Complex. J. Med. Chem. 2005, 48, 2336–2345. [DOI] [PubMed] [Google Scholar]
- Davies D. R.; Interthal H.; Champoux J. J.; Hol W. G. J. Explorations of Peptide and Oligonucleotide Binding Sites of Tyrosyl-DNA Phosphodiesterase Using Vanadate Complexes. J. Med. Chem. 2004, 47, 829–837. [DOI] [PubMed] [Google Scholar]
- Andoh T.; Ishii K.; Suzuki Y.; Ikegami Y.; Kusunoki Y.; Takemoto Y.; Okada K. Characterization of a Mammalian Mutant with a Camptothecin Resistant DNA Topoisomerase I. Proc. Natl. Acad. Sci. U.S.A. 1987, 84, 5565–5569. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.










